Abstract
Survival of bloodstream form Trypanosoma brucei, the agent of African sleeping sickness, normally requires mitochondrial gene expression, despite the absence of oxidative phosphorylation in this stage of the parasite's life cycle. Here we report that silencing expression of the α subunit of the mitochondrial F1-ATP synthase complex is lethal for bloodstream stage T. brucei as well as for T. evansi, a closely related species that lacks mitochondrial protein coding genes (i.e. is dyskinetoplastic). Our results suggest that the lethal effect is due to collapse of the mitochondrial membrane potential, which is required for mitochondrial function and biogenesis. We also identified a mutation in the γ subunit of F1 that is likely to be involved in circumventing the requirement for mitochondrial gene expression in another dyskinetoplastic form. Our data reveal that the mitochondrial ATP synthase complex functions in the bloodstream stage opposite to that in the insect stage and in most other eukaryotes, namely using ATP hydrolysis to generate the mitochondrial membrane potential.
Keywords: ATP synthase, dyskinetoplasty, mitochondria, RNA editing, trypanosome
Introduction
During its life cycle the unicellular eukaryote Trypanosoma brucei, an important pathogen of humans and livestock (http://www.who.int/tdr/diseases/tryp/default.htm), alternates between a mammalian host and an insect vector, the tsetse fly. The environmental changes encountered by the parasite require significant morphological and physiological adaptations, reflected, for example, by regulation of the activity of the organism's single mitochondrion (Schneider, 2001; Schnaufer et al, 2002; Matthews, 2005). Whereas the insect stage has a highly active mitochondrion and generates energy by both oxidative and substrate level phosphorylation, the long slender (LS) bloodstream stage in the mammalian host generates energy through glycolysis (Coustou et al, 2003; Hannaert et al, 2003). As a consequence, the LS stage mitochondrion is devoid of cristae and cytochrome-containing respiratory complexes and was thought to be largely inactive. Surprisingly, it was recently shown that replication of mitochondrial DNA (mtDNA) and editing of mitochondrial mRNAs, which is required for mitochondrial gene expression in these organisms, are essential processes, even in the LS stage (Timms et al, 2002; Stuart et al, 2005). These findings suggested the presence of unidentified mitochondrial functions in the bloodstream stage. They also represented a conundrum since dyskinetoplastic (Dk) bloodstream stage trypanosomes that lack mtDNA (kinetoplast DNA or kDNA in trypanosomatids) exist in the wild and have also been generated in the lab (Schnaufer et al, 2002). The Dk organisms are incapable of differentiating into the insect stage, underscoring the requirement for a functional respiratory chain in that stage of the life cycle, and rely on direct mechanical transmission between mammalian hosts (Schnaufer et al, 2002; Timms et al, 2002). Hence, the LS stage parasites may undergo adaptations that compensate for the loss of mtDNA or its expression.
The F0F1-ATP synthase complex (respiratory complex V) is partially encoded in the mtDNA and is expressed in the LS stage (Opperdoes et al, 1976; Bienen and Shaw, 1991; Williams, 1994). In eukaryotic cells, it is located in the inner mitochondrial membrane and participates in ATP generation through oxidative phosphorylation (Boyer, 1997). The F0 part is membrane embedded while the F1 part extends into the matrix. Each protozoan and yeast F0 complex is composed of one each of subunits 6 and 4 and a ring of multiple subunits 9. Subunit 6 is localized between the subunit 9 ring and subunit 4, which links F0 and F1 as a peripheral stalk. The function and location of a number of additional F0 subunits are less understood. F1 is comprised of a ring of six alternating α and β subunits that have ATP synthase catalytic sites at their interfaces. F1 is linked to the subunit 9 ring via a central stalk that is composed of subunits γ, δ, and ɛ. ATP synthesis is achieved by utilizing the proton motive force that is generated by the respiratory proton pumps. Protons are proposed to flow through a channel formed by subunits 6 and 9, inducing rotation of the subunit 9 ring, which forces the central stalk to rotate within the F1 ring. This in turn drives ATP synthesis by inducing conformational changes in the α and β subunits. Some inhibitors of ATP synthase are specific for the assembled F0F1 complex (e.g. oligomycin, which binds to the OSCP subunit of F0) while others also affect the isolated F1 moiety (e.g. azide).
The F0F1-ATP synthase can work in reverse and hydrolyze ATP to pump protons. Such a reverse function is well documented in prokaryotes, for example, in anaerobic bacteria (Futai and Kanazawa, 1983), but demonstrated cases are rare in eukaryotes and involve hypoxic or anoxic conditions. For instance, when ischemic conditions in mammalian tissues result in a loss of respiration, F0F1-mediated ATP hydrolysis leads to depletion of cellular ATP, with pathological consequences (St Pierre et al, 2000). For certain yeasts growing under anaerobic conditions, there is evidence that the complex functions as a proton pump in order to maintain a mitochondrial membrane potential (ΔΨm) (Chen and Clark-Walker, 1999; Clark-Walker, 2003; Lefebvre-Legendre et al, 2003). The requirement for maintaining a ΔΨm reflects the need to import the numerous essential mitochondrial proteins into the organelle (Neupert, 1997).
In the vast majority of eukaryotes, including trypanosomes, mtDNA encodes essential subunits of the respiratory proton pumps (complexes I, III, and IV) and of the F0 part of complex V (Burger et al, 2003). Hence, mtDNA loss would be expected to deprive the cell of both its usual mechanism for maintaining a ΔΨm and using an F0F1-ATP synthase working in reverse. Petite-positive yeasts and some mammalian cells can survive the complete (ρ0) or functionally complete (ρ−) loss of their mtDNA, apparently at least in part due to the electrogenic activity of the mitochondrial ATP/ADP carrier protein (AAC) (Giraud and Velours, 1997; Buchet and Godinot, 1998). In contrast, petite-negative yeasts cannot tolerate loss of mtDNA but mutations in F1 subunits can convert these yeasts into petite-positive forms (Clark-Walker et al, 2000).
These observations prompted us to investigate whether a similar scenario might occur in trypanosomes. Indeed, studies using the F0F1-ATP synthase complex inhibitor oligomycin had suggested that this complex is involved in maintaining a ΔΨm in the LS stage (Nolan and Voorheis, 1992; Vercesi et al, 1992; Divo et al, 1993; Bertrand and Hajduk, 2000). Here we present direct genetic evidence that the F0F1-ATP synthase complex fulfills an essential role in the LS stage of T. brucei by generating ΔΨm using ATP hydrolysis. This is a reversal of its physiological role in the insect stage of the parasite and in the vast majority of eukaryotic organisms. We also show that the F1 moiety of the complex is still required in Dk forms, whereas RNA editing is not. In addition, we identified a mutation in a subunit of F1 that appears to be involved in enabling trypanosomes to survive as Dk forms. Our findings provide insight into important questions regarding mitochondrial function in the disease-causing stage of trypanosomes.
Results
Knockdown of ATP synthase subunit α in LS stage T. brucei is lethal
We assessed the requirement for the F1 part of the ATP synthase complex in LS stage T. brucei by inducible silencing of its essential α subunit. A transgenic LS cell line, expressing the tet repressor protein (tetR) (Wirtz et al, 1999), was transfected with a construct containing inverted repeats of the first 530 bp of the T. brucei ATP synthase α gene placed downstream of a tetracycline (tet)-inducible promoter. Addition of tet to the culture medium, which results in expression of double-stranded RNA and RNAi-mediated degradation of the target mRNA (Shi et al, 2000), led to massive lysis of parasites after 56 h (Figure 1A). No live parasites were detected between 60 and 85 h after induction of RNAi by microscopic inspection of the culture (not shown), but culture growth resumed indicating escape from RNAi as commonly seen with RNAi in trypanosomes (Chen et al, 2003).
Figure 1.
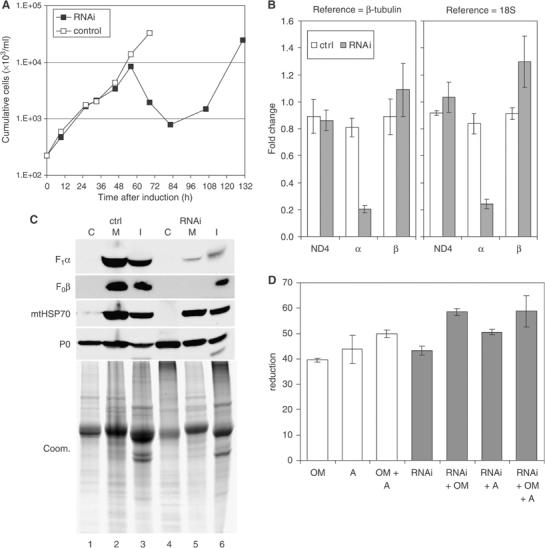
Knockdown of ATP synthase subunit α expression is lethal to LS stage T. brucei. (A) Culture growth shown as cumulative cell numbers after normalization for dilution during cultivation. Expression of subunit α was silenced using tet-inducible RNAi (solid squares); uninduced cells were maintained in parallel (open squares). (B) Determination of mRNA levels for ATP synthase subunits α and β and for the mitochondrial transcript ND4 by real-time RT–PCR (ΔΔCt method). Total RNA was isolated from induced cells after 43 h as well as from uninduced control cells. Primers for subunit α were located outside the region targeted by RNAi. Relative changes in mRNA levels with respect to the parental cell line are indicated, using β-tubulin mRNA or 18S rRNA for normalization. Average numbers for four amplifications are shown, using RNA preparations from two independent RNAi experiments. (C) Western blot analysis of crude cytosolic (C), soluble mitochondrial (M), and insoluble (I) fractions prepared from RNAi-induced cells after 45 h and from uninduced control cells. The blots were probed with reagents detecting the indicated proteins. Coomassie staining (bottom panel) revealed the protein loading. (D) Analysis of ATPase activity in crude mitochondrial fractions, generated as in (C) and assayed by measuring release of free phosphate. ATP synthase inhibitors oligomycin (OM; 2.5 μg/ml) and azide (A; 1 mM) were added where indicated. Average numbers for four assays are shown, using extract preparations from two independent RNAi experiments.
To assess the effectiveness and specificity of the knockdown, we performed quantitative real-time RT–PCR on RNA that was prepared from parasites 43 h after RNAi induction as well as from uninduced control and parental cell line parasites. The subunit α mRNA was reduced by ∼80% while the level of mitochondrially encoded ND4 mRNA, serving as an additional control, was not significantly changed compared to the parental cell line whether β-tubulin mRNA or 18S rRNA was used as RNA standard (Figure 1B). The slight increase in subunit β mRNA in the induced cells might be a response to loss of activity of the corresponding protein or complex (see below).
Western analysis of crude mitochondrial and cytosolic fractions prepared from 45 h RNAi-induced parasites revealed a substantial loss of F1 subunit β and F0 subunit 4 compared to uninduced control; reagents for the α subunit were not available (Figure 1C, compare lanes 2 and 5). A crude mitochondrial fraction was prepared by permeabilizing cells with 0.015% (w/v) digitonin, which leaves the mitochondrion intact (Tan et al, 2002), and was then lysed using 0.1% Triton X-100. Cells and other organelles that had escaped lysis together with insoluble proteins were pelleted in a subsequent centrifugation step. Analysis with an antibody specific for mitochondrial HSP70 as well as Coomassie staining of a parallel gel showed that comparable amounts of sample were loaded with perhaps slightly more for the uninduced samples. Analysis with an antiserum specific for the cytosolic ribosomal protein P0 showed cytosol was released by digitonin treatment (lanes 1 and 4). It also indicated that lysis was incomplete (as expected) since P0 was also present in the organellar and pellet fractions. Samples from recovered cultures (e.g. 130 h) had normal levels of β subunit, reflecting cell escape from RNAi (data not shown). The reduced levels of subunits 4 and β suggest that, as observed for yeast (Lai-Zhang et al, 1999; Lefebvre-Legendre et al, 2001), in the absence of the α subunit the F1F0 complex does not assemble properly and its unincorporated subunits degrade and/or mislocalize.
Mitochondrial ATPase activity was also substantially reduced upon RNAi induction (Figure 1D). ATP hydrolytic activity was measured via release of free phosphate (Law et al, 1995) in mitochondria prepared from 45 h RNAi-induced and uninduced cells using digitonin as described above. Oligomycin, a specific inhibitor of the F0F1 complex, and azide, an inhibitor of F1 (Buchet and Godinot, 1998), reduced activity by ∼40 and ∼44%, respectively. This is similar to the degree of inhibition observed with other crude mitochondrial fractions from T. brucei (Bienen and Shaw, 1991) while 50–60% inhibition was observed with purified preparations of T. brucei F0F1-ATP synthase (Opperdoes et al, 1976; Williams, 1994), indicating the presence of other ATP hydrolytic activities in these crude mitochondrial fractions. Induction of RNAi for 45 h reduced ATPase activity by ∼43%, which is comparable to the reduction obtained with oligomycin or azide. Addition of oligomycin and/or azide to these extracts resulted in minor further decreases of ATPase activity, presumably since none of these treatments alone is 100% effective. Together, these results demonstrate that the activity resulting from the mitochondrial ATP synthase complex in these extracts was reduced substantially, consistent with the results from the Western analysis. Overall, these results show that knockdown of ATP synthase subunit α by RNAi resulted in substantial and specific reduction of the amount and activity of the ATP synthase complex, which subsequently resulted in cell death.
Inactivation of the F1-ATP synthase complex results in loss of ΔΨm in LS stage T. brucei
We assayed ΔΨm in response to induction of RNAi since the ATP synthase complex in LS stage T. brucei was proposed to function in the generation of a ΔΨm (Nolan and Voorheis, 1992; Divo et al, 1993). Live T. brucei were stained with 0.25 μM rhodamine 123 (Rh123), a fluorescent dye that is a marker for energized mitochondria (Divo et al, 1993; Wilkes et al, 1997), and the resultant green fluorescence intensity was observed by epifluorescence microscopy and measured by flow cytometry (Figure 2A). Visual inspection by fluorescence microscopy showed bright fluorescence of the single tubular mitochondrion with very little background staining as reported (Divo et al, 1993). Inclusion of the uncoupler FCCP reduced the fluorescence substantially (bottom row). Induction of ATP synthase subunit α RNAi for 45.5 h resulted in fluorescence with an intensity that is comparable to that of FCCP-treated cells (center row; note that Dk T. evansi cells were included as controls for staining). Analysis of Rh123 fluorescence over the RNAi time course revealed continuously decreasing fluorescence for 56 h followed by emergence of a population of parasites with normal ΔΨm due to escape from RNAi (Figure 2B). Comparison of the percentage of normal Rh123 fluorescence (and thus ΔΨm) with the growth curve from Figure 1A showed that the decrease of ΔΨm set in just a few hours after induction of RNAi and reached 50% after ∼30 h, or more than 24 h before the onset of an effect on growth (Figure 2C). This suggests that the decrease in ΔΨm is a primary response to the inactivation of the F1-ATP synthase complex and not a consequence of other lethal events. In addition, the ΔΨm decrease is not immediately lethal but may result in lethality such as by loss of mitochondrial protein import as discussed below. In conclusion, these results strongly support an essential role of the F1 part of the ATP synthase in generation of a ΔΨm.
Figure 2.
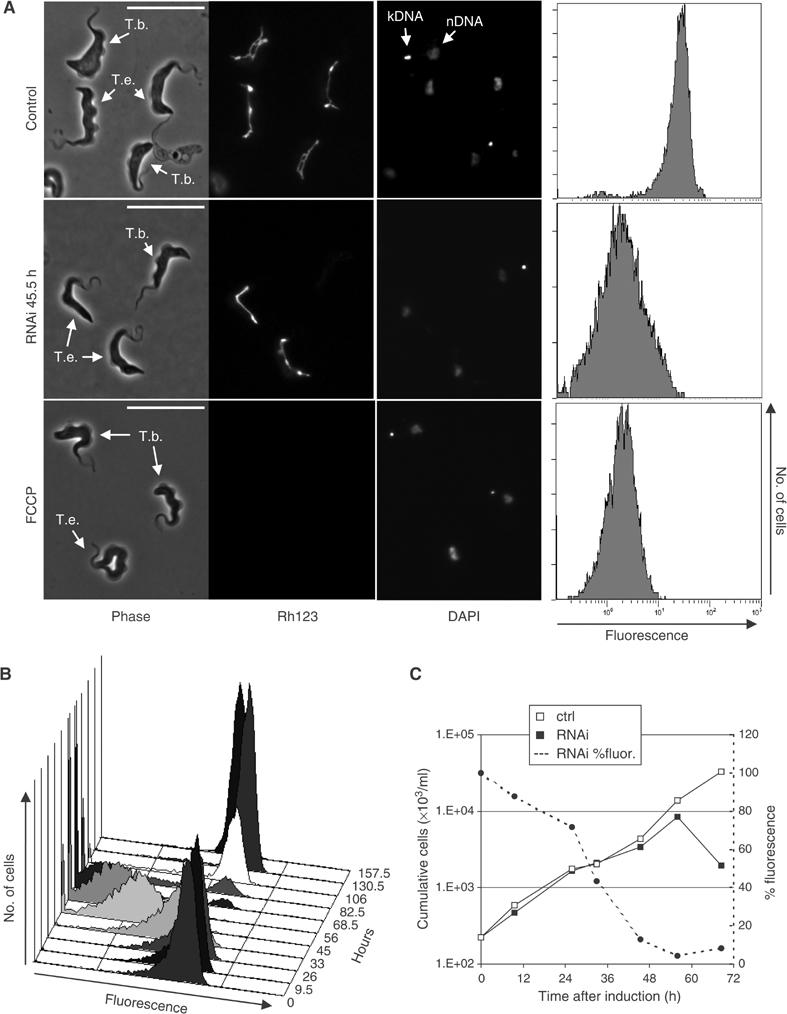
Knockdown of ATP synthase subunit α in LS T. brucei abolishes ΔΨm. (A) Live cells were stained with the fluorescent mitochondrial marker Rh123 (0.25 μM), immobilized, and analyzed by phase contrast and epifluorescence microscopy. ΔΨm-related fluorescence was also measured by flow cytometry (right column). Top row: uninduced T. brucei cells, mixed with Dk T. evansi, which can be distinguished by the absence of kDNA (DAPI staining; nDNA: nuclear DNA) and which serve as a control for Rh123 staining. Center panel: after induction of subunit α RNAi in T. brucei for 45.5 h. Bottom panel: cells treated with the uncoupler FCCP. In all panels, the bar represents 20 μm. (B) Change over time in ΔΨm-related fluorescence after induction of subunit α RNAi. (C) Change in ΔΨm-related fluorescence combined with growth analysis. Fluorescence is expressed as percent of average fluorescent signal compared to uninduced control cells.
Knockout of kinetoplastid RNA editing ligase 1 in Dk T. evansi
We investigated the requirement for the RNA editing complex in LS stage Dk trypanosomes since ATP synthase subunit 6 is encoded in the maxicircle component of mtDNA and its mRNA requires editing to be functional (Bhat et al, 1990). Analysis of two Dk strains showed that they contain functional editing complexes (Domingo et al, 2003). One of the strains, T. evansi Antat 3/3, has a single class of minicircles and lacks maxicircles (Borst et al, 1987; Songa et al, 1990) and thus lacks all mitochondrial protein coding genes, including that for ATP synthase subunit 6. We prepared null mutants of the kinetoplastid RNA editing ligase 1 (KREL1), which is essential in LS stage T. brucei (Schnaufer et al, 2001), using a gene replacement strategy (Wirtz et al, 1999). The first allele was replaced with the neomycin resistance marker and the second with a construct containing tetR and the hygromycin resistance marker. Clonal transfectants were obtained at normal frequency. Western analysis confirmed the absence of the KREL1 protein from these null mutants (Supplementary Figure S1A) and PCR analysis confirmed the absence of the KREL1 gene (data not shown). The growth rate of KREL1−/− cells was indistinguishable from that of wild-type and KREL1+/− cells (Supplementary Figure S1B). Thus, in contrast to T. brucei, the KREL1 enzyme and, by implication, the editing complex and RNA editing do not have a vital function in T. evansi Antat 3/3.
Knockdown of ATP synthase subunit α in Dk T. evansi Antat 3/3 is lethal
ATP synthase subunit α expression was silenced by inducible RNAi expression in the T. evansi KREL−/− mutant, taking advantage of the expression of tetR in these cells. This knockdown was lethal to the parasite and no escape was observed, implying a robust knockdown (Figure 3A). Real-time RT–PCR analysis of RNAs from cells induced for 24 h showed that the knockdown was specific for subunit α (Figure 3B). No signal was observed for ND4 mRNA as expected since mtDNA—and hence this gene—is absent (note that our analysis compared transcript levels of RNAi and uninduced control cells with levels in T. brucei). The relative amounts of α and β subunits in uninduced control cells were reduced by ∼50% when β-tubulin was used as a reference, which was not the case when 18S rRNA was used for normalization. This indicates that the relative amounts of the two reference RNAs differ between T. evansi and T. brucei.
Figure 3.
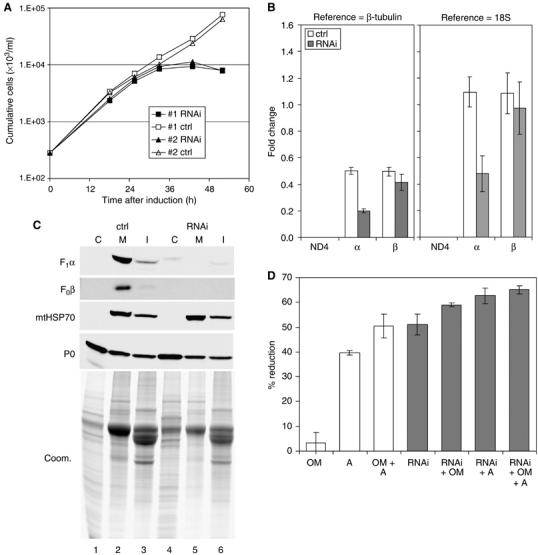
Knockdown of ATP synthase subunit α expression is lethal to Dk T. evansi Antat 3/3. (A) Culture growth shown as cumulative cell numbers after normalization for dilution during cultivation. Expression of subunit α was silenced using tet-inducible RNAi (solid squares), and uninduced cells were maintained in parallel (open squares). Growth curves for two individual clones are shown. (B) Determination of mRNA levels for ATP synthase subunits α and β and for the mitochondrial transcript ND4 as described for Figure 1B. Total RNA was isolated from induced cells after 24 h and from uninduced control cells. Indicated are relative changes in mRNA levels with respect to LS T. brucei. Average numbers for four real-time PCR experiments are shown, using RNA preparations from two independent RNAi experiments. (C) Western blot analysis of crude cytosolic (C), soluble mitochondrial (M), and insoluble (I) fractions prepared from RNAi-induced cells after 31 h and from uninduced control cells. The blots were probed with reagents detecting the indicated proteins. Coomassie staining (bottom panel) revealed the protein loading. (D) Analysis of ATP hydrolytic activity in crude mitochondrial fractions, generated as in (C) and assayed by measuring release of free phosphate. Average numbers for four assays are shown, using extract preparations from two independent RNAi experiments. See Figure 1D for abbreviations.
Subcellular fractions from the T. evansi parasites in which RNAi was induced for 31 h and from uninduced control cells were prepared by sequential lysis with digitonin and Triton X-100, as described above for T. brucei, and analyzed by immunoblotting (Figure 3C). Interestingly, abundance of F0 subunit 4 appeared to be reduced in T. evansi compared to T. brucei, possibly as a result of decreased stability of F0 in the absence of the mitochondrially encoded subunit 6. As in T. brucei, the amounts of F1 subunit β and F0 subunit 4 were substantially reduced upon RNAi induction. This again suggests that the ATP synthase complex cannot assemble in the absence of the α subunit and that unincorporated subunits are degraded. The RNAi knockdown resulted in reduction of ATP hydrolytic activity to a similar extent as in T. brucei using crude mitochondrial fractions obtained and assayed as described above (Figure 3D). Oligomycin had only a small effect, if any, which is consistent with the expected lack of a functional F0 part required to confer oligomycin sensitivity to the F0F1 complex (Boyer, 1997). Insensitivity of mitochondrial ATPase activity isolated from various Dk trypanosomes to oligomycin was reported previously (Opperdoes et al, 1976).
Knockdown of ATP synthase subunit α expression in T. evansi resulted in substantial reduction of mitochondrial staining with Rh123 (Figure 4A). ΔΨm decreased with a kinetic profile similar to T. brucei but the onset of cell death occurred sooner in T. evansi (Figure 4B). Cell growth had already slowed when the Rh123 fluorescence reached 50% in T. evansi (at ∼35 h) while growth was unaffected at this stage (50% fluorescence at ∼30 h) in T. brucei. These apparent differences in part may reflect a somewhat lower ΔΨm in T. evansi, judged from results obtained by fluorescence microscopy using alternative dyes (data not shown). However, they may also reflect an altered role of F1 in generating the ΔΨm (see below).
Figure 4.
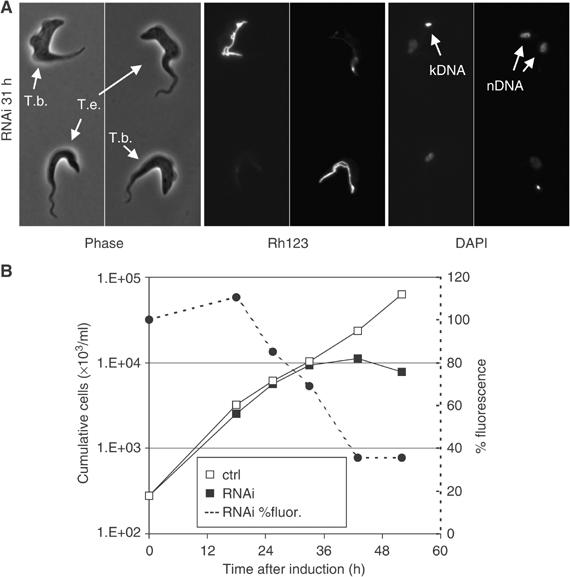
Knockdown of ATP synthase subunit α in T. evansi Antat 3/3 reduces ΔΨm. (A) Rh123 staining of immobilized cells as described for Figure 2A. T. brucei cells served as staining controls. Note that the T. evansi cell in the upper right corner has duplicated its nuclear genome. (B) Change in ΔΨm-related fluorescence combined with growth analysis as described for Figure 2C. Fluorescence is expressed as percent of average fluorescent signal compared to uninduced control cells.
Effect of ATP synthase inhibitors on growth and ΔΨm
One prediction from the above results is that growth of LS T. brucei and T. evansi Antat 3/3 should show very different susceptibilities to the F0F1 inhibitor oligomycin. Indeed, 125 ng/ml oligomycin was lethal to LS T. brucei while T. evansi was resistant to the inhibitor (Figure 5A and B). Growth of T. evansi did slow at concentrations above 500 ng/ml, perhaps due to nonspecific toxicity at high concentrations (data not shown). In contrast, T. evansi showed about two-fold higher sensitivity than T. brucei to the F1 inhibitor azide (Figure 5C and D). The effects of oligomycin and azide on growth were consistent with the effects of these inhibitors on ΔΨm: while 125 mg/ml oligomycin selectively abolished ΔΨm in T. brucei, T. evansi cells were unaffected (Supplementary Figure S2, center rows). In contrast, 0.5 mM azide abolished mitochondrial staining with Rh123 for both T. brucei and T. evansi. Insect form T. brucei, which, like most other organisms, generates ΔΨm using respiratory complexes III and IV, remained unaffected (Supplementary Figure S2, bottom rows).
Figure 5.
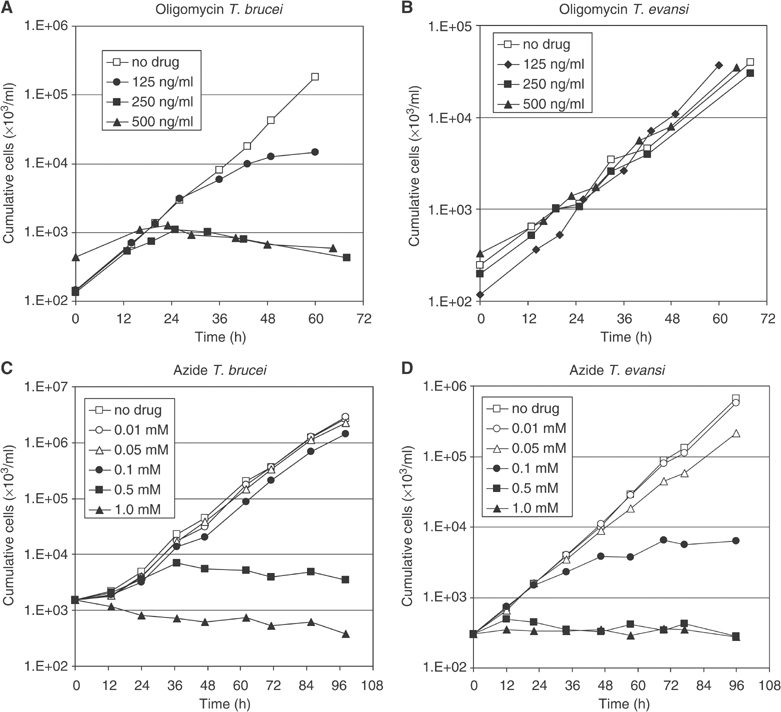
Comparison of the sensitivities of growth of LS form T. brucei and Dk T. evansi to the F0F1 inhibitor oligomycin (A, B) and the F1 inhibitor azide (C, D). Cells were grown in the presence of the indicated concentrations of inhibitor. Cumulative cell numbers reflect normalization for dilution during cultivation.
Incubation with 80 μM bongkrekic acid, an inhibitor of the AAC, also selectively decreased ΔΨm in LS T. brucei and T. evansi (Supplementary Figure S3). Hence, exchange of ATP and ADP between cytosol and mitochondrial matrix is involved in maintaining ΔΨm in the bloodstream forms, but not the insect form. This role of the AAC can be direct and/or indirect, as discussed below.
Identification of a potential compensatory mutation in a Dk trypanosome
In the petite-negative yeast Kluyveromyces lactis, several mutations that can compensate for the loss of mtDNA are located in subunits of F1 (Clark-Walker et al, 2000). To investigate whether a similar mechanism might be responsible for dyskinetoplasty in trypanosomes, we cloned and compared gene sequences for α, β, and γ subunits from normal T. brucei strains 427 and 164, the acriflavine-induced Dk strain 164 (Stuart, 1971), and T. evansi strain Antat 3/3. Among the three polymorphisms identified was a change from Leu to Pro at amino-acid residue 262 of the γ subunit of T. brucei Dk164 (Supplementary Figure S4). This mutation is in the C-terminal region of the γ subunit (in the matrix proximal region of F1), close to suppressor mutations identified in K. lactis (atp3-1 and atp3-2 in Supplementary Figure S4) (Clark-Walker et al, 2000). We therefore tested whether this mutation might be associated with increased tolerance to the loss of mtDNA. Conservation of this leucine between T. brucei and K. lactis enabled us to introduce the homologous amino-acid change into the yeast, where the γ subunit is encoded by the ATP3 gene. Yeast cells that harbor the Leu to Pro mutation (designated atp3-3) grew on standard glucose plates as well as wild type (Figure 6, top panel). Cells expressing atp3-2 grew a little slower. Cells expressing atp3-3 grew as well as wild type on a nonfermentable carbon source (glycerol), indicating that this mutation did not significantly affect the ability of the ATP synthase complex to function in oxidative phosphorylation (Figure 6, center panel). In contrast, as found before (Clark-Walker et al, 2000), yeast cells expressing atp3-2 grew poorly on glycerol. When plated on glucose medium containing ethidium bromide (EB), which causes deletions in mtDNA, the atp3-3 cells grew as well as the atp3-2 cells, while wild-type cells grew very poorly (Figure 6, bottom panel). Analysis of the individual EB-resistant atp3-3 colonies showed that they had lost mtDNA and did not grow on nonfermentable substrates (data not shown). The two EB-resistant colonies obtained for the wild-type strain are EB-induced mutants containing suppressors of ρ0/ρ− lethality (data not shown). Thus, the mutation identified in the ATP synthase subunit γ from T. brucei DK164 can convert the petite-negative yeast K. lactis into a petite-positive form. Consequently, it seems likely that the Leu to Pro change has a similar role in compensating the loss of kDNA in trypanosomes.
Figure 6.
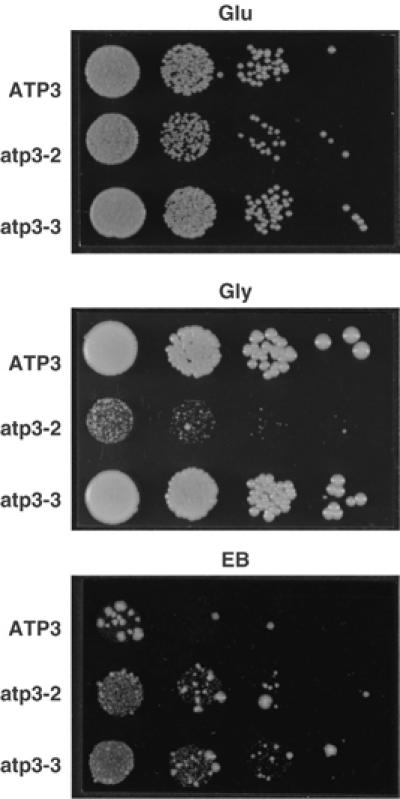
The Leu262Pro mutation in the gene encoding the γ subunit of the F1 moiety from a Dk strain of T. brucei converts the petite-negative yeast K. lactis into a petite-positive form. The Leu to Pro mutation was introduced into the conserved residue of the ATP synthase γ subunit of K. lactis (Leu265 in ATP3) and the mutant allele (atp3-3) expressed in a strain lacking the endogenous ATP3 gene. Control strains expressed the wild-type ATP3 gene or a suppressor mutation identified previously (atp3-2). Growth was analyzed after serial dilution on plates containing the carbon sources glucose (Glu), glycerol (Gly), or glucose medium with 16 μg/ml EB. Plates were incubated at 28°C for 2, 6, and 9 days, respectively.
Suppressor mutations in K. lactis reduce the Km of F1 for ATP (Clark-Walker, 2003). Similarly, we found that the atp3-3 mutation reduced the Km from 2.06±0.14 to 0.89±0.14 μmol/min mg protein (Supplementary Figure S5).
Discussion
The F1 part of the mitochondrial ATP synthase complex was found to be essential for generating ΔΨm and for survival of LS bloodstream form T. brucei as well as its Dk relative T. evansi Antat 3/3. We hypothesize that cell death is due to the collapse of critical transport processes between mitochondrion and cytoplasm, which depend on ΔΨm. These results indicate that the ATP synthase complex hydrolyzes ATP to generate the ΔΨm in bloodstream forms, hence functioning opposite to its ATP synthesis mode that occurs in the insect stage and in most eukaryotes. A mutation was found in the nuclearly encoded ATP synthase γ subunit of a Dk strain, which appears to help these cells evade the need for gene products encoded in mtDNA, such as subunit 6 of the ATP synthase complex. The RNA editing complex, which, like mtDNA, is essential in normal T. brucei, was shown not to be essential in T. evansi, suggesting that its function in T. brucei is limited to its role in the expression of mitochondrial genes.
The activity of the F0F1-ATP synthase complex is essential in LS T. brucei since both knockdown of expression of its α subunit and addition of oligomycin or azide, inhibitors of this complex, were lethal within a few days (Figures 1A and 6A). RNAi knockdown of the α subunit resulted in reduced levels of subunits β and 4, confirming that the complex cannot assemble in the absence of the α subunit (Lai-Zhang et al, 1999; Lefebvre-Legendre et al, 2001).
Collapse of ΔΨm appeared to be the primary consequence of subunit α knockdown since its decrease began shortly after induction of RNAi and reached background levels prior to inhibition of growth (Figure 2C). This finding supports the conclusion of others that oligomycin sensitivity of the ΔΨm reflects its generation by the ATP synthase complex in LS T. brucei (Nolan and Voorheis, 1992; Vercesi et al, 1992; Divo et al, 1993). ΔΨm is absolutely required for mitochondrial import of proteins encoded in the nucleus as well as for other transport processes and is therefore essential for the vast majority of mitochondrial activities, including biogenesis of the mitochondrion itself (Neupert, 1997). Some mitochondrial activities in LS T. brucei are repressed (Schneider, 2001), but recent findings have suggested important roles for the organelle in that stage of the life cycle (Schnaufer et al, 2002). For instance, respiration in LS T. brucei is mediated by a mitochondrial trypanosome alternative oxidase and this enzyme might be essential (Helfert et al, 2001). Collapse of ΔΨm is also a key event in apoptosis and although T. brucei lacks components of the classical apoptotic pathway (Esseiva et al, 2004), it appears to have some form of programmed cell death (Welburn et al, 1997), which may have been triggered by inactivation of the ATP synthase complex.
MtDNA and its expression with the help of RNA editing have recently been shown to be essential for survival of LS bloodstream form T. brucei (Timms et al, 2002; Stuart et al, 2005). The fact that Dk trypanosomes can survive as bloodstream forms therefore presents an intriguing conundrum (Schnaufer et al, 2002), which is underscored by our finding that the normally essential RNA editing enzyme KREL1 can be knocked out in Dk T. evansi without an obvious phenotype (Supplementary Figure S1). Findings presented in this paper help explain how LS Dk strains might generate the ΔΨm and survive the loss of the normally required mitochondrial gene products. The Dk strains require F1-ATP synthase activity, like the wild type, as evident from the loss of ΔΨm and growth inhibition resulting from knockdown of α subunit expression or addition of azide to the medium (Figures 3A, 4, and 5D). However, unlike the wild-type strain, ATP hydrolysis activity and cell growth of Dk T. evansi are essentially insensitive to oligomycin (Figure 5B; Opperdoes et al, 1976). This suggests that in T. evansi, the F1 part, which by itself is insensitive to oligomycin, still functions in the generation of the ΔΨm and that the F0F1 complex is altered. MtDNA of T. brucei encodes a protein with homology to ATP synthase subunit 6, the mRNA of which requires extensive RNA editing (Bhat et al, 1990). This subunit is an essential component of the proton channel of the F0 moiety (Boyer, 1997) and is therefore expected to be essential for the proton-pumping activity of F0F1-ATP synthase in LS T. brucei. This is also implied by collapse of the ΔΨm in T. brucei following treatment with acriflavine (Timms et al, 2002), which specifically deletes mtDNA. Based on our finding of a mutated ATP synthase γ subunit in a Dk strain of T. brucei, we suggest that mutations in subunits of the F0F1 complex may have altered the complex and compensated, at least in part, for the loss of mitochondrial gene products. Our finding resembles the identification of suppressor mutations in F1 subunits in the petite-negative yeast K. lactis, which, like T. brucei, does normally depend on the synthesis of mitochondrial gene products (Clark-Walker et al, 2000). Indeed, introduction of the identified mutation into the γ subunit of K. lactis suppressed the petite-negative phenotype and enabled growth under conditions that induce loss of mtDNA (Figure 6). Like previously identified suppressor mutations in K. lactis, the mutation identified here results in a lower Km for ATP (Supplemental Figure S5; Clark-Walker, 2003). In addition, the mutation may increase the stability of F1 in the absence of F0 and this possibility remains to be investigated. The situation in Dk cells therefore may be analogous to ρ0/ρ− mammalian cells and petite-positive yeasts, where ATP hydrolysis by F1 appears to result in a sufficiently low mitochondrial ATP concentration to allow the electrogenic exchange by the AAC of ADP3− for ATP4−. This exchange could generate a ΔΨm that would sustain essential organelle functions, even in the absence of processes that require mitochondrial gene products (Giraud and Velours, 1997; Buchet and Godinot, 1998). Hence, the AAC would have a more indirect role for generating ΔΨm in the normal LS form (by supplying ATP for proton pumping by the F0F1 complex) and a direct role in the Dk form. Indeed, ΔΨm of both forms (but not the insect form) was reduced by the AAC inhibitor bongkrekic acid (Supplementary Figure S3). Additional studies are needed to determine whether F1-ATPases from Dk trypanosomes in general have a lower Km for ATP and if the mutation that we identified is sufficient to compensate for the loss of mitochondrial gene products in T. brucei. Considering that a singular evolutionary event may have given rise to stable Dk trypanosomes in nature (Brun et al, 1998) and the difficulty of generating such strains in the laboratory (Stuart, 1971; Timms et al, 2002), it seems more likely that multiple mutations may be required.
Our model for the generation of ΔΨm in trypanosomes is summarized in Figure 7. The F0F1-ATP synthase complex has the conventional role of ATP generation during oxidative phosphorylation in the insect stage but it does not appear to be essential, since oligomycin does not affect intracellular ATP levels and only moderately affects growth (Coustou et al, 2003). In the LS bloodstream stage, by contrast, the complex is normally essential, although much less abundant than in the insect stage (Brown et al, 2001), and has a role opposite to that of the conventional role, namely generation of ΔΨm at the expense of ATP consumption. The shift between life cycle stages and the need to control ATP hydrolysis in insect stage mitochondria suggests the existence of a critical regulatory system. The identification of a peptide that inhibits the enzymes' ATP hydrolytic activity may be a component of such a regulatory system since its abundance appears to be upregulated in the insect stage (Chi et al, 1996). Intriguingly, our analysis of the T. brucei genome database (www.GeneDB.org) revealed three different genes for F0 subunit 9 (data not shown). Analysis of their expression or activity in different life cycle stages may indicate potential mechanisms that regulate the activity of the complex. The unusual but essential role of the mitochondrial ATP synthase complex in the disease-causing stage of T. brucei may open opportunities for drug development. An inhibitor selective for the ATP hydrolytic direction of the enzyme would be expected to be lethal to the parasite but not the host, and such inhibitors may be adapted from those under development to prevent tissue damage under ischemic conditions (St Pierre et al, 2000).
Figure 7.

Model for ATP synthase complex function in trypanosome mitochondria. The ATP synthase complex functions conventionally in the insect stage and generates ATP by utilizing the proton gradient produced by the respiratory chain. The ATP synthase complex works in reverse in the LS bloodstream stage, which lacks a cytochrome-mediated respiratory chain, and utilizes ATP to pump protons out of the matrix, generating a ΔΨm. In Dk trypanosomes, the F1 part is still essential for generating a ΔΨm, presumably for ATP hydrolysis as in the wild type, but the F0 part is absent or nonfunctional. The electrogenic ATP/ADP exchange that is catalyzed by AAC may be involved in the generation of a ΔΨm as has been proposed for ρ0 and ρ− mammalian and yeast cells (see text). A high rate of ATP hydrolysis by F1 may be required to keep the matrix ATP concentration sufficiently low, which may be achieved by mutations in F1 subunits like those identified here and in petite-negative yeasts. Note that each AAC catalyzes the exchange of one molecule of ADP for one molecule of ATP; two AAC proteins are shown solely to clarify ATP/ADP entry and exit.
Materials and methods
Trypanosome plasmid construction and transfection
The inducible RNAi plasmid for silencing ATP synthase subunit α was generated using the pQuadra system (Inoue et al, 2005). Briefly, the first 530 bp of the subunit α gene were amplified by PCR, using oligos with specifically designed BstXI sites. Ligation with BstXI-digested pQuadra1 and pQuadra3 plasmids generated pQuadra-ATPα, containing inverted repeats of the PCR product separated by a spacer region. Knockout plasmids for replacing the two KREL1 alleles with T7RNAP plus neomycin resistance marker and tetR plus hygromycin resistance marker, respectively, have been described previously (Schnaufer et al, 2001). Transfection of NotI-linearized constructs into the LS bloodstream form ‘single marker' cell line (Wirtz et al, 1999) or T. evansi Antat 3/3 (Borst et al, 1987) and selection of transgenic cell lines was carried out as described (Schnaufer et al, 2001).
Growth analysis of trypanosomes
Throughout growth analyses, cells were maintained at exponential growth (between 105 and 106 cells/ml). RNAi was induced by adding 1 μg/ml tet to the medium.
Quantitative real-time RT–PCR analysis
Mid-log trypanosomes were harvested at room temperature (10 min, 1300 g) and RNA was isolated using the Ultraspec RNA Reagent (Biotecx Laboratories Inc., Houston, TX). Quantitative real-time RT–PCR analysis was carried out as described (Carnes et al, 2005), using 10 μl of a mix of the specific primers at 1.5 μM (see Supplementary data). Reactions were analyzed with the ABI Prism 7000 software (Applied Biosystems). Relative amounts of RNA template in the preparations were calculated using the ΔΔCt method (Ingham et al, 2001). Parallel amplifications minus the reverse transcription step revealed only insignificant contaminations with genomic DNA.
Digitonin fractionation, ATPase assay, and Western blotting
Crude mitochondrial preparations were obtained by fractionation with digitonin (Tan et al, 2002). ATPase activity was measured based on release of free phosphate (Law et al, 1995). Briefly, ∼4 × 108 mid-log trypanosomes were harvested by centrifugation (1300 g, 10 min), washed, and permeabilized with 0.015% (w/v) digitonin. The crude cytosolic fraction was obtained as the supernatant from a 4000 g/3 min spin at 4°C. The pellet, representing the crude mitochondrial fraction, was resuspended in ATPase assay buffer (10 mM Tris–HCl, pH 8.2; 0.2 M KCl; 2 mM MgCl2). Where indicated, oligomycin or sodium azide was added to 25 μg/ml (∼10 μg/mg protein) and 1 mM, respectively. The reaction was started by addition of ATP to a final concentration of 5 mM. After 0, 10, and 20 min, 95 μl aliquots were added to 5 μl 3 M trichloric acid and kept on ice for 30 min. The precipitate was pelleted (16 000 g, 10 min), 90 μl of the supernatant was added to 0.5 ml Sumner reagent (Law et al, 1995), and absorbance was measured at 610 nm. For Western analysis, the crude mitochondrial fraction obtained above was lysed by addition of 0.1 volume of 10% Triton X-100. Soluble and insoluble fractions were obtained by centrifugation at 16 000 g for 10 min and separated by SDS–PAGE. Gels were either stained with Coomassie blue or transferred to PVDF membrane. Western blots were probed with reagents against the F1 moiety of Crithidia fasciculata (Speijer et al, 1997), which crossreacts only with the β subunit of the T. brucei complex, subunit 4 of Leishmania tarentolae (Nebohacova et al, 2003), mitochondrial HSP70 from T. brucei (Panigrahi et al, 2003), and ribosomal protein P0 from T. cruzi (Skeiky et al, 1994). For Western analysis of whole-cell lysates, cells were harvested by centrifugation and lysed in SDS sample buffer. Extracts corresponding to 5 × 107 cell equivalents were fractionated and transferred to PVDF membrane as described above and probed with monoclonal antibodies against editosome proteins KREPA1, KREPA2, KREL1, and KREPA3 (Panigrahi et al, 2001; Schnaufer et al, 2001).
Analysis of ΔΨm by microscopy and flow cytometry
A 1 ml portion of mid-log trypanosomes was incubated in the presence of 250 nM Rh123 (Molecular Probes; plus 1 μg/ml DAPI for microscopy) for 20 min at 37°C, harvested (1300 g, 10 min), and washed with 1 ml CytoMix (25 mM HEPES, pH 7.6; 120 mM KCl; 0.15 mM CaCl2; 10 mM K2HPO4/KH2PO4, pH 7.6; 2 mM EDTA; 5 mM MgCl2; 6 mM glucose). For microscopy, cells were resuspended in residual buffer, partially immobilized on HMI-9 medium containing 0.65% low-melting-type agarose, and analyzed using a Nikon Eclipse E600 fluorescence microscope. For flow cytometry, cells were resuspended in 0.5 ml CytoMix and analyzed for green fluorescence in a Beckman Coulter Epics XL-MCL flow cytometer.
Sequence comparison of ATP synthase subunits α, β, and γ
Genomic DNA was prepared from the following trypanosome strains: T. brucei brucei 427, procyclic and bloodstream stage (Cross, 1975); T. brucei brucei 164 and DK164 (Stuart, 1971); T. evansi Antat 3/3 (Borst et al, 1987). Complete coding sequences for ATP synthase subunits were amplified by PCR (see Supplementary data for primers), subcloned, and sequenced. In each case, sequences from at least three individual PCR reactions were analyzed to exclude PCR artifacts.
Yeast strains and plasmids; expression of mutagenized ATP3 in yeast
See Supplementary data for details.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary methods
Acknowledgments
We thank R Benne (University of Amsterdam) for the F1 antiserum, S Reed (Corixa Corp.) for the P0 antiserum, M Inoue (Kirume University) for the pQuadra plasmids, G Cross (Rockefeller University) for the single-marker cell line, A Schneider (University of Fribourg) for technical advice and David Pérez-Morga (Université Libre de Bruxelles) for advice on immobilizing trypanosomes. We thank J Carnes for advice on real-time PCR analysis and other members of the Stuart lab for helpful discussions. LiJun Ouyang (RSBS, ANU) is thanked for skilled technical assistance. We also thank H Interthal (University of Washington) and G Domingo (SBRI) for critical reading of the manuscript. This work was supported in part by NIH grant AI014102-28 (KS) and the MJ Murdock Charitable Trust (SBRI).
References
- Bertrand KI, Hajduk SL (2000) Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol 106: 249–260 [DOI] [PubMed] [Google Scholar]
- Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K (1990) An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61: 885–894 [DOI] [PubMed] [Google Scholar]
- Bienen EJ, Shaw MK (1991) Differential expression of the oligomycin-sensitive ATPase in bloodstream forms of Trypanosoma brucei brucei. Mol Biochem Parasitol 48: 59–66 [DOI] [PubMed] [Google Scholar]
- Borst P, Fase-Fowler F, Gibson WC (1987) Kinetoplast DNA of Trypanosoma evansi. Mol Biochem Parasitol 23: 31–38 [DOI] [PubMed] [Google Scholar]
- Boyer PD (1997) The ATP synthase—a splendid molecular machine. Annu Rev Biochem 66: 717–749 [DOI] [PubMed] [Google Scholar]
- Brown BSV, Chi TB, Williams N (2001) The Trypanosoma brucei mitochondrial ATP synthase is developmentally regulated at the level of transcript stability. Mol Biochem Parasitol 115: 177–187 [DOI] [PubMed] [Google Scholar]
- Brun R, Hecker H, Lun ZR (1998) Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship (a review). Vet Parasitol 79: 95–107 [DOI] [PubMed] [Google Scholar]
- Buchet K, Godinot C (1998) Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J Biol Chem 273: 22983–22989 [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF (2003) Mitochondrial genomes: anything goes. Trends Genet 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter J, Ernst N, Steinberg AG, Stuart K (2005) An essential RNA editing insertion specific endonuclease in Trypanosoma brucei. Proc Natl Acad Sci USA, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD (1999) Alpha and beta subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol Gen Genet 262: 898–908 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hung CH, Burderer T, Lee GS (2003) Development of RNA interference revertants in Trypanosoma brucei cell lines generated with a double stranded RNA expression construct driven by two opposing promoters. Mol Biochem Parasitol 126: 275–279 [DOI] [PubMed] [Google Scholar]
- Chi TB, Choi SY, Williams N (1996) The ATP synthase of Trypanosoma brucei is developmentally regulated by an inhibitor peptide. Arch Biochem Biophys 333: 291–297 [DOI] [PubMed] [Google Scholar]
- Clark-Walker GD (2003) Kinetic properties of F1-ATPase influence the ability of yeasts to grow in anoxia or absence of mtDNA. Mitochondrion 2: 257–265 [DOI] [PubMed] [Google Scholar]
- Clark-Walker GD, Hansbro PM, Gibson F, Chen XJ (2000) Mutant residues suppressing rho(0)-lethality in Kluyveromyces lactis occur at contact sites between subunits of F(1)-ATPase. Biochim Biophys Acta 1478: 125–137 [DOI] [PubMed] [Google Scholar]
- Coustou V, Besteiro S, Biran M, Diolez P, Bouchaud V, Voisin P, Michels PA, Canioni P, Baltz T, Bringaud F (2003) ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J Biol Chem 278: 49625–49635 [DOI] [PubMed] [Google Scholar]
- Cross GAM (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71: 393–417 [DOI] [PubMed] [Google Scholar]
- Divo AA, Patton CL, Sartorelli AC (1993) Evaluation of rhodamine 123 as a probe for monitoring mitochondrial function in Trypanosoma brucei spp. J Protozool 40: 329–335 [DOI] [PubMed] [Google Scholar]
- Domingo GJ, Palazzo SS, Wang B, Panicucci B, Salavati R, Stuart KD (2003) Dyskinetoplastic Trypanosoma brucei contain functional editing complexes. Eukaryot Cell 2: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseiva AC, Chanez AL, Bochud-Allemann N, Martinou JC, Hemphill A, Schneider A (2004) Temporal dissection of Bax-induced events leading to fission of the single mitochondrion in Trypanosoma brucei. EMBO Rep 5: 268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai M, Kanazawa H (1983) Structure and function of proton-translocating adenosine triphosphatase (F0F1): biochemical and molecular biological approaches. Microbiol Rev 47: 285–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud MF, Velours J (1997) The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho- yeast cells by a lack of assembly of the catalytic sector F1. Eur J Biochem 245: 813–818 [DOI] [PubMed] [Google Scholar]
- Hannaert V, Bringaud F, Opperdoes FR, Michels PA (2003) Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfert S, Estevez AM, Bakker B, Michels P, Clayton C (2001) Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem J 357: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. BioTechniques 31: 132–140 [DOI] [PubMed] [Google Scholar]
- Inoue M, Nakamura Y, Yasuda K, Yasaka N, Hara T, Schnaufer A, Stuart K, Fukuma T (2005) The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis and cell cycle. J Biol Chem 280: 14085–14096 [DOI] [PubMed] [Google Scholar]
- Lai-Zhang J, Xiao Y, Mueller DM (1999) Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J 18: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RH, Manon S, Devenish RJ, Nagley P (1995) ATP synthase from Saccharomyces cerevisiae. Methods Enzymol 260: 133–163 [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Balguerie A, Duvezin-Caubet S, Giraud MF, Slonimski PP, di Rago JP (2003) F1-catalysed ATP hydrolysis is required for mitochondrial biogenesis in Saccharomyces cerevisiae growing under conditions where it cannot respire. Mol Microbiol 47: 1329–1339 [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP (2001) Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem 276: 6789–6796 [DOI] [PubMed] [Google Scholar]
- Matthews KR (2005) The developmental cell biology of Trypanosoma brucei. J Cell Sci 118: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebohacova M, Maslov DA, Falick AM, Simpson L (2003) The effect of RNAi down regulation of RET1 3′ TUTase on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J Biol Chem 279: 7819–7825 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Nolan DP, Voorheis HP (1992) The mitochondrion in bloodstream forms of Trypanosoma brucei is energized by the electrogenic pumping of protons catalysed by the F1F0-ATPase. Eur J Biochem 209: 207–216 [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Borst P, De Rijke D (1976) Oligomycin sensitivity of the mitochondrial ATPase as a marker for fly transmissibility and the presence of functional kinetoplast DNA in African trypanosomes. Comp Biochem Physiol B 55B: 25–30 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Haynes PA, Gygi SP, Stuart K (2003) Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom 14: 728–735 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Carmean N, Igo RP Jr, Gygi S, Ernst N, Palazzo SS, Weston D, Aebersold R, Salavati R, Stuart KD (2001) Four related proteins of the T. brucei RNA editing complex. Mol Cell Biol 21: 6833–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Domingo GJ, Stuart KD (2002) Natural and induced dyskinetoplastid trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol 32: 1071–1084 [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP Jr, Salavati R, Stuart K (2001) An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2162 [DOI] [PubMed] [Google Scholar]
- Schneider A (2001) Unique aspects of mitochondrial biogenesis in trypanosomatids. Int J Parasitol 31: 1403–1415 [DOI] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E (2000) Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeiky Y, Benson DR, Elwasila M, Badaro R, Burns JM Jr, Reed SG (1994) Antigens shared by Leishmania species and Trypanosoma cruzi: immunological comparison of the acidic ribosomal P0 proteins. Infect Immun 62: 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songa EB, Paindavoine P, Wittouck E, Viseshakul N, Muldermans S, Steinert M, Hamers R (1990) Evidence for kinetoplast and nuclear DNA homogeneity in Trypanosoma evansi isolates. Mol Biochem Parasitol 43: 167–180 [DOI] [PubMed] [Google Scholar]
- Speijer D, Breek CK, Muijsers AO, Hartog AF, Berden JA, Albracht SP, Samyn B, Van Beeumen J, Benne R (1997) Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol Biochem Parasitol 85: 171–186 [DOI] [PubMed] [Google Scholar]
- St Pierre J, Brand MD, Boutilier RG (2000) Mitochondria as ATP consumers: cellular treason in anoxia. Proc Natl Acad Sci USA 97: 8670–8674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD (1971) Evidence for the retention of kinetoplast DNA in an acriflavin-induced dyskinetoplastic strain of Trypanosoma brucei which replicates the altered central element of the kinetoplast. J Cell Biochem 49: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK (2005) Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30: 97–105 [DOI] [PubMed] [Google Scholar]
- Tan TH, Bochud-Allemann N, Horn EK, Schneider A (2002) Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc Natl Acad Sci USA 99: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms MW, van Deursen FJ, Hendriks EF, Matthews KR (2002) Mitochondrial development during life cycle differentiation of African trypanosomes: evidence for a kinetoplast-dependent differentiation control point. Mol Biol Cell 13: 3747–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercesi AE, Docampo R, Moreno SNJ (1992) Energization-dependent Ca2+ accumulation in Trypanosoma brucei bloodstream and procyclic trypomastigotes mitochondria. Mol Biochem Parasitol 56: 251–258 [DOI] [PubMed] [Google Scholar]
- Welburn SC, Barcinski MA, Williams GT (1997) Programmed cell death in trypanosomatids. Parasitol Today 13: 22–26 [DOI] [PubMed] [Google Scholar]
- Wilkes JM, Mulugeta W, Wells C, Peregrine AS (1997) Modulation of mitochondrial electrical potential: a candidate mechanism for drug resistance in African trypanosomes. Biochem J 326: 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N (1994) The mitochondrial ATP synthase of Trypanosoma brucei: structure and regulation. J Bioenerg Biomembr 26: 173–178 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Simone L, Claudia O, Cross GAM (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary methods


