Abstract
We selected 125 candidate single nucleotide polymorphisms (SNPs) in genes belonging to the human type 1 interferon (IFN) gene family and the genes coding for proteins in the main type 1 IFN signalling pathway by screening databases and by in silico comparison of DNA sequences. Using quantitative analysis of pooled DNA samples by solid-phase mini-sequencing, we found that only 20% of the candidate SNPs were polymorphic in the Finnish and Swedish populations. To allow more effective validation of candidate SNPs, we developed a four-colour microarray-based mini-sequencing assay for multiplex, quantitative allele frequency determination in pooled DNA samples. We used cyclic mini-sequencing reactions with primers carrying 5′-tag sequences, followed by capture of the products on microarrays by hybridisation to complementary tag oligonucleotides. Standard curves prepared from mixtures of known amounts of SNP alleles demonstrate the applicability of the system to quantitative analysis, and showed that for about half of the tested SNPs the limit of detection for the minority allele was below 5%. The microarray-based genotyping system established here is universally applicable for genotyping and quantification of any SNP, and the validated system for SNPs in type 1 IFN-related genes should find many applications in genetic studies of this important immunoregulatory pathway.
INTRODUCTION
Single nucleotide polymorphisms (SNPs) represent the most abundant form of genetic variation, and they occur on average at every 1–2 kb in the human genome (1). Over 4 million SNPs have been identified (http://www.ncbi.nlm.nih.gov/SNP/), and of these SNPs over 1.2 million have been mapped to the human genome (http://snp.cshl.org/genome.shtml). Due to the abundant repetitive sequences in the genome, all of the SNPs contained in the databases are not necessarily true polymorphisms, or they may not be polymorphic in a specific population of interest (2,3). With the increasing attention towards whole-genome linkage studies and the interest in defining the haplotype structures across the genome (4–6), validation of the large collections of SNPs is a challenging task that requires universally applicable and efficient genotyping methods. Quantitative analysis of pooled DNA samples is a useful approach for increasing the throughput when large numbers of SNPs should be validated. Methods based on single nucleotide primer extension, mini-sequencing, have proven to be particularly well suited for this purpose (7,8). In this study we developed a ‘tag’-microarray system (9) for multiplex quantitative analysis of SNPs in pooled DNA samples (10) using four-colour fluorescence detection.
The human type 1 interferon (IFN) gene family consists of 13 IFN-α genes, the IFN-β gene, the IFN-ω gene and 11 pseudogenes (11). The main signalling pathways for the type 1 IFNs include at least two receptor subunits, two protein-tyrosine kinases and four signal transducers and activators of transcription (12–14). The type 1 IFNs are cytokines mediating both antiviral and growth-inhibiting effects. Despite the fact that the type 1 IFNs are part of the first line defence against various forms of infections (15) and may have a crucial role in the pathogenesis of several autoimmune diseases (16,17), the genetic variation of the type 1 IFNs and the components of their signalling pathway is still poorly characterised.
Of 125 candidate polymorphisms in IFN genes and genes belonging to the IFN signalling pathway, we assembled a panel of 25 SNPs that were polymorphic in the Swedish and Finnish populations and established the microarray-based genotyping system for this panel of SNPs. The system allows accurate quantitative determination of allele frequencies ranging from 5 to 95% in pooled DNA samples. As the format of our method permits analysis of each panel of SNPs in 56 samples per microscope slide, it is also a robust tool for high-throughput genotyping of SNPs in individual samples.
MATERIALS AND METHODS
DNA samples
DNA was extracted from blood samples of Swedish volunteer blood donors using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI). The DNA concentrations were measured spectrophotometrically at 260 nm. Equal amounts of DNA from 150 female and 100 male donors, respectively, were combined into two pooled DNA samples. The Finnish pooled DNA sample originated from a batch of pooled leukocytes from about 180 donors (18).
SNPs
One hundred and twenty five candidate SNPs were identified in silico by performing BLAST sequence comparisons against the NCBI human EST and the HTGS databases (http://www.ncbi.nlm.nih.gov/). Redundant matches were filtered out (19), and the potential SNPs were primarily selected at sites where the sequence variation was observed in at least one other sequence and was surrounded by good quality sequence with similarity scores of ≥98% and sequence lengths ≥1000 bases. SNPs were also identified in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and in HGVbase (http://hgvbase.cgb.ki.se/). Some of the SNPs in dbSNP were found using the SNPper (http://bio.chip.org:8080/bio/), a web-based tool to search for known SNPs in public databases. The RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) was used for excluding SNPs located in repetitive elements or sequences. In cases for which PCR primers that align outside the repetitive parts could be designed, a candidate SNP was studied further.
Primer design
The PCR primers were designed with the Primer3 program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Most of the PCR primers were designed to have the bases A and C at their 3′ end, and all primers had universal 5′ sequences to unify the kinetics of multiplex PCR. These sequences also facilitated a second amplification with universal biotinylated primers. The mini-sequencing primers contained 5′ 20-bp ‘tag’ sequences (Supplementary Material, Supplement 1) that were from the Affymetrix GeneChip® Tag collection. The SNPs included in the microarray panel were typed with mini-sequencing primers for both DNA strands. To avoid major hairpin–loop formations, the tagged mini-sequencing primers were tested with the Cybergene AB primer design program (http://www.cybergene.se/primer.html). The primers were synthesised by Thermo Hybaid Interactiva GmbH, (Ulm, Germany) or Sigma Genosys (Cambridgeshire, UK). The mini-sequencing primers are listed in Supplement 1, and the PCR and mini-sequencing primers used for analysing the SNPs that were found to be polymorphic in the populations studied are listed in Supplement 2.
PCR
PCR amplifications for the allele frequency quantification on the microarrays were done in multiplex reactions with three primer pairs per reaction. Additional SNPs were amplified in individual reactions. The PCR conditions were 95°C for 10 min, then 40 cycles of 95°C for 1 min, 55°C (in some cases 53°C) for 1 min, 72°C for 1.5 min and a final extension at 72°C for 5 min using 10 ng of genomic DNA with 0.02 or 0.035 U/µl (for the multiplex PCRs) AmpliTaq Gold DNA polymerase (Perkin-Elmer, Branchburg, NJ) and 200 µM of dNTPs in 50 µl of 10 mM Tris–HCl, pH 8.3, 50 mM KCl and 0.001% (w/v) gelatine. The MgCl2 concentration was 1.5 or 4 mM (for the multiplex PCRs) and the primer concentrations were 0.3 or 0.4 µM. For analysis on the microarrays the PCR products were pooled to contain roughly equal amounts of each amplicon and 7 µl of the pooled product was used further.
Solid-phase mini-sequencing in microtitre plates
To test if the initial 125 candidate SNPs, chosen using in silico search and from public databases, were polymorphic, they were analysed in pooled DNA samples. The DNA region spanning the SNP was amplified by PCR with primers containing universal 5′ sequences, followed by a second PCR amplification of 1 µl of a 1/100 dilution of the first amplification product using a universal biotinylated primer. The PCR products were captured in streptavidin-coated microtitre plate wells, and the variable nucleotides of the SNPs were detected using solid-phase mini-sequencing with 3H-labelled dNTPs according to a standard protocol (18). The signal ratios measured for the two alleles of an SNP in the pooled sample were normalised with respect to the corresponding signal ratios in a heterozygous sample, in which the two alleles are present in equal amounts, whenever there was a known heterozygote available (10). Alternatively, the specific activities of the 3H-labelled dNTPs (A 69 Ci/mmol, C 52 Ci/mmol, G 33 Ci/mmol, T 127 Ci/mmol; Amersham Pharmacia Biotech, Uppsala, Sweden) were used for calculating the allelic ratios in the pooled DNA samples (markers IFNAR2-g and IRF5-b) (10).
Preparation of microarrays
Oligonucleotides complementary to the ‘tag’ sequence of the mini-sequencing primers modified with NH2-groups in their 3′ ends, and containing a 3′-spacer sequence of 15 T residues were coupled covalently to Motorola Activated Slides (previously denoted 3D-Link™-slides; Motorola, Northbrook, IL). The attachment protocol given by the manufacturer was used, with the only exception that the ‘anti-tag’ oligonucleotides were dissolved in 400 mM sodium carbonate buffer, pH 9.0 at a 25 µM concentration, as this buffer was found to give better attachment efficiency in preliminary experiments. The oligonucleotides were applied onto the slides by contact printing using a ProSys 5510A instrument (Cartesian Technologies Inc., Irvine, CA) with four Stealth Micro Spotting Pins (SMP3) (TeleChem International Inc., Sunnyvale, CA). The oligonucleotide spots were 125–150 µm in diameter, and the centre-to-centre distance between two adjacent spots was 220 µm. Each slide contained 56 subarrays in a conformation of 14 rows and four columns at the same spacing as the wells of a 384-well microtitre plate (see Fig. 1). The ‘anti-tag’ olignucleotides were printed as duplicate spots in each subarray A Cy3-labelled oligonucleotide was included as a spotting control and an extra tag sequence was included as a control hybridising to a TAMRA-labelled complementary oligonucleotide added to the reaction mixture before the capturing step. After printing, the arrays were kept in a chamber with 75% relative humidity for a time period between 4 and 72 h (20), after which the excess of amine-reactive groups were deactivated by keeping the slides for 15 min at 50°C in a solution containing 50 mM ethanolamine, 0.1 M Tris–HCl, pH 9.0 and 0.1% sodium dodecyl sulphate (SDS). The slides were washed three times; first with dH2O, then with a solution containing 600 mM NaCl, 60 mM sodium citrate, pH 7.0, and 0.1% SDS at 50°C for 15–60 min, and finally with dH2O. The slides were stored desiccated at 20°C until use.
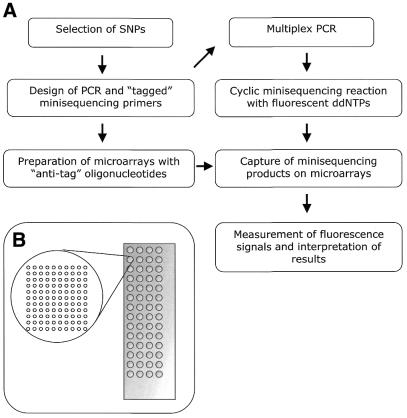
Figure 1.
(A) Steps of the genotyping procedure. (B) Schematic illustration of the ‘array of arrays’ format. Each microscope slide contains 56 subarrays in a 4 × 14 conformation, and each subarray carries ‘anti-tag’ oligonucleotides in a 10 × 11 spot conformation.
Cyclic mini-sequencing reactions
Pooled PCR products from each sample were treated with 0.5 U/µl Exonuclease I (USB Corporation, Cleveland, OH) and 0.1 U/µl shrimp alkaline phosphatase (USB Corporation) in 3.6 mM MgCl2, 50 mM Tris–HCl, pH 9.5, in a total volume of 10.5 µl. The mixture was heated to 37°C for 60 min, followed by inactivation of the enzymes at 99°C for 15 min. Each mini-sequencing reaction mixture contained 10.5 µl of PCR product treated with Exonuclease I and shrimp alkaline phosphatase, all the tagged mini-sequencing primers at a concentration of 2.5 nM each, 0.09 µM of each fluorescently-labelled ddNTP (Texas Red-ddATP 85 000 M–1 cm–1, TAMRA-ddCTP 91 000 M–1 cm–1, R110-ddGTP 78 000 M–1 cm–1, Cy5-ddUTP 250 000 M–1 cm–1, NEN™; Life Science Products, Brussels, Belgium), 0.017% Triton-X, 17 mM Tris–HCl, pH 9.5 and 0.07 U/µl of DynaSeq DNA polymerase (gift from Finnzymes OY, Helsinki, Finland) or ThermoSequenase (Amersham Pharmacia Biotech). Chemically synthesised single-stranded oligonucleotide templates were used at 1.5 nM concentration under identical reaction conditions. The total mini-sequencing reaction volume was 15 µl, and the cyclic reactions were performed in a Programmable Thermal Controller (MJ Tetrad Research) for 34 cycles of 95 and 55°C for 20 s each.
Capture by hybridisation
The array slides were pre-heated to 42°C in a custom-made aluminium reaction rack with a re-usable silicon rubber grid placed on the slides to form 56 separate reaction chambers on each slide (21). The hybridisation mixtures, containing 15 µl of mini-sequencing reaction product and a TAMRA-labelled hybridisation control oligonucleotide at 1 nM concentration in 23 µl of 900 mM NaCl, 90 mM sodium citrate, pH 7.0, were added to each reaction chamber on the microscope slide. Four control reactions without mini-sequencing reaction product were included on each slide. The hybridisation reaction time was 2.5–3 h at 42°C, after which the slides were briefly rinsed with 600 mM NaCl and 60 mM sodium citrate, pH 7.0, at 20–25°C. The slides were further washed twice for 5 min with 300 mM NaCl, 30 mM sodium citrate, pH 7.0, and 0.1% SDS that had been pre-heated to 42°C and twice for 1 min with 30 mM NaCl and 3 mM sodium citrate, pH 7.0, at 20–25°C. Finally, the slides were spin dried for 5 min at 140 g.
Signal detection and data interpretation
Fluorescence signals were measured with a ScanArray® 5000 instrument and the ScanArray® 3.1 software (Packard BioScience Ltd, UK) using the excitation lasers: Blue Argon, 488 nm; Green HeNe, 543.8 nm; Yellow HeNe, 594 nm; and Red HeNe, 632.8 nm. Due to the broad emission spectrum of the fluorophore TAMRA, the detector channel for Texas Red (emission maximum 612 nm) measures ∼10% of the signal from TAMRA (emission maximum 575 nm). The fluorescence signal intensities were quantified using the QuantArray® 3.0 software (ScanArray® 5000; Packard BioScience Ltd). The auto-balance/auto-range function of the ScanArray® 5000 instrument was used for normalisation of the signal intensities. The laser power was kept constant at 95%, whereas the photo-multiplier tube (PMT) gain varied between fluorophores and experiments. Typical settings for the PMT gain were 90, 80, 65 and 85% for the fluorophores Texas Red-ddA, TAMRA-ddC, R110-ddG and Cy5-ddU, respectively. The signal measured from each spot was corrected by subtracting the average background, measured from eight spots immediately below the primer spots, in each well. The genotypes for the individual SNPs were assigned using cluster analysis of the corrected signal intensities by a Microsoft Excel™ macro, using a cut-off value of 1000 fluorescence units for the sum of the signals for both alleles. Allele frequencies were determined from background- corrected signal intensity ratios after normalisation using a standard curve.
RESULTS
We selected 125 potential SNPs in the type 1 IFN genes and the genes coding for the proteins belonging to the main IFN signalling pathways by screening public databases and by in silico DNA sequence comparisons. The SNPs are denoted by acronyms that indicate the gene in which they are located (Supplement 1 and Table 1). The potential PCR products spanning the IFN regulatory factor 5 (IRF5-a) and IFN alpha (IFNA2-a, 4-d, -e, -f, 6-a, 7-a, -b) SNPs were found to align to multiple locations by BLAST analysis to the human working draft sequence (January 2001). Sixteen of the 125 candidate SNPs were located in an Alu repeat, mammalian-wide interspersed repeat (MIR), long interspersed element 1 (L1) or medium reiteration frequency 1 (MER1) repeat sequences, and the design of unique PCR assays was possible for eight of them. The design of primers for PCR amplification and genotyping by ‘mini-sequencing’ primer extension was successful for 101 of the 125 candidate SNPs.
Table 1. Allele frequencies of candidate SNPs in type 1 IFN-related genes determined by analysis of pooled DNA samples.
| SNPa,b |
Sequence variation |
Frequency of allele1c |
Frequency of allele1d |
|||
|---|---|---|---|---|---|---|
| Allele1/allele2 | Swedes | Finns | African Americans | Asians | Caucasians | |
| STAT1-b | C/T | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| STAT1-c | A/G | 1.00 | 1.00 | 0.78 | 0.92 | 0.95 |
| STAT1-d | C/G | 1.00 | 1.00 | 0.95 | 0.95 | 1.00 |
| STAT1-e | A/C | 0.88 | 0.90 | 0.71 | 0.84 | 0.96 |
| STAT1-f | C/G | 0.35 | 0.39 | n.a. | 0.93 | n.a. |
| STAT1-g | A/G | 0 | 0 | 0 | 0 | 0 |
| STAT1-h | T/C | 0.19 | 0.15 | 0.21 | n.a. | 0.25 |
| STAT1-i | A/G | 0.06 | 0.03 | 0 | 0.05 | 0 |
| STAT1-j | C/T | 0.52 | 0.53 | 1.00 | 1.00 | 1.00 |
| STAT3-a | C/A | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| STAT3-4 | G/A | 0 | 0 | 0 | 0 | 0 |
| STAT3-5 | A/G | 0 | 0 | 0 | 0 | 0 |
| STAT3-6 | A/G | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| STAT3-7 | G/A | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| STAT3-8 | T/A | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| STAT3-10 | C/G | n.a. | n.a. | 0.92 | n.a. | n.a. |
| STAT3-11 | G/C | n.a. | n.a. | 0 | n.a. | n.a. |
| STAT3-13 | A/G | 0.48 | 0.48 | 0.35 | 0.60 | n.a. |
| STAT3-14 | A/G | 0.38 | 0.38 | 0.28 | 0.56 | n.a. |
| IFNAR1-a | G/T | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| IFNAR1-e | C/G | 0.86 | 0.82 | n.a. | n.a. | n.a. |
| IFNAR1-t | G/T | 0.82 | 0.84 | 0.85 | 1.00 | 0.70 |
| IFNAR-1u | A/G | 0.82 | 0.80 | n.a. | n.a. | n.a. |
| IFNAR1-v | G/T | 0.60 | 0.67 | n.a. | n.a. | n.a. |
| IFNAR1-w | A/G | 0.18 | 0.16 | 0.13 | 0.26 | 0.16 |
| IFNAR2-g | G/T | 0.22 | 0.11 | n.a. | n.a. | n.a. |
| IFNAR2-k | G/T | 0.36 | 0.45 | 0.19 | 0.64 | 0.32 |
| IFNAR2-m | T/C | 0.34 | 0.34 | n.a. | n.a. | n.a. |
| IFI27 | C/G | 0.95 | 0.89 | 0.84 | 0.62 | 0.96 |
| IRF5-b | A/G | 0.48 | 0.60 | n.a. | n.a. | n.a. |
| IRF5-e | G/T | 0.84 | 0.83 | n.a. | n.a. | n.a. |
| IFNA21-a | G/A | 0.27 | 0.28 | 0.17 | 0.39 | 0.12 |
| IFNA21-b | G/A | 0.55 | 0.57 | 0.97 | 1.00 | 0.93 |
| IFNA21-d | G/A | 1.00 | 1.00 | 0.94e | 0.96e | 0.90e |
| JAK1-d | A/G | 0.77 | 0.75 | n.a. | n.a. | n.a. |
| TYK2-g | T/G | 0.44 | 0.30 | n.a. | n.a. | n.a. |
| TYK2-h | G/T | 0.16 | 0.10 | n.a. | n.a. | n.a. |
| TYK2-m | T/C | 0.74 | 0.74 | n.a. | P | P |
| TYK2-n | C/T | 0.88 | 0.91 | n.a. | 1.00 | 1.00 |
aThe SNPs included in the microarray panel are shown in bold.
bIFNAR, the IFN alpha, beta and omega receptor; JAK 1, Janus kinase 1; TYK 2, tyrosine kinase 2; STAT, signal transducer and activator of transcription; IRF, interferon regulatory factor; IFNA, interferon alpha; IFI27, interferon alpha-inducible protein 27.
cDetermined by solid-phase mini-sequencing in the current study. The average of the frequencies determined from a pool of Swedish women (n = 150) and a pool of Swedish men (n = 100) is given. Duplicate assays were performed for all SNPs.
dEstimated by R. Miller and P.-Y. Kwok (Washington University, St Louis, MO).
eFrequency of SNP in a repeat sequence according to the Kwok Laboratory; n.a., not available; P, polymorphic according to the Kwok Laboratory. The SNPs verified by the Kwok Laboratory have been included in dbSNP.
To assess if these candidate SNPs are polymorphic, and to determine their allele frequencies in the Swedish and Finnish populations, three pooled DNA samples containing equal amounts of DNA from 150 Swedish women, 100 Swedish men and 180 Finnish individuals, respectively, were analysed by solid-phase mini-sequencing in a microtitre plate format (10,18). This analysis revealed that only 25 of the candidate SNPs (20%) are polymorphic in the Finnish and Swedish populations as defined by allele frequencies >1% for the minor allele (Table 1). The allele frequencies for each SNP were identical for the Swedish women and men. The frequency of three SNPs (IFNAR2-g, IFNAR2-k and TYK2-g) differed from each other in the Swedish and Finnish populations by 10–15% (P < 0.05). A subset of 29 of the 125 SNPs was also validated by sequencing pooled samples with DNA from 42 individuals representing African Americans, Asians (10 Chinese, 32 Japanese) and Caucasians by the Kwok Laboratory at Washington University, St Louis, MO (3,22). For most of the SNPs, the polymorphic status was the same in these populations as in the Nordic populations, with the exception of two SNPs (STAT1-f and STAT1-j) that seemed to be specifically Nordic polymorphisms, and one SNP (IFNAR21-b) that was much more frequent in the Nordic populations (Table 1 and Supplement 1). The low yield of true polymorphic SNPs in the databases emphasises the requirement of efficient methods for verifying and genotyping SNPs. This need prompted us to develop a system for multiplex validations of SNPs by mini-sequencing in a microarray format to serve as a tool for multiplex quantitative analysis of SNPs in pooled DNA samples.
The 25 identified SNPs in candidate genes belonging to the IFN signalling and regulatory pathways (Table 1) were included in a panel for multiplex genotyping in a microarray format (Fig. 1). The genotyping system is based on cyclic mini-sequencing reactions in solution using primers with a 5′-tag sequence in the presence of dideoxynucleotides labelled with the four fluorescent dyes Texas Red, TAMRA, R110 and Cy5. The multiplex, fluorescent reaction products are captured and sorted on microarrays carrying tag sequences complementary to the 5′ part of the mini-sequencing primers. The tag oligonucleotides are positioned in an ‘array of arrays’ conformation (Fig. 1B) on the microscope slides to facilitate the genotyping of 56 samples for both strands of the 25 SNPs in duplicate on each microscope slide. Figure 2 shows the four fluorescence images of one row of ‘subarrays’ in which four individual DNA samples have been analysed.
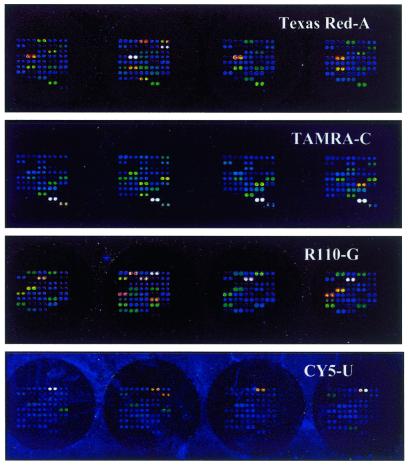
Figure 2.
Image at four wavelengths of a row of four ‘subarrays’ on an oligonucleotide array. Four samples have been genotyped for 25 IFN-related SNPs with primers for both DNA strands spotted in duplicate. The respective laser power and PMT gain settings of the array scanner were 90 and 75% for Texas Red-ddATP and R110-ddGTP, 100 and 75% for TAMRA-ddCTP and 100 and 80% for Cy5-ddUTP.
The DNA polymerase used for the mini-sequencing reactions has been engineered to efficiently incorporate dideoxy nucleotide analogues (23), but the incorporation efficiency varies between the four nucleotides, and is dependent on the sequence context of the SNP. Using synthetic oligonucleotides with identical sequences flanking one variable base as templates we compared the efficiency of incorporation of the four ddNTPs labelled with the same fluorophore (TAMRA). We found that TAMRA-ddCTP is most efficiency incorporated followed by ddATP, ddGTP and ddUTP (Table 2). Moreover, the four fluorophores exhibit different fluorescent properties (molar extinction coefficients, emission spectra and quantum yields), which are reflected as differences in signal intensities when measured in the array scanner. Table 2 illustrates that the relative fluorescence intensities between the fluorophores differ when they are measured directly, and after the four-colour mini-sequencing reactions using the synthetic template. Comparison of these fluorescence intensities indirectly shows that the efficiency of the DNA polymerase to incorporate a nucleotide is affected by the fluorophore attached to it. The differences in measured signal intensity between the fluorophores after mini-sequencing reactions can be partly compensated for by adjusting the laser power and PMT gain of the array scanner to avoid saturation of the signals or signals falling below the detection threshold. Most importantly, the sequence flanking an SNP affects the efficiency of nucleotide incorporation. The efficiency of incorporation as well as possible mis-incorporation of labelled ddNTP are unpredictable and thus difficult to correct for. Figure 3 shows two examples of cluster analysis obtained when 75 individual samples were genotyped for two SNPs, IFNAR1-w with an A to G transition, and STAT1-f with a C to G transversion. Due to the combined effect of the factors described above, the positions along the x-axis of the clusters corresponding to the heterozygous genotypes differ between the two SNPs, but the clear difference in ratios between the three clusters allows unequivocal genotype assignment for both SNPs.
Table 2. Relative signal intensities of the fluorophores and relative incorporation efficiencies of the fluorophores during mini-sequencing reactions in solution.
| |
Relative signal intensities |
|||
|---|---|---|---|---|
| A | C | G | U | |
| TAMRA ddNTPsa | 0.5 | 1.0 | 0.4 | 0.2 |
| Fluorophores onlyb | 0.5 | 1.0 | 1.0 | 0.7 |
| Four fluorophore ddNTPsc | 0.3 | 0.4 | 1.0 | 0.4 |
aRelative incorporation efficiency of the four nucleotides labelled with the TAMRA fluorophore during the mini-sequencing reaction.
bRelative signal intensities from equal amounts of ddNTP labelled with the four different fluorophores: Texas Red-A, TAMRA-C, R110-G, Cy5-U.
cRelative signal intensities measured from the four nucleotides labelled with the four different fluorophores during mini-sequencing reactions.
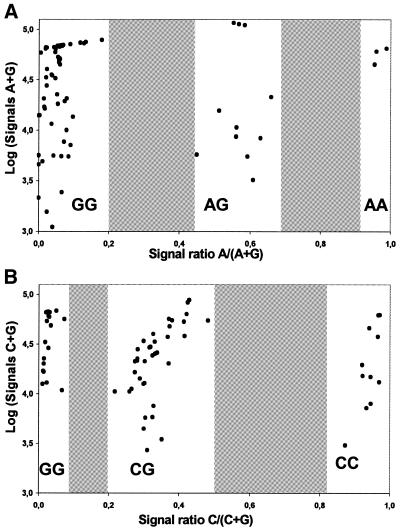
Figure 3.
Cluster analysis of the results from typing the SNPs IFNAR-w (A) and STAT1-f (B) in 75 samples using the IFN oligonucleotide array system. The values on the y-axes are logarithms of the sum of background corrected signals for both alleles. The allele ratios on the x-axes are calculated from the background corrected signal intensities for both alleles. The genotype frequencies conform to Hardy–Weinberg equilibrium.
The power of the microarray-based system for multiplex quantitative determination of allele frequencies in pooled DNA samples was evaluated. Quantification standard curves were prepared for nine SNPs by mixing DNA of different genotypes in ratios ranging from 100% of one allele to 100% of the other allele at 5 or 10% increments. The mixed samples were analysed by the four-colour fluorescence mini- sequencing method. Figure 4 shows the regression lines obtained when the measured fluorescence signal ratios were plotted as a function of the corresponding allelic ratios in the mixed samples. Those with correlation coefficients between 0.95 and 0.99 show that the allelic ratios determined by four-colour mini-sequencing are proportional to the original allelic ratios in the mixed samples. The detection sensitivity for the minority allele varied between the SNPs, owing to differences in the fluorescence intensities and incorporation efficiency for the fluorescent nucleotides. For half of the alleles included, the sensitivity of detecting a minority allele was below 5%, and for several alleles it was below 2%. The standard deviations (SDs) were calculated based on four parallel reactions and varied between the SNPs from 1 to 10%. Given this encouraging result, the allele frequencies for the nine SNPs were determined by analysing the pooled DNA samples representing the Swedish and Finnish populations. The allele frequencies were determined by comparing the fluorescence ratios obtained in the pooled samples with standard curves. The allele frequencies could also be determined by normalisation against a heterozygous sample, as was done for the individual SNPs. The allele frequencies determined by four-colour fluorescence mini-sequencing (Table 3) are concordant with those obtained by solid-phase mini-sequencing of individual SNPs using 3H-labelled dNTPs (Table 1), providing evidence for the accuracy of the four-colour multiplexed mini-sequencing method.
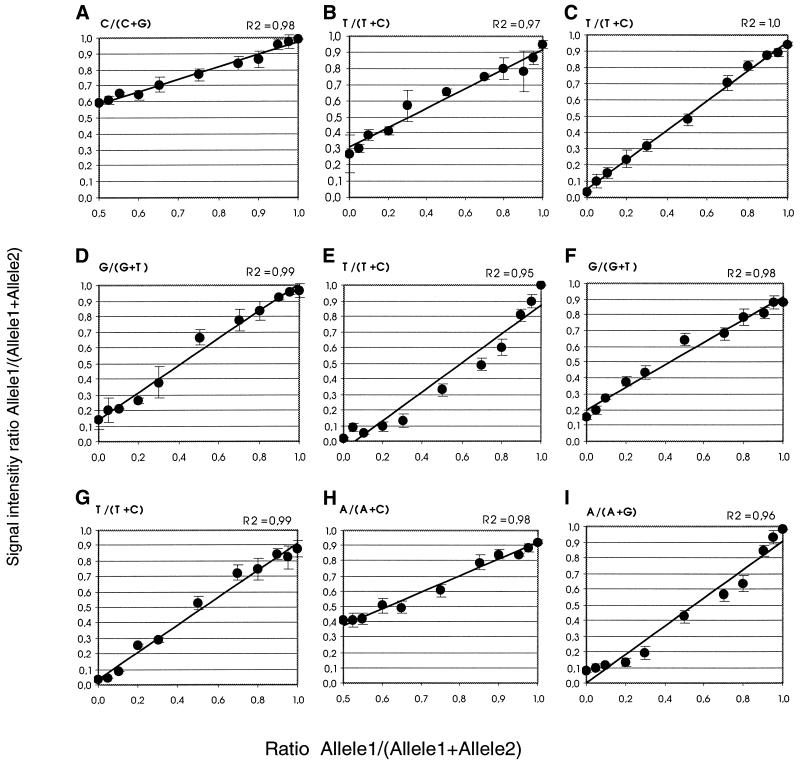
Figure 4.
Standard curves for nine SNPs. The SNPs are: (A) IFI27, (B) Jak1-d, (C) IFNAR1-e, (D) IFNAR1-v, (E) IFNAR1-w, (F) IFNAR2-k, (G) IFNAR2-m, (H) STAT1-e, (I) STAT3-14. Samples homozygous for both alleles of each SNP were mixed in appropriate ratios to obtain the allele ratios given on the x-axes. The correlation coefficient (R2) for the curves is given in each panel. The SDs indicated by vertical bars in the curves are calculated from four reactions performed in separate ‘subarrays’. No homozygotes to prepare mixtures of allelic ratios below 0.5 were available for the curves (A) and (H).
Table 3. Allele frequencies of polymorphic SNPs determined by multiplex, four-colour mini-sequencing in the microarray format.
| SNP | Variation | Average signalsa | Frequency of allele1b | |
|---|---|---|---|---|
| Allele1/allele2 | Allele1 | Allele2 | ||
| STAT1-e | A/C | |||
| Swedish pool | 5162 | 1591 | 0.87 ± 0.04 | |
| Finnish pool | 3997 | 961 | 0.91 ± 0.04 | |
| STAT3-14 | A/G | |||
| Swedish pool | 750 | 2149 | 0.24 ± 0.05 | |
| Finnish pool | 433 | 1037 | 0.30 ± 0.07 | |
| IFNAR1-e | C/G | |||
| Swedish pool | 8623 | 3591 | 0.74 ± 0.02 | |
| Finnish pool | 13 452 | 4525 | 0.77 ± 0.02 | |
| IFNAR1-v | G/T | |||
| Swedish pool | 638 | 391 | 0.60 ± 0.04 | |
| Finnish pool | 959 | 449 | 0.73 ± 0.04 | |
| IFNAR1-w | A/G | |||
| Swedish pool | 29 115 | 20 287 | 0.24 ± 0.05 | |
| Finnish pool | 23 435 | 19 580 | 0.18 ± 0.03 | |
| IFNAR2-k | G/T | |||
| Swedish pool | 5000 | 4969 | 0.34 ± 0.06 | |
| Finnish pool | 3281 | 4025 | 0.55 ± 0.06 | |
| IFNAR2-m | T/C | |||
| Swedish pool | 3886 | 23 425 | 0.26 ± 0.02 | |
| Finnish pool | 2556 | 14 465 | 0.28 ± 0.03 | |
| IFI27 | C/G | |||
| Swedish pool | 5031 | 294 | 0.93 ± 0.02 | |
| Finnish pool | 3041 | 324 | 0.89 ± 0.03 | |
| JAK1-d | T/C | |||
| Swedish pool | 17 729 | 4281 | 0.74 ± 0.06 | |
| Finnish pool | 7603 | 2134 | 0.73 ± 0.01 | |
aAverage values and SDs for four parallel reactions.
bDetermined from the standard curves in Figure 4.
DISCUSSION
In the present study we developed a system that multiplexes SNP analysis in two dimensions. First, the assay format facilitates genotyping of up to 100 SNPs in 56 samples per microscope slide using four-colour fluorescence detection and, secondly, the system allows quantitative analysis of SNPs in pooled samples containing DNA from hundreds of individuals (10). The flexible strategy for SNP genotyping based on cyclic mini-sequencing reactions in solution with 5′-tagged primers used here as a tool for quantification was first described for genotyping individual SNPs in a format based on microspheres detectable by flow cytometry (24,25). The tag-array concept has previously been combined with high-density GeneChips (Affymetrix) that use a double labelling with Cy5 and indirect detection of biotin with phycoerythrin-labelled streptavidin (9) and with low density microarrays using nucleotides labelled with three fluorophores (26). A system with four-colour detection has the advantage that it allows multiplex detection of both DNA strands of all possible SNPs in the same reaction (27). For quantitative application the use of four fluorophores in the same reaction is particularly advantageous because it eliminates variation between signals due to possible spot to spot variation originating from array printing, which would otherwise hamper the quantitative analysis on the microarrays.
The accuracy and sensitivity of the quantitative analysis is affected by the accuracy of the single nucleotide incorporation catalysed by the DNA polymerase. Using fluorescent ddNTPs and the ThermoSequenase DNA polymerase we were able to detect as little as 2% of the minority allele for some of the SNPs. Our original mini-sequencing method uses 3H-labelled dNTPs that are chemically similar to the natural dNTPs in combination with Taq DNA polymerase, and allows detection of as little as 1% of the minority allele for most SNPs (7,10). Other primer extension methods that should allow accurate and sensitive determination of allelic ratios for SNPs are pyrosequencing that uses unlabelled dNTPs followed by luminescence detection of released pyrophosphate (28) and assays with mass spectrometric detection of unlabelled dNTPs and ddNTPs (8,29). These methods cannot, however, be multiplexed for more than a few SNPs. A ligation assay based on a ‘zip-code’ strategy allowed detection of <1% of a single SNP allelic variant in a preliminary study (30), and could in principle be developed further towards a multiplexed microarray format. The quantitative performance of the four-colour mini-sequencing tag-array system established in our study compares favourably with detecting minority alleles by extension of immobilised allele-specific primers using a reverse transcriptase (21).
There is an evident lack of studies applying the generic tag-microarray format to high-throughput genotyping of SNPs. One obvious reason for this, shared by all current SNP-genotyping methods, is that the throughput of the methods is limited by the requirement of performing multiplex PCR amplification of the region spanning each SNP prior to genotyping. Another reason for the lack of high-throughput applications is that the microarray formats, in which one microscope slide is used for each sample, were originally designed for expression profiling of large numbers of RNA species in relatively few samples (9,26). This is impractical for SNP analysis where typically many DNA samples are analysed. The ‘array of arrays’ conformation that we have developed allows analysis of up to 80 individual samples per microscope slide and thus removes the latter limitation (21,31). The possibility of multiplex quantification of minority allelic variants of SNPs implies that the four-colour mini-sequencing system also performs robustly for high-throughput genotyping of multiplex SNPs in individual samples. Compared to mini-sequencing with specific primers immobilised on microarrays (20,32), or arrayed primer extension (27,33), the tag-array approach, in which the primer extension reactions are performed in solution, is less prone to false positive signals due to template independent extension of the primers. An additional advantage is that the production of the microarrays with generic anti-tag sequences is more convenient as the same arrays can be used for multiple applications.
Identification of SNPs in the type 1 IFN gene family and design of assays for potential SNPs in these genes proved to be a difficult task. The IFN type 1 gene family has a complex genomic organisation with 15 intronless genes and 11 pseudogenes mapping to the short arm of chromosome 9. It is likely that the complexity of the type 1 IFN gene family has led to the inclusion of an unusually large number of false SNPs in the databases. In our study, we identified altogether 23 potential SNPs in seven type 1 IFN genes through database searches, but out of these 23 candidate SNPs only two were found to be real polymorphisms in our populations. The draft sequence of the human genome published in February 2001 revealed that at least 50% of the sequence is comprised of repetitive sequences (34). The high number of repetitive elements and paralogous sequences complicate the identification of informative SNPs and the design of assays for them. In our study, 16 potential SNPs were located in repeated elements, and only one of them turned out to be a true polymorphism. As the draft sequence of the human genome is constantly being updated, the information on SNPs contained in the databases changes frequently as new data is gained.
Due to the important immunoregulatory functions of the IFN family and the IFN signalling pathway, the panel of SNPs for these specific genes that we have validated in the current study should find a variety of applications in genetic studies of cancer, autoimmune disorders and viral infections. A system for multiplex SNP validation is particularly useful for complex genes such as those of the type 1 IFN gene family for which the databases contain incomplete information. In the system established here for SNPs in type 1 IFN genes and genes of the IFN signalling pathway, 100 genotypes are generated per subarray, and each microscope slide holds 56 of these ‘subarrays’, which translates into 5600 genotypes per microscope slide. When pooled samples with DNA from 100 to 200 individuals are analysed, data corresponding to 0.5–1.0 million SNP alleles is extracted from a single microscope slide. The four-colour mini-sequencing system described here represents a significant improvement over existing microarray-based methods for SNP genotyping and is universally applicable to any SNP. Its multiplexed format opens prospects for performing high-throughput association studies with large numbers of SNP markers using multiple pooled samples from individuals with different phenotypic characteristics.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
Pui-Yan Kwok and Raymond D. Miller are thanked for analysing a subset of our SNPs in the African Americans, Asians and Caucasians. Raul Figueroa, Kristina Larsson and David Olsson are acknowledged for their excellent technical assistance. We thank one of the referees for his valuable suggestions for improvements to the manuscript. The study was made possible by grants from the Swedish Research Council, the Knut & Alice Wallenberg Foundation (WCN), Orchid BioSciences, Ltd, and the European Commission (Contract QLRT-2001-0004) (to A.-C. S), and the Swedish Rheumatism Foundation and the 80 years Foundation of King Gustaf V (to L.R.).
REFERENCES
- 1.Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 2.Douabin-Gicquel V., Soriano,N., Ferran,H., Wojcik,F., Palierne,E., Tamim,S., Jovelin,T., McKie,A.T., Le Gall,J.Y., David,V. et al. (2001) Identification of 96 single nucleotide polymorphisms in eight genes involved in iron metabolism: efficiency of bioinformatic extraction compared with a systematic sequencing approach. Hum. Genet., 109, 393–401. [DOI] [PubMed] [Google Scholar]
- 3.Marth G., Yeh,R., Minton,M., Donaldson,R., Li,Q., Duan,S., Davenport,R., Miller,R.D. and Kwok,P.Y. (2001) Single-nucleotide polymorphisms in the public domain: how useful are they? Nature Genet., 27, 371–372. [DOI] [PubMed] [Google Scholar]
- 4.Kruglyak L., (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet., 22, 139–144. [DOI] [PubMed] [Google Scholar]
- 5.Hoh J., Wille,A. and Ott,J. (2001) Trimming, weighting and grouping SNPs in human case–control association studies. Genome Res., 11, 2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,D.H., Marjoribanks,C., McDonough,D.P. et al. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science, 294, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 7.Syvanen A.C., (1999) From gels to chips: ‘minisequencing’ primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 8.Buetow K.H., Edmonson,M., MacDonald,R., Clifford,R., Yip,P., Kelley,J., Little,D.P., Strausberg,R., Koester,H., Cantor,C.R. et al. (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA, 98, 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J.B., Chen,X., Halushka,M.K., Berno,A., Huang,X., Ryder,T., Lipshutz,R.J., Lockhart,D.J. and Chakravarti,A. (2000) Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res., 10, 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsson C., Waldenstrom,E., Westermark,K., Landegren,U. and Syvanen,A.C. (2000) Determination of the frequencies of ten allelic variants of the Wilson disease gene (ATP7B), in pooled DNA samples. Eur. J. Hum. Genet., 8, 933–938. [DOI] [PubMed] [Google Scholar]
- 11.Strissel P.L., Dann,H.A., Pomykala,H.M., Diaz,M.O., Rowley,J.D. and Olopade,O.I. (1998) Scaffold-associated regions in the human type I interferon gene cluster on the short arm of chromosome 9. Genomics, 47, 217–229. [DOI] [PubMed] [Google Scholar]
- 12.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 13.Su L., and David,M. (2000) Distinct mechanisms of STAT phosphorylation via the interferon-alpha/beta receptor. Selective inhibition of STAT3 and STAT5 by piceatannol. J. Biol. Chem., 275, 12661–12666. [DOI] [PubMed] [Google Scholar]
- 14.Gauzzi M.C., Barbieri,G., Richter,M.F., Uze,G., Ling,L., Fellous,M. and Pellegrini,S. (1997) The amino-terminal region of Tyk2 sustains the level of interferon alpha receptor 1, a component of the interferon alpha/beta receptor. Proc. Natl Acad. Sci. USA, 94, 11839–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdan C., (2000) The function of type I interferons in antimicrobial immunity. Curr. Opin. Immunol., 12, 419–424. [DOI] [PubMed] [Google Scholar]
- 16.Ronnblom L., and Alm,G. (2001) A pivotal role for the natural interferon alpha producing cells (plasmacytoid dendritic cells) in the pathogenesis of lupus. J. Exp. Med., 194, F1–F6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronnblom L., and Alm,G.V. (2001) An etiopathogenic role for the type I IFN system in SLE. Trends Immunol., 22, 427–431. [DOI] [PubMed] [Google Scholar]
- 18.Syvanen A.C., Sajantila,A. and Lukka,M. (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am. J. Hum. Genet., 52, 46–59. [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnhammer E.L., and Durbin,R. (1994) A workbench for large-scale sequence homology analysis. Comput. Appl. Biosci., 10, 301–307. [DOI] [PubMed] [Google Scholar]
- 20.Lindroos K., Liljedahl,U., Raitio,M. and Syvanen,A.C. (2001) Minisequencing on oligonucleotide microarrays: comparison of immobilisation chemistries. Nucleic Acids Res., 29, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastinen T., Raitio,M., Lindroos,K., Tainola,P., Peltonen,L. and Syvanen,A.C. (2000) A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res., 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marth G.T., Korf,I., Yandell,M.D., Yeh,R.T., Gu,Z., Zakeri,H., Stitziel,N.O., Hillier,L., Kwok,P.Y. and Gish,W.R. (1999) A general approach to single-nucleotide polymorphism discovery. Nature Genet., 23, 452–456. [DOI] [PubMed] [Google Scholar]
- 23.Tabor S., and Richardson,C.C. (1995) A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA, 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai H., White,P.S., Torney,D., Deshpande,A., Wang,Z., Keller,R.A., Marrone,B. and Nolan,J.P. (2000) Flow cytometry-based minisequencing: a new platform for high-throughput single-nucleotide polymorphism scoring. Genomics, 66, 135–143. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Iannone,M.A., Li,M.S., Taylor,J.D., Rivers,P., Nelsen,A.J., Slentz-Kesler,K.A., Roses,A. and Weiner,M.P. (2000) A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res., 10, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschhorn J.N., Sklar,P., Lindblad-Toh,K., Lim,Y.M., Ruiz-Gutierrez,M., Bolk,S., Langhorst,B., Schaffner,S., Winchester,E. and Lander,E.S. (2000) SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc. Natl Acad. Sci. USA, 97, 12164–12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurg A., Tonisson,N., Georgiou,I., Shumaker,J., Tollett,J. and Metspalu,A. (2000) Arrayed primer extension: solid-phase four-color DNA resequencing and mutation detection technology. Genet. Test., 4, 1–7. [DOI] [PubMed] [Google Scholar]
- 28.Alderborn A., Kristofferson,A. and Hammerling,U. (2000) Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res., 10, 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauer S., Lechner,D., Berlin,K., Lehrach,H., Escary,J.L., Fox,N. and Gut,I.G. (2000) A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res., 28, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerry N.P., Witowski,N.E., Day,J., Hammer,R.P., Barany,G. and Barany,F. (1999) Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol., 292, 251–262. [DOI] [PubMed] [Google Scholar]
- 31.Pastinen T., Perola,M., Ignatius,J., Sabatti,C., Tainola,P., Levander,M., Syvanen,A.C. and Peltonen,L. (2001) Dissecting a population genome for targeted screening of disease mutations. Hum. Mol. Genet., 10, 2961–2972. [DOI] [PubMed] [Google Scholar]
- 32.Pastinen T., Kurg,A., Metspalu,A., Peltonen,L. and Syvanen,A.C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 33.Tonisson N., Zernant,J., Kurg,A., Pavel,H., Slavin,G., Roomere,H., Meiel,A., Hainaut,P. and Metspalu,A. (2002) Evaluating the arrayed primer extension resequencing assay of TP53 tumor suppressor gene. Proc. Natl Acad. Sci. USA, 99, 5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.