Abstract
We have shown previously that the human cytomegalovirus (HCMV) major immediate-early (MIE) distal enhancer is needed for MIE promoter-dependent transcription and viral replication at low multiplicities of infection (MOI). To understand how this region works, we constructed and analyzed a series of HCMVs with various distal enhancer mutations. We show that the distal enhancer is composed of at least two parts that function independently to coordinately activate MIE promoter-dependent transcription and viral replication. One such part is contained in a 47-bp segment that has consensus binding sites for CREB/ATF, SP1, and YY1. At low MOI, these working parts likely function in cis to directly activate MIE gene expression, thus allowing viral replication to ensue. Three findings support the view that these working parts are likely cis-acting elements. (i) Deletion of either part of a bisegmented distal enhancer only slightly alters MIE gene transcription and viral replication. (ii) Reversing the distal enhancer’s orientation largely preserves MIE gene transcription and viral replication. (iii) Placement of stop codons at −300 or −345 in all reading frames does not impair MIE gene transcription and viral replication. Lastly, we show that these working parts are dispensable at high MOI, partly because of compensatory stimulation of MIE promoter activity and viral replication that is induced by a virion-associated component(s) present at a high viral particle/cell ratio. We conclude that the distal enhancer is a complex multicomponent cis-acting region that is required to augment both MIE promoter-dependent transcription and HCMV replication.
The human cytomegalovirus (HCMV) major immediate-early (MIE) gene products, IE1 p72 and IE2 p86, are required for initiating viral replication (10, 16, 21, 30). Their transcription is controlled by the MIE regulatory region, which is composed of a promoter, enhancer, unique region, and modulator (reviewed in references 25 and 29). The 485-bp enhancer segment spans base positions −65 to −550 with respect to the +1 start site of MIE RNAs (25). The enhancer is recognized for its vigor in reliably activating transcription from MIE promoter constructs when put into widely diverse in vitro, transfection, and transgenic animal systems (25). However, enhancer strength depends on cell type, degree of cellular differentiation, and activity of certain signal transduction pathways (25, 28). This variable functioning is a result of changes in the amounts or activities of assorted cellular and viral proteins that act on the enhancer. Examples of several distinct sets of cellular transcription factors that bind to and stimulate the enhancer include NF-κB/rel, CREB/ATF, AP-1, SP-1, serum response factor, ELK-1, and liganded retinoic acid receptor (7, 25, 28). Some of these (NF-κB/rel, CREB/ATF, SP-1, and retinoic acid receptor) bind to multiple cognate sites, suggesting that they may interact cooperatively to influence enhancer function. Viral proteins pp71 and IE1 p72 can also act through cis-acting sites to augment enhancer activity (8, 18, 19, 39). While our understanding of the functional interplay between these transcription factors is quite limited, such interplay is likely complex and a distinguishing feature of this enhancer.
Only recently has the functional role of the MIE enhancer been examined in the natural setting of infection. For murine CMV (MCMV), deletion of the enhancer greatly impairs viral replication in culture but is rescued by expression of the MCMV MIE gene products from a separate vector (2). Replacement of MCMV’s enhancer with the paralogous HCMV enhancer does not appreciably alter viral replication in mouse livers and cultured fibroblasts (2, 12), although the enhancer change coincides with a decreased likelihood of infection at extrahepatic sites in mice (12). Rat CMV is less able to replicate in cultured rat fibroblasts and to establish infection of salivary tissue when its MIE enhancer is replaced with the paralogous murine enhancer (35). By analogy, the MIE enhancer of HCMV has likely evolved to ensure optimal MIE gene expression and viral replication in humans. The functional importance of HCMV’s enhancer in human cells is further supported by the findings of two recent studies. The first study showed that in HCMV-infected human embryonal NTera2 cells, enhancer silencing is linked to a block in both MIE gene transcription and viral replication, whereas reactivation of the enhancer by trichostatin A reverses the block (22). In the second study, the selective removal of the distal segment of the enhancer (−300 to −580) was found to greatly decrease both MIE gene transcription and viral replication in human fibroblasts at low multiplicities of infection (MOI) but not at high MOI (23). This finding was a surprise, given that transient transfection analyses of comparable enhancer segments in reporter plasmids revealed that such a mutation does not cause an appreciable decrease in MIE promoter activity in various differentiated cell types (17, 42). The reason for these contrasting findings is unknown, but the results underscore the importance of examining regulatory elements in the context of the viral genome.
The mechanism by which the 280-bp distal enhancer augments MIE transcription and viral replication at low MOI is open to question. One possibility is that removal of one or more specific cis-acting sites in the distal enhancer (e.g., serum response factor, ELK-1, NF-κB/rel, CREB/ATF, SP-1, YY-1, and/or retinoic acid receptor binding sites) directly impairs transcriptional activation of the MIE genes and, consequently, viral replication. Another possibility is that the distal enhancer deletion unexpectedly disrupts regulatory elements or coding regions of overlapping or neighboring genes that indirectly affect MIE gene expression or viral replication. The latter consideration is also plausible given that viral RNAs of low abundance cross the distal enhancer during lytic infection and a distal enhancer deletion lessens transcription of the distantly located viral immediate-early (IE) kinetic-class US3 gene (23). Thus, the distal enhancer could conceivably operate in a cis- or trans-acting manner.
In this report, we describe the initial characterization of the mechanisms underlying MIE distal enhancer function and elucidate the compensatory role of supernumerary viral particles in rendering the distal enhancer dispensable.
MATERIALS AND METHODS
Cells, viruses, and infections.
Primary human foreskin fibroblast (HFF) cells were isolated and grown as described previously (24). Hypoxanthine-guanine phosporibosyltransferase (HGPRT)-deficient fibroblasts (GM02291) were obtained from Coriell Institute for Medical Research (Coriell Cell Repositories, Camden, N.J.) and grown in Eagle’s minimal essential medium supplemented with 5% newborn bovine serum and 5% fetal bovine serum. HCMVs (Towne strain) were propagated, harvested, and titered as described previously (23, 24). Titers of HCMVs with distal enhancer mutations cannot be accurately determined by plaque assay. Therefore, titers of these viruses were normalized to known titers of replication-competent viruses on the basis of cytopathic effect (CPE) in HFF cells at 24 h postinfection (p.i.) and amount of viral DNA in HFF cells prior to viral replication (4 to 5 h p.i.) (23). For comparative studies, the viruses were propagated, harvested, and titered in parallel. Viral stocks with titers differing less than threefold were studied after adjustment by dilution with growth medium.
For inactivation of HCMV by UV light, extracellular HCMV particles were first isolated according to the method of Stinski (40, 41). Briefly, the growth medium overlying infected HFF cells at 100% CPE was centrifuged at 7,000 × g for 10 min, the resultant supernatant was centrifuged through a 20% d-sorbital cushion (17,500 × g) in 1× phosphate-buffered saline, and the viral pellet was resuspended in 4 ml of Eagle’s minimal essential medium. Two milliliters of viral suspension (∼108 PFU/ml) was placed in a lidless 30-mm2 cell culture dish and irradiated with UV light at a wavelength of 260 nm (13) for 60 to 70 min by use of a Mineralight Lamp model UVSL-25 (UVP, Inc., San Gabriel, Calif.) mounted 4.5 cm above the bottom of the culture dish. The minimum duration of UV irradiation needed to completely inactivate HCMV replication was determined by standard viral plaque assay and by monitoring for green fluorescent protein (GFP) expression from recombinant rΔ-582/-1108Egfp or rΔ-300/-1108Egfp (22) as a marker of viral lytic life cycle activity.
Plasmids.
Plasmids p1.6 and pIE1 have been reported previously (23). pΔ-300/-579, pΔ-521/-579, and pΔ-347/-579 were derived from pIE1 by multistep cloning. Briefly, a synthetic duplex oligonucleotide (5′-CTAGCCCGGGCCTAGG-3′) was inserted into the SpeI site of the HCMV MIE regulatory region contained in an intermediate shuttle vector to yield plasmid pLC12. This oligonucleotide insertion contains AvrII and SmaI sites. To construct pΔ-300/-579, pΔ-521/-579, and pΔ-347/-579, respectively, the BstN1 (blunted with Klenow fragment)-to-SmaI, BstUI-to-SmaI, and NdeI (blunted with Klenow fragment)-to-SmaI fragments of pLC12 were deleted. From each of the three resultant constructs, the EagI-to-BsrGI fragment was isolated and subcloned into corresponding sites in pIE1 to yield pΔ-300/-579, pΔ-521/-579, and pΔ-347/-579. All of these constructs contain an AvrII site at the site of the deletion. pΔ-300/-347 was derived from pΔ-300/-579 by deletion of the SmaI-to-NdeI fragment and insertion of a duplex oligonucleotide (5′-GGGCCTAGACCTAGCTACA-3′) containing an NheI site. pIE1.AvrII contains an AvrII site at −300 of pIE1. This was constructed by subcloning a PCR-generated fragment spanning nucleotides −300 to −640 of the MIE regulatory region into the AvrII (−300)-to-BsrGI (−640) sites of pΔ-300/-579, while maintaining an AvrII site at −300. A duplex oligonucleotide (5′-CTAGGTAGCTAGCTAGCTAC-3′) containing stop codons in all open reading frames (ORFs) was inserted into the AvrII site of pIE1.AvrII to yield pIE1.AvrII.stop. p-579/-300 was constructed by subcloning the AvrII-to-SpeI fragment of pIE1.AvrII into the AvrII site of pΔ-300/-579. All plasmids were sequenced to confirm the presence of the desired mutations.
HCMV recombination.
Recombinant HCMVs rΔ-300/-579, rΔ-521/-579, rΔ-347/-579, rΔ-300/-347, r-579/-300, r-300.A, and r-300.AS were derived from rΔMSVgpt by homologous recombination with pΔ-300/-579, pΔ-521/-579, pΔ-347/-579, pΔ-300/-343, p-580/-300, pIE1.AvrII, and pIE1.AvrII.stop, respectively (23). The method of cotransfection of plasmid (5 μg) and rΔMSVgpt (20 to 50 μg) DNAs has been detailed previously (23, 24). Recombinant viruses were selected in HGPRT-deficient fibroblasts (MOI, 0.3 to 0.5) exposed to 6-thioguanine (50 μg/ml) as described previously (23). All recombinant viruses were subjected to at least two rounds of plaque isolation. Genomes of plaque-purified viruses were analyzed by restriction endonuclease and Southern blot analyses. Two recombinant virus clones were obtained from independent transfection-recombination procedures to control for spurious genomic mutations.
DNA analysis.
HCMV genomic DNA was isolated as described previously (24). Viral genomes were digested with each of the EcoRI, PstI, and BamHI enzymes and fractionated on a 0.7% agarose gel. Ethidium bromide-stained restriction enzyme fragment profiles were analyzed prior to Southern blot analysis and autoradiography, as described previously (24). Probes were generated by either multiprime 32P labeling of indicated double-stranded DNA fragments or 32P end labeling of indicated oligonucleotides. Blots were stripped of probes by boiling in 0.2% sodium dodecyl sulfate prior to reuse.
HCMV DNA replication was analyzed by using the methods described previously (23, 24). Briefly, after virus absorption for 1.5 h, cells were exposed for 1 min to citrate buffer (50 mM sodium citrate and 4 mM KCl [pH 3]) to inactivate extracellular virus and were then washed three times with Hanks’ balanced salt solution without calcium and magnesium. Infected cell DNA was isolated, digested with HindIII, fractionated electrophoretically on a 0.5% agarose gel, and subjected to Southern blot analysis. Lambda DNA (2 μg) was added to each cell lysate sample to control for variation in processing, endonuclease digestion, and loading. Southern hybridization was carried out with a probe that is complementary to HCMV genomic termini containing TRL or IRL. This probe, termed T probe, was generated by multiprime 32P labeling of the 1.6-kbp BamHI-HindIII fragment of plasmid p1.6 (23). Hybridization signals were quantitated by image acquisition analysis (Hewlett-Packard Instant Imager).
Multiplex RT-PCR.
Whole-cell RNA was isolated according to the method of Chomczynski and Sacchi (9). SUPERSCRIPT II RNase H-negative reverse transcriptase (RT) (Life Technologies, Gaithersburg, Md.) was used according to the manufacturer’s directions to generate cDNA from 4 μg of RNA and 250 ng of random hexamers (Invitrogen, Carlsbad, Calif.) in a final volume of 20 μl. Thereafter, samples were heat inactivated at 70°C for 15 min and then diluted 1:5 with H2O. Multiplex real-time PCR was performed using the ABI PRISM 7700 Sequence Detection System (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.) according to the manufacturer’s directions for simultaneous detection of probes containing 6-carboxyfluorescein (FAM) and VIC reporter fluorophores. Amplifications were performed in a final volume of 25 μl of the PLATINUM Quantitative PCR SUPERMIX-UDG cocktail (Life Technologies) prepared according to the manufacturer’s directions. Each reaction mixture contained 2 μl of the test sample, 5 mM MgCl, 500 nM each MIE primer, 250 nM MIE probe, 12.5 nM each 18S primer, and 50 nM 18S probe. MIE primer sequences 5′-GCATTGGAACGCGGATTC-3′ and 5′-CAGGATTATCAGGGTCCATCTTTC-3′ (Life Technologies) are located in MIE exons 1 (forward) and 2 (reverse), respectively, which are separated by the 827-bp intron A. The MIE reporter probe, 5′-FAM-AGTGACTCACCGTCCTTGACACGATGG-tetramethyl rhodamine (TAMRA)-3′ (IDT, Coralville, Iowa), straddles the junction of MIE exons 1 and 2. Cellular 18S rRNA served as the endogenous control. The primers used to amplify the intronless ribosomal 18S cDNA and the VIC-labeled probe used for detecting this amplicon were obtained from PE Applied Biosystems. Thermal cycling conditions were an initial 50°C for 2 min and 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primer and template concentrations were chosen based on results of relative standard curves and validation experiments, as detailed by PE Applied Biosystems. Each experiment was performed in parallel with a relative standard curve and validation experiment, corroborating the use of the comparative threshold cycle (CT) method for relative quantitation of MIE RNA. Average CT values were simultaneously determined for MIE and 18S RNAs, and the difference between them is ΔCT. Standard deviations of the MIE and 18S CT values (SDMIE and SD18S, respectively) were also determined. The relative difference among samples, ΔΔCT, is ΔT − ΔCT,x, where the ΔCT,x value is that of the comparator (x). The standard deviation of the ΔΔCT value (SD) is equal to (SDMIE2 + SD18S2)1/2. The average quantity of MIE RNA normalized to 18S RNA and relative to comparator RNA is equal to 2−ΔΔCT, and the SD determines the values of the error bars (2−ΔΔCT+ SD and 2−ΔΔCT− SD). CT values of samples not treated with RT, but analyzed in parallel to the RT-treated samples, did not appreciably differ from the baseline.
RESULTS
The distal enhancer is composed of at least two functional parts that independently activate HCMV replication.
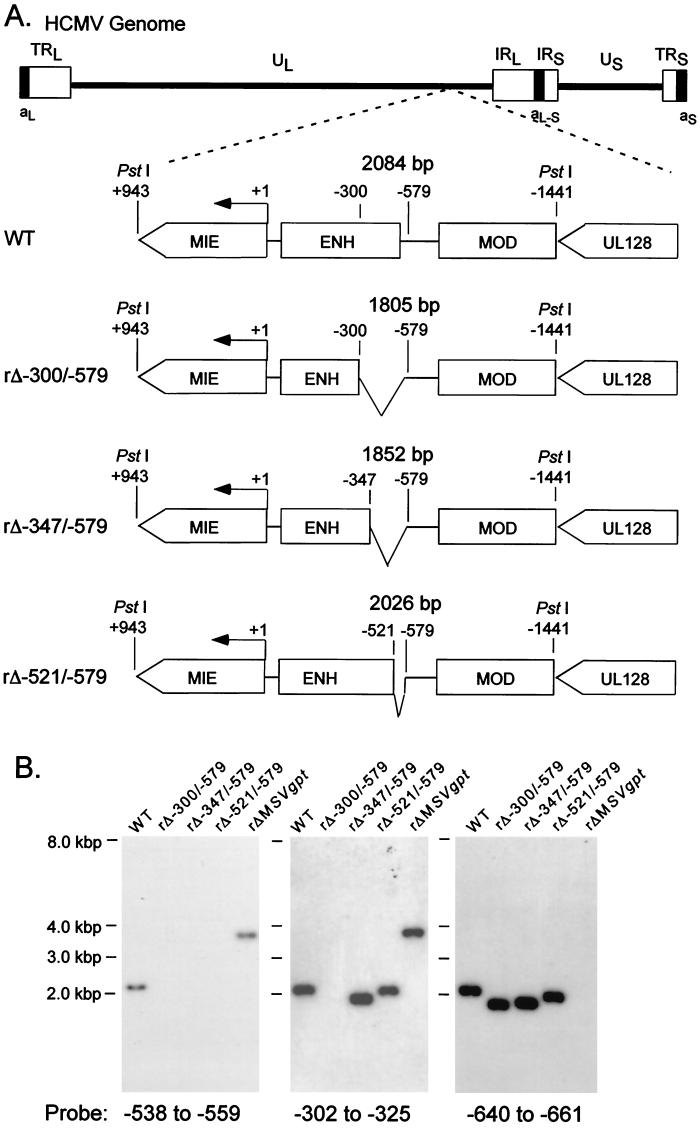
Our prior findings using a replacement mutagenesis strategy revealed that the distal enhancer segment from −300 to −582 possessed activity for augmenting MIE gene transcription and viral replication at low MOI. To determine whether the distal enhancer is composed of a single or multiple functional units, we subjected this region to sequential deletion analysis. Site-directed deletions were introduced into the HCMV genome by homologous recombination in fibroblasts, as described in Materials and Methods. We began by creating deletions of increasing size at the 5′ end of the distal enhancer. As depicted in Fig. 1A, the deletions are located at positions −521 to −579, −347 to −579, and −300 to −579 in recombinant viruses rΔ-521/-579, rΔ-347/-579, and rΔ-300/-579, respectively. These recombinant HCMVs differ from wild-type HCMV (WT) only by their respective deletions. Two distinct isolates of each recombinant type derived from independent recombination procedures were studied to control for spurious mutations. All of the recombinant viral genomes were analyzed in comparison to WT for restriction fragment length polymorphism (RFLP) by using three separate endonucleases (see Materials and Methods). Findings of Southern blot analyses of PstI RFLPs are shown in Fig. 1B for a representative set of viruses.
FIG. 1.
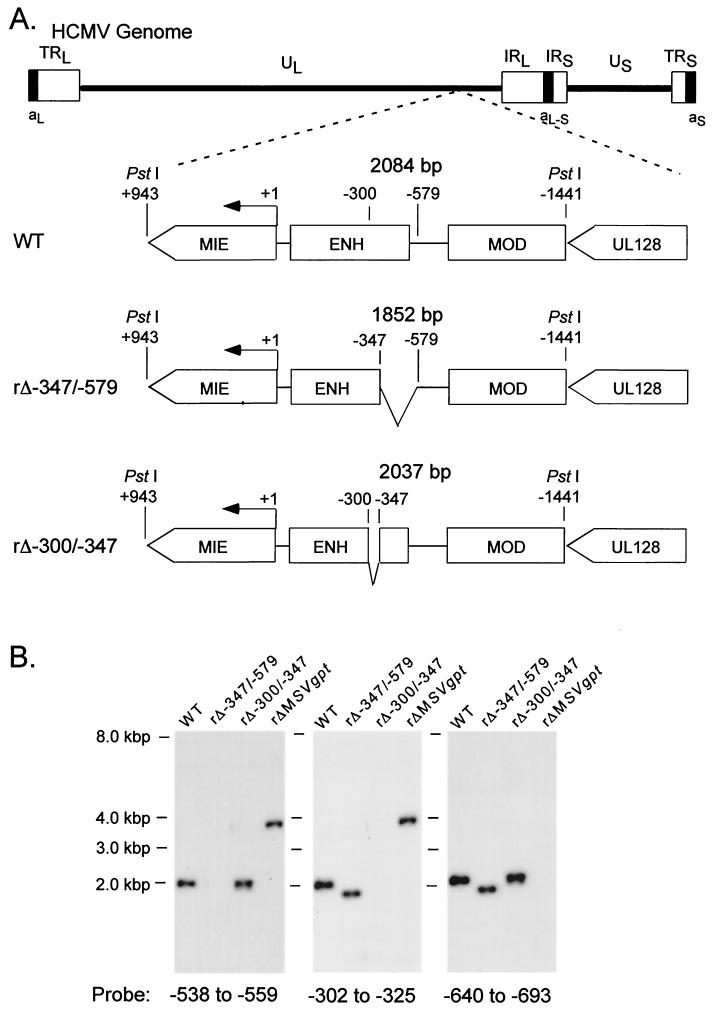
Effects on viral DNA replication of successively larger 5′-subsegmental distal enhancer deletions. (A) Schematic diagram of deleted distal enhancer segments in recombinant HCMVs rΔ-300/-579, rΔ-347/-579, and rΔ-521/-579. Base positions of deletions and PstI sites are depicted relative to the +1 RNA start-site of the MIE promoter. Predicted sizes of PstI RFLPs are shown. MIE, MIE transcription unit; ENH, enhancer; MOD, modulator; UL128, putative UL128 gene. (B) Southern blot analyses of PstI RFLPs of WT, rΔ-300/-579, rΔ-347/-579, rΔ-521/-579, and rΔMSVgpt. rΔ-300/-579, rΔ-347/-579, and rΔ-521/-579 were derived from rΔMSVgpt. Probe coordinates are given in base positions relative to the RNA start-site of the MIE promoter. (C) Abundances of WT, rΔ-347/-579, and rΔ-521/-579 genomes in HFF cells at MOI of 1.0 and 0.001. Infections were performed in parallel with equivalent input viral titers (see Materials and Methods). On the indicated day p.i. (DPI), infected-cell DNA was isolated, digested with HindIII, fractionated by gel electrophoresis, and subjected to Southern blot analysis. The 32P-labeled probe hybridizes to HCMV genomic fragments containing either the terminal repeat-long (TRL) or the internal repeat-long (IRL) region (23). The IRL is fused (Fused End) to the short genome segment, which is contained in the 17.2- and 13-kb fragments. The TRL is not fused (Free End) to the short genome segment and is contained in the 9.7-kb fragment. The blot was stripped and rehybridized to a λ-specific, 32P-labeled probe for detection of the lambda DNA internal control (Control). (D) Abundances of WT, rΔ-347/-579, and rΔ-300/-579 genomes in HFF cells at MOI of 1.0 and 0.001. The analysis was performed as described for Fig. 1C.
The recombinant viruses were first examined for their abilities to replicate in HFF cells at high and low MOI (1.0 and 0.001, respectively). Two independent methods were used to normalize input viral titers (see Materials and Methods). DNA from infected cells was isolated at the indicated times p.i. and subjected to HindIII digestion and Southern blot analysis. As shown in Fig. 1C, all of the recombinant viruses replicate DNA at high MOI at rates comparable to that of WT. However, only rΔ-300/-579 is substantially impaired in its ability to replicate viral DNA at low MOI compared to rΔ-521/-579, rΔ-347/-579, or WT (Fig. 1D).
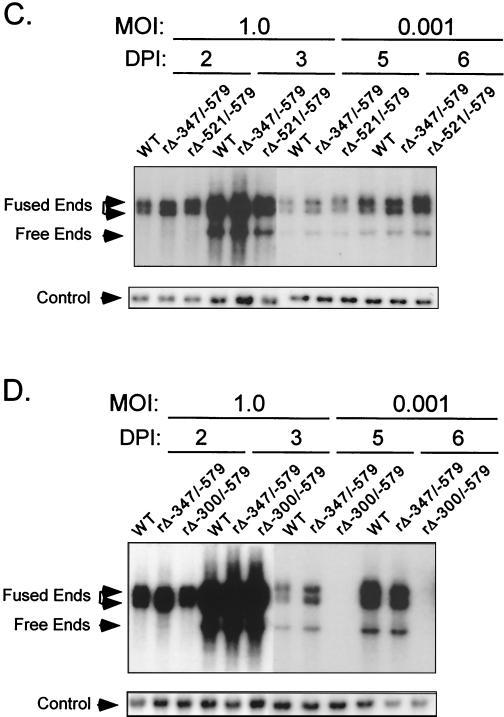
Based on these findings, we determined whether the remaining short segment from −300 to −347 of the distal enhancer had the functional activity needed for normal replication at low MOI. Therefore, recombinant rΔ-300/-347, lacking the −300 to −347 segment but with a genome otherwise matching that of WT, was made (Fig. 2A). This virus’s genome structure was verified by RFLP and Southern blot analyses (Fig. 2B). The replication rates of rΔ-300/-347 at high and low MOI (1.0 and 0.005, respectively) were compared to those of WT, rΔ-347/-579, and rΔ-300/-579 by using the same viral DNA replication assay described for Fig. 1. As expected, the viruses exhibited similar rates of viral DNA replication at high MOI (Fig. 2C). Remarkably, only rΔ-300/-579’s DNA replication rate was substantially impaired at the low MOI (five- to sixfold difference) in comparison to those of the other viruses (Fig. 2D). Thus, neither deletion of the −300 to −347 segment nor deletion of the −347 to −579 segment greatly lowers the replication rate of HCMV at low MOI. This finding indicates that the distal enhancer (−300 to −579) is made of at least two working parts that need not all be present to sufficiently stimulate HCMV replication at low MOI.
FIG. 2.
Multiple distal enhancer parts are involved in augmenting viral DNA replication. (A) Schematic diagram of deleted distal enhancer segments in HCMVs rΔ-347/-579 and rΔ-300/-347. Base positions of deletions and PstI sites are depicted relative to the RNA start site of the MIE promoter. Predicted sizes of PstI RFLPs are shown. (B) Southern blot analyses of PstI RFLPs of WT, rΔ-347/-579, rΔ-300/-347, and rΔMSVgpt. Probe coordinates are given relative to the RNA start site of the MIE promoter. (C) Abundances of WT, rΔ-300/-347, rΔ-347/-579, and rΔ-300/-579 genomes in HFF cells at an MOI of 1.0 on days 2 and 3 p.i. (DPI 2 and 3). (D) Abundances of WT, rΔ-300/-347, rΔ-347/-579, and rΔ-300/-579 genomes in HFF cells at an MOI of 0.005 on days 6 and 8 p.i.. Infections for which results are shown in panels C and D were performed in parallel, using methods described in the legend to Fig 1C.
The same working parts may be involved in activating MIE promoter-dependent transcription.
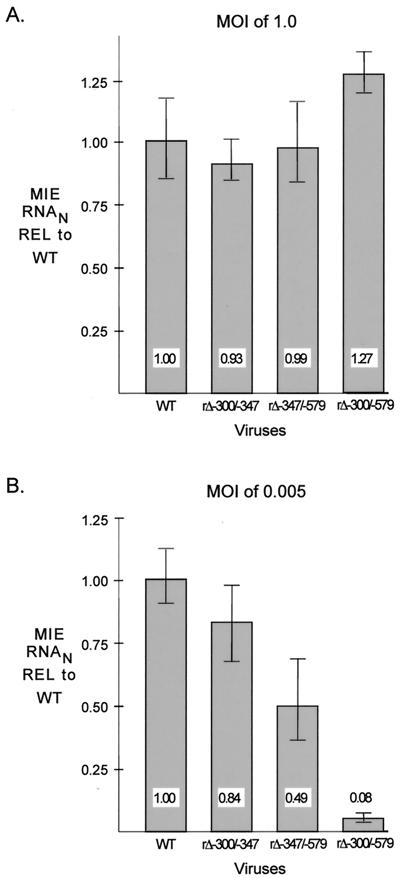
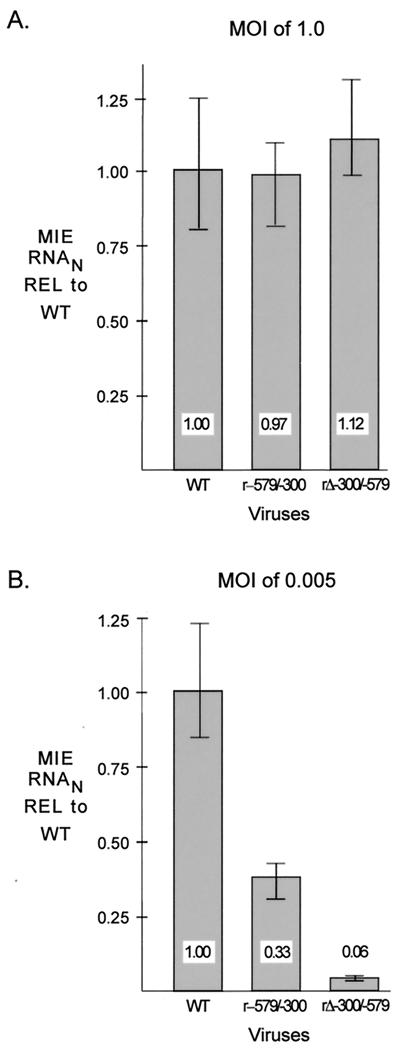
We determined whether the distal enhancer parts involved in stimulating HCMV replication are linked to transcriptional activation of the MIE promoter. Amounts of viral MIE RNAs produced by WT, rΔ-300/-347, rΔ-347/-579, and rΔ-300/-579 in HFF cells were determined at 6 h p.i. for both MOI of 1.0 and 0.005, using a multiplex real-time RT-PCR assay. Concomitant measurement of cellular 18S rRNA controlled for sample-to-sample variation. Input viral titers were normalized by methods described in Materials and Methods. The findings depicted in Fig. 3A reveal that WT, rΔ-300/-347, rΔ-347/-579, and rΔ-300/-579 make similar amounts of MIE RNAs at a high MOI; the mean amounts vary less than 28%. In contrast, rΔ-300/-579 produces 12.5-fold less MIE RNA at a low MOI than WT, whereas rΔ-300/-347 and rΔ-347/-579 yield only slightly less MIE RNA than WT (<2.1-fold difference). The results of this analytical method were verified by additional findings indicating that the abundance of IE1 protein and spliced exon 3/4 MIE RNA produced by rΔ-300/-579 was also markedly lower than that of WT at low MOI, but not at high MOI, when analyzed by semiquantitative Western blotting and RT-PCR methods, respectively (data not shown). Thus, the distal enhancer is composed of at least two working parts that function independently to activate MIE promoter-dependent transcription. The same working parts may be involved in activating both MIE promoter-dependent transcription and viral replication.
FIG. 3.
Multiple distal enhancer parts are involved in augmenting MIE promoter-dependent transcription. Relative quantitation of WT, rΔ-300/-347, rΔ-347/-579, and rΔ-300/-579 MIE RNAs at 6 h p.i. in HFF cells was performed for quadruplicate samples by multiplex real-time RT-PCR, using the CT method and validation experiments according to the manufacturer’s specifications. Viral MIE RNA abundance was normalized to cellular 18S RNA (MIE RNAN) and expressed relative (REL) to that of WT at an MOI of 1.0 (A) or 0.005 (B). Shown are means and standard deviations. Parallel determinations of CT values of corresponding samples lacking RT and of mock-infected samples revealed no appreciable difference from the baseline.
The distal enhancer is a cis-acting region that can operate in reverse orientation.
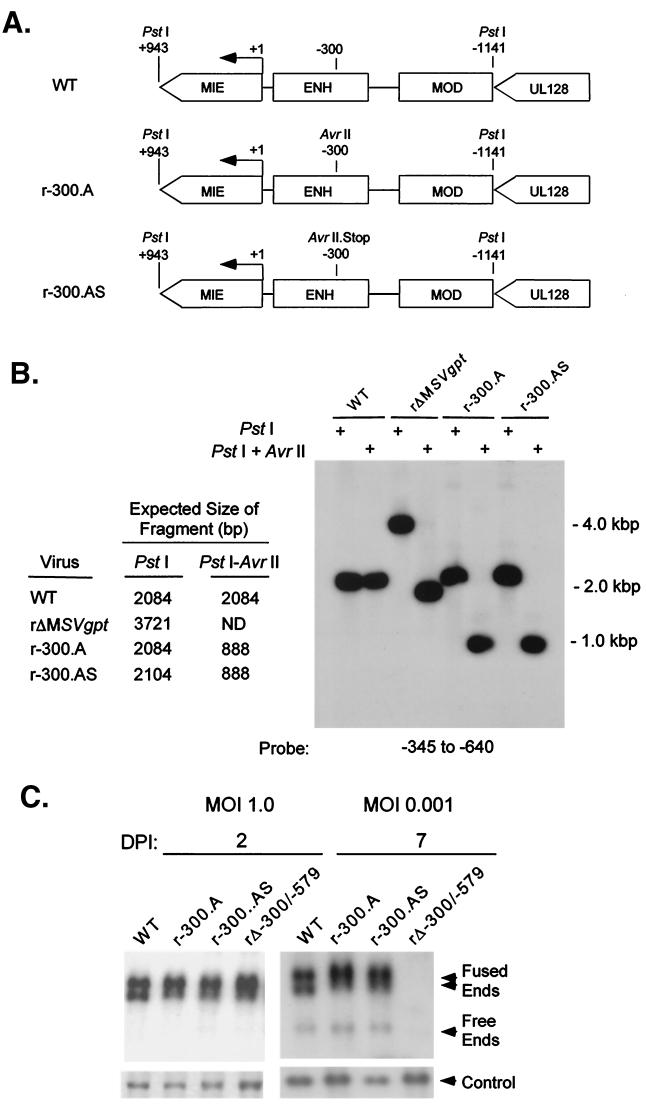
To further determine whether the distal enhancer operates in a cis- or trans-acting manner, the distal enhancer was subjected to insertion mutagenesis to disrupt overlapping hypothetical ORFs. Two types of insertions were introduced at position −300 of the distal enhancer to make recombinant viruses r-300.A and r-300.AS. This specific site was first targeted because it was ideally situated in two different hypothetical ORFs containing motifs showing limited homologies with those in a cellular tyrosine phosphatase and a G protein-coupled receptor. The insertion mutation of r-300.A adds a new AvrII site that disrupts the reading frames, whereas the insertion mutation of r-300.AS creates stop codons in all reading frames coded by each DNA strand (Fig. 4A and B). The DNA replication rates of these mutated viruses were analyzed at MOI of 1.0 and 0.001 on days 2 and 7 p.i., respectively. As shown in Fig. 4C, the rate of viral DNA synthesis is unaltered by either insertion mutation located at −300 in the distal enhancer. These mutations also had no appreciable effect on MIE promoter-dependent transcription under the same growth conditions (data not shown). Insertion mutations were also introduced at −345 of the distal enhancer, and they too did not impair viral replication at high or low MOI (data not shown). Thus, disruption of hypothetical ORFs overlapping the distal enhancer does not hinder viral transcription and replication, unlike a distal enhancer deletion.
FIG. 4.
Stop codons inserted into the distal enhancer do not alter viral DNA replication. (A) Schematic diagram of HCMVs r-300.A and rΔ-300.AS. (B) Southern blot analyses of PstI RFLPs of WT, r-300.A, rΔ-300.AS, and rΔMSVgpt. Probe coordinates relative to the RNA start site of the MIE promoter and predicted sizes of PstI RFLPs are given. (C) Abundances of WT, r-300.A, rΔ-300.AS, and rΔ-300/-579 genomes in HFF cells at MOI of 1.0 and 0.001 on days 2 and 7 p.i. (DPI 2 and 7). Infections were performed in parallel by methods described in the legend to Fig. 1C.
We next determined whether the distal enhancer could function in reverse orientation. A virus, designated r-579/-300, was made in which the distal enhancer (−300 to −579) was inverted at its original location in the MIE regulatory region (Fig. 5A and B). Two viruses derived independently from separate transfection-recombination procedures were subsequently studied. Their DNA replication rates at MOI of 1.0 and 0.005 were compared to those of WT and rΔ-300/-579. Figure 5C shows that r-579/-300, rΔ-300/-579, and WT exhibited similar replication rates at high MOI. At low MOI, both isolates of r-579/-300 replicated much better than rΔ-300/-579 (25- to 30-fold) but somewhat less well than WT (3- to 5-fold) (Fig. 5D and E). Hence, a distal enhancer in reverse orientation substantially compensates for the replication abnormality created by a distal enhancer deletion.
FIG. 5.
An inverted distal enhancer augments viral DNA replication. (A) Schematic diagram of WT and r-579/-300. Base positions of PstI, NdeI, and NcoI sites and probe (solid bar) coordinates are given relative to the RNA start site of the MIE promoter. INV, inverted distal enhancer. (B) Southern blot analyses of PstI and combined NdeI and NcoI RFLPs of WT, r-579/-300, and rΔMSVgpt. Predicted sizes of RFLPs are shown. (C) Abundances of WT, r-579/-300, and rΔ-300/-579 genomes in HFF cells at an MOI of 1.0 on days 2 and 3 p.i. (DPI 2 and 3). (D) DNA replication rates of WT, r-579/-300, and rΔ-300/-579 genomes in HFF cells at an MOI of 0.005 on days 6 and 7 p.i. Infections for which results are shown in panels C and D were performed in parallel by methods described in the legend to Fig. 1C. (E) Abundances of WT, r-579/-300a, r-579/-300b, and rΔ-300/-579 genomes in HFF cells at an MOI of 0.005 on day 7 p.i. Viruses r-579/-300a and r-579/-300b were derived from independent transfection-recombination procedures. The abundances of viral genomes at 4 h, 2 days, and 3 days p.i. at an MOI of 1 were equivalent for all four viruses (data not shown). Analysis was performed as described in the legend to Fig. 1C.
We examined whether the inverted distal enhancer yielded a level of MIE promoter-dependent transcription that is commensurate with viral replication. The amount of MIE RNA produced by r-579/-300 in HFF cells at 6 h p.i. was compared to that of WT and rΔ-300/-579 by the multiplex real-time RT-PCR method. Viral stocks tested in the experiments for which results are shown in Fig. 5C and D were used in these studies to confirm that input viral titers were equivalent. Figure 6A reveals that all three viruses produced similar amounts of IE1 RNA at high MOI. In contrast, r-579/-300 made 5.5-fold more MIE RNA than rΔ-300/-579 and 3-fold less MIE RNA than WT (Fig. 6B). Again, the amount of MIE RNA produced by WT was more than 12-fold that produced by rΔ-300/-579 for the infections carried out at an MOI of 0.005. The inverted distal enhancer replacement yields comparable levels of MIE-dependent transcription and viral replication, although neither of these viral activities is fully restored.
FIG. 6.
An inverted distal enhancer augments MIE promoter-dependent transcription. Relative quantitation of WT, r-579/-300, and rΔ-300/-579 MIE RNAs at 6 h p.i. in HFF cells was performed for quadruplicate samples by multiplex real-time RT-PCR, using the CT method and validation experiments according to the manufacturer’s specifications. The standard curve method was also used in parallel to validate results. Comparative input viral titers are shown in Fig. 5C and D. Viral MIE RNA abundance was normalized to that of cellular 18S RNA (MIE RNAN) and expressed relative (REL) to WT at an MOI of 1.0 (A) or 0.005 (B). Shown are means and standard deviations. Parallel determinations of CT values of corresponding samples lacking RT and of mock-infected samples revealed no appreciable difference from the baseline.
Taken together, the findings support a cis-acting, but not a trans-acting, role of the distal enhancer in stimulating MIE promoter-dependent transcription. This transcriptional regulatory activity is concordantly linked to viral replication.
Addition of UV-inactivated HCMV virions at a high viral-particle/cell ratio compensates for an absent distal enhancer at low MOI.
Because a distal enhancer deletion does not impair HCMV replication at high MOI, we determined whether the application of inactivated purified HCMV particles in numbers simulating a high MOI would compensate for a missing distal enhancer in a virus grown at a low MOI. The minimum amount of UV irradiation needed to fully inactivate HCMV particles isolated from extracellular medium was first determined using HCMV rΔ-582/-1108Egfp because GFP expression served as a convenient marker of lytic viral gene expression (22) (see Materials and Methods). This UV dose was then used to inactivate isolated WT particles (WTUV). HFF cells were infected in parallel with WT or rΔ-300/-579 at an MOI of 0.001 in the presence or absence of WTUV at a high viral-particle/cell ratio (∼3 to 5 PFU/cell prior to UV irradiation), and the viral plaques that formed were enumerated. Figure 7 shows the representative results of one of five experiments. Exposure of HFF cells to WTUV alone did not yield viral plaques. The combination of WTUV and rΔ-300/-579 produced 21.8-fold more plaques than rΔ-300/-579 alone. In contrast, WTUV was less able to increase the number of plaques produced by WT than to increase the number produced by rΔ-300/-579 (2.1- versus 21.8-fold, respectively), suggesting that a virus missing the distal enhancer benefits most from the stimulation. rΔ-300/-579’s ability to form plaques at low MOI was also greatly increased by addition of UV-inactivated rΔ-300/-579 but was not increased by addition of growth medium of infected cells that was cleared of viral particles (data not shown).
FIG. 7.
UV-inactivated HCMV virions applied at a high viral-particle/cell ratio greatly increase the ability of rΔ-300/-579 to produce plaques at low MOI. Purified WTUV (3 to 5 PFU/cell prior to UV inactivation) was applied at the time of infection of HFF cells with WT or rΔ-300/-579 at an MOI of 0.001. After viral absorption, cells were washed and covered with agarose. Viral plaques were visualized by light microscopy and enumerated. Mean plaque numbers and SDs were derived from triplicate experiments performed in parallel.
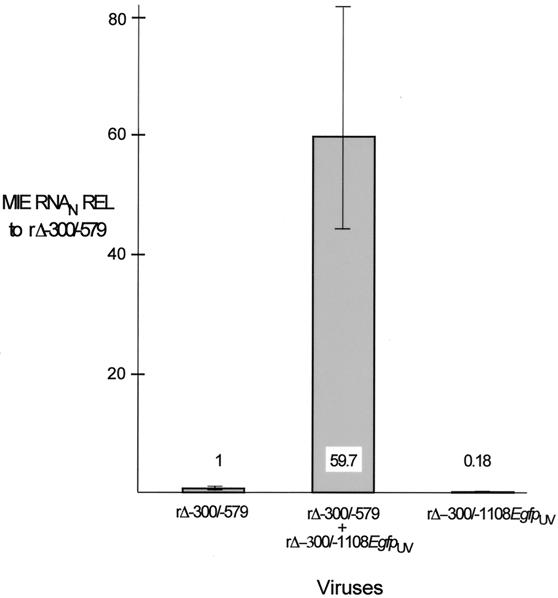
To determine whether an MIE promoter lacking a distal enhancer responds to stimulation provided by virion components, we applied UV-inactivated virions at a high viral-particle/cell ratio (∼5 PFU/cell prior to UV irradiation) to HFF cells infected with rΔ-300/-579 at an MOI of 0.005. For these studies, we inactivated recombinant rΔ-300/-1108Egfp because this virus’s MIE distal enhancer is replaced with an adenovirus E1b TATA box and GFP cassette that function with IE-class kinetics (22). The requisite demonstration that inactivated rΔ-300/-1108Egfp (rΔ-300/-1108EgfpUV) did not yield viral plaques or GFP fluorescence in exposed cells was assurance that HCMV lytic gene expression was successfully prevented. HFF cells were either mock infected or infected with rΔ-300/-579 in the presence or absence of rΔ-300/-1108EgfpUV. The relative MIE RNA abundance at 6 h p.i. was determined using multiplex real-time RT-PCR. As shown in Fig. 8, addition of the inactivated viral-particle preparation increased rΔ-300/-579 MIE RNA abundance 59.7-fold. A very small amount of MIE RNA was detected in cells exposed only to rΔ-300/-1108EgfpUV, a finding consistent with the recent report that HCMV particles themselves contain MIE RNAs (11). Such virion-associated RNAs would be anticipated to be detectable by the PCR-based method despite their inactivation by UV irradiation.
FIG. 8.
Augmentation of rΔ-300/-579 MIE promoter-dependent transcription by rΔ-300/-1108EgfpUV at a high viral-particle/cell ratio. HFF cells were infected with rΔ-300/-579 at an MOI of 0.005, and rΔ-300/-1108EgfpUV (MOI of 3 prior to UV inactivation) was added concomitantly to or omitted from the infection. Relative quantitation of viral MIE and cellular 18S RNAs was performed on quadruplicate samples by multiplex real-time RT-PCR, using the CT method and validation experiments according to the manufacturer’s specifications. Viral MIE RNA abundance was normalized to that of 18S RNA (MIE RNAN) and expressed relative (REL) to that of rΔ-300/-579 in the absence of rΔ-300/-1108EgfpUV. Shown are means and standard deviations. Parallel determinations of CT values of corresponding samples lacking RT and of mock-infected samples revealed no appreciable difference from the baseline.
Thus, the findings indicate that hyperexposure to virion-associated factors largely compensates for the distal enhancer’s absence at low MOI. These findings may partly explain why WT and a virus lacking a distal enhancer are similar at high MOI with regard to both viral MIE RNA production and DNA replication.
DISCUSSION
The requirement by HCMV of an MIE distal enhancer to activate both MIE gene expression and viral replication at a low MOI was shown previously (23), but the mechanism by which this 280-bp genome region confers these activities was unknown. In this report, we have shown that the distal enhancer is composed of at least two working parts that operate independently to activate MIE promoter-dependent transcription and viral replication. These parts likely function in cis to directly activate the MIE promoter, based on three sets of findings observed at low MOI. (i) Deletion of either part of a divided distal enhancer minimally alters viral MIE gene transcription and DNA replication (Fig. 1, 2, and 3). (ii) Reversing the distal enhancer’s orientation largely preserves MIE gene transcription and viral replication (Fig. 5 and 6). (iii) Insertion of stop codons in all potential reading frames at position −300 or −345 of the distal enhancer does not impair viral replication (Fig. 4) or MIE gene expression (data not shown).
Our results indicated that at least one of the cis-acting elements is located in a 47-bp segment (Fig. 1, 2, and 3) that has consensus-binding sites for CREB/ATF, SP1, and YY1 (25). Therefore, one or a combination of these specific cellular transcription factor binding sites may be needed to activate MIE gene transcription at low MOI, although we cannot exclude the possibility of an unrecognized binding site conferring the activation. Notably, each of the CREB/ATF, SP1, and YY1 recognition motifs is present in three copies distributed throughout the distal enhancer, and two additional CREB/ATF sites are located in the proximal enhancer (25). The distal enhancer also has binding sites for serum response factor, ELK-1, retinoic acid receptor, and NF-κB/rel, whereas the proximal enhancer has additional binding sites for AP-1, retinoic acid receptor, and NF-κB/rel (7, 25). Given the extent and variety of possible transcription factors that act on the enhancer, why should deletion of the entire 279-bp distal enhancer, but not its subsegments, have such a dramatic effect? The answer to this query is likely to be complex. It is conceivable that activation of MIE gene expression and viral replication at low MOI requires at least one cis-acting element in the distal enhancer in order to achieve a synergistic interaction that involves other regulatory elements located in neighboring regions. The synergism may derive from the coordinate assembly of multiple transcription factors on these different elements. Such a paradigm would resemble the multicomponent enhanceosome that forms to synergistically trans-activate the cellular beta interferon promoter (20, 31). This analogy may also apply to other viruses. The human T-cell leukemia virus type 1 (HTLV-1) enhancer of the long terminal repeat (LTR) promoter is one example. This enhancer contains three 21-bp repeats that are called TAX-responsive elements (TxREs) because they confer trans-activation by the viral TAX protein (26). CREB/ATF binds to each TxRE and is also required for its function. However, a single TxRE only weakly activates transcription, whereas two or three TxREs greatly activate the promoter (36). Thus, the synergistic or cooperative interaction between regulatory elements is important for activation of the HTLV-1 LTR promoter. Notably, the multiple CREB/ATF sites located in both proximal and distal portions of the HCMV MIE enhancer are functionally responsive to signaling induced by virion-associated components, mitogens, catecholamines, membrane depolarization, or forskolin when assayed in transfected or transduced cells (15, 33, 38, 42, 43). Whether these or other regulatory elements in the proximal and distal MIE enhancer regions (e.g., CREB/ATF sites) interact in a synergistic manner awaits the findings of future studies.
The effects of a distal enhancer deletion on MIE gene expression and viral replication have been validated previously using HCMVs in which the distal enhancer was replaced with heterologous sequence and subsequently reverted to WT (23). While the findings of several short subsegmental distal enhancer deletions have not been validated in this fashion, we believe it unlikely that the findings reflect artifactual idiosyncrasies of the viral construction. Nonetheless, additional mutagenesis of this complex region will be required in the future to identify and fully characterize the functional role of the cis-acting elements. Our studies have not discounted the possibility that distal enhancer regulatory elements may also have unanticipated effects on the expression of neighboring genes in a manner that could alter viral replication. There is at present no evidence to support this notion for HCMV, although swapping the MIE enhancer of rat CMV for the paralogous murine enhancer impairs rat CMV replication without altering MIE gene transcription for reasons that are unknown (35). In contrast, compelling evidence is mounting to support the idea that HCMV or MCMV replication requires sufficient activation of MIE gene expression (1, 2, 10, 16, 21–23, 30).
Another interesting characteristic of the distal enhancer’s cis-acting elements is that they are dispensable at high MOI. We have shown that UV-inactivated HCMV virions applied at a high viral-particle/cell ratio can largely offset the loss of a distal enhancer in a virus grown at a low MOI. This finding may explain why the distal enhancer is not needed at an MOI of 1 when the average infected cell may be exposed concomitantly to as many as 100 noninfectious or defective viral particles (41). We surmise that heightened exposure to a component(s) of the virion preparation functionally compensates for removal of these regulatory elements, since infected cell culture supernatant cleared of virus fails to restore function. Earlier studies showing trans-activation of transiently or stably transfected MIE promoter-enhancer segments by HCMV infection in the presence of protein synthesis inhibitors first suggested the role of virion-associated components in augmenting MIE gene expression (18, 37, 42). Transfection experiments have revealed that virion tegument proteins, such as UL82 (pp71) (18), UL69 (44), and TRS1/IRS1 (34) are able to augment MIE promoter activity. UL82 was also shown to activate an HCMV MIE promoter implanted in the herpes simplex virus genome (14) and to enhance the infectivity of HCMV DNA (3). Recently, Bresnahan and Shenk used a recombinant HCMV lacking UL82 to confirm the idea that virion-associated UL82 has an important role in activating MIE gene transcription from within the HCMV genome (6). The mechanism by which UL82 functions is poorly understood, although Liu and Stinski have found that this protein activates the MIE promoter-enhancer through CREB/ATF and AP-1 elements when assayed in a transient transfection system (18). Perhaps large numbers of UL82-containing virions can further stimulate remaining CREB/ATF and AP-1 elements located in the proximal enhancer to help compensate for the distal enhancer deletion.
We do not exclude the possibility that other virion components may be involved in activation of MIE gene expression. For example, viral gB and gH activate signal transduction pathways via ligand-receptor engagement to increase the binding of NF-κB trans-activators at multiple cognate sites in the MIE enhancer (4, 45). The virion also carries kinases and serine/threonine protein phosphatases that may regulate viral or cellular gene expression by modifying signal transduction pathways and activities of transcription factors (27). In addition, the virus has been reported to package the MIE gene product, IE1 p72 (45), but this association has not been detected by others (32). If IE1 p72 is provided by the virion, it would be predicted to stimulate MIE promoter activity and/or viral replication based on prior findings of transient transfection assays (8, 19, 39) and HCMVs lacking IE1 p72 (10, 30). Finally, an effect caused by a copurification contaminant or the translation of incompletely inactivated virion-associated RNA of viral or cellular origin (5, 11) is unlikely given the findings with our experimental controls. Notably, the inhibition of protein synthesis also restores activity to an MIE promoter lacking the distal enhancer (J. L. Meier, unpublished data), so that virion-mediated effects in the absence of de novo protein synthesis cannot be accurately discerned.
We infer from these studies that the MIE distal enhancer is complex and likely to contain multiple cis-acting elements that function together with other elements in the MIE regulatory region to confer promoter-dependent transcription. The pivotal role of these regulatory elements in HCMV lytic replication underscores the need to further determine their identity and mechanism of action.
Acknowledgments
We are grateful to Mark Stinski for critical reading of the manuscript. We thank members of the Stinski laboratory and Arnold Rabson for helpful discussions of this work. We also thank Kevin Knudtson of the DNA Core Facility for generously providing advice and help in performing the real-time PCR assays.
This work was supported by the National Institutes of Health (AI-40130), an American Cancer Society institutional research grant (IN-122R), and the March of Dimes (FY99-549).
REFERENCES
- 1.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene IE3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502–8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick, C. J., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within the human cytomegalovirus particles. Science 288:2373–2376. [DOI] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate-early genes in human cytomegalovirus infected cells. Proc. Natl. Acad. Sci. USA 97:14506–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, Y.-J., C.-J. Chiou, Q. Huang, and G. S. Hayward. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness or primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J. Virol. 70:8590–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159. [DOI] [PubMed] [Google Scholar]
- 10.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol 72:366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greijer, A. E., C. A. J. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during assembly process. J. Virol. 74:9078–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grzimek, N. K. A., J. Podlech, H. P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch, A. J., and T. Shenk. 1999. Human cytomegalovirus inhibits transcription of the CC chemokine MCP-1 gene. J. Virol. 73:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homer, E. G., A. Rinaldi, M. J. Nicholl, and C. M. Preston. 1999. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J. Virol. 73:8512–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunninghake, G. W., M. M. Monick, B. Liu, and M. F. Stinski. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP response elements. J. Virol. 63:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kothari, S., J. Baillie, J. G. Sissons, and J. H. Sinclair. 1991. The 21-bp repeat element of the human cytomegalovirus major immediate early enhancer is a negative regulator of gene expression in undifferentiated cells. Nucleic Acids Res. 19:1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609–620. [DOI] [PubMed] [Google Scholar]
- 21.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 deleted fails to express early lytic genes. J. Virol. 75:1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier, J. L., and J. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, J. L., and M. F. Stinski. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier, J. L., and M. F. Stinski. 1996. Regulation of cytomegalovirus immediate early genes. Intervirology 39:331–342. [DOI] [PubMed] [Google Scholar]
- 26.Mesnard, J. M., and C. Devaux. 1999. Multiple control levels of cell proliferation by human T-cell leukemia virus type 1 TAX protein. Virology 257:277–284. [DOI] [PubMed] [Google Scholar]
- 27.Michelson, S., P. Turowski, L. Picard, J. Goris, M. P. Landini, A. Topliko, B. Hemmings, C. Bessia, A. Garcia, and J. L. Virelizier. 1996. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and host cell-derived PP2A. J. Virol. 70:1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski, E., and C. Tan Courcelle. 2001. Cytomegaloviruses and their replication, p 2629–2674. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Mocarski, E. J. 1988. Biology and replication of cytomegalovirus. Transfus. Med. Rev. 2:229–234. [DOI] [PubMed] [Google Scholar]
- 30.Mocarski, E. S., G. Kemble, J. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491 aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321–11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munishi, N., J. Yie, M. Merika, K. Senger, S. Lomvardas, T. Agalioti, and D. Thanos. 1999. The IFN-β enhancer: a paradigm for understanding activation and repression of inducible gene expression. Cold Spring Harbor Symp. Quant. Biol. 64:149–159. [DOI] [PubMed] [Google Scholar]
- 32.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642–652. [DOI] [PubMed] [Google Scholar]
- 33.Prosch, S., C. E. C. Wendt, P. Reinke, C. Priemer, M. Oppert, D. H. Kruger, H. D. Volk, and W. D. Docke. 2000. A novel link between stress and human cytomegalovirus (HCMV) infection: sympathetic hyperactivity stimulates HCMV activation. Virology 272:357–365. [DOI] [PubMed] [Google Scholar]
- 34.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanford, G. R., L. E. Brock, S. Voight, C. M. Forester, and W. H. Burns. 2001. Rat cytomegalovirus major immediate-early enhancer switching results in altered growth characteristics. J. Virol. 75:5076–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimotohono, K., M. Takano, T. Teruuchi, and M. Miwa. 1986. Requirement of multiple copies of a 21-nucleotide sequence in the U3 regions of human T-cell leukemia virus type I and type II long terminal repeats for trans-acting activation of transcription. Proc. Natl. Acad. Sci. USA 83:8112–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spaete, R. R., and E. S. Mocarski. 1985. Regulation of cytomegalovirus gene expression: α and β promoters are trans-activated by viral functions in permissive human fibroblasts. J. Virol. 56:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamminger, T., H. Fickenscher, and B. Fleckenstein. 1990. The cell type-specific induction of the major immediate-early enhancer of human cytomegalovirus by cAMP. J. Gen. Virol. 71:105–113. [DOI] [PubMed] [Google Scholar]
- 39.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans-activation and repression by human cytomegalovirus immediate-early proteins involve common and unique protein domains. J. Virol. 64:1556–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stinski, M. F. 1976. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J. Virol. 19:594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stinski, M. F., E. S. Mocarski, and D. R. Thomsen. 1979. DNA of human cytomegalovirus: size heterogeneity and defectiveness resulting from serial undiluted passage. J. Virol. 31:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stinski, M. F., and T. J. Roehr. 1985. Activation of the major immediate-early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J. Virol. 55:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeler, D. G., and E. Cooper. 2001. Depolarization strongly induces human cytomegalovirus major immediate-early promoter/enhancer activity in neurons. J. Biol. Chem. 276:31978–31985. [DOI] [PubMed] [Google Scholar]
- 44.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yurochko, A. D., E.-S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E.-S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of SP1 and NF-κB during infection. J. Virol. 71:5051–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]