Abstract
A 15-nucleotide (nt) unstructured RNA with an initiation site but lacking a promoter could direct the initiation of RNA synthesis by the brome mosaic virus (BMV) replicase in vitro. However, BMV RNA with a functional initiation site but a mutated promoter could not initiate RNA synthesis either in vitro or in vivo. To explain these two observations, we hypothesize that RNA structures that cannot function as promoters could prevent RNA synthesis by the BMV RNA replicase. We documented that four different nonpromoter stem-loops can inhibit RNA synthesis from an initiation-competent RNA sequence in vitro. Destabilizing these structures increased RNA synthesis. However, RNA synthesis was restored in full only when a BMV RNA promoter element was added in cis. Competition assays to examine replicase–RNA interactions showed that the structured RNAs have a lower affinity for the replicase than do RNAs lacking stable structures or containing a promoter element. The results characterize another potential mechanism whereby the BMV replicase can specifically recognize BMV RNAs.
Keywords: RNA-dependent RNA polymerase, RNA promoter, repression of RNA synthesis, virus replication
INTRODUCTION
Viruses with genomes of positive-strand RNA include important pathogens of plants, animals, and humans, such as the causative agents of polio and hepatitis C (Buck 1996). The replication of these viruses is essential for virulence. Replication requires specific interaction between sequences within the viral genome and the viral replicase, a complex of viral and cellular proteins. How this specificity is achieved is only beginning to be elucidated.
Brome mosaic virus (BMV) is a model positive-strand RNA virus of plants that is widely used to study viral replication and gene expression (Kao and Sivakumaran 2000). Its genome is composed of three RNAs that share a common sequence at their 3′ 190 nucleotides (nt) that can fold into a tRNA-like structure containing the canonical 3′ CCA sequence (Fig. 1A ▶; Ahlquist et al. 1981; Fechter et al. 2001). During infection, the tRNA-like structure directs minus-strand RNA initiation at the 3′ CCA sequence (the preferred initiation cytidylate is underlined). The minus-strand RNAs then direct genomic plus-strand RNA synthesis. In addition, minus-strand RNA3 can direct the synthesis of a subgenomic plus-strand RNA4 from an internal initiation site. Specific recognition of the tRNA-like structure requires a structure named SLC (Fig. 1A ▶; Dreher and Hall 1988). The high-resolution structure of the terminal stem loop portion of SLC was determined by NMR (Kim et al. 2000) and has a well-defined conformation in which the 5′-most adenine protrudes from the loop and forms a clamped adenine motif (CAM). This adenine is important for RNA synthesis in vitro and in vivo, and can bind the BMV replicase (Chapman and Kao 1999; C.H. Kim and Kao 2001; Rao and Hall 1993).
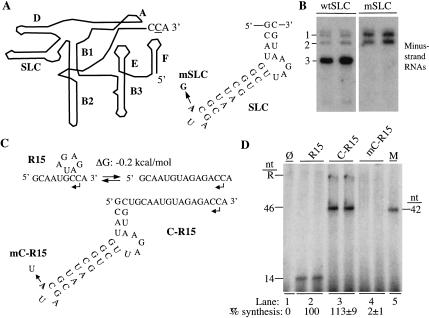
FIGURE 1.
BMV RNA synthesis in vivo and in vitro. (A) Schematic of the BMV 3′ ∼190 nt, which functionally mimics a tRNA-like sequence. The structure is modeled after the one in Fechter et al. (2001). The names of the various stem-loops are in bold letters from A to F, except for SLC. The sequence and secondary structure of SLC is shown to the right of the schematic. The name mSLC denotes an RNA3 with mutation in the CAM. (B) Northern blot result demonstrating that BMV RNA3 with a single-nucleotide change in the CAM of SLC is unable to direct RNA synthesis in transfected barley protoplasts despite having a wild-type initiation CCA sequence. As will be the case for all of the figures, most results will be shown in duplicate, independent reactions to demonstrate reproducibility. Lanes labeled “wtSLC” contain RNA from protoplasts transfected with wild-type transcripts of the three BMV RNAs. Lanes labeled “mSLC” were from protoplasts transfected with wild-type BMV RNA1 and RNA2 and RNA3 with the mSLC mutation. The RNAs were probed to detect minus-strand BMV RNAs. Probing with a probe recognizing cellular 18S rRNA revealed that all lanes contained comparable amounts of RNA (data not shown). The numbers to the left of the autoradiogram identifies the minus-strand RNAs in the Northern blot. (C) Schematics of R15, a relatively unstructured RNA that could potentially exist in two conformations in solution, and C-R15, which contains SLC fused 5′ of R15. mC-R15 contains a single nucleotide change at the CAM. (D) RNA synthesis by the BMV replicase from the templates shown in (C). Qualitative results for all reactions were run in duplicate in the autoradiogram to demonstrate reproducibility of the results. “φ” denotes a reaction performed in the absence of exogenously provided RNA. The products from C-R15 contain both the template-length product (46 nt) and a putative recombinational dimeric product (R) similar to the ones previously observed in M.J. Kim and Kao (2001). The lengths of the products were determined by comparison to the previously characterized products from SLC+8 (Kim et al. 2000) and −20/13 (Siegel et al. 1998). Quantifications of the 14- and 46-nt RNAs synthesized from three independent reactions are shown below the autoradiogram. All quantifications were normalized for the number of radiolabled cytidylates incorporated relative to the control and are from two independent assays.
Despite the apparent requirement for specificity determinants in BMV RNA synthesis, a sequence containing only an initiation site, such as a 5′CCA3′ can direct RNA synthesis in vitro. Deiman et al. (1998) found that disruption of the 5′-stem loop that directs turnip yellow mosaic virus (TYMY) minus-strand RNA synthesis only results in a twofold decrease of transcription efficiency. However, mutations that prevented access to the 3′ two cytidylates that serve as the TYMV initiation site abolished RNA synthesis completely. Yoshinari et al. (2000) confirmed these results and further showed that the replicases from turnip crinkle virus (TCV) phage Qβ can synthesize RNA from an RNA containing 3-nt repeats of the sequence 5′CCA3′. In fact, even the BMV replicase is capable of RNA synthesis from some single-stranded templates less than 20 nt long if the preferred initiation cytidylate is positioned as the penultimate nucleotide from the 3′ terminus (Siegel et al. 1998; Sivakumaran and Kao 1999; Tayon et al. 2001). Altogether, these results suggest that the elements for replicase binding and for the initiation of RNA synthesis can function independently of each other. Consistent with this, RNA synthesis in vitro by the BMV and cucumber mosaic virus (CMV) replicases can be inhibited by specificity elements that lack an initiation site (Siegel et al. 1998; Chapman and Kao 1999; Chen et al. 2000; Sivakumaran et al. 2002).
If the initiation site is sufficient to direct RNA synthesis, one would expect that some cellular RNAs, such as tRNAs with their canonical 3′ CCA sequence, could direct RNA synthesis by the BMV replicase. In fact, tRNATyr can not direct synthesis unless the BMV SLC was fused to it and 2 nt are inserted just 5′ of the CCA sequence (Ranjith-Kumar et al. 2003). The addition of the 2 nt alone is insufficient for RNA synthesis in vitro, and a mutation in the clamp adenine of SLC also decreased RNA synthesis to near background levels (Ranjith-Kumar et al. 2003).
The results showing that specificity determinants are required in longer RNAs but not in minimal length RNAs that, presumably, lack stable structure seemed incongruous. To reconcile both sets of results, we hypothesize that RNA structures that are incapable of serving as promoter elements will inhibit initiation of RNA synthesis by the BMV replicase. This model accounts for the observation that the BMV replicase is incapable of efficient RNA synthesis in vitro when the CAM is mutated (Chapman and Kao 1999; Kim et al. 2000). Nagy et al. (1999) also observed that nonpromoter stem-loops decreased RNA synthesis by the turnip crinkle virus replicase in vitro. Yoshinari et al. (2000) proposed that an RNA secondary structure could prevent initiation, but evidence in support of this model has been heretofore lacking.
In this work, we document that four different nonpromoter stem-loops can inhibit RNA synthesis from an initiation-competent RNA sequence, but that the inhibition is relieved by the presence of SLC. Competition assays indicate that replicase binds the structured RNAs poorly in comparison to RNAs that lack stable structures. Nonpromoter RNA structures can provide a new level of regulation to ensure the specific interaction between the replicase and viral RNA.
RESULTS
RNA secondary structures can inhibit minus-strand RNA synthesis
To demonstrate that an initiation sequence is insufficient to direct RNA synthesis in infected cells, we mutated the clamped adenine in BMV RNA3 to a guanine and transfected this RNA into barley protoplasts along with in vitro transcribed wild-type RNA1 and RNA2. The initiation site is unaffected in this mutant RNA3. Consistent with previous results that targeted portions of the BMV tRNA-like sequence, this mutation reduced minus-strand RNA accumulation to 13% of comparably transfected wild-type BMV RNA (Rao and Hall 1993; Fig. 1B ▶). A change of the clamped adenine to a uridylate reduced RNA synthesis to 10% of wild type (data not shown). These results demonstrate that the BMV initiation sequence is insufficient to direct RNA synthesis in vivo.
To examine the effects of nonpromoter RNA structure on RNA synthesis by the BMV replicase in vitro, we designed a 15-nt RNA (R15) that is predicted by the mfold computer program to form, at most, a 2 base-pair (bp) stem (ΔG: −0.2 kcal/mole) (Jaeger et al. 1989). R15 is derived from the 3′-terminus of BMV RNA3 and includes the CCA sequence at its 3′ terminus (Fig. 1C ▶). R15 directs the synthesis of a 14-nt RNA, suggesting that the penultimate cytidylate is required for initiation (Fig. 1C,D ▶, lane 2). Adding SLC to the 5′ end of R15 in an RNA named C-R15 resulted in 113% of the RNA synthesized by R15 (Fig. 1D ▶, lane 3) while adding a SLC with a mutated CAM (mC-R15) decreased synthesis to background level (Fig. 1D ▶, lane 4). Factors regulating initiation from R15 thus mimics results from protoplasts transfected with BMV RNA3 with a mutated CAM (Fig. 1B ▶) and in vitro characterization of SLC (Kim et al. 2000). Hence, R15 can be used for further analysis of the effects of nonpromoter RNA structure.
Three derivatives of R15 that differ in their predicted RNA secondary structures were made to test the effects of RNA structure on initiation (Fig. 2A ▶). The last five nucleotides of all three RNAs are identical to those in R15. However, R15m1 differed from R15 by having nucleotide changes that are predicted by mfold to prevent the formation of the 2-bp stem (Fig. 2A ▶). R15m2 and R15m3 should form structures that possess, respectively, a 4- and 5-bp stems (Fig. 2A ▶). The predicted ΔGs for R15m2 and R15m3 were −3.4 and −4.8 kcal/mole, respectively. At 25°C, R15 and R15m1 produced comparable amounts of RNA products while R15m2 and R15m3 resulted in synthesis at 4% of R15 (Fig. 2B ▶). These results demonstrate that structures within R15 can decrease its ability to direct RNA synthesis.
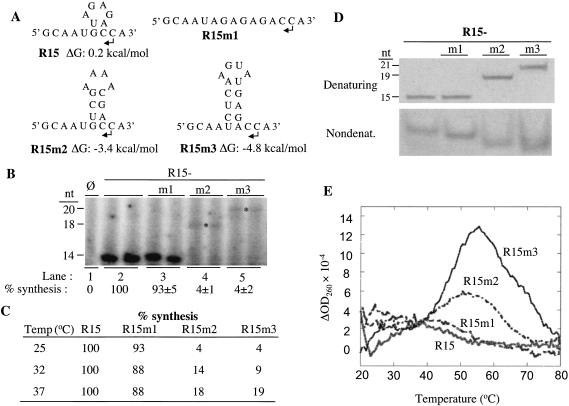
FIGURE 2.
Effects of R15 and its derivatives on RNA synthesis. (A) Sequence and predicted structures of R15 and its derivatives. Bent arrows denote the initiation cytidylates. Names and ΔG are shown below the sequences. (B) Effects of the structures on RNA synthesis by BMV replicase at 25°C. Qualitative results for all reactions are run in duplicate. “φ” denotes a reaction performed in the absence of exogenously provided RNA. Quantifications of at least three independent reactions for each RNA are shown below the autoradiogram. All quantifications were normalized for the number of radiolabled cytidylates incorporated relative to the control and are from two independent assays. The size of the replicase products, in nucleotides, is shown to the left of the autoradiogram. Asterisks are used to aid the identification of the faint bands from R15m1 and R15m2. (C) Quantified results of RNA synthesis assay performed at 25, 32, and 37°C. (D) RNAs visualized after electrophoresis of 40 pmole of RNAs in a 7 M urea PAGE (upper image) and 120 pmole of RNA in a nondenaturing PAGE (lower image). In our hands, RNAs in nondenaturing gels tend to appear as fuzzier bands, likely due to the RNAs retaining a greater degree of conformational flexibility in the absence of urea. (E) RNA thermodenaturation analysis. A first derivative of the absorbance at 260 nm is plotted as a function of temperature. The Tm of R15 and R15m1 are estimated to be 18°C, while the Tms of R15 m2 and R15m3, are 52 and 56°C, respectively.
If structured RNA represses RNA synthesis, higher temperature should decrease the stability of the structures in R15m2 and R15m3 and result in increased RNA synthesis. RNA synthesis reactions were performed with R15 and its derivatives at 32 and 37°C, instead of the usual 25°C (Fig. 2C ▶). At 32°C, RNA synthesis from R15m2 and R15m3 increased to 14 and 9%, relative to that from R15. At 37°C, synthesis from R15m2 and R15m3 increased further to 18 and 19% of the output of R15. The proportional increase of the RNA synthesis with temperature of R15 m2 and R15m3 is consistent with our hypothesis that a structured nonpromoter RNA can decrease RNA synthesis from an otherwise functional initiation site.
We examined the structures, or lack thereof, of R15 and its derivatives by analyzing their mobility in nondenaturing gels and their thermal denaturation profiles. In nondenaturing gels, only one predominant band was observed for all four RNAs, indicating that any structure that forms will be predominantly due to intramolecular interactions (Fig. 2D ▶). Furthermore, the 15-nt R15 and R15m1 migrated more slowly than the 19-nt R15m2 and 21-nt R15m3, indicating that R15 and R15m1 exist in less compact conformations than do R15m2 and R15m3 (Fig. 2D ▶). The thermal denaturation profiles showed that R15 and R15m1 have broad transitions that lack a distinct melting temperature (Tm; Fig. 2E ▶). In contrast, R15m2 and R15m3 have Tms of 52 and 56°C, confirming that they possess some stable structures. The results indicate that even minimally stable RNA secondary structure can decrease promoter-independent RNA synthesis by the BMV replicase.
To confirm that stable RNA structure is inhibitory to initiation, we attached a GNRA tetraloop and stem to the 5′ end of R15 and named the resultant RNA GNRAR15 (Fig. 3A ▶). The tetraloop sequence used is GCAA, which is commonly found in rRNAs (Haus and Pardi 1991). GNRAR15 is predicted by mfold to fold into the expected tetraloop-stem while the R15 portion of the RNA is predicted to lack stable structure (ΔG for GNRAR15 is −14.3 kcal/mole). Consistent with the results with R15 and its derivatives, GNRAR15 was a poor template for RNA synthesis by the BMV replicase, producing only 2% of the product that R15 did (Fig. 3B ▶, lane 3). In an attempt to resuscitate RNA synthesis, a number of changes to the loop were made but were found to have no effect. Among them is a loop sequence consisting of AAAA, resulting in AAAAR15 (Fig. 3A,B ▶, lane 6). We reason that despite having destabilized the interactions in a tetraloop, the RNAs remain sufficiently stable to inhibit RNA synthesis. Therefore, a series of truncations to the stem of AAAA-R15 were made to result in AAAAR15m1 (ΔG of −5.5 kcal/mole), AAAAR15m2 (ΔG of −3.7 kcal/mole), and AAAAR15m3 (ΔG of −1.7 kcal/mole), which are predicted to fold into RNAs with 4-, 3-, and 2-bp stems, respectively (Fig. 3A ▶). The RNA synthesis assay at 25°C showed that AAAAR15, AAAAR15m1, and AAAAR15m2 all produced less than 5% of the synthesis of R15, while AAAAR15m3 had synthesis at 30% (Fig. 3B ▶, lanes 6–9).
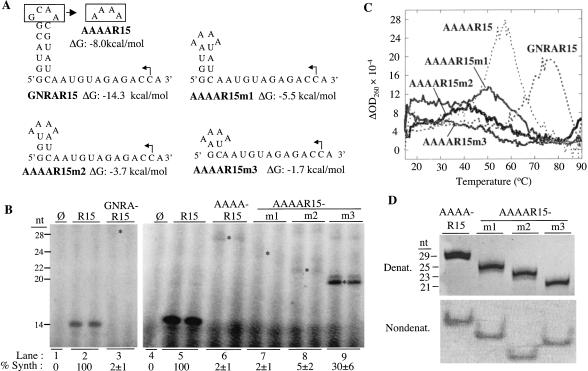
FIGURE 3.
Effects of GNRAR15 and AAAAR15 and its derivatives on RNA synthesis. (A) Sequences and predicted structures of GNRAR15, AAAAR15, and its derivatives. The names and predicted ΔG values for each RNA are shown below the sequences. The arrows show the initiation cytidylates. (B) RNA synthesis directed by GNRAR15, AAAAR15, and its derivatives. The size of the replicase products, in nucleotides, is shown to the left of the autoradiogram. Asterisks denote the expected positions of the 28-, 28-, 24-, 22-, and 20-nt products from GNRAR15, AAAAR15, AAAAR15m1, AAAAR15m2, and AAAAR15m3, respectively. The percentages of synthesis are normalized to number of radiolabeled cytidylates incorporated relative to the control; they are from two replicates. (C) Results from thermodenaturation analysis. A first derivative of the absorbance at 260 nm is plotted as a function of temperature. The Tm of GNRAR15 is 78°C, AAAAR15 is 57°C, AAAAR15m1 is 50°C, AAAAR15m2 is 40°C, and AAAA15m3 is 16°C. (D) RNAs visualized after electrophoresis of 40 pmole of RNA in a 7 M urea PAGE (upper image) and 120 pmole of RNA in a nondenaturing PAGE (lower image).
Thermal denaturation results showed that GNRAR15 had a Tm of ∼78°C. AAAAR15 had a Tm of ∼57°C (Fig. 3C ▶), consistent with our expectation that changes in the loop nucleotides should decrease the stability of the tetraloop. Although they showed broad transitions, suggesting flexibility in their conformations, AAAAR15m1 and m2 had Tms of ∼50 and 40°C, respectively (Fig. 3C ▶). AAAAR15m3, which produced 15-fold more product than did AAAAR15, had a broad transition in its thermal denaturation profile, with the peak being at ∼16°C (Fig. 3C ▶). The relative motilities of these RNAs in a 7.5 M urea PAGE corresponded to the lengths of the RNAs (Fig. 3D ▶, upper panel). However, in a nondenaturing gel, the 21-nt AAAAR15m3 migrated more slowly than did the 23-nt AAAAR15m2 (Fig. 3D ▶, lower panel). These results showed that AAAAR15m3 exists in a different conformation than does AAAAR15 and the other two RNAs. Altogether, these results demonstrate that RNAs with more stable structures are poor templates for RNA synthesis by the BMV replicase until their structures are nearly abolished.
Structured nonpromoter BMV RNA can inhibit RNA synthesis
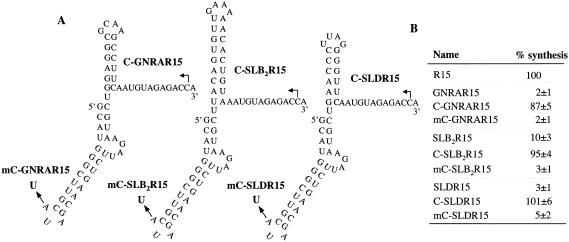
The RNAs tested thus far to inhibit RNA synthesis by the BMV replicase are all nonviral sequences. We wanted to determine whether nonpromoter BMV RNAs could also inhibit RNA synthesis. SLB2 consists of a 9-bp stem and a 1-nt loop from the 3′ terminal tRNA-like sequences of BMV RNAs that are not directly required for minus-strand RNA synthesis (Fig. 1A ▶; Fechter et al. 2001). When SLB2 (with a 9-bp stem) was fused to the 5′ of R15, the resultant RNA directed RNA synthesis at 10% of R15 (SLB2R15; ΔG of −17.0 kcal/mole, Fig. 4B ▶, lane 3). Derivatives of SLB2R15 with predicted stems of 3- (SLB2R15m1), or 2-bp (SLB2R15m2) increased RNA synthesis to 21 and 25%, respectively (Fig. 4B ▶, lanes 4,5). Thermal denaturation experiments showed that the Tms of SLB2R15, SLB2R15m1 and SLB2R15m2 were 60, 35, and 20°C, respectively, consistent with our observation that RNAs with less stable structures are better templates for RNA synthesis (Fig. 4C ▶).
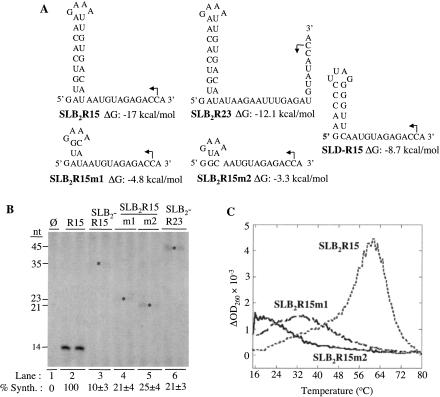
FIGURE 4.
Effects of SLB2R15 and its derivatives on RNA synthesis. (A) Sequences and structures of SLB2R15 and its derivatives. SLB2R23, has a 3′ 23-nt single-stranded region. (B) Results of RNA synthesis using the RNAs shown in (A). Two independent reactions are shown for each RNA tested and the quantified results shown are from these and other independently assayed reactions. Asterisks are used to aid the identification of the faint bands from RNAs containing stable nonpromoter structures. (C) A first derivative of the absorbance at 260 nm is plotted as a function of temperature. Tm of SLB2R15 is 60°C, SLB2R15m1 is 35°C, and SLB2R15m2 is 20°C.
We also tested the effects of SLD, another structure within the BMV 3′ tRNA-like sequence (Fig. 1A ▶; Fechter et al. 2001). SLD consists of a 4-bp stem with a 4-nt loop and is not believed to play a role in directing RNA synthesis (Chapman and Kao 1999). When fused to R15, the resulting RNA named SLDR15 directed 3% of the RNA product of R15 (Fig. 4A ▶; data not shown). Together with the results with SLB2, these results demonstrate that structured nonpromoter RNAs from BMV can inhibit viral RNA synthesis.
Previous research showed that the length of the 3′ single-stranded sequence affects RNA synthesis (Chapman and Kao 1999; Ranjith-Kumar et al. 2003). A 5-nt single-stranded sequence containing an initiation site is required for the initiation of minus-strand RNA synthesis in vitro. We tested whether a 23-nt sequence that was predicted by mfold to lack stable structure could affect RNA synthesis when fused to SLB2 (SLB2R23; Fig. 4A ▶). SLB2R23 produced 21% of the products that R15 did (Fig. 4B ▶, lane 6), indicating that a 23-nt sequence is unable to completely rescue RNA synthesis. However, this represents a twofold higher amount of product than that from SLB2R15, suggesting that a longer single-stranded initiation site may lessen the repressive effect of stable RNA structures. We attempted to design single-stranded sequences longer than 23 nt, but were unable to do so because all were predicted by mfold to contain some secondary structures even at 20°C.
SLC can overcome the inhibitory effects of a stable RNA structure
SLC was able to function at several positions relative to the initiation site (Ranjith-Kumar et al. 2003). We wanted to see whether the addition of SLC could rescue the inhibitory effects of RNA structures. SLC was fused to the 5′ termini of GNRAR15, SLB2R15, and SLDR15 to generate RNAs C-GNRAR15, C-SLB2R15, and C-SLDR15 (Fig. 5A ▶). In addition, SLC, with the clamped adenine changed to a uracil, was fused to the same RNAs to generate mC-GNRAR15, mC-SLB2R15, and mC-SLDR15. The addition of SLC increased RNA synthesis by a minimum of ninefold relative to the RNAs lacking SLC (Fig. 5B ▶), a level of synthesis comparable to that from R15. The CAM is required for this activity because changing it to a uridylate failed to increase RNA synthesis (Fig. 5B ▶).
FIGURE 5.
SLC can overcome the inhibitory effects of stable RNA. (A) Sequences and predicted structures of C-GNRAR15, C-SLB2R15, and C-SLDR15. Arrows show the mutation of the clamped adenine to a uracil. Bent arrows show the initiation sites. (B) RNA synthesis from the RNAs of (A). All quantified results are from more than three independent assays and reflect normalization to the number of radiolabled cytidylates incorporated relative to the R15.
Subgenomic RNA synthesis is also inhibited by an RNA secondary structure
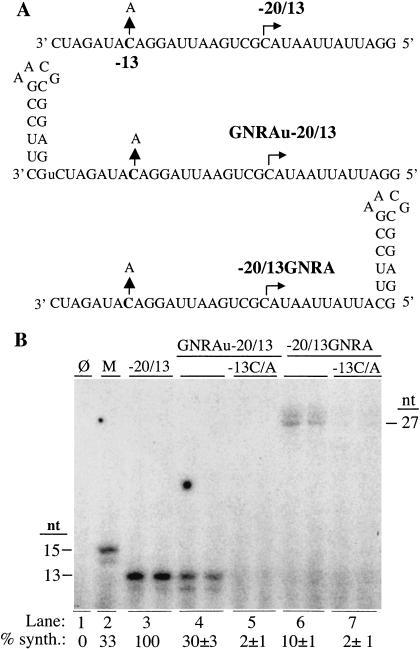
We wanted to determine whether RNA structure could affect the initiation of the BMV subgenomic RNA. Unlike minus-strand RNA synthesis, subgenomic RNA synthesis initiates within an RNA sequence, thus permitting the examination of stable RNA structure either 3′ or 5′ of the initiation site. RNA −20/13 is a well-characterized 33-nt RNA termed a “proscript” because it contains a 20-nt core subgenomic promoter and a 13-nt template (Siegel et al. 1998; Fig. 6A ▶). Within the promoter, nucleotides -17, -14, -13, and -11 are critical for RNA synthesis because substitutions at these positions decreased RNA synthesis to less than 20% (Siegel et al. 1998). We added a GNRA stem-loop along with an extra uridylate 3′ of proscript −20/13 to generate RNA GNRAU-20/13. The uridylate is intended to serve as a spacer to decrease the possibility of perturbing the crucial nucleotides. Additional uridylates 3′ of the subgenomic core promoter is known to not affect the initiation of the 13-nt product (Adkins et al. 1997). In addition, the GNRA stem-loop was added 5′ of −20/13 in RNA −20/13GNRA. Both RNAs directed product synthesis at 30% or less of the amount produced by −20/13, indicating that the tetraloop inhibited subgenomic RNA synthesis (Fig. 6A,B). We note that the effect of the tetraloop was more severe when it was present 5′ of the initiation site (Fig. 6B ▶, cf. lanes 4 and 6). For RNAs GNRAu-20/13 and −20/13GNRA, changing the −13 C to an A further decreased synthesis to 2% (Fig. 6A,B ▶), suggesting that the replicase recognition of the BMV subgenomic core promoter was not altered in these two constructs.
FIGURE 6.
Subgenomic RNA synthesis. (A) Sequences and structures of subgenomic RNAs. −20/13 is a 33-nt RNA containing a 20-nt core subgenomic promoter and a 13-nt template. Arrows show the mutations from the −13 C to an A, which should abolish RNA synthesis (Siegel et al. 1998). The GNRA tetraloop is shown as stems attached to the 3′ or 5′ termini of the subgenomic proscript. An extra uridylate (lower case “u”) is added at the 3′ terminus to provide a one-nucleotide spacer from the core promoter to the tetraloop stem. The prefix or suffix of GNRA indicates whether the tetraloop stem is placed at the 3′ or 5′ of the subgenomic proscript. Bent arrows show the initiation sites for RNA synthesis. (B) Autoradiogram of the RNAs synthesized by the BMV replicase from the templates shown in (A). Sizes of synthesis products are shown beside the figures. Quantified results (below the lanes) are normalized to number of radiolabled cytidylates incorporated relative to the control and are from three independent replicates.
RNA synthesis by the CMV replicase and flaviviral RdRps
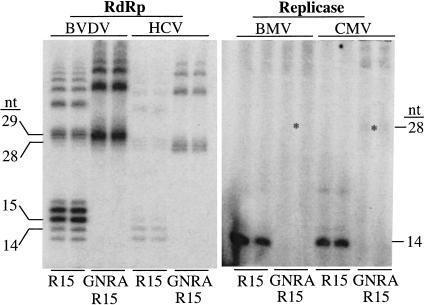
To determine whether the inhibitory effect of the structured nonpromoter RNAs is specific to the BMV replicase, we tested for RNA synthesis from R15 and GNRAR15 with the RdRps from hepatitis C virus (HCV) and bovine viral diarrhea virus (BVDV) that were expressed as recombinant proteins in Escherichia coli (Ranjith-Kumar et al. 2002b), and the CMV replicase (Fig. 7 ▶). The HCV and BVDV RdRps preferred an initiation cytidylate either as the 3′-terminal nucleotide or as the penultimate nucleotide, but they do not have promoter-specificity in vitro (Kao et al. 1999, 2000). Although the HCV and BVDV RdRps produced different amounts of RNA relative to each other, both synthesized comparable levels of RNA from R15 and GNRAR15 (Fig. 7 ▶, left panel), demonstrating that these RNAs are equally capable of directing RNA synthesis, despite the BMV replicase being unable to efficiently use GNRAR15 as a template. However, like the BMV replicase, the CMV replicase was able to direct a robust level of synthesis from only R15, and not from GNRAR15 (Fig. 7 ▶, right panel). Thus, structured nonpromoter RNA had similar effects on the two plant viral replicases, but not the HCV and BVDV RdRps.
FIGURE 7.
RNA synthesis by the CMV replicase and two flaviviral RdRps. The two template RNAs used are R15 and GNRAR15 (see Figs. 2A, 3A ▶ ▶). (Left panel) RNA synthesis assay by BVDV and HCV RdRps. Both RdRps have prominent nontemplated nucleotide addition activity that generates products that appear as a ladder of bands that differ by one nucleotide (Ranjith-Kumar et al. 2001). (Right panel) Synthesis from BMV and CMV replicases. Sizes of synthesis products are shown beside the figures and asterisks are used to aid the identification of the faint bands from GNRAR15.
A structured, nonpromoter RNA does not produce detectable prematurely terminated products
The inhibitory effects of structured nonpromoter RNAs could be either due to: (1) lack of replicase binding; (2) inability of the replicase to initiate RNA synthesis; or (3) premature termination of RNA synthesis due to the BMV replicase being unable to synthesize past the stable stem-loop. Should premature termination occur, we should be able to detect products that are 4 nt or longer, due to the use of CTP as the radiolabeled nucleotide in our reaction and the sequence of GNRAR15 (Fig. 3A ▶). We examined whether prematurely terminated products were made in transcription from GNRAR15. As a control for the sizes of RNAs produced, we used a template with the T7 promoter and a 14-nt template that is identical to the sequence in R15. T7 RNA polymerase transcribed the 14-nt RNA, RNAs 15- and 16-nt long that contain one or two 3′ nontemplated nucleotides, and an abundance of the 4- to 6-nt aborted products, all normal activity of the T7 RNA polymerase (Fig. 8 ▶, lane 5; Milligan et al. 1987). With the BMV replicase R15 was able to direct the synthesis of a 14-nt RNA that initiated from the penultimate cytidylate, because a change of the CCA initiation site to GGA abolished RNA synthesis (Fig. 8 ▶, lanes 2,4). The BMV replicase did not produce detectable amounts of product shorter than 14 nt from GNRAR15, suggesting that the inhibitory effect of structured nonpromoter RNA did not result in a prematurely terminated product; the effects of the RNA structure must therefore be exerted at or before the initiation of RNA synthesis.
FIGURE 8.
GNRAR15 does not cause premature termination of RNA synthesis by the BMV replicase. Products for RNA synthesis were separated on a 25% polyacrylamide–7 M urea gel. The templates for the reaction are shown above the autoradiogram. Sequences for R15 and GNRAR15 were presented in Figures 2A and 3A ▶ ▶. The sequence of the initiation-incompetent RNA, R15init-, is shown above the autoradiogram. “M” denotes products made by the T7 RNA polymerase from a DNA template that should generate identical products to the ones made by the BMV replicase from R15. The sizes of the products from the T7 RNA polymerase are indicated to the right of the autoradiogram. The sizes of the products that could be made by the BMV replicase are indicated to the left of the autoradiogram. The asterisk identifies the faint bands produced by the BMV replicase using GNRAR15 as a template.
Structured RNAs do not interact efficiently with the BMV replicase
To determine whether the structured nonpromoter RNAs can interact with the BMV replicase, we used a template competition assay. The assay compares the relative ability of different RNAs to prevent RNA synthesis by the BMV replicase from a functional reference template (Siegel et al. 1998). The reference RNA template, −1/13, can direct synthesis of a 13-nt RNA product that is easily distinguishable from any possible other products from the competitor RNAs (Tayon et al. 2001). The amount of product generated from −1/13 was quantified in the presence of increasing amounts of competitor RNA, and IC50 values were determined as the concentration of competitor that can reduce synthesis from −1/13 by 50%.
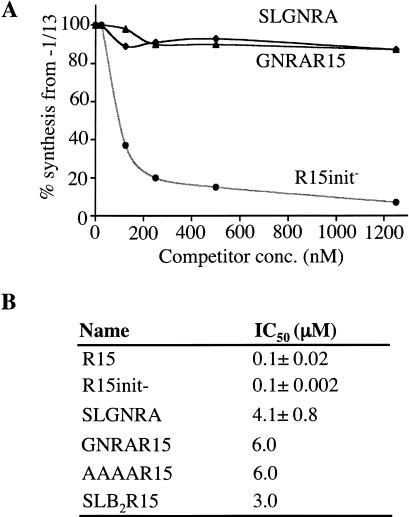
R15 had an IC50 value of 0.1 μM, a value similar to our previous IC50 measurement of another single-stranded RNA (0.2 μM; Siegel et al. 1999; Fig. 9 ▶). In addition, we designed the two competitor RNAs, SLGNRA, consisting of the 6-bp stem and the 4-nt GCAA loop (see Fig. 4A ▶) and R15 init- (Fig. 8 ▶). Both RNAs cannot direct RNA synthesis, and, therefore, will not compete for NTPs with −1/13, the functional template. Based on previous observations, the ability to direct RNA synthesis is not a prerequisite for binding to the replicase (Chapman and Kao 1999; Chen et al. 2000). Indeed, R15init- remains an efficient competitor with an IC50 value of 0.1 μM while SLGNRA had an IC50 of 4.1 μM, a 40-fold increase compared to R15 and R15init- (Fig. 9A,B ▶; data not shown).
FIGURE 9.
Structured RNAs do not interact efficiently with BMV replicase. (A) Quantitative result from competition assays in which the amount of products made from 25 nM of a reference template are plotted against the concentration of the competitor RNAs: SLGNRA, GNRAR15, and R15init-. (B) A summary of the results from several competition assays. Shown are the concentrations (IC50 values) of competitor RNAs needed to reduce RNA synthesis from the reference template to 50%. IC50s of R15init- and SLGNRA were from three independent assays, with the values for one standard deviation shown after the mean. Other values were the average from two independent assays that yielded consistent results.
Additional RNAs that are poor templates for RNA synthesis by the BMV replicase were also tested in the template competition assay. GNRAR15, AAAAR15, and SLB2R15 (Fig. 3A, 4A ▶ ▶) had IC50s of 6, 6, and 3 μM, respectively. We note that in another assay, SLC, the promoter element for minus-strand RNA synthesis had an IC50 of 0.025 μM (Chapman and Kao 1999) and the subgenomic promoter had an IC50 of 0.13 μM (Siegel et al. 1998). Altogether, these results show that relatively unstructured RNAs and promoter elements can interact with the BMV replicase approximately 30- to 60-fold better than structured nonpromoter RNAs.
DISCUSSION
As in transcription from DNA templates, viral RNA-dependent RNA synthesis requires the specific recognition of both the template to be replicated and the nucleotide that directs the initiation of RNA synthesis (Kao 2002). Specific structures/sequences that bind the BMV replicase have been characterized to act as positive regulators specifying the templates to be replicated by the viral replication proteins (Kao 2002). In this work, we document a negative regulator of RNA synthesis.
We found that an RNA containing an initiation sequence can direct promoter-independent RNA synthesis unless a portion of the RNA forms a stable structure such as stem-loops. Destabilizing the RNA structure will allow initiation of RNA synthesis. The presence of structured nonpromoter RNAs in cis of the initiation site decreased RNA synthesis by the BMV replicase. We have determined that SLGNRA cannot inhibit RNA synthesis from SLC+8 in trans, consistent with SLGNRA being unable to bind well to the BMV replicase (data not shown). Furthermore, adding lengths ranging from 3- to 23-nt of single-stranded sequence from the structured nonpromoter RNA did not overcome the inhibition (Figs. 2A,B, 4A,B ▶ ▶), suggesting that the inhibitory effect can be exerted at different distances relative to the initiation site. However, the level of inhibition did depend on the structure of the nonpromoter RNA elements (e.g., Fig. 2B ▶) and, to an extent, on the length of the single-stranded sequence (Fig. 4B ▶). Because RNAs tend to exist in a highly structured state, cellular RNAs should be repressed for replicase-mediated RNA synthesis unless specific promoter elements can overcome the inhibitory effect. This mechanism could allow the BMV replicase to selectively replicate BMV RNAs and not the vast majority of cellular RNAs.
Are the effects of structured nonpromoter RNAs likely to be used by other viral replicases? The effects of the GNRA stem-loop on BMV subgenomic RNA synthesis indicate that the inhibitory effects of structured nonpromoter RNA are not restricted to minus-strand RNA synthesis. Similarly, we demonstrate that the replicase of CMV is also affected by nonpromoter structured RNAs. RNA synthesis by the TCV replicase also appears to be affected when stem-loops with 10-bp stems were fused in cis to a 28-nt functional initiation element from a TCV satellite RNA. Furthermore, increased synthesis was observed when the stem was only 3-bp long (Nagy et al. 1999). These results suggest that a similar mechanism is at work in TCV, although the structures and syntheses by the TCV RNAs were not specifically studied for this effect and their promoter elements increased RNA synthesis more substantially than did the BMV SLC. Presently, this regulation may either not apply to flaviviral RdRps or require proteins in the replicase in addition to the RdRp (Fig. 7 ▶).
How can the replicase distinguish a single-stranded RNA from the same sequence attached to a structured sequence? Because a structured portion prevents efficient recognition of the RNA unless a promoter is also present in cis, we speculate that the structured nonpromoter RNA affects the scanning of the replicase along the RNA in a polarity-dependent manner during genomic RNA synthesis. That is, to recognize an initiation site, the replicase gains access to it by scanning in from the 5′ end of the RNA. Nonpromoter RNA structure would prevent the scanning of the replicase while unstructured RNA would not. The promoter element could overcome the impediment presented by stable RNA structure, perhaps by providing a tethering point to which the replicase remains attached or by permitting access to the RNA by the replicase in a scanning-independent mechanism. Although this model is highly speculative, it provides testable hypotheses for further experimentation. Furthermore, there is precedent for scanning being a mechanism for promoter recognition. The E. coli RNA polymerase has been observed via atomic force microscopy and optical traps, and it slides along nonspecific DNA until it encounters a promoter (Bustamante et al. 1999; Harada et al. 1999). Because viral RNA replication occurs in membrane-associated complexes (den Boon et al. 2001; Lyle et al. 2002), it is perhaps more realistic to envision the RNAs translocating along a relatively more stationary replicase complex.
What is the role of a core promoter for minus-strand BMV and CMV RNA synthesis? One common perception of a viral RNA promoter is that it actively recruits the replicase, and directs the initiation of RNA synthesis. We observed that the presence of a promoter element does not appear to significantly increase the amount of RNA synthesized relative to an unstructured RNA containing an appropriate initiation sequence (113 versus 100%, Fig. 1C ▶). Therefore, it is equally feasible to consider the BMV core promoter element as a factor to overcome the repressive effect of nonspecific secondary structure rather than to increase RNA initiation. Although this difference is partially one of semantics, the distinction can have important mechanistic implications. We note that SLC should be considered a core promoter directing basal levels of RNA synthesis and that additional promoter element(s) may regulate the frequency of initiation.
In summary, transcription from DNA templates is often subject to positive and negative regulation by factors such as enhancers, repressors, and nucleosomes (Weintraub 1984; Myers and Kornberg 2000; Strahl and Allis 2000). RNA-dependent RNA synthesis appears to share many parallels with DNA-dependent RNA synthesis (Adkins et al. 1998; Lai 1998), including potential negative regulation by a feature common to RNAs, their complex structure.
MATERIALS AND METHODS
Preparation of transcripts
RNAs were produced by transcription using the T7 RNA polymerase as described in Milligan et al. (1987). Following transcription, the RNAs were electrophoresed in a denaturing PAGE, and the bands of correct molecular mass were excised with a razor. The gel bands were then crushed to elute the RNA in a solution of 0.3 M ammonium acetate. Concentrations of RNA were determined by spectrometry and staining with Toluidine blue following denaturing PAGE.
RNA synthesis assay
BMV replicase was prepared from BMV-infected barley as previously described (Sun et al. 1996), and the CMV replicase was prepared as described in Sivakumaran et al. (2000). Replicase activity assays were carried out as described by Adkins et al. (1997). Each assay consisted of a 40-μL reaction containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP, 500 μM GTP, 200 μM UTP, 242 nM [α-32P]CTP (400 Ci/mmole, 10 mCi/mL [Amersham]), the desired amount of template and 8 μL of replicase preparation. The BVDV and HCV RdRps were purified from recombinant E. coli, and their use in RNA synthesis assays was as described by Ranjith-Kumar et al. (2002a). Following incubation for 60 min at 25°C, the reaction products were extracted with phenol/chloroform (1:1, vol/vol) and precipitated with six volumes of ethanol, 10 μg of glycogen, and a 0.4-M final concentration of ammonium acetate. Loading buffer (45% deionized formamide, 1.5% glycerol, 0.04% Bromophenol blue, and 0.04% xylene cyanol) was added to the products, and the samples were denatured by heating to 90°C for 3 to 5 min prior to electrophoresis on 20% acrylamide–7 M urea gels (unless noted otherwise). All gels were exposed to film at −80°C, and the amount of label incorporated into newly synthesized RNAs was quantified with a Phosphorimager (Molecular Dynamics).
Nondenaturing gel analysis
RNAs used for conformation analysis via nondenaturing gel electrophoresis were first heated to 90°C for 2 min in buffer A (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 0.1 mM EDTA) and placed on ice for 10 min. The samples were then adjusted to contain 0.5× TBE, 5% (v/v) glycerol, and bromophenol blue. Nondenaturing gel electrophoresis was performed in 15% polyacrylamide gels containing 0.5× TBE (Maniatis et al. 1982). After electrophoresis for 20 min without sample, 120 pmole samples of RNA were loaded. Electrophoresis was performed at a constant 200 volts until bromophenol blue migrated to the proper position according to the sizes of the samples. The gel was then stained with Toluidine blue to visualize the RNAs.
RNA UV melting analysis
The protocol used was as described by Puglisi and Tinoco (1989) using a Perkin-Elmer Lamdba 35 UV/VIS spectrometer with a PTP6 Peltier Temperature Programmer. Each RNA was dissolved in the same buffer (10 mM Tris, 25 mM NaCl, pH 7.0). Right before the melting experiments, each RNA was heated to 85°C and then cooled on ice to encourage proper RNA folding. The concentration of each sample was determined by calculations using the Beer-Lambert Law with a calculated extinction coefficient for each molecule. After adjusting the absorbance of the samples to between 0.1 and 0.4 at room temperature, data were collected with a rate of change in temperature of 0.5°C/min. Every experiment was repeated at least twice. The raw data were analyzed and plotted using a Kaleidagraph software package. The Tm for each molecule was calculated using a derivative of the raw data.
Acknowledgments
This work would not be possible without the generosity and guidance of Professor Ignacio Tinoco, Jr., in whose lab C.-H. Kim is a postdoctoral scientist. We thank C. Ranjith-Kumar and L. Kao for editing the manuscript. Funding was provided by a National Institute of Health Grant and Department of Energy Grant to I.T. and a National Science Foundation grant to C.K. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2190803.
REFERENCES
- Adkins, S., Siegel, R., Sun, J.H., and Kao, C.C. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3: 634–647. [PMC free article] [PubMed] [Google Scholar]
- Adkins, S., Stawicki, S., Faurote, G., Siegel, R.W., and Kao, C. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4: 455–470. [PMC free article] [PubMed] [Google Scholar]
- Ahlquist, P., Dasgupta, R., and Kaesberg, P. 1981. Near identity of 3′ RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell 23: 183–189. [DOI] [PubMed] [Google Scholar]
- Buck, K.W. 1996. Comparision of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 47: 159–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C., Guthold, M., Zhu, X., and Yang, G. 1999. Facilitated target location on DNA by individual Escherichia coli RNA polymerase molecules observed with the scanning force microscopy operating in liquid. J. Biol. Chem. 274: 16665–16668. [DOI] [PubMed] [Google Scholar]
- Chapman, M. and Kao, C.C. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286: 709–720. [DOI] [PubMed] [Google Scholar]
- Chen, M.H., Roossinck, M.J., and Kao, C.C. 2000. Efficient and specific initiation of subgenomic RNA synthesis by cucumber mosaic virus replicase in vitro requires an upstream RNA stem-loop. J. Virol. 74: 11202–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon, J.A., Chen, J., and Ahlquist, P. 2001. Identification of sequences in brome mosaic virus replication protein 1a that mediates association with endoplasmic reticulum membranes. J. Virol. 75: 12370–12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiman, B.A.L.M., Koenen, A.K., Verlaan, P.W.G., and Pleij, C.W.A. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 72: 3965–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, T.W. and Hall, T.C. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus-strand promoter activity. J. Mol. Biol. 201: 31–40. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Giegé, R., and Rudinger-Thirion, J. 2001. Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J. Mol. Biol. 309: 387–399. [DOI] [PubMed] [Google Scholar]
- Hans, A.H. and Pardi, A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194. [DOI] [PubMed] [Google Scholar]
- Harada, Y., Funatsu, T., Murakami, K., Nonoyama, Y., Ishihama, A., and Yanagida, T. 1999. Single molecule imaging of RNA polymerase–DNA interactions in real time. Biophys. J. 76: 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, J.A., Turner, D.H., and Zuker, M. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. 86: 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.C. 2002. Lessons learned from the core RNA promoters of Brome Mosaic Virus and Cucumber Mosaic Virus. Mol. Plant Pathol. 3: 53–59. [DOI] [PubMed] [Google Scholar]
- Kao, C.C. and Sivakumaran, K. 2000. Brome mosaic virus, good for an RNA virologists basic needs. Mol. Plant Pathol. 1: 91–98. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Del Vecchio, A., and Zhong, W. 1999. De novo initiation of RNA synthesis by a Flaviviridae RNA-dependent RNA polymerase. Virology 253: 1–7. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Yang, X., Kline, A., Wang, Q.M., Barket, D., and Heinz, B.A. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74: 11121–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H. and Kao, C.C. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA 7: 1476–1485. [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H., Kao, C.C., and Tinoco, I. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7: 415–423. [DOI] [PubMed] [Google Scholar]
- Kim, M.J. and Kao, C. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases. Proc. Natl. Acad. Sci. 98: 4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.M.C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription. Virology 244: 1–12. [DOI] [PubMed] [Google Scholar]
- Lyle, J.M., Bullitt, E., Bienz, K., and Kirkegaard, K. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296: 2218–2222. [DOI] [PubMed] [Google Scholar]
- Maniatis, S.T., Fritsch, E.F., and Sambrook, J. 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, L.C. and Kornberg, R.D. 2000. Mediators of transcriptional regulation. Annu. Rev. Biochem. 69: 729–749. [DOI] [PubMed] [Google Scholar]
- Nagy, P.D., Pogany, J., and Simon, A.E. 1999. RNA elements required for RNA recombination function as replication enhancer in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18: 5653–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi, J.D. and Tinoco, I. 1989. Absorbance melting curves of RNA. Methods Enzymol. 180: 304–325. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Gajewski, J., Maley, D., Sarisky, R.T., and Kao, C.C. 2001. Terminal nucleotidyl transferase activity of recombinant flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA synthesis. J. Virol. 75: 8615–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Gutshall, L., Kim, M.-J., Sarisky, R.T., and Kao, C.C. 2002a. Requirements for de novo initiation of viral RNA synthesis by recombinant Flaviviral RNA-dependent RNA polymerases. J. Virol. 76: 12526–12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Kim, Y.C., Gutshall, L., Silverman, C., Khandekar, S., Sarisky, R., and Kao, C.C. 2002b. Mechanism of de novo initiation of RNA synthesis by the hepatitis C virus RNA-dependent RNA polymerase: Role of divalent metals. J. Virol. 76: 12513–12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Zhang, X., and Kao, C.C. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77: 1830–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L.N. and Hall, T.C. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.W., Bellon, L., Beigelman, L., and Kao, C.C. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. 95: 11613–11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999. Use of DNA, RNA, and chimeric templates by a viral RNA-dependent RNA polymerase: Evolutionary implications for the transition from the RNA to the DNA world. J. Virol. 73: 6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K. and Kao, C.C. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 73: 6415–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K., Bao, Y., Roossinck, M.J., and Kao, C.C. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicase of brome mosaic virus and cucumber mosaic virus. J. Virol. 74: 10323–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K., Chen, M.H., Roossnick, M.J., and Kao, C.C. 2002. Minimal core promoter for correct and efficient initiation of cucumber mosaic virus subgenomic RNA4A. Mol. Plant Pathol. 3: 43–54. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Sun, J.H., Adkins, S., Faurote, G., and Kao, C.C. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226: 1–12. [DOI] [PubMed] [Google Scholar]
- Tayon, R., Kim, M.J., and Kao, C.C. 2001. Completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 29: 3576–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub, H. 1984. Histone-dependent chromatin superstructures and the suppression of gene activity. Cell 38: 17–27. [DOI] [PubMed] [Google Scholar]
- Yoshinari, S., Nagy, P.D., Simon, A.E., and Dreher, T.W. 2000. CCA initiation boxes without unique promoter elements support in vitro transcription by three viral RNA-dependent RNA polymerases. RNA 6: 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]