Abstract
Cell recognition molecules are involved in nervous system development and participate in synaptic plasticity in the adult brain. The close homolog of L1 (CHL1), a recently identified member of the L1 family of cell adhesion molecules, is expressed by neurons and glia in the central nervous system and by Schwann cells in the peripheral nervous system in a pattern overlapping, but distinct from, the other members of the L1 family. In humans, CHL1 (also referred to as CALL) is a candidate gene for 3p- syndrome-associated mental impairment. In the present study, we generated and analyzed CHL1-deficient mice. At the morphological level, these mice showed alterations of hippocampal mossy fiber organization and of olfactory axon projections. Expression of the mRNA of the synapse-specific neural cell adhesion molecule 180 isoform was upregulated in adult CHL1-deficient mice, but the mRNA levels of several other recognition molecules were not changed. The behavior of CHL1-deficient mice in the open field, the elevated plus maze, and the Morris water maze indicated that the mutant animals reacted differently to their environment. Our data show that the permanent absence of CHL1 results in misguided axonal projections and aberrant axonal connectivity and alters the exploratory behavior in novel environments, suggesting deficits in information processing in CHL1-deficient mice.
The L1 family of neural recognition molecules is a subgroup of the immunoglobulin superfamily and is characterized by six immunoglobulin-like and four to five fibronectin type III-like domains. L1 family members occur in species from invertebrates to mammals and 10 members have been identified thus far in the mouse. Four members, namely, L1, CHL1, neurofascin, and Nr-CAM, carry a transmembrane domain and a carboxy-terminal, intracellular domain, whereas the other members are glycosylphosphatidylinositol linked (reference 39 and references therein). The cytoplasmic domain of neurofascin, L1, and Nr-CAM have been shown to interact with ankyrin linking cell recognition to the cytoskeletal scaffold (23, 24). L1 family members are potent promoters of neurite outgrowth and are expressed relatively late in development, mostly at stages when axonogenesis occurs. L1 family members are predominantly expressed by neurons, although some family members are also present on glial cells (11, 17, 41, 51, 78). Several L1 family members can be expressed simultaneously in one set of neuronal cell types, whereas in other subsets of neurons only a particular L1 family member is expressed. It is likely that the structurally and functionally similar L1 family molecules subserve subtly distinct and finely tuned functions in neural cell interactions.
Expression of CHL1 appears to be restricted to the nervous system (39, 41). In mice, CHL1 expression is first detectable in the brain at embryonic day 13, reaches peak levels from embryonic day 18 to postnatal day 7, and subsequently declines to lower levels in the adult brain (39). Neurons in most brain areas express CHL1 mRNA, but its expression is often restricted to particular subsets of neurons, e.g., in the hippocampal formation, where CHL1 shows a stronger expression in the CA1 field and the dentate gyrus than in the CA2, CA3, and CA4 fields (39). CHL1 is also expressed by subpopulations of astrocytes and oligodendrocyte precursors in the central nervous system and by nonmyelinating Schwann cells and some neurons in the peripheral nervous system (39, 95), where prolonged upregulation of CHL1 mRNA by neuronal and glial cells during regeneration has been described (13, 14, 95). In vitro, CHL1 strongly promotes neurite outgrowth by hippocampal and cerebellar neurons via an as-yet-unidentified receptor or receptors on the neuronal surface (39).
CHL1 has been identified in mice, rats (41), and humans (90). Mapping to chromosome h3p26.1, the CHL1 gene, also referred to as CALL, may be associated with intelligence (3), and its loss may contribute to mental impairment associated with the “3p- syndrome” (4).
Here, we have investigated the functions of CHL1 in vivo by generation and characterization of mice deficient for CHL1. Our data reveal differences in hippocampal mossy fiber and olfactory sensory axon projections, upregulation of neural cell adhesion molecule 180 (NCAM180) mRNA in several brain areas, and behavioral abnormalities in CHL1-deficient mice.
MATERIALS AND METHODS
CHL1 targeting construct.
A genomic clone containing the 5′ part of the Chl1 gene was isolated from a mouse 129Sv genomic library (60) by hybridization with a mouse cDNA fragment, Pst159 to Pst645 of pX#2 (41), that corresponds to part of the 5′-untranslated region and the coding region for the signal sequence, the first immunoglobulin domain, and part of the second immunoglobulin domain of CHL1. Partial genomic sequences have been submitted to GenBank/EMBL (accession numbers AJ319655, AJ319656, and AJ319657). For homologous recombination, the targeting vector pDMCHL1ATGΔ (Fig. 1A ) was cloned containing the herpes simplex virus (HSV) thymidine kinase gene (tk) for selection against random integration (50), 1.6-kb 5′ homologous intronic sequence, a PGK-neo cassette (83) replacing 0.4 kb of the Chl1 gene, including exon 1, and 4 kb of the 3′ homologous region from intron 1 in pBluescript KS(−) (Stratagene, La Jolla, Calif.).
FIG. 1.
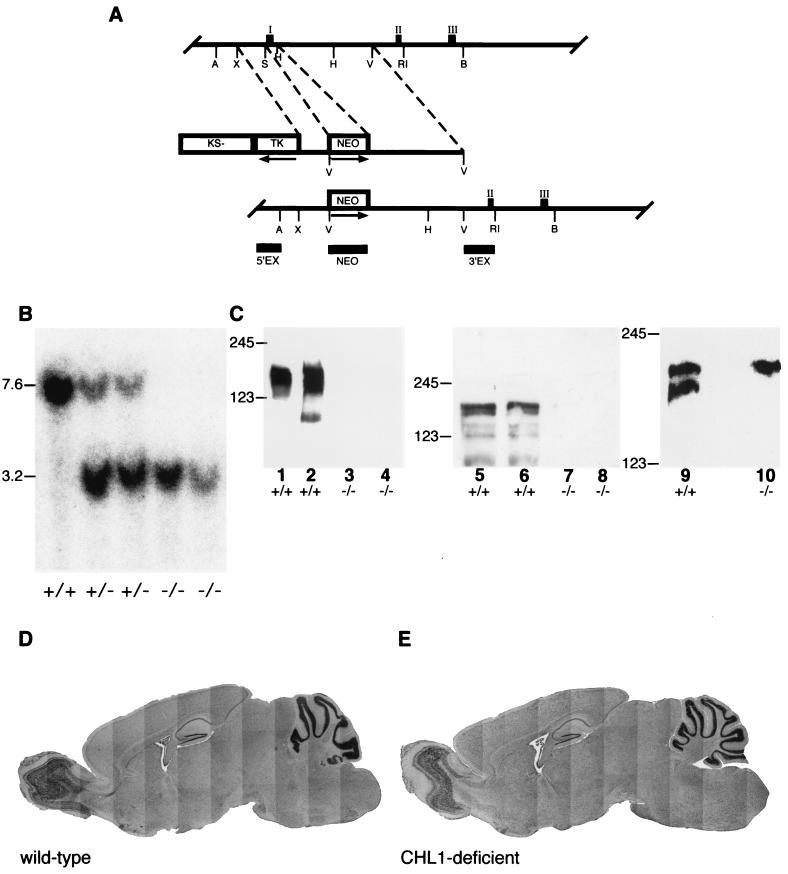
Generation and analysis of CHL1-deficient mice. (A) Restriction maps of part of the mouse Chl1 gene, targeting construct, and the expected and observed structures of the disrupted Chl1 gene after homologous recombination. Exons are represented by filled boxes and numbered with roman numerals. Exon 1 encodes part of the 5′-untranslated sequence, the translation initiation codon, and the signal sequence. Exon 2 encodes the first (IgIa) and exon 3 encodes the second (IgIb) half of the first immunoglobulin-like domain. The targeting construct contains 1.6- and 4-kb of homologous sequences on the 5′ and 3′ sites of the neomycin gene insertion, respectively. The replacement removes intronic sequences and exon 1 of CHL1. PGKneobpA and HSV-tk cassettes and the pBluescript KS(−) vector part are indicated by open boxes. Arrows indicate the transcriptional orientation of the respective genes. Horizontal bars indicate the localization of the hybridization probes 5′EX, 3′EX, and NEO. A, B, H, RI, S, X, and V represent cleavage sites for AccI, BamHI, HindIII, EcoRI, SpeI, XbaI, and EcoRV, respectively (only relevant sites are indicated). (B) Southern blot analysis. DNA from wild-type (+/+) heterozygous (+/−), and CHL1-deficient (−/−) mice that was digested with BamHI and EcoRV was separated by agarose gel electrophoresis, transferred to nylon membranes by Southern blotting, and hybridized with probe 5′EX, revealing bands of 7.6 and 3.2 kb for the wild-type and mutant alleles, respectively. (C) Western blot analysis. Proteins of detergent extracts from crude membrane fractions (lanes 2, 4, and 5 to 10) or the supernatant fraction (lanes 1 and 3) from the brains of 4-day-old (lanes 1 to 4, 9, and 10; 20 μg of protein per lane) or olfactory bulbs of adult (lanes 5 to 8; 22 μg of protein in lane 6, 33 μg in lanes 5 and 7, and 50 μg in lane 8) wild-type (+/+; lanes 1, 2, 5, 6, and 9) and CHL1-deficient (−/−; lanes 3, 4, 7, 8, and 10) mice were analyzed by Western blot with polyclonalantibodies directed to the sixth immunoglobulin domain and fibronectin domains (lanes 1 to 4) or the cytoplasmic domain of CHL1 (lanes 5 to 8) and monoclonal antibody 2C2 that recognizes the carboxy-terminal domain of L1 and CHL1 (lanes 9 and 10). In the wild-type, CHL1 is clearly detectable by polyclonal antibodies with the typical banding pattern (lanes 1, 2, 5, and 6), whereas no signal is obtained in CHL1-deficient mice (lanes 3, 4, 7, and 8). With monoclonal antibody 2C2 two bands corresponding to the full-length forms of L1 and CHL1 are detected in proteins from wild-type mice (lane 9), whereas in CHL1-deficient mice only L1 can be detected (lane 10). The position of molecular weight markers is indicated at the margin. (D and E) General brain anatomy. Wax-embedded sagittal sections of 8-month-old wild-type (D) and CHL1-deficient (E) mice stained with Mayer's hematoxylin. No obvious difference in the general anatomy and morphology could be detected between wild-type and mutant brains.
Embryonic stem cell culture.
A total of 2 × 107 embryonic stem cells of cell line E14.1 (42) were transfected by electroporation (Bio-Rad Gene Pulser; 230 V and 500 μF) with 20 μg of BamHI-linearized targeting construct, cultured on irradiated mouse primary embryonic fibroblasts neoR feeder cells (a gift of H. Blüthmann, F. Hofmann-La Roche, Basel, Switzerland), and selected with 0.2 μM 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU; Bristol-Myers, New York, N.Y.) and 300 μg of G418 (Gibco-BRL, Rockville, Md.)/ml for 3 and 6 days, respectively. Single colonies were expanded, and aliquots of the clones were frozen as described previously (15) or cultured in medium containing 60% buffalo rat liver cell-conditioned medium without feeder cells for DNA isolation.
Screening of recombinant clones and Southern blot analysis.
Embryonic stem cells were lysed, and DNA was isolated as described previously (69). The DNA of individual embryonic stem cell clones was digested with EcoRV and BamHI and then analyzed by Southern blotting as described previously (53) by using probe 5′EX (950-bp fragment the BamHI site resulting from construction of the genomic library to AccI in intron 0 and labeled to 108 cpm/μg by random priming [31]). Genomic DNA from positive embryonic stem cells was further analyzed after digestion with the appropriate restriction enzymes by Southern blot analysis with the probes 3′EX (1.3-kb fragment EcoRV intron 1 to EcoRI intron 1) and NEO (XhoI-SalI fragment of pPGKneobpA (83; also data not shown).
Blastocyst injection and mating of mice.
Blastocyst injections were performed by J. P. Julien and coworkers (McGill University, Montreal, Quebec, Canada) on a commercial basis. Male chimeras were mated with C57BL/6 females, and heterozygous offspring were crossed to obtain homozygous mice. The genotype of the mice was characterized by Southern blot analysis of DNA isolated from tail biopsies.
Protein analysis of brain extracts.
For analysis of proteins, detergent extracts of brains were prepared as described previously (53). Briefly, total brains of wild-type and CHL1-deficient mice were homogenized in buffer H (1 mM NaHCO3, 0.2 mM CaCl2, 0.2 mM MgCl2, 1 mM spermidine) complemented with protease inhibitors (10 μg of soybean trypsin inhibitor/ml, 10 μg of turkey egg white trypsin inhibitor/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM iodoacetamide). The homogenate was centrifuged at 4°C, first at 1,500 × g and then at 30,000 × g. The 30,000 × g pellet was then suspended in buffer (20 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.5% Triton X-100; pH 7.2) complemented with protease inhibitors as described above, and the protein concentrations of the resuspended pellet fraction (crude membrane fraction) and the 30,000 × g supernatant (soluble fraction) were determined (BCA assay; Pierce, Rockford, Ill.). After addition of 2× loading buffer and heat denaturation, the samples were analyzed under reducing conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (46) and Western blotting (88). Primary antibodies were visualized by using horseradish peroxidase-coupled antibodies to mouse or rabbit immunoglobulin G (IgG; diluted 1:10,000; Dianova, Hamburg, Germany) and enhanced chemiluminescence (Amersham Pharmacia, Freiburg, Germany).
Antibodies.
For immunoblot analysis, polyclonal antibodies against the recombinantly expressed domain of CHL1 comprising the sixth immunoglobulin-like domain and the fibronectin type III repeats (41) or against the cytoplasmic domain and monoclonal antibody 2C2 reacting with the cytoplasmic domain of L1 and CHL1 (gifts of M. Grumet, Rutgers University, Piscataway, N.J.) were used as first antibodies and detected by using horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence (Amersham Pharmacia).
For immunohistochemistry, synaptophysin was detected with mouse monoclonal anti-synaptophysin antibodies (diluted 1:200; Sigma-Aldrich, Taufkirchen, Germany), biotin-SP-conjugated goat anti-mouse secondary antibodies (diluted 1:200; Jackson Immunoresearch Laboratories, West Grove, Pa.), and Cy3-conjugated streptavidin (diluted 1:100; Dianova). For detection of calbindin, rabbit polyclonal anti-calbindin D-28k antibodies (diluted 1:10,000; Swant, Bellinzona, Switzerland) and Alexa 488 goat anti-rabbit secondary antibodies (diluted 1:100; Molecular Probes, Leiden, The Netherlands) were used.
General anatomy and histology.
For preparation of wax-embedded sections, deeply anesthetized animals were perfused with phosphate-buffered saline (PBS; pH 7.4), and the brains were removed and incubated overnight at 4°C in 70% ethanol-5% acetic acid, washed for 24 h in 70% ethanol at 4°C, dehydrated at room temperature in ascending concentrations of ethanol, and incubated three times for 12 h in wax at 38°C. Then, 20-μm sections were mounted on gelatine-coated slides, dried for at least 24 h, dewaxed in ascending concentrations of ethanol, and stained with Mayer's hematoxylin (Sigma-Aldrich).
Timm's staining.
Animals were deeply anesthetized with chloral hydrate (7% in saline, intraperitoneally) and perfused intracardially with PBS, followed by sodium sulfide solution (24.37 mM disodium sulfide, 43.11 mM sodium phosphate) and 4% paraformaldehyde in PBS. The brains were removed from the skull, postfixed overnight at 4°C in the same fixative, and cryoprotected in PBS containing 15% sucrose. Sagittal cryosections (20-μm thick) were processed as described by Cremer and coworkers (21). Briefly, sections were stained in the dark at 24°C with a freshly prepared solution of 1.2 mM gum arabic, 0.15 M hydroquinone, and 0.05 M silver nitrate in sodium citrate buffer (0.12 M citric acid, 0.08 M trisodium citrate) and then fixed in photofixative (Hypamfix; Ilford, Mobberly Knutsford, United Kingdom). The sections were then counterstained with neutral red (1%), dehydrated, mounted in Entellan (Merck Eurolab, Berlin, Germany), and placed under coverslips.
In situ hybridization.
In situ hybridization analysis with digoxigenin-labeled sense and antisense cRNA probes generated by in vitro transcription (27) was carried out as described previously (54). CHL1 and L1 cRNA probes were synthesized as described previously (41). Probes for NrCAM and neurofascin were generated from mouse cDNA clones containing fragments corresponding to the CHL1 and L1 probes (gifts of P. Dirks, Leibniz Institute, Magdeburg, Germany). Sense and antisense probes for NCAM180 and F3/contactin were synthesized by transcription from the plasmids pblueNCAM180 (a gift of J. Holm, Swiss Federal Institute of Technology, Zurich, Switzerland), carrying a 750-bp cDNA fragment specific for NCAM180, and pKSF3, containing a 3.2-kb cDNA fragment of mouse F3 (a gift of G. Rougon, CNRS Marseille) in pBluescript KS(−) (Stratagene). For quantitative analysis, a probe for NCAM180 was synthesized by using 35S-labeled UTP (Amersham Pharmacia).
Brain tissue was processed for in situ hybridization as described previously (54). Briefly, brains were quickly frozen by immersion in isopentane (−40°C). Frontal sections (14 μm thick) were collected on silane-coated glass slides. After fixation in 4% paraformaldehyde in PBS (pH 7.2), sections were acetylized, dehydrated, and prehybridized for 3 h at 37°C in prehybridization buffer. Hybridization was conducted for 12 to 15 h at 55°C in hybridization buffer. For the 35S-labeled NCAM180 probe, the sections were washed, dehydrated through a series of ethanol baths of ascending concentrations, and air dried before exposure on β-Max Hyperfilms (Kodak, Rochester, N.Y.) for 5 days at 4°C. For the digoxigenin-labeled probes, after a blocking step (1% Roche blocking reagent, 0.5% bovine serum albumin [BSA] fraction V; Sigma-Aldrich), sections were incubated overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (Roche, Mannheim, Germany). Sections were then washed and developed for several hours in the dark with 0.34 mg of 4-nitroblue tetrazolium chloride (Roche)/ml, 0.175 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate; Roche)/ml, and 0.25 mg of levamisole (Sigma-Aldrich)/ml. Sections were then mounted in Fluoromount. Sense probes (either 35S or digoxigenin labeled) at the same concentration as the antisense probes did not provide any significant labeling (data not shown).
Densitometric analysis of film autoradiograms was carried out as described previously (55) by using the public domain program NIH Image 1.62b7 developed at the U.S. National Institutes of Health (http://rsb.info.nih.gov/nih-image/). Uncalibrated optical densities were evaluated for several brain areas: the glomerular cell, mitral cell, and granular cell layers of the olfactory bulb, the cortex, the hippocampus (the CA1, CA2, and CA3 subfields and dentate gyrus), the thalamus, and the amygdala. These values were corrected by using as a reference the optical density of a control area (the central part of the olfactory bulb or the corpus callosum), allowing the comparison of the relative optical densities obtained for each brain area between the different animal groups without the need to measure optical density standards or use known optical density values (semiquantitative method). Sections taken from each brain were 4.28 mm anterior (olfactory bulb areas) and 1.46 to 1.70 mm posterior to the bregma (cortex, hippocampus, thalamus, and amygdala). Brain regions were defined according to the method of Franklin and Paxinos (34).
Immunocytochemistry for synaptophysin and calbindin.
Animals were deeply anesthetized with chloral hydrate (7% in saline, intraperitoneally) and perfused intracardially with PBS, followed by 4% paraformaldehyde in PBS. The brains were postfixed overnight at 4°C in the same fixative. Sections 60 μm thick were cut by using a vibratome and collected in PBS. The free-floating sections were incubated for 15 min in methanol-PBS (1:1) containing 1% H2O2 and blocked 45 min in a 5% BSA solution in PBS. Sections were then incubated with primary antibodies in PBS containing 0.25% BSA, 0.1% Triton X-100, and 0.05% NaN3 for two nights at 4°C with gentle shaking. For the detection of synaptophysin, sections were incubated for 90 min with biotinylated secondary antibody diluted in PBS, washed three times, and incubated for 45 min with Cy3-conjugated streptavidin. Then, sections were incubated for 1 h with secondary antibodies for the detection of calbindin, mounted on glass slides in PBS-glycerol (1:1) containing 0.1% Dapco (1,4-diazabicyclo[2.2.2.]octane; Sigma-Aldrich), and analyzed with a Leica Leitz DM RD microscope with a Kappa CF 20 DXC charge-coupled device color camera and a confocal microscope (TCS4D; Leica, Bensheim, Germany). An experimenter not aware of the genotype of the mice was able to identify 70 to 80% of the mutants according to the synaptophysin and calbindin immunostaining detected in the CA3 subfield of the hippocampus.
Lectin staining.
Animals were perfused as described for the immunocytochemistry of synaptophysin and calbindin. The brains were postfixed 4 to 6 h at 4°C in the same fixative, cryoprotected overnight in a solution of 30% sucrose in PBS, and frozen. Frontal olfactory bulb cryosections (60 μm thick) were collected in PBS. The free-floating sections were incubated 15 min in methanol-PBS (1:1) containing 1% H2O2, blocked with a 2% BSA solution in PBS for 30 min, and then incubated for 2 h with the plant lectin Dolichos biflorus agglutinin conjugated to biotin (DBA; Sigma) at 20 μg/ml in PBS and 0.25% Triton X-100. For the detection of the lectin, sections were incubated for 45 min with Cy3-conjugated streptavidin. After being washed in PBS, sections were mounted on glass slides and coverslipped with antifade media (Vectashield; Vector Labs) containing DAPI (4′,6′-diamidino-2-phenylindole; 1.5 μg/ml) and sealed. DAPI was used for nuclear staining.
Animals.
For all experiments, CHL1-deficient and matched littermate wild-type mice (referred to as “wild type”) derived from heterozygous matings were used. For behavioral experiments, heterozygous CHL1-deficient mice originating from strain 129Ola embryonic stem cells were backcrossed for at least eight generations to strain C57BL/6 (Charles River, Sulzfeld, Germany), including one generation with a C57BL/6 male, and then intercrossed to obtain homozygous CHL1-deficient mice. One week prior to behavioral testing, the animals were removed from the central facility and housed individually. CHL1-deficient and wild-type mice were tested alternately by an experimenter unaware of the genotype and during the light phase of the light-dark cycle.
General and neurological state.
General parameters indicative of the health and neurological state were addressed after the neurobehavioral examination of naive mice (appearance, sensorimotor behavior, immobility, and the animal's reflexes) as described by Wishaw et al. (91). In addition, the tests described in the primary screen of the SHIRPA protocol except for the startle response (71; www.mgc.har.mrc.ac.uk/mutabase/shirpa_summary.html) were carried out.
Grip strength measurement.
Grip strength was measured (52) with a high-precision force sensor (grip strength meter; TSE, Bad Homburg, Germany) to evaluate muscle strength and function of the neuromuscular junction. The animals previously used to determine the general and neurological state were used.
Rota-rod test.
An accelerating rota-rod (TSE) was used to analyze motor coordination (43). Mice were subjected to two training sessions (3-h intertrial interval) with accelerating speeds from 4 to 40 rpm over a 5-min period on the first day. Five days later, animals were tested at 16-, 24-, 32-, and 40-rpm constant speeds with a 5-min maximal duration of the trial. The duration that animals were able to maintain balance on the rod was measured. Animals previously tested in the water maze were used.
Open field test.
The open field test was performed in a square gray plastic arena (50 by 50 cm, 25 cm high; ca. 200 lx). Naive animals were placed in the middle of the arena, and their behavior was videotaped for 15 min. For evaluation, tracks were recorded by using the VideoMot 2 system (TSE), and the path length, relative time, visits, and walking speed in the central area (infield; square, 30 by 30 cm) in the area closer to the walls (outfield; within 10 cm) and in the four corners (10 by 10 cm each) of the arena were analyzed for the total test time (15 min) and for each 5-min interval. In addition, tracks were evaluated with the Wintrack analysis software, release 2000 (92).
Light-dark avoidance test.
Light-dark avoidance behavior (18, 19) was analyzed in a rectangular gray plastic arena with a dark (12.5 by 25 cm) and an illuminated compartment (25 by 25 cm) separated by a wall with a 5-cm-by-5-cm opening (85). Animals were placed in the middle of the illuminated compartment (ca. 250 lx), and their behavior was videotaped for 10 min. For analysis, the time spent in the dark versus the illuminated area, the number of transitions between the compartments, the duration of the stays, and the latency to enter the dark compartment were compared. Animals previously tested in the open field were used.
Elevated plus maze.
Naive animals were placed in the center of an elevated plus maze (48, 64) (6.5- by 45-cm arms, 75 cm above floor level, 22-cm high nontransparent side walls), and the number of entries into the central part or the closed and open arms and the time spent in these compartments during a 5-min testing period were recorded on videotape and analyzed by using the VideoMot 2 system.
Morris water maze.
The water maze (56) consisted of a dark-gray circular basin (130 cm in diameter) filled with water (24 to 26°C, 30 cm deep) made opaque by the addition of white paint. A circular platform (10 cm in diameter) was placed 1.5 cm below the water surface. Naive mice were subjected to six trials per day for 6 days. They were allowed to swim until they found the platform or until 120 s had elapsed. In the latter case, animals were guided to the platform and allowed to rest for 10 s. The hidden platform remained at a fixed position for the first 4 days (24 trials, aquisition phase) and was moved into the opposite quadrant for the 2 last days (12 trials, reversal phase). Trials 25 and 26 were defined as probe trials to analyze the precision of the spatial learning. All trials were videotaped and then analyzed by using the VideoMot 2 system and Wintrack analysis software.
Statistical analysis.
Statistical analysis was performed by using analysis of variance (ANOVA) and post hoc with Scheffe's test (Statview Program; SAS Institute, Inc., Cary, N.C.). Radioactive in situ hybridization for NCAM180 and behavioral experiments were examined by one-way ANOVA (with genotype as a factor). In addition, for the water maze experiment statistical analysis was performed by using repeated measures ANOVA (with between-subject factor genotype and within-subject factor acquisition day or retrieval day). For all analyses, a P value of <0.05 was considered significant.
RESULTS
Generation of CHL1-deficient mice.
Using a mouse cDNA fragment corresponding to the 5′-untranslated region and the region encoding the amino terminus of the CHL1 mRNA, a clone containing the first translated exons of the Chl1 gene was isolated from a mouse 129Sv genomic library. The structure of the partial Chl1 gene is shown in Fig. 1A. Exon 1 encodes the translational start codon, followed by the signal sequence; exon 2 codes for the amino-terminal part of the first immunoglobulin-like domain, and exon 3 encodes the second half of the first immunoglobulin-like domain. To inactivate the Chl1 gene, a targeting vector was constructed (Fig. 1A). This vector contains the HSV thymidine kinase gene (tk) for selection against random integration (50), a 1.6-kb 5′ homologous sequence, a PGK-neo cassette (83) replacing 0.4 kb of the Chl1 gene including exon 1, and 4 kb of 3′ homologous region. Homologous recombination with this targeting vector results in a replacement mutation (Fig. 1A), deleting the region encoding the ribosomal binding site, the translation initiation codon, and the amino terminus, including the signal sequence, and was thus expected to result in a null mutation.
After electroporation of the linearized targeting vector into strain 129Ola-derived embryonic stem cells and double selection with FIAU and G418, clones carrying the expected mutation were identified by Southern blot analysis with the external probe 5′EX. In addition to the wild-type band of ca. 7.6 kb, the appearance of a new 3.2-kb band was detected (due to the presence of a new EcoRV site introduced by insertion of the neomycin gene replacing exon I of the Chl1 gene) with a frequency of 1 positive clone in 11 clones analyzed. Further analysis with the internal probe NEO and 3′EX confirmed the pattern expected after homologous recombination. Two independently obtained targeted clones were further used to generate chimeric mice.
Highly chimeric mice were obtained after injection of targeted embryonic stem cells into blastocysts. Chimeric males showed germ line transmission of the disrupted Chl1 gene, as revealed by Southern blot analysis. Crossing of heterozygous offspring yielded homozygous CHL1-deficient mice with strictly Mendelian frequencies. Southern blot analysis of these mice with the probes 3′EX, NEO (data not shown), and 5′EX showed the pattern expected for a single integration by homologous recombination (Fig. 1B).
To confirm that the mutation generated a null allele, proteins from brains of wild-type and CHL1-deficient mice were analyzed by immunoblot analysis. In protein extracts from brains of wild-type mice, the typical banding pattern of the membrane-bound form and fragments of CHL1 was detected by using polyclonal antibodies against the extracellular domain (41), but no signal could be detected in protein extracts from brains of CHL1-deficient mice (Fig. 1C). Furthermore, CHL1 was not detected in protein extracts from brains of CHL1-deficient mice (Fig. 1C), whereas L1 was detectable with the monoclonal antibody 2C2 that recognizes the carboxy-terminal domains of CHL1 and L1 and thus detects only the full-length forms of both molecules.
Neither heterozygous nor homozygous CHL1-deficient mice showed any obvious, grossly abnormal neurological or behavioral phenotype up to an age of ca. 2 years, the latest time point investigated. Furthermore, we have established CHL1-deficient lines by intercrossing homozygous CHL1-deficient mice, proving that both sexes are fertile.
Morphological analysis of the central nervous system of CHL1-deficient mice.
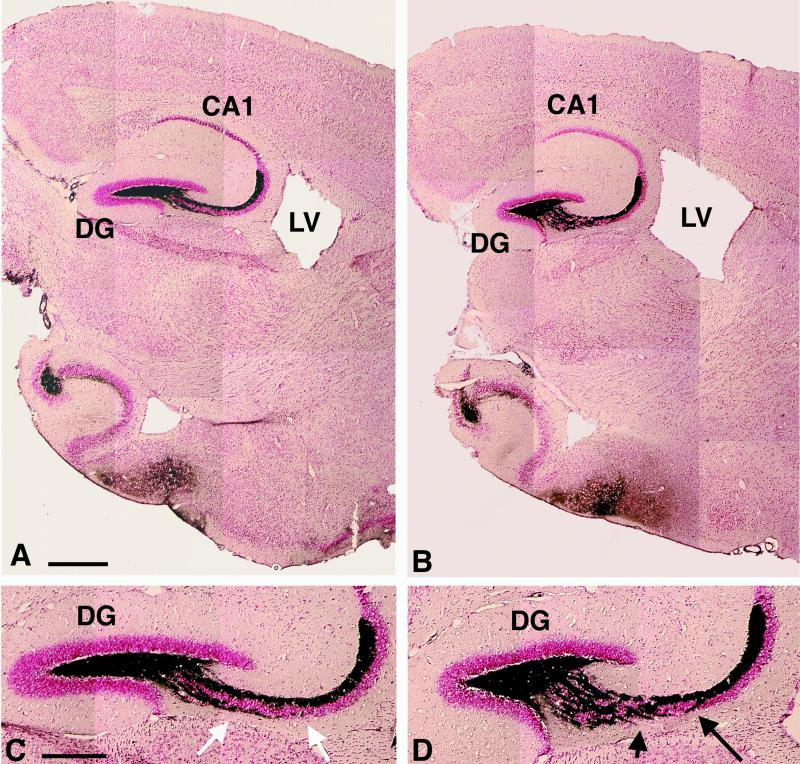
The anatomy of brains from CHL1-deficient mice and wild-type littermates was compared by using two complementary histological methods. Wax-embedded sections of 8-month-old mice were used to analyze the general morphology of the brain (Fig. 1D and E), and semithin sections were used to study the fine anatomy, as well as the cytology of different brain regions of 7-day-old and 4-month-old mice. Using these methods, the general morphology of brains from CHL1-deficient mice revealed a seemingly normal structure indistinguishable from that of wild-type littermates at the light microscopic level. No significant differences in either the shape or the size of the main brain structures could be observed and, in particular, the cerebellum, hippocampus, olfactory bulb, and cortical layers appeared to be morphologically unaffected in the CHL1-deficient mice. Similarly, the fine analysis of the anatomy and cytology of the brains did not show any major differences between mutant and wild-type mice (data not shown). However, after backcrossing to the mouse strain C57BL/6 for 10 generations, an enlargement of the lateral ventricles became apparent in CHL1-deficient mice (Fig. 2).
FIG. 2.
Mossy fiber distribution in adult CHL1-deficient mice. Sagittal sections of wild-type (A and C) and CHL1-deficient (B and D) mice stained by Timm's method to reveal the mossy fibers and counterstained with neutral red. (A and B) Low-magnification photomicrographs showing the general anatomy of the hippocampus and an enlarged lateral ventricle (LV) in CHL1-deficient (B) compared to the wild-type mice (A). (C and D) High-magnification photomicrographs showing the altered organization of the mossy fibers in CHL1-deficient (D) compared to wild-type mice (C); note the bundles of fibers crossing the CA3 region of the hippocampus in CHL1-deficient mice (D, arrows). CA1: hippocampal CA1 region; CA3: hippocampal CA3 region; DG: dentate gyrus of the hippocampus. Scale bars: 617 μm (A, B); 302 μm (C, D).
Mossy fiber organization.
The distribution of the hippocampal mossy fibers in CHL1-deficient mice was analyzed by using Timm's staining (Fig. 2). Wild-type mice showed a typical strong and ordered labeling of the mossy fibers, whereas in CHL1-deficient mice, mossy fibers invaded the CA3 pyramidal cell layer. No difference in the intensity of staining could be detected between mutant and control mice. Immunohistochemistry for two specific markers was used for detailed analysis of the organization of hippocampal mossy fibers and their terminals. Synaptophysin, a 38-kDa synaptic vesicle protein (also known as P38 or SVP38), is present in the presynaptic boutons of the mossy fibers (82) and has been successfully used by Cremer et al. to study the organization of mossy fiber terminals in NCAM-deficient mice (21). The calcium-binding protein calbindin D28K shows a characteristic spatial pattern of expression in the hippocampal formation and identifies the axons of dentate granular cells, the mossy fibers (5, 75, 81, 87).
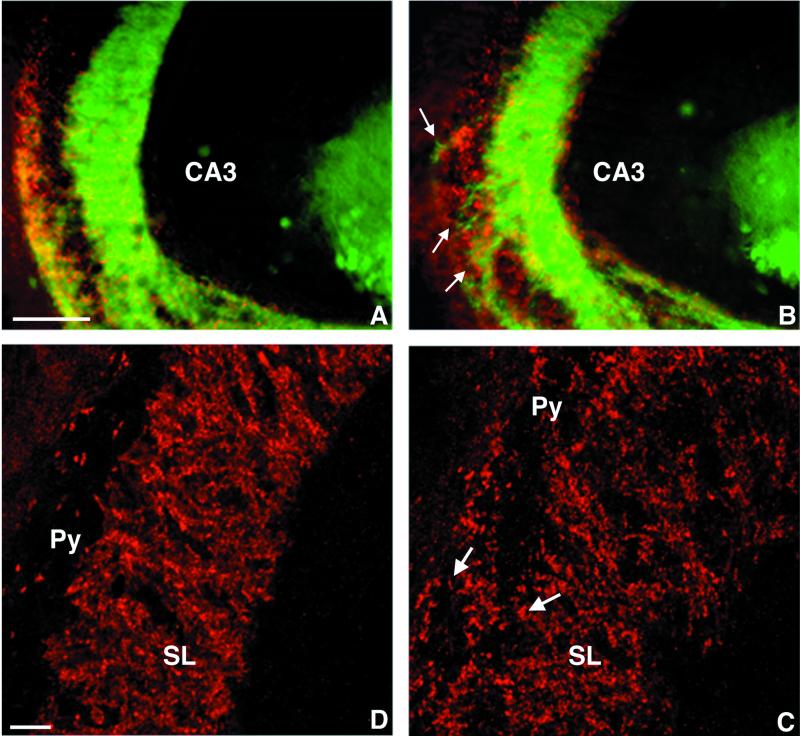
In the CA3 subfield of the hippocampus of adult wild-type mice (n = 10), immunocytochemistry for calbindin revealed that the mossy fibers were oriented strictly parallel to the direction of the pyramidal cell layer and formed a suprapyramidal and an infrapyramidal bundle (Fig. 3A). In CHL1-deficient mice (n = 10), this configuration of the mossy fibers was altered (Fig. 3B). Most of the axons still traveled along the CA3 pyramidal cell layer, and a suprapyramidal and an infrapyramidal bundle could be distinguished, but many small bundles or individual thin mossy fibers projected through the CA3 region, forming a network between the suprapyramidal and infrapyramidal bundles. The distribution of the mossy fibers terminals, as revealed by immunocytochemistry for synaptophysin (Fig. 3C and D), confirmed these observations. In wild-type mice, the mossy fiber synaptic boutons are organized in a laminated pattern (Fig. 3C) as described by Amaral and Witter (2). Only a few synapses are detectable within the pyramidal cell layer of the CA3 subfield, and the pyramidal cell layer and the stratum lucidum can be clearly distinguished. In contrast, in CHL1-deficient mice this laminated organization was lost (Fig. 3D). Mossy fiber terminals appeared to be distributed not only in the suprapyramidal and infrapyramidal bundles but also throughout the CA3 pyramidal cell body layer. Numerous boutons surrounding the pyramidal cell soma were detected (see arrows in Fig. 3D), suggesting that the terminals were formed not only on the proximal part of the pyramidal cell dendrites but also on their cell bodies.
FIG. 3.
Mossy fiber organization in adult CHL1-deficient mice. Sagittal sections stained for the presence of calbindin (green in panels A and B; mossy fiber axons) and synaptophysin (red in panels A to D; mossy fiber synaptic boutons). Conventional fluorescence microscopy (A and B) showing that in wild-type mice (A) the mossy fibers are orientated strictly parallel to the pyramidal cell layer; in contrast, in CHL1-deficient mice (B) many small bundles or individual thin mossy fibers travel through the CA3 region. Higher-magnification confocal microscopy showing the distribution of the mossy fiber terminals in the CA3 subfield of the hippocampus. (C) In wild-type mice the mossy fiber synaptic boutons are organized in a laminated pattern, only few synapses are detectable within the pyramidal cell layer (Py); the stratum lucidum (SL) can be clearly distinguished from the stratum pyramidale. (D) In CHL1-deficient mice, mossy fiber terminals are also detected throughout the CA3 pyramidal cell body layer surrounding the pyramidal cell soma (arrows). Scale bars, 200 μm (A and B) and 50 μm (C and D).
Projections of olfactory neurons.
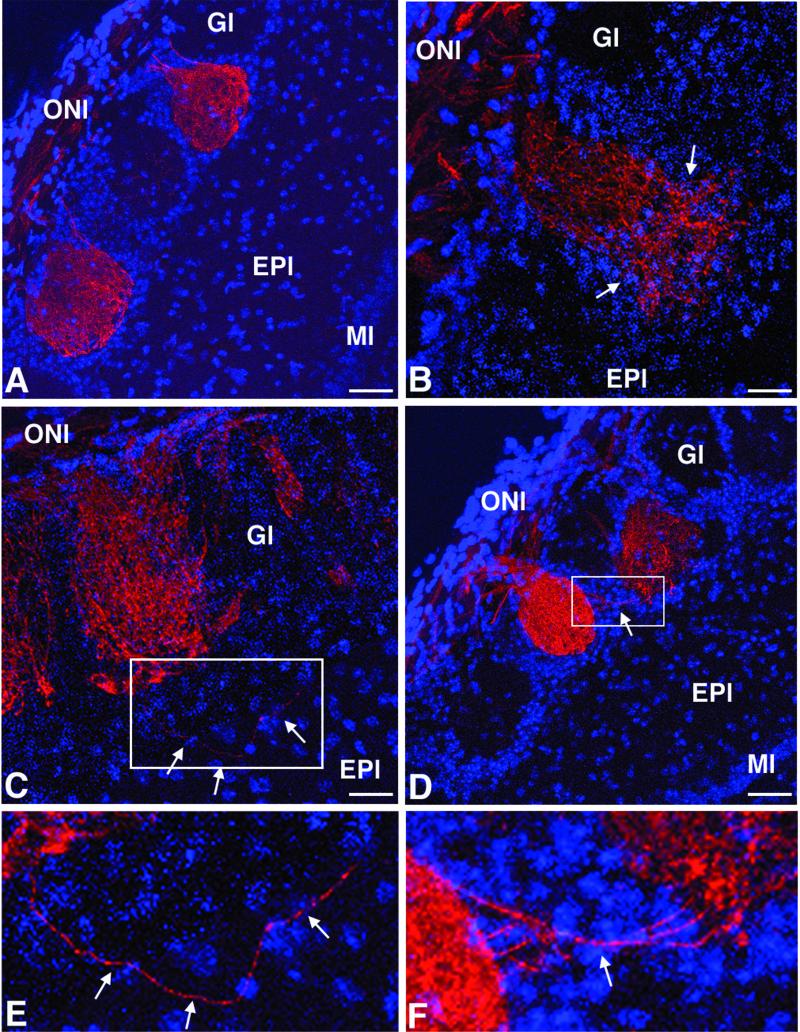
The projection pattern of the primary sensory olfactory neurons in CHL1-deficient mice was analyzed by using as a marker the plant lectin DBA. This lectin labels a subpopulation of primary sensory olfactory neurons representative of the total population of these cells (86). In adult wild-type mice (n = 5), DBA-labeled olfactory neurons terminated exclusively in the olfactory bulb glomeruli, and one axon contacted always selectively one glomerulus (Fig. 4A). In contrast, in CHL1-deficient mice (n = 5), some axons were observed to project to two glomeruli (Fig. 4D) or passing through the glomerular layer and terminating in the external plexiform layer (Fig. 4C). In addition, as shown in Fig. 4B, the arborizations of some olfactory axons were not restricted to a glomerulus, delimited by the periglomerular cell populations, but extended outside of this cellular border. As indicated by DAPI staining of the cell nuclei, no other major morphological differences between wild-type and CHL1-deficient mice were detected in the olfactory bulb (Fig. 4A to D).
FIG. 4.
Olfactory nerve axons in adult CHL1-deficient mice. Confocal microscopy pictures of olfactory bulb frontal sections stained for the detection of the olfactory nerve axons labeled with the plant lectin DBA conjugated to biotin (in red). Cell nuclei labeled with DAPI appear in blue. (A) In wild-type mice, olfactory nerve terminals generally project to only one olfactory glomerulus. In contrast, in CHL1-deficient mice, some olfactory axons terminate in two glomeruli (D) or pass through the glomerular layer and terminate in the external plexiform layer (C). (B) In addition, the arborizations of some olfactory axons extend outside a particular olfactory glomerulus. (E and F) Higher-magnification photomicrographs of the ares indicated in panels C and D, respectively. Scale bars, 50 μm (A and D) and 25 μm (B and C).
Expression of L1, NrCAM, neurofascin, F3/contactin, and NCAM180 mRNA in CHL1-deficient mice.
Levels of L1, Nr-CAM, neurofascin, F3/contactin, and NCAM180 mRNAs were examined in tissue sections of brains from adult mice by using in situ hybridization of digoxigenin-labeled antisense cRNA probes. No significant differences in L1, NrCAM, neurofascin, and F3/contactin mRNA expression were detected in adult CHL1 mutant mice (n = 5) compared to wild-type littermates (n = 4) (Fig. 5 and data not shown). In contrast, CHL1-deficient mice showed an elevated expression of NCAM180 mRNA in comparison to wild-type littermates (Fig. 5). Quantitative in situ hybridization with a 35S-labeled probe revealed an increase of NCAM180 mRNA in CHL1-deficient mice (n = 5) (Fig. 6) in the glomerular (33.03%; F(1,34) = 26.80 and P < 0.0001), mitral cell (31.63%; F(1,34) = 17.15 and P = 0.0002), and granule cell (39.92%; F(1,34) = 12.05 and P = 0.0014) layers of the olfactory bulb; in the cortex (39.1%; F(1,34) = 7.12 and P = 0.0116); in the CA1, CA2, and CA3 fields of the hippocampus (57.45%; F(1,34) = 11.79 and P = 0.0016); in the dentate gyrus (46.98%; F(1,34) = 8.53 and P = 0.0062); and in the amygdala (33.04%; F(1,34) = 5.63 and P = 0.0234) in comparison to wild-type littermate mice (n = 4). In the thalamus of mutant mice, NCAM180 mRNA expression was also increased (25.83%) compared to wild-type mice, but this difference did not reach statistical significance.
FIG. 5.
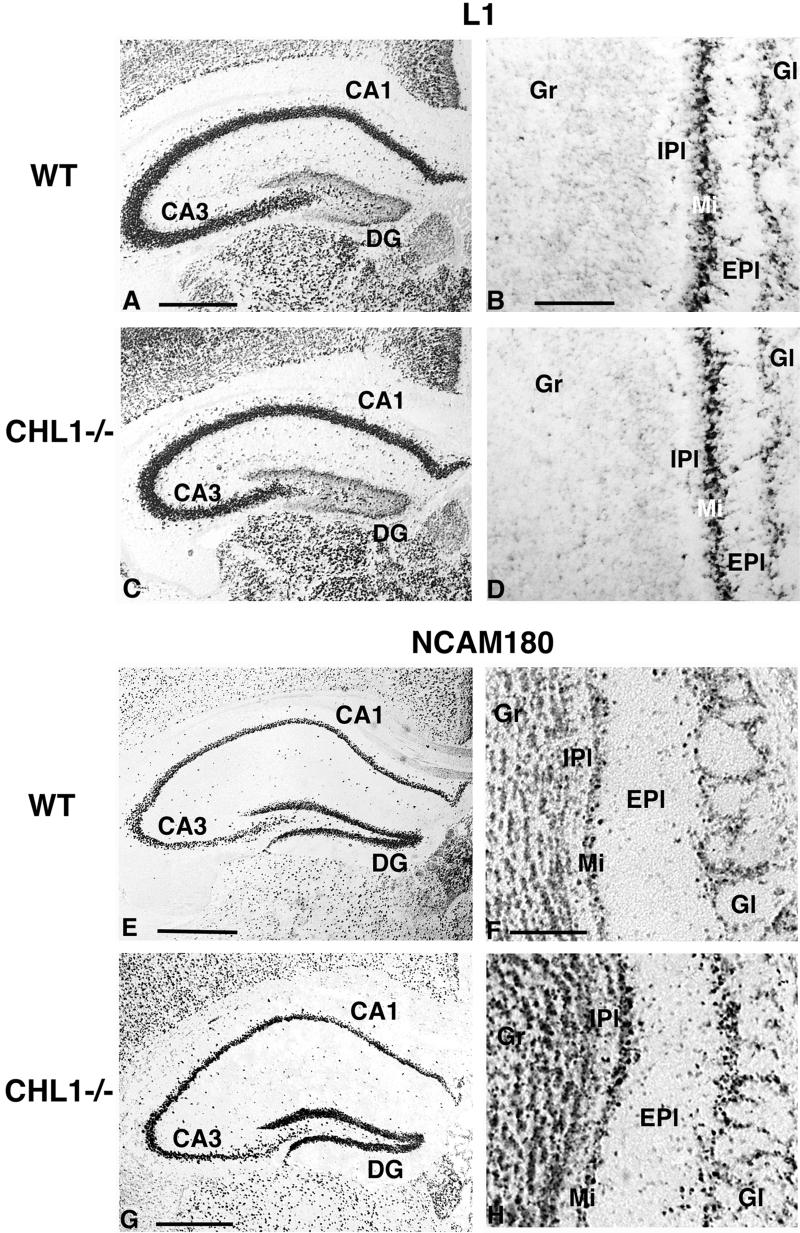
L1 and NCAM180 mRNA expression in adult wild-type and CHL1-deficient mice. Frontal brain sections showing the expression of L1 (A to D) and NCAM180 (E to H) mRNA detected by in situ hybridization with digoxigenin-labeled probes in the hippocampus (A, C, E, and G) and olfactory bulb (B, D, F, and H) of wild-type (A, B, E, and F) and CHL1-deficient (C, D, G, and H) mice. The spatial pattern and the staining intensity for L1 mRNA are similar for wild-type and mutant mice. Note the characteristic spatial expression pattern of L1 in the hippocampus (high in the CA1 to CA3 regions and low in the dentate gyrus of the hippocampus) and in the olfactory bulb (high in the mitral cell layer and glomerular layer of the olfactory bulb and extremely low in the granular cell layer). The staining intensity and number of NCAM180 mRNA-expressing cells are increased in mutant compared to wild-type mice. Abbreviations: CA1, hippocampal CA1 region; CA3, hippocampal CA3 region; DG, dentate gyrus of the hippocampus; Gr, granular cell layer, IPl, internal plexiform layer, Mi, mitral cell layer, EPl, external plexiform layer; Gl, glomerular layer of the olfactory bulb. Scale bars: 565 μm (A and C), 85 μm (B and D), 700 μm (E), 180 μm (F and H), and 600 μm (G).
FIG. 6.
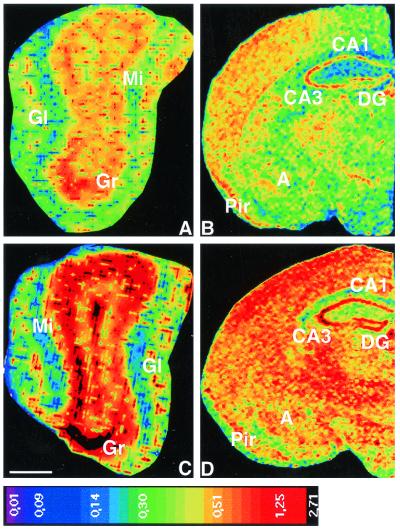
NCAM180 mRNA expression in adult CHL1-deficient mice. Calibrated pseudocolor images of film autoradiograms showing densities of hybridization in the main olfactory bulb (A and C) and the brain (B and D) of wild-type (A and B) and CHL1-deficient (C and D) mice. Compared to wild-type mice, CHL1-deficient mice showed a strong increase in NCAM180 mRNA expression in the glomerular (Gl), mitral cell (Mi), and granular cell (Gr) layers of the olfactory bulb; in the cortex; in the CA1, CA2, and CA3 fields of the hippocampus; in the dentate gyrus (DG); and in the amygdala (A). Pir, piriform cortex. The values of the color calibration bar are uncorrected optical densities (NIH program). Scale bar (in panel C), 500 μm (A and C) and 1,000 μm (B and D).
Behavioral analysis.
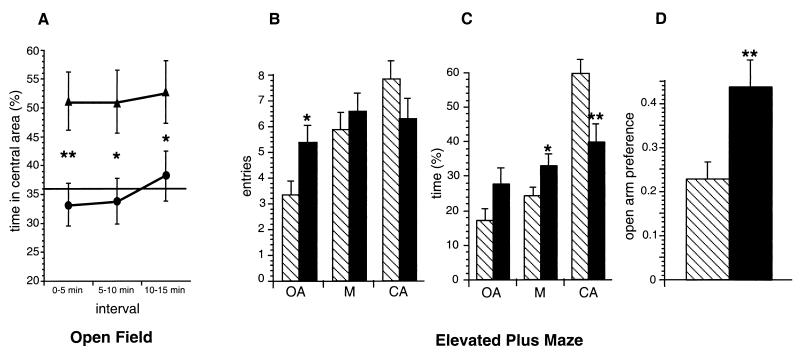
A behavioral analysis of CHL1-deficient mice in comparison to wild-type littermates was performed after backcrossing to strain C57BL/6 for eight generations. Assessment of the general state, gross sensory functions, reflexes, and motor abilities based on grip strength and rota-rod tests did not reveal any significant difference between adult CHL1-deficient (n = 12) and wild-type littermate control animals (n = 12) (data not shown). In the open field test, CHL1-deficient animals (n = 17) spent significantly more time (52%) in the central area of the maze during the entire test time compared to control animals (36%, n = 20) (F(1,35) = 8.12 and P = 0.0073) and also during each 5-min interval (Fig. 7A), which could indicate changes in the exploratory behavior or reduced anxiety in the novel environment. During the 15 min of the test, the CHL1-deficient mice moved a shorter total distance (72.0 m) than the controls (85.6 m; F(1,35) = 6.302 and P = 0.0168), but this was not caused by inactivity since the percentages of active times, the times resting, and the durations of the rests were similar. Repetition of the experiment with a different group of animals (n = 8 for each genotype) yielded very similar results (data not shown). The elevated plus maze (48, 64) and the light-dark avoidance test (18, 19, 85) were performed as paradigms indicative of the anxiety level. In the elevated plus maze, CHL1-deficient mice (n = 35) showed significant differences to their littermates (n = 34) (Fig. 7B to D). Mutant mice entered the open arm more often (F(1,67) = 5.45 and P = 0.0226), spent less time in the closed arm (F(1,67) = 9.21 and P = 0.0034), and stayed longer in the central part of the maze (F(1,67) = 4.11 and P = 0.0467). The preference for the open arm (time on open arm divided by the total time on closed and open arms) as defined by Fernandes and File (32) was significantly higher for mutants than for controls (F(1,67) = 8.29; P = 0.0054). In contrast, the light-dark avoidance paradigm revealed no significant differences either in the number of entries into or in the time spent in the dark compartment for CHL1-deficient (n = 12) compared to wild-type (n = 12) littermate mice.
FIG. 7.
Behavior of CHL1-deficient and wild-type littermate mice. (A) CHL1-deficient (▴, n = 17) and wild-type (•, n = 20) littermate mice were placed for 15 min in an open field (50 by 50 cm). The percentage of the time ± the standard error of the mean (SEM) spent in the central area of the open field maze during 5-min intervals was significantly higher for CHL1-deficient mice compared to controls (✽✽, P < 0.01; ✽, P < 0.05 [according to ANOVA]). The horizontal bar (36%) indicates the percentage of the time expected to be spent by chance in the central area according to the proportions of the maze (chance level). (B to D) CHL1-deficient (solid bars, n = 35) and wild-type (hatched bars, n = 34) littermate mice were placed for 5 min on an elevated plus maze. In panel B, the bars represent the average number of entries ± the SEM into the open arms (OA), the closed arms (CA), and the center (M) (✽, P < 0.05 [according to ANOVA]). The number of entries into the open arm were significantly higher for the mutants compared to the controls. In panel C, the bars represent the average time as a percentage of the total time ± the SEM spent on the open arms (OA), in the closed arms (CA), and in the center (M). CHL1-deficient mice spent significantly more time in the center (✽, P < 0.05 [according to ANOVA]) and less time in the closed arms (✽✽, P < 0.01 [according to ANOVA]). In panel D, the bars represent the preference for the open arm ± the SEM (time on open arm divided by the total time on closed and open arms), which was significantly higher for mutant compared to control mice (✽✽, P < 0.01 [according to ANOVA]).
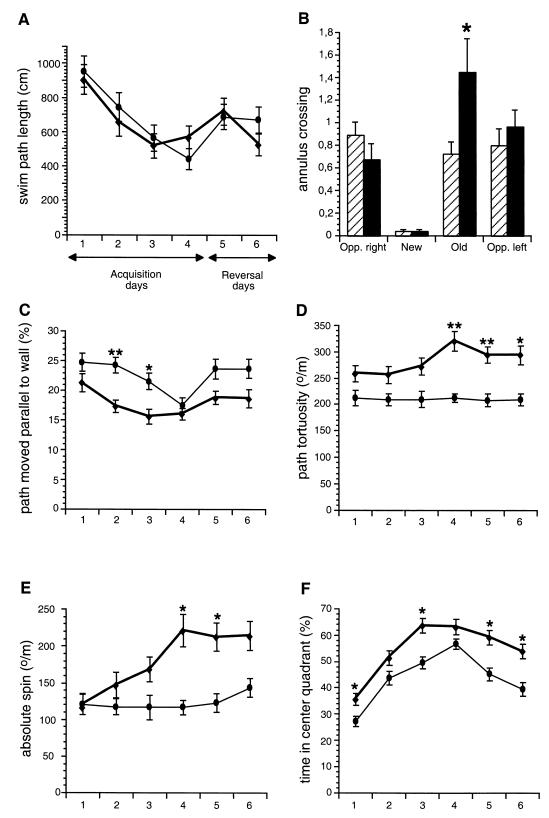
Hippocampal deficits are often reflected by reduced performance and an altered search strategy in the Morris water maze (57, 58, 59, 94). We subjected CHL1-deficient (n = 12) and wild-type littermates (n = 11) to an intensive training in the hidden platform version of the water maze. In 24 training trials, the mutant mice showed similar acquisition of the task, as measured by escape latency (data not shown) and path length (Fig. 8A). During the probe trials after platform reversal, escape latency (data not shown) and path length were similar for both groups. However, the number of crossings over the old goal was significantly increased for CHL1-deficient mice at day 5 (one-way ANOVA, genotype: F(1,21) = 6.95 and P = 0.0154; Fig. 8B). In subsequent trials, escape latency and swim path length for the new platform position did not differ significantly between wild-type and mutants. Analysis of the swim patterns by use of the Wintrack analysis program revealed differences between CHL1-deficient and wild-type control animals (Fig. 8C to F). The percentage of the time spent swimming parallel to the wall of the pool was reduced for CHL1-deficient mice compared to controls during acquisition (repeated-measures ANOVA, genotype: F(1,21) = 9.70 and P = 0.0052) and reversal days (not significant; P = 0.0667). As shown in Fig. 8C, the mutant mice spent significantly less time swimming parallel to the wall of the pool at days 2 and 3 (one-way ANOVA, genotype: F(1,21) = 12.95 and P = 0.0017 and F(1,21) = 6.60 and P = 0.0179, respectively). In addition, the swim path tortuosity was significantly elevated for CHL1-deficient mice compared to wild-type during both acquisition and reversal (repeated measures ANOVA, genotype, acquisition: F(1,21) = 13.57 and P = 0.0014; reversal: F(1,21) = 10.87 and P = 0.0034; Fig. 8D). Furthermore, the absolute spin was elevated for the mutants compared to controls (repeated measures ANOVA, genotype, reversal: F(1,21) = 5.34 and P = 0.0311; Fig. 8E). Finally, the time spent in the center quadrant was also elevated for the CHL1-deficient mice compared to controls during acquisition and reversal days (repeated-measures ANOVA, genotype, acquisition: F(1,21) = 5.91 and P = 0.0241; reversal: F(1,21) = 6.90 and P = 0.0158; Fig. 8F).
FIG. 8.
Performance of CHL1-deficient and wild-type littermate mice in the spatial version of the water maze. CHL1-deficient (♦, n = 12) and wild-type (•, n = 11) littermate mice were trained in the hidden-platform version of the water maze for four consecutive days with six trials per day (Acquisition). On day 5 the platform was moved into the opposite quadrant for the 2 last days (12 trials, Reversal). (A) Average swim path length ± the SEM. CHL1-deficient and wild-type mice showed similar acquisitions of the task. (B) Bars represent average number of annulus crossings ± the SEM at day 5 (six trials). The number of annulus crossings over the old goal (Old) was significantly higher for CHL1-deficient (solid bars) in comparison to wild-type mice (hatched bars). New, new platform position; Opp. right and Opp. left, two virtual platform positions. (C) Percentage of the path swimming parallel to the wall ± the SEM. At days 2 and 3, CHL1-deficient mice compared to control mice moved for shorter distances parallel to the wall. (D) Swim path tortuosity ± the SEM. In comparison to wild-type mice, CHL1-deficient mice showed more absolute direction changes per path length. (E) Absolute spin ± the SEM. Signed direction changes per path length were elevated for CHL1-deficient mice. (F) Time spent in the center quadrant ± the SEM. CHL1-deficient mice spent more time in the center quadrant, corresponding to half the diameter of the water maze, than their wild-type littermates (for all graphs: ✽, P < 0.05; ✽✽, P < 0.01; ✽✽✽, P < 0.001 [according to ANOVA]).
DISCUSSION
In this study, CHL1-deficient mice were generated and analyzed at the molecular, anatomical, and behavioral levels. At the light microscopic level, brains from CHL1-deficient mice showed a normal morphology of the cerebellum, hippocampus, olfactory bulb, and cortical layers indistinguishable from wild-type littermates. Only after backcrossing to C57BL/6 mice for 10 generations, enlargement of the lateral ventricles in CHL1-deficient mice became apparent. Interestingly, two independent strains of L1-deficient mice display enlarged ventricles in the C57BL/6 but not on a 129 background (22, 72), whereas another independent L1-deficient strain (16) shows an enlarged ventricular system in the 129 background, indicating the influence of unidentified modifier genes (25, 36). Severely enlarged ventricles may be the primary cause for the development of a hydrocephalus observed in some L1-deficient mice (72) and in humans affected with the CRASH syndrome caused by mutations in L1 (for reviews, see references 35 and 44).
We investigated potential functions of CHL1 in axon guidance and synapse formation in the hippocampal formation and in the olfactory bulb. In the hippocampus, subsets of neurons express CHL1 mRNA (39, 41). Mossy fibers represent the axonal projections of dentate gyrus granule cells on pyramidal cells in the hippocampus proper and their terminals form en passant synapses in the stratum lucidum with the proximal portion of the apical dendrites of CA3 pyramidal cells. Timm's staining and immunocytochemistry for calbindin and synaptophysin as markers for mossy fibers and their terminals, respectively, showed that the mossy fiber organization in CHL1-deficient mice is altered, irrespective of their strain backgrounds. Some axons formed small bundles or traveled as individual thin mossy fibers through the CA3 region, forming a network between the suprapyramidal and infrapyramidal bundles. Numerous synaptic boutons surrounding the pyramidal cell soma indicate that terminals were formed not only on the proximal part of pyramidal cell dendrites but also on their cell bodies. In the olfactory bulb, CHL1 protein is detectable despite the lack of CHL1 mRNA-expressing cells (M. Montag-Sallaz, unpublished observations), suggesting a localization of the protein on the axons and terminals of the olfactory nerves and/or of centrifugal afferents. In adult wild-type mice, olfactory axons terminate exclusively in the glomeruli, with each axon connecting to a single specific glomerulus. In contrast, the projection pattern of some sensory olfactory neurons is altered in the olfactory bulb of CHL1-deficient mice. Using the plant lectin DBA as a marker, we show here that some axons (ca. 1 to 5% of labeled axons) either cross the glomerular layer without branching and terminate in the underlying bulbar layer, the external plexiform layer, or clearly connect to more than one glomerulus. Our findings are in agreement with the proposed role of CHL1 in neurite extension and pathfinding. The abnormal anatomy detected in adult mice could be attributed to the lack of CHL1 during development and/or to later events. Noteworthy, neurogenesis persists in the hippocampal formation during adulthood. New neurons being continuously added to the population of dentate granule cells (1, 7) have to establish correct synaptic contacts with their dendritic targets throughout the animal's lifetime (37). Neurogenesis in the dentate gyrus has been related to the behavioral trait of reactivity to novelty (47). Furthermore, granule cells have been shown to be more numerous in the dentate gyrus of adult mice living in an enriched environment (45), and mossy fiber terminals in the CA3 stratum oriens region were significantly increased in rats trained in a water maze (68). Therefore, correct integration of newly born granule cells into functional neuronal networks is certainly crucial for the processing of environmental information. Continuous expression of CHL1 may thus be central to this integration in both the developing and mature hippocampus.
Like dentate granule cells, primary sensory olfactory neurons are generated throughout life in the mature olfactory neuroepithelium of the nasal cavity. Their axons grow continually along the olfactory nerve, enter the olfactory bulb, and make connections with second-order mitral and tufted cells in synaptic complexes, the glomeruli. Our results demonstrate that in the abscence of CHL1 some axons or axonal ensembles do not find their proper targets, supporting a possible role for CHL1 in the establishment of connections between sensory and second-order olfactory neurons. Although CHL1-deficient mice actively respond to olfactory stimulation in a way similar to that of wild-type mice (A. Baarke and D. Montag, unpublished results), detailed characterization of their olfactory capabilities will require future studies.
Aberrant targeting of axons has also been observed in mice deficient for the L1 family members L1 (12, 16), contactin (9), and Nr-CAM (T. Sakurai, M. Lustig, and M. Grumet, Proc. 29th Annu. Meet. Soc. Neurosci., abstr. 9.3, p. 8, 1999) and for NCAM (20, 21), netrin-1 (6, 80), and the L1 family-related netrin-1 receptor DCC (30). As in CHL1-deficient mice, not all axons in a pathway fail to reach their proper target in these mutants, indicating that a particular cell recognition molecule is not essential for, but optimizes proper targeting. Alternatively, loss of one cell recognition molecule may be partially compensated or circumvented by others and/or may result in deregulation of their expression. Therefore, we analyzed the expression of mRNAs for NCAM180 and several L1 family members by in situ hybridization. In adult CHL1-deficient and littermate mice, L1, Nr-CAM, neurofascin, and F3/contactin mRNA expression did not appear to differ. In contrast, NCAM180 mRNA was upregulated in the mutants in many brain regions, e.g., olfactory bulb, cortex, hippocampus, and amygdala. In the olfactory bulb, NCAM180 mRNA was strongly upregulated by mitral and granule cells. These neurons do not express CHL1 mRNA in wild-type mice (Montag-Sallaz, unpublished) but are the targets of sensory and centrifugal afferents, which are likely to express CHL1 (see above). In the hippocampus, NCAM180 is enriched at postsynaptic sites (65, 66, 77) and CHL1 is expressed by dentate granule and pyramidal cells (41). Furthermore, the hippocampus is a brain area well known for its plasticity and participation in learning and memory (70, 93). Evidence for the involvement of cell recognition molecules in neuronal plasticity has been accumulated during recent years (8, 26, 33, 40, 49, 61, 74). It is thus tempting to speculate that CHL1 and NCAM180 may act in concert to support synaptic plasticity and elaboration of neuronal networks.
Interestingly, phenotypically very similar alterations of mossy fiber organization and formation of aberrant synaptic boutons on CA3 pyramidal cells that we observed in CHL1-deficient mice have been reported for NCAM-deficient mice (21); in mice with an ablation of the NCAM180 isoform, which is the predominant polysialic acid-carrying molecule in the CNS; and after enzymatic removal of polysialic acid (79). Upregulation of NCAM180 mRNA in CHL1-deficient mice may be the cellular response to the absence of CHL1; however, it is evidently not sufficient to compensate for the phenotype. This is supported by work of Stork et al. (84), who showed that transgenic expression of NCAM180 does not suppress the mossy fiber lamination deficits in NCAM-deficient mice but rescues some behavioral deficits.
In a variety of tests, we analyzed whether the alterations observed at the molecular and cellular levels in the absence of CHL1 bear consequences for the animal's behavior. CHL1-deficient mice did not show abnormalities with respect to general behavior, neurological reflexes, and sensory functions and did not differ from wild-type littermates in life span, viability, or fertility. Grip strength and rota-rod performance did not indicate motor impairments. In contrast, significant differences were revealed by the open field paradigm. The mutants spent much more time in the central area, which may indicate reduction of anxiety or a different exploratory behavior. Anxiety was assessed by using the elevated plus maze and the light-dark avoidance paradigms. The performance of CHL1-deficient mice in the elevated plus maze is compatible with reduced anxiety, whereas the light-dark avoidance did not reveal such a trait. Although these paradigms address different forms of anxiety, it appears more likely that the behavior of CHL1-deficient mice in the novel plus maze environment reflects a different exploratory reaction rather than reduced anxiety. In support of this view is the observation that CHL1-deficient mice showed significant differences in the Morris water maze compared to littermates with respect to the path swimming parallel to the wall and increased swim path tortuosity, absolute spin, and time spent in the center quadrant. This may also indicate a different exploratory response toward the environment, whereas the acquisition of the task by CHL1-deficient mice, as measured by escape latency and swim path length, was similar to that of the littermates, indicating that spatial learning was not impaired. However, during the first reversal day the number of crossings over the old goal was significantly increased for mutant compared to wild-type mice, suggesting reduced flexibility. In conclusion, the observed behavioral abnormalities of CHL1-deficient mice indicate an alteration of exploratory behavior.
A dentate gyrus-selective colchicine lesion results in a loss of behavioral flexibility (94), and connectivity and morphology of mossy fibers have been correlated with the spatial behavior of mice (10, 76; for discussion, see reference 67). The alterations of hippocampal mossy fiber connectivity in CHL1-deficient mice may correlate with their different behavior, suggesting that the abnormalities of intrahippocampal neuronal networks may alter the processing of spatial information. However, environmental stimuli are complex and require the integration of sensory and emotional information at multiple levels. The crucial involvement of the hippocampus in these integrative mechanisms is well documented (29, 38), and the altered connectivity in the absence of CHL1 could result in a more general disturbance of this capacity.
In humans, distal deletions of chromosome 3 in the vicinity of the CHL1 (CALL) gene are associated with 3p- syndrome (28, 62, 63), which is characterized by multiple congenital anomalies and mental retardation (89). Recently, it has been hypothesized that CHL1 haploinsufficiency may underlie some of the mental impairment characteristics of 3p- syndrome patients (4). The behavioral and morphological abnormalities of CHL1-deficient mice revealed by our study lend support to a possible contribution of CHL1 deletions to mental deficits of 3p- patients. In addition, an association between a missense polymorphism in the CHL1 (CALL) gene and schizophrenia has been described (73).
Acknowledgments
We thank M. Grumet for providing monoclonal antibodies 2C2. We are grateful to H. Blüthmann and Y. Lang for providing feeder cells and to J. P. Julien and his coworkers for generation of chimeric mice. We thank J. Holm, G. Rougon, and P. Dirks for the donation of cDNA clones and T. Ahmed, C. Wotjak, and P. Beesley for critically reading the manuscript.
REFERENCES
- 1.Altman, J., and G. D. Das. 1965. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124:319-335. [DOI] [PubMed] [Google Scholar]
- 2.Amaral, D. G., and M. P. Witter. 1989. The three-dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience 31:571-591. [DOI] [PubMed] [Google Scholar]
- 3.Angeloni, D., M. H. Wei, and M. I. Lerman. 1999. Two single nucleotide polymorphisms (SNPs) in the CALL gene for association studies with IQ. Psychiatr. Genet. 9:165-167. [DOI] [PubMed] [Google Scholar]
- 4.Angeloni, D., N. M. Lindor, S. Pack, F. Latif, M. H. Wei, and M. I. Lerman. 1999. CALL gene is haploinsufficient in a 3p- syndrome patient. Am. J. Med. Genet. 86:482-485. [DOI] [PubMed] [Google Scholar]
- 5.Baimbridge, K. G., and J. J. Miller. 1982. Immunohistochemical localization of calcium-binding protein in the cerebellum, hippocampal formation, and olfactory bulb of the rat. Brain Res. 245:223-229. [DOI] [PubMed] [Google Scholar]
- 6.Barallobre, M. J., J. A. Del Rio, S. Alcantara, V. Borrell, F. Aguado, M. Ruiz, M. A. Carmona, M. Martin, M. Fabre, R. Yuste, M. Tessier-Lavigne, and E. Soriano. 2000. Aberrant development of hippocampal circuits and altered neural activity in netrin 1-deficient mice. Development 127:4797-4810. [DOI] [PubMed] [Google Scholar]
- 7.Bayer, S. A., J. W. Yackel, and P. S. Puri. 1982. Neurons in the granule cell layer substantially increase during juvenile and adult life. Science 216:890-892. [DOI] [PubMed] [Google Scholar]
- 8.Benson, D. L., L. M. Schnapp, L. Shapiro, and G. W. Huntley. 2000. Making memories stick: cell-adhesion molecules in synaptic plasticity. Trends Cell Biol. 10:473-482. [DOI] [PubMed] [Google Scholar]
- 9.Berglund, E. O., K. K. Murai, B. Fredette, G. Sekerkova, B. Marturano, L. Weber, E. Mugnaini, and B. Ranscht. 1999. Ataxia and abnormal cerebellar microorganization in mice with ablated contactin gene expression. Neuron 24:739-750. [DOI] [PubMed] [Google Scholar]
- 10.Bernasconi-Guastalla, S., D. P. Wolfer, and H.-P. Lipp. 1994. Hippocampal mossy fibers and swimming navigation in mice: correlations with size and left-right asymmetries. Hippocampus 4:53-64. [DOI] [PubMed] [Google Scholar]
- 11.Bixby, J. L., J. Lilien, and L. F. Reichardt. 1988. Identification of the major proteins that promote neuronal process outgrowth on Schwann cells in vitro. J. Cell Biol. 107:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellani, V., A. Chedotal, M. Schachner, C. Faivre-Sarrailh, and G. Rougon. 2000. Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signalling pathways in axonal guidance. Neuron 27:237-249. [DOI] [PubMed] [Google Scholar]
- 13.Chaisuksunt, V., G. Campbell, Y. Zhang, M. Schachner, and A. R. Lieberman. 2000. The cell recognition molecule CHL1 is strongly upregulated by injured and regenerating thalamic neurons. J. Comp. Neurol. 425:382-392. [DOI] [PubMed] [Google Scholar]
- 14.Chaisuksunt, V., Y. Zhang, P. N. Anderson, G. Campbell, E. Vaudano, M. Schachner, and A. R. Lieberman. 2000. Axonal regeneration from CNS neurons in the cerebellum and brainstem of adult rats: correlation with the patterns of expression and distribution of messenger RNAs for L1, CHL1, c-jun and growth-associated protein-43. Neuroscience 100:87-108. [DOI] [PubMed] [Google Scholar]
- 15.Chan, S. Y., and M. J. Evans. 1991. In situ freezing of embryonic stem cells in multiwell plates. Trends Genet. 7:76.. [DOI] [PubMed] [Google Scholar]
- 16.Cohen, N. R., J. S. Taylor, L. B. Scott, R. W. Guillery, P. Soriano, and A. J. Furley. 1998. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr. Biol. 8:26-33. [DOI] [PubMed] [Google Scholar]
- 17.Collinson, J. M., D. Marshall, C. S. Gillespie, and P. J. Brophy. 1998. Transient expression of neurofascin by oligodendrocytes at the onset of myelinogenesis: implications for mechanisms of axon-glial interaction. Glia 23:11-23. [DOI] [PubMed] [Google Scholar]
- 18.Crawley, J. N. 1981. Neuropharmacologic specificity of a simple animal model for the behavioral actions of benzodiazepines. Pharm. Biochem. Beh. 15:695-699. [DOI] [PubMed] [Google Scholar]
- 19.Crawley, J. N., and F. K. Goodwin. 1980. Preliminary report of a simple animal behavior model for the anxiolytic effects of benzodiazepines. Pharm. Biochem. Beh. 13:167-170. [DOI] [PubMed] [Google Scholar]
- 20.Cremer, H., G. Chazal, A. Carleton, C. Goridis, J. D. Vincent, and P. M. Lledo. 1998. Long-term but not short-term plasticity at mossy fiber synapses is impaired in neural cell adhesion molecule-deficient mice. Proc. Natl. Acad. Sci. USA 95:13242-13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cremer, H., G. Chazal, C. Goridis, and A. Represa. 1997. NCAM is essential for axonal groth and fasciculation in the hippocampus. Mol. Cell. Neurosci. 8:323-335. [DOI] [PubMed] [Google Scholar]
- 22.Dahme, M., U. Bartsch, R. Martini, M. Anliker, M. Schachner, and N. Mantei. 1997. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 17:346-349. [DOI] [PubMed] [Google Scholar]
- 23.Davis, J., and V. Bennett. 1994. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 269:27163-27166. [PubMed] [Google Scholar]
- 24.Davis, J. Q., T. McLaughlin, and V. Bennett. 1993. Ankyrin-binding proteins related to nervous system cell adhesion molecules: candidates to provide transmembrane and intercellular connections in adult brain. J. Cell Biol. 121:121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demyanenko, G. P., A. Y. Tsai, and P. F. Maness. 1999. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 19:4907-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dityatev, A., G. Dityatev, and M. Schachner. 2000. Synaptic strength as a function of post- versus presynaptic expression of the neural cell adhesion molecule NCAM. Neuron 26:207-217. [DOI] [PubMed] [Google Scholar]
- 27.Doerries, U., U. Bartsch, C. Nolte, J. Roth, and M. Schachner. 1993. Adaptation of a non-radiactive in situ hybridization method to electron microscopy: detection of tenascin messenger RNAs in mouse cerebellum with digoxigenin-labelled probes and gold-labelled antibodies. Histochemistry 99:251-262. [DOI] [PubMed] [Google Scholar]
- 28.Drumheller, T., B. C. McGillivray, D. Behrner, P. MacLeod, D. E. McFadden, J. Roberson, C. Vendittie, M. Chorney, and D. I. Smith. 1996. Precise localisation of 3p25 breakpoints in four patients with the 3p- syndrome. J. Med. Genet. 33:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichenbaum, H., G. Schoenbaum, B. Young, and M. Bunsey. 1996. Functional organization of the hippocampus memory system. Proc. Natl. Acad. Sci. USA 93:13500-13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fazeli, A., S. Dickinson, M. L. Hermiston, R. V. Tighe, R. G. Steen, C. G. Small, E. T. Stoeckli, K. Keino-Masu, M. Masu, H. Rayburn, J. Simons, R. T. Bronson, J. L. Gordon, M. Tessier-Lavigne, and R. A. Weinberg. 1997. Phenotype of mice lacking functional deleted in colorectal cancer (DCC) gene. Nature 386:796-804. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes, C., and S. E. File. 1996. The influence of open arm ledges and maze experience in the elevated plus-maze. Pharm. Biochem. Beh. 54:31-40. [DOI] [PubMed] [Google Scholar]
- 33.Fields, R. D., and K. Itoh. 1996. Neural cell adhesion molecules in activity-dependent development and synaptic plasticity. Trends Neurosci. 19:473-480. [DOI] [PubMed] [Google Scholar]
- 34.Franklin, K. B. J., and G. Paxinos. 1997. The mouse brain in stereotaxic coordinates. Academic Press, Inc., San Diego, Calif.
- 35.Fransen, E., G. Van Camp, L. Vits, and P. J. Willems. 1997. L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 6:1625-1632. [DOI] [PubMed] [Google Scholar]
- 36.Fransen, E., R. D'Hooge, G. Van Camp, M. Verhoye, J. Sijbers, E. Reyniers, P. Soriano, H. Kamiguchi, R. Willemsen, S. K. Koekkoek, C. I. De Zeeuw, P. P. De Deyn, A. Van der Linden, V. Lemmon, R. F. Kooy, and P. J. Willems. 1998. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum. Mol. Genet. 7:999-1009. [DOI] [PubMed] [Google Scholar]
- 37.Hastings, N. B., and E. Gould. 1999. Rapid extension of axons into the CA3 region by adult-generated granule cells. J. Comp. Neurol. 413:146-154. [DOI] [PubMed] [Google Scholar]
- 38.Henke, K., A. Buck, B. Weber, and H. G. Wieser. 1997. Human hippocampus establishes associations in memory. Hippocampus 7:249-256. [DOI] [PubMed] [Google Scholar]
- 39.Hillenbrand, R., M. Molthagen, D. Montag, and M. Schachner. 1999. The close homologue of the neural adhesion molecule L1 (CHL1): patterns of expression and promotion of neurite outgrowth by heterophilic interactions. Eur. J. Neurosci. 11:813-826. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann, K. B. 1998. The relationship between adhesion molecules and neuronal plasticity. Cell. Mol. Neurobiol. 18:461-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm, J., R. Hillenbrand, V. Steuber, U. Bartsch, M. Moos, H. Lübbert, D. Montag, and M. Schachner. 1996. Structural features of a close homologue of L1 (CHL1) in the mouse: a new member of the L1 family of neural recognition molecules. Eur. J. Neurosci. 8:1613-1629. [DOI] [PubMed] [Google Scholar]
- 42.Hooper, M., K. Hardy, A. Handsyde, S. Hunter, and M. Monk. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326:680-685. [DOI] [PubMed] [Google Scholar]
- 43.Jones, B. J., and D. J. Roberts. 1968. The quantitative measurement of motor inco-ordination in naive mice using an accelerating rota-rod. J. Pharm. 20:302-304. [DOI] [PubMed] [Google Scholar]
- 44.Kamiguchi, H., M. L. Hlavin, M. Yamasaki, V. Lemmon. 1998. Adhesion molecules and inherited diseases of the human nervous system. Annu. Rev. Neurosci. 21:97-125. [DOI] [PubMed] [Google Scholar]
- 45.Kempermann, G., H. G. Kuhn, and F. H. Gage. 1997. More hippocampal neurons in adult mice living in an enriched environment. Nature 386:493-495. [DOI] [PubMed] [Google Scholar]
- 46.Lämmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 47.Lemaire, V., C. Aurousseau, M. Le Moal, and D. N. Abrous. 1999. Behavioral trait of reactivity to novelty is related to hippocampal neurogenesis. Eur. J. Neurosci. 11:4006-4014. [DOI] [PubMed] [Google Scholar]
- 48.Lister, R. G. 1987. The use of a plus-maze to measure anxiety in the mouse. Psychopharmology 92:180-185. [DOI] [PubMed] [Google Scholar]
- 49.Lüthi, A., J. P. Laurent, A. Figurov, D. Muller, and M. Schachner. 1994. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature 372:777-779. [DOI] [PubMed] [Google Scholar]
- 50.Mansour, S. L., K. R. Thomas, and M. R. Capecchi. 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336:348-352. [DOI] [PubMed] [Google Scholar]
- 51.Martini, R., and M. Schachner. 1986. Immunoelectron microscopic localization of neural cell adhesion molecules (L1, N-CAM, and MAG) and their shared carbohydrate epitope and myelin basic protein in developing sciatic nerve. J. Cell Biol. 103:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer, O. A., H. A. Tilson, W. C. Byrd, and M. T. Riley. 1979. A method for the routine assessment of fore- and hindlimb grip strength of rats and mice. Neurobehav. Toxicol. 1:233-236. [PubMed] [Google Scholar]
- 53.Montag, D., K. P. Giese, U. Bartsch, R. Martini, Y. Lang, H. Bluethmann, J. Karthigasan, D. A. Kirschner, E. S. Wintergerst, K. A. Nave, J. Zielasek, K. V. Toyka, H.-P. Lipp, and M. Schachner. 1994. Mice deficient for the myelin-associated glycoprotein show subtle abnormalities in myelin. Neuron 13:229-246. [DOI] [PubMed]
- 54.Montag-Sallaz, M., H. Welzl, D. Kuhn, D. Montag, and M. Schachner. 1999. Novelty-induced increased expression of the immediate early genes c-fos and arg 3.1 in the mouse brain. J. Neurobiol. 38:234-246. [PubMed] [Google Scholar]
- 55.Montag-Sallaz, M., and N. Buonviso. 2002. Altered odor-induced expression of c-fos and arg 3.1 immediate early genes in the olfactory system after familiarization with an odor. J. Neurobiol. 52:61-72. [DOI] [PubMed]
- 56.Morris, R. G. M. 1984. Development of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11:47-60. [DOI] [PubMed] [Google Scholar]
- 57.Morris, R. G. M., and U. Frey. 1997. Hippocampal synaptic plasticity: role in spatial learning or the automatic recording of attended experience? Phil. Trans. R. Soc. Lond. B 352:1489-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris, R. G. M., P. Garrud, J. N. P. Rawlins, and J. O'Keefe. 1982. Place navigation impaired in rats with hippocampal lesions. Nature 297:681-683. [DOI] [PubMed] [Google Scholar]
- 59.Morris, R. G. M., F. Schenk, F. Tweedie, and L. E. Jarrard. 1990. Ibotenate lesions of hippocampus and/or subiculum: dissociating components of allocantric spatial learning. Eur. J. Neurosci. 2:1016-1028. [DOI] [PubMed] [Google Scholar]
- 60.Müller, U., N. Christina, Z. W. Li, D. P. Wolfer, H.-P. Lipp, T. Rüllicke, S. Brandener, A. Aguzzi, and C. Weissmann. 1994. Behavioral and anatomical deficits in mice homozygous for a modified beta-amyloid precursor protein gene. Cell 79:755-765. [DOI] [PubMed] [Google Scholar]
- 61.Murphy, K. J., and C. M. Regan. 1998. Contributions of cell adhesion molecules to altered synaptic weightings during memory consolidation. Neurobiol. Learning Memory 70:73-81. [DOI] [PubMed] [Google Scholar]
- 62.Narahara, K., K. Kikkawa, M. Murakami, K. Hiramotot, H. Namba, K. Tsuji, Y. Yokoyama, and H. Kimoto. 1990. Loss of the 3p25.3 band is critical in the manifestation of del(3p) syndrome: karyotype-phenotype correlation in cases with deficiency of the distal portion of the short arm of chromosome 3. Am. J. Med. Genet. 35:269-273. [DOI] [PubMed] [Google Scholar]
- 63.Nienhaus, H., U. Mau, and K. D. Zhang. 1992. Infant with del(3) (p25-pter): karyotype-phenotype correlation and review of previously reported cases. Am. J. Med. Genet. 44:573-575. [DOI] [PubMed] [Google Scholar]
- 64.Pellow, S., P. Chopin, S. E. File, and M. Briley. 1985. Validation of open:closed arm entries in an elevated plus-maze as a measure of anxiety in the rat. J. Neurosci. Methods 14:149-167. [DOI] [PubMed] [Google Scholar]
- 65.Persohn, E., G. E. Pollerberg, and M. Schachner. 1989. Immunoelectron-microscopic localization of the 180 kD component of the neural cell adhesion molecule N-CAM in postsynaptic membranes. J. Comp. Neurol. 288:92-100. [DOI] [PubMed] [Google Scholar]
- 66.Persohn, E., and M. Schachner. 1990. Immunohistological localization of the neural adhesion molecules L1 and N-CAM in the developing hippocampus of the mouse. J. Neurocytol. 19:807-819. [DOI] [PubMed] [Google Scholar]
- 67.Pleskacheva, M. G., D. P. Wolfer, I. F. Kupriyanova, D. L. Nikolenko, H. Scheffrahn, G. Dell'Omo, and H.-P. Lipp. 2000. Hippocampal mossy fibers and swimming navigation learning in two vole species occupying different habitats. Hippocampus 10:17-30. [DOI] [PubMed] [Google Scholar]
- 68.Ramirez-Amaya, V., M. L. Escobar, V. Chao, and F. Bermúdez-Rattoni. 1999. Synaptogenesis of mossy fibers induced by spatial water maze overtraining. Hippocampus 9:631-636. [DOI] [PubMed] [Google Scholar]
- 69.Ramirez-Solis, R., J. Rivera-Perez, J. D. Wallace, M. Wims, H. Zheng, and A. Bradley. 1992. Genomic DNA microextraction: a method to screen numerous samples. Anal. Biochem. 201:331-335. [DOI] [PubMed] [Google Scholar]
- 70.Riedel, G., J. Micheau, A. G. M. Lam, E. V. L. Roloff, S. J. Martin, H. Bridge, L. de Hoz, B. Poeschel, J. McCulloch, and R. G. M. Morris. 1999. Reversible neural innactivation reveals hippocampal participation in several memory processes. Nat. Neurosci. 2:898-905. [DOI] [PubMed] [Google Scholar]
- 71.Rogers, D. C., E. M. Fisher, S. D. Brown, J. Peters, A. J. Hunter, and J. E. Martin. 1997. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome 8:711-713. [DOI] [PubMed] [Google Scholar]
- 72.Rolf, B., M. Kutsche, and U. Bartsch. 2001. Severe hydrocephalus in L1-deficient mice. Brain Res. 891:247-252. [DOI] [PubMed] [Google Scholar]
- 73.Sakurai, K., O. Migita, M. Toru, and T. Arinami. 2002. An association between a missense polymorphism in the close homologue of L1 (CHL1. CALL) gene and schizophrenia. Mol. Psychiatr. 7:412-415. [DOI] [PubMed] [Google Scholar]
- 74.Schachner, M. 1997. Neural recognition molecules and synaptic plasticity. Curr. Opin. Cell Biol. 9:627-634. [DOI] [PubMed] [Google Scholar]
- 75.Scharfmann, H. E., J. H. Goodman, and A. L. Sollas. 2000. Granule-like neurons at the hilar/CA3 border after status epilepticus and their synchrony with area CA3 pyramidal cells: functional implications of seizure-induced neurogenesis. J. Neurosci. 20:6144-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schöpke, R., D. P. Wolfer, H.-P. Lipp, and M.-C. Leisinger-Trigona. 1991. Swimming navigation and structural variations of the infrapyramidal mossy fibers in the hippocampus of the mouse. Hippocampus 1:315-328. [DOI] [PubMed] [Google Scholar]
- 77.Schuster, T., M. Krug, H. Hassan, and M. Schachner. 1998. Increase in proportion of hippocampal spine synapses expressing neural cell adhesion molecule NCAM180 following long-term potentiation. J. Neurobiol. 15:359-372. [DOI] [PubMed] [Google Scholar]
- 78.Seilheimer, B., and M. Schachner. 1988. Studies of adhesion molecules mediating interactions between cells of peripheral nervous system indicate a major role for L1 in mediating sensory neuron growth on Schwann cells in culture. J. Cell Biol. 107:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seki, T., and U. Rutishauser. 1998. Removal of polysialic acid-neural cell adhesion molecule induces aberrant mossy fiber innervation and ectopic synaptogenesis in the hippocampus. J. Neurosci. 18:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serafini, T., S. A. Colamarino, E. D. Leonardo, E. Wang, R. Beddington, W. C. Skarnes, and M. Tessier-Lavigne. 1996. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87:1001-1014. [DOI] [PubMed] [Google Scholar]
- 81.Seress, L., A. I. Gulyas, I. Ferrer, T. Tunon, E. Soriano, and T. F. Freund. 1993. Distribution, morphological features, and synaptic connections of parvalbumin- and calbindin D28k-immunoreactive neurons in the human hippocampal formation. J. Comp. Neurol. 337:208-230. [DOI] [PubMed] [Google Scholar]
- 82.Smith, T. D., M. M. Adams, M. Gallagher, J. H. Morrison, and P. R. Rapp. 2000. Circuit-specific alterations in hippocampal synaptophysin immunoreactivity predict spatial learning impairment in aged rats. J. Neurosci. 20:6587-6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 84.Stork, O., H. Welzl, D. Wolfer, T. Schuster, N. Mantei, S. Stork, D. Hoyer, H. Lipp, K. Obata, and M. Schachner. 2000. Recovery of emotional behavior in neural cell adhesion molecule (NCAM) null mutant mice through transgenic expression of NCAM180. Eur. J. Neurosci. 12:3291-3306. [DOI] [PubMed] [Google Scholar]
- 85.Stork, O., H. Welzl, C. T. Wotjak, D. Hoyer, M. Delling, H. Cremer, and M. Schachner. 1999. Anxiety and increased 5-HT1A receptor response in NCAM null mutant mice. J. Neurobiol. 40:343-355. [DOI] [PubMed] [Google Scholar]
- 86.Tenne-Brown, J., and B. Key. 1999. Errors in lamina growth of primary olfactory axons in the rat and mouse olfactory bulb. J. Comp. Neurol. 410:20-30. [DOI] [PubMed] [Google Scholar]
- 87.Toth, K., and T. F. Freund. 1992. Calbindin D28k-containing nonpyramidal cells in the rat hippocampus: their immunoreactivity for GABA and projection to the medial septum. Neuroscience 49:793-805. [DOI] [PubMed] [Google Scholar]
- 88.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verjaal, M., and M. B. De Nef. 1978. A patient with a partial deletion of the short arm of chromosome 3. Am. J. Dis. Child 132:43-45. [DOI] [PubMed] [Google Scholar]
- 90.Wei, M. H., I. Karavanova, S. V. Ivanov, N. C. Popescu, C. L. Keck, S. Pack, J. A. Eisen, and M. I. Lerman. 1998. In silico-initiated cloning and molecular characterization of a novel human member of the L1 gene family of neural cell adhesion molecules. Hum. Genet. 103:355-364. [DOI] [PubMed] [Google Scholar]
- 91.Wishaw, I. Q., F. Haun, and B. Kolb. 1999. Analysis of behavior in laboratory rodents, p. 1243-1275. In U. Windhorst and H. Johansson (ed.), Modern techniques in neuroscience. Springer-Verlag, Berlin, Germany.
- 92.Wolfer, D., and H.-P. Lipp. 1992. A computer program for detailed off-line analysis of Morris water maze behavior. J. Neurosci. Methods 41:65-74. [DOI] [PubMed] [Google Scholar]
- 93.Wood, E. R., P. A. Dudchenko, R. J. Robitsek, and V. Eichenbaum. 2000. Hippocampal neurons encode information about different types of memory episodes occurring in the same location. Neuron 27:623-633. [DOI] [PubMed] [Google Scholar]
- 94.Xavier, G. F., I. Oliveira, F. J. B. Filho, and A. M. G. Santos. 1999. Dentate gyrus-selective colchicine lesion and disruption of performance in spatial tasks: difficulties in “place strategy” because of a lack of flexibility in the use of environmental cues? Hippocampus 9:668-681. [DOI] [PubMed] [Google Scholar]
- 95.Zhang, Y., R. Roslan, D. Lang, M. Schachner, A. R. Lieberman, and P. N. Anderson. 2000. Expression of CHL1 and L1 by neurons and glia following sciatic nerve and dorsal root injury. Mol. Cell Neurosci. 16:71-86. [DOI] [PubMed] [Google Scholar]