Abstract
An important consideration in the design of oligonucleotide probes for homogeneous hybridization assays is the efficiency of energy transfer between the fluorophore and quencher used to label the probes. We have determined the efficiency of energy transfer for a large number of combinations of commonly used fluorophores and quenchers. We have also measured the quenching effect of nucleotides on the fluorescence of each fluorophore. Quenching efficiencies were measured for both the resonance energy transfer and the static modes of quenching. We found that, in addition to their photochemical characteristics, the tendency of the fluorophore and the quencher to bind to each other has a strong influence on quenching efficiency. The availability of these measurements should facilitate the design of oligonucleotide probes that contain interactive fluorophores and quenchers, including competitive hybridization probes, adjacent probes, TaqMan probes and molecular beacons.
INTRODUCTION
The ability of fluorescent compounds to transfer energy absorbed from light to nearby molecules has been exploited for the development of homogeneous methods of nucleic acid detection (1–5). In these assays, it is not necessary to isolate the probe–target hybrids from the non-hybridized probes to detect the presence of target nucleic acids. Instead, alterations in the emission of fluorescent labels that are attached to the probes are measured to determine the number of target strands that are present. These techniques are being used to monitor the progress of nucleic acid amplification reactions in real time in sealed reaction tubes, resulting in more accurate quantification of nucleic acid targets (6–9). These methods are also useful for imaging gene expression in living cells (10,11).
In homogeneous hybridization assays, a pair of interactive fluorophores is attached to the ends of two different oligodeoxyribonucleotide probes or to the two ends of the same oligodeoxyribonucleotide probe. A target nucleic acid reveals itself by either bringing the donor fluorophore and the acceptor fluorophore close to each other, permitting energy transfer between them to occur, or by separating them from each other, precluding the transfer of energy. The earliest formats for homogeneous hybridization assays utilized a pair of oligodeoxyribonucleotide probes labeled at their respective 5′ and 3′ ends that were designed to bind to adjacent sites on a target strand, thereby bringing a donor and acceptor moiety close to each other (1,2). A second approach utilizes a pair of mutually complementary oligodeoxyribonucleotides, in which one of the oligodeoxyribonucleotides serves as a probe for a single-stranded target sequence. The 5′ end of one oligodeoxyribonucleotide is labeled with a donor fluorophore and the 3′ end of the other oligodeoxyribonucleotide is labeled with an acceptor fluorophore, such that when the two oligodeoxyribonucleotides are annealed to each other, the two labels are close to one another. Since small complementary oligodeoxyribonucleotides bind to each other in a dynamic equilibrium, target strands compete for binding to the probe, causing the separation of the labeled oligodeoxyribonucleotides (3). In a third approach, the donor and acceptor fluorophores are attached to the ends of the same oligodeoxyribonucleotide, which serves as the probe. Since an oligodeoxyribonucleotide in solution behaves like a random coil, its ends occasionally come close to one another, resulting in a measurable change in energy transfer. However, when the probe binds to its target, the rigidity of the probe–target helix keeps the two ends of the probe apart from each other, precluding interaction between the donor and the acceptor moieties (12,13). In the fourth approach, single-stranded oligodeoxyribonucleotides called molecular beacons possess short additional sequences at either end of a probe sequence that are complementary to one another, enabling terminal labels to be in close proximity through the formation of a hairpin stem. Binding of this probe to its target creates a relatively rigid probe–target hybrid that causes the disruption of the hairpin stem and the removal of the donor moiety from the vicinity of the acceptor moiety, thus restoring the fluorescence of the donor (5,8). In addition to these hybridization-based schemes and their variations, dual-labeled randomly coiled probes that bind to template strands during PCR, can be enzymatically cleaved by the 5′→3′ endonuclease activity of DNA polymerase (TaqMan probes), separating the donor and acceptor moieties and enabling nucleic acid synthesis to be monitored in real time (6). Furthermore, oligodeoxyribonucleotides labeled with a pair of interactive fluorophores have proven to be useful in real-time monitoring of a number of other reactions involving nucleic acids (14–17).
If an acceptor fluorophore is brought closer to a donor fluorophore within the range 20–100 Å, the intensity of the fluorescence of the acceptor fluorophore increases, whereas the intensity of the fluorescence of the donor fluorophore decreases. This is due to an increase in the efficiency of fluorescence resonance energy transfer (FRET) from the donor to the acceptor fluorophore. However, if the two moieties are brought any closer, the intensity of the fluorescence of both the donor fluorophore and the acceptor fluorophore is reduced. At these intimate distances, most of the absorbed energy is dissipated as heat and only a small amount of energy is emitted as light, a phenomenon sometimes referred to as static or contact quenching (8,18,19). In adjacent probes and in randomly coiled probes, the donor and the acceptor moieties remain at such a distance from each other that FRET is the predominant mechanism of quenching. On the other hand, in competitive hybridization probes and in molecular beacons, when they are not hybridized to targets, the two moieties are very close to each other and contact quenching is the predominant mechanism of quenching. One of the useful features of contact quenching is that all fluorophores are quenched equally well, irrespective of whether the emission spectrum of the fluorophore overlaps the absorption spectrum of the quencher, one of the key conditions that determines the efficiency of FRET (8).
A further simplification of homogeneous assays that utilize fluorescently labeled probes is the use of non-fluorescent dyes as acceptors or quenchers (5,20). Quenching by non- fluorescent dyes enables changes in the intensity of fluorescence to be measured directly, rather than as an alteration in the shape of the emission spectrum, which is more difficult to monitor. This improvement has also led to a higher degree of multiplexing, as the part of the spectrum that would have been occupied by the fluorescence of the quencher can instead be reserved for the fluorescence of additional fluorophores for the detection of more targets (21). Recently, a number of unique non-fluorescent quenchers, ranging from nucleotides to gold particles, have been introduced for use in fluorogenic probes (14,22–25). Quenching efficiencies up to several thousand-fold have been reported for some of these quenchers (8,23). Although most of this work concentrated on the labeling of molecular beacons, non-fluorescent quenchers are useful in both the FRET mode and in the contact mode of fluorescence quenching. A number of new fluorophores and fluorescence measurement instruments have also become available. Thus, there is a great need to carry out a comprehensive determination of the quenching efficiencies for many different combinations of fluorophores and quenchers. We employed a simple strategy to measure the efficiency of quenching of 22 different fluorophores by five different quenchers, both in the FRET mode and in the contact mode of quenching. We also measured the efficiency of quenching by nucleotides for each of the fluorophores.
MATERIALS AND METHODS
Synthesis of labeled oligodeoxyribonucleotides
Labeled oligodeoxyribonucleotides were synthesized by direct incorporation of either a 5′-amino group or a 5′-sulfhydryl group during automated synthesis. Iodoacetamide or maleimide derivatives of the fluorophores Alexa 350, Alexa 568, Alexa 594, Alexa 633, Alexa 680 or fluorescein (FAM) were then coupled to the 5′-sulfhydryl group. Succinimidyl ester derivatives of the fluorophores Pacific Blue, coumarin, Alexa 430, Alexa 488, Alexa 532, Alexa 546, Alexa 660, Cy2, Cy3, Cy3.5, Cy5, Cy5.5, tetramethylrhodamine (TMR) or Texas red were coupled to the 5′-amino group. Tetrachlorofluorescein (TET) or hexachlorofluorescein (HEX) was directly incorporated into the 5′ end of oligodeoxyribonucleotides through the use of a phosphoramidite containing the fluorophore. In order to introduce quenchers at the 3′ ends of the oligodeoxyribonucleotides, we utilized controlled pore glass columns covalently linked to either amino groups, dabcyl, black hole quencher 1 or black hole quencher 2 (Biosearch Technologies). Succinimidyl ester derivatives of TMR or QSY-7 quencher were coupled to the 3′ amino groups. Excess dyes were removed by passage through a NAP-5 gel exclusion column (Amersham Biosciences) and the oligodeoxyribonucleotides were then purified by high pressure liquid chromatography. The concentration of each oligodeoxyribonucleotide was determined using a microvolume spectrophotometer (Nanodrop). DNA synthesis reagents were purchased from Glen Research and the dyes were obtained from Amersham Biosciences and Molecular Probes. The sequences of the oligodeoxyribonucleotides are shown in Figure 1.
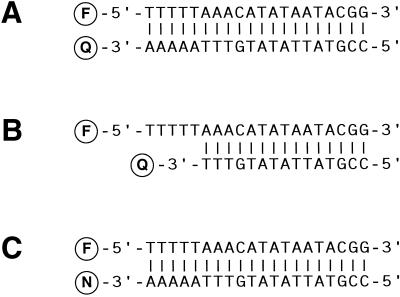
Figure 1.
Structure of oligodeoxyribonucleotide hybrids used to measure the efficiencies of quenching. (A) Blunt-ended hybrids that place the fluorophore (F) and the quencher (Q) so close together that contact quenching occurs. (B) Staggered hybrids that place the fluorophore and quencher at a distance at which FRET is the predominant mode of quenching. (C) Hybrids used to study the quenching effect of nucleotides (N).
Determination of optimum overhang length
The optimal hybrid overhang for measuring FRET efficiencies was determined with fluorescent oligodeoxyribonucleotides whose sequence was TET-5′-(T)nAAACATATAATACGG-3′, where n was either 0, 5 or 10, and with a quencher oligodeoxyribonucleotide whose sequence was 5′-CCGTA TTATATGTTT-3′-TMR. The absorption spectra of the hybrids and of the individual oligodeoxyribonucleotides (7.5 µM) dissolved in hybridization buffer (4 mM MgCl2, 15 mM KCl and 10 mM Tris–HCl, pH 8.0) were obtained using a Nanodrop spectrophotometer. The emission spectra of the hybrids and of the individual oligodeoxyribonucleotides (0.03 µM) dissolved in hybridization buffer were obtained using a QuantaMaster spectrofluorometer (Photon Technology International).
Measurement of quenching efficiencies
The fluorescence of a 0.03 µM solution (150 µl) of each fluorophore-labeled oligodeoxyribonucleotide was measured in hybridization buffer at the optimal excitation and emission wavelengths of the fluorophore (see Table 3). The quencher-labeled oligodeoxyribonucleotide was added in a small volume (5 µl) so that its final concentration was 0.15 µM. The decrease in fluorescence was monitored over time until it reached a stable plateau. Quenching efficiency was determined by dividing the fluorescence intensity of the hybrid by the fluorescence intensity of the fluorophore-labeled oligodeoxyribonucleotide, multiplying the result by 100 and then subtracting the result from 100.
Table 3. Quenching efficiencies (%) of nucleotides.
| Fluorophore | Excitation/emission maximum (nm) | Adenosine | Cytidine | Guanosine | Thymidine |
|---|---|---|---|---|---|
| Alexa 350 | 351/441 | 5 | –7 | 20 | 3 |
| Pacific Blue | 406/457 | 6 | 2 | 47 | –4 |
| Coumarin | 436/475 | 13 | 14 | 17 | 15 |
| Cy2 | 493/507 | 35 | 51 | 40 | 35 |
| Alexa 488 | 494/517 | –8 | –13 | -6 | 3 |
| FAM | 494/517 | 23 | 8 | 32 | 8 |
| Alexa 430 | 430/535 | 18 | 31 | 46 | 25 |
| TET | 523/540 | 16 | 16 | 35 | 14 |
| Alexa 532 | 526/551 | 9 | 3 | 29 | 3 |
| HEX | 538/554 | 62 | 56 | 69 | 65 |
| Cy3 | 550/564 | –47 | –45 | –46 | –97 |
| Alexa 546 | 556/570 | 39 | 35 | 57 | 34 |
| TMR | 555/577 | 9 | 11 | 20 | 11 |
| Cy3.5 | 581/593 | 15 | 9 | 37 | 17 |
| Alexa 568 | 577/599 | 4 | 4 | –60 | 3 |
| Texas red | 584/603 | 22 | 11 | 8 | 11 |
| Alexa 594 | 591/612 | 12 | 16 | 26 | 14 |
| Alexa 633 | 629/645 | 18 | 21 | 30 | 15 |
| Cy5 | 651/663 | 15 | 15 | 19 | 10 |
| Cy5.5 | 675/687 | 15 | 15 | 17 | 18 |
| Alexa 660 | 663/690 | 18 | 20 | 20 | 17 |
| Alexa 680 | 679/702 | 15 | 16 | 17 | 17 |
Negative quenching efficiencies indicate that the fluorescence of the fluorophore increased upon hybridization.
Determination of melting temperatures
Thermal denaturation profiles for hybrids labeled with fluorophores emitting in the range 507–603 nm were obtained using a 7700 Prism spectrofluorometric thermal cycler (Applied Biosystems), and for fluorophores emitting in the range 612–702 nm they were obtained using an IQ iCycler (Bio-Rad). We could not measure thermal denaturation profiles for hybrids that contained Alexa 350, Pacific Blue, coumarin and Alexa 430. All measurements were made in hybridization buffer. The concentration of the fluorophore-labeled oligodeoxyribonucleotides was 0.3 µM and the concentration of the quencher-labeled oligodeoxyribonucleotides was 1.5 µM. The temperature was increased in steps of 1°C, from 20 to 80°C, with each step lasting 60 s. Fluorescence was measured during the final 30 s.
RESULTS
Design of labeled complementary oligodeoxyribonucleotides
In order to measure quenching efficiencies, we designed two complementary oligodeoxyribonucleotides that would bring a fluorophore close to a quencher when the oligodeoxyribonucleotides hybridized to each other. A fluorophore was linked to the 5′ end of one oligodeoxyribonucleotide and a quencher was linked to the 3′ end of the other oligodeoxyribonucleotide. Quenching efficiency could then be measured by the drop in the intensity of emission of the fluorophore that occurs upon hybridization of the two oligodeoxyribonucleotides. In order to compare the efficiencies of quenching that occurs when a fluorophore and a quencher interact by FRET to the efficiencies of quenching that occurs when the same fluorophore and the same quencher interact by contact, two different types of quencher-labeled oligodeoxyribonucleotides were prepared for each of the quenchers. The first type of quencher-labeled oligodeoxyribonucleotide was of the same length as each fluorophore-labeled oligodeoxyribonucleotide, enabling the quencher and the fluorophore to interact by contact when the oligodeoxyribonucleotides were hybridized to each other (Fig. 1A). The second type of quencher-labeled oligodeoxyribonucleotide was shorter at its 3′ end, yielding hybrids containing a quencher and a fluorophore that were separated from one another by a distance that was optimal for them to interact by FRET (Fig. 1B).
In order to study FRET-mediated quenching, we carried out preliminary experiments to determine the optimal difference in length between a quencher-labeled oligodeoxyribonucleotide and a fluorophore-labeled oligodeoxyribonucleotide that would produce a hybrid in which quenching by FRET was maximal and quenching by contact was minimal. In these experiments, TET was used as the donor on the fluorophore-labeled oligodeoxyribonucleotide and another fluorophore, TMR, was used as the acceptor on the quencher-labeled oligodeoxyribonucleotide. The following characteristics were used to distinguish the contact mode of quenching from the FRET mode of quenching: (i) in contact quenching, the emission of both the donor and the acceptor fluorophore is diminished, whereas in the FRET mode of quenching, the emission of the donor fluorophore is decreased and the emission of the acceptor fluorophore is increased; (ii) in the contact mode of quenching the visible absorption spectrum of the fluorophores is substantially altered, whereas in the FRET mode of quenching, the visible absorption spectrum of the fluorophores is unchanged.
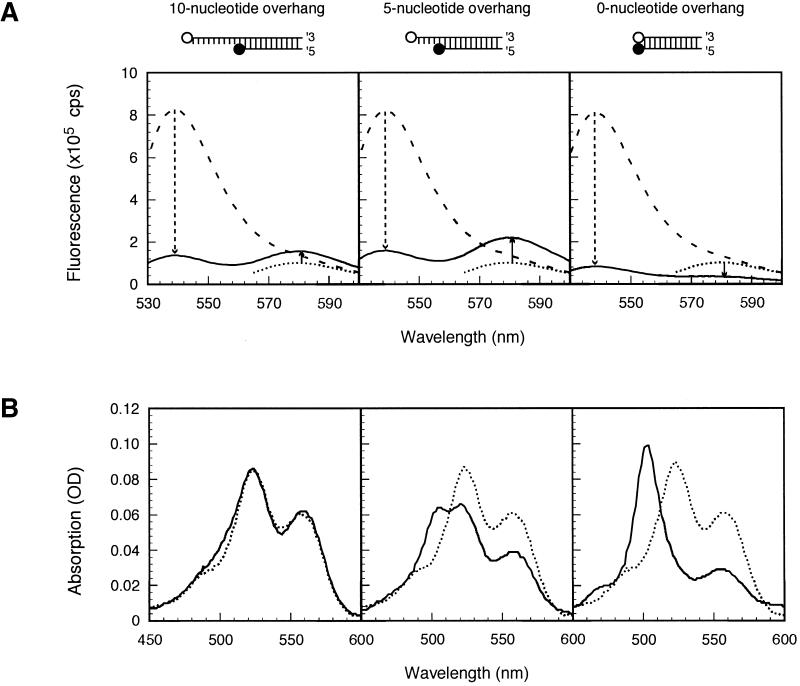
In these experiments, we varied the length of the oligodeoxyribonucleotide possessing the donor fluorophore, so that the resulting hybrids would contain an overhanging 5′-terminal segment that was either 0, 5 or 10 nt long (Fig. 2). We excited solutions of the hybrids at the wavelength that was optimal for the excitation of TET (522 nm) and we monitored the emission spectrum in the wavelength range that encompasses the fluorescence of both TET and TMR (530–600 nm). The increase in acceptor fluorescence (TMR) that occurs upon hybridization (indicated by the solid arrows in Fig. 2A) was used to determine the optimal overhang length for assessing the FRET mode of quenching. The results show that a 5 nt overhang produces a stronger increase in FRET than a 10 nt overhang. When there was no overhang, the fluorescence of both TET and TMR decreased, indicating that contact quenching was occurring.
Figure 2.
Spectral characteristics of hybrids with an overhang of 10, 5 or 0 nt. (A) Emission spectra of the TET-labeled oligodeoxyribonucleotides alone (dashed lines) and emission spectra of the hybrids they form with the TMR-labeled oligodeoxyribonucleotide (solid line), when they are both stimulated by 522 nm light, which is the optimal excitation wavelength for TET. A dotted line shows the emission spectrum of the TMR-labeled oligodeoxyribonucleotide alone, when stimulated by 555 nm light, which is the optimal excitation for TMR. Dashed arrows indicate the magnitude of the decrease in TET fluorescence that occurs when the TET-labeled strands are hybridized to the TMR-labeled strand, and solid arrows indicate the magnitude and direction of the change in TMR fluorescence that occurs when the TET-labeled strands are hybridized to the TMR-labeled strand. (B) Solid lines show the absorption spectra of the hybrids and dotted lines show the absorption spectra obtained by combining the absorption spectra of the individual oligodeoxyribonucleotides.
We next measured the changes that occur upon hybridization in the absorption spectra of the dyes. No change occurred in the absorption spectrum of the two fluorophores in hybrids possessing a 10 nt overhang, small changes occurred with hybrids possessing a 5 nt overhang and very large changes occurred in the blunt-ended hybrids (Fig. 2B). Although the data is shown only for the TET–TMR pairs, similarly notable changes in the absorption spectra were observed for all blunt-ended hybrids. These results indicate that while the mechanism of quenching in a 10 nt overhang is purely FRET and in the blunt-ended hybrid it is purely contact based, in the hybrids with a 5 nt overhang, both mechanisms may be involved. Giving more weight to the efficiency of energy transfer, rather than to the changes in the absorption spectrum, we decided to use a 5 nt rather than 10 nt overhang for the determination of the quenching efficiencies.
In the staggered hybrids designed to measure FRET-mediated quenching, we chose to place the fluorophores at the tip of the overhang and the quenchers at the recessed end (rather than vice versa) to avoid any quenching of the fluorophore by the nucleotides in the complementary strand (Fig. 1B). On the other hand, the hybrids designed to measure the quenching effect of nucleotides were blunt-ended, in which the test nucleotide occupied the position of the quencher (Fig. 1C).
Efficiencies of quenching
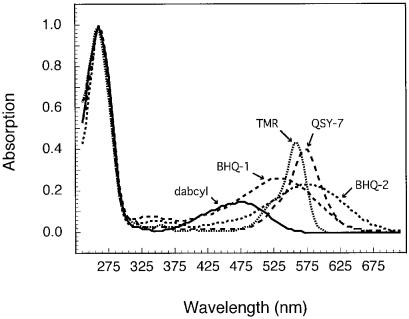
We measured quenching efficiency for 22 different fluorophores whose emission wavelength maxima ranged from 441 to 702 nm. Five different quenchers were tested with each of these fluorophores. Of the five quenchers, one (TMR) was fluorescent and the rest were non-fluorescent. The results are shown in Table 1. In order to relate the observed quenching efficiencies to the absorption characteristics of the quenchers, we determined the absorption spectrum of each of the quencher oligodeoxyribonucleotides (Fig. 3). The quenching efficiency that was observed with staggered hybrids (primarily attributable to quenching by FRET) decreased as the extent of overlap between the absorption spectrum of the quencher and the emission spectrum of the fluorophore decreased. On the other hand, the quenching efficiency that was observed with blunt-ended hybrids (due to contact quenching) did not decrease with the decrease in spectral overlap. This confirms and extends our previous observation that, in the contact mode of quenching, a good spectral overlap between the absorption spectrum of the quencher and the emission spectrum of the fluorophore is not necessary for efficient quenching (8). For each fluorophore–quencher pair that we examined, the efficiency of contact quenching was always greater than the efficiency of quenching by FRET. In the FRET mode of quenching, quenchers that exhibited a broader absorption spectrum efficiently quenched a wider range of fluorophores than quenchers with a narrow absorption spectrum. In addition, quenching efficiency was greater with quenchers that had higher extinction coefficients.
Table 1. Quenching efficiency (%) of different fluorophore–quencher combinations.
| Fluorophore | Emax (nm) | Dabcyl (Amax 475 nm) | BHQ-1 (Amax 534 nm) | TMR (Amax 558 nm) | QSY-7 (Amax 571 nm) | BHQ-2 (Amax 580 nm) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| QFRET | Qcontact | QFRET | Qcontact | QFRET | Qcontact | QFRET | Qcontact | QFRET | Qcontact | ||
| Alexa 350 | 441 | 82 | 95 | 87 | 97 | 81 | 95 | 81 | 97 | 87 | 96 |
| Pacific Blue | 457 | 83 | 96 | 93 | 98 | 90 | 98 | 86 | 97 | 93 | 96 |
| Coumarin | 475 | 84 | 95 | 92 | 97 | 87 | 97 | 84 | 98 | 92 | 97 |
| Cy2 | 507 | 78 | 95 | 92 | 98 | 84 | 96 | 76 | 96 | 91 | 97 |
| Alexa 488 | 517 | 86 | 94 | 91 | 95 | 89 | 94 | 86 | 95 | 91 | 93 |
| FAM | 517 | 80 | 91 | 88 | 93 | 86 | 92 | 85 | 93 | 89 | 92 |
| Alexa 430 | 535 | 74 | 76 | 88 | 92 | 83 | 82 | 66 | 77 | 89 | 96 |
| TET | 540 | 67 | 89 | 84 | 91 | 81 | 90 | 72 | 93 | 83 | 91 |
| Alexa 532 | 551 | 80 | 93 | 93 | 95 | 81 | 90 | 87 | 96 | 92 | 93 |
| HEX | 554 | 60 | 88 | 76 | 92 | 68 | 88 | 73 | 93 | 84 | 93 |
| Cy3 | 564 | 69 | 94 | 92 | 97 | – | – | 78 | 95 | 97 | 93 |
| Alexa 546 | 570 | 64 | 93 | 93 | 98 | – | – | 89 | 98 | 94 | 96 |
| TMR | 577 | 78 | 83 | 89 | 87 | – | – | 84 | 87 | 88 | 86 |
| Cy3.5 | 593 | 53 | 89 | 88 | 96 | – | 66 | 79 | 95 | 87 | 95 |
| Alexa 568 | 599 | 60 | 91 | 96 | 98 | – | 57 | 83 | 99 | 97 | 98 |
| Texas red | 603 | 88 | 96 | 97 | 98 | – | 86 | 89 | 98 | 97 | 97 |
| Alexa 594 | 612 | 62 | 90 | 92 | 95 | – | 66 | 78 | 95 | 93 | 94 |
| Alexa 633 | 645 | 79 | 96 | 96 | 98 | – | 95 | 54 | 97 | 97 | 97 |
| Cy5 | 663 | 43 | 84 | 89 | 96 | – | 71 | 63 | 79 | 91 | 96 |
| Cy5.5 | 687 | 44 | 82 | 81 | 96 | – | 77 | 61 | 74 | 84 | 95 |
| Alexa 660 | 690 | 31 | 81 | 84 | 96 | – | 77 | 37 | 94 | 92 | 95 |
| Alexa 680 | 702 | 44 | 81 | 88 | 94 | – | 79 | 70 | 90 | 91 | 93 |
Emax is the emission maximum of the fluorophore, Amax is the absorption maximum of the quencher, QFRET is the efficiency of quenching due to FRET, and Qcontact is the efficiency of quenching due to contact between the fluorophore and the quencher. In the case of the fluorescent quencher TMR, the quenching efficiencies could not be determined for a number of fluorophores (indicated by –) as the fluorescence of TMR interfered with the measurement.
Figure 3.
Absorption spectra of each of the five quencher-labeled oligodeoxyribonucleotides normalized for absorption at 260 nm (which is due to the nucleotides), in order to compare the absorption spectra of the quenchers.
Effects of affinity between the fluorophore and the quencher
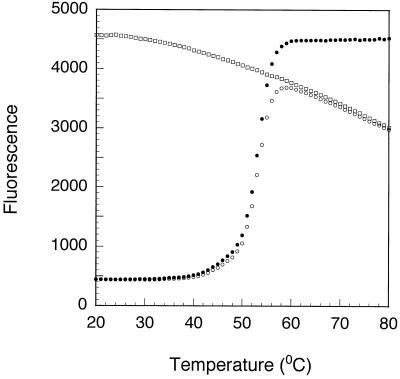
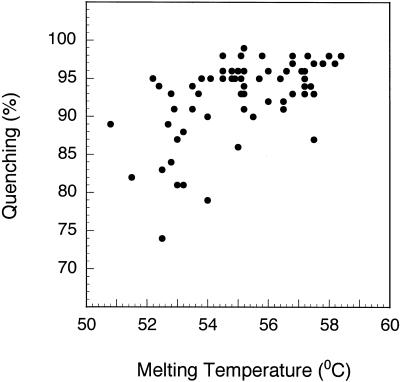
Even though the major force that brings the fluorophore and the quencher together in our experimental system is the binding affinity of the complementary oligodeoxyribonucleotides, the possibility exists that the two label moieties will attract or repel each other once they are in close proximity. Although the affinity between each quencher and fluorophore cannot be predicted a priori, an attraction between the fluorophore and the quencher would stabilize the hybrid and a repulsion would destabilize it. We therefore measured the melting temperature of each blunt-ended hybrid by monitoring its fluorescence as a function of temperature (Fig. 4). We observed that the melting temperatures ranged from 51 to 59°C, even though the same pair of complementary oligodeoxyribonucleotides was used to form each hybrid (see Table 2). The melting temperature of the same hybrids without any labels was determined to be 49°C by hypochromicity measurements. These results indicate that strong bonds can form between some fluorophore–quencher pairs and nucleic acid parameters alone are insufficient to accurately predict the thermodynamic behavior of oligodeoxyribonucleotides labeled with these fluorophores and quenchers. In order to determine the effect of the affinity between the fluorophore and quencher on the quenching efficiency, we plotted the melting temperature obtained for each blunt-ended hybrid as a function of its quenching efficiency (Fig. 5). Despite the great photochemical variation that exists between the different fluorophore–quencher pairs (which are the main determinants of quenching efficiency), a correlation was observed between quenching efficiency and melting temperature. Fluorophore– quencher pairs that stabilize the hybrid have higher quenching efficiencies. A similar trend was observed in staggered hybrids.
Figure 4.
Determination of the melting temperature of a hybrid possessing a 5 nt overhang that was labeled with FAM and dabcyl. The fluorescence of the hybrid (open circles) was divided by the fluorescence of the FAM-labeled oligodeoxyribonucleotide alone (open squares), which decreases as the temperature is increased, in order to obtain the corrected fluorescence of the hybrid as a function of temperature (filled circles). The corrected values were used to calculate the melting temperature of the hybrid.
Table 2. Melting temperatures (°C) for blunt-ended hybrids containing different fluorophore–quencher combinations.
| Fluorophore | Dabcyl (Amax 475 nm) | BHQ-1 (Amax 534 nm) | TMR (Amax 558 nm) | QSY-7 (Amax 571 nm) | BHQ-2 (Amax 580 nm) |
|---|---|---|---|---|---|
| Cy2 | 55 | 59 | 56 | 55 | 59 |
| Alexa 488 | 52 | 55 | 54 | 55 | 55 |
| FAM | 52 | 56 | 54 | 55 | 56 |
| TET | 52 | 55 | 56 | 56 | 57 |
| Alexa 532 | 54 | 56 | 54 | 56 | 55 |
| HEX | 52 | 55 | 54 | 56 | 56 |
| Cy3 | 54 | 58 | 54 | 54 | 56 |
| Alexa 546 | 53 | 55 | 53 | 55 | 55 |
| TMR | 53 | 56 | 53 | 53 | 55 |
| Cy3.5 | 51 | 55 | 52 | 53 | 55 |
| Alexa 568 | 53 | 57 | 54 | 55 | 57 |
| Texas red | 55 | 58 | 54 | 55 | 57 |
| Alexa 594 | 54 | 57 | 53 | 55 | 57 |
| Alexa 633 | 55 | 57 | 56 | 56 | 57 |
| Cy5 | 52 | 57 | 53 | 55 | 57 |
| Cy5.5 | 51 | 55 | 52 | 53 | 54 |
| Alexa 660 | 52 | 56 | 53 | 53 | 56 |
| Alexa 680 | 52 | 57 | 52 | 55 | 58 |
The melting temperature of the blunt-ended hybrid without a fluorophore or quencher is 49°C.
Amax, absorption maximum of the quencher.
Figure 5.
Correlation between quenching efficiency and melting temperature for blunt-ended hybrids containing different fluorophore–quencher pairs. Even though all of the hybrids were formed from the same complementary oligodeoxyribonucleotides, they exhibited different melting temperatures, due to the degree of attraction or repulsion of the fluorophore and the quencher.
Quenching by nucleotides
Several investigators have observed that nucleotides can quench the fluorescence of fluorophores (26,27). We systematically measured the quenching efficiency of each of the four deoxyribonucleotides in combination with each of the 22 fluorophores, utilizing the hybrid structure shown in Figure 1C. The results (Table 3) show that nucleotides do not quench as efficiently as the moieties discussed above. Moreover, the nucleotides exhibited a variable degree of quenching, with guanosine being the most efficient, followed by adenosine, cytidine and thymidine. In general, fluorophores that fluoresce in the green and yellow wavelengths were quenched more efficiently by nucleotides than fluorophores that fluoresce in the blue and red wavelengths. When oligoguanosine was used as a quencher in place of a single guanosine, quenching efficiency was higher (results not shown). The interpretation of the results obtained with nucleotides is more complex than the interpretation of the results obtained with the quencher moieties discussed above. First, the magnitude of nucleotide quenching efficiencies is rather small and, secondly, the influence of neighboring nucleotides in both strands of the hybrid cannot be isolated from the influence of the nucleotide that is immediately opposite the fluorophore.
DISCUSSION
The quenching efficiencies that we measured can be used to aid the design of different kinds of probes for homogeneous hybridization assays. The contact quenching efficiencies apply to molecular beacons and competitive hybridization probes, and the FRET quenching efficiencies apply to adjacent probes and TaqMan probes. The values for the quenching efficiencies that we obtained in these experiments are lower than the values we normally obtain in dual-labeled molecular beacon and TaqMan probes, because there is always a small fraction of dissociated oligodeoxyribonucleotides present at room temperature that lowers the measured efficiencies. In the case of TaqMan probes, another factor to consider in the prediction of the quenching efficiency is the length of the probe. Our measurements are for the smallest possible distance (5 nt) for FRET to occur. In practice, TaqMan probes are much longer and therefore may yield a lower degree of quenching than reported here. Our results also suggest that when using a pair of labeled probes that are designed to hybridize close to one another on a target sequence and to interact by FRET (Light Cycler probes), the donor and the acceptor fluorophores should not be in immediate contact with each other, as this will decrease the emission intensity of both fluorophores. Instead, the target sequences for each probe should be at least 5 nt apart, to prevent contact quenching and to maximize emission from the acceptor.
The stabilizing effect of some fluorophore–quencher pairs on the hybrid has important consequences for the design of probes. For example, a molecular beacon labeled with Cy3.5 and BHQ-2 exhibited a stem melting temperature that was 5°C higher than the same molecular beacon labeled with Cy3.5 and dabcyl. This factor should be considered when predicting their thermodynamic behavior. In the case of probes that form a random coil, such as TaqMan probes, it has been thought that the fluorophore and the quencher wander into a FRET radius by thermal motion (12,13). If the fluorophore and the quencher (including the nucleotides) have substantial attraction for each other, then the two moieties will stay within that radius more or less permanently, creating highly quenched probes. For example, when the fluorophore Cy2 is in contact with the BHQ-1, the melting temperature of the hybrid is 59°C and the fluorescence is quenched by 97%, whereas when the fluorophore Cy3.5 is in contact with dabcyl, the melting temperature of the hybrid is 51°C and the quenching of the fluorescence is 89%. The results suggest that the enhanced quenching ability of black hole quenchers in FRET mode is due to their propensity to bind to fluorophores. This has also been observed by another group in a recent paper (24).
In this report, we have characterized five quenchers in combination with 22 different fluorophores, and as new pairs become available, they can similarly be tested. Our results confirm that in probe systems that place two label moieties within an appropriate distance for FRET to occur, spectral overlap between the emission spectrum of the fluorophore and the absorption spectrum of the quencher is an important determinant of quenching efficiency. Spectral overlap is not a significant determinant of quenching efficiency in probes that cause the fluorophore and the quencher to come within contact distance of each other, and quenching efficiencies remain very high in the absence of spectral overlap.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Diana P. Bratu and Musa M. Mhlanga for critical review of the manuscript. This work was supported by National Institutes of Health grants HL-43521 and ES-10536.
REFERENCES
- 1.Heller M.J., Morrison,L.E., Prevatt,W.D. and Akin,C. (1983) Patent no. EPO070685.
- 2.Cardullo R.A., Agrawal,S., Flores,C., Zamecnik,P.C. and Wolf,D.E. (1988) Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 85, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison L.E., Halder,T.C. and Stols,L.M. (1989) Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal. Biochem., 183, 231–244. [DOI] [PubMed] [Google Scholar]
- 4.Livak K.J., Flood,S.J., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 5.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 6.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 7.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–138. [DOI] [PubMed] [Google Scholar]
- 8.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 9.Li Q., Luan,G., Guo,Q. and Liang,J. (2002) A new class of homogenous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res., 30, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuji A., Koshimoto,H., Sato,Y., Hirano,M., Sei-Iida,Y., Kondo,S. and Ishibashi,K. (2000) Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J., 78, 3260–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkhurst K.M. and Parkhurst,L.J. (1995) Kinetic studies by fluorescence resonance energy transfer employing a double-labeled oligonucleotide: hybridization to the oligonucleotide complement and to single-stranded DNA. Biochemistry, 34, 285–292. [DOI] [PubMed] [Google Scholar]
- 13.Parkhurst K.M. and Parkhurst,L.J. (1995) Donor-acceptor distance distributions in a double-labeled fluorescent oligonucleotide both as a single strand and in duplexes. Biochemistry, 34, 293–300. [DOI] [PubMed] [Google Scholar]
- 14.Walter N.G. and Burke,J.M. (1997) Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA, 3, 392–404. [PMC free article] [PubMed] [Google Scholar]
- 15.Ledford M., Friedman,K.D., Hessner,M.J., Moehlenkamp,C., Williams,T.M. and Larson,R.S. (2000) A multi-site study for detection of the factor V (Leiden) mutation from genomic DNA using a homogeneous invader microtiter plate fluorescence resonance energy transfer (FRET) assay. J. Mol. Diagn., 2, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggins J.B., Prudent,J.R., Marshall,D.J., Ruppen,M. and Thorson,J.S. (2000) A continuous assay for DNA cleavage: the application of “break lights” to enediynes, iron-dependent agents and nucleases. Proc. Natl Acad. Sci. USA, 97, 13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamaguchi N., Ellington,A. and Stanton,M. (2001) Aptamer beacons for the direct detection of proteins. Anal. Biochem., 294, 126–131. [DOI] [PubMed] [Google Scholar]
- 18.Lakowicz J.R. (1999) Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Publishers, New York, NY.
- 19.Bernacchi S. and Mely,Y. (2001) Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Res., 29, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matayoshi E.D., Wang,G.T., Krafft,G.A. and Erickson,J. (1990) Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science, 247, 954–958. [DOI] [PubMed] [Google Scholar]
- 21.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 22.Knemeyer J.P., Marme,N. and Sauer,M. (2000) Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem., 72, 3717–3724. [DOI] [PubMed] [Google Scholar]
- 23.Dubertret B., Calame,M. and Libchaber,A.J. (2001) Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol., 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 24.Johansson M.K., Fidder,H., Dick,D. and Cook,R.M. (2002) Intramolecular dimers: a new strategy to fluorescence quenching in dual-labeled oligonucleotide probes. J. Am. Chem. Soc., 124, 6950–6956. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell D.J., Taylor,J.R. and Nie,S. (2002) Self-assembled nanoparticle probes for recognition and detection of biomolecules. J. Am. Chem. Soc., 124, 9606–9612. [DOI] [PubMed] [Google Scholar]
- 26.Seidel C.A.M., Schulz,A. and Sauer,M.M.H. (1996) Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem., 100, 5541–5553. [Google Scholar]
- 27.Crockett A.O. and Wittwer,C.T. (2001) Fluorescein-labeled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem., 290, 89–97. [DOI] [PubMed] [Google Scholar]