Abstract
The Génolevures online database (http://cbi.labri.fr/Genolevures/) provides tools and data relative to 4 complete and 10 partial genome sequences determined and manually annotated by the Génolevures Consortium, to facilitate comparative genomic studies of hemiascomycetous yeasts. With their relatively small and compact genomes, yeasts offer a unique opportunity for exploring eukaryotic genome evolution. The new version of the Génolevures database provides truly complete (subtelomere to subtelomere) chromosome sequences, 25 000 protein-coding and tRNA genes, and in silico analyses for each gene element. A new feature of the database is a novel collection of conserved multi-species protein families and their mapping to metabolic pathways, coupled with an advanced search feature. Data are presented with a focus on relations between genes and genomes: conservation of genes and gene families, speciation, chromosomal reorganization and synteny. The Génolevures site includes an area for specific studies by members of its international community.
INTRODUCTION
Comparative analysis of genomes is greatly facilitated when their sequences are complete, fully assembled and carefully annotated. Detailed analysis of species- and clade-specific gain or loss of function, and expansions or contractions of gene families, provide useful insight into the mechanisms of molecular evolution and can be performed with confidence when data are complete. The Génolevures online database provides such data for complete genomes of four species from the class of Hemiascomycete yeasts, search and analysis tools for comparing these genomes and community pages for ongoing developments. New complete genomes will be added in 2006.
With their relatively small and compact genomes, yeasts offer a unique opportunity to explore eukaryotic genome evolution by comparative analysis of several species. Yeasts are widely used as cell factories, for the production of beer, wine and bread and more recently of various metabolic products such as vitamins, ethanol, citric acid, lipids, etc. Yeasts can assimilate hydrocarbons (genera Candida, Yarrowia and Debaryomyces), depolymerise tannin extracts (Zygosaccharomyces rouxii) and produce hormones and vaccines in industrial quantities through heterologous gene expression. For review see Ref. (1). Several yeast species are pathogenic for humans. Among the most frequent disease agents are the Hemiascomycetes Candida albicans, Candida glabrata, Candida tropicalis and the Basidiomycete Cryptococcus neoformans. Even Saccharomyces cerevisiae may be pathogenic in immunocompromised patients (2). The most well known yeast in the Hemiascomycete class is S.cerevisiae (3), widely used as a model organism for molecular genetics and cell biology studies, and as a cell factory. As the most thoroughly-annotated genome of the small eukaryotes, it is a common reference for the annotation of other species. The hemiascomycetous yeasts represent a homogeneous phylogenetic group of eukaryotes with a relatively large diversity at the physiological and ecological levels. Comparative genomic studies within this group have proved very informative (4–7).
The Génolevures program is devoted to large-scale comparisons of yeast genomes from various branches of the Hemiascomycete class, with the aim of addressing basic questions of molecular evolution such as the degrees of gene conservation, the identification of species-specific, clade-specific or class-specific genes, the distribution of genes among functional families, the rate of sequence and map divergences and mechanisms of chromosome shuffling.
COMPLETE SEQUENCING AND ANNOTATION OF YEAST GENOMES
The Genoscope and the Institut Pasteur provide high-quality sequence data at 10× or better coverage, assembled into complete chromosomes from subtelomere to subtelomere, usually with no more than one gap per chromosome. Protein-coding and tRNA genes are identified using a variety of in silico methods reported elsewhere and are manually annotated by a network of volunteer experts. Comparative analysis of four genomes was reported in (8). Ongoing Génolevures sequencing projects are reported on and included in the online database as data are released. Currently the database contains 55 693 317 nt comprising 24 147 protein-coding genes and 1124 tRNA or snRNA genes.
The focus of the Génolevures database is to describe the relations between genes and genomes. We curate relations of orthology and paralogy between genes, as individuals or as members of protein families, chromosomal map reorganization and gain and loss of genes and functions. We do not provide detailed annotations of individual genes and proteins of S.cerevisiae which are already carefully maintained by MIPS and CYGD (http://mips.gsf.de/projects/fungi) (9) and SGD (http://www.yeastgenome.org/) (10) as well as in general-purpose databases such as UniProt (11) and EMBL (12).
GÉNOLEVURES PROTEIN FAMILIES
While extensive chromosomal rearrangements combined with segmental and massive duplications make comparisons of yeast genome sequences difficult (13), relations of homology between protein-coding genes can be identified despite their great diversity at the molecular level (8). Families of homologous proteins provide a powerful tool for appreciating conservation, gain and loss of function within yeast genomes. Génolevures provides a unique collection of paralogous and orthologous protein families, identified using a novel consensus clustering algorithm (M. Nikolski, manuscript submitted) applied to a complementary set of homeomorphic [sharing full-length sequence similarity and similar domain architectures, see (14)] and nonhomeomorphic systematic Smith–Waterman (15) and Blast (16) sequence alignments. Similar approaches are developed on a wider scale (14) and are complementary to these yeast-specific families.
EXPLORING GENOLEVURES DATA
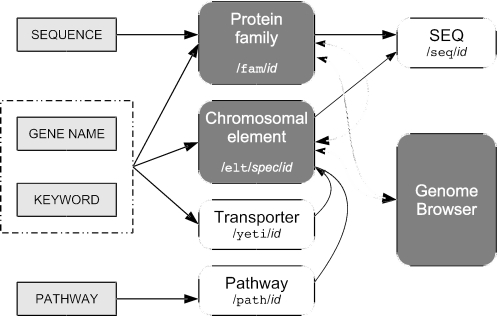
The Génolevures online database is designed to help scientists gain insight into the mechanisms of eukaryotic molecular evolution by asking specific questions about the relationships between DNA and protein sequences (Figure 1; examples are shown in online Supplementary Data).
Figure 1.
Links between Génolevures data and tools showing the principal workflows used by scientific users. Dark gray boxes represent dynamic, database-backed web pages, white boxes represent static web pages. Shorthand URL prefixes for these pages are shown in a monospaced font.
What genes exist, as orthologs for my favorite gene or as members of a functional class? (URL prefixes /concordance and /blast) Génolevures data can be searched by keyword, S.cerevisiae gene name, alignment to an arbitrary DNA or protein sequence and protein family identifier. A query simultaneously searches for and can return genes that have or may have a translation product, RNA and other genes that may have a transcription product only, cis-active elements and cross-genome protein families.
What is known about a given chromosomic element? (URL prefixes /elt) Each element, coding or not, has a summary page with a linkable URL that presents what is known about that element: annotation, chromosomic neighborhood and inter-genome alignments (with a clickable map), membership in a protein family, sequence data and domain architecture when known. Protein family membership is indicated both with the phyletic pattern and the phylogenetic profile of the family, which provides an immediate impression of the degree of conservation of that gene in hemiascomycete yeast species.
What relations exist in a protein family? (URL prefixes/fam) A protein family contains proteins with an observable evolutionary relationship that generally speaking lets one infer functional similarity. Each protein family is described on a summary page with a linkable URL that shows a cartoon of the pairwise relations between family members, linked annotations of the individual genes and a decorated multiple alignment of the family members computed with T-COFFEE (17). Links are provided to a pairwise distance matrix, a FastA file of protein sequences and a position-specific scoring matrix; the latter can be used to jump-start an iterative PSI-BLAST (16) search in public databanks for proteins similar to family members.
How are the individual genomes organized? (URL prefixes/elt and /perl/gbrowse) Chromosomal maps can be explored starting from the species page (e.g. /elt/CAGL for C.glabrata or /elt/YALI for Y.lipolytica) or directly through the genome browser (18), which provides a zoomable view of a chromosomal neighborhood with annotation tracks for different gene types and sequence features, and relations to orthologs in protein families showing conservation of function and synteny (when observable).
How are metabolic pathways conserved? (URL prefixes/path) Conservation of genes participating in KEGG (19) metabolic pathways may be explored, which makes it possible to emit hypotheses concerning the conservation of those pathways or the necessity of a particular gene for a given enzymatic function. Pathway conservation in a species is computed by coloring S.cerevisiae KEGG pathways with orthologs identified by Génolevures protein families. Each colored pathway contains both a summary and a detailed table of orthologs for each enzyme with useful information such as gene deletion effects.
How are membrane proteins and transporters classified? (URL prefixes /yeti) The YETI classification of these proteins from André Goffeau's lab (20), which indicates evolutionary relationships traced using non-ambiguous functional and phylogenetic criteria derived from the TCDB (21) classification system, can be explored and searched across the sequenced species.
Can I obtain sequence data? (URL prefixes /seq and /download) The latest release of annotated sequence data and protein family classification may be downloaded for local analysis. All Génolevures DNA and protein sequence data are also publicly available in EMBL and UniProt.
ONGOING DEVELOPMENTS
The Consortium is currently sequencing other yeast genomes from the Hemiascomycete class which will benefit from the same annotation pipeline. These genomes will be particularly helpful in refining Génolevures protein families, and in ongoing work on the construction of comparative views of cell function through inference of networks of protein–protein and protein–ligand interactions. Consortium member laboratories will continue to contribute results from a variety of focused studies, e.g. (22–24).
TECHNICAL NOTES
The Génolevures database uses a straightforward object model mapped to a relational database. Flexibility in the design is guaranteed through the use of controlled vocabularies: the Sequence Ontology (25) for DNA sequence features and GLO, our own ontology for comparative genomics (D. Sherman, unpublished data). Browsing of genomic maps and sequence features is provided by the Generic Genome Browser (18). The Blast service is provided by NCBI Blast 2.2.6 (16). The Génolevures web site uses a REST architecture internally (26) and extensively uses the BioPerl package (27) for manipulation of sequence data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We wish to thank all our colleagues from the Génolevures Consortium for numerous, friendly and creative discussions and for their devoted contributions to the sequencing, assembly and annotations of the yeast genomes. Ingrid Lafontaine, Emmanuel Talla and Hélène Ferry-Dumazet significantly contributed to the refinement of Génolevures protein families. The YETI classification of transporters and membrane proteins is kindly provided by Benoît De Hertogh, Frédéric Hancy, Philippe Baret and André Goffeau. Comparative analysis of metabolic pathways is developed and maintained by F.I. Lionel Frangeul developed and maintains the CAAT-Box annotation system. Special thanks to Jean Weissenbach and Patrick Wincker of the Génoscope and Christiane Bouchier of the Institut Pasteur who have made the sequencing of these yeasts possible. Hardware and technical support for Génolevures is provided by the Laboratoire Bordelais de Recherche en Informatique (LaBRI, CNRS UMR 5800) for the Bordeaux Center for Bioinformatics (CBiB) and is made possible by funding from the University Bordeaux 1, the Aquitaine Région through the program ‘Génotypage et Génomique comparée’ and the ACI IMPBIO ‘Génolevures En Ligne.’ Génolevures is supported by CNRS (GDR 2354), various sources from host institutions of participating laboratories and by CNRG through Génoscope and the Réseau National des Génopoles. Funding to pay the Open Access publication charges for this article was provided by the CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kurtzmann C.P., Fell J.W., editors. The Yeasts, A Taxonomic Study, 4th edition. Amsterdam: Elsevier; 1998. [Google Scholar]
- 2.Tawfik O.W., Papasian C.J., Dixon A.Y., Potter L.M. Saccharomyces cerevisiae pneumonia in a patient with acquired immune deficiency syndrome. J. Clin. Microbiol. 1989;27:1689–1691. doi: 10.1128/jcm.27.7.1689-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. 563–567. [DOI] [PubMed] [Google Scholar]
- 4.Souciet J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Casaregola S., de Montigny J., Duchateau-Nguyen G., et al. Génolevures: Genomic exploration of the hemiascomycetous yeasts. FEBS Lett. 2000;487:1–149. doi: 10.1016/s0014-5793(00)02292-4. [DOI] [PubMed] [Google Scholar]
- 5.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 6.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 8.Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuveglise C., Talla E., et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 9.Mewes H.W., Frishman D., Guldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry J.M., Adler C., Ball C., Chervitz S.A., Dwight S.S., Hester E.T., Jia Y., Juvik G., Roe T., Schroeder M., et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro M., Cochrane G., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llorente B., Malpertuy A., Neuveglise C., de Montigny J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., et al. Genomic exploration of the hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. [DOI] [PubMed] [Google Scholar]
- 14.Wu C.H., Nikolskaya A., Huang H., Yeh L.S., Natale D.A., Vinayaka C.R., Hu Z.Z., Mazumder R., Kumar S., Kourtesis P., et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 18.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arwa A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Hertogh B., Hancy F., Goffeau A., Baret P.V. Emergence of species-specific transporters during evolution of the Hemiascomycete phylum. Genetics. 2005 doi: 10.1534/genetics.105.046813. doi:10.1534/genetics.105.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier M.H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuveglise C., Chalvet F., Wincker P., Gaillardin C., Casaregola S. Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot. Cell. 2005;4:615–624. doi: 10.1128/EC.4.3.615-624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richard G.F., Kerrest A., Lafontaine I., Dujon B. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol. Biol. Evol. 2005;22:1011–1023. doi: 10.1093/molbev/msi083. [DOI] [PubMed] [Google Scholar]
- 24.Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 25.Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fielding R., Taylor R.N. Principled design of the modern Web architecture. ACM Trans. Internet Technol. 2002;2:115–150. [Google Scholar]
- 27.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G.R., Korf I., Lapp H., et al. The Bioperl Toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.