Abstract
XPC DNA repair gene mutations result in the cancer-prone disorder xeroderma pigmentosum. The XPC gene spans 33 kb and has 16 exons (82–882 bp) and 15 introns (0.08–5.4 kb). A 1.6 kb intron was found within exon 5. Sensitive real- time quantitative reverse transcription–polymerase chain reaction methods were developed to measure full-length XPC mRNA (the predominant form) and isoforms that skipped exons 4, 7 or 12. Exon 7 was skipped in ∼0.07% of XPC mRNAs, consistent with the high information content of the exon 7 splice acceptor and donor sites (12.3 and 10.4 bits). In contrast, exon 4 was skipped in ∼0.7% of the XPC mRNAs, consistent with the low information content of the exon 4 splice acceptor (–0.1 bits). A new common C/A single nucleotide polymorphism in the XPC intron 11 splice acceptor site (58% C in 97 normals) decreased its information content from 7.5 to 5.1 bits. Fibroblasts homozygous for A/A had significantly higher levels (∼2.6-fold) of the XPC mRNA isoform that skipped exon 12 than those homozygous for C/C. This abnormally spliced XPC mRNA isoform has diminished DNA repair function and may contribute to cancer susceptibility.
INTRODUCTION
Xeroderma pigmentosum (XP) is an autosomal recessive disease with defective nucleotide excision repair (NER) (1,2). XP patients have a high incidence of skin cancers (∼1000 times that of the general population) which occur at an average age of <10 years (3,4). The defects in XP patients fall into seven NER complementation groups: XP-A to XP-G and a separate group XP variant (XP-V) with a defect in DNA polymerase η. XP-C is one of the more common forms in the USA (1). The XPC gene encodes a 940 amino acid protein uniquely involved in global genome repair (5–7).
We examined the structure of the human XPC gene in order to determine its arrangement and splicing signals. Extremely sensitive real-time quantitative reverse transcription–polymerase chain reaction (QRT–PCR) assays were developed to measure full-length XPC message as well as alternatively spliced isoforms that skip exon 4, 7 or 12. We found that a splice acceptor with high information content had a low level of alternatively spliced isoform, while a splice acceptor with low information content was associated with a higher level of exon skipping. A common single nucleotide polymorphism (SNP) in a splice acceptor reduced its information content and was associated with increased skipping of exon 12. Utilizing a post-UV host cell reactivation assay we have demonstrated that cells with increased levels of the XPC isoform that skips exon 12 have reduced DNA repair function.
MATERIALS AND METHODS
Cell lines, culture conditions and DNA/RNA isolation
Normal primary fibroblasts (AG04349, AG13145, AG13153, AG04659, AG05186, AG13354, AG13129, AG06239 and AG04438) and lymphoblastoid cells (AG10107) were obtained from the Human Aging Cell Repository (Camden, NJ) through the assistance of Dr J. Metter (National Institute on Aging, Baltimore, MD). XP heterozygote lymphoblast line KR04491 (XPH310BE, mother of XP-C patient XP25BE) was obtained from the Human Mutant Cell Repository (Camden, NJ). SV40-transformed XP-C (XP4PA-SV-EB) cells (5) were a gift from Dr R. Legerski (M.D. Anderson Hospital, Houston, TX). Fibroblast cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) containing 40 mM glutamine, and 15% fetal calf serum (Hyclone Laboratories Inc., Logan, UT). Lymphoblastoid cells were grown in RPMI 1640 medium with 10% fetal calf serum. Exposure to emetine (300 µg/ml) (Sigma-Aldrich Chemical Co., St Louis, MO) in serum-free DMEM for 2 h was used to reverse the premature termination codon-mediated mRNA decay in some experiments (8). Total cytoplasmic RNA was isolated from cells using the RNAqueous small scale phenol-free total RNA isolation kit (Ambion, Austin, TX) as per the vendor’s protocol. DNA was isolated utilizing DNAzol reagent as per the vendor’s protocol (Invitrogen). Measurement of mRNA in 24 normal human tissues utilized Rapid-Scan gene expression panels (catalogue no. HSCA-101; Origene Technologies, Rockville, MD), using the real-time QRT–PCR assays described below.
PCR amplification and sequencing of XPC exons and introns including splice donor and acceptor sites
In order to determine the DNA sequences at the splice donor and acceptor sites of all of the 16 XPC exons, each exon and their respective splice donor and acceptor sites were PCR amplified using a total of 17 intronic primers flanking these sequences, as shown in supplementary Table A. The primers listed in supplementary Table B were used to amplify the XPC introns. The primers were designed on the basis of the XPC cDNA and the sequences we submitted (GenBank accession nos AF261892–AF261901) or the GenBank reference sequence (accession no. AC090645). Some of the primers were previously reported (5). PCR amplification and DNA sequencing were performed using the conditions described in the Supplementary Material. A total of 16 primer pairs (in either an exon or an intron) were employed using the PCR conditions described in the Supplementary Material to amplify the entire ∼33 kb XPC gene.
DNA sequence information analysis
Sequences were scanned with the donor and acceptor individual information weight matrices and the identified sites were displayed as described previously (9,10). These analyses can be performed on a Web server (http://www.lecb.ncifcrf.gov/∼toms/delilaserver.html).
Real-time QRT–PCR for XPC mRNA
XPC mRNA isoforms were quantitated using isoform-specific real-time QRT–PCR as described in the Supplementary Material. Real-time QRT–PCR assays were carried out on a Bio-Rad iCycler iQ system (Bio-Rad, Hercules, CA) using intercalation of SYBR Green as the fluorescence reporter. Reactions were carried out using the SYBR Green kit from PE Applied Biosystems following the manufacturer’s protocol. Isoform-specific real-time QRT–PCR assays were designed to discriminate between RNAs including or excluding specific exons (see supplementary Table C and Fig. S1). Primer sequences were based on the GenBank sequence for full-length XPC mRNA (accession no. D21089) and the exon junction information in Table 1. All primer pairs showed at least a 20 cycle difference between cDNA clones for exon inclusion and exon exclusion, indicating an ∼106-fold specificity. In preliminary experiments we determined that use of a primer that spans the junction of adjacent exons (supplementary Fig. S1) can eliminate the possibility of amplification of intron-containing sequences, thereby greatly reducing interference due to cross contamination of cDNA with genomic DNA (data not shown).
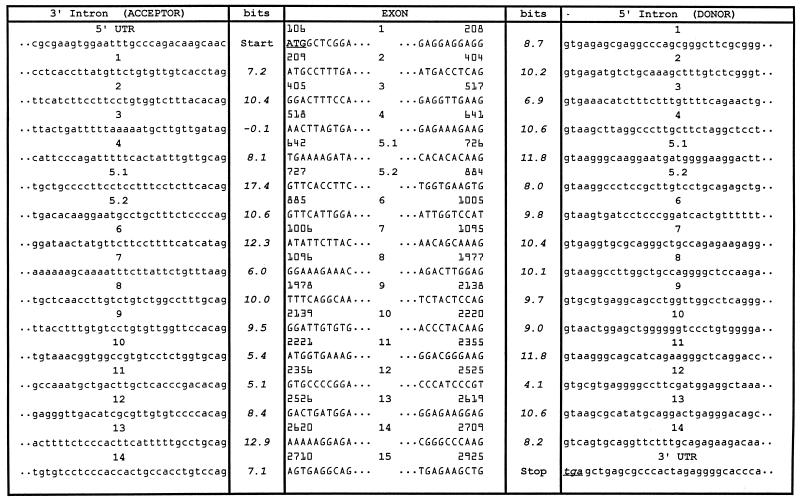
Table 1. The human XPC gene: sequence and information content of splice sites.
XPC cDNA standards for real-time QRT–PCR were generated by end-point PCR with the primers shown in supplementary Table C and subcloned with the TOPO TA cloning kit (Invitrogen) as per the vendor’s protocol. The concentration of each standard was determined using picogreen (Molecular Probes) and also by real-time quantitative polymerase chain reaction using a primer pair for the ampicillin resistance gene (supplementary Table C). This approach allows data from all assays to be expressed in terms of fg XPC clone pXPC-3 (5) (this clone contains the partial XPC cDNA sequence GenBank accession no. NM_004628, which does not contain the first 272 nt of XPC cDNA GenBank accession no. D21089), thus facilitating comparison between assays. For reference purposes, there are ∼150 single-stranded DNA molecules per fg clone pXPC-3.
Determination of XPC intron 11 (–5 C→A) acceptor site SNP
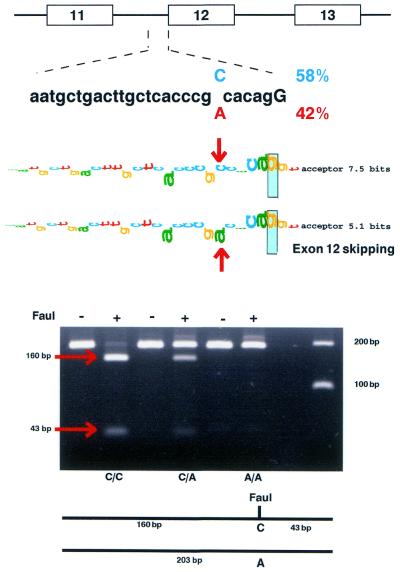
The intron 11 –5 C→A SNP was detected by PCR amplification of this region using the Advantage cDNA PCR kit (Clontech, CA) with forward primer E1 and reverse primer H2 (supplementary Table B) as per the manufacturer’s protocol. The PCR steps were conducted as follows: 94°C for 3 min, then 35 cycles of amplification [94°C for 20 s (denaturation) and 67°C for 3 min (annealing/extension)], ending with 67°C for 3 min. The polymorphic position was analyzed by FauI restriction endonuclease (SibEnzyme) digestion of the 203 bp PCR products amplified from donor DNA using primer pairs E1 and H2. The C at position –5 of XPC intron 11 creates a FauI site. FauI digestion converts the 203 bp PCR product into two fragments of 43 and 160 bp. The fragments were resolved on 2% agarose gels.
Host cell reactivation assay and functional analysis of XPC mRNA isoforms
The pCMVLuc reporter gene plasmid (a generous gift from M. Hedayati and L. Grossman, Johns Hopkins University, Baltimore, MD) was used to measure post-UV host cell reactivation as described previously (11). Briefly, 4 µl (200 ng) of CsCl-purified pCMVLuc, either UV-irradiated (1000 J/m2) or unirradiated, was transfected into cells using Lipofectamine (Invitrogen). After 48 h, the luciferase activity was measured with a luminometer (Monolight 2010; Analytical Luminescence Laboratory, San Diego, CA) using luciferase assay reagent (Promega) as per the vendor’s protocol. Relative luciferase activities are presented as a percentage of activities obtained with UV-irradiated versus unirradiated control plasmids.
In order to assess the function of the XPC mRNA isoform that skips exon 12, an expression vector plasmid containing the XPC cDNA with deletion of exon 12 (pcDNA3/HA-XPC-deleted-exon12) was constructed. The heterozygous XP-C cell line KR04491 was used as a source of cDNA with deletion of exon 12. This material was used to replace the same region of wild-type XPC cDNA in the expression vector plasmid pcDNA3/HA-XPC (a gift from Dr Fumio Hanaoka, Osaka University, Osaka, Japan).
RESULTS
Structure of the normal XPC gene
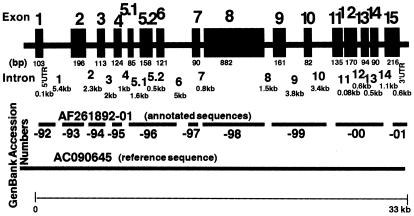
Detailed knowledge of the genomic structure of the human XPC gene is crucial for identifying the genotype of individuals with inherited defects and increased cancer risk. Primer pairs and sequencing conditions were developed to amplify 16 regions of the XPC gene spanning ∼33 kb (Fig. 1 and supplementary Tables A and B). The human XPC gene was previously reported to have 15 exons (12). We found an additional 1.6 kb intron within exon 5 (between nucleotides 726 and 727) that splits this region into exons 5.1 and 5.2. The 16 exons of the human XPC gene range in size from 82 bp (exon 10) to 882 bp (exon 8) (Fig. 1 and Table 1). The 15 introns vary from 0.08 kb (intron 11) to 5.4 kb (intron 1) (Fig. 1). The nucleotide sequence in 10 regions was determined and submitted to GenBank (accession nos AF261892–AF261901). The GenBank reference sequence AC090645 from high-throughput sequencing completes the gaps in the intron sequences.
Figure 1.
Structural map of the human XPC gene. The 16 exons and 15 introns of the 33 kb XPC gene are numbered and their size in bp or kb is indicated below each segment. The parts of the genomic DNA sequence that we determined were submitted to GenBank (accession nos AF261892– AF261901). AC090645 is the chromosome 3 reference sequence that was later posted in GenBank.
Analysis of the XPC exon–intron boundaries
Intron primers were developed to permit amplification of each of the 16 exons separately (supplementary Table A). All of the exon–intron junctions in the XPC gene were sequenced and some of the previously reported exon–intron boundaries were corrected (12). The human XPC gene intronic splice donor and acceptor site sequences and their location in the coding XPC sequence are listed in Table 1.
The information contents of the XPC splice donor and acceptor sites were evaluated using an information theory-based approach incorporating information weight matrices that reflect features of nearly 2000 published donor and acceptor sites (9,10). Information is an additive measure of sequence conservation and describes the degree to which a member contributes to the conservation of an entire sequence family in contrast to examining only the consensus sequence (9,13). Information content is defined as the number of choices needed to describe a sequence pattern, using a logarithmic scale in bits. The magnitude of the information content indicates how strongly conserved a base is in natural splice junction binding sites, with ∼2.4 bits being the apparent minimal functional value (14). All but one of the XPC splice sites demonstrate information contents that range from 4.1 bits (donor splice site in intron 12) to 17.4 bits (acceptor splice site in intron 5.1), consistent with functional activity but with considerable variation in strength (Table 1). The exception is the acceptor splice site of intron 3, which possesses a very low information content (–0.1 bits).
Measurement of XPC mRNA levels by real-time QRT–PCR
Real-time QRT–PCR is an extremely sensitive and specific technique for measurement of specific mRNA levels in human cells (for a review see 15). For this study isoform-specific primers were designed using the same principles that have been developed for allele-specific PCR (16,17). We first created an assay for XPC cDNA that includes exon 12 by designing a sense primer that recognizes the exon 11–exon 12 junction (oCCB-331) and pairing it with an antisense primer that is contained in exon 12 (oCCB-337, supplementary Fig. S1 and Table C). This assay was applied to RT reactions from duplicate RNA samples from nine normal skin fibroblast cultures (Table 2). Close agreement was found between duplicate samples. In addition, similar results were obtained using a second primer pair that detects the exon 12–exon 13 junction (oCCB-333 and oCCB-335 in supplementary Fig. S1; data not shown). The normal level of XPC cDNA was 294 ± 15.7 fg pXPC-3 with a range of 197–348 fg based on measurement of message that contained exon 12 (Table 2). Assuming 100% efficiency of the RT reaction, this corresponds to an average of ∼10 XPC mRNA molecules/cell. However, since we do not know the efficiency of the RT reaction, all real-time QRT–PCR results will be expressed in terms of fg pXPC-3.
Table 2. Full-length (FL) and alternatively spliced XPC mRNA levels in normal fibroblasts.
| Cell strain | Intron 11 genotype | FL XPC (mean ± SDa) (fgb) | Exon 7 skipping (fgb) | Exon 7 skip/FL XPC (%) | Exon 4 skipping (mean ± SDa) (fgb) | Exon 4 skip/FL XPC (%) |
|---|---|---|---|---|---|---|
| AG04349 | CC | 244 ± 10 | 0.3 | 0.10 | 1.7 ± 0.1 | 0.75 |
| AG13145 | CC | 335 ± 14 | 0.2 | 0.06 | 2.4 ± 0.2 | 0.75 |
| AG13153 | CC | 258 ± 7 | 0.2 | 0.08 | 1.0 ± 0.1 | 0.42 |
| AG04659 | AA | 206 ± 12 | 0.1 | 0.07 | 1.4 ± 0.5 | 0.53 |
| AG05186 | AA | 315 ± 6 | 0.2 | 0.05 | 2.1 ± 0.04 | 0.66 |
| AG13354 | AA | 336 ± 18 | 0.2 | 0.07 | 2.7 ± 0.2 | 0.76 |
| AG13129 | AC | 332 ± 9 | 0.3 | 0.08 | 3.0 ± 0.8 | 0.73 |
| AG06239 | AC | 304 ± 13 | 0.2 | 0.07 | 3.1 ± 0.7 | 0.87 |
| AG04438 | AC | 322 ± 24 | 0.2 | 0.06 | 1.6 ± 0.3 | 0.55 |
| Average | 295 | 0.2 | 0.07 | 2.1 | 0.67 |
aMean of two independent cultures analyzed for each cell strain.
bUnits are expressed in terms of fg control plasmid pXPC-3 (see Materials and Methods for details).
Relationship of splice acceptor information content to extent of exon 4 and 7 skipping
The information theory-based approach along with the exon recognition model of vertebrate splicing predicts that splice acceptors with high information content would be less likely to be associated with exon skipping than splice acceptors with low information content (9,14,18). We found that the splice acceptor for exon 7 had an information content of 12.3 bits, while that for exon 4 had an information content of –0.1 bits (Table 1). This predicts that normal cells would contain a smaller amount of alternatively spliced XPC mRNA that skipped exon 7 than XPC mRNA that skipped exon 4. Isoform-specific primers were developed for QRT–PCR to measure mRNA that skipped exon 7 (using oCCB-382, a primer that includes the junction of exons 6 and 8) and that skipped exon 4 (using oCCB-364, a primer that includes the junction of exons 3 and 5.1). These primers are designed only to detect exon skipping. For example, primer oCCB364 spans the exon 3–exon 5.1 junction and has a 3′ two base mismatch with exon 4, which allows us to discriminate between the two isoforms (supplementary Fig. S1 and Table C). These were paired with primers oCCB-384 and oCCB-368, respectively.
RT reactions from normal fibroblasts contained extremely little detectable XPC cDNA that skipped exon 7 (nine normal fibroblast lines had a mean of 0.2 ± 0.01 fg; Table 2). This represents <0.07% of the full-length isoform that contains exon 12 (Table 2). In contrast, the same RT reactions had a 10-fold greater amount of alternatively spliced XPC cDNA that skipped exon 4 (mean 2.0 ± 0.2 fg; Table 2). This represents 0.7% of the full-length isoform that contains exon 12 (Table 2).
A common SNP in the intron 11 splice acceptor
A C/A SNP in the 3′ end of XPC intron 11 at the –5 position was found where the C creates a new FauI restriction site (Fig. 2). We examined buccal DNA from 97 normal subjects, randomly selected among NIH employees (19), to determine the frequencies of this polymorphism by restriction fragment length polymorphism (RFLP) analysis (Fig. 2 and Table 3). This was a common polymorphism, with the C being present in 58% of the alleles and the A present in 42%. The observed genotype distribution was not significantly different from that expected by the Hardy–Weinberg theory (Table 3).
Figure 2.
XPC intron 11 splice acceptor polymorphism. (Top) A C/A SNP is located at the –5 position of the XPC intron 11 splice acceptor. (Middle) The walkers indicate the change in splice acceptor information content from 7.5 to 5.1 bits by the polymorphism. (Lower) RFLP assay for detection of the C/A polymorphism by use of digestion with FauI. The 203 bp sequence generated by PCR is cut into fragments of 160 and 43 bp by FauI if the C is present in the sequence, permitting detection of C/C, C/A and A/A genotypes.
Table 3. XPC intron 11 splice acceptor polymorphism allele frequencies and genotype distribution.
| |
NIH donors |
Alleles |
Genotype distribution observed [expected] |
||||
|---|---|---|---|---|---|---|---|
| Total | –5 C | –5 A | C/C | C/A | A/A | ||
| p | q | p2 | 2pq | q2 | |||
| Number | 97 | 194 | 112 | 82 | 37 | 38 | 22 |
| Frequency | 100% | 100% | 58% | 42% | 38% [33%] | 39% [49%] | 23% [18%] |
The distribution of the XPC intron 11 splice acceptor polymorphism was compared with that of two other polymorphisms in the XPC gene: a poly(AT) insertion/deletion polymorphism in intron 9 (PAT) and a SNP in XPC exon 15 (A2920C, Lys939Gln) (19). These markers are ∼9 kb apart in the XPC gene (Figs 1 and 3). Among the 97 normal donors, 59 (61%) were homozygous for either C or A in the XPC intron 11 splice acceptor (Table 3). These same 59 donors were also homozygous for the PAT polymorphism and 56 of the donors were homozygous for the XPC exon 15 polymorphism (Fig. 3, upper and middle). These three markers are in linkage disequilibrium and are consistent with a haplotype of PAT–/intron 11 C/exon 15 A in ∼60% of the donors and PAT+/intron 11 A/exon 15 C in ∼40% of the donors (Fig. 3, lower).
Figure 3.
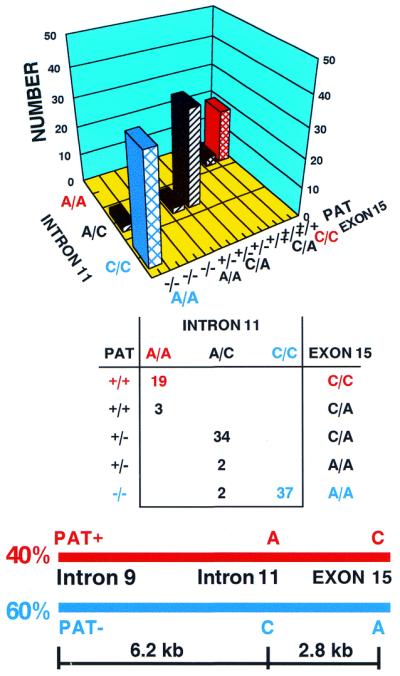
Linkage disequilibrium among three XPC polymorphisms in 97 normal donors. (Upper) The number of donors with the XPC intron 9 PAT+/–, intron 11 A/C and exon 15 C/A genotypes is indicated on the bar graph. (Middle) The table shows the number of individuals homozygous for all three polymorphisms (in red or blue) or heterozygous for at least one of the polymorphisms (in black). (Lower) Schematic diagram of the physical location and haplotypes of three XPC polymorphisms. The XPC intron 11 splice acceptor A at position –5 is linked to C2920 in exon 15 and PAT+ in intron 9 in a haplotype covering ∼9 kb and occurring at a genotype frequency of ∼40% (in red). The XPC intron 11 splice acceptor C at position –5 is linked to A2920 in exon 15 and PAT– in intron 9 in a haplotype covering ∼9 kb and occurring at a genotype frequency of ∼60% (in blue).
Effect of a splice acceptor polymorphism on exon 12 skipping
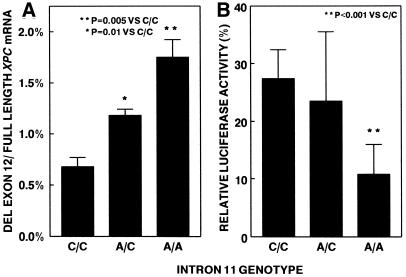
Information theory analysis indicated that an intron 11 splice acceptor site with an A at position –5 would have a lower information content (5.1 bits) than a splice acceptor site with a C at that position (7.5 bits) (Fig. 2). This analysis would predict that XPC mRNA from cells with the A at –5 would have a greater amount of alternatively spliced message that skipped exon 12 in comparison to cells with C at that position. A real-time QRT–PCR assay was developed utilizing an isoform-specific primer that spans the exon 11–exon 13 junction (oCCB-330; supplementary Fig. S1 and Table C) to measure XPC mRNA with deletion of exon 12. Skipping of exon 12 creates a premature stop codon which may trigger nonsense-mediated decay. Treatment of the fibroblasts with emetine prior to harvesting RNA stabilized this alternatively spliced product (8,20). Emetine treatment elevated the level of alternatively spliced XPC mRNA isoforms that deleted exon 4, 7 or 12 approximately 4- to 7-fold (data not shown). Normal fibroblasts with the A/A genotype had a 2.6-fold (P = 0.005) greater proportion of alternatively spliced XPC mRNA with deletion of exon 12 in comparison with cells from donors with the C/C genotype (Fig. 4A). The A/C heterozygotes had an intermediate level (1.8-fold, P = 0.01) of increase in the proportion of XPC mRNA with deletion of exon 12 (Fig. 4A). Similar results were seen in the absence of emetine (data not shown).
Figure 4.
Relation of XPC intron 11 genotype to deletion of exon 12 in XPC mRNA and DNA repair function. (A) Real-time QRT–PCR was used to measure the amount of XPC mRNA deleting exon 12 and retaining exon 12. The proportion of deleted exon 12 XPC mRNA was significantly greater in cells with the A/A and A/C genotypes compared with the C/C genotype (the nine fibroblast lines assayed in duplicate are listed in Table 2). (B) Post-UV host cell reactivation was used to measure DNA repair function. The relative luciferase activity was significantly greater in the cells with the C/C genotype than in cells with the A/A genotype (the nine fibroblast lines assayed in triplicate are listed in Table 2).
The real-time QRT–PCR assays were used to examine the relative proportion of XPC mRNA with deletion of exon 12 in a panel of 24 normal human tissues. While most tissues showed 0.1–0.5% skipping of exon 12, there was greater skipping (2–5% skipping) of exon 12 in muscle, bone marrow, fetal brain, plasma blood leukocytes and stomach (data not shown).
Function of XPC cDNA with deletion of exon 12
A host cell reactivation assay (21) was employed to assess the function of wild-type XPC mRNA and alternatively spliced mRNA that skipped exon 12. This assay measures the ability of cells to repair DNA damage in a UV-treated plasmid (which contained a luciferase reporter gene) 2 days after transfection in vivo. The three normal fibroblast strains that were homozygous for the XPC intron 11 C/C genotype had significantly higher post-UV relative luciferase activity than that of three normal fibroblasts with the A/A genotype (P < 0.001) (Fig. 4B). The three normal fibroblasts with the A/C genotype had intermediate post-UV relative luciferase activity (Fig. 4B). Post-UV host cell reactivation was also measured in a lymphoblastoid cell line (XPH310BE) from the parent of an XP patient who is heterozygous for a mutation in intron 11 that leads to a elevated level (15.6%) of exon 12 skipping and from a normal donor (AG10107) with a low level (1%) of exon 12 skipping. There was a significantly lower level of relative luciferase activity in the XP heterozygote cells (8.0%) than in the normal cells (13.4%) (n = 6, P = 0.04, one tailed t-test) (data not shown). Both the fibroblast and lymphoblast results indicate that an increasing proportion of alternatively spliced XPC mRNA that skips exon 12 is associated with decreased DNA repair activity in normal cells.
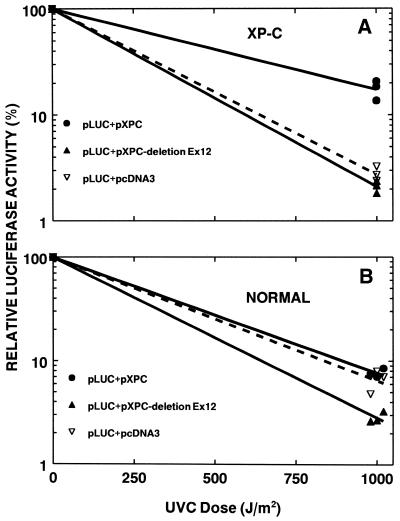
In order to directly assess the function of the XPC mRNA that skips exon 12 we co-transfected expression vectors containing either wild-type XPC mRNA or XPC mRNA with deletion of exon 12 along with UV-treated luciferase plasmid (Fig. 5). Repair-deficient XP-C cells show very low post-UV luciferase expression relative to the untreated plasmid (Fig. 5A). Co-transfection of the XP-C cells with UV-treated plasmid along with another plasmid expressing wild-type XPC mRNA results in increased luciferase activity reflecting complementation of the DNA repair defect by the wild-type XPC cDNA. In contrast, co-transfection of the XP-C cells with a UV-treated luciferase plasmid along with a plasmid expressing XPC mRNA with deletion of exon 12 showed no significant complementation. These results indicate a lack of function of the alternatively spliced XPC mRNA isoform with deletion of exon 12.
Figure 5.
Post-UV host cell reactivation assay assessment of DNA repair activity of XPC cDNA deleting exon 12. (A) Increased expression of luciferase activity in XPC cells [XP4PA (SV40)] by co-transfection of full-length XPC cDNA expression vector (pcDNA3/HA-XPC) (solid circle) was not seen on co-transfection with the XPC cDNA expression vector deleted of exon 12 (pcDNA3/HA-XPC-deleted-exon 12) (solid triangle) or with the empty vector (pcDNA3/HA) (open triangle). (B) Reduction in expression of luciferase in normal cells (AG13129) by co-transfection with XPC cDNA expression vector deleted for exon 12 (solid triangle). Each data point represents an independent transfection. Data points at 1000 J/m2 are displaced for clarity.
The same assay was performed on wild-type cells to look for possible dominant negative effects of the alternatively spliced XPC isoform with deletion of exon 12 on DNA repair in normal cells. Normal cells were fully able to repair UV-induced DNA damage to the luciferase plasmid (Fig. 5B). There was no additional repair seen with co-transfection with wild-type XPC cDNA expression vector. In contrast, co-transfection with XPC cDNA plasmid expressing XPC mRNA with deletion of exon 12 showed lower activity, suggesting a dominant negative effect.
DISCUSSION
Rapid, sensitive, real-time QRT–PCR assays were developed to measure XPC message levels. This technique combines real-time quantitative PCR with reverse transcription of mRNA into cDNA. The advantages of this technique include a very large dynamic range and the capability for absolute quantitation of cDNA by the use of a standard curve. This technique is extremely sensitive, with the potential to measure <1 molecule/cell (15). The mean value of 295 fg pXPC-3 for the amount of normally spliced XPC cDNA derived from 50 ng total RNA (Table 2) corresponds to an average of ∼10 XPC mRNA molecules/cell, assuming 100% efficiency of the RT reaction. This number is similar to the 5–8 transcripts/cell estimated for another NER gene, XPA (22), and is in line with average transcript levels for other genes. Warrington et al. (23) looked at the expression of 7000 genes in human adult and fetal tissues and found that most transcripts were detected at ≤5 copies/cell. Lockhart and Winzeler (24) found that the peak of the expression profile for 40 000 human genes lies between 5 and 10 copies/cell.
Information theory is an important tool to rank normal and mutant splice junctions (9,14). Some of the exon and intron junctions were corrected based on genomic DNA sequencing and employing information theory. Most splice junctions that are fully functional have an information content of at least 2.4 bits (14). The XPC splice acceptor and donor sites all had very high information contents, ranging from 4.1 to 17.4 bits. The only exception was the exon 4 splice acceptor site, which had an information content of –0.1 bits.
We attempted to determine if a splice acceptor site with low information content was more likely to result in increased skipping of the following exon in comparison with an exon with high information content. The splice acceptors at exons 4 (–0.1 bits) and 7 (12.3 bits) were chosen for study since they had markedly different information contents. Accordingly, real-time QRT–PCR assays were developed to measure the level of XPC cDNA with deletion of exon 7 or 4. The level of XPC cDNA skipping exon 7 was very low, with an average of 0.07%. In contrast, the XPC cDNA skipping exon 4 was ∼10 times greater at 0.7% (Table 2). This indicates a relationship between information content and extent of exon skipping. The very low information content of the acceptor at exon 4 (–0.1 bits) would suggest that this would be a very poor splice site and result in even greater levels of exon 4 skipping than we observed. Examination of the exon 4 splice acceptor sequence (Table 1) reveals a run of five uridines upstream of five adenines. In spite of the low information content, the five uridines should be a strong binding site for U2AF65, the protein that recognizes polypyrimidine tracts (25,26). In addition, a strong splice donor (10.6 bits) lies 124 nt downstream at the 3′ end of exon 4 (Table 1) and most likely functions as a splicing enhancer sequence (27). Additional intronic or exonic splicing enhancer elements may also be present and help to compensate for the poor splice acceptor site (28).
A new SNP was discovered in the XPC intron 11 splice acceptor. This polymorphism results in lower information content in an acceptor with A (5.1 bits) than with C (7.5 bits). We predicted that more exon 12 skipping would be found in cells with the A/A genotype than in cells with the C/C genotype. Using a real-time QRT–PCR assay to measure the extent of skipping of exon 12 there was a genotype-dependent increase in skipping of exon 12.
The presence of premature termination codons in the message decreases the levels of most (but not all) known mRNAs, a process called ‘nonsense-mediated message decay’ (29–31). Multiple alternatively spliced mRNA isoforms of the XPG gene have been described in normal tissues (32). Alternative splicing that eliminates the ATG start site in the XP-V gene (DNA polymerase η) has been reported (33). Multiple splice variants of the human base excision repair protein, DNA polymerase β, are common in normal bladder tissues and bladder cancer and appear to be related to the information content of the splice donor and acceptor sequences (34). Recent analyses of the EST database and the sequence of the human genome suggest that at least 38% of human mRNAs have alternatively spliced forms with an average of 2.75 splice forms per spliced gene (35). Thus alternative splicing significantly increases the size of the proteome.
Two common polymorphisms were reported in the XPC gene: an intron 9 insertion/deletion polymorphism (PAT) and an exon 15 A/C polymorphism (19) ∼9 kb apart in the XPC gene (Fig. 3). There was non-disjunction between these polymorphisms and the intron 11 SNP. Thus the C of intron 11 appears to be linked to the PAT– allele and the exon 15 A allele in one haplotype covering at least 9 kb on chromosome 3. PAT+ is linked to the exon 15 C allele and the A in intron 11 in another haplotype. Haplotypes covering 3–84 kb on chromosome 5 have been reported in the human genome (36,37). There was equal DNA repair function of both the exon 15 A and the exon 15 C alleles as measured in the host cell reactivation assay (19). In contrast, deletion of exon 12 results in loss of function of the XPC cDNA in correcting the defect in XP-C cells with the host cell reactivation assay and also results in dominant negative inhibition of function in normal cells (Fig. 5A and B). We also found that increasing amounts of exon 12 skipping are associated with reduced DNA repair in normal cells (Fig. 4A and B). There is evidence that HHR23B protein binds to the exon 12 region of XPC protein (6,38).
Epidemiologic study of cancer patients showed that the PAT+/+ genotype was associated with a 1.85-fold increase in squamous cell carcinoma of the head and neck (39). The mechanism of this increased cancer risk has not been determined. The present study suggests a possible explanation. The PAT+ allele is in linkage disequilibrium with the A allele of intron 11 splice acceptor, which has a lower information content than the C allele and is associated with a higher frequency of deletion of exon 12. This exon 12 deleted XPC mRNA isoform has reduced DNA repair activity. While cells with the A/A genotype in intron 11 have normal levels of full-length XPC mRNA, there is evidence of a dominant negative suppression of DNA repair in normal cells by the XPC exon 12 deleted cDNA and reduced DNA repair in cells with the XPC intron 11 A/A genotype. Thus the increased cancer susceptibility may be related to a reduced level of DNA repair in PAT+/+ patients. This linkage analysis establishes a correlation between the genotype of the XPC gene and cancer predisposition.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
DDBJ/EMBL/GenBank accession nos AF261892–AF261901
REFERENCES
- 1.Van Steeg H. and Kraemer,K.H. (1999) Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol. Med. Today, 5, 86–94. [DOI] [PubMed] [Google Scholar]
- 2.Kraemer K.H. (1999) Heritable diseases with increased sensitivity to cellular injury. In Freedberg,I.M., Eisen,A.Z., Wolff,K., Austen,K.F., Goldsmith,L., Katz,S.I. and Fitzpatrick,T.B. (eds), Fitzpatrick’s Dermatology in General Medicine. McGraw Hill, New York, NY, pp. 1848–1862.
- 3.Kraemer K.H., Lee,M.M. and Scotto,J. (1987) Xeroderma pigmentosum. Cutaneous, ocular and neurologic abnormalities in 830 published cases. Arch. Dermatol., 123, 241–250. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer K.H., Lee,M.-M., Andrews,A.D. and Lambert,W.C. (1994) The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer: the xeroderma pigmentosum paradigm. Arch. Dermatol., 130, 1018–1021. [PubMed] [Google Scholar]
- 5.Legerski R. and Peterson,C. (1992) Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature, 359, 70–73. [Erratum (1992) Nature, 360, 610.] [DOI] [PubMed] [Google Scholar]
- 6.Masutani C., Sugasawa,K., Yanagisawa,J., Sonoyama,T., Ui,M., Enomoto,T., Takio,K., Tanaka,K., Van der Spek,P.J., Bootsma,D. et al. (1994) Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J., 13, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K., Ng,J.M., Masutani,C., Iwai,S., Van der Spek,P.J., Eker,A.P., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- 8.Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- 9.Schneider T.D. (1997) Information content of individual genetic sequences. J. Theor. Biol., 189, 427–441. [DOI] [PubMed] [Google Scholar]
- 10.Schneider T.D. (1997) Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res., 25, 4408–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slor H., Batko,S., Khan,S.G., Sobe,T., Emmert,S., Khadavi,A., Frumkin,A., Busch,D.B., Albert,R.B. and Kraemer,K.H. (2000) Clinical, cellular and molecular features of an Israeli xeroderma pigmentosum family with a frameshift mutation in the XPC gene: sun protection prolongs life. J. Invest. Dermatol., 115, 974–980. [DOI] [PubMed] [Google Scholar]
- 12.Li L., Peterson,C. and Legerski,R. (1996) Sequence of the mouse XPC cDNA and genomic structure of the human XPC gene. Nucleic Acids Res., 24, 1026–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mount S.M. (1982) A catalogue of splice junction sequences. Nucleic Acids Res., 10, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogan P.K., Faux,B.M. and Schneider,T.D. (1998) Information analysis of human splice site mutations. Hum. Mutat., 12, 153–171. [DOI] [PubMed] [Google Scholar]
- 15.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 16.Kwok S., Kellogg,D.E., McKinney,N., Spasic,D., Goda,L., Levenson,C. and Sninsky,J.J. (1990) Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res., 18, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottema C.D. and Sommer,S.S. (1998) Seletive amplification of specific alleles. In Cotton,R.G.H., Edkins,E. and Forrest,S. (eds), Mutation Detection: A Practical Approach. Oxford Univerisity Press, Oxford, UK, pp. 161–187.
- 18.Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 19.Khan S.G., Metter,E.J., Tarone,R.E., Bohr,V.A., Grossman,L., Hedayati,M., Bale,S.J., Emmert,S. and Kraemer,K.H. (2000) A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis, 21, 1821–1825. [DOI] [PubMed] [Google Scholar]
- 20.Dietz H.C. (1997) Nonsense mutations and altered splice-site selection. Am. J. Hum. Genet., 60, 729–730. [PMC free article] [PubMed] [Google Scholar]
- 21.Gozukara E.M., Khan,S.G., Metin,A., Emmert,S., Busch,D.B., Shahlavi,T., Coleman,D.M., Miller,M., Chinsomboon,N., Stefanini,M. et al. (2001) A stop codon in xeroderma pigmentosum group C families in Turkey and Italy: molecular genetic evidence for a common ancestor. J. Invest. Dermatol., 117, 197–204. [DOI] [PubMed] [Google Scholar]
- 22.Layher S.K. and Cleaver,J.E. (1997) Quantification of XPA gene expression levels in human and mouse cell lines by competitive RT-PCR. Mutat. Res. DNA Repair, 383, 9–19. [DOI] [PubMed] [Google Scholar]
- 23.Warrington J.A., Nair,A., Mahadevappa,M. and Tsyganskaya,M. (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics, 2, 143–147. [DOI] [PubMed] [Google Scholar]
- 24.Lockhart D.J. and Winzeler,E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- 25.Roscigno R.F., Weiner,M. and Garcia-Blanco,M.A. (1993) A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem., 268, 11222–11229. [PubMed] [Google Scholar]
- 26.Zamore P.D., Patton,J.G. and Green,M.R. (1992) Cloning and domain structure of the mammalian splicing factor U2AF. Nature, 355, 609–614. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman B.E. and Grabowski,P.J. (1992) U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev., 6, 2554–2568. [DOI] [PubMed] [Google Scholar]
- 28.Blencowe B.J. (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 29.Culbertson M.R. (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- 30.Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 31.Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- 32.Emmert S., Schneider,T.D., Khan,S.G. and Kraemer,K.H. (2001) The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res., 29, 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakur M., Wernick,M., Collins,C., Limoli,C.L., Crowley,E. and Cleaver,J.E. (2001) DNA polymerase eta undergoes alternative splicing, protects against UV sensitivity and apoptosis, and suppresses Mre11-dependent recombination. Genes Chromosom. Cancer, 32, 222–235. [DOI] [PubMed] [Google Scholar]
- 34.Thompson T.E., Rogan,P.K., Risinger,J.I. and Taylor,J.A. (2002) Splice variants but not mutations of DNA polymerase beta are common in bladder cancer. Cancer Res., 62, 3251–3256. [PubMed] [Google Scholar]
- 35.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 36.Daly M.J., Rioux,J.D., Schaffner,S.F., Hudson,T.J. and Lander,E.S. (2001) High-resolution haplotype structure in the human genome. Nature Genet., 29, 229–232. [DOI] [PubMed] [Google Scholar]
- 37.Reich D.E., Cargill,M., Bolk,S., Ireland,J., Sabeti,P.C., Richter,D.J., Lavery,T., Kouyoumjian,R., Farhadian,S.F., Ward,R. et al. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 38.Li L., Lu,X.Y., Peterson,C. and Legerski,R. (1997) XPC interacts with both HHR23B and HHR23A in vivo. Mutat. Res. DNA Repair, 383, 197–203. [DOI] [PubMed] [Google Scholar]
- 39.Shen H., Sturgis,E.M., Khan,S.G., Qiao,Y., Shahlavi,T., Eicher,S.A., Xu,Y., Wang,X., Strom,S.S., Spitz,M.R. et al. (2001) An intronic poly(AT) polymorphism of the DNA repair gene XPC and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Res., 61, 3321–3325. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.