Abstract
To investigate the role of chromatin remodeling in nucleotide excision repair, we prepared mononucleosomes with a 200-bp duplex containing an acetylaminofluorene-guanine (AAF-G) adduct at a single site. DNase I footprinting revealed a well-phased nucleosome structure with the AAF-G adduct near the center of twofold symmetry of the nucleosome core. This mononucleosome substrate was used to examine the effect of the SWI/SNF remodeling complex on the activity of human excision nuclease reconstituted from six purified excision repair factors. We found that the three repair factors implicated in damage recognition, RPA, XPA, and XPC, stimulate the remodeling activity of SWI/SNF, which in turn stimulates the removal of the AAF-G adduct from the nucleosome core by the excision nuclease. This is the first demonstration of the stimulation of nucleotide excision repair of a lesion in the nucleosome core by a chromatin-remodeling factor and contrasts with the ACF remodeling factor, which stimulates the removal of lesions from internucleosomal linker regions but not from the nucleosome core.
Nucleotide excision repair (excision repair) is a multistep and all-purpose repair system which removes all DNA lesions, including UV photoproducts and alkylated and oxidized bases, from DNA (32, 52). The basic steps of this repair system include damage recognition, dual incision, excision (12), repair synthesis, and ligation. The excision of damage in human cells by dual incision is carried out by six repair factors, RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1 (4, 29, 30). Similarly, the Saccharomyces cerevisiae homologs of these six factors are necessary and sufficient for dual incision (8). The basic enzymology of excision repair in both mammalian and yeast cells has been determined in considerable detail with naked DNA substrates (4, 8, 29, 30, 48). However, the natural substrate of this repair system in vivo is chromatin, and there have been only limited studies on the molecular mechanisms of excision repair of DNA damage in the nucleosome or chromatin in vitro (9, 17, 23, 45).
The nucleosome is the fundamental repeating unit of chromatin and constitutes the first order of DNA compaction in the nucleus. A nucleosome consists of two structurally different parts, the core particle and the linker. The nucleosome core particle consists of about 145 bp of DNA wrapped around the histone octamer. The nucleosome core particles are joined by the linker, consisting of approximately 50 bp of DNA associated with a linker histone to form the “beads-on-a-string” structure (16, 50). Packaging of DNA into the nucleosome has strong negative effects on essentially all DNA transactions, including replication, recombination, repair, and transcription (16, 41, 44); these effects have been best characterized with respect to transcriptional regulation. The development of an in vitro excision repair assay (12) and the reconstitution of the excision reaction in a defined six-factor system (29) have provided the opportunity to investigate the effect of DNA compaction in chromatin on excision repair in defined in vitro systems. With this strategy, it was found that a DNA lesion within the nucleosome core particle was repaired 5- to 10-fold less efficiently than a lesion in naked DNA (9). Molecular analysis of the inhibitory effect of the nucleosome on excision repair revealed that the binding of damage recognition factors RPA, XPA, and XPC was severely hampered in nucleosomal DNA and suggested that the accessibility of damage in chromatin to these factors may be the rate-limiting step in excision repair and that factors which increase this accessibility may modulate excision repair (9).
Most of the factors which increase DNA accessibility in chromatin have been identified by genetic and biochemical tests which affect transcription. Such studies have revealed two major classes of chromatin-modifying (and hence transcription-modulating) factors (for reviews, see references 1, 15, 44, and 46). One class of factors alters DNA-histone interactions through covalent modification of histones by acetylation, phosphorylation, and methylation (36). The other class encompasses several multisubunit complexes which utilize the energy of ATP hydrolysis to alter DNA-histone interactions. ATP-dependent chromatin-remodeling complexes are further classified into three groups based on the ATPase subunit of the complex: SWI2/SNF2, ISWI, and Mi (1, 15, 46). At least for certain genes, SWI/SNF is recruited to the particular promoter by a transcriptional activator and, upon arrival, remodels the local chromatin structure to facilitate the formation of a preinitiation complex (54). Although the two general classes of chromatin-remodeling factors, in combination with transcriptional activators, regulate transcription by a variety of methods (5), the details of the mechanisms are beyond the scope of this work. Of relevance to our study are two recent reports which implicate ATP-dependent chromatin-remodeling complexes in recombination and in repair. In one study, it was found that SWI/SNF enhanced cleavage of the V(D)J recombination signal sequence in the mononucleosome by the RAG1/RAG2 recombinase (19). In the second study, it was found that the ACF chromatin-remodeling complex (an ISWI family member) from Drosophila enhanced the excision of a DNA lesion by human excision nuclease only when the lesion was in the linker region of a dinucleosome and had no effect on lesions in the nucleosome core (45). Since the majority of DNA lesions are in the nucleosome core simply because it is larger than the linker region, it is important to know whether any chromatin-remodeling factor alters the accessibility of nucleosomal core DNA to human excision nuclease.
In the present study, we used a mononucleosome substrate containing an acetylaminofluorene-guanine (AAF-G) adduct to test the effect of the SWI/SNF remodeling complex on human excision nuclease. We found that the 200-bp “random-sequence” DNA is assembled into the mononucleosome with a unique rotational setting and in a manner which places the lesion in the center of the nucleosome core particle. Damage recognition factors RPA, XPA, and XPC facilitate the remodeling activity of SWI/SNF, which in turn enhances the overall excision activity of the six-factor excision nuclease. These findings constitute the first evidence for the participation of chromatin-remodeling factors in the removal of DNA damage from the nucleosome core particle by the human nucleotide excision repair system.
MATERIALS AND METHODS
DNA substrate.
The substrate was a 200-bp duplex containing a single AAF-G adduct and a radiolabel either at the 11th phosphate 5′ to the lesion or at the 5′ terminus of the damaged strand (Fig. 1). The substrate was prepared as follows. A 20-nucleotide-long oligomer containing a single G residue was treated with N-acetoxy-2-acetylaminofluorene (Chemsyn Science Laboratories, Lenexa, Kans.) as described previously (22), and the modified oligomer was purified from the unmodified duplex through a nondenaturing polyacrylamide gel (20% in 1× Tris-borate-EDTA [TBE]). The 20-mer with the AAF-G adduct was labeled at the 5′ terminus with [γ-32P]ATP and polynucleotide kinase and used to prepare circular plasmid DNA with AAF-G as described previously (10, 12). The plasmid DNA was then digested with HinPI and HindIII restriction endonucleases to generate a 200-bp duplex containing AAF-G in a central location (Fig. 1). The adducted fragment was separated from the other restriction fragments on a nondenaturing polyacrylamide gel (5% in 1× TBE). The control DNA was prepared by the same procedure with an unmodified 20-mer as the starting material.
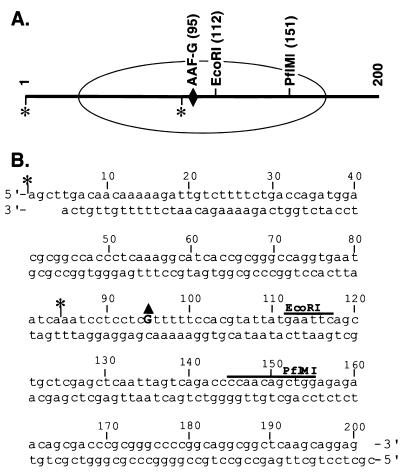
FIG. 1.
Nucleosome substrate for excision nuclease. The substrate is 200 bp long. (A) Site of AAF-G damage (diamond), restriction sites used for testing remodeling, and approximate location of the area covered by the nucleosome core (ellipse). The substrate was radiolabeled with 32P at either of two sites (asterisk). The internally labeled substrate was used for excision and remodeling assays, and the terminally labeled duplex was used for footprinting analysis. (B) Sequence with appropriate landmarks. The triangle indicates AAF-G.
For footprinting experiments, the AAF-G-containing 20-mer was phosphorylated with cold ATP and used to prepare plasmid DNA. The plasmid was then digested with HindIII and dephosphorylated, and the 5′ terminus was labeled with [γ-32P]ATP by standard procedures. Following labeling, the plasmid was digested with HinPI, and the resulting 200-bp HindIII-HinPI fragment was purified through a nondenaturing polyacrylamide gel (5% in 1× TBE) and isolated by electroelution.
Proteins.
The core histone proteins H2A, H2B, H3, and H4 (53) and the core human excision nuclease factors RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1 (2, 27, 30, 31) were purified as described previously. The yeast SWI/SNF protein complex was a generous gift from Craig L. Peterson (University of Massachusetts, Worcester) and was prepared as described previously (25).
Nucleosome assembly.
To prepare nucleosomes, 100 fmol of the 200-bp AAF-G-containing DNA fragment and 1 μg of salmon sperm DNA were incubated with core histone proteins at a 1:1 molar ratio of octamer to nucleosome unit of DNA and assembled into nucleosomes as described previously (35). Thus, the nucleosome substrate prepared in this manner is a mixture of 32P-labeled nucleosomes containing AAF-G and unlabeled nucleosomes from salmon sperm DNA. Nucleosome assembly was monitored by gel mobility shift experiments according to published methods (18); more than 95% of the AAF-G DNA was assembled into nucleosomes.
Footprinting.
DNase I footprinting was carried out according to published methods (51). Briefly, 20 fmol of nucleosomal or naked DNA substrate was treated with DNase I (Promega) and directly loaded onto a nondenaturing polyacrylamide gel (5% in 1× TBE). The nucleosome bands were located by autoradiography of the wet gel and excised, and the DNA was purified from the gel slice. The DNA was then analyzed by denaturing polyacrylamide gel (5% in 2× TBE) electrophoresis.
Excision assay.
The excision assay was carried out as described previously (12, 30) with minor modifications. Briefly, 0.16 nM 32P-labeled nucleosomal or naked DNA, 1.6 μg of unlabeled salmon sperm DNA nucleosomes/ml, purified excision repair factors (42 nM RPA, 6.5 nM XPA, 2.2 nM XPC, 16 nM TFIIH, 3 nM XPG, and 6 nM XPF-ERCC1) and, when needed, 0.48 nM SWI/SNF were mixed in excision buffer (32 mM HEPES-KOH [pH 7.9], 64 mM KCl, 6.4 mM MgCl2, 0.24 mM EDTA, 0.8 mM dithiothreitol, 2 mM ATP, 200 μg of bovine serum albumin/ml, 5.5% glycerol, and 0.05% NP-40 in a 12.5-μl volume) and incubated at 30°C for various times. The reaction products were extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a denaturing polyacrylamide gel (8% in 2× TBE). The level of excision was determined by measuring the amount of radioactivity in the bands corresponding to the excision products and unexcised substrate with PhosphorImager analysis and the ImageQuant system (Molecular Dynamics) and was plotted as the percentage of excision or the amount of excision products.
Coupled remodeling-restriction assay.
The coupled remodeling-restriction assay measures the effect of nucleosome-remodeling factors on the accessibility of nucleosomal DNA by digestion of the nucleosome with a restriction enzyme in the absence or in the presence of a particular remodeling factor (24). To test the accessibility of the nucleosomal DNA used in our experiments, 0.16 nM radiolabeled nucleosome substrate and 1.6 μg of unlabeled nucleosomes/ml were incubated with either EcoRI or PflMI restriction endonuclease and, when needed, 0.48 nM SWI/SNF in excision buffer at 30°C for 1 h. EcoRI cuts near the center and PflMI cuts near the end of nucleosomal core DNA; hence, the levels of incisions obtained with these restriction enzymes are indicative of the accessibility of nucleosomal DNA in these two locations (Fig. 1).
To test the effect of repair factors implicated in damage recognition on SWI/SNF remodeling activity, a 0.16 nM concentration of 32P-labeled nucleosomes was first incubated with EcoRI for 50 min at 30°C to eliminate the high-DNA-accessibility subpopulation of nucleosomes (24). Then, the reaction mixture was diluted twofold with excision buffer; 16 pM SWI/SNF, 6.5 nM XPA, 42 nM RPA, and 2.2 nM XPC were added; and the reaction mixture was further incubated at 30°C for various times. The reaction products were deproteinized by phenol-chloroform extraction, ethanol precipitated, and analyzed on a denaturing polyacrylamide gel (5% in 2× TBE). The level of restriction enzyme digestion was quantified with PhosphorImager analysis and the ImageQuant system.
RESULTS
Characterization of the nucleosome substrate.
The nucleosome substrate was prepared by the salt dilution method with a 200-bp duplex DNA containing AAF-G and core histones isolated from HeLa cells (35). The DNA contained 32P label either at the 11th phosphodiester bond 5′ to the AAF-G or at the 5′ terminus of the AAF-G-containing strand (Fig. 1). After assembly into nucleosomes, more than 95% of the DNA was found to be in the nucleosomes, as determined by nondenaturing polyacrylamide gel electrophoresis (data not shown).
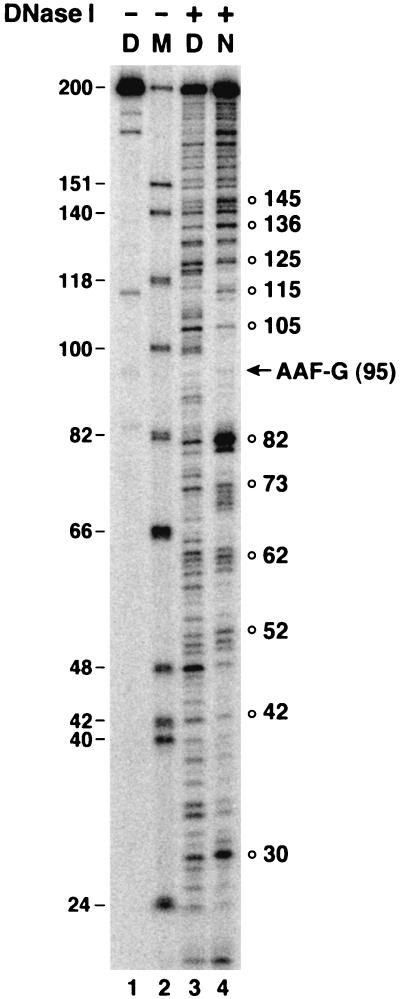
In reconstituting nucleosomes with DNA fragments substantially longer than 146 bp, there is a concern of obtaining a heterogeneous nucleosome population with multiple nucleosome positions along the DNA unless the DNA contains specific nucleosome-positioning sequences, such as the TG motif and the Xenopus 5S rRNA gene (11, 33, 51). Our DNA substrate was not specifically designed to have such positioning sequences. However, when the nucleosomes prepared with this DNA were analyzed by DNase I footprinting, the cleavage pattern revealed a strong 10-base periodicity over the region of bp 30 to 145 (Fig. 2), indicating a rather homogeneous nucleosome population in which the histone octamer is more or less symmetrically positioned relative to the two ends of the duplex. At the boundaries of the 10-base periodic region, the DNase I cleavage pattern of nucleosomal DNA is different from that of naked DNA, although the 10-base periodicity is not that obvious, indicating weaker interactions with the nucleosome core in the area close to the linker DNA. Importantly, these data show that we have obtained an essentially homogeneous nucleosome population in which AAF-G is “buried” within the nucleosome core particle and that this preparation is suitable for testing the effect of a remodeling factor on the activity of excision nuclease on damage within the nucleosome core.
FIG. 2.
DNase I footprint of AAF-G mononucleosome. Naked DNA (D) and nucleosome DNA (N) were treated with DNase I for 3 min and separated on a denaturing polyacrylamide gel (5% in 2× TBE) along with size markers (M). The position of AAF-G is indicated by an arrow, and the DNase I-hypersensitive sites in nucleosomal DNA with the 10-nucleotide periodicity are indicated by open circles.
SWI/SNF stimulates the activity of human excision nuclease on a lesion within the nucleosome core particle.
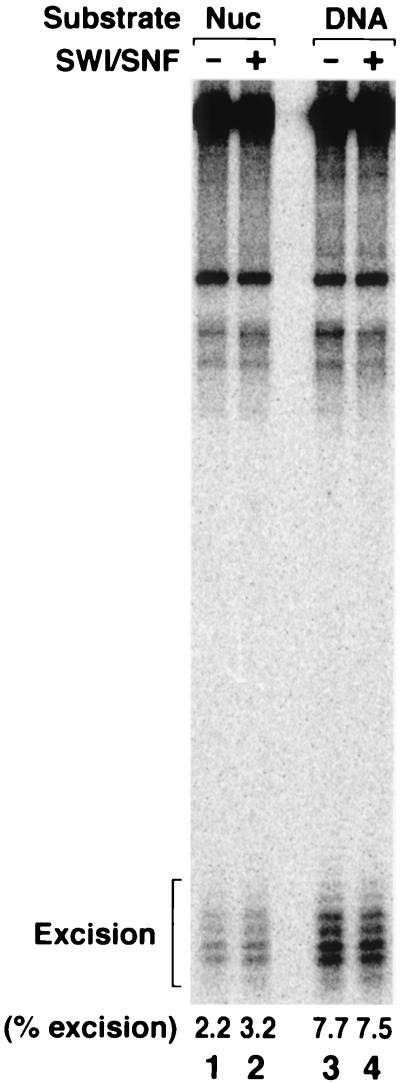
The positioning of the AAF-G lesion within the nucleosome drastically inhibited its excision (Fig. 3), as was previously observed for a (6-4) photoproduct (9, 45), although this inhibition was somewhat less severe than that for the (6-4) photoproduct. These two lesions are equally good substrates for human excision nuclease (14, 28). Therefore, the differential inhibition was likely due to other factors. First, it is possible that the nucleosomes used in this study and a previous study (9) have different levels of quality. Second, the lengths of the substrates and the sequences used in the two studies are different. Finally, it is conceivable that, for structural reasons, the AAF-G adduct is more accessible to excision nuclease than the (6-4) photoproduct in nucleosomal DNA but not in naked DNA. Importantly, however, the addition of SWI/SNF to the reaction mixture stimulated excision from the nucleosomal substrate (Fig. 3, lanes 1 and 2) but had no effect or had a slightly inhibitory effect on naked DNA (Fig. 3, lanes 3 and 4).
FIG. 3.
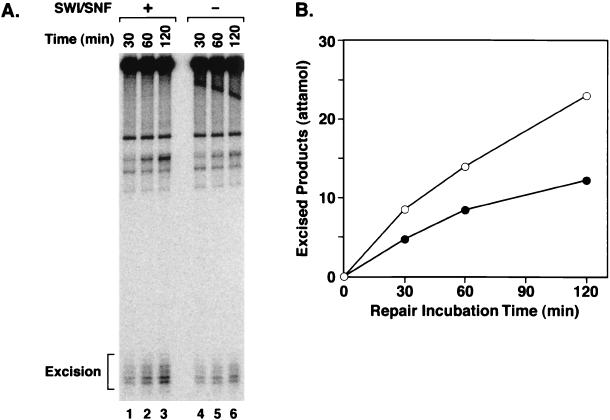
Effect of the SWI/SNF remodeling factor on excision nuclease. AAF-G nucleosome (Nuc) or naked DNA was incubated with human excision nuclease for 90 min at 30°C in the absence or presence of 0.48 nM SWI/SNF. The reaction products were analyzed on a denaturing polyacrylamide gel (8% in 2× TBE). The positions of the excision products and the percentage of the damage excised are indicated.
The stimulating effect of SWI/SNF on the excision of damage from nucleosomal DNA was analyzed in more detail by performing a kinetic assay. As shown in Fig.4, under our assay conditions, SWI/SNF stimulated human excision nuclease activity for AAF-G in the nucleosome core particle by about a factor of 2, indicating that this remodeling factor, in contrast to ACF (45), may play an important role in nucleosomal DNA repair.
SWI/SNF remodeling of the AAF-G nucleosome core substrate.
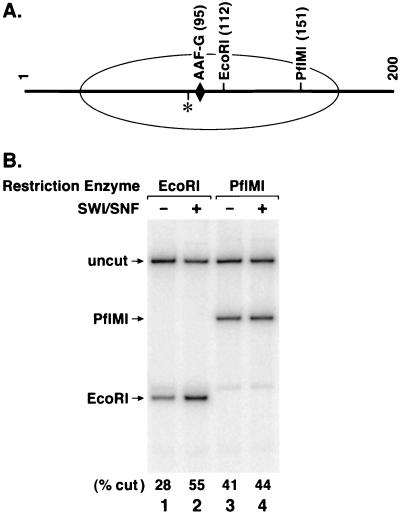
A likely explanation for the stimulation of human excision nuclease by SWI/SNF is that this remodeling factor increases the accessibility of damage within the nucleosome core to excision nuclease. To test the effect of SWI/SNF on the AAF-G nucleosome core substrate, we used a coupled remodeling-restriction enzyme digestion assay commonly used to monitor nucleosome-remodeling activity (24). In this assay, the effect of a remodeling factor on nucleosome structure is monitored by the accessibility of the nucleosomal DNA to restriction enzymes which digest DNA at various positions along the nucleosome core DNA (24). With our particular substrate, EcoRI and PflMI were convenient enzymes for probing DNA accessibility within the center of the core particle and near the linker region, respectively (Fig. 5A). As shown in Fig. 5B, digestion of nucleosomal DNA with EcoRI was severely inhibited relative to digestion with PflMI (Fig. 5B, lanes 1 and 3). Importantly, the addition of SWI/SNF strongly stimulated EcoRI digestion but had only a marginal effect on PflMI digestion (Fig. 5B, lane 1 versus lane 2 and lane 3 versus lane 4).
FIG. 5.
Remodeling of the AAF-G nucleosome by SWI/SNF. An internally labeled AAF-G nucleosome was incubated with either EcoRI or PflMI in excision buffer in the absence or presence of 0.48 nM SWI/SNF for 1 h at 30°C. Following deproteinization, the reaction products were separated on a denaturing polyacrylamide gel (5% in 2× TBE). (A) Site of AAF-G damage (diamond), restriction sites for EcoRI and PflMI, location of 32P label (asterisk), and approximate location of the area covered by the nucleosome core (ellipse). (B) Autoradiogram of a denaturing polyacrylamide gel. Positions of uncut substrate and restriction enzyme digestion products are indicated by arrows. The levels of digestion as percentages of input substrate are given at the bottom of the gel.
These findings are consistent with the known properties of SWI/SNF, which increases the accessibility of DNA within the nucleosome core to enzymes but which has a lesser effect on DNA near the entry and exit from the nucleosome core; this DNA appears to have weaker interactions than DNA in the center of the core particle (24). It must be noted, however, that under the reaction conditions used for the experiment shown in Fig. 5B, we observed essentially a similar effect on unmodified DNA (data not shown); these results indicate that SWI/SNF may increase the accessibility of nucleosomal DNA to excision nuclease whether or not the DNA is damaged. However, the difference between damaged DNA and undamaged DNA, from the perspective of excision nuclease, is quantitative and not qualitative, as excision nuclease attacks even undamaged DNA at a low but significant efficiency (3). Thus, to search for a damage-specific interaction of SWI/SNF with nucleosomes and the potential effect of repair factors on this interaction, we performed the coupled remodeling-restriction assay with limiting concentrations of SWI/SNF in the following experiments.
Effects of repair factors on SWI/SNF remodeling activity.
During transcription, transcriptional activators, histone-modifying enzymes, and ATP-dependent remodeling enzymes are recruited to the promoter region in various orders of assembly, depending on the particular gene, to promote the formation of the preinitiation complex by general transcription factors and RNA polymerase II (5, 54). Thus, we wished to determine the effects of repair factors involved in damage recognition on chromatin remodeling by SWI/SNF in order to gain insight into the mechanism of stimulation of excision repair by SWI/SNF. We conducted remodeling studies in the presence of the repair factors XPA, RPA, and XPC with 1/30 the concentration of SWI/SNF used in the experiment shown in Fig. 5B. This level of SWI/SNF itself was determined empirically to have no measurable effect on nucleosomal DNA accessibility, as analyzed by the coupled remodeling-restriction enzyme assay.
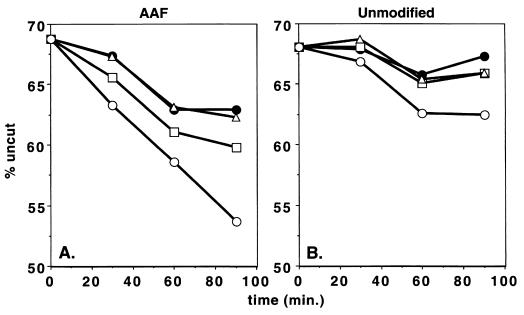
When either control (undamaged) or AAF-G nucleosomes were incubated with EcoRI, biphasic digestion kinetics were observed, as has been reported for a nucleosomal array substrate; this result is indicative of two populations of nucleosomes (24), in one of which the restriction site is not occluded by the nucleosome and hence the DNA is rapidly cleaved. In agreement with an earlier report (24), we found this rapid cleavage phase to be essentially complete in 50 min, after which the rate of digestion was reduced to about 10% the initial rate. Thus, we limited our analysis of the effects of SWI/SNF plus repair factors on EcoRI digestion to the slower phase of the biphasic digestion to detect any damage-specific effects of these two classes of factors on the accessibility of nucleosomal DNA. In the fast phase of the EcoRI digestion, the AAF-G nucleosome and the control nucleosome were digested at identical rates, with a first-order rate constant (k1) of 0.6 min−1. In the second, slow phase, however, there was a marked difference in the rates of digestion of the two substrates (Fig. 6): the k1 for the AAF-G nucleosome was 0.07 min−1, and the k1 for the control nucleosome was 0.02 min−1. These results indicate that AAF-G by itself causes a structural change in the nucleosome so as to make it more accessible to a restriction endonuclease.
FIG. 6.
Effect of damage recognition factors on SWI/SNF remodeling activity. AAF-G (AAF) and control (Unmodified) nucleosomes were incubated for 50 min at 30°C with EcoRI before the addition of 16 pM SWI/SNF at time zero and three repair factors, RPA, XPA, and XPC, as indicated below, and incubation was continued for the indicated time periods. Reaction products were analyzed on denaturing polyacrylamide gels (5% in 2× TBE). The data points are averages of three independent experiments, and the standard errors for each data point were less than 11% the value for each data point. Symbols: •, no SWI/SNF or repair factors; ▵, SWI/SNF but no repair factors; □, repair factors (RPA, XPA, and XPC) but no SWI/SNF; ○, SWI/SNF and repair factors.
When SWI/SNF, the combination of the three repair factors implicated in damage recognition (RPA, XPA, and XPC), and the ensemble of the remodeling and damage recognition factors were tested for their effects on nucleosomal DNA accessibility, the following results were obtained (Fig. 6). With AAF-G nucleosomes, repair factors alone increased k1 from 0.07 to 0.10 min−1; SWI/SNF at the concentration used had no effect; but the combination of repair factors plus SWI/SNF increased k1 to 0.20 min−1. Thus, it appears that the repair factors increase the remodeling activity of SWI/SNF in the presence of limiting concentrations of the remodeling factor. With control nucleosomes, the overall rate of the slow phase was slow (k1, 0.02 min−1) relative to that with the damaged nucleosomes and, unlike that with the AAF-G nucleosomes, it was not affected by the repair factors alone or SWI/SNF alone but was enhanced from 0.02 to 0.07 min−1 by the combination of the two under these experimental conditions. Thus, it appears that the three repair factors involved in damage recognition recruit SWI/SNF to both damaged and undamaged nucleosomes but may do so more efficiently with damaged nucleosomes and, as a consequence, have a greater effect on remodeling and repair of damaged DNA than remodeling and futile repair of undamaged DNA.
Finally, we wished to determine which of the three repair factors was responsible for stimulating SWI/SNF-independent and -dependent increases in nucleosomal DNA accessibility by using a single factor or two-factor combinations in these experiments. The results obtained under these conditions were rather minor and variable from experiment to experiment (data not shown), preventing us from drawing a firm conclusion. We suspect that all three factors aid in SWI/SNF remodeling activity in a cooperative manner.
DISCUSSION
Chromatin structure affects every aspect of DNA-protein interactions in eukaryotes, including transcription, replication, recombination, and repair. In general, chromatin inhibits these processes by interfering with enzymes which mediate these reactions (7, 16, 20, 21, 41, 44). Recent work in the transcription field has identified many factors which increase the accessibility of DNA in chromatin to the transcription machinery involved in initiation and elongation. No such specific factors which promote other DNA transactions have been described. However, it has been found that some of the factors identified as transcription accessibility factors have more general roles in promoting the functions of other DNA-acting enzymes as well. For example, SWI/SNF has been found to enhance V(D)J cleavage by RAG1/RAG2 recombinase (19) by weakening the DNA-protein interactions within the nucleosome. Thus, it is possible that the other histone-modifying and nucleosome-remodeling complexes are general accessibility factors for all DNA-acting enzymes. Indeed, in this study, we show that SWI/SNF, which was originally identified as a chromatin-remodeling transcription factor, in addition to stimulating RAG1/RAG2 recombinase, stimulates human excision nuclease as well. To put our findings in the broader context of chromatin remodeling and repair, in the following we briefly discuss the effects of DNA damage on nucleosome stability, summarize current knowledge on nucleotide excision repair of nucleosomal DNA in vitro, and present a plausible model for the cooperation of nucleosome-remodeling factors and excision repair proteins during the repair of damaged chromatin.
Effect of AAF-G on nucleosome stability.
When we tested the AAF-G and control nucleosomes for incision by EcoRI, which cuts near the center of nucleosomal core DNA, we observed biphasic digestion kinetics for both. In the rapid, first phase, the rates for damaged and undamaged nucleosomes were essentially identical. However, in the slower, second phase, the damaged nucleosomes were digested at a 3.5-fold higher rate than the control nucleosomes. It has been reported that DNA damage can alter the stability of nucleosomes, and it may decrease or increase the stability depending on the type of lesion; it was found that while UV damage made nucleosomes unstable, benzo[a]pyrene-G adducts increased nucleosome stability (26). Thus, it appears that the AAF-G adduct, at least at the specific position at which it is located within our nucleosomes, has more of a UV lesion-type destabilizing effect, as measured by accessibility to a restriction endonuclease. It is interesting, however, that in contrast to EcoRI digestion, the excision repair of nucleosomes exhibited linear kinetics, indicating that nucleosome instability is a relative term and may or may not be apparent, depending on the method used to probe it. In a related matter, the homogeneous phasing of our 200-bp random-sequence DNA within the nucleosome core deserves some comments. Traditionally, uniformly phased nucleosomes are assembled in vitro with DNAs containing specific nucleosome-positioning sequences, such as a TG motif and the popular Xenopus 5S RNA gene fragment (11, 33, 51). The fact that we obtained strong phasing with our random sequence suggests that the local denaturation caused by AAF-G might have contributed to the uniform positioning of our substrate within the nucleosome. Another possible explanation is that the 200-bp fragment that we chose for our study exhibits certain features which are not obvious but which nevertheless exert a strong positioning effect.
Excision repair of nucleosomal DNA in vitro.
Elegant in vivo work with a variety of methods showed that packaging of DNA into chromatin had an inhibitory effect on excision repair (34, 39) and that the kinetics of damage removal depended on the location of the damage relative to the nucleosome, with excision, in general, being slower in the nucleosome core particle than in the linker region (42). However, in vivo studies could not address the effect of chromatin-remodeling factors on excision repair. Insight into this question can be unambiguously gained only by in vitro studies. Early in vitro studies with randomly damaged minichromosomes and human cell extracts essentially confirmed the in vivo data by revealing an overall inhibition of repair (37, 49). Some insight into the effect of nucleosome structure on eukaryotic excision repair has been gained by using mono- or dinucleosomes with either a single lesion or multiple lesions. In studies with nuclear extracts from Xenopus oocytes and either nucleosomes containing random UV damage (23) or a single cyclobutane-thymine dimer (17), it was found that even though there was some variability in the efficiencies of repair of photoproducts at different sites within the mononucleosome core particle, repair was in general very efficient. When compared to naked DNA, the level of inhibition by nucleosome DNA at different sites varied from fivefold to negligible; moreover, at a majority of the sites, all of the naked DNA and about 50% of the nucleosomal DNA were repaired within 2 h (17, 23). These values represent extremely high repair efficiencies never achieved with mammalian cell extracts, even though the basic mechanisms of excision repair in Xenopus oocytes and mammalian cells are the same and DNA microinjected into Xenopus oocytes is repaired with a low efficiency comparable to that seen with mammalian cell extracts (38). Despite their significant contribution to the understanding of nucleosome repair, Xenopus oocyte nuclear extracts so far have not been amenable to addressing the role of remodeling factors in excision repair.
The development of the highly specific and sensitive excision assay (12) and the reconstitution of human excision nuclease from six general repair factors (30) made it possible to investigate the effects of specific factors on nucleosomal DNA in a completely defined system (9). With this system, it was found that a (6-4) photoproduct in the nucleosome core was excised at about 10% the rate of naked DNA (9). This finding was confirmed and extended in a subsequent study which also showed that the excision of a (6-4) photoproduct in a dinucleosome substrate was equally inhibited whether the lesion was in the nucleosome core or the linker region (45). Significantly, it was found that the ACF remodeling factor stimulated excision from the linker region by about a factor of 2 but had no effect on excision from the nucleosome core (45). In this study, we show that another member of the group of ATP-dependent remodeling enzymes, SWI/SNF, stimulates the excision of AAF-G from the nucleosome core particle. The fact that about 30% of the EcoRI site in our nucleosome preparation is relatively accessible to the restriction enzyme suggests that this fraction may be more accessible to excision nuclease as well; in this case, the “real” inhibition of excision of the AAF-G adduct by the nucleosome is more severe than we have estimated and therefore the stimulatory effect of SWI/SNF is more drastic than we have calculated based on the results obtained with the “heterogeneous” nucleosome populations. It should be noted that the stimulatory effect of SWI/SNF on damage excision from the nucleosome core is not restricted to the AAF-G adduct. We saw similar stimulation with the nucleosome substrate assembled with a 136-bp duplex containing a (6-4) photoproduct in another study (data not shown).
We do not know the reason for the differential effects on excision repair of these two remodeling enzymes. However, there are significant differences between the subunit compositions and biochemical properties of the two remodeling factors. ACF is a member of the ISWI group of chromatin-remodeling factors; it is made up of 140-kDa (ISWI) and 180-kDa (ACF1) subunits (13, 43) and is thought to loosen histone-DNA contacts in a manner that allows the nucleosome to slide. In fact, ACF was identified as a factor that creates an ordered array of nucleosomes with uniform spacing between the nucleosomes (13, 43). It is therefore quite likely that the stimulatory effect of ACF on the excision repair of a dinucleosome substrate is due to the increased size of the linker between the two nucleosomes (45). SWI/SNF is a member of the SWI2 group of remodeling factors; it is made up of 11 or 12 subunits and is thought to convert a nucleosome into a stably remodeled status, or active form, without actually dissociating DNA and histones. Thus, it is possible that the conversion of a nucleosome into this stably remodeled active form provides the time necessary for the assembly of excision nuclease and the repair of damage in the core particle. In summary, it appears that the location of damage along chromatin may determine which remodeling factor facilitates excision repair, with ACF working at the linker region and SWI/SNF working at the core particle. It must be noted, however, that SWI/SNF may stimulate excision at the linker region as well. This possibility was not tested in our study. Moreover, repair may be significantly affected by histone-modifying enzymes, acetylases, and methyltransferases, which will be the subjects of future studies.
Model for repair of nucleosomal DNA.
The simplest model for the activation of transcription by chromatin-remodeling factors posits that transcriptional activators bind upstream of target genes and then recruit histone-modifying and ATP-dependent chromatin-remodeling enzymes to facilitate the assembly of the transcription machinery and the subsequent transcription initiation and elongation processes (54). However, recent studies have shown that activators, chromatin-remodeling factors, general transcription factors, and RNA polymerase II may assemble at the target site in a variety of orders of assembly, depending on the particular gene (5). It is conceivable that the same may also be applicable to excision repair. However, the results reported in this study are more consistent with the prototypical mechanism of remodeling factor action in ensuring DNA accessibility. We found that the three proteins involved in damage recognition facilitate the remodeling activity of SWI/SNF and thus may be assumed to be the functional analogs of transcriptional activators which recruit remodeling factors to target genes. Admittedly, in our study, the damage recognition factors facilitated the action of SWI/SNF for both undamaged and damaged nucleosomes; therefore, a question might be raised as to whether such an effect of damage recognition factors has enough specificity to facilitate the repair of nucleosomal DNA. However, we do not think that this is a serious concern, because excision nuclease is an all-purpose repair enzyme which excises lesions ranging from bulky adducts to minor modifications to unmodified DNA (3); hence, damage recognition factors which recruit SWI/SNF to damaged nucleosomes are expected to recruit the remodeling factor to undamaged nucleosomes as well.
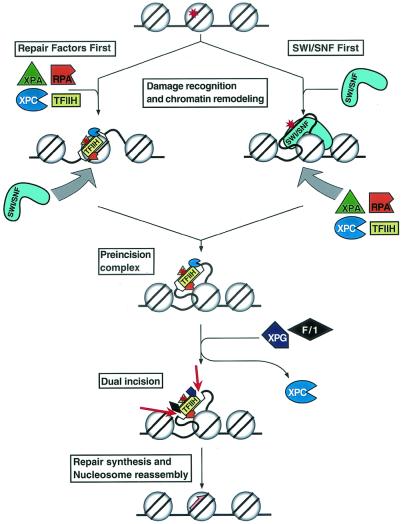
Within these general parameters of the functions of excision nuclease and remodeling factors, then, we propose the following model for the repair of nucleosomal DNA (Fig. 7). Damage recognition factors RPA, XPA, and XPC locate the damage and may facilitate the recruitment of SWI/SNF individually or after forming preincision complex 1 (PIC1) with TFIIH. A complex of the three damage binding proteins has not been detected (29, 48), and inclusion of TFIIH in the remodeling reaction had a marginal effect above the level achieved with the three factors alone (data not shown). Thus, recruitment could be mediated by RPA, XPA, RPA-XPA, and XPC in isolation or after forming PIC1. This intermediate is rather unstable (30a, 48a) and may be stabilized by SWI/SNF, which locally remodels the nucleosome and, in doing so, allows for the tighter binding of PIC1 and facilitates the entry of XPG and XPF-ERCC1 to form PIC2 and PIC3, respectively, leading to dual incision and excision of the 24- to 32-mer carrying the damaged base. However, our data do not eliminate the possibility that damaged nucleosomes, especially those with reduced stability, are more accessible to SWI/SNF which remodels the nucleosome and subsequently recruits the repair factors. During repair synthesis, nucleosomes may disassemble and then reassemble on repaired DNA with the aid of chromatin assembly factor (6). It must be noted, however, that this is a working model which does not encompass all the known factors of excision repair. For example, damage-specific binding proteins, such as DDB (damaged DNA binding protein) and high-mobility group proteins, may be important factors acting in the initial stages of damage recognition in chromatin (40, 47, 55) and hence may play a role in the recruitment of remodeling factors. Further studies are needed to address this issue as well as the very important question of the role of histone-modifying enzymes in excision repair.
FIG. 7.
Model for the role of SWI/SNF in excision repair. Our data suggest that repair factors recruit SWI/SNF to lesion sites to remodel the nucleosome (left arm) but do not exclude the alternative pathway, in which remodeling by SWI/SNF first accelerates the assembly of the repair factors to form PIC1 (right arm). SWI/SNF may contribute to the subsequent steps, PIC2 and PIC3 formation, as well. Circles indicate histones, asterisks indicate DNA damage, and the half arrow indicates repair synthesis. F/1, XPF/ERCC1.
FIG. 4.
Effect of SWI/SNF on the kinetics of excision of AAF-G from the nucleosome core by human excision nuclease. The AAF-G nucleosome was incubated with six-factor reconstituted human excision nuclease in the absence or presence of 0.48 nM SWI/SNF for the indicated times at 30°C. The reaction products were separated on a denaturing polyacrylamide gel (8% in 2× TBE). The level of excision was quantified with PhosphorImager analysis. (A) Autoradiogram of a kinetics experiment. (B) Plot of average data from three independent experiments, including the one shown in panel A. Symbols: •, absence of SWI/SNF; ○, presence of SWI/SNF. The standard errors were less than 15% the value for each data point.
Acknowledgments
This work was made possible through a generous gift of SWI/SNF from C. L. Peterson and P. Horn (University of Massachusetts, Worcester). We thank Joyce T. Reardon and Mark E. Branum for comments on the manuscript.
This work was supported by NIH grant GM32833.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ′chromatin remodeling' mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Bessho, T., A. Sancar, L. H. Thompson, and M. P. Thelen. 1997. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J. Biol. Chem. 272:3833-3837. [DOI] [PubMed] [Google Scholar]
- 3.Branum, M. E., J. T. Reardon, and A. Sancar. 2001. DNA repair excision nuclease attacks undamaged DNA. A potential source of spontaneous mutations. J. Biol. Chem. 276:25421-25426. [DOI] [PubMed] [Google Scholar]
- 4.Evans, E., J. G. Moggs, J. R. Hwang, J. M. Egly, and R. D. Wood. 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry, C. J., and C. L. Peterson. 2002. Unlocking the gates to gene expression. Science 295:1847-1848. [DOI] [PubMed] [Google Scholar]
- 6.Gaillard, P.-H. L., E. M.-D. Martini, P. D. Kaufman, B. Stillman, E. Moustacchi, and G. Almouzni. 1996. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86:887-896. [DOI] [PubMed] [Google Scholar]
- 7.Green, C. M., and G. Almouzni. 2002. When repair meets chromatin. EMBO Rep. 3:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270:12973-12976. [DOI] [PubMed] [Google Scholar]
- 9.Hara, R., J. Mo, and A. Sancar. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20:9173-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara, R., C. P. Selby, M. Liu, D. H. Price, and A. Sancar. 1999. Human transcription release factor 2 dissociates RNA polymerase I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 274:24779-24786. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, J. J., and A. P. Wolffe. 1992. Transcription factor interaction with nucleosomal DNA. Bioessays 14:597-603. [DOI] [PubMed] [Google Scholar]
- 12.Huang, J. C., D. L. Svoboda, J. T. Reardon, and A. Sancar. 1992. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. USA 89:3664-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 14.Kazantsev, A., D. Mu, A. F. Nichols, X. Zhao, S. Linn, and A. Sancar. 1996. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc. Natl. Acad. Sci. USA 93:5014-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingston, R. E., and G. L. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 17.Kosmoski, J. V., E. J. Ackerman, and M. J. Smerdon. 2001. DNA repair of a single UV photoproduct in a designed nucleosome. Proc. Natl. Acad. Sci. USA 98:10113-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosmoski, J. V., and M. J. Smerdon. 1999. Synthesis and nucleosome structure of DNA containing a UV photoproduct at a specific site. Biochemistry 38:9485-9494. [DOI] [PubMed] [Google Scholar]
- 19.Kwon, J., K. B. Morshead, J. R. Guyon, R. E. Kingston, and M. A. Oettinger. 2000. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell 6:1037-1048. [DOI] [PubMed] [Google Scholar]
- 20.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 21.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 22.Lindsley, J. E., and R. P. P. Fuchs. 1994. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry 33:764-772. [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., and M. J. Smerdon. 2000. Nucleotide excision repair of the 5S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem. 275:23729-23735. [DOI] [PubMed] [Google Scholar]
- 24.Logie, C., and C. L. Peterson. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16:6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logie, C., and C. L. Peterson. 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304:726-741. [DOI] [PubMed] [Google Scholar]
- 26.Mann, D. B., D. L. Springer, and M. J. Smerdon. 1997. DNA damage can alter the stability of nucleosome: effects are dependent on damage type. Proc. Natl. Acad. Sci. USA 94:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga, T., C. H. Park, T. Bessho, D. Mu, and A. Sancar. 1996. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 271:11047-11050. [DOI] [PubMed] [Google Scholar]
- 28.Mu, D., E. Bertrand-Burggraf, J.-C. Huang, R. P. P. Fuchs, and A. Sancar. 1994. Human and E. coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 22:4869-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 30.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 30a.Mu, D., M. Wakasugi, D. S. Hsu, and A. Sancar. 1997. Characterization of reaction intermediates of human excision repair nuclease. 1997. J. Biol. Chem. 272:28971-28979. [DOI] [PubMed] [Google Scholar]
- 31.Reardon, J. T., D. Mu, and A. Sancar. 1996. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J. Biol. Chem. 271:19451-19456. [DOI] [PubMed] [Google Scholar]
- 32.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 33.Shrader, T. E., and D. M. Crothers. 1989. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 86:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smerdon, M. J., and F. Thoma. 1990. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell 61:675-684. [DOI] [PubMed] [Google Scholar]
- 35.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferase complex stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strahl, B. D., and D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 37.Sugasawa, K., C. Masutani, and F. Hanaoka. 1993. Cell-free repair of UV-damaged simian virus 40 chromosome in human cell extracts. I. Development of a cell-free system detecting excision repair of UV-irradiated SV40 chromosomes. J. Biol. Chem. 268:9098-9104. [PubMed] [Google Scholar]
- 38.Svoboda, D. L., J-. S. Taylor, J. E. Hearst, and A. Sancar. 1993. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 268:1931-1936. [PubMed] [Google Scholar]
- 39.Tanaka, S., M. Livingston-Zatchej, and F. Thoma. 1996. Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosome in the chromosomal context. J. Biol. Chem. 257:919-934. [DOI] [PubMed] [Google Scholar]
- 40.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoma, F. 1999. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18:6585-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tjisterman, M., R. De Pril, J. G. Tasseron-DeJong, and J. Brouwer. 1999. RNA polymerase II transcription suppresses nucleosomal modification of UV-induced (6-4) photoproducts and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol. 19:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 44.Tyler, J. K., and J. T. Kadonaga. 1999. The “dark side”of chromatin remodeling: repressive effects on transcription. Cell 99:443-446. [DOI] [PubMed] [Google Scholar]
- 45.Ura, K., M. Araki, H. Saeki, C. Masutani, T. Ito, S. Iwai, T. Mizukoshi, Y. Kaneda, and F. Hanaoka. 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 20:2004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakasugi, M., A. Kawashima, H. Morioka, S. Linn, A. Sancar, T. Mori, O. Nikaido, and T. Matsunaga. 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277:1637-1640. [DOI] [PubMed] [Google Scholar]
- 48.Wakasugi, M., and A. Sancar. 1999. Order of assembly of human DNA excision repair nuclease. J. Biol. Chem. 274:18759-18768. [DOI] [PubMed] [Google Scholar]
- 48a.Wakasugi, M., and A. Sancar. 1998. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl. Acad. Sci. USA 95:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Z., X. Wu, and E. C. Friedberg. 1991. Nucleotide excision repair by human cell extract is suppressed in reconstituted nucleosomes. J. Biol. Chem. 266:22472-22478. [PubMed] [Google Scholar]
- 50.Wolffe, A. P. 1997. Chromatin: structure and function. Academic Press, Inc., New York, N.Y.
- 51.Wolffe, A. P., and J. J. Hayes. 1993. Transcription factor interactions with model nucleosomal templates. Methods Mol. Genet. 2:314-329. [Google Scholar]
- 52.Wood, R. D. 1996. DNA repair in eukaryotes. Annu. Rev. Biochem. 65:135-167. [DOI] [PubMed] [Google Scholar]
- 53.Workman, J. L., I. C. A. Taylor, and R. E. Kingston, R. G. Roeder. 1991. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 35:419-447. [DOI] [PubMed] [Google Scholar]
- 54.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zamble, D. B., D. Mu, J. T. Reardon, A. Sancar, and S. J. Lippard. 1996. Repair of cisplatin-DNA adducts by the mammalian excision nuclease. Biochemistry 35:10004-10013. [DOI] [PubMed] [Google Scholar]