Abstract
Essential isoprenoid compounds are synthesized using the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway in many gram-negative bacteria, some gram-positive bacteria, some apicomplexan parasites, and plant chloroplasts. The alternative mevalonate pathway is found in archaea and eukaryotes, including cytosolic biosynthesis in plants. The existence of orthogonal essential pathways in eukaryotes and bacteria makes the MEP pathway an attractive target for the development of antimicrobial agents. A system is described for identifying mutations in the MEP pathway of Salmonella enterica serovar Typhimurium. Using this system, point mutations induced by diethyl sulfate were found in the all genes of the essential MEP pathway and also in genes involved in uptake of methylerythritol. Curiously, none of the MEP pathway genes could be identified in the same parent strain by transposon mutagenesis, despite extensive searches. The results complement the biochemical and bioinformatic approaches to the elucidation of the genes involved in the MEP pathway and also identify key residues for activity in the enzymes of the pathway.
It was recently discovered that bacteria synthesize isoprenoids by a pathway that differs from that found in eukaryotes. In bacteria, pyruvate and glyceraldehyde 3-phosphate are converted, through the 2-C-methyl-d-erythritol phosphate (MEP) pathway, to isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). This has focused recent research on the elucidation of the biosynthetic steps and genes encoding the catalytic enzymes in bacteria. To date, all synthetic steps of the MEP pathway and the genes encoding the biosynthetic enzymes have been identified. Isoprenoid biosynthesis begins with the condensation of pyruvate and glyceraldehyde 3-phosphate by 1-deoxy-d-xylulose 5-phosphate (DXP) synthase, encoded by dxs (26). Next, in the first committed step of the pathway, DXP reductoisomerase, encoded by dxr, converts DXP to MEP (16, 32). In the following three steps, a cytidyl monophosphate moiety is attached to the phosphate of MEP; the resulting diphosphodiester is phosphorylated at the C-2 hydroxyl in the methylerythritol moiety and then cyclized to form an eight-membered cyclic diphosphate diester, and these steps are catalyzed by the proteins encoded by ispD (25), ispE (18), and ispF (11), respectively. The enzyme encoded by ispG, 1-hydroxy-2-methyl-2-(E)-butenol 4-diphosphate (HDMAPP) synthase, catalyzes the ring opening and reduction of 2-C-methyl-d-erythritol-2,4-cyclodiphosphate to yield HDMAPP (10). Finally, HDMAPP is converted to a mixture of IPP and DMAPP by HDMAPP reductase encoded by ispH (1).
Mutants blocked in the MEP pathway of Salmonella spp. or Escherichia coli are expected to be lethal, since these organisms are unable to utilize exogenously supplied IPP, DMAPP, or their corresponding alcohols. To allow viability of such mutants, genes of the alternative mevalonate pathway were introduced into bacteria, either on plasmids or in the chromosome (2, 5, 9, 15, 24). Strains containing the genes of the mevalonate pathway can synthesize isoprenoids using the eukaryotic pathway when mevalonate is provided. This allows identification of mutants blocked in the alternative bacterial pathway by their requirement for mevalonate. An approach similar to that described here was used recently in an E. coli study (28) and revealed point mutations for all MEP pathway genes except ispH.
The system described here for Salmonella bacteria employs a strain with genes of the MVA pathway of yeast inserted in the bacterial gene dxs, which encodes deoxyxylulose phosphate synthase in the bacterial MEP pathway. This strain uses the MEP pathway when supplied with methylerythritol and the MVA pathway when supplied with MVA. Extensive attempts to isolate insertion mutations in the MEP pathway of Salmonella bacteria by this means were unsuccessful. However, following mutagenesis with diethyl sulfate (DES), mutants were recovered for all steps in the MEP pathway downstream of dxr that were identified by alternative biochemical or bioinformatic methods. Mutants with mutations in the srl operon, which has been implicated in the uptake and phosphorylation of exogenously supplied ME, were also recovered.
The failure of the genetic approach using transposons was surprising, since constructed Salmonella mutations for each of the genes showed the expected phenotype when introduced into the parental strain—an absolute requirement for mevalonate (33). The evidence described here demonstrates the efficacy of the genetic system and raises the question of why transposon mutagenesis failed. This genetic system promises to be a useful adjunct to other methods in further analysis of the bacterial MEP pathway, which remains an attractive target for the design of antibiotic, antimalarial, and herbicidal compounds, as demonstrated by the use of fosmidomycin, a MEP pathway inhibitor used in treatment of bacterial infections and malaria (13).
Structural information about the active sites of the MEP pathway proteins can be an invaluable tool for the rational design of inhibitors. While the crystal structures have been reported for the proteins encoded by dxr (21, 39), ispD (13, 23), ispE (20, 36), and ispF (13, 23, 31), little structural data are available for the other MEP pathway proteins. In addition, the oxygen sensitivity of the iron sulfur proteins encoded by ispG and ispH (30, 38) presents special problems when work is performed with the purified enzymes. In such cases, alternative methods, such as random mutagenesis, are useful for identifying residues in the enzymes essential for function and a method for testing the in vivo efficacy of potential inhibitors.
MATERIALS AND METHODS
Genetic media and methods.
Chloramphenicol (Cam), kanamycin (Kan), tetracycline (Tet), l-arabinose (l-ara), mevalonolactone, and DES were purchased from Sigma. Klentaq-LA polymerase was purchased from Clontech. Luria-Bertani (LB) full medium was used with or without supplementation for all growth conditions (27). E minimal medium was prepared without carbon as described by Vogel and Bonner (35). Cam was used at a final concentration of 20 μg/ml, Kan at 40 μg/ml, and Tet at 30 μg/ml unless otherwise noted. Methylerythritol was synthesized using the method of Duvold (8) and supplemented at a final concentration of 50 μg/ml. l-Arabinose was used at a final concentration of 0.02%. Mevalonic acid was prepared by hydrolysis of 1 volume of 1 M mevalonolactone with 1.02 volumes of 1 M KOH, followed by incubation at 37°C for 30 min, and used at a final concentration of 5 mM.
Transductions were mediated by the high-frequency P22 mutant HT105/1 int-201 as previously described (29). Phage P22 lysates were prepared as previously described (7). All DNA sequencing was performed at the Health Sciences Center Sequencing Facility, Eccles Institute of Human Genetics, University of Utah.
Cloning and sequencing methods.
Genomic DNA was isolated using Easy DNA kits (Invitrogen). PCR was performed using a Perkin-Elmer GeneAmp PCR system 2400 DNA thermal cycler as directed by the polymerase vendor. PCR nucleotide mix was purchased from Roche Molecular Biochemicals. All oligonucleotides were synthesized by the Protein/DNA Core Facility of the Utah Regional Cancer Center. Agarose gel purifications and PCR purifications of DNA were performed using QIAquick gel extraction kits and PCR purification kits (QIAGEN), respectively. Sequence searches were performed on the National Center for Biotechnology Information (NCBI) and Washington University School of Medicine Genome Sequencing Center BLAST servers. Gene sequences were downloaded from NCBI. Sequence alignments were performed using Vector NTI Align-X software. Strains used in this work are listed in Table 1, and oligonucleotides are described in Table 2.
TABLE 1.
Salmonella enterica serovar Typhimurium LT2 strains used in this study
TABLE 2.
Oligonucleotide sequences used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| sAMPygbPB | GCACAGCCACTGCTCGGTAA |
| asAMPygbPB | AGCCGCGTCAACGATGAT |
| sAMPychB | GGTAAAAACTGGAAAGTGG |
| asAMPychB | AAACCCGAATGCGTTAGA |
| sAMPgcpE | TTCGCCAGGCAGATAATC |
| asAMPgcpE | CGCGTCTGACCCTTAATG |
| sAMPlytB | CGGGCATACCGTTCACTTTGA |
| asAMPlytB | CAACGTAGCGTCATCAGGCA |
| sAMPsrlAEB | GAAGCGGCACAAGAGAAT |
| asAMPsrlAEB | CGACATTCGCGGCTTTAT |
| sSEQygbPB1 | CGCAGCAAAATGTGCTCGCCTT |
| sSEQygbPB2 | ACGCCACATCGCCATCGGAA |
| sSEQygbPB3 | GTTCGCCGCGTTTCATGGTG |
| sSEQychB1 | TATGACAGCAAAACGCAGCC |
| sSEQychB2 | TTGGCAATGCGGGCTTTCC |
| aSEQgcpE | TGCCACGTTTATTCTCTTTC |
| asSEQgcpE | GCAGTAACAGACGGGTAA |
| sSEQlytB1 | TGATATTGAAGTGCTGGAAA |
| sSEQlytB2 | GGATGTACCTGGTGGAGTCG |
| sSEQsrlAEB1 | TGATTACAATGAACAGGAAA |
| sSEQsrlAEB2 | GCTGGTAGGTCTGGTGACGA |
| sSEQsrlAEB3 | CCAGCCTGCCAAAACGACAT |
| sSEQsrlAEB4 | TCGGCGTACCTTCTGTGCTG |
Identifying and classifying mutations.
Mutants were generated by exposing stationary-phase cultures of RMC26 to a saturated solution of DES in E medium. A saturated DES solution was prepared by placing two drops of DES in 5 ml of E medium. The solution was mixed and incubated at 37° for 10 min. To the saturated DES solution, 0.1 ml of RMC26 stationary-phase culture was added. After the cells were incubated at 37° for 30 min, 0.1 ml aliquots were used to inoculate 5 ml overnight cultures of LB-MVA-l-ara-Kan medium. The following day, full-cell suspensions were diluted 106-fold, and 0.1 ml of diluted cells was plated to LB-MVA-l-ara-Kan plates. A separate overnight culture was used for each plate in an effort to reduce the isolation of sibling mutants. After 2 days growth at 37°C, colonies were replica printed to LB-MVA-l-ara-Kan and LB-ME-Kan media. Mutants were isolated that demonstrated viability in the presence of MVA-l-ara but not in the presence of ME. Mutants were verified by patching to media supplemented with MVA-l-ara and printing to both MVA-l-ara media and ME-l-ara media.
Identification of isolated mutants.
Strains CR5, CR33, CR34, and CR35 (see Table 1) were constructed from parental strain RMC26 to contain a chloramphenicol acetyltransferase (CAT) cassette in place of individual MEP pathway genes or srlE by using linear recombination as previously described (33). Isolated mutants were used as recipients in P22-mediated transductional crosses with donor strains having known MEP genes or genes within the srl operon replaced by a CAT cassette. The following strains, all with the MVA operon inserted in the chromosomal copy of dxs, were used as donors in the P22 crosses: CR4 for the ispD replacement, CR33 for the ispE replacement, CR34 for the ispG replacement, CR35 for the ispH replacement, and CR5 for the srlE replacement. RMC26 was also used as a donor in a control cross. DES-generated random mutants were used as recipients in the transductional crosses and were plated to LB-ME-Kan media. The number of transductants from each cross was counted. Any recipient point mutant that could not generate recombinants able to use 2-C-methyl-d-erythritol (ME) when crossed with a particular donor mutant was likely to have a mutation in the same gene that was lacking in the donor strain.
Once identified by cross, the mutant allele was PCR amplified from the genomic template by use of KlenTaq LA polymerase mix, followed by sequencing of the amplificate to identify the position of mutation. In cases where the results of complementation by strains with disruptions in MEP pathway genes were ambiguous, the implicated genes were amplified and sequenced. PCRs were performed in duplicate, followed by sequencing in duplicate. Primers used to amplify the target genes were as follows: for ispD and ispF, primers sAMPygbPB and asAMPygbPB; for ispE, primers sAMPychB and asAMPychB; for ispG, primers sAMPgcpE and asAMPgcpE; for ispH, primers sAMPlytB and asAMPlytB; and for srlA, srlB, and srlE, primers sAMPsrlAEB and asAMPsrlAEB. The following sequencing primers were used for each target gene: primers sSEQygbPB1, sSEQygbPB2, and sSEQygbPB3 were used to sequence ispD and ispF amplificates; primers sSEQychB1 and sSEQychB2 were used for ispE amplificates; primers sSEQgcpE and asSEQgcpE were used for ispG; sSEQlytB1 and sSEQlytB2 were used for ispH; and sSEQsrlAEB1-sSEQsrlAEB4 were used for srlA, srlE, and srlB.
A subset of mutant strains that failed to grow on ME as an isoprenoid precursor did not appear to have mutations in any known MEP pathway gene, as determined by the recombination test described above. To test their dependence on the MVA operon, these mutants were used as recipients in transductional crosses with donor DM269 (37), which harbors a Tn10d-Tet insertion in the chromosomal copy of thiI, a thiamine biosynthesis gene in close proximity to dxs. As a control, RMC26, CR4, and CR5 were also used as recipients in the same cross. Recombinants were selected on LB-MVA-l-ara-Tet media. After 2 days growth at 37°C, transductants were replica printed to LB-MVA-l-ara-Kan media and scored for the presence of the MVA operon as indicated by Kan resistance.
RESULTS
Structure of the parental stain RMC26.
Strain RMC26 has a synthetic operon encoding the Saccharomyces cerevisiae genes ERG12, ERG8, and ERG19 (for the conversion of MVA to IPP) inserted into the chromosomal copy of dxs and expressed from an arabinose-inducible promoter (1). This strain has an absolute growth requirement for an early compound of either the yeast MVA pathway or for the bacterial MEP pathway. When either 1-deoxy-d-xylulose or ME is supplied, the bacterial pathway is used. When MVA and the inducer l-ara are provided, the yeast pathway is used. Mutants with disruptions in the MEP pathway were identified by their viability on media supplemented with MVA-l-ara but not on medium with ME.
Isolating mutants in strain RMC26.
Random mutants were generated in Salmonella serovar Typhimurium strain RMC26 by exposure to DES, a DNA-alkylating agent that results in primarily G:C to A:T transitions (12). Approximately 20,000 colonies were screened for mutations that allow growth in the presence of MVA-l-ara but not in the presence of ME. A total of 28 mutants were isolated that grew less well on media containing ME than on media supplemented with MVA-l-ara. Of the total, 23 exhibited a complete lack of growth on ME and were further characterized. The remaining five mutants showed only slightly impaired growth on ME, suggestive of partial loss of functions, and were not pursued.
Characterization of mutations.
To identify the mutated genes, each of the mutants was used as recipient in a transduction cross with individual donor strains bearing CAT replacements in one of the known MEP pathway genes downstream of dxr and srlE (Fig. 1). Selection was made for ability to grow on ME. A failure to yield recombinants indicated that the recipient point mutation affected the gene that was deleted in the donor. The implicated region of the mutant chromosome was amplified from genomic DNA using KlenTaq LA polymerase mix followed by sequencing of the amplificate. Each mutant allele was PCR amplified and sequenced in duplicate to reduce the chance of reporting a mutation introduced by PCR or an error in sequencing. This possibility was further reduced by using KlenTaq LA polymerase mix (3), which contains a small amount of editing 3′ exonuclease which provides proofreading and reduces the probability of introducing mutations by PCR.
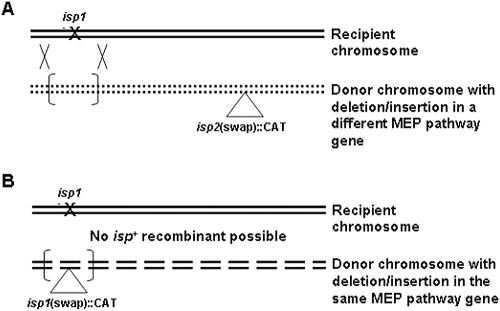
FIG. 1.
Identification of DES-generated mutations by complementation. A) Bracketed region suggests the presence of a 44-kb recombination fragment. A donor chromosome with a mutation in a MEP pathway gene, different (isp2) than the unidentified mutants (isp1), produces a recombination fragment able to repair the unidentified mutation. B) A donor chromosome with a mutation in the same gene as the recipient chromosome does not produce a recombination fragment able to repair the lethal mutation.
All of the sequenced mutations were G:C-to-A:T transitions, as expected by the use of the ethylating agent DES (12). Of the 23 mutants characterized, four independent mutations were found in ispD, one in ispE, two in ispF, four in ispG, one in ispH, and six in the srl operon (Table 3). The remaining six mutants were recombinationally repaired in crosses with any of the donor strains lacking one of the known genes in the MEP pathway, suggesting that their failure to grow on ME was not due to lack of a known MEP enzyme.
TABLE 3.
Mutants of RMC26 unable to utilize ME for the biosynthesis of isoprenoids
| Mutant | Amino acid substitutionc
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ispD | ispE | ispFa | ispG | ispH | srlAb | srlE | None | |
| PM1 | unk | |||||||
| PM3 | R237C | |||||||
| PM5 | R133H | |||||||
| PM6 | G78A | |||||||
| PM7 | G295A | |||||||
| PM8 | unk | |||||||
| PM9 | G281A | |||||||
| PM10 | D38N | |||||||
| PM11 | E196K | |||||||
| PM12 | unk | |||||||
| PM13 | unk | |||||||
| PM14 | W11Stp | |||||||
| PM15 | G20Q | W315Stp | ||||||
| PM16 | A33V | |||||||
| PM17 | G130E | |||||||
| PM18 | Q100Stp | |||||||
| PM19 | G309R | |||||||
| PM21 | C96Y | |||||||
| PM23 | G92Q | |||||||
| PM24 | G239R | |||||||
| PM25 | G206R | |||||||
| PM26 | unk | |||||||
| PM28 | unk | |||||||
Complementation deficiency mapped with deletion insertion in ispD.
Complementation deficiency mapped with deletion insertion in srlE.
Mutations noted are amino acid substitutions. Numbering is consistent with the enzymes from S. typhimurium. unk, not identified; Stp, stop codons.
If these strains carried some novel block in the MEP pathway, they would be expected to require their MVA operon to supply isoprenoids. To test this, the MVA operon was removed from each mutant by a cross with donor strain DM269 (37), which carries a Tn10 insertion near the dxs gene in which the MVA operon was inserted. In all six cases, more than 90% of Tetr transductants lost the MVA operon (and became dsx+), as determined by their Kan sensitivity. This is the same loss frequency observed in crosses between donor DM269 and recipient RMC26, with no additional blocks in the MEP pathway, and recipient CR5, which has the insertion of a CAT cassette in the chromosomal copy of srlE. These dsx+ transductants grew normally with without added MVA or MEP, demonstrating that their de novo MEP pathway is unaffected by any of the six unlinked mutations. When this cross was done using a recipient with a block in MEP (e.g., CR4 which has a CAT insertion in the chromosomal copy of ispD), none of the transductants were Kan sensitive, suggesting the MVA operon is essential when the endogenous MEP pathway is blocked. We conclude that these six mutants have an intact MEP pathway and failed to grow on ME in the original background because they interfere with assimilation of ME (outside the normal MEP pathway).
DISCUSSION
A system is described for isolation of mutants with blocks in the MEP pathway of S. typhimurium. Using this screen, approximately 20,000 DES-mutagenized colonies were tested, and 23 mutants were isolated that exhibited a strong growth defect on media containing ME compared to the results seen with media containing MVA-l-ara. The gene affected by each of the 23 mutations was identified by failure to recombine with deletions of one of the known genes in the MEP pathway, and their base changes were determined by DNA sequencing. The resulting mutations included blocks in each of the genes of the MEP pathway downstream of dxr. In the study of E. coli reported by Sauret-Gueto et al. (28), a screen of ∼27,000 strains yielded no mutations in ispH, when the expected frequency of hits was ∼2.7. Those authors suggested that the region of the E. coli chromosome might be poorly accessible to the mutagen. This now appears unlikely, and the absence of an ispH mutant more likely reflects the small number of mutations isolated. In addition, we isolated several strains bearing mutations in the sorbitol operon, in agreement with previous work suggesting its involvement in the import and phosphorylation of exogenous ME (33).
The results reported here for a chemical mutagen contrast with extensive earlier mutant screens looking for insertional inactivation of MEP pathway genes. These hunts employed a MutJ(Cm) element, a derivative of phage Mu carrying chloramphenicol resistance (6). No insertion mutations were found in any gene of the MEP pathway. The only genes detected by insertions were in the sorbitol phosphotransferase system (PTS) (33). Sufficient insertion mutations (150,000) were screened to assure complete coverage—on the order of 30 hits per gene. At this point we have no explanation for this discrepancy but suggest several possibilities below.
The puzzle posed by failure to identify insertion mutations might reflect any of the following possibilities. (i) The isp genes could be poor targets for the insertion element used. This seems unlikely, since this transposon has been used extensively in other systems and shows very little target specificity. While rare genes might be poor targets, it seems unlikely that all of the scattered genes for one particular pathway would be poor targets. (ii) The chloramphenicol resistance might be unexpressed in strains using the MVA instead of the MEP pathway, or chloramphenicol might interfere with functioning of the MVA pathway. This seems unlikely, since successfully constructed MEP mutants carried a Camr gene very similar to that cloned into the various MEP genes and all these mutants showed simultaneous resistance to chloramphenicol and ability to use MVA. (iii) Intermediates in the MEP or MVA pathways might affect transposition. We have shown that transposition occurs in cells of the parent strain grown on mevalonate both with and without added methylerythritol. Thus, no intermediate in the MVA pathway interferes with transposition and no intermediate in the MEP pathway is required for transposition. Any methylerythritol phosphate accumulated in the parent (dxs) strain does not inhibit transposition. The idea that a particular internal MEP intermediate inhibits transposition does not explain the results, since the mutational block would not be imposed until after transposition had occurred and methylerythritol was provided. (iv) The explanation we favor is that polar insertions in the MEP genes are nonviable. While it seems unlikely that essential genes are located downstream of each MEP gene, this possibility fits our data quite well, based on the evidence summarized below.
The insertion element used is strongly polar, and such polarity has been shown to be sufficient to eliminate expression of downstream genes in other systems. The constructed MEP pathway mutants received a Camr cassette that is known to have outward-directed promoters at both sides and would thus provide for expression of downstream genes. The idea of lethal polar effects is consistent with the rarity of nonsense mutations among the recovered MEP point mutations; such mutations would be expected to show polarity and may also be counterselected. The general idea predicts that all isoprenoid biosynthetic genes are in multigene operons with essential distal genes. This prediction has not yet been tested, but such mixed-function operons are known (17, 19, 34).
Of the genes downstream of dxr in the MEP pathway, crystal structures are reported for the proteins encoded by ispD (14, 22), ispE (20, 36), and ispF (13, 23, 31). Four mutants, defective in ispD, were isolated. PM18 had a nonsense mutation (Q100). The remaining three had missense mutations in PM11 (E196K), PM17 (G130E), and PM23 (G92Q). The crystal structure of E. coli 4-diphosphocytidyl-2-C-methyl-d-erythritol (CDP-ME) synthase, encoded by ispD, suggests that the protein is a homodimer (22). Each subunit is comprised of two structurally distinct domains, a larger core domain, comprised of residues 1 to 136 and 160 to 236, and a smaller lobe domain, comprised of residues 137 to 159. The core domain is globular in structure and includes a distinctive parallel β-sheet motif. The lobe domains of the two subunits resemble “curved arms” which interlock to mediate dimer formation and organize parts of the catalytic site (22). The truncated IspD protein resulting from the nonsense mutation in PM18 lacks both the small lobe domain and a portion of the large globular domain, making dimer formation and catalysis unlikely. The three missense mutations do not appear in amino acids thought to be involved directly in substrate binding or catalysis as predicted by the crystal structure. In addition, none of the three mutated residues are strictly conserved, as determined on the basis of sequence alignments of the proteins from 35 organisms. However, in all three cases a change in polarity of the residue occurs that may translate into altered protein structure. Negatively charged Glu196 is changed to positively charged lysine, while nonpolar Gly92 and Gly130 are mutated to larger polar glutamine and negatively charged glutamate, respectively. Aside from polarity, the protein structure may be compromised by steric interactions in which small buried glycine residues are replaced by the much larger glutamine and glutamate residues.
A single missense mutation was isolated in ispE (G239R). This same mutation was isolated by Sauret-Güeto and coworkers (28) in ethyl methanesulfonate-generated E. coli mutants containing a MVA operon. They identified a strictly conserved motif characterized by the consensus sequence G-[S,T]-G that is critical for enzyme function. The crystal structure of the IspE protein from E. coli implicates Gly239 as an important residue in substrate binding. The Gly239 amide is thought to aid in positioning Asn12 through hydrogen bonding interactions, which allow the residue to in turn hydrogen bond with the O4 hydroxyl group in CDP-ME (20). The substitution of the much larger and charged Arg residue may disrupt the secondary structure of the protein backbone sufficiently to inhibit formation of the stabilizing hydrogen bond between the amide and Asn12, which in turn likely disrupts substrate binding.
Two missense mutations were isolated in the protein encoded by ispF. The crystal structures of E. coli 2-C-methyl-d-erythritol-2,4-cyclodiphosphate synthase show that the protein is a bell-shaped homotrimer (13, 23, 31). Each monomer is comprised of a large and a small β-sheet and four α-helices. The catalytic sites are located at the interfaces of each of the three subunits and are characterized by three distinct binding pockets: a central pocket binds the sugar and diphosphate of the cytidyl moiety; a second pocket binds the nucleoside; and a third pocket binds the 2-phosphate and carbon chain of the 2C-methyl-d-erythritol moiety (31). Both mutations, A33V and D38N, appear to be very near residues involved in substrate binding by the third pocket. The hydrophilic side chains of Ser35, Ser73, and Asp63 are thought to be important for binding and positioning the 2C-methyl-d-erythritol moiety (31). Additionally, His34 is thought to stabilize a flexible loop which caps the active site (31). The pentapeptide sequence starting at His34, including fully conserved residues Ser35 and Asp38, is highly conserved and is positioned along the side of the active site, suggesting a role in substrate binding (13). Though Asp38 is not directly implicated in substrate binding or catalysis by the current crystal structures, its involvement in enzyme activity is not surprising given the close proximity to the binding site and fully conserved nature of the residue. Similarly, the close proximity of Ala33 may suggest that it is important for correct positioning of the conserved pentapeptide within the active site.
Though crystal structure data are not available for the remaining enzymes of the MEP pathway, some structural information can be inferred based on function and homology to enzymes with better characterized motifs. The amino acid sequence of the IspG protein has strong homology to enzymes possessing a [4Fe-4S] cluster (30), and a conserved ferrodoxin motif spanning residues 300 to 319 in E. coli has been identified (38). However, alignments have failed to identify additional conserved motifs that would give an insight into the mechanism of catalysis (2, 4). IspG possesses three fully conserved cysteines, residues 269, 272, and 305 in the S. typhimurium enzyme, which have been suggested to participate in iron binding (10). The current screen identified four missense mutations in ispG: R237C (PM3), R133H (PM5), G309R (PM19), and G206R (PM25). Of these, only G309R is not in a fully conserved residue. However, Gly309 is highly conserved, located within the ferrodoxin motif and near a conserved Cys305. The other three residues, Arg133, Arg237, and Gly206, are fully conserved among bacteria, plants, and the malaria parasite Plasmodium falciparum. The polar-charged nature of Arg133 and Arg237 allows the possibility of involvement in substrate binding or catalysis, while the small size and nonpolar aliphatic nature of Gly206 would suggest a structural role. Nonetheless, identification of these residues by the current screen confirms that they are essential for enzyme function.
A single missense mutation in ispH was isolated. Like IspG, IspH is a [4Fe-4S] protein and possesses three fully conserved cysteine residues at positions 12, 96, and 197 in Salmonella serovar Typhimurium. It has been suggested that these cysteines are involved in iron binding (2). Therefore, it is not surprising that the missense mutation isolated in ispH (C96Y) results in a loss of function phenotype, which may be attributed to the inability of the protein to form the [4Fe-4S] cluster.
Five DES-generated mutants, showing MVA auxotrophy, were isolated in the sorbitol operon, which was previously implicated in the import and phosphorylation of exogenous ME (33). While these mutations are not directly relevant to the function of the MEP pathway, isolation of these mutations supports the current model in which exogenous ME is imported and phosphorylated by the sorbitol phosphoenolpyruvate:phosphotransferase system. While use of an insertion element to generate gene disruptions in previous screens for MVA auxotrophic mutants resulted in the isolation of only srlE, the current screen also identified a random mutation in srlA. Both srlA and srlB had been previously implicated in import and phosphorylation by directed disruption (28). A single mutant, PM15, carried two independent mutations, a missense mutation in srlA (G20Q) and a nonsense mutation in srlE (W315), as determined by sequencing. It is unclear at this time which mutation results in the MVA-dependent phenotype. One additional nonsense mutation was isolated in srlA at W11. Thus, inactivation of the SrlA protein is not surprising given the early translation termination. Three additional point mutations were isolated in srlE, G78A, G281A, and G295A. The substitution of glycine to alanine is conservative. However, two of these glycines, G78 and G295, are fully conserved among sorbitol PTSs, which may explain the loss of function for the similar substitution. It is unclear why G281A causes a loss-of-function phenotype. Although glycine 281 is highly conserved, alanine substitutes for glycine at that position in S. mutans.
Six mutants isolated in the screen failed to use ME but were not completely characterized, because they do not affect function of the MEP pathway. All six were used as recipients in a transductional cross with donor DM269, which has a Tn10 insertion in the thiI gene closely linked to the dxs insertion (containing MVA pathway genes) in the parent strain. Many Tetr recombinants lost the dxs insertion and the MVA genes and regained a functional dxs gene. These transductants still carried the new mutation but were clearly able to use the bacterial MEP pathway. All of the six mutants thus failed to grow on ME in the parent dxs background due to some defect in uptake or phosphorylation of methylerythritol. They have no impairment in the MEP pathway. The behavior of these Dxs+ transductants contrasts with that for the same recombinants obtained using recipient strain CR4, which bears a dysfunctional copy of the MEP pathway gene ispD. We suggest (but have not directly tested) that these six mutations affect the sorbitol PTS, which requires proteins in addition to SrlA, SrlE, and SrlB. The six mutations could thus block uptake of ME without blocking the MEP pathway or affecting the srl operon. Essential genes outside of the known MEP pathway were not identified in this screen or that described by Sauret-Güeto (28). This agrees with biochemical and bioinformatic studies indicating that all genes required for biosynthesis of IPP and DMAPP by MEP pathway have been identified. Both IspG (GcpE) and IspH (LytB) enzymes require regeneration systems such as flavodoxin, flavodoxin reductase, and NADPH for activity in vitro (30). Regeneration systems specific to the [4Fe-4S] proteins, IspG and IspH, have been hypothesized to exist in vivo, but this has not been conclusively established. Genes encoding enzymes for the specific reduction of either IspG or IspH are unlikely to exist. Similarly, screens performed with E. coli failed to isolate strains with mutations in additional genes (28).
Six of the mutants recovered that failed to grow on ME were shown to possess an intact MEP pathway. We suggest that these mutants are likely to interfere with some early step in uptake and metabolism of ME or to make strains sensitive to growth inhibition by ME.
Acknowledgments
The work was supported by National Institutes of Health grants GM25521 (C.D.P.) and GM23408 (J.R.R.). R.M.C. was supported by a chemistry/biology interface predoctoral training grant (GM08537).
REFERENCES
- 1.Altincicek, B., E. C. Duin, A. Reichenberg, R. Hedderich, A. K. Kollas, M. Hintz, S. Wagner, J. Wiesner, E. Beck, and H. Jomaa. 2002. LytB protein catalyzes the terminal step of the 2-C-methyl-d-erythritol-4-phophate pathway of isoprenoid biosynthesis. FEBS Lett. 532:437-440. [DOI] [PubMed] [Google Scholar]
- 2.Altincicek, B., A. K. Kollas, S. Sanderbrand, J. Wiesner, M. Hintz, E. Beck, and H. Jomaa. 2001. GcpE is involved in the 2-C-methyl-d-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol. 183:2411-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, W. M. 1992. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene 112:29-35. [DOI] [PubMed] [Google Scholar]
- 4.Campos, N., M. Rodrigues-Concepcion, M. Seemann, M. Rohmer, and A. Boronat. 2001. Identification of gcpE as a novel gene of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 488:170-173. [DOI] [PubMed] [Google Scholar]
- 5.Campos, N., M. Rodriguez-Concepcion, S. Sauret-Gueto, F. Gallego, L. Lois, and A. Boronat. 2001. Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate: a novel system for the genetic analysis of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem. J. 353:59-67. [PMC free article] [PubMed] [Google Scholar]
- 6.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Duvold, T. P., P. Cali, J. Bravo, and M. Rohmer. 1997. Incorporation of 2-C-methyl-d-erythritol, a putative precursor in the mevalonate-independent pathway, into ubiquinone and menaquinone of Escherichia coli. Tetrahedron Lett. 38:6181-6184. [Google Scholar]
- 9.Hahn, F. M., L. M. Eubanks, C. A. Testa, B. S. J. Blagg, J. A. Baker, and C. D. Poulter. 2001. 1-Deoxy-d-xylulose 5-phosphate synthase, the gene product of open reading frame (ORF) 2816 and 2895 in Rhodobacter capsulatus. J. Bacteriol. 183:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht, S., W. Eisenreich, P. Adam, S. Amslinger, K. Kis, A. Bacher, D. Arigoni, and F. Rohdich. 2001. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA 26:14837-14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herz, S., J. Wungsintaweekul, C. A. Schurh, S. Hecht, H. Luttgen, S. Sagner, M. Fellermeier, W. Eisenreich, M. H. Zenk, A. Bacher, and F. Rohdich. 2000. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate to 2C-methyl-d-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. USA 97:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann, G. R. 1980. Genetic effects of dimethyl sulfate Hoffmann, diethyl sulfate, and related compounds. Mutat. Res. 75:63-129. [DOI] [PubMed] [Google Scholar]
- 13.Kemp, L. E., C. S. Bond, and W. N. Hunter. 2002. Structure of 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthetase: an essential enzyme for iso-prenoid biosynthesis and target for antimicrobial drug development. Proc. Natl. Acad. Sci. USA 99:6591-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemp, L. E., C. S. Bond, and W. N. Hunter. 2001. Crystallization and early X-ray diffraction studies of recombinant Escherichia coli 4-diphosphocytidyl-2-C-methyl-d-erythritol synthetase. Acta Crystallogr. Sect. D Biol. Crystallogr. 57:1189-1191. [DOI] [PubMed] [Google Scholar]
- 15.Kuzuyama, T., M. Takagi, K. Kaneda, H. Watanabe, T. Dairi, and H. Seto. 2000. Formation of 4-(cytidine 5-diphospho)-2-C-methyl-d-erythritol from 2-C-methyl-d-erythritol by 2-C-methyl-d-erythritol 4-phosphate cytidyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett. 41:2925-2928. [Google Scholar]
- 16.Kuzuyama, T., S. Takahashi, H. Watanabe, and H. Seto. 1998. Direct formation of the 2-C-methyl-d-erythritol 4-phosphate from 1-deoxy-d-xylulose 5-phosphate by 1-deoxy-d-xylulose 5-phosphate reductoisomerase, a new enzyme in the non-mevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett. 39:4509-4512. [Google Scholar]
- 17.Lam, H. M., and M. E. Winkler. 1992. Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J. Bacteriol. 174:6033-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luttgen, H., F. Rohdich, S. Herz, J. Wungsintaweekul, S. Hecht, C. A. Schuhr, M. Fellermeier, S. Sagner, M. H. Zenk, A. Bacher, and W. Eisenreich. 2000. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-d-erythritol. Proc. Natl. Acad. Sci. USA 97:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Man, T. K., A. J. Pease, and M. E. Winkler. 1997. Maximization of transcription of the serC (pdxF)-aroA multifunctional operon by antagonistic effects of the cyclic AMP (cAMP) receptor protein-cAMP complex and Lrp global regulators of Escherichia coli K-12. J. Bacteriol. 179:3458-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miallau, L., M. S. Alphey, L. E. Kemp, G. A. Leonard, S. M. McSweeney, S. Hecht, A. Bacher, W. Eisenreich, F. Rohdich, and W. N. Hunter. 2003. Biosynthesis of isoprenoids: crystal structure of 4-diphosphocytidyl-2C-methyl-d-erythritol kinase. Proc. Natl. Acad. Sci. USA 100:9173-9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter, K., S. Sanderbrand, H. Jomaa, J. Wieser, I. Steinbrecher, E. Beck, M. Hintz, G. Klebe, and M. T. Stubbs. 2002. Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis. J. Biol. Chem. 277:5378-5384. [DOI] [PubMed] [Google Scholar]
- 22.Richard, S. B., M. E. Bowman, W. Kwiatkowski, I. Kang, C. Chow, A. M. Lillo, D. E. Cane, and J. P. Noel. 2001. Structure of E. coli 4-diphosphocytidyl-2-C-methyl-d-erythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nat. Struct. Biol. 8:641-648. [DOI] [PubMed] [Google Scholar]
- 23.Richard, S. B., J. L. Ferrer, M. E. Bowman, A. M. Lillo, C. N. Tetzlaff, D. E. Cane, and J. P. Noel. 2002. Structure and mechanism of 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway. J. Biol. Chem. 277:8667-8672. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Concepcion, M., N. Campos, L. M. Lois, C. Maldonado, J. Hoeffler, C. Grosdemange-Billiard, M. Rohmer, and A. Boronat. 2000. Genetic evidence of branching in the isoprenoid pathway for the production of isopentenyl diphosphate and dimethylallyl diphosphate in Escherichia coli. FEBS Lett. 473:328-332. [DOI] [PubMed] [Google Scholar]
- 25.Rohdich, F., J. Wungsintaweekul, M. Fullermeier, S. Sagner, S. Herz, K. Kis, W. Eisenreich, A. Bacher, and M. H. Zenk. 1999. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphoctidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA 96:11758-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohmer, M., M. Seemann, S. Horbach, S. Bringer-Meyer, and H. Sahm. 1996. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoids biosynthesis. J. Am. Chem. Soc. 118:2564-2566. [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sauret-Gueto, S., A. Ramos-Valdivia, E. Ibanez, A. Boronat, and M. Rodriguez-Concepcion. 2003. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem. Biophys. Res. Commun. 307:408-415. [DOI] [PubMed] [Google Scholar]
- 29.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:9-12. [DOI] [PubMed] [Google Scholar]
- 30.Seemann, M., B. T. S. Bui, M. Wolff, D. Tritsch, N. Campos, A. Boronat, A. Marquet, and M. Rohmer. 2002. Isoprenoid biosynthesis through the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) is a [4Fe-4S] protein. Angew. Chem. Int. Ed. Engl. 41:4337-4339. [DOI] [PubMed] [Google Scholar]
- 31.Steinbacher, S., J. Kaiser, J. Wungsintaweekul, S. Hecht, W. Eisenreich, S. Gerhardt, A. Bacher, and F. Rohdich. 2002. Structure of 2C-methyl-d-erythritol-2,4-cyclodiphosphate synthase involved in mevalonate-independent biosynthesis of isoprenoids. J. Mol. Biol. 316:79-88. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, S., T. Kuzuyama, H. Watanabe, and H. Seto. 1998. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95:9879-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Testa, C. A., R. M. Cornish, and C. D. Poulter. 2004. The sorbitol phosphotransferase system is responsible for transport of 2-C-methyl-d-erythritol into Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsui, H. C., G. Feng, and M. E. Winkler. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eσ32-specific promoters during heat shock. J. Bacteriol. 178:5719-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel, H. J., and D. M. Bonner. 1965. Acetyl-ornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 36.Wada, T., T. Kuzuyama, S. Satoh, S. Kuramitsu, S. Yokoyama, S. Unzai, J. R. H. Tame, and, S. Y. Park. 2003. Crystal structure of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. J. Biol. Chem. 278:30022-30027. [DOI] [PubMed] [Google Scholar]
- 37.Webb, E., F. Febres, and D. M. Downs. 1996. Thiamine pyrophosphate (TPP) negatively regulates transcription of some thi genes of Salmonella typhimurium. J. Bacteriol. 178:2533-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff, M., M. Seemann, B. T. S. Bui, Y. Frapart, D. Tritsch, A. G. Estrabot, M. Rodriguez-Concepcion, A. Boronat, A. Marquet, and M. Rohmer. 2003. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett. 541:115-120. [DOI] [PubMed] [Google Scholar]
- 39.Yajima, S., T. Nonaka, T. Kuzuyama, H. Seto, and K. Ohsawa. 2002. Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase complexed with cofactors: implications of a flexible loop movement upon substrate binding. J. Biochem. 131:313-317. [DOI] [PubMed] [Google Scholar]