Abstract
Localization of mRNA is an important way of generating early asymmetries in the developing embryo. In Drosophila, Staufen is intimately involved in the localization of maternally inherited mRNAs critical for cell fate determination in the embryo. We show that double-stranded RNA-binding Staufen proteins are present in the oocytes of a vertebrate, Xenopus, and are localized to the vegetal cytoplasm, a region where important mRNAs including VegT and Vg1 mRNA become localized. We identified two Staufen isoforms named XStau1 and XStau2, where XStau1 was found to be the principal Staufen protein in oocytes, eggs, and embryos, the levels of both proteins peaking during mid-oogenesis. In adults, Xenopus Staufens are principally expressed in ovary and testis. XStau1 was detectable throughout the oocyte cytoplasm by immunofluorescence and was concentrated in the vegetal cortical region from stage II onward. It showed partial codistribution with subcortical endoplasmic reticulum (ER), raising the possibility that Staufen may anchor mRNAs to specific ER-rich domains. We further showed that XStau proteins are transiently phosphorylated by the MAPK pathway during meiotic maturation, a period during which RNAs such as Vg1 RNA are released from their tight localization at the vegetal cortex. These findings provide evidence that Staufen proteins are involved in targeting and/or anchoring of maternal determinants to the vegetal cortex of the oocyte in Xenopus. The Xenopus oocyte should thus provide a valuable system to dissect the role of Staufen proteins in RNA localization and vertebrate development.
Keywords: RNA localization, translational control, Xenopus oocyte, RNA-binding protein, phosphorylation, MAPK
INTRODUCTION
Staufen is a double-stranded RNA-binding protein involved in RNA localization and the control of translation. During Drosophila oogenesis, Staufen is necessary to anchor bicoid transcripts to the anterior pole of the oocytes and to localize oskar mRNA to the posterior pole (for review, see Palacios and St. Johnston 2001). The correct localization of these mRNAs is critical for establishing the protein gradients that ultimately control transcription of zygotic target genes in a concentration-dependent manner. Staufen is further involved in the derepression of translation of oskar mRNA when it is localized at the posterior pole (Kim-Ha et al. 1995; Micklem et al. 2000). Later, during neurogenesis, Staufen asymmetrically localizes prospero mRNA to the apical crescent of dividing neuroblasts (Li et al. 1997; Broadus et al. 1998). Distinct domains of Staufen mediate microtubule- and actin-based mRNA transport (Micklem et al. 2000). Thus, Drosophila Staufen is a common component of RNA transport in oocytes and neurons (Roegiers and Jan 2000), and is also involved in translational regulation.
Mammals, including human, mouse, and rat, have two genes encoding Staufen, both of which undergo alternative splicing to generate several isoforms (Marion et al. 1999; Wickham et al. 1999; Duchaine et al. 2002). Staufens 1 and 2 are highly related at the sequence level (Fig. 1 ▶) and possess several copies of the double-stranded RNA binding domain (dsRBD), of which there are five in Drosophila Staufen. All isoforms contain dsRBD2, 3, and 4, of which dsRBD3 and 4 are the principal RNA-binding domains, and the most highly conserved regions of the protein. Staufens 1 lack dsRBD1, while Staufens 2 appear to have a truncated dsRBD5. The dsRBDs do not recognize particular nucleotide sequences in vitro, but bind nonspecifically to dsRNAs and ssRNAs with extensive secondary structure, suggesting that target specificity may generally be governed through interactions with other proteins (St Johnston et al. 1992; Saunders and Barber 2003). Lying between dsRBD4 and dsRBD5 is the so-called tubulin-binding domain (TBD), unique to the mammalian Staufens, and absent in Drosophila. The TBD bears partial homology to the tubulin-binding domain of MAP1B, and binds tubulin in vitro (Zauner et al. 1992; Wickham et al. 1999).
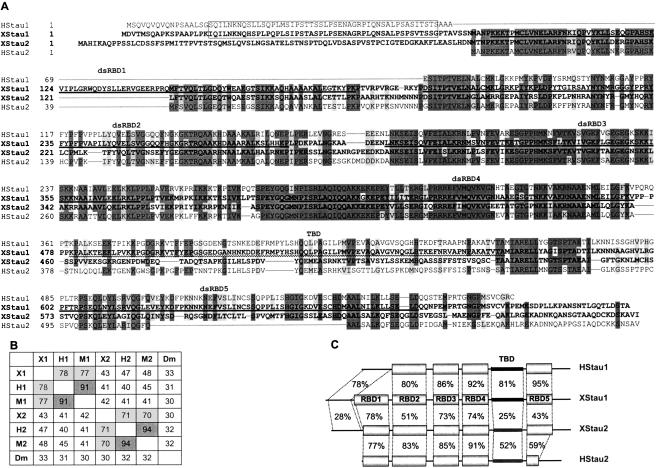
FIGURE 1.
Xenopus Staufen 1 and 2 sequences. (A) XStau1 (AY705672) and XStau2 (BC046732) sequences, in bold, were aligned with human Stau163 (AL133174) and Stau262 (AK002152) using Clustal 1.82. Boundaries of gaps longer than 10 residues were refined by comparison to exon/intron junctions in human Stau genomic sequences. Dark gray shadowing indicates identity in at least three of the four sequences, and light gray indicates conserved changes. dsRBD1–5 domains in Xenopus Staufens, based on Drosophila dsRBD domains (St Johnston et al. 1992) are shown as bold lines, and the tubulin-binding domain (TBD) (Wickham et al. 1999) is shown as a wavy line. The N-terminal region conserved between XStau1 and human Stau163 is boxed. (B) Xenopus Staufens were aligned with the longest available versions of mammalian Stau proteins, namely, human Stau163 (H1, AL133174), mouse Stau155 (M1, BC012959), human Stau262 (H2, AK002152), and mouse Stau262 (M2, AF459099), as well as Drosophila Staufen (Dm, M69111) using Clustal 1.82, and scores of % homology between any two sequences are tabulated. (C) Schematic representation of Xenopus Stau1 and 2 and human Stau163 and Stau262. Identity scores for the domains indicated in A were obtained using the default settings of the GAP algorithm (GCG); the drawing is not to scale.
Mammalian isoforms comprise Stau163 and Stau155, containing 577 and 496 amino acids, respectively (Wickham et al. 1999), and Stau262, Stau259, and Stau252, containing 570, 538, and 479 amino acids, respectively (Duchaine et al. 2002). These differ at their N- (Stau163 and Stau155; Stau262 and Stau259) and C-termini (Stau259 and Stau252). Due to the considerable homology between human, mouse, and rat proteins, we will refer to them, collectively, as either Stau1 or Stau2, while Staufen will be reserved for the Drosophila protein, and XStau1 and XStau2 for the Xenopus laevis homologs.
Stau1 and Stau2 transcripts appear to be fairly ubiquitously expressed in all species studied to date, with evidence of enhanced expression in mouse, rat and human of Stau1 in the ovary, and Stau2 in brain (Marion et al. 1999; Wickham et al. 1999; Saunders et al. 2000; Duchaine et al. 2002). In cultured cells, human Stau1 (hStau1) colocalizes with markers of the rough endoplasmic reticulum (RER), but not with endosomes, cytoskeleton, or Golgi apparatus (Marion et al. 1999; Wickham et al. 1999). Furthermore, sedimentation analyses indicate that hStau1 associates with ribosomes (Marion et al. 1999; Wickham et al. 1999), via protein–protein interactions mediated by dsRBD4 and TBD domains, in conjunction with the RNA-binding activity of dsRBD3 (Luo et al. 2002), consistent with a possible role in translation.
In neurons, Stau1 is found in the soma and dendrites, and is absent from axons. Mammalian Stau1 localizes to the somatodendritic domain of cultured hippocampal neurons, in which it concentrates in the vicinity of ER and microtubules (MTs) (Kiebler et al. 1999; Kohrmann et al. 1999; Krichevsky and Kosik 2001; Monshausen et al. 2001). Indeed, hStau1-GFP granules move bidirectionally along the dendritic shaft in an MT-dependent manner (Kohrmann et al. 1999). Stau2 also localizes to the somatodendritic domain of neurons, but, importantly, Stau1 and 2 particles do not colocalize in distal dendrites (Duchaine et al. 2002). A dominant negative form of Stau2 reduced, while overexpression of wild-type Stau2 increased, dendritic mRNA content in cultured hippocampal neurons, suggesting that Stau2 plays an important role in the transport of RNA to dendrites (Tang et al. 2001). RNAs are transported in motile granules, which are highly enriched in Staufen, and contain ribosomes (Krichevsky and Kosik 2001). Biochemical characterization of Staufen-containing complexes from rat brain confirmed their association with ER and ribosomes, and indicated differences between Stau1- and Stau2-containing complexes (Ohashi et al. 2002; Mallardo et al. 2003).
With the exception that mouse stau1 and stau2 genes are expressed in germ cells during oogenesis and spermatogenesis (Saunders et al. 2000), little is known of Stau1 and 2 proteins in early vertebrate development, when regulation at the level of RNA localization and translation is critical. In the frog oocyte in particular, vegetal localization of maternally inherited determinants such as VegT mRNA is required for establishing the correct formation of germ layers and subsequent patterning of the developing embryo. Here we report that the Xenopus laevis genome encodes homologs to the mammalian Staufen genes, XStau1 and XStau2. We characterize the expression, localization, and phosphorylation of the Xenopus Staufen proteins in oocytes, eggs, embryos, and adult tissues. We show that the two proteins, which have distinct properties, are present at a time and place consistent with roles in vegetal transport and anchoring of certain mRNAs to the vegetal cortex in Xenopus oocytes.
RESULTS
Cloning and sequence analysis of Xenopus Stau1 and 2
A Xenopus Staufen cDNA was obtained by screening an ovary cDNA library with a probe spanning the dsRBD3 and dsRBD4 region, highly conserved between fly, mouse, and man. The ~ 3.3-kb-long cDNA contained an open reading frame (ORF) with 718 amino acids and a predicted molecular weight of 79 kDa. Comparison with mammalian Stau proteins revealed this protein to correspond to the Stau1 family (XStau1, Fig. 1 ▶). Xenopus Stau2 (XStau2) was identified by a BLAST search using the human Stau2 sequence against the available Xenopus laevis database. A full-length clone from the I.M.A.G.E. consortium encoded a 75-kDa polypeptide of 685 amino acids with an extensive homology to XStau2 (Materials and Methods). XStau1 shows > 75% homology to mammalian Stau1 isoforms, but only ~45% to their Stau2 proteins, while, conversely, XStau2 shows ~ 70% homology to mammalian Stau2 isoforms, but only ~ 40% to their Stau1 proteins (Fig. 1B ▶).
Xenopus Staufens are significantly longer than their mammalian counterparts, largely due to N-terminal extensions, though they are not as long as the Drosophila protein. Interestingly, both Xenopus Staufens appear to contain five dsRBDs as originally described in Drosophila Staufen (St Johnston et al. 1992). Human Stau163 and XStau1 share a unique N-terminal extension that does not map to any conserved domain (Fig. 1A ▶). By inspection of the sequence alignments, it seems apparent that Stau163 has the entire dsRBD1 deleted, while still retaining this unique N-terminal domain. The XStau2 protein contains a unique N-terminal extension that is not present in any of the mammalian Stau2 group, and comparison with a zebrafish XStau2 homolog in available databases (acc. no. AAH45292, data not shown) indicates that this extension may be unique to amphibians. In comparison to the Stau2 and XStau2 proteins, the XStau1 dsRBD1 has an insert of 23 amino acids in loop 2, separating the β1 and β2 strands as determined from the structure of Drosophila dsRBD3 (Ramos et al. 2000). Xenopus tropicalis Stau1 expressed sequence tags (ESTs) contain the same dsRBD insert sequence as their laevis counterparts (e.g., Tegg050024 at http://www.sanger.ac.uk/Projects/X_tropicalis/; and see Discussion). The dsRBD2–4 regions are highly conserved between Xenopus and human Staufen proteins (Fig. 1A,C ▶). In dsRBD5, however, XStau2 differs from the mammalian Stau2 group in that XStau2 contains a complete dsRBD5, as can be found in the Stau1 proteins. This complete dsRBD5 is also conserved in the zebrafish homolog (data not shown, see above). This observation and the sequence alignment with XStau2 indicates that human, mouse, and rat Stau2 proteins harbor a large internal deletion within the dsRBD5 domain, since the proteins align well on either side of this deletion (Fig. 1A ▶; data not shown). One final sequence feature of note is that the putative TBD (Zauner et al. 1992; Wickham et al. 1999), is considerably less well conserved in the Xenopus proteins than the dsRBD regions (Fig.1A,C ▶).
Expression of XStau1 and 2 proteins peaks in mid-oogenesis
Xenopus Staufens 1 and 2 were expressed in Escherichia coli as full-length His-tagged proteins and purified by Ni-NTA chromatography for immunization of rabbits. XStau1 antibodies recognized a protein of about 80 kDa in Xenopus oocytes, slightly smaller than the recombinant protein extended by a C-terminal 6His tag, indicating that the maternal protein corresponds in size to the predicted cDNA-encoded ORF (Fig. 2A ▶). XStau2 antibodies detected multiple oocyte bands of ~ 80 kDa, the largest of which was slightly shorter than the recombinant protein, extended by an RGS-6His tag at its N-terminus, again indicating that this maternal protein corresponds in size to the predicted cDNA-encoded ORF (Fig. 2B ▶). The multiple bands most likely correspond to the three isoforms of mammalian Stau2 (Duchaine et al. 2002). To ensure that our reagents are specific for each of the two isoforms, XStau1 antibody was neutralized with XStau2 protein, and XStau2 antibodies were depleted of XStau1 epitopes (Materials and Methods). Comparison with the untreated antibodies shows that both treatments resulted in essentially isoform-specific reagents (Fig. 2A,B ▶). Unless otherwise specified, the data in this study were obtained with these isoform-specific reagents.
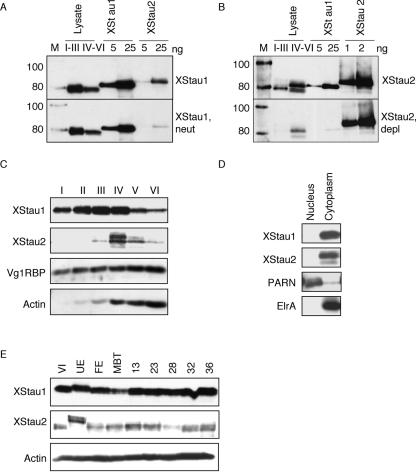
FIGURE 2.
The levels of Xenopus Staufens peak in mid-oogenesis. (A,B) Validation of enrichment in XStau1- and XStau2-specific antibodies. Two cell equivalent mixed-stage oocytes lysates (I–III) and (IV–VI) and indicated amounts of recombinant Xenopus Staufen proteins were analyzed by Western blotting. (A) Top panel, using XStau1 antibody (1:10,000 dilution) and bottom panel XStau1 antibody (1:10,000) neutralized with XStau2 protein (XStau1, neutr). (B) Top panel, using XStau2 antibody (1:2500) and bottom panel XStau2 antibody (1:2500) depleted of XStau1 epitopes (XStau2, depl). Lane M, Magic Mark Western Standard molecular weight standards, in kDa. Donkey antirabbit secondary antibodies and the molecular weight standards were detected by ECL. (C) The levels of Xenopus Staufens peak in mid-oogenesis. Lysates were prepared from staged oocytes (I–VI) and two cell equivalents were analyzed by Western blotting with specific XStau1 and XStau2 antibodies. The oocyte samples were also probed with control Vg1RBP and actin antibodies. (D) XStau1 and 2 are cytoplasmic proteins. Nuclear and cytoplasmic fractions were prepared from stage VI oocytes and equivalent cell samples analyzed by Western blotting. The fractions were verified by PARN antibody, which detects the nuclear poly(A) nuclease and HuR antibody, which detects the cytoplasmic elrA protein. In C,D, XStau antibodies were used and verified against recombinant proteins as in panels A,B (data not shown). (E) XStau protein expression in oocytes, eggs, and embryos. Lysates from two cell equivalents of stage VI oocytes, unfertilized and fertilized eggs, as well as MBT and embryonic stages 13–36 were analyzed by Western blotting with specific XStau1 and XStau2 antibodies, verified by comparison with indicated amounts of recombinant proteins. A control blot of the same samples with actin antibodies confirmed consistent protein loading.
To monitor levels of XStau1 and 2 during oogenesis, S10 lysates from the equivalent of two oocytes of each stage were analyzed by Western blotting. XStau1 was detectable in stage I oocytes; the levels then increased to a peak at stage III/IV of oogenesis, of about 10 ng/oocyte, and then declined to ~ 2 ng per stage VI oocyte (Fig. 2C ▶). XStau2, unlike XStau1, was more abundant in late-stage than early-stage oocytes (Fig. 2A–C ▶). It was not detectable in stage I oocytes, peaked at stage IV at about 1 ng/oocyte, and also declined subsequently (Fig. 2C ▶). XStau1 and XStau2 were completely absent from the insoluble pellet fractions (data not shown). In contrast, Vg1RBP, an RNA-binding protein that has been implicated in localizing Vg1 mRNA in the Xenopus oocyte (Deshler et al. 1998; Havin et al. 1998; Bubunenko et al. 2002; Kwon et al. 2002), is present at approximately constant amounts per cell throughout oogenesis (Zhang et al. 1999), while the levels of actin rise during oogenesis as the oocyte grows (Fig. 2C ▶). These controls validate the declining levels of XStau proteins from stage IV onward. We examined nuclear and cytoplasmic fractions from equivalent numbers of stage VI oocytes, and found that both XStau1 and 2 proteins were entirely cytoplasmic (Fig. 2D ▶). The fractions were verified by reprobing the blots with PARN antibodies that detect a nuclear poly(A) nuclease (Copeland and Wormington 2001) and HuR antibodies which detect cytoplasmic elrA (A. Devaut and N. Standart, unpubl.). Levels of XStau1 and 2 remain relatively constant from stage VI through embryogenesis, up to at least stage 36. In two independent experiments, a slight decline in the levels of XStau 1 and 2 proteins around the mid-blastula transition (MBT) was observed, followed by a slight increase, when equivalent numbers of eggs and embryos were compared (Fig. 2E ▶; data not shown). The level of XStau1 mRNA, assessed by RNAse protection, remained essentially constant from fertilization and throughout embryogenesis, but starts to increase from around stage 13 onward (data not shown). We conclude that XStau1 is more abundant than XStau2 in oocytes and embryos of all stages, both being cytoplasmic proteins. Levels of both XS-tau1 and 2 proteins are highest during mid-oogenesis, with XStau2 expression essentially restricted to stage IV. Our data also suggest that maternal transcripts and proteins are replaced by zygotic versions after MBT.
Xenopus Staufen proteins are principally expressed in ovary and testis
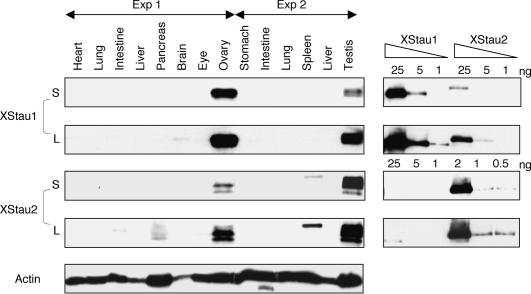
To determine Staufen protein expression in adult Xenopus, lysates made from tissues in two independent experiments were analyzed by Western blotting (Fig. 3 ▶). Approximately 40 μg of total protein was loaded in each lane; Coomassie Blue staining confirmed approximately equal protein loading in all tissues, and duplicate samples showed very similar protein patterns (data not shown). Staufen protein expression was largely confined to ovary and testis; with XStau1 more highly expressed in ovary than in testis and XStau2, in contrast, more highly expressed in testis than in ovary. Longer exposures of the same blots showed weak expression of XStau1 in brain, and of XStau2 in spleen, pancreas, and intestine. In the case of XStau2, different tissues appeared to contain varying ratios of the different isoforms. No significant XStau expression was observed in heart, lung, liver, stomach, and eye. Reprobing the blot with an actin antibody demonstrated that protein was present in lanes without XStau1 or 2 signal. We conclude that Staufen expression is largely confined to reproductive tissues and embryonic cells.
FIGURE 3.
Xenopus Staufen expression in adults is largely confined to reproductive tissues. Samples were prepared from indicated adult tissues, in two independent experiments as detailed in Materials and Methods, and 40 μg of total protein was analyzed by Western blotting, alongside indicated amounts of recombinant proteins. Short (S, XStau1 10 sec, XStau2, 30 sec) and Long (L, XStau1 and XStau2, 10 min) exposures are shown. The blots were overprobed with anti-actin antibody.
Xenopus Staufens are vegetally localized during oogenesis
To assess the distribution of Staufens in the Xenopus oocyte, immunofluorescence was performed with the anti-XStaufen antibodies (Fig. 4 ▶). Neutralized/depleted antibodies and affinity-purified antibodies gave equivalent results (see Materials and Methods for details). Both proteins were cytoplasmic in all regions of early-stage (I–III) and late-stage (VI) oocytes, the XStau1 signal being stronger than the XStau2 signal in accordance with the relative levels of the proteins detected in the cytoplasm by Western blotting (Fig. 2 ▶).
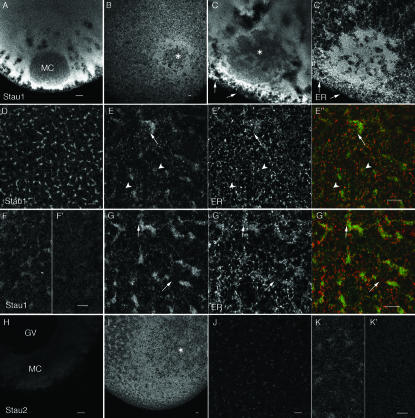
FIGURE 4.
Localization of XStau1 and XStau2 proteins in oocytes. Confocal images of oocytes stained with anti-XStau1 (A–G) and anti-XStau2 (H–K) antibodies. Stage I oocytes are shown in A,H; Stage II oocytes in B,C,I. All other images are of stage VI oocytes. Staining with both anti-Staufen antibodies was excluded from the mitochondrial cloud (MC) at stage I and from the site where it has joined the cortex at stage II–III (*). The images in D,F–I are confocal sections through a vegetal subcortical layer of stage VI oocytes containing distinctive patches of ER, which lies about 5 μm beneath the oocyte surface. E is a more superficial confocal section of the same oocyte as G, taken at the level of the cortical ER network. C′, E′, and G′ show the ER distribution in oocytes costained with anti-GRP94. E″ and G″ are corresponding overlays of XStau1 (green) and GRP94 (red). Arrows in C,E,G point to sites of XStau1-ER colocalization; arrowheads in E indicate absence of localization at the level of the outer cortex. F′ and I′ are images of oocytes stained with antibodies pre-incubated with the corresponding protein, acquired with the same confocal settings as those of the oocytes stained with BSA-incubated antibodies in F and I. Scale bars 10 μm throughout.
In stage I–II oocytes, XStau1 staining was mainly cytoplasmic (Fig. 4A ▶). There was no concentration in the mitochondrial cloud (MC), a structure in which a number of RNAs involved in germ plasm formation become localized (King et al. 1999); indeed the protein appeared partially excluded from the MC region. In stage II and III oocytes, XStau1 staining was concentrated in a cortical region surrounding, but excluded from, the site where the MC had joined the cortex (* in Fig. 4B,C ▶). Double-immunofluorescence with an antibody recognizing the ER luminal protein GRP-94 (Beckhelling et al. 2003) revealed that XStau1 staining did not colocalize with the dense ER found within the MC (Fig. 4C ▶′ Chang et al. 2004). Some codistribution with ER was, however, detectable in the surrounding region of the vegetal cortex (arrows in Fig. 4C ▶). In stage VI oocytes, XStau1 staining showed a striking concentration in irregular-shaped patches, measuring tens of micrometers in length, beneath the vegetal cortex (Fig. 4D ▶). Note that in confocal sections the size and morphology of these patches are variable, depending on their precise depth and location within the vegetal hemisphere. The staining of the patches was severely reduced or abolished by pre-incubation with purified XStau1 protein, demonstrating the specificity of the staining (Fig. 4F ▶). The patches stained by XStau1 clearly coincided with similarly shaped patches of concentrated ER below the vegetal cortex (arrows in Fig. 4G ▶). The colocalization was not, however, perfect, and there was little colocalization of XStau1 and ER outside these dense patches. This was particularly obvious in confocal sections taken at superficial levels containing the cortical ER layer and the surface of some dense XStau1 patches (Fig. 4E ▶). We hypothesize that XStau1 becomes associated with ER in the vegetal cortical region from stage II onward. The imprecise colocalization of XStau1 and ER suggests that their association is indirect, and may depend on the presence of other structural components.
Anti-XStau2 staining in stage VI oocytes was weaker than that of anti-XStau1. It was more uniform, but also showed a weak concentration in subcortical vegetal patches from stage II onward (Fig. 4H,I ▶). This pattern was abolished by preabsorption of the antibody with XStau2 protein (Fig. 7I ▶). At earlier stages, very weak cytoplasmic staining was obtained with XStau2 antibodies (Fig. 4J ▶). Staining was again stronger in the vegetal cortex around the site of MC fusion at stages II–III (Fig. 4K ▶). These data indicate that both Xstau1 and 2 proteins are widely distributed in the oocyte, but that their distributions are not identical. Their distribution first becomes polarized during Stage II, both proteins becoming enriched in the vegetal cortical region from this stage onward. Their concentration in this region coincides temporally and spatially with that of RNAs that localize by the “late pathway” such as Vg1 and VegT mRNAs (Kloc and Etkin 1998).
FIGURE 7.
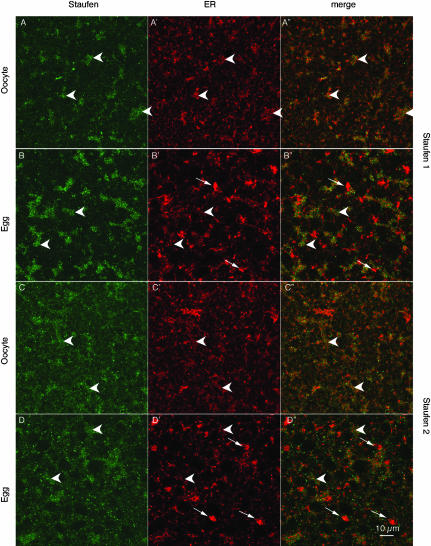
Staufen distribution in the vegetal region of oocytes and eggs. Confocal images of oocytes and progesterone-matured eggs stained with anti-XStau1 (A,C) and anti-XStau2 (B,D) antibodies. All samples were costained with anti-GRP 94 to visualize ER (images A′–D′). A′–D′ are the corresponding overlays of XStau (green) and GRP94 (red). These confocal sections were taken about 5 μm below the vegetal surface in a layer containing distinctive patches (arrowheads) containing XStau1, and to a lesser extent XStau2. In oocytes, these patches coincide closely with domains rich in ER. During maturation the ER reorganizes to form distinctive, brightly stained whorls of ER (white arrows in B,D), while ER is partially lost from the Staufen-containing patches. Scale bar, 10 μm.
Characterization of Staufen complexes
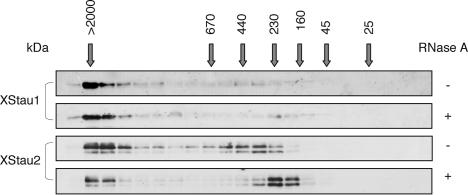
To reinforce the finding that XStau1 and 2 proteins localize independently within the oocyte, we compared their distribution among fractions produced by gel filtration of oocyte lysates from stage V/VI oocytes, and RNase-treated or mock-treated prior to fractionation (Fig. 5 ▶). Interestingly, differences between XStau1- and 2-containing complexes were detected. In mock-treated extracts, the majority of XStau1 eluted very early, with the molecular weight markers indicating that it is found in granules of a size ≥ 2 MDa, with a very small proportion trailing evenly throughout the rest of the elution up to the 160-kDa marker. XStau2 eluted as two peaks, the first one also very early (> 2 MDa); however, a second peak eluted later in the profile, just after the 440-kDa molecular weight marker. The very large XStau1 and XStau2 granules were less sensitive to RNase treatment, with the small XStau2 400-kDa particles being reduced in size by nuclease digestion. The gel filtration data suggest that the different distributions of XStau1 and XStau2 in oocytes reflect their inclusions in different-sized complexes.
FIGURE 5.
XStau granules and particles in Xenopus oocyte lysates: 200 μL stage V/VI S10 extract was fractionated through a Superose 6 HR 10/30 column at a flow rate of 0.5 mL/min, and 0.3- mL fractions collected. Alternate fractions were separated by SDS-PAGE and visualized by Western blotting using antibodies as shown. Where indicated, lysate was treated with RNase A (167 μg/mL, 10 min, 20°C) prior to loading on the column. This treatment did not significantly affect the fractionation behavior of the Coomassie Blue-stainable proteins (data not shown). Elution volumes of size markers [Blue Dextran (> 2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), ovalbumin (43 kDa), and chymotryp-sinogen A (25 kDa)] are marked by arrows.
XStau1 and 2 proteins are phosphorylated during meiotic maturation
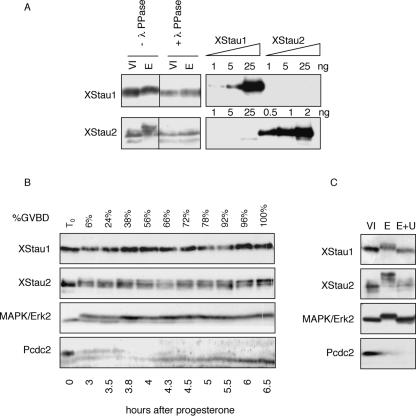
Interestingly, the migration of Xenopus Staufen proteins on SDS-PAGE in samples prepared from meiotically mature eggs was retarded relative to their migration in oocyte or embryo samples, particularly notable in the case of XStau2 (Fig. 6A ▶). We observed these mobility changes in both in vitro progesterone-matured oocytes, and in laid unfertilized eggs (Figs. 2 ▶, 6 ▶). The apparent size shift in XStau1 and XStau2 proteins was abolished by treatment of progesterone-matured eggs with λ-phosphatase prior to Western blot analysis, while control incubations lacking the phosphatase left migration unaffected, indicating that this size shift is due to phosphorylation (Fig. 6A ▶). Moreover, immunoprecipitation experiments demonstrated that Xenopus Staufen proteins can directly incorporate 33P phosphate in eggs (data not shown).
FIGURE 6.
XStau1 and XStau2 are phosphorylated during meiotic maturation. (A) Lysates were prepared from stage VI oocytes and progesterone-matured eggs, and treated (+) or left untreated (−) with λ-phosphatase, and subsequently analyzed by Western blotting with specific XStau1 and XStau2 antibodies as in Figures 2 ▶ and 3 ▶. (B) Timing of phosphorylation during meiotic maturation. Random groups of 20 cells out of ~ 250 stage VI oocytes treated with progesterone were taken at the time of progesterone addition (T0) and when the indicated proportion of cells has undergone GVBD (%). Oocytes were then subjected to Western blot analysis with the indicated antibodies. (C) XStau1 and XStau2 are phosphorylated by the MAPK kinase pathway. Stage VI oocytes, some of which were preincubated with 50 μM U0126 for 4 h, were matured with progesterone. Samples of untreated oocytes (O), control matured eggs (E), and eggs matured in the presence of UO126 (E+U) were analyzed by western blotting with the indicated antibodies.
To examine the timing of phosphorylation during meiotic maturation, a large population of oocytes was matured with progesterone, and cells were withdrawn at frequent intervals for Western blot analysis (Fig. 6B ▶). In this culture, 50% germinal vesicle breakdown (GVBD) occurred around 4 h after hormone addition, as monitored by appearance of the “white spot.” Phosphorylation of XStau1 and XStau2 are early events in meiotic maturation, as retarded bands were observed in the 3-h sample, with 6% GVBD. We note that not all XStau2 isoforms are phosphorylated early, as the higher-molecular-weight one in particular appears to be modified later in maturation. The timing of the early phosphorylation of Xenopus Staufens coincides temporally with the activation of MAP and cdc2 kinases, as shown by Western blotting with antibodies raised against Erk2 and phospho-cdc2, which undergo phosphorylation and dephosphorylation reactions, respectively, during their activation. To identify which of these two pathways was responsible for Staufen phosphorylation, oocytes were matured in the presence of UO126, a specific MAPK inhibitor (Gross et al. 2001). Western blotting confirmed that this reagent prevented Erk2 kinase activation, while allowing cdc2 kinase activation (Fig. 6C ▶). The phosphorylation of both XStau1 and XStau2 was abolished by U0126, indicating that the kinase responsible lies on the MAPK pathway.
Distribution of XStau proteins during meiotic maturation
To assess whether the meiotic phosphorylation of the XStau proteins might correlate with a change in distribution concurrent with the release of Vg1 and other RNAs from their vegetal localization (King et al. 1999), the localization of XStau1 and XStau2 was compared in oocytes and progesterone-matured eggs by immunofluorescence (Fig. 7 ▶). Particular attention was paid to their relation with ER, revealed using anti-GRP94 antibodies, as ER has been implicated in Vg1 RNA anchoring (Alarcon and Elinson 2001). The prominent large patches of XStau1 detected in the vegetal subcortical region of Stage VI oocytes (Fig. 7B ▶; see also Fig. 4 ▶) were also detected in eggs (Fig. 7B ▶); however, the ER associated with these patches became noticeably less dense during maturation. Thus the colocalization of XStau1 and the ER was significantly reduced (Fig. 7 ▶, cf. A″ and B″). Extensive reorganization of the ER network in parallel to the meiotic divisions has been described previously (Kume et al. 1997; Terasaki et al. 2001), with dense ER whorls forming close to the egg surface (arrows in Fig. 7B ▶′ and D′).
XStau2 also appeared to maintain its slight concentration in subcortical patches during maturation while the ER concentration in these patches was lost (Fig. 7C,D ▶). As mentioned above, the patchy distribution of XStau2 was much less marked than that of XStau1, reflecting the lower abundance of the protein and the presence of additional staining in the cytoplasm. These observations are consistent with the idea that a direct or indirect association between Staufen proteins and ER is weakened during maturation.
DISCUSSION
Conserved features of Staufen proteins
Xenopus Staufen 1 and 2 proteins are highly homologous to Drosophila Staufen, and to their mouse, human (Fig. 1 ▶), and rat counterparts (Kiebler et al. 1999; Monshausen et al. 2001). Oocytes contain one major form of XStau1, corresponding in size with the cDNA-encoded protein of 79 kDa, and most closely similar to the mammalian Stau163 alternatively spliced isoform. Three maternal XStau2 proteins of ~ 80 kDa were detected, presumably corresponding to mammalian Stau252, Stau259, and Stau262 alternatively spliced isoforms, although we cannot rule out allelic variation or posttranslational modification being responsible for the different-sized proteins.
The only features of Staufen that appear to have been conserved throughout animal evolution are five dsRBDs and an insertion that splits dsRBD2 into two halves. Drosophila dsRBDs 1, 3, and 4 bind dsRNA in vitro, but dsRBDs 2 and 5 do not (St Johnston et al. 1992; Micklem et al. 2000). Alignments of Staufen homologs from multiple vertebrates with Drosophila Staufen indicate that the sequences of dsRBD3 and 4 demonstrate the highest conservation across this large evolutionary distance (data not shown), suggesting that these form the conserved core of the Staufen family. XStau1 contains a dsRBD1, in addition to dsRBD2–5 domains, somewhat surprisingly, as no isoform of the mammalian Stau 1 group contains this feature. This observation seems to indicate that this dsRBD has been lost by internal deletion of this domain from the mammalian Stau1 family. XStau1 dsRBD1 contains an insert of 23 amino acids in loop2, separating the β1 and β2 strands based on the αβββα dsRBD3 NMR structure (Ramos et al. 2000). The similarly located insert in Drosophila dsRBD2 prevents its potential to bind dsRNA in vitro (Micklem et al. 2000). Indeed, all known Staufen proteins contain an insert in the same location of dsRBD2, albeit of somewhat different sequence and length, leading to lack of RNA-binding (Duchaine et al. 2002; Wickham et al. 1999). In a novel rat and mouse isoform of Stau1, a six-aminoacid extension of the β1 strand in dsRBD3 also abrogates RNA-binding (Kiebler et al. 1999; Monshausen et al. 2001). Thus the insert-containing XStau1 dsRBD1 is very likely to be inactive in RNA binding; indeed preliminary NorthWestern experiments indicate that the only region of XStau1 capable of binding bcd and Vg1 VLE RNA contains dsRBD3+dsRBD4, and that regions containing dsRBD1+dsRBD2 or dsRBD5 are inactive (E. Adegbenro and N. Standart, unpubl.).
Implication of XStau proteins in oocyte RNA localization
Maximum levels of both sets of XStau proteins were detected during mid-oogenesis, with XStau1 present at ~ 10-fold higher levels than XStau2 in oocytes, eggs, and embryos (Fig. 2 ▶). In comparison, Vg1RBP is ~ 5–10× more abundant than XStau1, and is present at approximately equal levels throughout oogenesis (Fig. 2 ▶; Zhang et al. 1999; Git and Standart 2002). Different again, the temporal expression of PrrP, a proline-rich protein which binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization, increases during oogenesis (Zhao et al. 2001). Thus, of the proteins implicated in Vg1 mRNA localization, the expression profile shown by the Xenopus Staufens is unique, and peak levels temporally correlate with the timing of the vegetal cortical anchoring step of the so-called late pathway mRNAs, including Vg1 mRNA (King et al. 1999). The spatial distribution of these proteins, determined by immunofluorescence, is also consistent with a role in this process (see below).
XStau1 and 2 proteins are expressed throughout embryogenesis, up to at least stage 36 (Fig. 4 ▶), suggesting that they continue to function in the cells of the early embryo. However, the proteins are absent in most tissues, and even when protein was detectable, the levels were considerably less than in embryos. This indicates that expression of these proteins is down-regulated in adult differentiated tissues, except in the reproductive tissues of testis and ovary. In germ cells, a surprising difference in XStau1 and XStau2 protein expression was revealed, with XStau1 being preferentially expressed in ovary, and XStau2, unexpectedly, being preferentially expressed in testis. Although we are not aware of reports of localized RNAs in testis, posttranscriptional control at the level of translation during spermatogenesis is well documented (Braun 1998). Characterization of mammalian Stau proteins in human, mouse, and rat adults has been largely confined to determination of mRNA expression, rather than protein levels (see Introduction). Further analysis will be required to determine whether this unexpected pattern of protein expression is also demonstrated in the mammalian germ line.
For the first time for any Staufen protein, we have shown that both sets of Staufen proteins are phosphorylated, by MAP kinase, during meiotic maturation. At this time RNAs such as Vg1 RNA are released from their tight localization at the vegetal cortex (Figs. 2 ▶, 6 ▶; King et al. 1999). The transient Xenopus Staufen phosphorylation in eggs, and by inference, dephosphorylation in embryos (Fig. 2 ▶) may relate to the report that Stau1 interacts with, and colocalizes with, protein phosphatase-1 (PP-1) in rat neurons (Monshausen et al. 2002). Indeed, the short motif in rat Stau1 shown to bind PP1 (RKVTF; located between dsRBD4 and dsRBD5) is conserved in XStau1 (Fig. 1 ▶). It will clearly be of interest to determine whether and how phosphorylation modifies Staufen functions. For example, phosphorylation may modulate RNA-binding or Staufen-protein interactions, or its interactions with cytoskeleton and ER.
Association of Xenopus Staufen proteins with vegetal ER
Confocal imaging of stage I–III oocytes stained using anti-XStau antibodies showed that both XStau1 and 2 are largely excluded from the mitrochondrial cloud (MC) (Fig. 4 ▶). The MC is a macroscopic structure lying close to the nucleus where germ plasm mRNAs such as Xcat-2 localize during very early stages of oogenesis, using the “early” or “METRO” pathway (King et al. 1999; Kloc et al. 2002). Vg1RBP/Vera, a protein implicated in targeting Vg1 and VegT mRNAs to the vegetal cortex during oogenesis, using the “late” localization pathway (Deshler et al. 1998; Havin et al. 1998; Bubunenko et al. 2002; Kwon et al. 2002), is also cytoplasmic and largely excluded from the MC in early-stage oocytes (Chang et al. 2004). The exclusion of XStau proteins and Vg1RBP from the MC region suggests that these proteins are not involved in the “early” RNA localization pathway. From stage II onward, however, both XStau1 and Vg1RBP concentrate in a region of vegetal cortex, around the site where the MC has joined it. Within this region, double immunofluorescence with an ER marker indicated that the proteins partially colocalize with ER, although they clearly do not colocalize with the dense ER originating from within the MC (Fig. 4 ▶; Chang et al. 2004). In later-stage oocytes, irregular-shaped XStau1 patches were detected beneath the vegetal cortex, and were found to coincide with similar patches of concentrated ER (Figs. 4 ▶, 7 ▶). These observations suggest that a direct or indirect association between Staufen proteins and part of the ER network beneath the vegetal cortex is established during stage II and maintained during the vitellogenic stages of oogenesis. In eggs, this association is lost, as the ER undergoes extensive restructuring (Fig. 7 ▶; Kume et al. 1997; Terasaki et al. 2001), and the Staufen proteins are phosphorylated. We cannot yet determine to what extent one or both of these processes disrupts the association between ER and XStau proteins. It is noteworthy that the formation of the characteristic dense ER whorls during maturation is probably mediated by cdc2 (Terasaki et al. 2001), whereas XStau phosphorylation is due to the MAPK pathway (Fig. 6 ▶).
There is precedent for an association between Stau proteins and ER. Mammalian Stau1 proteins colocalize with RER in tissue culture cells (Marion et al. 1999; Wickham et al. 1999), and with ER near synaptic contacts in hippocampal neurons (Kiebler et al. 1999), and copurify with ER markers upon gel filtration fractionation of rat brain lysates (Mallardo et al. 2003). Vg1RBP cofractionates with ER membranes following centrifugation of undiluted oocyte lysates (Deshler et al. 1997) and as we have observed for XStau1, colocalizes with vegetal ER in mid-late-stage oocytes (Chang et al. 2004). Both proteins may be components of mRNPs that associate directly or indirectly with ER in a distinct region of the oocyte vegetal hemisphere. This ER is located around the MC remnants during stages I and II, initially occupying a “wedge” between the germinal vesicle and the vegetal cortex (Deshler et al. 1997; Kloc and Etkin 1998). As the oocyte grows this wedge flattens into a thin subcortical layer containing the characteristic dense ER patches. It will be important to establish what other components associate with the ER, XStau, and Vg1RBP in these structures. Their location beneath the vegetal cortex makes them good candidates for a role in (late pathway) RNA anchoring. ER as well as cytokeratin filaments have previously been implicated in the vegetal anchoring of Vg1 mRNA (Alarcon and Elinson 2001).
XStau2 staining in oocytes was considerably weaker than that of XStau1, reflecting its lower abundance, and making it harder to draw strong conclusions concerning its localization. XStau2 in oocytes appeared to be distributed more uniformly than XStau1, though some enrichment in subcortical vegetal patches was detected in later-stage oocytes (Fig. 4 ▶). The more cytoplasmic distribution of XStau2 may reflect its participation in a distinct population of small (size) particles from which XStau1 is largely absent (Fig. 5 ▶). A recent study of Stau-containing complexes in rat brain also analyzed by gel filtration (Mallardo et al. 2003) revealed a similar distribution of Stau1 and 2. Stau1 and 2 granules were very large (> 2 MDa) and were RNase-insensitive, while Stau1 particles (~ 440–670 kDa) cofractionated with kinesin and were nuclease-sensitive (Mallardo et al. 2003). In oocytes, large XStau1- and 2-containing complexes were largely RNase-insensitive, while smaller XStau2-containing particles were more nuclease-sensitive (Fig. 5 ▶). Based on these findings, we can speculate that in the Xenopus oocyte, granules containing (most) XStau1 and (some) XStau2 preferentially associate with the vegetal cortical ER-rich domains, accounting for their strong staining with XStau1 and weaker staining with XStau2. While the present report was under review, Yoon and Mowry (2004) reported the identification of a Xenopus Staufen (XStau) protein, essentially identical to XStau1. The two proteins differ only in the inclusion of six amino acids, GERKYP, located between dsRBD1 and 2, in the XStau1 isoform reported here (see Fig. 1 ▶). In agreement with our results, Yoon and Mowry found that XStau is expressed in oocytes and localizes to the vegetal cortex in mid-oogenesis, using an antibody that probably detects both XStau1 and XStau2. In addition they show that XStau interacts with late-pathway RNAs, and functions in vegetal RNA localization, as shown by overexpression of a dominant negative form containing dsRBD2–4 (Yoon and Mowry 2004). Given that dsRBD2–4 are highly conserved between XStau1 and 2, it remains to be established whether both or just one of the XStau isoforms that we have identified functions in vegetal RNA localization. Likewise, the identity of the isoform that interacts with kinesin remains to be established, although given the presence of this motor protein in smaller, RNase-sensitive complexes (Yoon and Mowry 2004), our results suggest that it may be XStau2.
In summary, this characterization of Xenopus Staufens in early development, particularly in terms of expression and distribution during oogenesis, implicates them in localizing RNAs to the vegetal cortex. They could be involved in motor-based transport of RNPs through the cytoplasm (Betley et al. 2004; Yoon and Mowry 2004) and/or anchoring by association with cytoskeletal elements or specific regions containing ER. It will clearly be of interest to identify the mRNAs that associate with these proteins, and to test the role of Xenopus Staufens in different phases of RNA localization in the oocyte.
METHODS AND MATERIALS
cDNAs and constructs
Xenopus Staufen 1 cDNA
Degenerate oligonucleotides were designed to regions highly conserved in Drosophila, mouse, and human staufen, encoding HMKNFV/IT and GTNKKVA in domains 3 and 4, respectively (forward: 5′-CAC/TATGAAGAAAC/TTTT/CG/ATIAC-3′ and reverse 5′-GCIACCTTC/TTTG/ATTIGTICC-3′ where I = Inosine). These primers amplified a 388-bp product from stage III/IV Xenopus oocyte cDNA under low-stringency conditions (annealing 45°C, 1 min; extension 72°C, 1 min; 40 cycles) which was then used to screen a Xenopus ovary cDNA library (Lemaire et al. 1995) at high stringency for full-length clones. A 3.3-kb clone was isolated with multiple stop codons upstream of a predicted initiator methionine and large ORF encoding 718 amino acid residues. Northern blot analysis of total oocyte RNA showed a single band of ~ 3.6 kb using a 1.2-kb EcoR1/Not1 staufen cDNA fragment encoding the N-terminal half of the protein as a probe (data not shown).
The XStau1 ORF (AY705672) was amplified using pfu DNA polymerase and oligonucleotides 2737 (5′-TGGCTAGCATGGAC GTCACCATGTCTC-3′) and 2738 (5′-TTGCGGCCGCACTC GAGGGCTGTGCTGTCCAGTGTT-3′). The NheI-XhoI-digested ORF was subcloned into NheI/XhoI sites of pGIT, a plasmid based on pET21 (Minshall and Standart 2004).
Xenopus Staufen 2 cDNA
Xenopus laevis Staufen 2 (BC046732) was available as a full-length I.M.A.G.E. clone (IMAGp998A1412240Q3, https://www.rzpd.de) in the pCMV-SPORT6 vector. The cDNA was prepared with RNA from embryonic stages 31/32. The XStau2 coding region from BC046732 was amplified using pfu polymerase using (forward 5′-GCGCGAGATCTATGGCACACATTAAGGCACAGCCACCATC-3′, reverse 5′-GCGCGCCCGGGATGACAGCCTTTGAATCCTTGCA GTC-3′) and cloned into BamHI-SmaI sites of pQE32 (QIAGEN).
Expression of XStau1 and XStau2 in E. coli
Xstaul His6 in pGIT was expressed in E. coli BL21* cells, and purified by Ni-NTA chromatography as described for Vg1RBP (Git and Standart 2002) with the following modifications. First, the HNTA buffer was supplemented with 10% v/v glycerol, 7 mM β-mercaptoethanol, 0.5 M NDSB 201 (pyridinium propane sulfonate), 2 mM benzamidine, 10 mM imidazole, and 2× Complete Protease Inhibitors for the cell lysis step, and secondly, the Ni-NTA resin was washed with HNTA buffer, then with PBS containing 10% v/v glycerol, 7 mM β-mercaptoethanol, 2 mM benzamidine, and with PBS+ containing 50 mM imidazole prior to elution of the recombinant protein with PBS+ containing 1 M imidazole pH 7.5, and dialysis against buffer M (150 mM NaCl, 50 mM NaPi, pH 7.8, 1 mM DTT, 10% v/v glycerol, 0.1% v/v Triton X-100).
BL21pREP4 cells containing pQE32XStau2 were grown at 30°C to A600 of 0.8; IPTG was added to 0.5 mM, and cells were incubated for 6 h at 24°C. Cells were lysed by French press in ice-cold 20 mM Tris, pH 8.0, 500 mM NaCl, 0.5 mM PMSF. Triton X-100 was added to 0.1% v/v, and the lysate was centrifuged for 30 min at 14,000 rpm in a Sorval SS-34 rotor. The supernatant was bound to Ni-NTA resin (1 mL per liter cell culture) for 1 h in the presence of 10 mM imidazole. The resin was washed with lysis buffer containing 0.01% Triton X100, and bound protein was eluted with 0.3 M imidazole in lysis buffer with 0.01% Triton X100 and dialyzed against 20 mM Tris, pH 8.0, 50 mM NaCl, 0.01% v/v Triton X-100, and 0.5 mM DTT. The supernatant obtained following centrifugation for 15 min at 10,000 rpm was applied to Heparin Sepharose (Pharmacia) equilibrated in dialysis buffer. Heparin-Sepharose was washed with dialysis buffer and then eluted with a linear 50–1000 mM NaCl gradient in eight column volumes. The XStau2 peak was concentrated and dialyzed against PBS-G-500 (PBS with 20% v/v glycerol and NaCl to 500 mM), PBS-G-300. and then PBSG. Part of the XStau2 protein precipitated in PBSG and was resuspended in PBS by sonication.
Both recombinant proteins were quantitated by Bradford assays, and verified by comparing titrations on a Coomassie Blue-stained 10% SDS-PAGE gel and known amounts of BSA.
Antibody preparation and purification
Soluble XStau1 protein was used to raise rabbit antibodies by CovalAB. To prepare rabbit polyclonal antisera against the XStau2 protein, 500 μg of the precipitated XStau2 protein pellet from the Heparin-Sepharose column was used to immunize New Zealand White rabbits using the Titermax Gold Adjuvant system (Stratech Scientific). Rabbits were boosted using the soluble XStau2 protein.
XStau1 and XStau2 pre-immune antibodies did not detect any oocyte protein (data not shown). To neutralize XStau2 epitopes in XStau1 antibody, XStau2 protein was added to XStau1 antibody to a final concentration of 100 ng/μL and incubated at room temperature for 15 min to allow neutralization prior to Western blotting. To deplete XStau1 epitopes from XStau2 antibody, ~ 20 μg of XStau1 protein was Western-blotted and the membrane then stained with Ponceau S (0.2% Ponceau in 0.3% TCA) to allow excision of the recombinant protein slice. The membrane was blocked overnight at 4°C in 5% BSA in PBS with rotation. Four hundred μL of XStau2 antisera was mixed with an equal volume of 5% BSA in PBS and incubated with the membrane overnight at 4°C with rotation. The depleted antisera was removed and tested for XStau1 and XStau2 cross-reactivity by Western blot analysis. This depletion was then repeated another three times until the cross-reactivity was such that the majority of XStau1 cross-reacting epitopes had been removed.
To affinity-purify antibodies for immunostaining, the depletion protocol was followed until the initial antibody membrane-binding step. Following the removal of the depleted antisera, the membrane was washed three times in PBS and the XStau1- or XStau2-specific antibodies were eluted from the membrane in 100 μL 0.1 M glycine, pH 2.5 for 10 min at room temperature with rotation. The solution was then neutralized with 10 μL 1 M Tris, pH 8.0 and finally diluted in 5% BSA in PBS with the addition of 0.03% NaAzide.
Xenopus oocyte, egg, embryo, and tissue sample preparation
Ovarian lobes were removed from Xenopus laevis females and oocytes were manually staged (Dumont 1972) following collagenase treatment to remove follicle cells. Meiotic maturation of stage VI oocytes was induced by the addition of 10 μg/mL progesterone for 16 h. Where indicated, oocytes were pre-incubated with U0126 (Promega V1121) for 4 h prior to addition of progesterone. Dry frozen oocytes, progesterone-matured eggs, or dejellied laid eggs and staged embryos (Nieuwkoop and Faber 1956) were lysed in cold BFH [20 mM Tris, pH 7.4, 50 mM NaCl, 12.5 mM β-glycerophosphate, 15 mM NaF, 10 mM EGTA, 2.2 mM MgCl2, 6 mM DTT, Complete EDTA-free Protease Inhibitors (Roche)] to a final 5–20 μL/cell. The extract was cleared from yolk and debris for 10 min in a minifuge, and the supernatants were denatured in protein sample buffer.
To obtain nuclear and cytoplasmic fractions, nuclei were dissected out of 10 stage VI oocytes stored in J buffer (10 mM HEPES KOH, pH 7.2, 70 mM KCl, 1 mM MgCl2, 2 mM DTT, 0.1 mM EDTA pH 8.0, Complete EDTA-free Protease Inhibitors, 5% glycerol), and the nuclei and the cytoplasmic fractions were lysed in a volume of 2.5 μL J buffer per oocyte by vigorous pipetting. The samples were centrifuged for 10 min at 10,000 rpm, and the supernatant was denatured with protein SDS sample buffer. Lysates from stage VI oocytes and progesterone-matured eggs were treated with λ-phosphatase (New England BioLabs) as follows. Twenty frozen cells were lysed in λ-phosphatase buffer supplemented with Complete EDTA-free Protease Inhibitors and cleared for 10 min in a microfuge. Next, 250 ng of λ-phosphatase and 2 mM MnCl2 were added to two cell equivalents of lysate in a total volume of 10 μL, then incubated for 1 h at 30°C, and an equal volume of 2× protein sample buffer was added to each sample prior to gel electrophoresis and Western blotting. Gel filtration of stage V/VI oocyte lysates (prepared in column buffer, 34.2 mM Na2HPO4, 15.8 mM NaH2PO4, 150 mM NaCl) and treatment with RNase A were described previously (Minshall and Standart 2004).
Lysates from adult tissues in Experiment 1 were prepared in the following manner. Pieces of tissue were frozen in liquid nitrogen immediately after dissection, and then ground in a tissue grinder in RIPA buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% v/v NP40, 0.1% w/v SDS, with a protease inhibitor cocktail). The homogenates were allowed to sit for 10 min before a 10-min spin in a microcentrifuge. The protein concentration of the twice-cleared supernatant was determined using the Bio-Rad Dc Assay system. In Experiment 2, lysates were made as described previously (Cosson et al. 2002). In both experiments, 40 μg of total lysate protein was analyzed by Western blotting.
Protein gel electrophoresis and Western blotting
Two cell equivalents of protein lysates, unless otherwise stated, were resolved on 10% acrylamide SDS-PAGE gels and electrotransferred onto Immobilon-P membrane (Millipore). The membrane was blocked and probed following standard protocols using TBST (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.1% Tween-20) supplemented at the blocking and probing stages with 2% skim milk powder (Marvel). Antibodies were visualized using enhanced chemiluminescence (ECL). Magic Mark Western Molecular weight standards (Invitrogen), run alongside, were also visualized by ECL. The following antibodies were used: rabbit XStau1 (XStau2-neutralized, 1:10,000), XStau2 (XStau1-depleted, 1:2500), Vg1RBP (1:10,000; Zhang et al. 1999), PARN (1:2000; Copeland and Wormington 2001), actin (1:2000; Sigma A2066), MAPK/Erk2 (K-23, 1:25,000; Santa Cruz), phospho-cdc2 (1:1000; Cell Signalling), mouse anti-elrA (HuR 3A2, 1:2000; Santa Cruz) and mouse anti-His (1:1000; Abcam). Secondary antibodies were obtained from Jackson Immunoresearch, all used at 1:10,000.
Immunofluorescence
Oocytes and eggs were fixed in methanol/1% formaldehyde at −20°C for a minimum of 2 h. After stepwise rehydration to PBS containing 0.05% Triton X-100, oocytes were extracted for 10 min in PBS/0.25% Triton X-100, rinsed in PBS, and blocked for at least 1 h in 1% BSA/PBS at 4°C. Samples were incubated with primary antibodies overnight at 4°C and secondary antibodies for 2 h at room temperature, washing in PBS/0.1% Tween-20 and mounted in Citifluor (Citifluor). XStau1 and 2 antibodies were affinity-purified as described above; the ER marker rat monoclonal anti-GRP-94 was obtained from StressGen Biotechnologies, and preabsorbed rhodamine or FITC-labeled anti-Ig antibodies were from Jackson Immunoresearch. Controls in which primary antibodies were omitted confirmed that no cross-reaction between antibodies occurred in double-stain experiments. To control for specificity of anti-XStau1 and 2 staining, affinity-purified antibodies diluted 1/50 and 1/5 respectively were incubated prior to staining with 100 μg/mL of the corresponding XStau protein or BSA overnight at 4°C, then centrifuged for 15 min at 4°C in a microfuge prior to further 1/10 dilution in PBS. Images were acquired on an inverted Leica SP2 confocal microscope equipped with Argon ion and Helium-Neon lasers, and processed using ImageJ software.
Acknowledgments
We thank Francis von Horck (University of Cambridge) for eggs and staged embryos, Michel Philipe (Univ. of Rennes) for tissue samples (Exp. 2), Mike Wormington (University of Virginia) for PARN antibody, and Lucy Otero, Nicola Minshall, and Patrick Chang for assistance and advice. This work was funded by the Wellcome Trust and a BBSRC studentship (R.A.), as well as ACI #167 from the French MRT to E.H. K.C. was supported by fellowships from the NIH and DFG during the course of this work and is grateful to Iain Mattaj for assistance. F.S. was supported by a Howard Florey Fellowship of the Royal Society and NH&MRC of Australia and thanks John Gurdon for support and advice, and Josie McConnell for helpful discussions.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7450204.
REFERENCES
- Alarcon, V.B. and Elinson, R.P. 2001. RNA anchoring in the vegetal cortex of the Xenopus oocyte. J. Cell Sci. 114: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Beckhelling, C., Chang, P., Chevalier, S., Ford, C., and Houliston, E. 2003. Pre-M Phase-promoting factor associates with annulate lamellae in Xenopus oocytes and egg extracts. Mol. Biol. Cell 14: 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley, J.N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., and Deshler, J.O. 2004. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14: 219–224. [DOI] [PubMed] [Google Scholar]
- Braun, R.E. 1998. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 9: 483–489. [DOI] [PubMed] [Google Scholar]
- Broadus, J., Fuerstenberg, S., and Doe, C.Q. 1998. Staufen-dependent localisation of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391: 792–795. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M., Kress, T.L., Vempati, U.D., Mowry, K.L., and King, M.L. 2002. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 248: 82–92. [DOI] [PubMed] [Google Scholar]
- Chang, P., Torres, J., Lewis, R., Mowry, K., Houliston, E., and King, M.L. 2004. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes by entrapment and association with endoplasmic reticulum. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Copeland, P.R. and Wormington, M. 2001. The mechanism and regulation of deadenylation: Identification and characterisation of Xenopus PARN. RNA 7: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, B., Couturier, A., Le Guellec, R., Moreau, J., Chabelskaya, S., Zhouravleva, G., and Philippe, M. 2002. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol. Cell 94: 217–231. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., and Schnapp, B.J. 1997. Localisation of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276: 1128–1131. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Abramson, T., and Schnapp, B.J. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8: 489–496. [DOI] [PubMed] [Google Scholar]
- Duchaine, T.F., Hemraj, I., Furic, L., Deitinghoff, A., Kiebeler, M.A., and DesGroseillers, L. 2002. Staufen2 isoforms localise to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 115: 3285–3295. [DOI] [PubMed] [Google Scholar]
- Dumont, J.N. 1972. Oogenesis in Xenopus laevis (Daudin) I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136: 153–180. [DOI] [PubMed] [Google Scholar]
- Git, A. and Standart, N. 2002. The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA 8: 1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S.D., Schwab, M.S., Taieb, F.E., Lewellyn, A.L., Qian, Y.W., and Maller, J.L. 2001. The critical role of the MAP kinase pathway in meiosis II in Xenopus oocytes is mediated by p90(Rsk). Curr. Biol. 10: 430–438. [DOI] [PubMed] [Google Scholar]
- Havin, L., Git, A., Elisha, Z., Oberman, F., Schwartz, S.P., Standart, N., and Yisraeli, J.K. 1998. RNA binding protein conserved in both microtubule and microfilament-based RNA localization. Genes & Dev. 12: 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler, M.A., Hemraj, I., Verkade, P., Kohrmann, M., Fortes, P., Marion, R.M., Ortin, J., and Dotti, C.G. 1999. The mammalian staufen protein localises to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J. Neurosci. 19: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha, J., Kerr, K., and Macdonald, P. 1995. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- King, M.L., Zhou, Y., and Bubunenko, M. 1999. Polarizing genetic information in the egg: RNA localization in the frog oocyte. BioEssays 21: 546–557. [DOI] [PubMed] [Google Scholar]
- Kloc, M. and Etkin, L.D. 1998. Apparent continuity between the messenger transport organizer and late RNA localization pathways during oogenesis in Xenopus. Mech. Dev. 73: 95–106. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Zearfoss, N.R., and Etkin, L.D. 2002. Mechanisms of subcellular mRNA localization. Cell 108: 533–544. [DOI] [PubMed] [Google Scholar]
- Kohrmann, M., Luo, M., Kaether, C., DesGroseillers, L., Dotti, C.G., and Kiebler, M.A. 1999. Microtubule-dependent recruitment of staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10: 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M. and Kosik, K.S. 2001. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696. [DOI] [PubMed] [Google Scholar]
- Kume, S., Yamamoto, A., Inoue, T., Muto, A., Okano, H., and Mikoshiba, K. 1997. Developmental expression of the inositol 1,4,5-trisphosphate receptor and structural changes in the endoplasmic reticulum during oogenesis and meiotic maturation of Xenopus laevis. Dev. Biol. 182: 228–239 [DOI] [PubMed] [Google Scholar]
- Kwon, S., Abramson, T., Munro, T.P., John, C.M., Kohrmann, M., and Schnapp, B.J. 2002. UUCAC and Vera dependent localisation of VegT RNA in Xenopus oocytes. Curr. Biol. 12: 558–564. [DOI] [PubMed] [Google Scholar]
- Lemaire, P., Garrett, N., and Gurdon, J. 1995. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell 81: 85–94. [DOI] [PubMed] [Google Scholar]
- Li, P., Yang, X., Wasser, M., Cai, Y., and Chia, W. 1997. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90: 437–447. [DOI] [PubMed] [Google Scholar]
- Luo, M., Duchaine, T.F., and DesGroseillers, L. 2002. Molecular mapping of the determinants involved in human staufen-ribosome association. Biochem. J. 365: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo, M., Deitinghoff, A., Muller, J., Goetze, B., Macchi, P., Peters, C., and Kiebler, M.A. 2003. Isolation and characterisation of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. 100: 2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, R.M., Fortes, P., Beloso, A., Dotti, C., and Ortin, J. 1999. A human sequence homologue of staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell Biol. 19: 2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem, D.R., Adams, J., Grunert, S., and St. Johnston, D. 2000. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19: 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall, N. and Standart, N. 2004. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 32: 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen, M., Putz, U., Rehbein, M., Schweizer, M., DesGroseillers, L., Kuhl, D., Richter, D., and Kindler, S. 2001. Two rat brain staufen isoforms differentially bind RNA. J. Neurochem. 76: 155–165. [DOI] [PubMed] [Google Scholar]
- Monshausen, M., Rehbein, M., Richter, D., and Kindler, S. 2002. The RNA-binding protein staufen from rat brain interacts with protein phosphatase-1. J. Neurochem. 81: 557–564. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P.D. and Faber, J. 1956. Normal tables of Xenopus laevis Daudin. Elsevier-North Holland, Amsterdam.
- Ohashi, S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T.-A., and Anzai, K. 2002. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, Fragile X protein, and Myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277: 37804–37810. [DOI] [PubMed] [Google Scholar]
- Palacios, I.M. and St. Johnston, D. 2001. Getting the message across: The intracellular localisation of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17: 569–614. [DOI] [PubMed] [Google Scholar]
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St. Johnston, D., and Varani, G. 2000. RNA recognition by a staufen double-stranded RNA- binding domain. EMBO J. 19: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers, F. and Jan, Y.N. 2000. Staufen: A common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 10: 220–224. [DOI] [PubMed] [Google Scholar]
- Saunders, L.R. and Barber, G.N. 2003. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 17: 961–983. [DOI] [PubMed] [Google Scholar]
- Saunders, P.T., Pathirana, S., Maguire, S.M., Doyle, M., Wood, T., and Bownes, M. 2000. Mouse staufen genes are expressed in germ cells during oogenesis and spermatogenesis. Mol. Hum. Reprod. 6: 983–991. [DOI] [PubMed] [Google Scholar]
- St Johnston, D., Brown, N.H., Gall, J.G., and Jantsch, M. 1992. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. 89: 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S.J., Meulemans, D., Vazquez, L., Colaco, N., and Schuman, E. 2001. A role for a rat homologue of staufen in the transport of RNA to neuronal dendrites. Neuron 32: 463–475. [DOI] [PubMed] [Google Scholar]
- Terasaki, M., Runft, L.L., and Hand, A.R. 2001. Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol. Biol. Cell 12: 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, L., Duchaine, T., Luo, M., Nabi, I.R., and DesGroseillers, L. 1999. Mammalian staufen is a double-stranded RNA-and tubulin-binding protein which localises to the rough endoplasmic reticulum. Mol. Cell. Biol. 19: 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y.J. and Mowry, K.L. 2004. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131: 3035–3045. [DOI] [PubMed] [Google Scholar]
- Zauner, W., Kratz, J., Staunton, J., Feick, P., and Wiche, G. 1992. Identification of two distinct microtubule binding domains on recombinant rat MAP1B. Eur. J. Cell Biol. 57: 66–74. [PubMed] [Google Scholar]
- Zhang, Q., Yaniv, K., Oberman, F., Wolke, U., Git, A., Fromer, M., Taylor, W.L., Meyer, D., Standart, N., Raz, E., et al. 1999. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88: 101–106. [DOI] [PubMed] [Google Scholar]
- Zhao, W.M., Jiang, C., Kroll, T.T., and Huber, P. 2001. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 20: 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]