Abstract
We have discovered a novel cortical patch structure in Saccharomyces cerevisiae defined by a family of integral plasma membrane proteins, including Sur7p, Ynl194p, and Ydl222p. Sur7p-family patches localized as cortical patches that were immobile and stable. These patches were polarized to regions of the cell with a mature cell wall; they were absent from small buds and the tips of many medium-sized buds. These patches were distinct from other known cortical structures. Digestion of the cell wall caused Sur7p patches to disassemble, indicating that Sur7p requires cell wall-dependent extracellular interactions for its localization as patches. sur7Δ, ydl222Δ, and ynl194Δ mutants had reduced sporulation efficiencies. SUR7 was originally described as a multicopy suppressor of rvs167, whose product is an actin patch component. This suppression is probably mediated by sphingolipids, since deletion of SUR7, YDL222, and YNL194 altered the sphingolipid content of the yeast plasma membrane, and other SUR genes suppress rvs167 via effects on sphingolipid synthesis. In particular, the sphingoid base length and number of hydroxyl groups in inositolphosphorylceramides were altered in sur7Δ, ydl222Δ, and yne194Δ strains.
The importance of membrane domains and local differences in membrane composition and structure has been described for several processes, including endocytosis, signaling through caveoli, and protein trafficking (17, 32, 34). For the yeast Saccharomyces cerevisiae, two reports have presented evidence for detergent-insoluble plasma membrane lipid rafts, which are important for protein sorting through the endoplasmic reticulum and Golgi (2, 30). Also, septins maintain the daughter cell plasma membrane as a domain with distinct markers from the mother’s plasma membrane (4, 49). However, most yeast plasma membrane proteins, whether integral, peripheral, or glycosylphosphatidylinositol anchored, are dispersed evenly throughout the plasma membrane. With the notable exception of actin patch components, relatively few proteins are known to localize as patches or domains associated with the plasma membrane.
SUR7 was originally identified as a multicopy suppressor of mutations in rvs161 and/or rvs167 (45). rvs161 and rvs167 mutants have reduced viability upon starvation (6, 13) and are also defective in actin polarization, bipolar bud site selection (6, 19, 44), endocytosis (35), and sporulation (12, 15). Overexpression of SUR7 suppressed rvs161 and rvs167 defects in actin polarization, bud site selection, and growth (45). rvs161 and rvs167 are also suppressed by loss-of-function mutations in the nonessential genes sur1, sur2, and sur4 (15). Sur1p, Sur2p, and Sur4p are a mannosyl-transferase, a hydroxylase, and an acyl chain elongation protein, respectively, all involved in sphingolipid biosynthesis (see references 16 and 42).
The Sur7p family includes three members in S. cerevisiae: Sur7p, Ynl194p, and Ydl222p. They are predicted to be integral membrane proteins; each has a signal sequence and three transmembrane helices (45). These proteins contain 301 to 309 residues, with the N-terminal one-third predicted to be extracellular, the middle third largely transmembrane, and the C-terminal third cytoplasmic. Pairwise comparisons of their sequences show 27 to 34% identity and 42 to 49% similarity. The extracellular portions are more conserved than the intracellular portions (33.6% of extracellular residues are identical in all three protein sequences, in contrast to 5.2 and 16% of intracellular and transmembrane residues, respectively). Overexpression of YNL194 did not suppress rvs161 and rvs167, and no additional phenotypes were described for strains with a sur7 disruption alone or in combination with a ydl222 or rvs167 mutation (45). SUR7, YDL222, and YNL194 are differentially expressed: SUR7 expression is increased in late G2/M phase (48), YDL222 is induced during pseudohyphal growth or osmotic shock (33, 38), and YNL194 is induced during the shift to anaerobic metabolism, carbon starvation, or osmotic shock (14, 37).
Since Rvs167p is a component of actin patches (3) and SUR7 interacts genetically with rvs167, we hypothesized that Sur7p was an integral membrane actin patch component. No integral membrane or lipidated proteins have yet been localized to actin patches or implicated in tethering actin patches to the membrane. We report here that Sur7p is not a component of actin patches and that Sur7p-family members are not necessary for actin- or Rvs167p-associated functions. Instead, we found that Sur7p-family proteins localize as cortical patches that are distinct from any previously described structure or domain in yeast. We also found that these patches interact with the cell wall, are involved in sporulation, and probably suppress rvs161 and rvs167 through sphingolipids.
MATERIALS AND METHODS
Construction of fusion proteins.
Strains used in this study are listed in Table 1. C-terminal GFP(S65T) and 9myc fusions were constructed as described previously (25). DNA fragments carrying green fluorescent protein (GFP) and HIS3 or 9myc and HIS5 from Schizosaccharomyces pombe, flanked by the genomic sequences homologous to sequences downstream and upstream of the appropriate stop codon, were generated by PCR. The products for SUR7-GFP, YNL194-GFP, and SUR7−9myc were integrated into the genome of YJC1193 yielding YJC2054, YJC2032, and YJC2009, respectively; YNL194-9myc was integrated into YJC1092, yielding YJC2080. GFP tagging was confirmed by PCR, and 9myc tagging was confirmed by Western hybridization with the mouse anti-c-myc antibody 9E10 (BAbCO/Covance). YJC2054 was crossed with YJC2106 and YJC2080 to produce YJC2124 and YJC2126, respectively. YJC2054 and YJC2032 were diploidized by HO induction, yielding YJC2210 and YJC2629. Plasmids containing STD1 tagged with hemagglutinin (3HA) or GFP on 2μm plasmids under the ADH2 promoter were a generous gift of Martin Schmidt (University of Pittsburgh School of Medicine) and were transformed into YJC2054 and YJC2009.
TABLE 1.
Yeast strains
| Strain | Relevant genotype (all strains are leu2 ura3 his3-Δ200) | Source or reference |
|---|---|---|
| YJC1092 | MATa lys2 ade2 trp1 | YPH499 (P. Hieter) |
| YJC1193 | MATα | 24 |
| YJC1411 | MATa/MATα | 24 |
| YJC1423 | MATα CAP1-GFP-HIS3 | 26 |
| YJC2004 | MATasur7Δ::HIS3 lys2 ade2 trp1 | This study |
| YJC2009 | MATα SUR7-9myc-SpHIS5 | This study |
| YJC2019 | MATα NUM1-GFP-HIS3 | 23 |
| YJC2032 | MATα YNL194C-GFP-HIS3 | This study |
| YJC2041 | MATα ydl222cΔ::HIS3 | This study |
| YJC2042 | MATaydl222cΔ::HIS3 lys2 ade2 trp1 | This study |
| YJC2044 | MATα ynl194cΔ::HIS3 | This study |
| YJC2054 | MATα SUR7-GFP-HIS3 | This study |
| YJC2080 | MATaYNL194c-9myc-SpHIS5 lys2 ade2 trp1 | This study |
| YJC2106 | MATaNUM1-9myc::SpHIS5 ade2 trp1 | 23 |
| YJC2122 | MATasur7Δ::HIS3 ydl222cΔ::HIS3 ynl194cΔ::HIS3 | This study |
| YJC2124 | MATα SUR7-GFP-HIS3 NUM1-9myc-SpHIS5 trp1 | This study |
| YJC2126 | MATα SUR7-GFP-HIS3 YNL194c-9myc-SpHIS5 lys2 trp1 | This study |
| YJC2191 | MATa/MATα sur7Δ::HIS3/sur7Δ::HIS3 lys2/lys2 ade2/ade2 trp1/trp1 | This study |
| YJC2197 | MATa/MATα sur7Δ::HIS3/sur7Δ::HIS3 ydl222c::HIS3/ydl222c::HIS3 ynl194cΔ::HIS3/ynl194cΔ::HIS3 | This study |
| YJC2204 | MATa/MATα ynl194cΔ::HIS3/ynl194cΔ::HIS3 | This study |
| YJC2208 | MATa/MATα ydl222c::HIS3/ydl222c::HIS3 lys2/lys2 ade2/ade2 trp1/trp1 | This study |
| YJC2210 | MATa/MATα SUR7-GFP-HIS3/SUR7-GFP-HIS3 | This study |
| YJC2212 | MATα rvs161Δ::HIS3 | This study |
| YJC2213 | MATα rvs167Δ::HIS3 | This study |
| YJC2629 | MATa/MATα YNL194C-GFP-HIS3/YNL194C-GFP-HIS3 | This study |
| YJC2631 | MATα sur7Δ::HIS3 | This study |
| YJC2632 | MATa/MATα ydl222c::HIS3/ydl222c::HIS3 | This study |
| YJC2636 | MATα ydl222cΔ886-927-LACZ-URA3 | This study |
| YJC2638 | MATa/MATα YDL222C-885-3HA/YDL222C-885-3HA | This study |
| YJC2661 | MATa/MATα sur7Δ::HIS3/sur7Δ::HIS3 | This study |
| YJC2700 | MATα ynl194cΔ412-903-LACZ-URA3 | This study |
| YJC2701 | MATa/MATα YNL194C-411-3HA/YNL194C-411-3HA | This study |
Plasmids containing transposon-generated tags of YNL194 and YDL222 were a generous gift from M. Snyder’s TRIPLES database (31, 39). Plasmids V133H1 and V157A8 were cut with NotI and transformed into YJC1193, resulting in the presence of the URA3-containing transposon with LACZ in frame at codons 295 of YDL222 (YJC2636) and 137 of YNL194 (YJC2700). These strains were then transformed with pSH62, a CEN6 HIS3 plasmid containing CRE under the GAL1 promoter, a generous gift from J. Hegemann (22). The majority of the transposable element was then removed by galactose-induced lox/cre excision, leaving only an in-frame, 93-codon, 3HA and no selectable marker. These strains were then diploidized by HO induction, resulting in YDL222-885HA (YJC2638) or YNL194-412HA (YJC2701).
Gene disruptions.
Deletions of entire coding sequences of genes were performed as described previously (5). SUR7 and YDL222 were disrupted in YJC1092, creating YJC2004 and YJC2042. YDL222, YNL194, RVS161, RVS167, and SUR7 were disrupted in YJC1193, creating YJC2041, YJC2044, YJC2212, YJC2213, and YJC2631. All disruptions were confirmed by PCR; disruption of YNL194 in YJC2044 was also confirmed by sequencing. Strain YJC2122 with disruptions of SUR7, YNL194, and YDL222 was obtained by sequential mating and tetrad dissection of YJC2004 with YJC2041, followed by YJC2044. Strains YJC2191, YJC2197, YJC2204, YJC2208, YJC2210, YJC2632, and YJC2661 were generated by HO-induced diploidization of YJC2004, YJC2122, YJC2044, YJC2042, YJC2054, YJC2041, and YJC2631.
Growth on plates.
Strains were grown from fresh overnight cultures to an optical density at 600 nm (OD600) of 0.6. Tenfold serial dilutions of each strain were transferred to yeast-peptone or synthetic dextrose agar plates containing glucose, raffinose, galactose, or glycerol as a carbon source. Yeast-peptone-dextrose (YPD) or synthetic dextrose agar plates were also tested with the following: 2 μg of antimycin A/ml; 5 or 20 mg of 3-amino-1,2,4-triazole/ml; 2.5, 5, 7.5, or 10% NaCl, KCl, or CaCl2; 2.5 to 10 μg of calcofluor/ml; 0.75 to 3 mg of caffeine/ml; 0.003 or 0.001% sodium dodecyl sulfate; and 0.001% methylene blue with or without 1 M sorbitol.
Growth in liquid.
Fresh overnight cultures were diluted to an OD600 of 0.2 in YPD with carbenicillin. The OD600 was monitored over time until it reached at least 4.0.
Electron microscopy.
Mid-log-phase cultures were fixed by addition of 2% glutaraldehyde, followed by suspension of cells in 2% glutaraldehyde−0.1 M cacodylate for 3 h at 4°C. The cells were rinsed and then treated for 1 h at room temperature with 1.25% osmium−0.1 M cacodylate. Cells were rinsed with buffer, rinsed with 15% ethanol, stained for 1 h in 4% uranyl acetate, and dehydrated. They were embedded in a pure polybed, incubated at 60°C overnight, thin-sectioned, poststained in 4% uranyl acetate and lead citrate, and viewed on a Zeiss 902 electron microscope.
Cell wall digestion.
Mid-log-phase cultures were suspended in 0.1 M KPO4−1 M sorbitol, pH 7.0, and incubated with 0.5 mg of zymolyase/ml for 2 h at 37°C. β-glucuronidase (Sigma Chemical Co.) was added to a concentration of 1,000 U/ml, and cultures were incubated for an additional 15 h at 37°C. The cells were washed, and Sur7-GFP (YJC2054), Cap1-GFP (YJC1423), or Num1-GFP (YJC2019) was imaged.
Microscopy.
Movies of GFP fluorescence and rhodamine-phalloidin staining of mid-log-phase cultures were performed as described (51). Latrunculin A treatment consisted of adding 0.01 volumes of 50 mM Latrunculin A in dimethyl sulfoxide to the culture. Latrunculin A was from Philip Crews, Department of Chemistry, UCSC, NIH grant CA47135. Rhodamine-phalloidin staining confirmed that all filamentous actin was depolymerized. Immunofluorescence of strains containing a 9myc, 3HA, or β-galactosidase tag was performed as described with monoclonal 9E10 or HA11 antibodies (BAbCO/Covance) or rabbit anti-β-galactosidase immunoglobulin Gs (Cappel/ICN) and rhodamine-labeled secondary antibodies (1).
Mass spectrometry.
Plasma membranes were isolated, and their lipids were prepared and analyzed by nano-electrospray ionization tandem mass spectrometry as described previously (9, 43), including positive ion scans specific for phosphatidylserines, phosphatidylcholines, and phosphatidylethanolamines, and a negative ion precursor scan for lipids containing inositol phosphate (m/z 241).
Other assays.
FM4-64 and lucifer yellow uptake endocytosis assays were performed on YJC1092, YJC1193, YJC2004, YJC2042, YJC2044, YJC2122, YJC2212, and YJC2213 as described previously (18, 50). Analysis of bipolar bud site selection was performed on YJC1411, YJC2191, YJC2197, YJC2204, and YJC2208 as described previously (10). Cells with three or more bud scars were scored as having a random or bipolar budding pattern. Osmotically induced, calcofluor-accumulating domains were induced and imaged as described previously (47).
Functional genomics data.
Data from gene chip studies were obtained from the corresponding websites (http://cellcycle-www.stanford.edu [48], http://cmgm.stanford.edu/pbrown/explore/diauxsearch.html [14], http://staffa.wi.mit.edu/fink_public/mapk [33], and http://CC.uab.es/∼ivbq3/Hoja_WEB_chips.html [36]). Strains from the TRIPLES database are described online (http://ygac.med.yale.edu/).
RESULTS
Sur7p, Ynl194p, and Ydl222p define novel cortical patch structures.
Sur7p, Ynl194p, and Ydl222p are homologous proteins that define a family of predicted integral membrane proteins in S. cerevisiae (45). We found proteins with similar sequences in the fungi Aspergillus nidulans (AI211325), Mycosphaerella graminicola (AW180313), Neurospora crassa (AI397953, AA898805), and Magnaporthe grisea (AI069195) and in the higher plant barley (Hordeum vulgare [BE455024]) and the budding yeast Candida albicans (AL033391). Database searches revealed no homologues in Arabidopsis or in any organism lacking a cell wall.
We localized Sur7p by GFP tagging its C terminus, which is predicted to be intracellular (45). Sur7-GFP was readily detected in vegetative cells growing in rich media and localized as patches at the cell cortex (Fig. 1A), consistent with the prediction that it is an integral plasma membrane protein. The localization was verified by immunofluorescence staining of a strain containing Sur7p-9myc (YJC2009) (data not shown). Sur7-GFP was able to rescue the null sporulation phenotype, as described below.
FIG. 1.
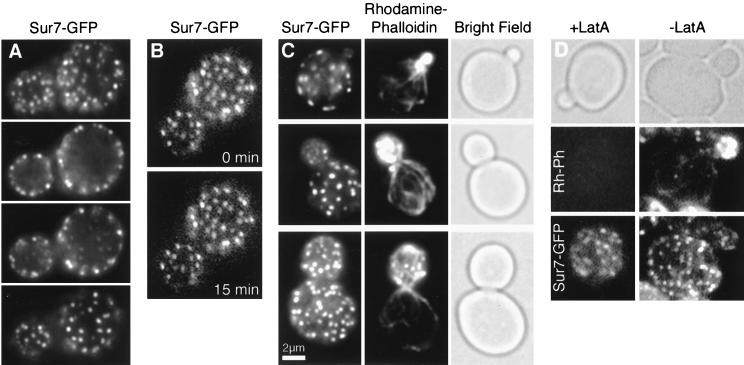
Sur7p localizes as cortical, stationary, polarized patches. (A) Fluorescence microscopy of the top, middle, and bottom of a haploid cell containing Sur7-GFP (YJC2054) reveals that Sur7p localizes as patches at the cortex. (B) Time-lapse imaging shows Sur7-GFP patches are stationary and stable. A 15-min interval separates the two single focal plane frames. (C). Fixation and costaining of Sur7-GFP and actin patches, stained with rhodamine-phalloidin, shows that the two signals do not colocalize and that Sur7p patches are not present in small buds. (D) Sur7-GFP patches remain after actin has been depolymerized by latrunculin A (LatA), which was confirmed by rhodamine-phalloidin staining.
We further characterized these cortical Sur7p patch structures in living and fixed cells. A typical cell contained 40 to 80 Sur7p patches during vegetative growth, which were stationary and stable over a wide range of time scales (from 4 frames/s to 15 frames/h) in time-lapse movies of GFP fluorescence in living cells (Fig. 1B). In contrast, actin patches move rapidly in time intervals of <1 s (51).
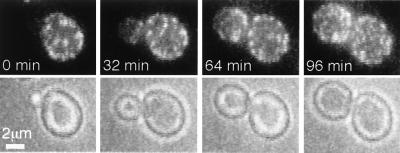
The distribution of Sur7p patches within vegetatively growing cells was polarized. Sur7p patches were not present in small buds but were found in medium to large buds and in mother cells (Fig. 1C). This distribution was confirmed by movie analysis, in which Sur7p patches were initially absent from the small bud and subsequently appeared in the older proximal region of the medium-sized bud (Fig. 2). Eventually, patches were present throughout the large bud. Patches did not move from the mother into the bud. Sur7p’s localization did not require filamentous actin. Latrunculin A treatment under conditions that completely depolymerized F-actin did not delocalize Sur7-GFP patches (Fig. 1D).
FIG. 2.
Sur7p patches assemble during bud growth. Selected frames are shown from time-lapse fluorescence microscopy of Sur7-GFP cells (YJC2054). Each frame is a two-dimensional projection of a z-series. The interval between frames is 4 min. The movie is available online (http://www.cooperlab.wustl.edu).
We performed similar analysis on the Sur7p homologue Ynl194p. Endogenous Ynl194p tagged C-terminally with GFP was only faintly visible in a small proportion of vegetative cells. The addition of 0.4 M NaCl or the absence of carbon sources induces YNL194 transcription (37), and we found that these conditions also increased Ynl194-GFP expression to a readily detectable level. Ynl194-GFP patches were similar in number and distribution to Sur7p patches, also being cortical and absent from small buds and some medium buds (Fig. 3A). Ynl194-GFP was also able to rescue the null sporulation phenotype, and its localization was confirmed by immunofluorescence staining of Ynl194-9myc (Fig. 3B). Movie analysis of Ynl194-GFP revealed that Ynl194p patches, like Sur7p patches, were stationary and stable. Comparison of Sur7-GFP and Ynl194-9myc patches after growth in salt revealed partial colocalization of the two proteins (Fig. 3B). Forty-six percent of 149 Ynl194p patches in 22 YJC2126 cells colocalized with Sur7-GFP patches. This partial colocalization is more than expected from random distribution because it is significantly more than the 12% overlap between another protein found in patches, Num1p, and Sur7-GFP (n = 218 Num1p-9myc patches in 23 YJC2124 cells).
FIG. 3.
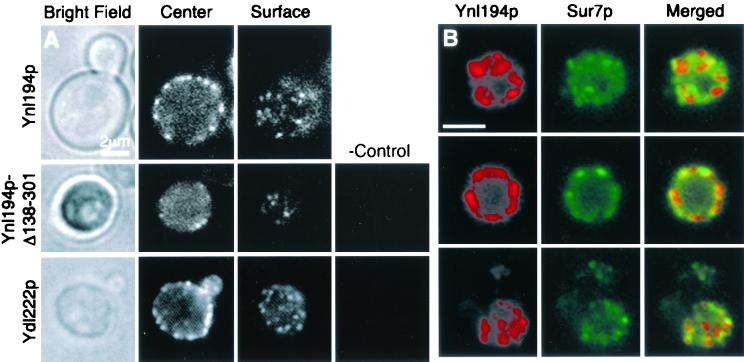
Ynl194p and Ydl222p localize similarly to Sur7p. All strains were incubated in YPD with 0.4 M NaCl for 75 min prior to viewing to induce YNL194 and YDL222 expression. (A) Single focal planes of Ynl194-GFP (YJC2032) in a living cell show Ynl194-GFP as cortical patches that are absent from the bud (top panels). A Ynl194p construct truncated at the first intracellular residue and tagged with β-galactosidase (YJC2700) localizes similarly to the full-length protein (middle panels). A Ydl222p construct tagged with 3HA before its 14 C-terminal residues (YJC2638) localizes similarly to Sur7p and Ynl194p. The rightmost panels are immunostaining of untagged cells (YJC1193). (B) Cells containing Sur7-GFP and Ynl194p-9myc (YJC2126) were fixed and stained with anti-myc antibodies. Depicted are representative single focal planes showing little (top panels), extensive (middle panels), or partial (bottom panels) colocalization of Sur7p and Ynl194p. Scale bars are 2 μm.
The first extracellular domain of Ynl194p was sufficient for its localization as cortical patches. Insertion of a transposon from M. Snyder’s TRIPLES library at codon 137 of Ynl194p results in a protein containing only the first extracellular loop, the following transmembrane helix, and a β-galactosidase tag. Immunofluorescence with anti-β-galactosidase antibody revealed staining in cortical patches, although there were fewer patches than in the strain with the full-length tagged protein (Fig. 3A). This indicates that the localization of Ynl194p, and probably of Sur7p and Ydl222p, as patches in the plasma membrane is largely independent of cytosolic interactions.
Another plasmid from the TRIPLES database contains YDL222 with a transposon after codon 295, 42 nucleotides before the stop codon. We integrated this plasmid into a haploid strain and then induced the transposon’s excision by CRE induction; this leaves a 93-amino-acid 3HA insertion in Ydl222p. After this strain was diploidized and grown in the same salt conditions that induced YNL194 expression, staining of HA showed Ydl222p localization to cortical patches that appeared similar to Sur7p and Ynl194p patches (Fig. 3A). However, the tagged Ydl222p was not functional in the sporulation assay, indicating that the intracellular C terminus of Ydl222p is important for its function. Our attempts with several primer pairs to tag endogenous YDL222 by PCR product integration were unsuccessful.
The characteristics of Sur7p, Ynl194p, and Ydl222p patches were not similar to those of other described cortical structures. However, we performed localization experiments with several proteins known to localize as patches. First, we compared Sur7-GFP patches to actin patches. Since Sur7p patches were polarized almost inversely to actin patches, which polarize to the bud tip and at the neck (27), these two types of patches were usually not found in the same region of the cell. In large buds, which do contain both types of patches, Sur7p patches and actin patches did not colocalize (Fig. 1C, bottom frames). We also compared Sur7p-GFP patches with Num1p-9myc patches because Num1p patches are stationary and more prominent in mother cells (20, 23), but Sur7p and Num1p patches did not colocalize (see above). Finally, we compared the localization of Sur7-GFP or Sur7-9myc patches with Std1-3HA or Std1-GFP patches. Std1p is found in the nucleus and in cortical patches (41). No colocalization was observed (data not shown), although the Std1p signal was weak in conditions that also preserved the Sur7p signal. However, the size and number of Std1p patches in most cells and Sur7p patches were clearly different.
Sur7p requires extracellular interactions for its localization.
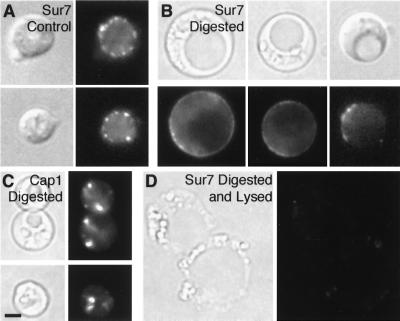
Since the N-terminal third of Sur7p-family proteins is highly conserved and the first extracellular portion of Ynl194p was sufficient for its patched localization, we examined whether Sur7p’s localization depends on extracellular interactions. First, we treated cells with zymolyase for 2 h, which digests β1-3 glucan; Sur7-GFP patch localization was not affected (data not shown). However, longer incubations, and especially the addition of a crude solution of glucuronidase, which provides a less specific and more effective digestion of the cell wall, caused dissipation of most Sur7-GFP patches, and in some cells diffuse cortical fluorescence was observed (Fig. 4B). We were concerned that cells might have lysed during the digestion, allowing proteases or other enzymes into the cell interior. However, in control experiments, actin patches labeled with Cap1-GFP (Fig. 4C) and Num1-GFP patches (not shown) did not disappear under these digestion conditions, indicating that plasma membranes were intact. Furthermore, occasional cells that were clearly lysed based on their bright field morphology lost the diffuse cytoplasmic fluorescence present in most Sur7-GFP cells (Fig. 4D).
FIG. 4.
Disassembly of Sur7p patches upon cell wall digestion. Sur7-GFP cells (YJC2054) were incubated for 15 h at 37°C in the absence (A) or presence (B) of zymolyase and glucuronidase. Cap1-GFP patches (in YJC1423) were retained in the presence of enzymes (C). Sur7-GFP fluorescence was lost from cells that appeared lysed by bright-field microscopy (D).
The Sur7p family is involved in sporulation.
SUR7 was identified as a multicopy suppressor of mutations in the gene encoding the actin patch component Rvs167p, so we hypothesized that the function of Sur7p patches might be related to actin patches or Rvs167p. To test this hypothesis, we generated strains carrying null mutations of sur7, ydl222, and ynl194, singly and in combination. All three single mutants and the triple mutant (YJC2004, YJC2042, YJC2044, YJC2122) were viable and grew well on plates and in liquid media. In a previous study, a disruption leading to a truncation of the C-terminal two-thirds of ynl194 was inviable (45). We confirmed that our ynl194 deletion was correct by PCR testing and by sequencing genomic DNA. The reason for this difference may be the nature of the disruption or differences in genetic backgrounds.
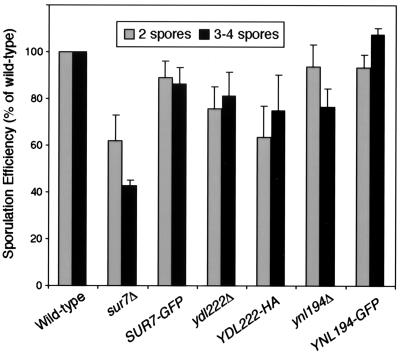
One phenotype of rvs161 and rvs167 mutants is a reduced frequency of sporulation. We found that the sporulation efficiency of sur7Δ, ydl194Δ, and ydl222Δ strains was reduced to various degrees (Fig. 5). Relative to wild-type cells, sur7Δ/sur7Δ strains showed almost a 60% reduction in sporulation, while ydl222Δ/ydl222Δ and ydl194Δ/ydl194Δ strains showed reductions of only 20 and 25%. Strains homozygous for SUR7-GFP or YNL194-GFP showed rescue of the null phenotypes, while the YDL222-3HA strain did not. The ydl222Δ/ydl222Δ strain had a particularly variable phenotype, but within experiments, the YDL222-3HA strain was always the same as the null strain. The lack of YDL222-3HA rescue is probably due to the position of the tag, which is inserted within Ydl222p’s last intracellular loop, while Sur7p and Ynl194p were tagged at their C termini. A construct with 3HA inserted into Ynl194p at the N terminus of the first intracellular loop (YJC2701) also rescued the null phenotype (data not shown).
FIG. 5.
Sur7p and its homologues function in sporulation. Strains homozygous for sur7Δ, SUR7-GFP, ydl222Δ, YDL222-3HA, ynl194Δ, or YNL194-GFP (YJC2661, YJC2210, YJC2632, YJC2638, YJC2204, YJC2629) were sporulated on an MSPO plate for 7 days at 30°C. Well-formed asci with three to four clearly visible spores, as well as incomplete asci containing only two spores, were counted. Percentages shown are of the total number of cells and asci. Error bars show the standard error of the mean for 4 to 13 independent experiments, each of which counted 500 to 2,000 cells. Data from each experiment were normalized to results for a wild-type strain (YJC1411) from the same plate, in which 4 to 10% of cells became well-formed asci.
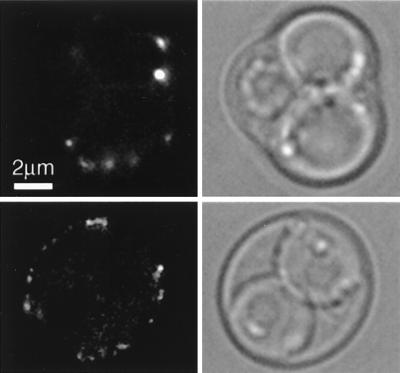
We then examined Sur7-GFP localization in sporulating cells (YJC2210). Sur7p patches were visible in the outer ascus membrane, which is consistent with the patches remaining intact in the plasma membrane throughout sporulation. However, sporulating cells contained fewer Sur7p patches than did vegetative cells (Fig. 6). Sur7p patches were not observed to assemble in the ascospore membrane, which is synthesized de novo around each spore, starting at the spindle pole body, although these membranes may contain diffuse Sur7-GFP not visible above the spore’s autofluorescence.
FIG. 6.
Sur7p patches during sporulation. A diploid strain homozygous for SUR7-GFP (YJC2210) was sporulated and viewed by fluorescence microscopy. Sur7p patches are visible in the membrane of the ascus, but not the ascospores of mature (top panels) or incomplete (bottom panels) asci. A single focal plane is shown.
We tested the single and triple sur7, ydl222, and ynl194 mutants for other actin- and Rvs161p-dependent phenotypes, including defective endocytosis, actin depolarization, and a loss of bipolar bud site selection. Endocytosis was assessed by FM4-64 and lucifer yellow uptake under conditions where rvs161Δ and rvs167Δ strains were defective. We examined the polarization of actin patches and cables by staining with rhodamine-phalloidin. Finally, we examined diploid cells stained with calcofluor to determine if they displayed bipolar or random budding patterns. Endocytosis, actin polarization, and bipolar bud site selection were normal in all the mutants. These results indicate that SUR7, YDL222, and YNL194 are not necessary for actin function.
The original phenotype of rvs161 and rvs167 mutants was reduced viability upon starvation, and expression of YNL194 and YDL222 is induced by osmotic stress (38). Therefore, we tested the growth of Sur7p-family mutants under various starvation and stress conditions. Unlike rvs161 or rvs167 mutants, sur7Δ, ynl194Δ, and ydl222Δ strains grew normally on plates containing high levels of salt (NaCl, KCl, or CaCl2), with poor carbon sources, or at elevated temperatures. Similarly, all the mutants grew normally in liquid medium with or without 0.4 M NaCl.
YDL222 has a minor role in cell wall function.
The requirement of a cell wall for Sur7p’s localization as patches suggests a role in cell wall function. We examined the ultrastructure of the cell wall in mutant strains by thin-section electron microscopy. The cell walls of a ydl222Δ strain (YJC2042) were slightly thinner than normal (data not shown), but the layered structure appeared normal, and we did not observe any significant differences between wild-type strains and the other, single-mutant or triple-mutant strains (YJC2004, YJC2044, YJC2122). Another indicator of cell wall function is inhibition of growth on plates containing calcofluor or sodium dodecyl sulfate. sur7Δ, ynl194Δ, and ydl222Δ strains grew as well as wild-type strains on these plates. While there was not a noticeable decrease in colony growth, we did detect a slight sorbitol-remediated susceptibility to lysis at 37°C in the ydl222Δ single strain and sur7Δ ynl194Δ ydl222Δ triple strain, by including methylene blue in the plates. This result is consistent with a slightly defective cell wall.
Hyperosmotic shock has also been shown to induce calcofluor-accumulating domains in the cell wall (47). We found that the majority of these domains had disappeared before Ynl194-GFP patches were induced to a visible level, and these cell wall domains did not colocalize with Ynl194-GFP or Sur7-GFP patches (data not shown).
The Sur7p family affects the sphingolipid composition of the plasma membrane.
sur1, sur2, and sur4 are null suppressors of rvs167 and encode enzymes involved in sphingolipid synthesis. Therefore, we hypothesized that SUR7 might also suppress rvs167 through effects on sphingolipid metabolism.
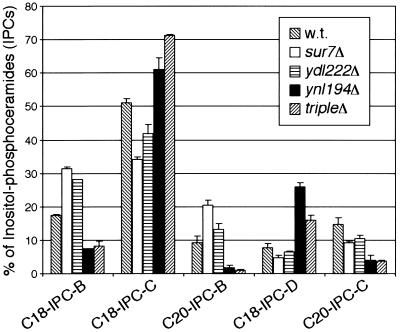
We analyzed the lipid composition of plasma membranes from Sur7p-family mutants by nano-electrospray ionization tandem mass spectrometry. The composition of inositolphosphorylceramide (IPC) was altered in the mutants (Fig. 7). IPCs, like all ceramides and other sphingolipids in yeast, have variations in the sphingoid base length (C18 or C20) and in the number of hydroxyl groups (subclasses A to D contain 1 to 4 hydroxyl groups, respectively). In ynl194Δ single- and sur7Δ ynl194Δ ydl222Δ triple-mutant strains, the fraction of IPCs containing C20 sphingoid bases was reduced from 25 to 5%. sur7Δ and ydl222Δ single mutants had IPCs with normal levels of C20 sphingoid bases. sur7Δ and ydl222Δ strains showed a decrease in IPC hydroxylation, having relatively more IPC-Bs, whereas the ynl194Δ single mutant and sur7Δ ynl194Δ ydl222Δ triple mutant showed an increase in IPC hydroxylation, having relatively more IPC-Cs and IPC-Ds (Fig. 7). The composition of glycerol-based lipids, phosphatidylinositol, phosphatidylserine, phosphatidylcholine, and phosphatidylethanolamine, was not altered in any of the mutants (data not shown).
FIG. 7.
Altered sphingolipid content of sur7Δ, ydl222Δ, and ynl194Δ plasma membranes. Mass spectrometry of plasma membrane lipids from the mutant strains showed differences in the composition of IPC sphingolipids. The IPCs varied in the number of hydroxyl groups (subclasses A to D) and in the length of their sphingoid base component (C18 or C20). Error bars are the standard error of proportion. m/z values are as follows: C18-IPC-B, 936; C18-IPC-C, 952; C20-IPC-B, 964; C18-IPC-D, 968; C20-IPC-C, 980.
DISCUSSION
We found that integral membrane proteins of the Sur7p family define novel domains in the yeast plasma membrane. Plasma membrane domains in mammalian cells have been defined and studied, but their existence and importance in yeast have not been clearly established. The Sur7p-defined plasma membrane domains do not correspond to actin patches, Num1p patches, Std1p patches, or chitin domains induced by osmotic shock. Sur7p-family patches are stationary and stable and have a polarized distribution. They are present throughout the cortex of the mother and the older portions of the bud, but they are absent in small buds and the tips of some medium buds. The reason Sur7p patches are not seen in sites of growth could be that Sur7p is absent from these regions or, more likely, that assembly of Sur7p into patches requires a certain amount of time or the presence of an additional component.
Several pieces of evidence indicate that Sur7p patches interact with the cell wall. First, cell wall digestion caused Sur7-GFP patches to dissipate, often resulting in diffuse fluorescence throughout the plasma membrane. Second, a Ynl194p truncation, including its major extracellular domain and a transmembrane domain but not intracellular portions of the protein, was able to localize as cortical patches. Third, ydl222Δ cells had minor cell wall morphology and cell lysis defects. Interactions with the cell wall are also consistent with observations that extracellular regions of Sur7p-family proteins have the highest sequence conservation and that Sur7p homologues are present in several fungi and a higher plant but not in organisms lacking a cell wall. A requirement for a cell wall structure or domain that is not yet present in the growing areas of the cell or in the spore wall could account for the absence of Sur7p patches from these regions and Sur7p’s polarized distribution. The cell wall is important for maintaining polarization in Fucus zygotes, because their polarization is lost upon cell wall digestion (8, 29).
SUR7 was discovered and defined as a multicopy suppressor of rvs161 and rvs167, and our results indicate that this suppression may be mediated by sphingolipids. Other rvs161 and rvs167 suppressors, sur1, sur2, and sur4 mutants, are defective in sphingolipid biosynthesis (15), and the plasma membranes of sur7Δ, ynl194Δ, and ydl222Δ mutants all displayed altered sphingolipid compositions, particularly in IPCs. The IPC compositions in sur7Δ and ydl222Δ single mutants showed decreases in IPC hydroxylation, while ynl194Δ single mutants and sur7Δ ynl194Δ ydl222Δ triple mutants showed an increase in IPC hydroxylation and a relative decrease in IPCs with longer (C20) sphingoid bases.
The mechanism of sphingolipid suppression of rvs167Δ may be through intermediates of sphingolipid metabolism that serve as cellular signals. Sphingoid bases, sphingoid base phosphates, ceramides, and the C subclass of IPCs affect processes including endocytosis (21, 52), the heat shock response (11, 46), growth inhibition (28), and Ca2+ sensitivity (7). However, we did not find phenotypes associated with any of these processes in single or triple deletions of SUR7, YNL194, and YDL222.
Sur7p, Ydl222p, and Ynl194p may affect sphingolipid metabolism in several ways. They are probably not components of the major sphingolipid synthesis pathway, which does includes Sur1p, Sur2p, and Sur4p (16, 42); IPC is probably synthesized in the endoplasmic reticulum and modified in the Golgi, but Sur7p-family proteins are at the plasma membrane. Alternatively, the sphingolipid composition in sur7Δ, ydl222Δ, and ynl194Δ strains may be altered in response to the loss of Sur7p-family patches. Sphingolipid derivatives are present in the same places as Sur7p’s extracellular interactions; they are in both the outer leaflet of the plasma membrane and the cell wall, where they are part of many glycosylphosphatidylinositol-anchored proteins.
We found that SUR7 and, to a lesser extent, YDL222 and YNL194 contribute to efficient sporulation. The Sur7p family could influence a novel ceramide-based signaling pathway that affects sporulation, similar to the glycerol-based lipid pathway including Spo14p (40). Alternatively, interactions of Sur7p with the ascospore wall may be important.
Acknowledgments
This work was supported by the Peter and Traudel Engelhorn Foundation, the German Research Foundation, SFB 352, and the Human Frontiers Science Program to B. Brügger and F. T. Wieland; the Austrian Science Foundation P13767 to R. Schneiter; and NIH grant GM47337 to J. A. Cooper.
We thank Martin Schmidt, Valerie Brachet, Michael Snyder, Johannes Hegemann, and Richard Heil-Chapdelaine for generous sharing of reagents; Neil Adames, Wei-Lih Lee, and Ken Blumer for helpful discussions; and Lori LaRose for assistance with electron microscopy.
REFERENCES
- 1.Amatruda, J. F., and J. A. Cooper. 1992. Purification, characterization and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J. Cell Biol. 117:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., S. Keränen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balguerie, A., P. Sivadon, M. Bonneu, and M. Aigle. 1999. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112:2529–2537. [DOI] [PubMed] [Google Scholar]
- 4.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841–851. [DOI] [PubMed] [Google Scholar]
- 5.Baudin, A., O. Ozierkalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, F., M. Urdaci, M. Aigle, and M. Crouzet. 1993. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13:5070–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeler, T., D. Bacikova, K. Gable, L. Hopkins, C. Johnson, H. Slife, and T. Dunn. 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J. Biol. Chem. 273:30688–30694. [DOI] [PubMed] [Google Scholar]
- 8.Brownlee, C., and F. Y. Bouget. 1998. Polarity determination in Fucus: from zygote to multicellular embryo. Semin. Cell Dev. Biol. 9:179–185. [DOI] [PubMed] [Google Scholar]
- 9.Brügger, B., G. Erben, R. Sandhoff, F. T. Wieland, and W. D. Lehmann. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 94:2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chant, J., and J. R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, N., G. Jenkins, Y. A. Hannun, J. Heitman, and L. M. Obeid. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275:17229–17232. [DOI] [PubMed] [Google Scholar]
- 12.Colwill, K., D. Field, L. Moore, J. Friesen, and B. Andrews. 1999. In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics 152:881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crouzet, M., M. Urdaci, L. Dulau, and M. Aigle. 1991. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast 7:727–743. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. [DOI] [PubMed] [Google Scholar]
- 15.Desfarges, L., P. Durrens, H. Juguelin, C. Cassagne, M. Bonneu, and M. Aigle. 1993. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast 9:267–277. [DOI] [PubMed] [Google Scholar]
- 16.Dickson, R. C., and R. L. Lester. 1999. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1438:305–321. [DOI] [PubMed] [Google Scholar]
- 17.Dobrowsky, R. T. 2000. Sphingolipid signalling domains floating on rafts or buried in caves? Cell Signal 12:81–90. [DOI] [PubMed] [Google Scholar]
- 18.Dulic, V., M. Egerton, I. Elguindi, S. Raths, B. Singer, and H. Riezman. 1991. Yeast endocytosis assays. Methods Enzymol. 194:697–710. [DOI] [PubMed] [Google Scholar]
- 19.Durrens, P., E. Revardel, M. Bonneu, and M. Aigle. 1995. Evidence for a branched pathway in the polarized cell division of Saccharomyces cerevisiae. Curr. Genet. 27:213–216. [DOI] [PubMed] [Google Scholar]
- 20.Farkasovsky, M., and H. Kuntzel. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friant, S., B. Zanolari, and H. Riezman. 2000. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 19:2834–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heil-Chapdelaine, R. A., J. R. Oberle, and J. A. Cooper. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karpova, T. S., J. G. McNally, S. L. Moltz, and J. A. Cooper. 1998. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142:1501–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpova, T. S., S. L. Moltz, L. E. Riles, U. Gueldener, J. H. Hegemann, S. Veronneau, H. Bussey, and J. A. Cooper. 1998. Depolarization of the actin cytoskeleton is a specific phenotype in Saccharomyces cerevisiae. J. Cell Sci. 111:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpova, T. S., S. L. Reck-Peterson, N. B. Elkind, M. S. Mooseker, P. J. Novick, and J. A. Cooper. 2000. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell 11:1727–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilmartin, J. V., and A. E. M. Adams. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S., H. Fyrst, and J. Saba. 2000. Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156:1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kropf, D. L., B. Kloareg, and R. S. Quatrano. 1988. Cell wall is required for fixation of the embryonic axis in Fucus zygotes. Science 239:187–190. [DOI] [PubMed] [Google Scholar]
- 30.Kübler, E., H. G. Dohlman, and M. P. Lisanti. 1996. Identification of Triton X-100 insoluble membrane domains in the yeast Saccharomyces cerevisiae. Lipid requirements for targeting of heterotrimeric G-protein subunits. J. Biol. Chem. 271:32975–32980. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., K. H. Cheung, P. Ross-Macdonald, P. S. Coelho, P. Miller, and M. Snyder. 2000. TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res. 28:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurzchalia, T. V., and R. G. Parton. 1999. Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11:424–431. [DOI] [PubMed] [Google Scholar]
- 33.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muniz, M., and H. Riezman. 2000. Intracellular transport of GPI-anchored proteins. EMBO J. 19:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6, and end7: Mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1721–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posas, F., J. R. Chambers, J. A. Heyman, J. P. Hoeffler, E. de Nadal, and J. Arino. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275:17249–17255. [DOI] [PubMed] [Google Scholar]
- 37.Puig, S., and J. E. Pérez-Ortín. 2000. Stress response and expression patterns in wine fermentations of yeast genes induced at the diauxic shift. Yeast 16:139–148. [DOI] [PubMed] [Google Scholar]
- 38.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290–8300. [DOI] [PubMed] [Google Scholar]
- 39.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413–418. [DOI] [PubMed] [Google Scholar]
- 40.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wölfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneiter, R. 1999. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays 21:1004–1010. [DOI] [PubMed] [Google Scholar]
- 43.Schneiter, R., B. Brügger, R. Sandhoff, G. Zellnig, A. Leber, M. Lampl, K. Athenstaedt, C. Hrastnik, S. Eder, G. Daum, F. Paltauf, F. T. Wieland, and S. D. Kohlwein. 1999. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivadon, P., F. Bauer, M. Aigle, and M. Crouzet. 1995. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol. Gen. Genet. 246:485–495. [DOI] [PubMed] [Google Scholar]
- 45.Sivadon, P., M. F. Peypouquet, F. Doignon, M. Aigle, and M. Crouzet. 1997. Cloning of the multicopy suppressor gene SUR7: evidence for a functional relationship between the yeast actin-binding protein Rvs167 and a putative membranous protein. Yeast 13:747–761. [DOI] [PubMed] [Google Scholar]
- 46.Skrzypek, M. S., M. M. Nagiec, R. L. Lester, and R. C. Dickson. 1999. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol. 181:1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slaninová, I., S. Ŝesták, A. Svoboda, and V. Farkaŝ. 2000. Cell wall and cytoskeleton reorganization as the response to hyperosmotic shock in Saccharomyces cerevisiae. Arch. Microbiol. 173:245–252. [DOI] [PubMed] [Google Scholar]
- 48.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takizawa, P. A., J. L. DeRisi, J. E. Wilhelm, and R. D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341–344. [DOI] [PubMed] [Google Scholar]
- 50.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waddle, J. A., T. S. Karpova, R. H. Waterston, and J. A. Cooper. 1996. Movement of cortical actin patches in yeast. J. Cell Biol. 132:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanolari, B., S. Friant, K. Funato, C. Sütterlin, B. J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19:2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]