Abstract
Recent developments in single molecule force spectroscopy have allowed investigating the interaction between two redox partners, Azurin and Cytochrome C 551. Azurin has been directly chemisorbed on a gold electrode whereas cytochrome c has been linked to the atomic force microscopy tip by means of a heterobifunctional flexible cross-linker. When recording force-distance cycles, molecular recognition events could be observed, displaying unbinding forces of ∼95 pN for an applied loading rate of 10 nN/s. The specificity of molecular recognition was confirmed by the significant decrease of unbinding probability observed in control block experiments performed adding free azurin solution in the fluid cell. In addition, the complex dissociation kinetics has been here investigated by monitoring the unbinding forces as a function of the loading rate: the thermal off-rate was estimated to be ∼14 s−1, much higher than values commonly estimated for complexes more stable than electron transfer complexes. Results here discussed represent the first studies on molecular recognition between two redox partners by atomic force microscopy.
INTRODUCTION
Over the last decade, redox metalloproteins have gained particular interest for possible applications in the challenging field of molecular bio-nanoelectronics (1–4). Their nanoscale dimension, as well as the variety of exploitable biological functions—such as recognition capability, catalysis, and electron transfer (ET)—may be combined with the progressing power of modern microelectronics for implementation of hybrid nanodevices. The possible integration of metalloproteins with electronic transducers has been recently explored with the aim to “wire them up” in efficient ET chains for biosensing applications at the level of the single molecule (1,5).
Planar gold surfaces appear as attractive solid supports for such applications, thanks to the ability to stably bind various kinds of proteins without affecting their biological activity (5) as well as for enabling both optical and electrical transduction schemes (6,7). Several groups have addressed this issue exploiting the high affinity of protein disulphides and thiols for gold, resulting in efficient conduction through the molecule toward the electrode (8–13).
Once the metalloprotein is successfully coupled with the electrode, a further step toward the applications in nano-biosensing devices may be represented by the recognition of the biomolecule physiological partner. Indeed, intermetalloprotein ET occurs through several steps (14,15) including protein recognition, which implies discriminating between reaction partners and other proteins, and after protein-specific binding with the formation of a (transient) complex (16). Once the specific complex is formed, ET can take place with optimal efficiency; the complex subsequently dissociates to yield the products (14,17).
Even if the ET process between two metalloproteins has been subject of investigation by many groups, most experiments were focused on the study of the free proteins in liquid. In this article we focus on the individual recognition between two redox partners after one of them has been immobilized on a gold electrode. The recognition of a metalloprotein by its redox partner directly linked to a metal electrode may, in principle, be detected as an electric signal, with potential applications for ultrasensitive biosensing at the single-molecule level.
The coupling of the redox protein Azurin (AZ) (from Pseudomonas aeruginosa) to gold electrodes has been extensively studied by our group (9,18). This small metalloprotein (MW = 14.6) bears a single copper ion that switches between Cu(I) and Cu (II) oxidation states during the ET process. Opposite to the active site there is an exposed disulfide moiety (Cys3-Cys26) which is capable to provide a stable binding of the protein to a gold substrate (19,20), resulting in efficient conduction through the molecule toward the electrode (9,11). The retention of AZ redox functionality upon chemisorption on gold has been clearly assessed by differential pulse voltammetry experiments (12) as well as by cyclic voltammetry measurements and electrochemical scanning tunneling microscopy (9,11,18).
P. aeruginosa cytochrome c 551 (C551) is a redox partner of AZ. This small protein (MW = 9) has a heme-iron redox center, the iron switching its oxidation state between +2 and +3 in biological function. Both C551 and AZ are involved in the anaerobic respiratory chain of P. aeruginosa, and several experimental evidences of ET between these two metalloproteins have been provided (21–25). In particular, the observed efficiency of this ET process—in both directions—is consistent with the formation of an ET complex (17).
Even if, to the best of our knowledge, crystallographic evidence of the AZ/C551 complex is not yet available, protein residues involved in the interaction have been explored experimentally by means of site-directed mutagenesis strategy by Cutruzzolà et al. (24): these authors established that the interface between the two partners, during mutual interaction, comprises the hydrophobic patches surrounding residues Val23, Ile59 for C551 and Met44, Met64 for AZ.
A possible configuration of the AZ/C551 complex has been provided by computational docking methods (Brunori, E., A. R. Bizzarri, B. Bonanni, and S. Cannistraro, unpublished). Indeed, starting from the structures of component proteins and applying the docking method known as GRAMM (26,27), we were able to determine a configuration for the AZ/C551 complex which is consistent with experimental data about residues involved in AZ/C551 interaction (24). The best complex provided by our docking procedure (Brunori, E., A. R. Bizzarri, B. Bonanni, and S. Cannistraro, unpublished) has been selected by optimizing a variety of complex characteristic quantities (distance Fe-Cu, accessible surface area, H-bond, etc.) and is shown in Fig. 1.
FIGURE 1.
Schematic representation of best C551/AZ complex as obtained by GRAMM computational docking method (Brunori, E., A. R. Bizzarri, B. Bonanni, and S. Cannistraro, unpublished). The active sites (Cu for AZ, Fe for C551) are represented with small spheres. The disulfide group Cys3-Cys26 of AZ is represented in golden-yellow. Residues involved in protein interaction (Met44 and Met64 for AZ, Val23 and Ile59 for C551, as from Ref. 24) are evidenced as well.
The molecular interaction between AZ and C551 can, in principle, be investigated by atomic force microscopy (AFM). Undoubtedly, AFM has been proven to be an ideal tool for measuring intermolecular forces, thanks to force sensitivity of the order of no more than a few picoNewtons, contact areas as small as 10 nm2, displacement sensitivity of 0.1 nm, and the ability to operate in physiological conditions. By using a measuring tip with ligands bound and recording force versus distance cycles on surface-bound receptors, unbinding forces of ligand-receptor pairs can be probed at the level of single-molecule (28–32). Ligands have been often covalently coupled to the tip via long flexible spacer molecules (33–35). By this linkage method, the ligand on the tip is free to move and orient for unconditioned recognition of the surface-bound receptor, thus favoring complex formation. The characteristic stretching of the spacer (36), preceding complex dissociation, allows us to better discriminate specific unbinding events from unspecific adhesion, with great advantages for the study of low-affinity physiological partners (37).
In this article the molecular interaction between C551 and gold-immobilized AZ has been investigated by force spectroscopy. For optimal interaction between the two partners, proteins have been linked to substrate and tip with proper orientation for mutual interaction. More specifically, AZ has been immobilized on gold via Cys3 and Cys26 residues, thus directing the protein to a configuration in which the hydrophobic patch, involved in interaction with C551 (24), is oriented up (38)—away from the electrode surface—i.e., facing the tip of an AFM (see Fig. 1). Conversely, C551 molecules have not been adsorbed directly on the AFM tip, but by employing a few-nm-long cross-linker that allows protein reorientation over the AZ sample, thus facilitating mutual interaction.
The two redox partners here investigated display a relative low affinity (the estimated value of equilibrium constant for complex dissociation being in the range 10−6–10−4 M; Ref. 22)—that is, lower than values for ligand-receptor pairs commonly studied by force spectroscopy, but still comparable to that for cadherin dimers, which have been successfully investigated by this technique (37). It is also important to keep in mind that the likely transient nature of the ET complex here investigated (16) renders such ligand-receptor pairs very different from more stable complexes commonly studied by force spectroscopy experiments. To the best of our knowledge, this article represents the first study of molecular recognition of two redox partners by means of force spectroscopy. Here, the specificity of molecular recognition has been clearly demonstrated at the level of single molecule even after immobilization on gold of one of the partners. Additionally, by exploring the unbinding force of the complex for different values of loading rate, the dissociation kinetics has been investigated.
MATERIALS AND METHODS
Sample preparation
The AZ was purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification, after dissolving in 50 mM sodium phosphate buffer solution, pH 7. The substrates used in our experiments consist of vacuum-evaporated thin gold films (thickness 250 nm) on borosilicate glass (using the product arrandee, Dr. Dirk Schröer, Werther, Germany). Before incubation with 35 μM AZ solution, the gold substrates have been flame-annealed, to obtain recrystallized Au(111) terraces. The annealed Au(111) substrates were then incubated overnight at 4°C. Samples were gently rinsed with buffer solution to remove any unadsorbed material and immediately immersed in buffer for fluid imaging and force spectroscopy.
Tip functionalization
Cytochrome c 551 from P. aeruginosa (C551) was purchased from Sigma-Aldrich and used without further purification. The cross-linker used to bind C551 to the AFM tip is a heterobifunctional polyethylene-glycol derivative (PEG) which has been synthesized by some of the authors (33). The linker is 10-nm-long and bears an aldehyde (ALD) moiety at one end and n-hydroxy-succinimide (NHS) at the other end. A schematic representation of tip functionalization (with a success rate of ∼40%) is shown in Fig. 2 a. The Si3N4 cantilevers (Veeco Instruments, Santa Barbara, CA) were contact microlevers with backside gold coating and an oxide-sharpened tip. Probes have been first cleaned in three changes of chloroform and then treated with amine groups by overnight incubation with ethanolamine-HCl dissolved in dimethylsulfoxide. Subsequent incubation of tips with PEG solution results in stable binding of the cross-linker NHS-end with the amino-treated tip surface (being that the amine-NHS reaction is faster than the amine aldehyde reaction). PEG solution has been adjusted to ensure low density of cross-linkers on the Si3N4 surface, and therefore single-molecule detection by the tip. Tips functionalized with the PEG were finally incubated with the C551 solution. The cytochrome molecules stably bind to the ALD-end of the flexible PEG via one of its eight lysine residues, homogeneously distributed on the molecule surface (see Fig. 2 b). Before tip incubation with cytochrome, the C551 solution was passed through a Sephadex G-25 desalting column (PD10, Pharmacia, Peapack, NJ), to change the initial ammonium acetate pH 4.7 buffer (which may interfere with final aldehyde-lysine binding) to 50 mM sodium phosphate buffer solution, pH 7. Functionalized tips were stored in buffer solution at 4°C until use. The cantilever spring constant of the functionalized tips (nominal value 0.030 N/m) was experimentally determined by thermal noise analysis (39) with uncertainty of ∼7% on each cantilever. Measured values for used cantilevers ranged from 0.022 to 0.038 N/m.
FIGURE 2.
(a) Schematic representation of the AFM tip functionalized with C551 via the ALD-PEG-NHS cross-linker. (b) Schematic representation of C551, with the heme group in red. Binding of C551 to the ALD-PEG is achieved via one of the eight lysine residues (highlighted in light blue) homogeneously distributed on the molecule surface. Residues Val23 and Ile59, belonging to the protein surface involved in interaction with AZ (as from Ref. 24), are represented as well.
AFM imaging and force measurements
Measurements were carried out by using alternatively a PicoSPM (Molecular Imaging, Phoenix, AZ) or a Nanoscope IIIa/Multimode AFM (Digital Instruments, Santa Barbara, CA).
For the characterization of Au(111) substrates, images were taken in air in contact mode configuration, by using silicon nitride contact microlevers (Veeco Instruments) with backside gold coating and an oxide-sharpened tip. The nominal spring constant was 0.3 N/m.
Imaging of AZ/Au(111) samples was performed in buffer solution by intermittent contact AFM, either by magnetic AC mode (MAC mode by PicoSPM) or by conventional Tapping Mode (TMAFM by Nanoscope IIIa). In this case, the silicon nitride cantilevers (Veeco Instruments) had nominal spring constant of 0.1–0.5 N/m and typical correspondent resonant frequency of 8–30 KHz were used. The amplitude set point was selected to be ∼85% of free amplitude value.
To perform force spectroscopy experiments, the imaging tip was then replaced with the PEG-C551 functionalized tip. With such tips no images were taken, but only force-distance cycles were recorded over the AZ sample in buffer solution (50 mM sodium phosphate, pH 7) at constant lateral position. To limit the force that the tip applied to the surface, as well as to minimize deviations between experiments, a relative trigger of 15–30 nm was used in all deflection-distance curves. Approach and pulling velocity were identical in all experiments and varied from 30 nm/s to 3000 nm/s. All force-distance cycles here presented have been recorded setting the encounter time equal to zero.
RESULTS AND DISCUSSION
The quality of the annealed gold surface was assessed by contact-mode AFM measurements, which showed the presence of atomically flat (111) terraces over hundreds of nanometers, with a typical roughness of ∼0.05–0.1 nm.
The AZ/Au(111) sample was characterized in buffer solution by intermittent contact AFM. A typical MAC-Mode image of such a sample is reported in Fig. 3, which shows a stable and dense layer of molecules over the gold surface. The presence of a stable AZ monolayer on the gold surface after overnight incubation is also assessed by further AFM and scanning tunneling microscopy analysis performed by our group on analogous samples (9,18). In particular, samples prepared following such a protocol display reproducible good-quality scanning tunneling microscopy imaging, even after repetitive scans, indicative of full coverage with molecules stably bound to gold, as for covalent binding (9). The functionality of adsorbed AZ has been demonstrated by cyclic voltammetry experiments, shown elsewhere (9,11). The high concentration of molecules on the substrate, the stable linkage to gold and more importantly the preserved AZ functionality, are characteristics which make the sample suitable for force spectroscopy analysis. In particular, the retention of ET capability is indicative of redox center, and residues close to it (likely involved in interaction with C551; Ref. 24) being not significantly affected by immobilization.
FIGURE 3.
MAC-Mode AFM image of AZ monolayer over Au(111) substrate. The image has been recorded in buffer solution. Scale bar: 100 nm.
The imaging tip was then replaced with the PEG-C551 functionalized tip for force spectroscopy experiments in contact mode. A representative force versus distance cycle recorded with the functionalized tip is plotted in Fig. 4. The retrace curve displays the characteristic stretching of the flexible PEG-linker, which shows a typical nonlinear behavior of the force versus distance retraction curve (36), and following C551-AZ recognition and unbinding. Similar curves have been recorded, at a scan rate of 3000 nm/s, over different sites on the sample surface. Many specific recognition events (characterized by the delayed nonlinear course in the retraction curve) were observed in every lateral position probed with same tip, indicative of reproducible behavior during force-distance cycles, whereas very few adhesion events (characterized by linear retrace force slope) were observed. Reproducibility of unbinding events with the same tip is indicative of molecules not being pulled off the surfaces of the probe or the substrate during the process, since if AZ (or C551) molecules had remained attached to the probe (sample) surface, no repeatability would have been observed. Thousands of curves have been recorded and analyzed to evaluate the unbinding probability as well as the most probable unbinding force and typical unbinding length (see Fig. 4 for their definition). The unbinding force was measured from the pull-off jump of the retraction curve; in the case of multi-event retraces, only the last pull-off jump was considered. Values below the noise level, namely the standard deviation of the flat portion of sampled force-distance curve (for instance, 10 pN for the force-distance curve plotted in Fig. 4), were not taken into account. Statistical distribution of unbinding forces was found to be reproducible when hundreds of curves were recorded over different sites on the sample surface. In Fig. 5 a, the histogram of pull-off forces measured in AFM force-distance curves between C551-PEG tip and AZ substrate (scan rate = 3000 nm/s) is represented. Data come from the analysis of 268 unbinding events over 1500 force-distance cycles, which correspond to a binding probability of ∼18%. From the force histogram we estimate for the most probable unbinding force a value of 74 pN. To ensure that the most probable values are independent of the histogram arrangement, different classes of variable width were compared, and no significant differences in the maxima were found.
FIGURE 4.
Typical force-distance curve recorded with a PEG-C551 functionalized tip over an AZ/Au(111) sample showing a rupture event during retraction. A graphical representation of unbinding length (L) and unbinding force (F) is provided.
FIGURE 5.
(a) Histogram of the unbinding forces as measured from 268 rupture events observed over 1500 force-distance cycles. (b) Unbinding length statistical distribution evaluated for same series of curves providing the force distribution shown in a.
Regarding the binding probability, 18% is a bit lower than binding probabilities usually experienced by other complexes. For instance, many antigen-antibody investigated by force spectroscopy experiments show a binding probability close to 50% (40% for Human Serum Albumin and its antibody, Ref. 34; 60% for single-chain Fv antibody and antigen, Ref. 40; and 50% for γ-glutamyltransferase and anti-γ-glutamyltransferase AFM tip, Ref. 41). On the other hand, a binding probability as low as 15% is also found for some complexes such as P-selectin/ligand complexes (10–15% at a loading rate close to 10 nN/s; Ref. 42), or in some experiments on streptavidin-biotin complexes (25%; Ref. 43), as well as for cadherin dimers (44). In the latter case, the binding probability is found to be ∼10%, even if such a value significantly increases when the tip is allowed to rest on the sample hundreds of milliseconds before retracting. As a matter of fact, a similar increase in the binding probability is also observed for our complex, the frequency of recognition events almost doubling for encounter time varying between 0 and 500 ms, even if in the latter case multiple interactions cannot be ruled out (37). Undoubtedly, the steric hindrance of the binding process, due to the densely packed AZ-monolayer as well as to the low flexibility of the molecules on the gold substrate, is likely to have an effect in lowering binding probability (45) and, on the other hand, in increasing it for longer tip-surface contact times. In this respect, a different immobilization strategy (namely with lower AZ concentration and/or with the introduction of a spacer between AZ and gold) might somehow help in protein recognition. Nevertheless, it is also worth noticing that the relative low binding probability we observe may be a particular aspect of AZ/C551 interaction, since the not-high-specificity is thought to be a characteristic of ET complexes (16).
From the unbinding curves, a distribution of unbinding length was calculated, by evaluating the extension corresponding to the nonlinear portion of the force versus distance retraction curve (see Fig. 4). The resulting length statistical distribution, correspondent to force distribution of Fig. 5 a, is reported in the histogram of Fig. 5 b. The most probable unbinding length is ∼18 nm, a value which well compares to typical values found in other force spectroscopy experiments using similar PEG cross-linkers (34). Moreover, this value is in good agreement with the extension expected for similar PEG upon stretching (36). Such finding provides further evidence that the corresponding pull-off events, selected as specific unbinding events, derive from the stretching of the PEG linker and following protein unbinding (46), whereas a possible unfolding of one of the interacting proteins is unlikely.
To test the reproducibility of the experiment, thousands of force-versus-distance curves were also recorded with different C551-PEG tips over AZ/Au(111) samples (data not shown here). The statistical distributions found for unbinding force and unbinding length provided similar peak values to those shown in Fig. 5, a and b, previously discussed, indicative of the good reproducibility of our results.
As already mentioned, the selection of curves displaying specific unbinding events is facilitated by the introduction of the PEG cross-linker, since its stretching preceding the pull-off and characterized by nonlinear behavior of force versus distance curve allows us to discard aspecific adhesion curves. Nevertheless, to sustain the specificity of rupture events, control experiments were performed to test inhibition of AZ/C551-tip recognition. To this aim, force-distance curves were recorded over the AZ/Au(111) sample using the same C551 functionalized tip which provided the 18% probability of unbinding events, but after the addition of free AZ solution in the fluid cell. To ensure that most of the C551 on the tip was in complex with free AZ, the added AZ solution concentration was 15 μM, that is, comparable to Kd for C551/AZ complex (Kd ≈ 10−6–10−4 M; see Ref. 22). After free AZ addition, the tip and substrate were incubated 30 min in the AZ solution before starting to record force-versus-distance cycles. A corresponding increase of number of curves displaying no rupture events was observed, the unbinding probability progressively changing from the initial average value of 18% down to a final stable value of 5.5%, therefore resulting in 70% overall decrease of number of unbinding events. The significant decrease of binding probability is indicative of blocking recognition between C551 on the tip and the AZ adsorbed on the Au(111) surface, likely due to binding of free AZ to the C551 proteins on the tip (i.e., tip-blocking).
The unbinding force distribution before and during tip-blocking is represented in Fig. 6. The similarity of the two histograms seems to indicate that the observed 5.5% pertaining unbinding events are related to C551/AZ specific recognition not-blocked. The 70% decrease in unbinding events is consistent with choosing blocking agent concentration close to Kd (as also observed for other complexes; Ref. 37), Kd representing the concentration of ligand that—at equilibrium—causes binding to half receptors (47). It is worth noting that the persistence of a 3–6% residual unbinding activity is, however, observed in most blocking experiments, even in very effective blocking conditions (34,41), and that such residual activity is likely to be induced by the forced interaction between the two molecules, due to the experimental setup. Nevertheless, the lack of full blocking may be ascribed, to some extent, also to the transient nature of the complex, which may determine the occurrence of a sort of dynamic exchange of binding between the cytochrome on the tip and the free AZ in solution.
FIGURE 6.
Comparison of unbinding force statistical distribution before (dashed) and after (solid) tip-blocking. After tip-blocking, the total number of unbinding events significantly decreases, corresponding to a final 70% reduction of unbinding probability.
After the tip-blocking experiment, the measuring tip was washed out in copious buffer solution. By recording force-distance cycles in temporal sequence, a progressive recovering in the number of unbinding events was observed, up to a final stable unbinding probability of 17%. Unbinding probability during tip-wash is shown in Fig. 7, together with values before and during tip-blocking (note that reported values refer to statistical analysis of force-distance curve series recorded in temporal sequence). The recovering of unbinding events during the washout is indicative of a reversible blocking mechanism, and reminds us of the weak nature of the ET complex here investigated (16).
FIGURE 7.
Unbinding probability before (shaded) and during (solid) tip-blocking, as well as after tip-washing (dashed shaded line). The reported values refer to a statistical analysis of a force-distance curve series recorded in a temporal sequence.
Additional experiments have been performed to investigate the unbinding kinetics of C551/AZ complex. In principle, from loading rate-dependent measurements of the unbinding force (dynamic force spectroscopy), it is possible to determine the dissociation rate of a bond (48,49). More specifically, the unbinding process of a ligand-receptor pair under the influence of an external loading force can be treated, in terms of the Bell model (50), as a kinetic problem of escape from a potential well: the effect of the external load is then to tilt the interaction potential and facilitate thermally activated escape from the bound state (48,51,52). As a consequence, the dissociation rate constant koff of a bond is amplified by the application of an external force F as
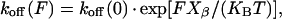 |
(1) |
where koff(0) is the natural thermal off-rate for dissociation in the absence of applied force, KB the Boltzmann's constant, and Xβ has the dimension of a length and can be interpreted as the difference in separation between ligand and receptor in the bound and transition states. Assuming that the bond dissociation is a random process, and in the case of load on complex increasing with constant rate ν (F = ν · t), the most probable unbinding force F* is given by
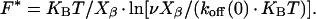 |
(2) |
Therefore by plotting F* versus ν and fitting data using Eq. 2 it is possible to estimate the length scale Xβ (describing the difference in separation of C551 and AZ between the bound and the transition state) and, more importantly, an AFM-measured off-rate koff(0) can be determined. The nominal loading rate (calculated as the product of the cantilever nominal spring constant and the scan rate, i.e., ν = Knominal × scan rate) has been varied between 7 nN/s and 150 nN/s. The value of nominal loading rate is then substituted with the real loading rate, which has been evaluated by taking into account the PEG spacer elasticity (36), namely the cantilever effective spring constant. For every value of applied loading rate, 1000 force-distance cycles have been recorded. For increasing loading rate, we observed a progressive clear upward shift of measured forces, as shown in the histograms of Fig. 8 a corresponding to minimum and maximum values of applied loading rate. The most probable unbinding force F*, resulting from a Gaussian fit to the histogram distribution, has been plotted in Fig. 8 b as a function of the loading rate ν. A nearly linear increase of the unbinding force with the loading rate on the half-logarithmic scale can be observed over almost two orders of magnitude in ν, which suggests that our data may be described in terms of the Bell model, as for rupture forces characteristic for a single energy-barrier in the thermally activated regime (48,50,51). Experimental data have been thus fitted with Eq. 2; from fitting procedure, an estimate of koff (0) and Xβ has been obtained. Best-fitting value for koff in absence of applied force is found to be koff (0) = (14 ± 2) s−1.
FIGURE 8.
(a) Statistical distributions of unbinding force for maximum and minimum values of applied loading rate. At every loading rate, 1000 force-distance curves have been recorded. (b) Loading-rate dependence of the most probable unbinding force F* resulting from a Gaussian fit to the histogram distribution. Force statistical errors are given by standard deviation (2σ/N1/2 for 95.4% confidence level). Error on loading rate is evaluated from uncertainty in the effective spring constant of the cantilever. The solid line is a numerical fit of experimental data to the Bell model (see Eq. 2 in the text). Best-fitting parameters are koff (0) = (14 ± 2) s−1 and Xβ = (0.14 ± 0.01) nm.
It is commonly accepted that ET complexes, such as AZ/C551, have high off-rates (as well as on-rates) with typical koff up to 103 s−1 (whereas kon is the range 107–109 M−1 s−1) (16). Such peculiarity renders the AZ/C551 complex very different from most complexes usually studied by force spectroscopy experiments, which are more stable. As a matter of fact, for the well-known biotin-avidin complex, the typical timescale of the binding process is of the order of months (koff = 9–10−8 s−1 and kon = 7–107 M−1 s−1; see Ref. 53), whereas antigen-antibody complexes have higher off-rates, but are still in the range of 0.01 s−1.
Even if the value of 14 s−1 we estimate is not as high as typical koff expected for ET transient complexes (16), it is, however, much higher than off-rates typically estimated by dynamic force spectroscopy for “stable” ligand-receptor pairs. For these, conversely, dynamic force spectroscopy provided koff (0) values in the range of 10−3–10−1 s−1 (as for antigen-antibody; Ref. 54) or even lower (for the well-known biotin-avidin, koff (0) ∼10−5 s−1; Ref. 43); still, for the low-affinity complex of cadherin dimers a value not larger than 1.8 s−1 has been provided experimentally (37).
The best-fitting value for Xβ, describing the difference in separation of C551 and AZ between the bound and the transition state, is (0.14 ± 0.01) nm—which is comparable to values found for other ligand-receptor pairs.
As widely discussed, the unbinding force may strongly depend on loading rate; thus the comparison of the AZ/C551 unbinding forces with values from literature for other complexes can be made only if considering the data recorded at same loading rate. Therefore, to complete the comparison of data from our experiments with similar studies—available from literature—on other complexes, we focus our attention on a particular value of the real loading rate, namely 10 nN/s. As a matter of fact, many force spectroscopy experiments on several ligand-receptor pairs have been performed applying such loading-rate value. From the plot shown in Fig. 8 b, we can provide an estimate for the most probable unbinding force F* of ∼95 pN when a real loading rate of 10 nN/s is applied. This value can be now compared to unbinding forces of other ligand-receptor pairs, as summarized in Table 1 from literature. From the table, we may conclude that the force we measure for AZ/C551 is in the range of forces typically measured for many ligand-receptor complexes, varying between 20 and 240 pN.
TABLE 1.
Unbinding forces for different complexes, as measured by force spectroscopy experiments at loading rate of 10 nN/s
| Complex | Reference | Force (pN) |
|---|---|---|
| α5β1 integrin ligand/GRGDSP peptide receptor | (55) | 20 ± 7 |
| Cadherin/cadherin | (35,37) | 35 ± 16 |
| Selectin/sialyl Lewis X tetrasaccharide | (56) | 50 ± 15 |
| Lysozyme/anti-lysozyme | (35) | 52 ± 18 |
| Anti-lysozyme Fv fragment/lysozyme | (57) | 55 ± 10 |
| Regulatory protein (His)6ExpG/DNA promoter regions | (58) | 75 ± 15 |
| Biotin/avidin | (59) | 80 ± 15 |
| AZ/C551 | This study | 95 ± 15 |
| ICAM-1 (Intracellular Adhesion Molecule)/anti-ICAM-1 | (35) | 100 ± 50 |
| P-selectin/ligand | (42) | 115 ± 40 |
| Anti-Sendai-antibody/Sendai bacteriorhodopsin | (60) | 126 ± 15 |
| Single-chain Fv fragment of fluorescein binding antibody/fluorescein antigen | (54) | 160 ± 15 |
| HSA (Human Serum Albumin)/anti-HSA | (35) | 244 ± 22 |
CONCLUSION
The study of individual recognition between C551 and gold-immobilized AZ has been, for the first time, accomplished by atomic force spectroscopy and the specificity of this molecular interaction has been clearly demonstrated by control block experiments. The dissociation kinetics of the complex has been explored and an estimate of the complex off-rate has been provided, which (as expected) results much higher than typical values for more stable complexes usually investigated by this technique.
Importantly, the effectiveness of molecular recognition between two redox partners when one of them is directly linked to a metal electrode is a good starting point for possible detection of single recognition events as a recordable electric signal. Our results thus confirm the great potentiality of redox proteins for implementation of ultrasensitive bio-nanodevices designed for biological screening at the single-molecule level.
Acknowledgments
We thank Elena Brunori for generating Fig. 1.
This work has been partially supported by the FIRB-MIUR project “Molecular Nanodevices” and a PRIN-MIUR 2004 project. The work was also supported by Austrian Science Foundation project Nos. P14549 (C.R. and P.H.) and P15295 (H.J.G.).
References
- 1.Gilardi, G., and A. Fantuzzi. 2001. Manipulating redox systems: application to nanotechnology. Trends Biotechnol. 19:468–476. [DOI] [PubMed] [Google Scholar]
- 2.Willner, I., and B. Willner. 2001. Biomaterials integrated with electronic elements: en route to bioelectronics. Trends Biotechnol. 19:222–230. [DOI] [PubMed] [Google Scholar]
- 3.Gorton, L., A. Lindgren, T. Larsson, F. D. Munteanu, T. Ruzgas, and I. Gazaryan. 1999. Direct electron transfer between heme-containing enzymes and electrodes as basis for third generation biosensors. Anal. Chim. Acta. 400:91–108. [Google Scholar]
- 4.Adams, D. M., L. Brus, C. E. D. Chidsey, S. Creager, C. Creutz, C. R. Kagan, P. V. Kamat, X. M. Lieberman, S. Lindsay, R. A. Marcus, R. M. Metzger, M. E. Michel-Beyerle, J. R. Miller, M. D. Newton, D. R. Rolison, O. Sankey, K. S. Schanze, J. Yardley, and X. Zhu. 2003. Charge transfer on the nanoscale: current status. J. Phys. Chem. B. 107:6668–6697. [Google Scholar]
- 5.Willner, I., and E. Katz. 2000. Integration of layered redox-proteins and conductive supports for bioelectronic applications. Angew. Chem. Int. Ed. Engl. 39:1180–1218. [DOI] [PubMed] [Google Scholar]
- 6.Ghindilis, A. L., P. Atansov, M. Wilkins, and E. Wilkins. 1998. Immunosensors: electrochemical sensing and other engineering approaches. Biosens. Bioelectron. 13:113–131. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura, H., and I. Karube. 2003. Current research activity in biosensors. Anal. Bioanal. Chem. 377:446–468. [DOI] [PubMed] [Google Scholar]
- 8.Bonanni, B., D. Alliata, A. R. Bizzarri, and S. Cannistraro. 2003. Topological and electron transfer properties of yeast cytochrome c adsorbed on bare gold electrode. Chem. Phys. Chem. 4:1183–1188. [DOI] [PubMed] [Google Scholar]
- 9.Bonanni, B., D. Alliata, L. Andolfi, A. R. Bizzarri, and S. Cannistraro. 2004. Redox metalloproteins on metal surface as hybrid system for bio-nanodevices: an extensive characterization at the single molecule level. In Surface Science Research Developments. C.P. Norris, editor. Nova Science Publishers, Commack, NY.
- 10.Andolfi, L., B. Bonanni, G. W. Canters, M. P. Verbeet, and S. Cannistraro. 2003. Scanning probe microscopy characterization of gold-chemisorbed poplar plastocyanin mutants. Surf. Sci. 530:181–194. [Google Scholar]
- 11.Andolfi, L., D. Bruce, S. Cannistraro, G. W. Canters, J. J. Davis, H. A. O. Hill, J. Crozier, M. Ph. Verbeet, C. L. Wrathmell, and Y. Aster. 2004. The electrochemical characteristics of blue copper protein monolayers on gold. J. Electroanal. Chem. 565:21–28. [Google Scholar]
- 12.Zhang, J., Q. Chi, J. U. Nielsen, A. G. Hansen, J. E. T. Andersen, H. Wackerbarth, and J. Ulstrup. 2002. Organized monolayers of biological macromolecules on Au(111) surfaces. Russ. J. Electrochem. 38:68–76. [Google Scholar]
- 13.Cavalleri, O., C. Natale, M. E. Stroppolo, A. Relini, E. Cosulich, S. Thea, M. Novi, and A. Gliozzi. 2000. Azurin immobilization on thiol covered Au(111). Phys. Chem. Chem. Phys. 2:4630–4635. [Google Scholar]
- 14.Marcus, R. A., and N. Sutin. 1985. Electron transfer in chemistry and biology. Biochim. Biophys. Acta. 811:265–322. [Google Scholar]
- 15.Bizzarri, A. R., and S. Cannistraro. 2005. Electron transfer in metalloproteins. In Encyclopedia of Condensed Matter Physics. Elsevier, Dordrecht, The Netherlands.
- 16.Crowley, P. B., and M. Ubbink. 2003. Close encounters of the transient kind: protein interactions in the photosynthetic redox chain investigated by NMR spectroscopy. Acc. Chem. Res. 36:723–730. [DOI] [PubMed] [Google Scholar]
- 17.Hervas, M., J. A. Navarro, A. Diaz, H. Bottin, and M. A. De la Rosa. 1995. Laser-flash kinetic analysis of the fast electron transfer from plastocyanin and cytochrome c-6 to photosystem I. Experimental evidence on the evolution of the reaction mechanism. Biochemistry. 34:11321–11326. [DOI] [PubMed] [Google Scholar]
- 18.Facci, P., D. Alliata, and S. Cannistraro. 2001. Potential-induced resonant tunneling through a redox metalloprotein investigated by electrochemical scanning probe microscopy. Ultramicroscopy. 89:291–298. [DOI] [PubMed] [Google Scholar]
- 19.Chi, Q., J. Zhang, J. U. Nielsen, E. P. Friis, I. Chorkendor, G. W. Canters, J. E. T. Andersen, and J. Ulstrup. 2000. Molecular monolayers and interfacial electron transfer of Pseudomonas aeruginosa azurin on Au(111). J. Am. Chem. Soc. 122:4047–4055. [Google Scholar]
- 20.Friis, E. P., J. E. T. Andersen, Y. I. Kharkats, A. M. Kuznestov, R. J. Nichols, J.-D. Zhang, and J. Ulstrup. 1999. An approach to long-range electron transfer mechanisms in metalloproteins: in situ scanning tunneling microscopy with submolecular resolution. Proc. Natl. Acad. Sci. USA. 96:1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zannoni, D. 1989. The respiratory chains of pathogenic pseudomonads. Biochim. Biophys. Acta. 975:299–316. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, M. T., C. Greenwood, M. Brunori, and E. Antonini. 1975. Electron transfer between azurin and cytochrome c-551. Biochem. J. 145:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corin, A. F., R. Bersonhn, and P. E. Cole. 1983. pH Dependence of the reduction-oxidation reaction of azurin with cytochrome c-551: role of histidine-35 of azurin in electron transfer. Biochemistry. 22:2032–2038. [DOI] [PubMed] [Google Scholar]
- 24.Cutruzzolà, F., M. Arese, G. Ranghino, G. van Pouderoyen, G. Canters, and M. Brunori. 2002. Pseudomonas aeruginosa cytochrome C-551: probing the role of the hydrophobic patch in electron transfer. J. Inorg. Biochem. 88:353–361. [DOI] [PubMed] [Google Scholar]
- 25.Rosen, P., and I. Petch. 1976. Conformational equilibria accompanying the electron transfer between cytochrome (P551) and azurin. Biochemistry. 15:775–786. [DOI] [PubMed] [Google Scholar]
- 26.Vajda, S., and C. J. Camacho. 2004. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 22:110–116. [DOI] [PubMed] [Google Scholar]
- 27.Vakser, I. A., and C. Aflalo. 1994. Hydrophobic docking: a proposed enhancement to molecular recognition techniques. Proteins. 20:320–329. [DOI] [PubMed] [Google Scholar]
- 28.Rief, M., and H. Grubmuller. 2002. Force spectroscopy of single biomolecules. Chem. Phys. Chem. 3:255–261. [DOI] [PubMed] [Google Scholar]
- 29.Lee, G. U., D. A. Kidwell, and R. J. Colton. 1994. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir. 10:354–357. [Google Scholar]
- 30.Florin, E.-L., V. T. Moy, and H. E. Gaub. 1994. Adhesion forces between individual ligand-receptor pairs. Science. 264:415–417. [DOI] [PubMed] [Google Scholar]
- 31.Moy, V. T., E.-L. Florin, and H. E. Gaub. 1994. Intermolecular forces and energies between ligands and receptors. Science. 266:257–259. [DOI] [PubMed] [Google Scholar]
- 32.Lo, Y.-S., N. D. Huefner, W. S. Chan, F. Stevens, J. M. Harris, and T. P. Beebe, Jr. 1999. Specific interactions between biotin and avidin studied by atomic force microscopy using the Poisson statistical analysis method. Langmuir. 15:1373–1382. [Google Scholar]
- 33.Haselgrübler, T., A. Amerstorfer, H. Schindler, and H. J. Gruber. 1995. Synthesis and applications of a new poly(ethylene glycol) derivative for the crosslinking of amines with thiols. Bioconjug. Chem. 6:242–248. [DOI] [PubMed] [Google Scholar]
- 34.Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinterdorfer, P., F. Kienberger, A. Raab, H. J. Gruber, W. Baumgartner, G. Kada, C. Riener, S. Wielert-Badt, C. Borken, and H. Schindler. 2000. Poly(ethylene glycol): an ideal spacer for molecular recognition force microscopy/spectroscopy. Single Mol. 1:99–103. [Google Scholar]
- 36.Kienberger, F., V. P. Pastushenko, G. Kada, H. J. Gruber, C. Riener, H. Schindler, and P. Hinterdorfer. 2000. Static and dynamical properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol. 1:123–128. [Google Scholar]
- 37.Baumgartner, W., P. Hinterdorfer, W. Ness, A. Raab, D. Vestweber, H. Schindler, and D. Drenckhahn. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 97:4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nar, H., A. Messerschmidt, R. Huber, M. van de Kamp, and G. W. Canters. 1991. X-ray crystal structure of the two site-specific mutants His35Gln and His35Leu of azurin from Pseudomonas aeruginosa. J. Mol. Biol. 218:427–447. [DOI] [PubMed] [Google Scholar]
- 39.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 40.Ros, R., F. Schwesinger, D. Anselmetti, M. Kubon, R. Schäfer, A. Plückthun, and L. Tiefenauer. 1998. Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. USA. 95:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wielert-Badt, S., P. Hinterdorfer, H. Gruber, J. T. Lin, D. Badt, B. Wimmer, H. Schindler, and R. K.-H. Kinne. 2002. Single molecule recognition of protein binding epitopes in brush border membranes by force microscopy. Biophys. J. 82:2767–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritz, J. A., G. Katopodis, F. Kolbinger, and D. Anselmetti. 1998. Force mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 95:12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan, C., A. Chen, P. Kolb, and V. T. Moy. 2000. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 39:10219–10223. [DOI] [PubMed] [Google Scholar]
- 44.Baumgartner, W., H. J. Gruber, P. Hinterdorfer, and D. Drenckhahn. 2000. Affinity of trans-interacting VE-cadherin determined by atomic force microscopy. Single Mol. 1:119–122. [Google Scholar]
- 45.Eckel, R., R. Ros, B. Decker, J. Mattay, and D. Anselmetti. 2005. Supramolecular chemistry at the single-molecule level. Angew. Chem. Int. Ed. Engl. 44:484–488. [DOI] [PubMed] [Google Scholar]
- 46.Ratto, T. V., K. C. Langry, R. E. Rudd, R. L. Balhorn, M. J. Allen, and M. W. McElfresh. 2004. Force spectroscopy of the double-tethered concanavalin-a mannose bond. Biophys. J. 86:2430–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bongrand, P. 1999. Ligand-receptor interactions. Rep. Prog. Phys. 62:921–968. [Google Scholar]
- 48.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strunz, T., K. Oroszlan, I. Schumakovitch, H.-J. Güntherodt, and M. Hegner. 2000. Model energy landscapes and the force-induced dissociation of ligand-receptor bonds. Biophys. J. 79:1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 51.Evans, E. 1998. Energy landscapes of biomolecular adhesion and receptor anchoring at interfaces explored with dynamic force spectroscopy. Faraday Discuss. 111:1–16. [DOI] [PubMed] [Google Scholar]
- 52.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 53.Green, N. M., and E. J. Toms. 1973. The properties of subunits of avidin coupled to sepharose. Biochem. J. 133:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwesinger, F., R. Ros, T. Strunz, D. Anselmetti, H.-J. Guntherodt, A. Honegger, L. Jermutus, L. Tiefenauer, and A. Pluckthun. 2000. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA. 97:9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kokkoli, E., S. E. Ochsenhirt, and M. Tirrell. 2004. Collective and single-molecule interactions of α5β1 integrins. Langmuir. 20:2397–2404. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, X., D. F. Bogorin, and V. T. Moy. 2004. Molecular basis of the dynamic strength of the sialyl Lewis X-selectin interaction. Chem. Phys. Chem. 5:175–182. [DOI] [PubMed] [Google Scholar]
- 57.Berquand, A., N. Xia, D. G. Castner, B. H. Clare, N. L. Abbott, V. Dupres, Y. Adriaensen, and Y. F. Dufrêne. 2005. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir. 21:5517–5523. [DOI] [PubMed] [Google Scholar]
- 58.Bartels, F. W., B. Baugmarth, D. Anselmetti, R. Ros, and A. Becker. 2003. Specific binding of the regulatory protein ExpG to promoter regions of the galactoglucan biosynthesis gene cluster of Sinorhizobium meliloti—a combined molecular biology and force spectroscopy investigation. J. Struct. Biol. 143:145–152. [DOI] [PubMed] [Google Scholar]
- 59.De Paris, R., T. Strunz, K. Oroszlan, H.-J. Güntherodt, and M. Hegner. 2000. Force spectroscopy and dynamics of the biotin-avidin bond studied by scanning force microscopy. Single Mol. 1:285–290. [Google Scholar]
- 60.Kienberger, F., G. Kada, H. Mueller, and P. Hinterdorfer. 2005. Single molecule studies of antibody-antigen interaction strength versus intra-molecular antigen stability. J. Mol. Biol. 347:597–606. [DOI] [PubMed] [Google Scholar]










