Abstract
HinP1I recognizes and cleaves the palindromic tetranucleotide sequence G↓CGC in DNA. We report three structures of HinP1I–DNA complexes: in the presence of Ca2+ (pre-reactive complex), in the absence of metal ion (binary complex) and in the presence of Mg2+ (post-reactive complex). HinP1I forms a back-to-back dimer with two active sites and two DNA duplexes bound on the outer surfaces of the dimer facing away from each other. The 10 bp DNA duplexes undergo protein-induced distortions exhibiting features of A-, B- and Z-conformations: bending on one side (by intercalation of a phenylalanine side chain into the major groove), base flipping on the other side of the recognition site (by expanding the step rise distance of the local base pair to Z-form) and a local A-form conformation between the two central C:G base pairs of the recognition site (by binding of the N-terminal helix in the minor groove). In the pre- and post-reactive complexes, two metals (Ca2+ or Mg2+) are found in the active site. The enzyme appears to cleave DNA sequentially, hydrolyzing first one DNA strand, as seen in the post-reactive complex in the crystalline state, and then the other, as supported by the observation that, in solution, a nicked DNA intermediate accumulates before linearization.
INTRODUCTION
Type II restriction endonucleases (REases) are a diverse and fascinating group of proteins. With the REBASE database currently listing more than 3600 Type II REases possessing more than 250 distinct DNA sequence specificities, they constitute one of the largest known families of enzymes (1). These DNA-cleaving enzymes combine very high catalytic efficiencies (kcat/kuncat = ∼1016) with exquisite DNA sequence selectivity. Restriction enzymes classified as Type II cleave DNA specifically within or close to their recognition sites and require Mg2+ for activity [reviewed in (2)]. DNA cleavage by these enzymes produces fragments with either 5′ or 3′ overhangs or with blunt ends.
Most Type II REases are homodimeric or tetrameric enzymes that recognize symmetrical DNA sequences (2). Dimerization results in a clamp-shaped molecule with two internal active sites facing each other (face-to-face), allowing the enzyme to wrap around a DNA duplex and cleave the two strands symmetrically and simultaneously. The first exception to this rule came from the MspI structure (3). An MspI monomer, rather than a dimer, binds to the palindromic DNA sequence and makes direct or water-mediated contacts with all four base pairs in the CCGG recognition sequence.
HinP1I is a Type II restriction enzyme from Haemophilus influenzae P1 (4) that recognizes and cleaves the palindromic DNA sequence G↓CGC. Structurally, HinP1I is strikingly similar to the monomeric MspI (5). Unlike MspI, however, HinP1I can dimerize through interactions at the ‘back’ of the enzyme such that the dimer has two external active sites and two DNA binding surfaces facing away from each other on the outside of the dimer. Here, we show that this back-to-back dimer of HinP1I binds to two copies of duplex DNA. Soaking Mg2+ into the pre-formed HinP1I–DNA co-crystals induced cleavage of one DNA strand in each bound duplex, indicating that the back-to-back HinP1I dimer is catalytically active in the crystalline state. In solution, HinP1I-digestion of supercoiled plasmid DNA results in the accumulation of nicked DNA intermediate before linearization, indicating that the two DNA strands are hydrolyzed sequentially, rather than simultaneously. In the crystal, we observed three major protein-induced DNA distortions: bending on one side of the recognition sequence and base flipping on the other, and local A-form conformation between the two central C:G base pair of the recognition site. Together, these structural and biochemical data provide snapshots of HinP1I–DNA specific interactions during pre- and post-reactive states of the enzyme and grant unique insights into the DNA recognition and cleavage mechanism of this endonuclease.
MATERIALS AND METHODS
HinP1I was expressed and purified as described previously (5). The enzyme was concentrated to 40–50 mg/ml in 10 mM Tris, pH 7.5, 1 mM DTT, 0.1 mM Na2EDTA, 5% glycerol and 400 mM NaCl. Following mixing with the oligodeoxynucleotide in an approximate 1:1 molar ratio, the protein–DNA complex was allowed to stand on ice for at least 1 h before crystallization. Crystals appeared within hours to several days in hanging drop experiments. We obtained two crystal forms of the pre-reactive complex of HinP1I–DNA–Ca2+. The P3221 crystal grew under conditions of 7.5% (v/v) ethanol, 1.5 M NaCl, 100 mM Bis-Tris propane, pH 7.8 and 20 mM CaCl2 and was very reproducible. The P6522 crystal grew under conditions of 30% PEG 4000, 100 mM Tris, pH 8.5, 0.2 M Li2SO4 and 20 mM CaCl2. Because of the high similarity between the two structures (the root-mean-square deviation of ∼0.3 Å) and the higher resolution of the P3221 structure (Table 1), we described in text the ternary structure in the P3221 space group. Using the protein coordinates of a free HinP1I monomer structure (5) as a search model (PDB 1YNM), the program REPLACE (6) determined the structural solutions by molecular replacement. DNA models were built manually into densities of difference maps using the program O (7). Refinement proceeded with the program CNS (8).
Table 1.
Statistics of X-ray data reduction and refinement
| Complex | Pre-reactive complex | Binary complex | Post-reactive complexa | |
|---|---|---|---|---|
| HinP1I/DNA/Ca2+ | HinP1I/DNA/Ca2+ | HinP1I/DNA | HinP1I/DNA/Mg2+ | |
| PDB ID | 2FKC | 2FKH | 2FL3 | 2FLC |
| Beamline (wavelength Å) | APS 22-ID (0.97179) | APS 22-BM (0.99997) | APS 22-ID (1.0) | APS 22-ID (1.0) |
| Space group | P3221 | P6522 | P6522 | P6522 |
| Unit cell dimensions (Å) | a = b = 105.1, c = 130.4 | a = b = 109.1, c = 113.3 | a = b = 100.9, c = 139.4 | a = b = 100.4, c = 139.4 |
| Resolution range (Å) (highest resolution shell) | 19.89–2.39 (2.48–2.39) | 31.51–3.09 (3.20–3.09) | 34.17–2.39 (2.48–2.39) | 34.10–2.59 (2.65–2.59) |
| Measured reflections | 225 720 | 130 658 | 325 661 | 106 424 |
| Unique reflections | 31 659 | 7757 | 15 524 | 13 460 |
| <I/σ> | 19.0 | 17.1 | 15.2 | 17.6 |
| Completeness (%) | 94.7 (97.2) | 99.9 (99.9) | 89.0 (50.2) | 99.2 (95.7) |
| Rlinear = Σ|I − 〈I〉|/Σ〈I〉 | 0.108 (0.368) | 0.071 (0.278) | 0.079 (0.374) | 0.070 (0.401) |
| R-factor = Σ|Fo − Fc|/Σ|Fc| | 0.222 (0.309) | 0.220 (0.255) | 0.246 (0.427) | 0.275 (0.475)a |
| R-free (5% data) | 0.260 (0.362) | 0.313 (0.338) | 0.284 (0.419) | 0.324 (0.504)a |
| Non-hydrogen atoms | ||||
| Protein | 3981 (two molecules) | 1958 (one molecule) | 1939 (one molecule) | 1925 (one molecule) |
| DNA | 808 (two duplexes) | 404 (one duplex) | 404 (one duplex) | 404 (one duplex) |
| Metal ions | 4 (Ca2+) | 2 (Ca2+) | – | 2 (Mg2+) |
| Water molecules | 45 | 12 | 10 | 1 |
| Root-mean-square deviation from ideality | ||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 | 0.011 |
| Bond angles (°) | 1.4 | 1.3 | 1.4 | 1.6 |
| Dihedral (°) | 23.0 | 22.6 | 22.7 | 22.7 |
| Improper (°) | 2.9 | 2.9 | 2.8 | 2.8 |
| Estimated coordinate error | ||||
| From Luzzati plot (Å) | 0.33 | 0.38 | 0.44 | 0.49 |
| From SIGMAA (Å) | 0.29 | 0.17 | 0.51 | 0.55 |
aThe refinement of the post-reactive complex was a challenge.
Analysis of reflection data indicated that Mg2+ soaking may lead the crystal to form a superlattice, which probably contains two alternating sublattices: a short one similar to the original native crystal and a long one with doubled a and b axes (see Supplementary Data). Efforts were made to separate the contributions from the two sublattices and the dominant short sublattice was finally used for the refinement of the post-reactive complex. Due to the error introduced by the contributions from the long sublattice (∼12% in the crystal), the R-factor and R-free values are relatively high although electron density maps appear suitable. Assignment of solvent molecules to discontinued densities would further reduce the R-factor, but not the R-free. We took a conservative approach without including such solvent molecules, and carefully examined omit difference electron density maps particularly in the area of the cleaved scissile bond and the Mg2+ binding sites.
The crystals of the binary HinP1I–DNA complex were grown with no added metal under conditions of 20% PEG 1000, 400 mM NaCl, 100 mM Bis-Tris propane (pH 6.2–6.8) and 5% ethylene glycol. These crystals were used in soaking experiments with compounds containing 1–10 mM MgCl2. Cation-containing solutions were added to the mother liquor with a cryoprotectant (25% ethylene glycol) and crystals were transferred to this mixture. Addition of Mg2+ caused the crystals to crack often within 10 s but occasionally after ∼45–60 s. Thus, in an attempt to capture a catalytic or product complex, the soaked crystals were flash frozen within these time limits. Crystals were acquired with a nylon loop (Hampton) and then quickly flash frozen directly in liquid nitrogen or in a cold nitrogen gas stream at 100K before being stored frozen and later data collection. X-ray data werecollected, from a crystal soaked in 5 mM MgCl2 for ∼45 s, at the beamline 22-ID at the Advanced Photon Source (0.5° oscillations, 221 images, 8 s per image, 1.0 Å wavelength).
RESULTS AND DISCUSSION
We crystallized a ternary complex containing HinP1I, a 10 bp oligonucleotide containing a single GCGC site, and a divalent metal ion, Ca2+, in the P3221 space group (PDB ID 2FKC). The crystallographic asymmetric unit contains two HinP1I molecules (A and B), forming a back-to-back dimer as observed in the structure of free HinP1I (5), and two DNA duplexes (Figure 1A)—strands C and D are bound to molecule A and strands E and F are bound to molecule B.
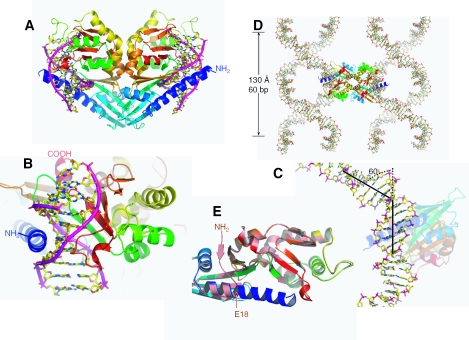
Figure 1.
HinP1I–DNA–Ca2+ structure. (A) The back-to-back HinP1I dimer bound with two DNA duplexes. The HinP1I monomer is colored using a rainbow spectrum with blue for the N-terminus and red for the C-terminus. (B) A HinP1I monomer bound with one DNA duplex, with the N-terminal helix in the minor groove and the central β sheet in the major groove. (C) The DNA is kinked ∼60° where it is bound by the HinP1I enzyme. (D) The 10 bp DNA self-assembles to generate a continuous superhelix. The back-to-back HinP1I dimer links two superhelices together. (E) Structural superimposition of DNA-bound HinP1I (in a rainbow spectrum) with that of free HinP1I (in salmon; PDB 1YNM). The N-terminal residues (amino acids 1–17, colored in blue) become part of a long helix in the DNA-bound structure.
DNA superhelix
The 10 bp DNA duplex is encircled by a single HinP1I molecule (Figure 1B); with the long 7-turn N-terminal helix in the minor groove and the β-strands and their associated loops in the major groove. Thus, ∼1900 Å2 of surface area per protein is buried upon DNA binding, comparable with what is observed for the monomeric mismatch repair endonuclease MutH when it binds DNA (9). Also, the bound DNA is substantially bent, ∼60° (Figure 1C), by its interaction with the enzyme (see below). The bent duplex stacks with one neighboring DNA molecule at each end forming a superhelix. The axis of the superhelix formed by the assembly of the oligonucleotide is parallel to the crystal c-axis and the DNA obeys the 32 screw-symmetry of the space group with a radius of ∼50 Å (half of the unit cell a-axis) and a rise of 130 Å (the length of the unit cell c-axis), giving a repeat of 60 bp. The assembled superhelices are connected via two HinP1I molecules through the back-to-back dimer interface (Figure 1D), defining the packing of the protein–DNA complexes that complete the formation of the crystal. The HinP1I–DNA packing is very similar to the DNA condensation induced by some DNA bridge factors, which use two DNA binding motifs located on opposite surfaces of a bridge dimer (10).
HinP1I changes conformation upon DNA binding
DNA molecules bound by face-to-face dimeric REases are almost all accommodated in a tight binding cleft formed by both monomers (2,11). In the cases of HinP1I and MspI (3), the DNA binding cleft lies within a single protein molecule, which raises the question whether a conformational change in the protein is necessary for tight binding. Comparing the structures of free and bound HinP1I, the largest change involves in the N-terminal 17 residues, which become part of a long helix upon binding to cognate DNA (Figure 1E). In the structure of free HinP1I, the first six N-terminal residues that are invisible in the electron density map are followed by a β-strand (residues 9–11) and a loop (residues 12–17), which is connected to the helix αA (5). Upon binding to DNA, helix αA is extended to the very first residue and the resulting longer helix clamps down along the minor groove of the DNA (Figure 1B). This results in a local A-form geometry around the two central C:G base pairs of the recognition site: a wider minor groove (∼10 Å width versus 6 Å in B-form), a shorter base rise distance (∼2.4–2.9 Å versus 3.4 Å in B-form) and a tilting of the bases (∼10–20°) rather than lying perpendicular to the helix axis (Table 2). Interestingly, the residue adjacent to this helix extension, E18, forms part of the catalytic center (see below). Comparing the two structures, with and without DNA bound, there is very little change in the catalytic center, suggesting that it is pre-formed and relatively rigid.
Table 2.
Local inter-base parameters of DNA
| Strand C | Strand D | Shift (C/D) | Slide (C/D) | Rise (C/D) | Tilt (C/D) | Roll (C/D) | Twist (C/D) |
|---|---|---|---|---|---|---|---|
| C1/C2 | G-1/G-2 | 0.87/0.34 | −0.43/−0.10 | 3.39/3.39 | 3.79/1.74 | 12.86/5.27 | 36.77/37.23 |
| C2/A3 | G-2/T-3 | 0.09/−0.06 | −0.72/−0.45 | 3.05/3.47 | 5.78/−2.77 | 8.87/10.73 | 23.88/27.90 |
| A3/G4 | T-3/C-4 | 2.04/1.57 | −2.23/−2.17 | 4.26/5.25 | −8.74/21.42 | −8.01/−6.69 | 33.70/29.96 |
| G4/C5 | C-4/G-5 | −0.15/0.70 | −2.91/−1.56 | 3.46/3.35 | −1.30/−1.57 | 2.10/−0.97 | 28.48/25.83 |
| C5/G6 | G-5/C-6 | 0.88/1.31 | −1.39/−1.08 | 2.94/2.74 | 17.51/9.23 | 5.35/15.01 | 23.54/21.34 |
| G6/C7 | C-6/G-7 | −1.54/−1.46 | −0.22/−1.22 | 3.21/3.34 | 2.97/−1.00 | −8.77/4.26 | 34.99/40.43 |
| C7/T8 | G-7/A-8 | 1.29/0.39 | 0.24/0.04 | 4.73/4.70 | −3.06/7.23 | −13.31/−41.27 | 45.28/41.35 |
| T8/G9 | A-8/C-9 | −1.12/−0.88 | 0.92/0.25 | 3.41/2.99 | −1.87/−12.39 | −14.96/9.07 | 35.32/33.41 |
| G9/G10 | C-9/C-10 | 0.61/−0.67 | −0.37/−0.88 | 3.26/3.58 | 5.50/−2.59 | 9.28/2.00 | 36.11/17.13 |
| A-DNA | 2.6 | 20 | 33 | ||||
| B-DNA | 3.4 | −6 | 36 | ||||
| Z-DNA | 4.5 | 7 | −30 | ||||
| MspI (averaged) (PDB 1SA3) | 3.37 | 0.42 | 32.74 | ||||
| Strand E | Strand F | Shift (E/F) | Slide (E/F) | Rise (E/F) | Tilt (E/F) | Roll (E/F) | Twist (E/F) |
| C1/C2 | G-1/G-2 | 0.73/0.28 | 1.42/1.33 | 3.21/3.37 | 13.51/−1.68 | 13.44/−2.46 | 31.68/35.63 |
| C2/A3a | G-2/T-3 | 5.40/−0.81 | 1.99/0.58 | a/3.42 | −18.28/−5.48 | 53.13/10.29 | 73.15/37.11 |
| A3a/G4 | T-3/C-4 | −7.00/2.23 | −0.49/−2.75 | a/5.69 | −25.75/22.24 | −54.97/−19.64 | −12.89/26.40 |
| G4/C5 | C-4/G-5 | −0.17/0.79 | −3.23/−1.74 | 3.41/3.43 | 1.26/0.67 | 4.54/1.62 | 27.99/24.62 |
| C5/G6 | G-5/C-6 | 0.85/1.74 | −1.35/−1.34 | 2.83/2.47 | 19.80/−11.28 | 4.61/14.11 | 21.54/17.80 |
| G6/C7 | C-6/G-7 | −1.41/−1.79 | −0.43/−1.03 | 3.41/3.37 | 0.72/2.10 | −8.14/3.78 | 34.25/49.46 |
| C7/T8 | G-7/A-8 | 0.89/0.77 | −0.04/−0.21 | 4.73/4.85 | −3.56/6.48 | −16.75/−37.91 | 41.96/33.41 |
| T8/G9 | A-8/C-9 | −0.60/−0.47 | 0.38/0.30 | 3.10/3.00 | 5.57/−15.27 | 5.28/8.77 | 31.90/31.39 |
| G9/G10 | C-9/C-10 | 0.56/−0.47 | −0.79/−1.80 | 3.55/3.55 | −2.16/1.79 | −1.00/2.95 | 28.46/38.42 |
aA3 in strand E is flipped.
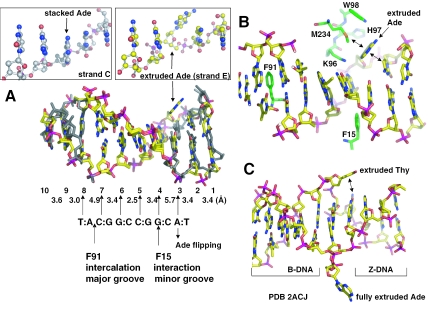
DNA distortion by side chain intercalation and base flipping
The major disruption to the base stacking and linearity of the DNA occurs at the two junctions of the recognition site. On one distal side of the recognition site, between the outer base pair of the recognition site (C:G at position 7) and the first base pair of the flanking DNA (T:A at position 8) (Figure 2A), the hydrophobic side chain of F91 intercalates the DNA from the major groove causing the DNA to be kinked by ∼60° (Figure 2B). The intercalation expands the base rise distance to ∼4.7 Å between the base pairs at positions 7 and 8 (Table 2). On the proximal side of the recognition site, the Gua of the outer G:C base pair (at position 4) is in van der Waals contact with the phenyl ring of F15 of the N-terminal helix αA, which approaches the DNA from the minor groove (Figure 2C). This asymmetric intercalation is similar to the intercalation of the repair proteins MutS and Vsr (which function as monomers) with DNA mismatches (12–15), but differs from the symmetric intercalation of the HincII restriction endonuclease, a face-to-face dimer, in which a Gln side chain intercalates between 2 bp on either side of the recognition site (16).
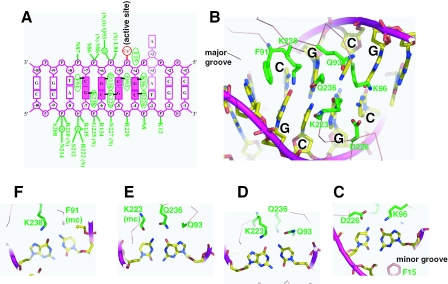
Figure 2.
HinP1I–DNA interactions. (A) Summary of the protein–DNA contacts of HinP1I (green). Backbone mediated interactions are indicated with main chain amine (N) or carbonyl (O). For simplicity, only single water (w) molecule mediated interactions are shown. The A(3) base can be in either an intrahelical or extrahelical location (see text). (B) HinP1I side chains (green) in the major groove of the DNA duplex with a central GCGC site. The side chain of F91 intercalates DNA immediately outside of the recognition sequence, between the outer base pair of the recognition site (G:C) and the first base pair of the flanking DNA (A:T). (C–F) Detailed interactions with the recognition sequence GCGC. All interactions occur from the major groove side, except for the side chain of F15, which approaches the DNA from the minor groove.
The most striking finding in the structure is that Ade3, immediately outside of the proximal side, can adopt at least two conformations, extrahelical and intrahelical. Shown in Figure 3A is a least-squares superimposition of two DNA duplexes (strands C and D versus strands E and F), using the protein component only to determine the superimposition (a root-mean-square deviation of ∼0.6 Å comparing 247 pairs of Cα atoms of two HinP1I molecules A and B). The two duplexes show high concordance in the interaction pattern of the recognition base pairs (at positions 4–7), and the distal T:A base pair (at position 8) immediately adjacent to the F91 intercalation site. On the other side of the recognition sequence, the helix conformation of the first flanking base pair, A:T (at position 3 of Figure 3A), is markedly different in the two duplexes. In molecule A, the A:T base pair at position 3 remains stacked and hydrogen bonded in the duplex (left boxed enlargement in Figure 3A). In molecule B, the Ade of the A:T base at position 3 is flipped out of the DNA helix (right boxed enlargement), where it is stabilized extrahelically by residues H97, W98, M234, and the 5′ Cyt base at position 2 (Figure 3B). Interestingly, the local DNA structure adopts features characteristic of Z-form DNA between the base pairs at positions 3 and 4 (Table 2), reminiscent of the recent finding of base flipping at the junction between B- and Z-DNA (17). An important parameter is a relatively long rise distance of 5.7 Å between the T(-3) and C(-4) bases (strand F in Table 2), which may be attributable to the flipping of the A(3) base of strand E, which has a twist angle of −13°, an indication of a transition from B-form (36°) to Z-form (−30°). The corresponding rise distance between the base pairs 3 and 4 in strand C and D is slightly smaller, one possible reason why A(3) base of strand C stays stacked. Compared with the two extruded bases at the junction between B-DNA and Z-DNA (Figure 3C) (17), the A(3) base lies in a position similar to the flipped thymine at the B-to-Z junction. Both flipped bases, A(3) in HinP1I and the thymine at the B-to-Z junction, have a stabilizing force via a face-to-edge van der Waals contact with an intrahelical base (comparing Figure 3B and C), while the fully extended adenine at the B-to-Z junction has neither contacts with the protein nor the DNA (17).
Figure 3.
Base flipping outside of the recognition sequence. (A) Superimposition of the two DNA duplexes, bound with molecule A (colored in grey) or molecule B (colored with yellow for carbon atoms, blue for nitrogen atoms, red for oxygen atoms and magenta for phosphate atoms). Base pairs are numbered from 1 to 10, as shown in Figure 2A. The distances between the stacked bases on strand F are indicated. For comparison, the flipped Ade, A(3) of strand E, and its corresponding A(3) of strand C are enlarged and boxed. (B) The flipped Ade, A(3) of strand E, lies against the HinP1I protein surface of molecule B. The double arrows indicate van der Waals contacts. (C) A view of DNA structure containing a junction between Z-DNA and B-DNA [PDB 2ACJ (17)]. The two bases of a A:T base pair at the junction are extruded. Note the van der Waals interaction (double arrow) between the extruded Thy and an intrahelical base.
We also obtained a second crystal form of HinP1I–DNA–Ca2+ (see Materials and Methods) where the A(3) base in every duplex in the crystal is flipped out (data not shown). The new crystal form can be indexed in a higher symmetry space group P6522 where the crystallographic asymmetric unit is reduced to containing one Hin1PI molecule and one DNA duplex (PDB ID 2FKH). The protein component of pre-reactive complex in the space group P6522 is more similar in structure to the extrahelical-A(3)-containing molecule B (root-mean-square deviation of ∼0.3 Å) than the intrahelical-A(3)-containing molecule A (root-mean-square deviation of ∼0.6 Å) in the space group P3221. The extrahelical-A(3) in both space groups, P3221 and P6522, is not involved in any crystal packing contacts. We suggest that the degree of deviation of local structure around A(3), away from B-DNA toward Z-DNA, determines whether A(3) flips.
Enzyme-induced DNA base flipping has been characterized structurally in DNA methyltransferases and glycosylases (18–20) and in a DNA polymerase (21). In these cases, the flipped nucleotide is either the target of modification or repair or it is the template nucleotide for DNA synthesis. In solution, incorporation of the nucleotide analog 2-aminopurine (2AP) into synthetic oligodeoxynucleotide duplexes has been used to probe conformational changes, such as base flipping (22–26), because 2AP fluorescence increases dramatically when it is removed from the stacking environment of double helical DNA (27). However, in some cases, the 2AP fluorescence change does not correlate with the target of modification or repair (28–30), when the 2AP was positioned at a non-target site or outside of recognition sequence. The ambiguity calls into questions whether 2AP fluorescence is a reliable diagnostic technique for DNA base flipping in solution. While the technique is being fine tuned, our structure of a HinP1I–DNA complex provides the first example of a nucleotide outside of a recognition sequence that can be either intrahelical or extrahelical upon binding of the DNA to protein. It should be noted that when MspI binds to an oligonucleotide of the same length as used here, it retains the typical B-form DNA structure (3). There are no equivalents in MspI to F15 and F91 in HinP1I.
Direct major groove protein–DNA contacts
In addition to phosphate interactions, which span 6 bp (Figure 2A), all eight bases of the tetranucleotide recognition sequence GCGC have direct hydrogen bond interaction with one HinP1I molecule. The interactions with the four guanines are all through their O6 atoms, while the four cytosines are contacted through their N4 atoms (Figure 2C–F). Three lysine side chains (K96, K223 and K238) and one glutamine side chain (Q236) are involved in the interactions with the guanines. Two side chain oxygen atoms (D226 and Q93) and two main chain carbonyl oxygen atoms (K223 and F91) are involved in the interactions with the cytosines.
Pre-reactive ternary complex of HinP1I–DNA–Ca2+
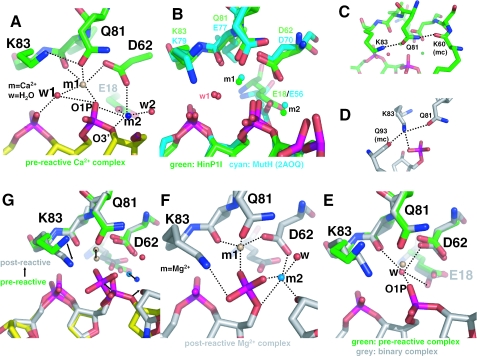
Two Ca2+ ions (m1 and m2, separated by 4.2 Å distance) were identified in the active site: both have octahedral coordination (Figure 4A). Metal m1 is coordinated by the side chain oxygen atoms of D62 (Oδ2, 2.5 Å) and Q81 (Oε1, 2.5 Å), the main chain carbonyl oxygen of V82 (2.4 Å), the oxygen O1P of the scissile phosphate (2.1 Å) and a water molecule w1 (2.4 Å). Besides making contact with the metal m1, the water molecule w1 is hydrogen bonded to the O1P oxygen of the scissile phosphate (3.0 Å) and the O2P oxygen of the 3′ nucleotide (2.6 Å). The water molecule w1 is well positioned, 3.4 Å from the phosphorous atom and makes an angle of ∼167° with the P-O3′ bond of the scissile phosphate group, to act as the attacking nucleophile for an in-line attack opposite the O3′ leaving group. It is unclear which component of the catalytic site is responsible for activating the nucleophilic water molecule. The side chain amino group of K83, a highly conserved catalytic site residue among restriction enzymes but is replaced by Glu in BamHI or Gln in BglII, appears to be too far removed from the water (4.0 Å) to accept the proton. If the Lys has any role in activating the water molecule, as proposed for many restriction enzymes, deprotonation of K83 would be critical. Because the typical pKa of a free Lys side chain is 10.8, the proper local environment (such as an immediately proximal positive charge or a hydrophobic microenvironment) might lead to a significantly depressed pKa value for K83. In the bacteriophage T5 flap endonuclease, the positively charged metal-ion cofactor lowered the pKa value of the ternary complex to 8.3 (with Mg2+ as a cofactor), 7.0 (Mn2+) or 6.0 (Co2+) (31). As an alternative, the negatively charged pro-Rp oxygen 3′ to the scissile bond might play a substrate-assisted catalytic role by accepting the proton [as proposed for EcoRI and EcoRV (32), MutH (9) and RNase H (33)], although its pKa, conversely, would ordinarily be too low.
Figure 4.
Two-metal mechanism. (A) In the pre-reactive complex, two Ca2+ ions (m1 and m2) are bound in the active site. The dashed lines indicate hydrogen bonds. (B) Structural superimposition of the active site of HinP1I (green) with that of MutH [cyan; PDB 2AOQ (9)]; both contain the DNA substrate and two Ca2+ ions. (C) In the pre-reactive complex, Q81 (a metal coordinator) interaction with the K83 side chain and the K60 main chain. (D) In the post-reactive complex, K83 links Q81 (a metal coordinator), Q93 (a base recognition residue) and the cleaved phosphate group. (E) In the metal-free binary HinP1I–DNA structure, a water molecule occupies the position near the m1 metal site. (F) In the post-reactive complex, two Mg2+ ions (m1 and m2) are bound in the active site. (G) Superimposition of the pre-reactive and post-reactive complexes, with arrows indicating the movements from pre-reactive to the post-reactive states.
Metal m2 is bound by the side chain oxygen atoms of D62 (Oδ1, 2.5 Å) and E18 (Oε1, 2.6 Å), the leaving group O3′ oxygen of 5′ Gua (2.6 Å), the oxygen O1P of the scissile phosphate (2.9 Å) and a water molecule w2 (2.6 Å). The metal-associated water molecule w2 is 3.3 Å away from the 3′-oxygen leaving group and could function as a general acid by protonating the leaving group. The geometries of the two metal ions and their associated water molecules support the two-metal catalytic mechanism (2,34).
A common catalytic site motif among restriction enzymes is characterized as PDXn(D/E)XK, with the consensus residues clustering around the scissile phosphate (2). The corresponding catalytic motif in HinP1I and MspI is (S/T)DX17-18(Q/N)XK (5). The active site of HinP1I is most similar to that of MutH (Figure 4B), all elements involved in the active-site formation are superimposable including the two Ca2+ metal ions, the attacking water molecule, the ion coordination, the scissile phosphate and the 3′ pro-Rp oxygen. One major difference is the replacement of E77 of MutH (EXK) by Q81 of HinP1I (QXK). The Nε2 atom of side chain of Q81 hydrogen bonds to the main chain carbonyl oxygen of K60 (Figure 4C), an interaction that might confer additional stability to the active site. A similar interaction is observed between the side chain of N117 of MspI (NXK) and the main chain carbonyl of K97. However, no corresponding side chain–main chain interaction exists in MutH or in the active sites of REases involving (E/D)XK.
DNA cleavage in solution
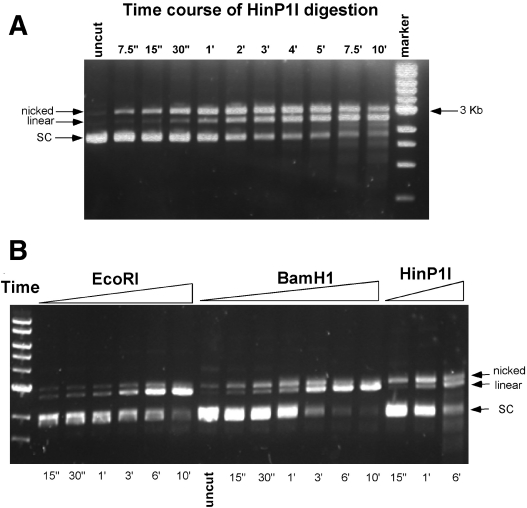
The back-to-back dimer of HinP1I bound to two DNA duplexes (Figure 1), with only one active site for each duplex (Figure 2A), raises the interesting question of whether HinP1I cleaves the two strands of duplex DNA separately. To answer this question, we tried to cleave DNA both in the crystal (see below) and in solution to see whether HinP1I generates a nicked intermediate. We used supercoiled pUC19 plasmid DNA (2.7 kb, with 17 HinP1I sites) as template. Figure 5A shows a time course of digestion with a 1:4 molar ratio of HinP1I to pUC19 DNA. The earliest product was a nicked open circle intermediate, which accumulated before being converted to a linear product, consistent with HinP1I cleaving DNA one strand at a time. In comparison, very little nicked open circle was produced by EcoRI (Figure 5B), while more nicked intermediate was produced by BamHI, an enzyme known to act on the two strands asymmetrically (35). Although HinP1I clearly formed a nicked intermediate, the amount of it seemed to be less, and the appearance of the linear product was earlier, than one would expect from a completely random nicking reaction, considering that pUC19 contains 17 HinP1I sites. This might suggest that the likelihood of strand-hydrolysis at any HinP1I site increases greatly once the other strand is already hydrolyzed. Either HinP1I remains in the vicinity of its site after hydrolysis and can rebind it with high probability, or hydrolysis of the second strand at any site proceeds more rapidly than does hydrolysis of the first.
Figure 5.
Digestion of supercoiled (SC) DNA by HinP1I in solution. (A) Each 5 µl reaction in the New England Biolabs (NEB) buffer 3 (50 mM Tris, pH 7.9, 10 mM MgCl2, 100 mM NaCl and 1 mM DTT) contains 0.16 µg of pUC19 DNA and 1 ng of HinP1I (a 1:4 molar ratio of HinP1I to pUC19 DNA). The reaction was carried out at room temperature for the indicated times. (B) Same as (A), except the NEB enzymes (EcoRI and BamHI) were used.
Metal-free binary complex of HinP1I–DNA
To investigate DNA cleavage in the crystal, we first crystallized the HinP1I–DNA complex in the absence of metals, in the space group P6522 (PDB ID 2FL3). One HinP1I (molecule C, to distinguish it from the A and B molecules in the pre-reactive complex) and one DNA duplex were present in the asymmetric unit. However, the HinP1I monomer interacts with its neighboring protein molecule using the same back-to-back dimer interface via a crystallographic 2-fold symmetry. The binary structure is highly similar to that of the pre-reactive ternary complex. Shown in Figure 4E is a superimposition of the two active sites both bound with DNA, using the protein component only to determine the superimposition (root-mean-square deviation of ∼0.35 Å comparing 247 pairs of Cα atoms between molecules A and C). A single water molecule was found in the active site,occupying a position ∼1.5 Å away from the first metal site m1. The water molecule is coordinated via three strong hydrogen bond interactions (∼2.7 Å) with the side chain oxygen of D62 (Oδ2), the main chain carbonyl oxygen of V82, the oxygen O1P of the scissile phosphate and a weak hydrogen bond interaction (3.3 Å) with the side chain oxygen of E18 (Oε1).
Post-reactive complex of HinP1I: two-metal catalytic mechanism
We soaked pre-formed crystals of the metal-free binary complex of HinP1I–DNA in mother liquors containing Mg2+ ions. Crystals usually cracked or disintegrated. After much trial and error, and careful control of metal concentration, soaking times and the size of crystals (see Materials and Methods), we were able to collect a dataset at 2.6 Å resolution (Table 1) (PDB ID 2FLC).
Remarkably, the DNA is cleaved in the crystal showing that the back-to-back dimer maintains a catalytically active conformation in the crystalline state. DNA cleavage occurs only in one strand and the active site contains two Mg2+ ions (Figure 4F). The overall root-mean-square deviation between the Cα positions of the pre-reactive Ca2+ complex (molecule A) and the post-reactive Mg2+ complex is only ∼0.5 Å. The main difference is the location of the scissile phosphate group and the O3′ leaving group after cleavage: each group is displaced by ∼1.3 Å from the position it occupied when the phosphodiester bond was intact (Figure 4G). Following cleavage, the phosphate group is hydrogen bonded with K83 (3.0 Å) (Figure 4F). The two Mg2+ ions are positioned similarly to the Ca2+ ions although they are slightly closer (3.6 Å as opposed to 4.2 Å). The smaller Mg2+ may shift the two metal sites closer together or a small shift in metal-ion coordination could be associated with activation of the enzyme, as suggested for the two Mg2+ ions in the RNase H–substrate complex (33). Because the scissile phosphate group and the O3′ leaving group are involved in the metal ion coordination both before and after cleavage, the distance between the two metal ions is determined by the configuration of the active-site residues (E, D, Q or N) as well as the substrate.
Mg2+ at the metal m1 position is coordinated by the side chain oxygen atoms of D62 (Oδ2) and Q81 (Oε1), the main chain oxygen atom of V82 and one of the oxygen atoms of the cleaved phosphate group, all within a coordination distance of 1.9–2.1 Å. The coordination of the second Mg2+ at the m2 position is, however, somewhat looser: ranging from 2.0 Å (the side chain oxygen Oδ1 of D62), 2.15 Å (the O3′ leaving oxygen group), 2.3 Å (a water molecule), to ∼2.6 Å (the oxygen atoms of the cleaved phosphate group). E18 is no longer coordinated to the metal at site m2 as in the Ca2+ complex. Overall, it appears that the metal at the m2 site is less tightly bound after the cleavage reaction.
K83 couples metal-ion coordination, sequence-specific recognition and DNA cleavage
The amino group of the side chain of K83, which is unlikely to be involved in the deprotonation of the nucleophilic water, is hydrogen bonded (3.2 Å) with the side chain oxygen Oε1 of Q81 (a metal coordinator) in the pre-reactive complex (Figure 4A). This hydrogen bond, however, is absent in the binary complex, where the amino group is re-oriented and hydrogen bonded (2.8 Å) instead with the carbonyl oxygen of Q93 (data not shown), a base recognition residue (Figure 2D). In the post-reactive complex, the amino group is within hydrogen bonding distance with three groups, the carbonyl oxygen of Q93 (2.85 Å), the side chain oxygen of Q81 (3.25 Å) and the scissile phosphate group (3.0 Å), after cleavage, in addition to its electrostatic interaction (Figure 4D). Thus, it appears that K83 couples metal-ion coordination and DNA cleavage with sequence-specific recognition, a linchpin model proposed recently for the function of the corresponding K79 in MutH (9).
CONCLUSION
HinP1I recognizes DNA in an asymmetric manner, in which a single protein molecule binds a DNA duplex with the palindromic tetranucleotide sequence. The conformationally induced long N-terminal helix binds in the minor groove of the recognition sequence, causing a local A-form conformation, while the conserved catalytic domain containing β-strands and their associated loops lies in the major groove. In the pre- and post-reactive complexes, two metals (Ca2+ or Mg2+) are found in the single active site. The enzyme cleaves first one DNA strand (as seen in the post-reactive complex in the crystalline state) and then the other (in solution, a nicked DNA intermediate is accumulated before linearization). The enzyme's specificity and cleavage apparently involves not only the amino acids that interact directly with base-specific groups and metal ions, but also those residues that control the conformational changes necessary to provide binding complementarity to the distorted DNA including bending by intercalation and base flipping (seen here for the first time for a restriction endonuclease) by transition to a local Z-conformation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Professor Stephen E. Halford FRS for discussion. The study was partly supported by US Public Health Service grants GM068680 and GM49245 (J.R.H., X.Z., Z.Y. and X.C.) and New England Biolabs (R.J.R., R.M. and G.G.W.). Data for this study were measured at the beamline SERCAT-22 of the Advanced Photon Source at Argonne National Laboratory. Financial support for the beamline operation of Emory's shares comes from the Dean's Office of Emory University School of Medicine. Figures were drawn using the program Pymol, a user-sponsored molecular modeling system with an OPEN-SOURCE foundation (http://pymol.sourceforge.net). Atomic coordinates have been deposited in the Protein Data Bank with accession numbers 2FKC and 2FKH (pre-reactive complexes), 2FL3 (binary complex) and 2FLC (post-reactive complex). Funding to pay the Open Access publication charges for this article was provided by New England Biolabs.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Q.S., Kucera R.B., Roberts R.J., Guo H.C. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Shen S., Li Q., Yan P., Zhou B., Ye S., Lu Y., Wang D. Restriction endonucleases from three strains of Haemophilus influenzae. Scientia Sinica. 1980;23:1435–1442. [PubMed] [Google Scholar]
- 5.Yang Z., Horton J.R., Maunus R., Wilson G.G., Roberts R.J., Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong L., Rossmann M.G. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 7.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallog. sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 8.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 9.Lee J.Y., Chang J., Joseph N., Ghirlando R., Rao D.N., Yang W. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Bradley C.M., Ronning D.R., Ghirlando R., Craigie R., Dyda F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nature Struct. Mol. Biol. 2005;12:935–936. doi: 10.1038/nsmb989. [DOI] [PubMed] [Google Scholar]
- 11.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamers M.H., Perrakis A., Enzlin J.H., Winterwerp H.H., de Wind N., Sixma T.K. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 13.Obmolova G., Ban C., Hsieh P., Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 14.Tsutakawa S.E., Jingami H., Morikawa K. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell. 1999;99:615–623. doi: 10.1016/s0092-8674(00)81550-0. [DOI] [PubMed] [Google Scholar]
- 15.Bunting K.A., Roe S.M., Headley A., Brown T., Savva R., Pearl L.H. Crystal structure of the Escherichia coli dcm very-short-patch DNA repair endonuclease bound to its reaction product-site in a DNA superhelix. Nucleic Acids Res. 2003;31:1633–1639. doi: 10.1093/nar/gkg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton N.C., Dorner L.F., Perona J.J. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nature Struct. Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- 17.Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 18.Klimasauskas S., Kumar S., Roberts R.J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 19.Roberts R.J., Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 20.Cheng X., Roberts R.J. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair D.T., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 22.Allan B.W., Reich N.O. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 23.Allan B.W., Beechem J.M., Lindstrom W.M., Reich N.O. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem. 1998;273:2368–2373. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- 24.Holz B., Klimasauskas S., Serva S., Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stivers J.T. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebert K., Hermann A., Schlickenrieder M., Jeltsch A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Ward D.C., Reich E., Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 28.Reddy Y.V., Rao D.N. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H., Jeltsch A. Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 30.Bernards A.S., Miller J.K., Bao K.K., Wong I. Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J. Biol. Chem. 2002;277:20960–20964. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 31.Tock M.R., Frary E., Sayers J.R., Grasby J.A. Dynamic evidence for metal ion catalysis in the reaction mediated by a flap endonuclease. EMBO J. 2003;22:995–1004. doi: 10.1093/emboj/cdg098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA. 1993;90:8499–8503. doi: 10.1073/pnas.90.18.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Horton J.R., Blumenthal R.M., Cheng X. Restriction endonucleases: structure of the conserved catalytic core and the role of metal ions in DNA cleavage. Nucleic Acids and Mol. Biol. 2004;14:361–392. [Google Scholar]
- 35.Vanamee E.S., Viadiu H., Likacs C.M., Aggarwal A.K. Two of a kind: BamHI and BglII. Nucleic Acids and Mol. Biol. 2004;14:215–236. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.