Abstract
The sequence of much of the human genome is now available and the next goal is to identify functional genes and to clarify their roles. We have recently developed a novel system for isolation of genes in the Fas- and TNF-α-mediated pathways to apoptosis using poly(A)-connected hammerhead ribozyme libraries with randomized substrate-binding arms at both the 5′ and 3′ ends of ribozymes. The transcripts of these hybrid ribozymes have a poly(A) motif that can recruit RNA helicases and, thus, they can effectively attack target sites. In the previous studies, hybrid ribozymes were stably expressed. In order to save selection times, in this study we adopted transiently expressed hybrid ribozymes. In the case of Fas-mediated apoptosis, when we transiently introduced these hybrid-ribozyme libraries into Fas-expressing HeLa cells, we were able to isolate surviving clones that were resistant to or exhibited a delay in Fas-mediated apoptosis. We identified many pro-apoptotic genes and novel genes using this strategy with these transiently expressed hybrid-ribozyme libraries. In contrast, we identified significantly smaller numbers of candidate genes using conventional ribozyme libraries that were expressed transiently. Thus, when changes of a particular phenotype occur within a short period of time, our gene discovery system based on transiently expressed hybrid-ribozyme libraries should also be useful for the rapid identification of functional genes in the post-genome era.
INTRODUCTION
The sequence of the human genome is now available (1,2) and it is important to identify functional human genes and to clarify their functions that are involved in particular signal transduction pathways such as growth factor-mediated pathways, a cell cycle or apoptosis. The shorter the time that is required for the identification of functional genes, the better it is as a method for the identification (we will describe the transient strategy later). The Fas protein is a member of the TNFR family of the tumor necrosis factor receptors and it induces apoptosis when associated with Fas ligand or a Fas-specific antibody (α-Fas) (3,4). Upon activation of Fas by the ligand or antibody, Fas induces formation of the death-inducing signaling complex, which consists of FADD and caspase 8 (5,6). Release of active caspase 8 from the complex initiates apoptosis. The major aspects of the mechanism in the Fas-mediated apoptosis are well known, but many factors that participate in this pathway remain to be characterized. Therefore, we tried to identify genes that function in this pathway using a novel method with randomized poly(A)-connected hammerhead ribozyme libraries (7,8). When the phenotype of cells changes upon introduction of a ribozyme library, genes that are responsible for the changes in phenotype can be identified by sequencing the active ribozyme clones.
Hammerhead ribozymes are small RNA molecules that can catalyze cleavage of an RNA in a sequence-specific manner (9–14). These ribozymes can be designed to cleave any RNA substrate that contains an NUX triplet, where N can be any nucleotide and X can be C, A or U and, thus, they have been used successfully to knock out the intracellular expression of a variety of specific viral and cellular targets (15–20). The major limitations to effective intracellular ribozyme use are target site accessibility and co-localization of ribozymes with target RNAs (21–25). Since transcripts of target RNAs form secondary and tertiary structures, they may interfere with accessions from ribozymes. To determine the best target site on a target RNA, an in vitro approach that utilizes the binding of randomized DNA oligonucleotides to an in vitro synthesized RNA and/or a computer prediction such as MulFold are generally used (20,26). To overcome this target site accessibility problem, we recently constructed two novel hybrid ribozymes, a poly(A)-connected ribozyme (tRNAVal-Rz-A60; 7,8) and a constitutive transport element-connected ribozyme (27), and demonstrated that ribozymes of both types have strong cleavage activity and substrate-unwinding activity, regardless of the putative secondary or tertiary structure of the target in the vicinity of the actual target site (27).
We also demonstrated recently that a rapid identification of functional genes is possible, using the RNA helicase-associated hybrid-ribozyme libraries, which were expressed stably, with randomized substrate-binding arms (7,8,28). We identified many pro-apoptotic genes and novel genes using this strategy with these hybrid-ribozyme libraries. In using this method, inhibition of the expression of a particular gene was reflected by a change in a particular phenotype; hence, the method allowed the easy and rapid identification of functional genes. However, in the previous studies, as we stably expressed the hybrid-ribozyme libraries, it took a few weeks for each selection cycle (7,8,28). As we felt that it would be possible, depending on the speed of appearance of a specific phenotype, to transiently express the hybrid-ribozyme libraries, we tried in this study to take such an approach in the Fas-mediated pathway to apoptosis. We demonstrate in this work that our gene discovery system based on transiently expressed hybrid-ribozyme libraries is indeed useful for the rapid identification of functional genes in the post-genome era.
MATERIALS AND METHODS
Construction of randomized hammerhead ribozyme libraries
Chemically synthesized oligonucleotides encoding a randomized ribozyme sequence (5′-TCC CCG GTT CGA AAC CGG GCA CTA CAA AAA CCA ACT TCN NNN NNN NNN CTG ATG AGG CCG AAA GGC CGA ANN NNN NNN NNG GTA CCC CGG ATA TCT TTT TTT-3′) with a total of 20 randomized nucleotides in the substrate-binding arm and a pol III termination sequence (TTTTTT) were converted to double-stranded DNAs by PCR. After digestion with Csp45I and KpnI, the fragments were cloned downstream of the tRNA promoter of pUC-dt (16). To generate a poly(A)-connected ribozyme expression library, we inserted a poly(A) sequence of 60 nt between the ribozyme and the pol III termination sequence.
Culture and transfection of HeLa-Fas cells
HeLa cells that stably expressed the human gene for Fas (HeLa-Fas cells) were prepared as described elsewhere (29,30). They were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Trans fections were performed with the Lipofectin™ reagent (Gibco-BRL, Rockville, MD) as described elsewhere (16). HeLa-Fas cells were selected by incubation with G418 for 3 weeks.
Analysis by RT–PCR
Total RNA was isolated from HeLa-Fas cells with Isogen™ (Nippon Gene, Toyama, Japan) according to the manufacturer’s protocol. RT–PCR was performed using an RNA PCR Kit version 2 (Takara, Kyoto, Japan) with Bik upstream (nucleotides 1–24) and downstream (nucleotides 421–445) primers, CAD upstream (nucleotides 20–44) and downstream (nucleotides 990–1014) primers, CBP upstream (nucleotides 442–467) and downstream (nucleotides 1660–1684) primers or GADPH upstream (nucleotides 230–254) and downstream (nucleotides 642–666) primers as a control. Unknown gene (UG) mRNAs (UG1–UG43) were detected by RT–PCR with up primers specific for the respective genes (see Table 1) and oligo dT down primer. The products of PCR were analyzed by electrophoresis on a 2% agarose gel.
Table 1. Genes identified by the gene discovery system.
Detection of apoptosis
Percentages of apoptotic cells were determined by TUNEL analysis as described elsewhere (16). Cells were fixed for 15 min in 4% paraformalaldehyde, permeabilized by treatment with 0.1% Triton X-100, washed with PBS and incubated in 1× terminal deoxy-nucleotidyltransferase (TdT) buffer that contained 300 U/ml TdT and 40 µM biotin-dUTP for 60 min at 37°C according to the protocol from the manufacturer of the TUNEL kit (Boehringer-Mannheim, Mannheim, Germany). Cells were then washed with PBS. TUNEL-positive cells were detected by incubation with fluorescein isothiocyanate-conjugated streptavidin for 30 min at 37°C.
Identification of functional genes using the randomized poly(A)-connected ribozyme library
Randomized poly(A)-connected ribozyme libraries were introduced transiently into HeLa-Fas cells (2 × 106 cells) using Lipofectin™ (16). After 24 h, HeLa-Fas cells that expressed the randomized library were treated with Fas-specific antibody (α-Fas). After 36 h, surviving cells were picked up manually and plasmid DNA was purified from these survival cells. Then we introduced isolated plasmids into Escherichia coli and these plasmids were amplified in E.coli. After purification of amplified plasmids, we re-introduced these plasmids into HeLa-Fas cells and repeated this cycle five times in all. Finally, we introduced these isolated plasmids into E.coli and these E.coli were plated on a LB plate. Each independent plasmid was isolated from clones of E.coli cells. The sequences of active ribozymes included in these plasmids were analyzed with an automated DNA sequencer. Finally, we searched for the target genes of these ribozymes and identified them using the BLAST search program (31), as we did previously (7,8,28).
RESULTS AND DISCUSSION
At first, we prepared two libraries of 1 × 107 different poly(A)-connected or unconnected ribozymes with randomized substrate-binding arms (Fig. 1A). The intracellular activities of ribozymes depend on the level of expression of the ribozyme and the secondary structure of the target mRNA in cells (15–20,24,25). Therefore, we chose to express ribozyme libraries under the control of the promoter of a human gene for tRNAVal, which can be expected to produce high-level expression of transcripts in cells. We then introduced these ribozyme libraries transiently into HeLa-Fas cells. A schematic diagram of the transient gene discovery system for the Fas-mediated pathway to apoptosis is shown in Figure 1B. After transfection with libraries, cells were treated with Fas-specific antibodies (α-Fas, C-11; MBL, Nagoya, Japan). After 36 h, surviving cells were picked up manually and the plasmid DNA was purified from these surviving cells. Then we introduced isolated plasmids into E.coli and these plasmids were amplified to a limited extent in E.coli. After purification of the amplified plasmids, we re-introduced these plasmids into HeLa-Fas cells and repeated this cycle five times in all. Finally, we introduced these isolated plasmids into E.coli and the transformed E.coli cells were plated on a LB plate. Each independent plasmid was isolated from clones of E.coli cells. The sequences of active ribozymes included in these plasmids were analyzed with an automated DNA sequencer. Finally, we searched for the target genes of these ribozymes and identified them using the BLAST search program (31), as we did previously (7,8,28).
Figure 1.
The rationale for the system for discovery of genes that act in Fas-mediated apoptosis using poly(A)-connected ribozyme libraries. (A) Construction of plasmids that contained randomized ribozyme (Rz) libraries with a poly(A) motif. Ampr, ampicillin; Puror, puromycin; Terminator, terminator of a pol III transcription. (B) Schematic diagram of the gene discovery system.
We realized that this transient gene discovery system would function optimally with the delivery of one type of ribozyme-encoding plasmid per cell. However, in transient transfection systems, in many cases, multiple plasmids are introduced into the same cell. Thus, background clones exhibiting Fas-mediated apoptosis can be observed when a cell contains a specific anti-apoptosis ribozyme and other unrelated background ribozymes. To overcome this problem, we performed multi-cycle selection, and determined numbers of surviving clones after each cycle (Fig. 2B). We also compared the effects of conventional ribozyme libraries and poly(A)-connected ribozyme libraries (ribozyme-A60 library) in order to confirm the effectiveness of the unwinding activity of RNA helicase eIF4AI [we have confirmed previously that the poly(A)-connected ribozymes can make a complex with eIF4AI (7,8)].
Figure 2.
The number of surviving clones after treatment of HeLa-Fas cells with Fas-specific antibody (α-Fas) in each selection cycle. (A) Expression of the various ribozymes in HeLa-Fas cells, as detected by RT–PCR. (B) The number of surviving clones after treatment of HeLa-Fas cells. Pink bars indicate a poly (A)-connected ribozyme library and light green bars indicate results for the conventional ribozyme library.
We first examined the levels of tRNAVal-Rz or tRNAVal-Rz-A60 transcripts in cells by RT–PCR. As shown in Figure 2A, the levels of expression were nearly identical for both ribozyme libraries, within the limits of experimental errors. Since a hybrid ribozyme with an attached helicase can attack the target site effectively (7,8,27), when we used the poly(A)-connected ribozyme library, the number of surviving clones and, thus, the number of genes identified was much greater than when we use the conventional ribozyme library (Fig. 2B). It should be noted that this is the third example that indicates that poly(A)-connected ribozyme libraries were significantly more effective than their conventional counterparts in the gene discovery system (7,8).
Although several genes were recently identified by randomized ribozyme libraries (7,8,28,32–35), our present method is the first transient method and our data clearly demonstrate that multi-cycle selection can be used to rapidly identify functional genes. In our previous strategies based on stably expressed hybrid-ribozyme libraries, it usually took 2 weeks for each selection cycle (7,8), whereas the time required for each cycle in this transient strategy was 4 days. Using our newly devised transient method, as shown in Table 1, we identified many genes that are known to have pro-apoptotic functions during Fas-induced apoptosis, such as genes for FADD, caspases 3, 8 and 9, Bik (36,37), Apaf-1 and DNase CAD (38). Moreover, we also isolated many novel genes (indicated by UG in Table 1). To confirm that the active ribozymes that were targeted to the various genes had, in fact, cleaved the respective mRNAs in cells, we performed RT–PCR analysis using primers specific for Bik and for CAD, which were not identified by our previous stable expression system (7,8).
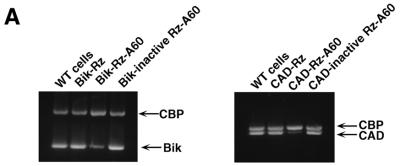
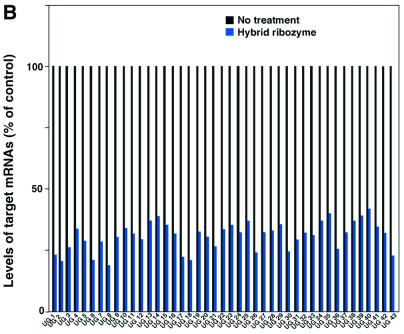
As shown in Figure 3A, the level of Bik mRNA in HeLa-Fas cells that expressed a poly(A)-connected Bik ribozyme was 5-fold lower than levels in wild-type HeLa-Fas cells and in cells that expressed inactive ribozymes (‘Bik-inactive Rz-A60’, with a single G-to-A mutation at a catalytically important, conserved nucleotide). Similar results were obtained in the case of CAD-Rz-A60 (Fig. 3A). The level of CAD mRNA in HeLa-Fas cells that expressed a poly(A)-connected CAD ribozyme was more than 10-fold lower than levels in wild-type HeLa-Fas cells and in cells that expressed inactive ribozymes. It should be emphasized that, in both cases, transiently expressed conventional ribozymes without the poly(A) tail failed to effectively cleave the mRNAs, indicating that we would not have identified these genes if we had not used the RNA helicase-associated ribozymes. Importantly, when the novel target genes (UG1–UG43) were analyzed using RT–PCR with specific up primers and oligo dT down primers, the level of respective mRNA was also reduced dramatically in HeLa-Fas cells that expressed a poly(A)-connected specific ribozyme, as compared with levels in wild-type HeLa-Fas cells (Fig. 3B).
Figure 3.
The determination of the levels of target genes expression. (A) The levels of expression of target genes in cells that expressed poly(A)-connected (A60) or unconnected Bik-ribozyme, or CAD-ribozyme. Bik mRNA and CAD mRNA were detected by RT–PCR with primers specific for the respective genes (see Materials and Methods). CBP is an endogenous control. The levels of expression of all target genes that were identified using the gene discovery system in cells that expressed a poly(A)-connected ribozyme. WT, wild-type. (B) Target mRNAs were detected by RT–PCR with up primers specific for the respective genes and oligo dT down primer. UG, unknown gene.
We next investigated whether apoptotic functions were correlated with phenotype in our gene discovery system. We examined the relative numbers of apoptotic cells that expressed various ribozymes during apoptosis that was induced by α-Fas using the TUNEL method (16). We counted TUNEL-positive cells 36 h after the start of treatment with α-Fas. As shown in Figure 4A, apoptosis occurred in wild-type cells after treatment with α-Fas. In contrast, cells that expressed the poly(A)-connected ribozymes Bik-Rz-A60 or CAD-Rz-A60 exhibited significantly reduced progress towards apoptosis. Control cells that expressed inactive ribozymes targeted to Bik mRNA or CAD mRNA underwent apoptosis. These results suggest that the phenotypes of cells that expressed either Bik-Rz or CAD-Rz were correlated with those of positive clones obtained by our gene discovery method, demonstrating the usefulness and validity of this transient method. In agreement with the results shown in Figure 3, the conventional ribozymes without the poly(A) motif did not slow down the apoptotic process. Therefore, it should be emphasized again that, in the absence of the poly(A) tail, we would not have identified Bik and CAD in our screening with the conventional ribozyme libraries (Fig. 4A). Moreover, cells that expressed poly(A)-connected ribozymes against the respective novel genes (UG1–UG43) exhibited a significant delay in apoptosis after treatment with α-Fas (Fig. 4B). We are now cloning and analyzing them.
Figure 4.
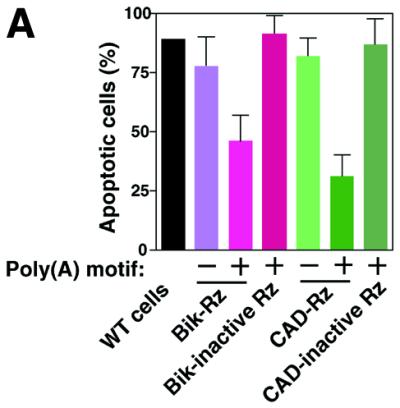
The levels of apoptotic functions of target genes. (A) The extent of apoptosis (%) 36 h after treatment with Fas-specific antibody in populations of cells that expressed poly(A)-connected or unconnected ribozymes, or an inactive ribozyme directed against the gene for Bik or the gene for CAD. Values are means with SD of results from three replicates in each case. WT, wild-type. (B) The extent of apoptosis (%) 36 h after treatment with α-Fas in populations of cells that expressed poly(A)-connected ribozymes directed against each gene. Values are means with SD of results from three replicates in each case. UG, unknown gene.
In this study, we developed a transient system for the rapid identification of functional genes using hybrid-ribozyme libraries. Progress with differential display and microarray technologies has recently been considerable. These technologies allow identification of genes that are turned on or off in association with a particular phenotype. However, genes identified using these systems might simply represent downstream effects (outcomes caused by other upstream genes) and it might prove difficult to identify the genes that are really responsible for a change in phenotype, in other words, the genes that are directly responsible for the specific phenotype. However, since genes identified in our gene discovery system are directly related to a specific change in phenotype, we can directly identify functional genes that are directly responsible for the phenotype (7,8,28,32–35,39). However, these systems depend heavily on the efficiency of ribozymes and it is important to use our hybrid ribozymes, such as poly(A)-connected ribozymes (7,8,28) that recruit the RNA helicase eIF4AI (40), which are significantly more effective than the corresponding unmodified conventional ribozymes (Figs 2 and 3).
We clearly demonstrate here that the time required for each selection cycle can be shortened from 2 weeks for stably expressed hybrid-ribozyme libraries to 4 days if we use the transient expression system. Moreover, since conventional ribozymes cannot cleave target sites that form stable secondary structures, our novel functional gene screening system using hybrid-ribozyme libraries, which can be expressed either stably (7,8) or transiently (as demonstrated here depending on the time required to observe a specific phenotype), should be a powerful tool for the rapid identification of functional genes in the post-genome era.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Professor S. Yonehara (Kyoto University) for a gift of human Fas cDNA. This research was supported by grants from the Ministry of Economy, Trade and Industry (METI) of Japan, by a grant from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Culture (MEXT) of Japan.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Trauth B.C., Klas,C., Peters,A.M., Matzku,S., Moller,P., Falk,W., Debatin,K.M. and Krammer,P.H. (1989) Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science, 245, 301–305. [DOI] [PubMed] [Google Scholar]
- 4.Yonehara S., Ishii,A. and Yonehara,M. (1989) A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J. Exp. Med., 169, 1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnaiyan A.M., O’Rourke,K., Tewari,M. and Dixit,V.M. (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell, 81, 505–512. [DOI] [PubMed] [Google Scholar]
- 6.Muzio M., Chinnaiyan,A.M., Kischkel,F.C., O’Rourke,K., Shevchenko,A., Ni,J., Scaffidi,C., Bretz,J.D., Zhang,M., Gentz,R., Mann,M., Krammer,P.H., Peter,M.E. and Dixit,V.M. (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell, 85, 817–827. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H. and Taira,K. (2002) Identification of genes by hybrid-ribozymes that couple cleavage activity with the unwinding activity of an endogenous RNA helicase. EMBO Rep., 3, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Kawasaki H., Ohnuki,R., Suyama,E. and Taira,K. (2002) Identification of genes that function in the TNF-α-mediated apoptotic pathway using randomized hybrid ribozyme libraries. Nat. Biotechnol., 20, 376–380. [DOI] [PubMed] [Google Scholar]
- 9.Uhlenbeck O.C. (1987) A small catalytic oligoribonucleotide. Nature, 328, 596–600. [DOI] [PubMed] [Google Scholar]
- 10.Haseloff J. and Gerlach,W.L. (1988) Simple RNA enzymes with new and highly specific endonuclease activities. Nature, 334, 585–591. [DOI] [PubMed] [Google Scholar]
- 11.Symons R.H. (1992) Small catalytic RNAs. Annu. Rev. Biochem., 61, 641–671. [DOI] [PubMed] [Google Scholar]
- 12.Slim G. and Gait,M.J. (1991) Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res., 19, 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grasby J.A., Jonathan,P., Butler,G. and Gait,M.J. (1993) The synthesis of oligoribonucleotides containing O6-methylguanosine: the role of conserved guanosine residues in hammerhead ribozyme cleavage. Nucleic Acids Res., 21, 4444–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou D.M. and Taira,K. (1998) The hydrolysis of RNA: From theoretical calculations to the hammerhead ribozyme-mediated cleavage of RNA. Chem. Rev., 98, 991–1026. [DOI] [PubMed] [Google Scholar]
- 15.Rossi J.J. (1999) Ribozymes, genomics and therapeutics. Chem. Biol., 6, R33–R37. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H., Eckner,R., Yao,T.P., Taira,K., Chiu,R., Livingston,D.M. and Yokoyama,K.K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- 17.Scanlon K.J. (1998) Therapeutic Applications of Ribozymes. Humana Press, Totowa, NJ, USA.
- 18.Rossi J.J. and Couture,L.A. (1999) Intracellular Ribozyme Applications. Horizon Scientific Press, Norfolk, UK.
- 19.Tanabe T., Kuwabara,T., Warashina,M., Tani,K., Taira,K. and Asano,S. (2000) Oncogene inactivation in a mouse model: tissue invasion by leukaemic cells is stalled by loading them with a designer ribozyme. Nature, 406, 473–474. [DOI] [PubMed] [Google Scholar]
- 20.Krupp G. and Gaur,R.K. (2000) Ribozyme: Biochemistry and Biotechnology. Eaton Publishing, Natick, MA, USA.
- 21.Sullenger B.A. and Cech,T.R. (1993) Tethering ribozymes to a retroviral packaging signal for destruction of viral RNA. Science, 262, 1566–1569. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand E., Castanotto,D., Zhou,C., Carbonnelle,C., Lee,N.S., Good,P., Chatterjee,S., Grange,T., Pictet,R., Kohn,D., Engelke,D. and Rossi,J.J. (1997) The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA, 3, 75–88. [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y., Kuwabara,T., Warashina,M., Toda,H. and Taira,K. (2001) Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem., 276, 15378–15385. [DOI] [PubMed] [Google Scholar]
- 24.Kuwabara T., Warashina,M., Koseki,S., Sano,M., Ohkawa,J., Nakayama,K. and Taira,K. (2001) Significantly higher activity of a cytoplasmic hammerhead ribozyme than a corresponding nuclear counterpart: engineered tRNAs with an extended 3′ end can be exported efficiently and specifically to the cytoplasm in mammalian cells. Nucleic Acids Res., 29, 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuwabara T., Warashina,M., Sano,M., Tang,H., Wong-Staal,F., Munekata,E. and Taira,K. (2001) Recognition of engineered tRNAs with an extended 3′ end by exportin-t (Xpo-t) and transport of tRNA-attached ribozymes to the cytoplasm in somatic cells. Biomacromolecules, 3, 1229–1242. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger J.A., Turner,D.H. and Zuker,M. (1989) Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol., 183, 281–306. [DOI] [PubMed] [Google Scholar]
- 27.Warashina M., Kuwabara,T., Kato,Y., Sano,M. and Taira,K. (2001) RNA–protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc. Natl Acad. Sci. USA, 98, 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taira K., Warashina,M., Kuwabara,T. and Kawasaki,H. (1999) Functional hybrid molecules with sliding ability. Japanese Patent Application H11-316133.
- 29.Goltsev Y.V., Kovalenko,A.V., Arnold,E., Varfolomeev,E.E., Brodianskii,V.M. and Wallach,D. (1997) CASH, a novel caspase homologue with death effector domains. J. Biol. Chem., 272, 19641–19644. [DOI] [PubMed] [Google Scholar]
- 30.Wajant H., Johannes,F.J., Haas,E., Siemienski,K., Schwenzer,R., Schubert,G., Weiss,T., Grell,M. and Scheurich,P. (1998) Dominant-negative FADD inhibits TNFR60-, Fas/Apo1- and TRAIL-R/Apo2-mediated cell death but not gene induction. Curr. Biol., 8, 113–116. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beger C., Pierce,L.N., Kruger,M., Marcusson,E.G., Robbins,J.M., Welcsh,P., Welch,P.J., Welte,K., King,M.C., Barber,J.R. and Wong-Staal,F. (2001) Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl Acad. Sci. USA, 98, 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q.X., Robbins,J.M., Welch,P.J., Wong-Staal,F. and Barber,J.R. (2000) A novel functional genomics approach identifies mTERT as a suppressor of fibroblast transformation. Nucleic Acids Res., 28, 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruger M., Beger,C., Li,Q.X., Welch,P.J., Tritz,R., Leavitt,M., Barber,J.R. and Wong-Staal,F. (2000) Identification of eIF2Bγ and eIF2γ as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl Acad. Sci. USA, 97, 8566–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch P.J., Marcusson,E.G., Li,Q.X., Beger,C., Kruger,M., Zhou,C., Leavitt,M., Wong-Staal,F. and Barber,J.R. (2000) Identification and validation of a gene involved in anchorage-independent cell growth control using a library of randomized hairpin ribozymes. Genomics, 66, 274–283. [DOI] [PubMed] [Google Scholar]
- 36.Boyd J.M., Gallo,G.J., Elangovan,B., Houghton,A.B., Malstrom,S., Avery,B.J., Ebb,R.G., Subramanian,T., Chittenden,T., Lutz,R.J. et al. (1995) Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene, 11, 1921–1928. [PubMed] [Google Scholar]
- 37.Chittenden T., Flemington,C., Houghton,A.B., Ebb,R.G., Gallo,G.J., Elangovan,B., Chinnadurai,G. and Lutz,R.J. (1995) A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein-binding functions. EMBO J., 14, 5589–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukae N., Enari,M., Sakahira,H., Fukuda,Y., Inazawa,J., Toh,H. and Nagata,S. (1998) Molecular cloning and characterization of human caspase-activated DNase. Proc. Natl Acad. Sci. USA, 95, 9123–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onuki R., Nagasaki,A., Kawasaki,H., Baba,T., Ueda,T.Q.P. and Taira,K. (2002) Confirmation by FRET in individual living cells of absence of significant amyloid-β-mediated caspase 8 activation. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craig A.W., Haghighat,A., Yu,A.T. and Sonenberg,N. (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature, 392, 520–523. [DOI] [PubMed] [Google Scholar]