Abstract
Among all types of RNA, tRNA is unique given that it possesses the largest assortment and abundance of modified nucleosides. The methylation at N1 of adenosine 58 is a conserved modification, occurring in bacterial, archaeal, and eukaryotic tRNAs. In the yeast Saccharomyces cerevisiae, the tRNA 1-methyladenosine 58 (m1A58) methyltransferase (Mtase) is a two-subunit enzyme encoded by the essential genes TRM6 (GCD10) and TRM61 (GCD14). While the significance of many tRNA modifications is poorly understood, methylation of A58 is known to be critical for maintaining the stability of initiator tRNAMet in yeast. Furthermore, all retroviruses utilize m1A58-containing tRNAs to prime reverse transcription, and it has been shown that the presence of m1A58 in human tRNA3 Lys is needed for accurate termination of plus-strand strong-stop DNA synthesis during HIV-1 replication. In this study we have identified the human homologs of the yeast m1A Mtase through amino acid sequence identity and complementation of trm6 and trm61 mutant phenotypes. When coexpressed in yeast, human Trm6p and Trm61p restored the formation of m1A in tRNA, modifying both yeast initiator tRNAMet and human tRNA3 Lys. Stable hTrm6p/hTrm61p complexes purified from yeast maintained tRNA m1A Mtase activity in vitro. The human m1A Mtase complex also exhibited substrate specificity—modifying wild-type yeast tRNAi Met but not an A58U mutant. Therefore, the human tRNA m1A Mtase shares both functional and structural homology with the yeast tRNA m1A Mtase, possessing similar enzymatic activity as well as a conserved binary composition.
Keywords: modified nucleoside, methyltransferase, tRNA modification, HIV-1 replication
INTRODUCTION
The cellular translational machinery is capable of converting nucleotide sequences into amino acid chains because of the distinctive structure of transfer RNA (tRNA) molecules, making tRNAs crucial for proper gene expression and protein biosynthesis. In order to reach their mature forms, tRNAs undergo a series of post-transcriptional processing events, including end trimming, splicing of intervening sequences, CCA addition to the 3′-end, and base or ribose modification (Hopper and Phizicky 2003). Modified nucleosides have been detected in most cellular RNAs; however, tRNAs are more abundantly and diversely modified than are other RNAs (Björk 1995). In many cases, the exact role modified nucleosides play in tRNA function remains unknown due to the finding that tRNAs lacking modified nucleotides retain their normal function. For example, deletion of genes from the yeast Saccharomyces cerevisiae encoding TRM2 (generates m5U54), TRM4 (generates m5C34, 40, 48, 49), or PUS6 (generates Ψ31) exhibit no visible growth defect (Wu et al. 1998; Nordlund et al. 2000; Ansmant et al. 2001).
The modified nucleoside 1-methyladenosine (m1A) is found at position 58 in tRNAs from organisms in each of the three domains of life. In S. cerevisiae, 23 of the 34 tRNA sequences that have been determined contain m1A58 (Sprinzl and Vassilenko 2005), and modification of tRNAGln(CTG) has recently been discovered by using a microarray (Hiley et al. 2005). We previously showed that a two-subunit methyltransferase (Mtase) encoded in yeast by the essential TRM6 (GCD10) and TRM61 (GCD14) genes is responsible for this post-transcriptional base modification (Anderson et al. 2000). In keeping with the conventional nomenclature of tRNA modification enzymes from yeast, GCD10 and GCD14 are now referred to as TRM6 and TRM61, respectively (Kadaba et al. 2004). While structural and functional homologs of Trm61p have been identified in Archaea and Eubacteria (Bujnicki 2001; Gupta et al. 2001; Droogmans et al. 2003; Roovers et al. 2004; Varshney et al. 2004), homologs of Trm6p have not been found in the sequenced genomes of organisms from these same domains (Bujnicki 2001). It has been suggested that a gene duplication of TRM61 appeared early in eukaryotic evolution to yield the progenitor of the TRM6 gene that is presently found in eukaryotes (Bujnicki 2001).
Previously, we reported that tRNAs from a yeast strain bearing a trm6 deletion (trm6Δ) lacked m1A58 and that its absence led to the selective reduction of steady-state initiator methionine tRNA (tRNAi Met) (Anderson et al. 1998). Accordingly, the lethality of this defect is suppressed by the overexpression of tRNAi Met (Anderson et al. 1998). Mutations in TRM61 that were predicted to interfere with S-adenosylmethionine (AdoMet) binding exhibited a null phenotype but did not disrupt formation of enzyme complexes. In addition, purified Trm61p alone failed to bind tRNA in an in vitro assay, leading to the hypothesis that Trm6p binds the tRNA substrate while Trm61p binds AdoMet and catalyzes the N1-methylation of adenosine 58 in tRNA (Anderson et al. 2000).
Along with the function of m1A58 in tRNAi Met stability, it has been reported that m1A58 in human tRNA3 Lys is important for efficient replication of human immunodeficiency virus type 1 (HIV-1) (Gilboa et al. 1979; Burnett and McHenry 1997; Renda et al. 2001). All retroviruses contain a primer binding site (PBS) of 18 nucleotides that is complementary to the 3′-end of a host tRNA, allowing the tRNA to be used as a primer for – strand DNA synthesis and as a template for replication of the PBS on the + strand. Past work has shown that the presence of m1A58 leads to proper termination of + strand strong-stop DNA synthesis during HIV replication, presumably because m1A can not base pair correctly (Burnett and McHenry 1997; Renda et al. 2001). The significance of m1A58 is further demonstrated by in vivo studies in which HIV replication is inhibited in T cells expressing tRNA3 Lys with A58 changed to U (Renda et al. 2001, 2004). Since tRNA3 Lys is directly involved in HIV replication and because HIV exhibits a preference for this tRNA (Das et al. 1995; Wakefield et al. 1995, 1996; Moore et al. 2004), preventing the modification of tRNA3 Lys has been suggested as a possible anti-HIV therapy (Burnett and McHenry 1997; Renda et al. 2001, 2004). While m1A58 is essential for yeast viability, a rat mammary adenocarcinoma with reduced m1A in tRNA has been described, demonstrating that decreased m1A methyltransferase activity is not lethal (Salas et al. 1982). Consequently, the human m1A58 methyltransferase may be one possible target for an HIV therapeutic agent.
Having established the identity of the yeast m1A58 Mtase in previous work, we sought to exploit the yeast system to expand our studies to include the apparent human homolog. Because this enzyme may represent a target for HIV treatment, it is important to determine if the human homologs of the yeast enzyme encode a functional m1A58 Mtase. Here we show the results of several experiments that reveal that human TRM6 (hTRM6) and human TRM61 (hTRM61) together encode an active tRNA m1A Mtase capable of modifying both yeast tRNAi Met and human tRNA3 Lys in vivo. In addition, we report that hTrm6p/ hTrm61p complexes purified from yeast modify an in vitro transcribed tRNA. Furthermore, we provide evidence that the human enzyme synthesizes m1A at position 58. The human m1A58 methyltransferase is now the fourth tRNA methyltransferase from humans to be cloned and characterized, and only the second to be purified (Liu and Straby 2000; Alexandrov et al. 2002; Brule et al. 2004).
RESULTS
Identification of human homologs of the yeast tRNA m1A methyltransferase
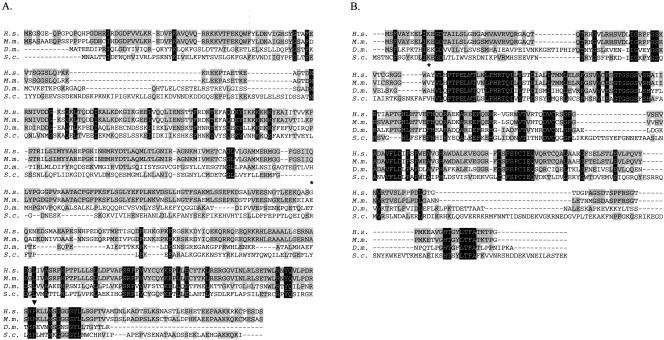
The presence of m1A in tRNA from the three kingdoms of life (Björk 1995) suggested that an enzyme with homologous function exists in all organisms, including humans. We began our search for the human homolog by conducting BLAST searches of the human EST database (Altschul et al. 1990) using the polypeptide sequences of Trm6p (previously Gcd10p) and Trm61p (previously Gcd14p). Several EST clones containing partial sequences that encoded proteins with significant similarity to Trm6p or Trm61p were identified (data not shown). The most complete human cDNA identified contained an open reading frame (ORF) with significant similarity (20% identity) to yeast Trm6p, with the greatest homology found near the C terminus (Fig. 1A). Of particular note is a conserved dipeptide near the C terminus, consisting of histidine 431 and proline 432. In yeast it has been found that substitution of proline 431 by arginine or leucine (trm6-504 or trm6-505, respectively) (J. Anderson, unpubl.) causes temperature-sensitive growth at 36°C, suggesting this conserved region has functional importance. This histidine/proline dipeptide is found in the sequence of the apparent human homolog as well as in putative homologs from other eukaryotes such as Mus musculus and Drosophila melanogaster (Fig. 1A; Bujnicki 2001). In the case of Trm61p, there is 30% identity overall with the apparent human homolog (Fig. 1B). In addition, the hTrm61p sequence shares identity with the Trm61p in specific regions that represent known motifs common among S-adenosylmethionine–dependent methyltransferases (Kagan and Clarke 1994; Fauman et al. 1999). From these preliminary structural comparisons, two cDNAs were obtained for further investigation.
FIGURE 1.
Eukaryotic orthologs of Trm6p and Trm61p. (A) Alignment of predicted polypeptide sequences of Trm6p from Homo sapiens (H.s.) with homologous proteins from Mus musculus (M.m.), Drosophila melanogaster (D.m.), and Saccharomyces cerevisiae (S.c.). The arrowhead highlights proline 431, the amino acid changed in the temperature-sensitive stains trm6-504 and trm6-505. The asterisk indicates the substitution of glycine for glutamic acid at amino acid 299. (B) Alignment of Trm61p from H.s. with homologous proteins from eukaryotes (M.m, D.m., S.c.) as in A. The alanine at position 66, which is found as valine in sequence from the human genome, is indicated by the asterisk. Shading designates the amino acids conserved between two or more homologs.
To study the function of hTrm6p, a cDNA (accession no. AK000613, plasmid p198) containing the full-length hTRM6 ORF was obtained. When compared to the human genome, this cDNA has two consecutive nucleotide changes, resulting in the substitution of glycine for glutamic acid at position 299 (Fig. 1A). As the amino acid at this position is not conserved, this substitution is not likely to affect hTrm6p function. A plasmid containing a full-length hTRM61 cDNA (plasmid p218) was obtained. When this cDNA was sequenced and the sequence was compared with both the human genome and another cDNA (BC010167), two nucleotide changes were noted. Only one of these changes alters the amino acid sequence, causing the substitution of alanine for valine at position 66 (Fig. 1B). Given the presence of alanine in this same position in the Trm61p homolog from D. melanogaster (Fig. 1B; Bujnicki 2001), it is unlikely that this change would alter hTrm61p function.
Complementation of temperature-sensitivity in yeast mutants by hTRM6/hTRM61
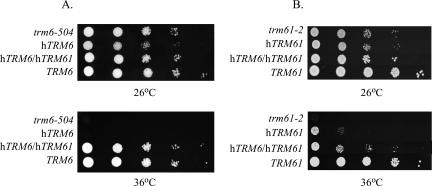
Temperature-sensitive trm6 and trm61 mutants exhibit a reduced steady-state level of tRNAi Met, which we proposed and demonstrated is due to a lack of m1A58 that leads to pre-tRNAi Met instability and degradation (Anderson et al. 1998; Calvo et al. 1999; Kadaba et al. 2004). These mutant strains grow at 26°C but are unable to grow at 36°C. Two yeast strains bearing point mutations, trm6-504 or trm61-2 (Y190 or Hm296, respectively), were used to determine whether expression of hTRM6 and/or hTRM61 could complement their temperature-sensitive growth defects. Both hTRM6 and hTRM61 cDNA sequences were amplified by PCR and cloned into pESC-URA (Stratagene), a vector that allows for the individual or simultaneous expression of two exogenous proteins in yeast using the galactose-inducible/ glucose-repressible GAL1-10 promoter. A TRM6 strain (Y200) was transformed with pESC-URA, while trm6-504 was transformed with pESC-URA or pESC-URA containing hTRM6 (p244), or both hTRM6 and hTRM61 (p230). Transformants were grown in synthetic complete liquid media supplemented with galactose and raffinose (SCgal/ raf) and lacking uracil to select for plasmids. Serial dilutions of these cultures were spotted onto selective SCgal/raf or SC glucose plates at permissive (26°C) and nonpermissive (36°C) temperatures. While colonies readily formed in all of the transformants at the permissive temperature, colony formation at the nonpermissive temperature was seen only for the wild-type TRM6 strain or the trm6-504 mutant strain containing both hTRM6 and hTRM61 (Fig. 2A). Expression of hTRM6 alone was unable to complement the trm6-504 temperature-sensitive growth defect at 36°C, demonstrating that hTrm6p is likely to be insufficient for m1A58 formation in yeast tRNAs. Moreover, this result provided the first indication that hTRM6 and hTRM61 together encode an active tRNA m1A Mtase. On SC glucose media (pESC-URA repressing condition), only the wild-type TRM6 strain exhibited growth at 36°C, demonstrating that complementation is dependent on expression of hTRM6 and hTRM61 (S. Ozanick and J. Anderson, unpubl.).
FIGURE 2.
hTrm6p/hTrm61p expression in yeast complements the temperature-sensitivity of yeast strains trm6-504 and trm61-2. (A) A trm6-504 strain was transformed with pESC-URA (shown as trm6-504) or pESC-URA containing hTRM6 (p244) or hTRM6/hTRM61 (p230). These transformants and an isogenic wild-type strain bearing pESC-URA (shown as TRM6) were grown to saturation at room temperature in liquid medium, SCgal/raf lacking uracil. Serial 10-fold dilutions of these cultures were spotted (5 μL) onto SCgal/raf plates lacking uracil and incubated at the indicated temperature for 2–4 d. (B) A trm61-2 strain was transformed with pESC-URA (shown as trm61-2) or with plasmids containing hTRM61 (p231), hTRM6/hTRM61 (p230), or TRM61 (pRC56). Growth of the transformants on SCgal/raf plates was as described in A at the indicated temperatures for 2–4 d.
Similarly, strain trm61-2 (Hm296) was transformed with a single-copy plasmid bearing TRM61 (pRC56) (Calvo et al. 1999), pESC-URA or pESC-URA containing hTRM61 (p231), or both hTRM6 and hTRM61 (p230) and tested for growth at permissive and nonpermissive temperatures on SCgal/raf plates. In each case, colonies formed at the permissive temperature (Fig. 2B). At the nonpermissive temperature, the formation of colonies by transformants expressing both hTRM6 and hTRM61, albeit somewhat smaller than TRM61 transformants, indicated that expression of hTrm6p and hTrm61p partially complements the trm61-2 temperature-sensitive growth defect. Interestingly, a trm61-2 mutant expressing hTRM61 alone exhibited some growth at 36°C (Fig. 2B). One possible explanation for the low level of complementation of trm61-2 by hTrm61p is that this subunit functions to some degree as an m1A Mtase in yeast independently of hTrm6p. However, Western blot analysis has illustrated that hTrm61p is expressed at very low levels in the absence of hTrm6p (S. Ozanick and J. Anderson, unpubl.). This is not unprecedented, as a similar situation exists for members of another two-subunit tRNA methyltransferase found in yeast, Trm8p and Trm82p. In a trm82 deletion strain, the level of Trm8p is reduced, even though TRM8 mRNA levels do not change (Alexandrov et al. 2005). Alternatively, hTrm61p may be capable of interacting with Trm6p to create an inefficient, yet active, enzyme. To address the possibility that hTrm61p restores some m1A formation in tRNA, total tRNA was purified from the trm61-2 strain expressing hTrm61p alone and analyzed by HPLC. The level of m1A in this strain was not found to be greater than that of the trm61-2 strain containing only pESC-URA (see below and Table 1). Therefore, the growth seen for the trm61-2 strain expressing hTrm61p is not due to a detectable increase in the amount of m1A in total tRNA.
TABLE 1.
HPLC analysis of tRNA
| Relative m1A | Relative m5C | |||
| Yeast strain/plasmid | m1A/Ψ | % Wild type | m5C/Ψ | % Wild type |
| Wild type/pESC-URA | 0.29 | 100 | 0.22 | 100 |
| trm6-504/pESC-URA | 0.02 | 6 | 0.21 | 98 |
| trm6-504/hTRM6/hTRM61 | 0.31 | 107 | 0.22 | 100 |
| trm61-2/TRM61 | 0.21 | 100 | 0.16 | 100 |
| trm61-2/pESC-URA | 0.02 | 10 | 0.16 | 100 |
| trm61-2/hTRM61 | 0.02 | 10 | 0.15 | 94 |
| trm61-2/hTRM6/hTRM61 | 0.15 | 71 | 0.18 | 113 |
The quantitative determination of m1A and 5-methylcytidine (m5C) levels was done by normalizing the percentage area of the m1A and m5C peaks to the percentage area of the pseudouridine (Ψ) peak in each sample. The amount of m1A and m5C found in the wild-type strain containing only pESC-URA and the trm61-2 strain complemented with TRM61 was set at 100%.
Human Trm6p/Trm61p expression restores m1A formation in tRNA in vivo
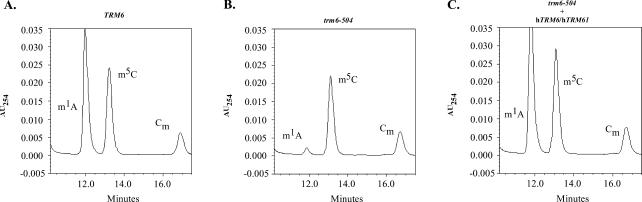
Complementation of the temperature-sensitivity exhibited by trm6-504 or trm61-2 mutants can occur by increasing the copy number of the IMT genes, encoding tRNAi Met, or LHP1, encoding the tRNA 3′-end processing factor Lhp1p (Yoo and Wolin 1997; Anderson et al. 1998; Calvo et al. 1999). This genetic complementation results in an increase in mature tRNAi Met but does not restore tRNA m1A formation in a strain lacking a functional m1A Mtase (Anderson et al. 1998). To determine whether complementation of trm6-504 and trm61-2 mutants by expression of the cloned human cDNAs is accompanied by formation of m1A in tRNA and not merely an increase in tRNAi Met levels, we determined the modified nucleoside content of total tRNA from a number of strains grown in selective SCgal/raf at 30°C (Fig. 3; Table 1). Total tRNA was purified from the wild-type (Y200) strain containing pESC-URA, the trm6-504 mutant containing pESC-URA, and the trm6-504 mutant expressing hTRM6/hTRM61 (p230). The quantitative determination of m1A and m5C levels was accomplished by normalizing the percentage area of the m1A or 5-methylcytidine (m5C) peaks with the percentage area of the pseudouridine (Ψ) peak in each sample. We chose to use Ψ to normalize the level of m1A and m5C because of its ubiquitous nature (Grosjean et al. 1995), and formation of Ψ does not depend on the presence of m1A58 in tRNA (Jiang et al. 1997). In contrast to the nearly equivalent levels of m5C detected in tRNAs from all three strains (Fig. 3A–C; Table 1), the trm6-504 mutant contained a low level of m1A (6% compared with wild type) (Table 1; Fig. 3B). Remarkably, the strain expressing hTRM6 and hTRM61 exhibited a level of m1A similar to, if not slightly higher than, the wild-type strain (107% vs. 100%) (Table 1; Fig. 3A, C). It is possible that the slightly increased level of m1A in this strain is due to modification of tRNAs by hTrm6p/Trm61p that are not normally modified by the yeast m1A Mtase (see below). The HPLC data corroborate our interpretation of the growth studies, demonstrating that expression of both hTRM6 and hTRM61 repairs the primary biochemical defect in the mutant strain by restoring the formation of m1A in tRNA in vivo. We propose from these observations that hTRM6 and hTRM61 encode proteins that form an active m1A Mtase in yeast and that complementation of trm6-504 and trm61-2 mutants is through m1A58 formation in tRNA.
FIGURE 3.
Expression of hTrm6p/hTrm61p restores wild-type levels of m1A in tRNA. Total tRNA purified from wild-type yeast (Y200) containing pESC-URA (A), trm6-504 (Y190) containing pESC-URA (B), and trm6-504 (Y190) containing hTRM6/hTRM61(p230) (C) was digested to single nucleosides and analyzed by HPLC. The absorbance at 254 nm (AU254) was measured and plotted against retention time in minutes, as previously described (Anderson et al. 1998). For clarity, only a portion of the chromatograph—the region where m1A, 5-methylcytidine (m5C), and 2′-O-methylcytidine (Cm) elute from the column—is shown.
To address whether the inability of hTRM6 and hTRM61 to fully complement a trm61-2 mutant is due to incomplete restoration of m1A formation in tRNA, and to investigate the partial complementation provided by hTrm61p alone, we conducted the same HPLC analysis on tRNA purified from trm61-2 strains containing pESC-URA, TRM61 (pRC56), hTRM61 alone (p231), or both hTRM6 and hTRM61 (p230). Similar to what we had observed for trm6-504, the level ofm1A in tRNA fromtrm61-2 containing only pESC-URA was found to be 10% of that found in trm61-2 containing TRM61 on a plasmid (Table 1). Furthermore, the trm61-2 strain expressing only hTRM61 did not contain an increased amount of m1A. Consistent with the partial growth complementation of trm61-2 by hTRM6 and hTRM61 expression, we determined that the m1A levels in tRNA isolated from that strain were restored to only 71% of the wild-type control (Table 1). This result, along with the complementation analysis, indicates that partial complementation of trm61-2 by hTRM6 and hTRM61 expression is due to an inability of the human m1A Mtase to fully restore m1A formation in the presence of a defective Trm61p protein. Although this result is not understood and is beyond the scope of this study, one possible explanation is that inactive complexes form from interactions between yeast and human subunits. Alternatively, differences in hTrm6p/hTrm61p expression due to the strain background may explain the full complementation seen in the trm6-504 strain but only partial complementation seen in the trm61-2 strain. It should also be noted that the trm61-2 mutation is located in the start codon of TRM61 and results in low levels of Trm61p (Calvo et al. 1999).
Modification of htRNA3 Lys by hTrm6p/hTrm61p in yeast
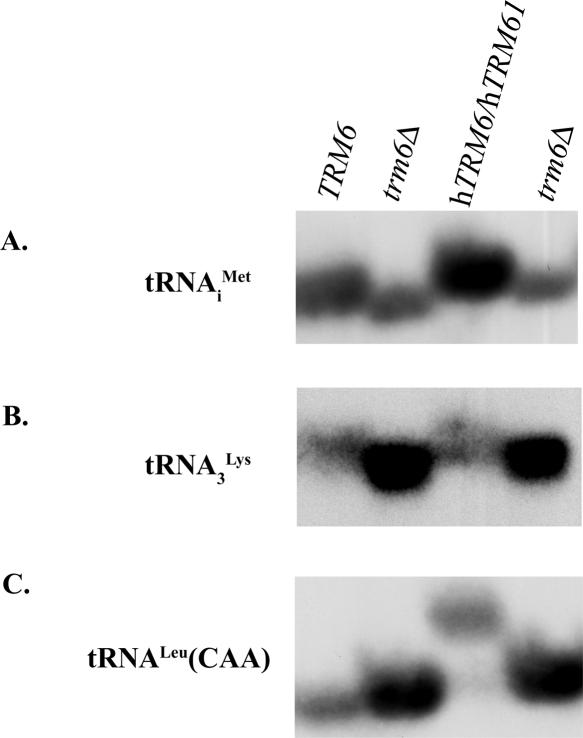
While HPLC analysis demonstrates that expression of hTRM6 and hTRM61 leads to the formation of m1A in tRNA in vivo, it does not reveal the m1A content of specific tRNAs. To establish the identity of tRNAs modified by hTrm6/hTrm61p, Northern blot analysis was performed. A plasmid containing the htRNA3 Lys gene was obtained as a gift from Anders Byström (Umeå University, Umeå, Sweden). Three strains were created starting with a trm6 deletion (trm6Δ): trm6Δ bearing htRNA3 Lys and pESC-URA; trm6Δ bearing htRNA3 Lys and pESC-URA containing hTRM6 and hTRM61; and trm6Δ bearing htRNA3 Lys and a plasmid borne copy of yeast TRM6. Total RNA prepared from these strains was subjected to acidic denaturing polyacrylamide gel electrophoresis (Varshney et al. 1991) and transferred to a nylon membrane for Northern hybridization. The blot was probed for the presence of tRNAi Met, htRNA3 Lys, or tRNALeu(CAA) using a radiolabeled oligo-nucleotide (Fig. 4). Northern hybridization using the probe specific for tRNAi Met illustrates that the mobility of this tRNA from yeast containing either TRM6 or hTRM6 and hTRM61 is altered compared with tRNA from the trm6Δ strain (Fig. 4A). This mobility shift is presumably due to the presence of m1A58 in this tRNA. Similarly, a change in the mobility of htRNA3 Lys is also seen when purified from strains expressing either TRM6 or hTRM6 and hTRM61 (Fig. 4B), suggesting that both the yeast and human m1A Mtases can modify this tRNA. Using a probe specific for tRNALeu(CAA), a tRNA that does not contain m1A in yeast, no difference was seen in the mobility of this tRNA when purified from the strain expressing TRM6 or the trm6Δ strain (Fig. 4C). However, a noticeable change was seen in the migration of tRNALeu(CAA) from cells expressing hTRM6 and hTRM61 (Fig. 4C), suggesting that hTrm6p/hTrm61p modifies this tRNA. It is known that tRNALeu with an anti-codon consisting of an unknown nucleotide followed by two adenosines is modified in humans (Sprinzl and Vassilenko 2005), and the results of this Northern blot analysis suggest that yeast tRNALeu(CAA) is recognized as a substrate for modification by hTrm6p/ hTrm61p, but not the yeast m1A Mtase. This is consistent with the small increase in m1A levels seen by HPLC analysis of total tRNA isolated from a trm6Δ strain expressing both hTRM6 and hTRM61 (Table 1). The ability of the human m1A Mtase to modify yeast tRNALeu(CAA) while the yeast enzyme does not implies that the yeast and human enzyme have different substrate specificities. This knowledge should be useful as we begin our studies aimed at defining protein determinants required for substrate specificity of the yeast and human enzymes.
FIGURE 4.
Modification of specific tRNAs by hTrm6p/hTrm61p. Northern blot analysis of total RNA (5 μg) from a trm6Δ strain expressing htRNA3 Lys and either TRM6 (TRM6), pESC-URA (trm6Δ), or hTRM6/hTRM61 (hTRM6/hTRM61). RNAs were separated on an acidic, denaturing 5% acrylamide/bis-acrylamide, 19:1, sequencing gel and transferred to a nylonmembrane for hybridization with radiolabeled probes specific for tRNAi Met (A), htRNA3 Lys (B), or tRNALeu(CAA) (C). Results were visualized by autoradiography.
hTrm6p/hTrm61p form a stable complex when expressed in yeast
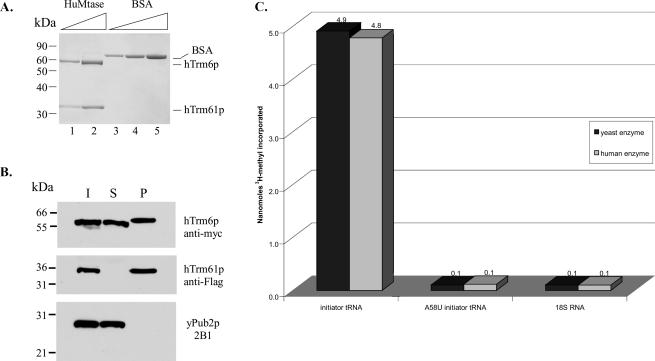
Since full complementation of trm6-504 or trm61-2 mutants required concomitant expression of hTrm6p and hTrm61p, we wanted to determine whether the human proteins also form complexes in vivo. To avoid the possibility of hTrm61p interacting with Trm6p (see above), hTrm6p and hTrm61p were coexpressed in a trm6Δ strain (Y146). Whole-cell extracts were prepared and the human m1A Mtase complexes were purified under nondenaturing conditions by affinity chromatography using anti-Flag antibody (Materials and Methods), which is directed against a single Flag epitope on hTrm61p. The purified proteins were separated by SDS-PAGE and analyzed by Coomassie blue staining (Fig. 5A). The Coomassie-stained immunoprecipitate showed the presence of two predominant proteins with apparent molecular weights of 57 kDa and 33 kDa. These apparent molecular weights are in good agreement with the predicted molecular weights of hTrm6p (55 kDa) and hTrm61p (31 kDa) based on the cDNA sequences and the apparent molecular weights of hTrm6p and hTrm61p detected by immunoblot analysis (Fig. 5B). Western blot analysis of whole-cell extract and the supernatant and pellet fractions following affinity purification was performed by using antibodies against a c-myc epitope in hTrm6p and the Flag epitope in hTrm61p. This analysis confirmed the presence of both hTrm6p and hTrm61p in affinity-purified complexes, and we concluded that hTrm6p was expressed to higher levels than was hTrm61p in yeast due to its presence and hTrm61p absence in the supernatant after affinity purification (Fig. 5B). The specificity of our affinity purification procedure was demonstrated by the inability to detect an abundant ribosomal protein, Pub2p (Anderson et al. 1993), in the immunoprecipitate (Fig. 5B) and the relative purity of the immunoprecipitate as shown by Coomassie blue staining (Fig. 5A). It is clear from these data that hTrm6p forms a stable complex with hTrm61p in vivo.
FIGURE 5.
Purified hTrm6p/hTrm61p complexes possess tRNA (m1A) Mtase activity in vitro. Whole-cell extract (~50 mg) from a trm6Δ strain (Y146) grown in SCgal/raf and containing hTRM6 and hTRM61 (p230) was used as starting material for purification of Flag-tagged hTrm61p using anti-Flag antibody bound to Sepharose beads (Sigma-Aldrich). Protein samples were separated by SDS-PAGE and stained with Coomassie blue (A) or transferred to a nitrocellulose membrane for immunoblotting (B). (A) Lanes 1 and 2 contain 5 and 15 μL of purified enzyme complex, respectively; lanes 3, 4, and 5 contain 0.25, 0.5, and 1.0 μg of BSA, respectively (included for quantitative purposes). (B) Lanes I (input) and S (supernatant) contain 50 μg each of whole-cell extract or the supernatant after anti-Flag affinity purification, respectively. Lane P (pellet) contains 0.5 μL (1/300) of the complex purified from 500 mL of yeast cells. hTrm6p, hTrm61p, and Pub2p were detected by ECL (Amersham) using anti-c-myc, anti-Flag, and 2B1 monoclonal antibodies, respectively. Pub2p is an abundant yeast large ribosomal subunit protein, which serves as a control for specificity. (C) Purified yeast or human Trm6p/Trm61p complexes were combined with S-adenosyl-[3H-methyl]- methionine and in vitro transcribed tRNAi Met, A58U tRNAi Met, or 18S rRNA (Ambion) under optimal reaction conditions (see Materials and Methods). The acid insoluble material was collected on nitrocellulose filters, dried, and counted by liquid scintillation. The amount of 3H-methyl incorporated is shown as the average of two identical, independent reactions. Each reaction contained 15 nmol tRNA or 18S RNA substrate.
Purified hTrm6p/hTrm61p complexes modify yeast tRNAi Met in vitro
We next examined whether the purified hTrm6p/hTrmp61p complexes possessed m1A Mtase activity in vitro. Initially, tRNAi Met purified to homogeneity from yeast cells lacking a functional m1A Mtase was used as substrate for yeast m1A Mtase complexes to optimize conditions for the methyltransferase activity in vitro (data not shown). To test the ability of hTrm6p/hTrm61p complexes to modify tRNA, yeast tRNAi Met was transcribed from a linearized plasmid (HG300) using T7 RNA polymerase. Unlabeled T7 transcripts were renatured and combined with purified hTrm6p/ hTrm61p and S-adenosyl-l-[3H-methyl]methionine under conditions optimized for m1A activity when using purified yeast m1A Mtase. Both human and yeast Trm6p/Trm61p complexes were able to incorporate 3H-methyl into the tRNAi Met substrate, demonstrating the methyltransferase activity of these enzymes. By using purified hTrm6p/ hTrm61p complexes from yeast, we observed the incorporation of 4.8nmol of 3H-methyl in reactions containing 15nmol tRNA (Fig. 5C). Similar results (4.9 nmol incorporated) were obtained when yeast Trm6p/Trm61p complexes were tested (Fig. 5C), indicating that the human and yeast m1A Mtases can modify approximately one third of the tRNAi Met substrate. Increasing the incubation time or concentration of enzyme in a reaction did not lead to a significant increase in the amount of substrate modified.
One caveat of these in vitro assays is the possibility of modification at an adenosine in the tRNA at a position other than A58 of the TΨC loop. To address this limitation, a variant of tRNAi Met in which A58 was changed to U58 was synthesized as a T7 transcript. While wild-type tRNAi Met contained a significant amount of radioactive methyl incorporated, the amount of 3H-methyl incorporated by both human and yeast Trm6p/Trm61p complexes into the A58U mutant tRNAi Met was the same as seen for a negative control, 18S rRNA (0.1 nmol) (Fig. 5C). This result illustrates not only the substrate specificity possessed by both the human and yeast m1A Mtases but also their ability to carry out position-specific modification of tRNA.
We conclude that the cDNAs identified do encode a human tRNA m1A Mtase that functions to modify tRNAs at position 58 of the TΨC loop. This conclusion supports the idea that the heteromeric structure of the yeast tRNA m1A58 Mtase has been conserved throughout eukaryotic evolution, further suggesting that subunit diversity is important for the function of this enzyme in eukaryotes. The exact role that each subunit plays in the formation of m1A in tRNA is presently unknown, but studies are ongoing to define specific enzymatic functions for each subunit using the yeast enzyme as a model to provide insight into how this and other eukaryotic tRNA m1A Mtases function. Finding that the human cDNAs identified in this study encode an active m1A Mtase that can replace a nonfunctional yeast m1A Mtase presents the possibility of using S. cerevisiae as a model system to identify potential in vivo inhibitors of the human m1A Mtase, which ultimately may have clinical significance in treating HIV-1 infections.
MATERIALS AND METHODS
Yeast strains and plasmid construction
The genotypes of yeast strains used in this work are listed in Table 2, and the construction of strains Y190 and Y200 has been described (Kadaba et al. 2004). Yeast transformations were done as described (Ito et al. 1983). To clone hTRM6, the PCR was used to amplify a 1511-bp fragment from AK000613, clone KAT06232 (NEDO Human cDNA-Sequencing Project, Institute of Medical Science, University of Tokyo), using oligos JA87 (5′- GAACGCGTCGACAGAGTCAGACTCTGGGCATT) and JA88 (5′-CGGGGATCCATGGAGGGCTCAGGGGAG). After SalI/BamHI digestion, the fragment was ligated into SalI/BamHI-digested pESC-URA, a yeast epitope-tagging vector (Stratagene) to create plasmid p244. A plasmid with both hTRM6 and hTRM61 was created by using oligos JA100 (5′-GACTAGTATGAGCTTCGTGGCATACGAG) and JA110 (5′-CCATCGATGCCTGGGGTCTTGGTGG) to amplify hTRM61 as a ~900-bp PCR product from p218 (Dr. Michael Tschudi, Yale University, New Haven, CT). After SpeI/ClaI digestion, the fragment was ligated into SpeI/ClaI-digested p244. The resulting plasmid was digested with ClaI, ClaI ends were blunt-ended using Klenow, and the resulting product ligated to form p230. To construct a plasmid containing only hTRM61, p230 was digested with SalI/ BamHI, the SalI/BamHI ends were blunt-ended using Klenow, and the plasmid containing only hTRM61 was gel purified and ligated to form p231.
TABLE 2.
Yeast strains used in this work
| Name | Description | Reference |
| H2457 | MATα, trm6-504, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | (Anderson et al. 1998) |
| Y200 | MATa, TRM6, gcn2-101, his1-29, ura3-52, ino1, (HIS4-lacZ, ura3-52) | (Kadaba et al. 2004) |
| Y190 | MATa, trm6-504, gcn2-101, his1-29, ura3-52m ubi1m (HIS4-lacZ, ura3-52) | (Kadaba et al. 2004) |
| Y146 | MATα, trm6Δ, ura3-52, trp1, leu2Δ1, his3Δ200, pep::HIS4, prb1Δ1′1.6, can1, Gal+ | (Anderson et al. 1998) |
| Hm296 | MATα, trm61-2, ura3-52, gcn2-101, gcn3- 101, his1-29, ino1, (HIS4-lacZ, ura3-52) | (Cuesta et al. 1998) |
Protein purification and detection
Purification of yeast Trm6p/Trm61p complexes was previously described (Anderson et al. 2000) Flag-hTRM61p/hTRM6p complexes were affinity purified from yeast whole-cell extracts. All steps in the enzyme purification process were carried out at 4°C. A trm6Δ strain containing a high-copy plasmid bearing the IMT4 gene (Y146) (Anderson et al. 1998) and transformed with p230 was grown in 2 L of SCgal/raf (synthetic complete media supplemented with 5% galactose and 2% raffinose) lacking uracil and leucine at 30°C to OD600 = 1. Cells were harvested by centrifugation at 7000g for 10 min, washed with 1× Tris buffered saline (TBS), and resuspended in breaking buffer consisting of 1× TBS/ 1 mM DTT/1× protease inhibitor cocktail (Roche), using 1 mL/g wet-weight cells. Cells were broken by using a French press (Thermo IEC) at a pressure of 20,000 psi. The whole-cell extract was centrifuged at 30,000g for 20 min, and the supernatant was then spun again at 75,000g for 20 min. The supernatant from this centrifugation step (20 mL, 10 mg/mL whole-cell extract) was incubated with anti-Flag M2-agarose affinity gel (Sigma) over-night on a nutator at 4°C. After washing the anti-Flag resin five times with breaking buffer, bound complexes were eluted by using breaking buffer containing 10% glycerol and 100 μg/mL Flag peptide (Sigma), and the eluate was concentrated to 100 μL by using a microcentricon YM-30 filter (Millipore). Purified proteins were separated by SDS-PAGE and either stained with Coomassie blue or transferred to nitrocellulose. Nitrocellulose filters were blocked in 5% milk in 1× TBS, 0.1% Tween20 prior to incubation with anti c-myc 1:1000 (Roche), anti-Flag 1:600 (Sigma- Aldrich), or 2B1 1:10,000 (gift of Dr. Maurice Swanson, University of Florida, Gainesville), in 5% milk/1× TBS 1 h to overnight at 4°C. Secondary antibody, goat anti-mouse conjugated to horseradish peroxidase (Amersham), was incubated at 1:10,000 for 0.5–1 h in 5% milk in 1× TBS at room temperature. Blots were washed after primary and secondary antibodies for 5 min, three times, using 1× TBS, 0.1% Tween20 prior to detection using ECL (Amersham). The blot was stripped by using a protocol suggested by the manufacturer (Amersham) prior to immunoblotting with additional antibodies.
Northern blot analysis
A plasmid containing the htRNA3 Lys gene was obtained as a gift from Anders Byström. The gene was cloned into the vector YEplac112 (Gietz and Sugino 1988) as a HindIII/PstI fragment, creating plasmid p241. Y146 was transformed with both p241 and pESC-URA (Y274) or p230 (Y275). The pESC-URA plasmid was selected against in Y274 on 5-fluoroorotic acid (5-FOA) and the 5-FOA–resistant colonies transformed with TRM6 in YCplac33 (p129) (Anderson et al. 1998). Yeast strains were grown in 50 mL selective SCgal/raf at 30°C to OD600 1.2–1.6. Total RNA was prepared as described (Varshney et al. 1991) but adjusted for yeast cultures. tRNA was deacylated with 2 M Tris (pH 8) at 37°C for 30 min, precipitated, and resuspended in diethylpyro-carbonate (DEPC)-treated water. Sample concentrations were determined by measuring the absorbance at 260 nm, and 5 μg of samples was separated on a 5% polyacrylamide gel as described (Varshney et al. 1991). RNA was transferred to Biodyne B (Pall Corp.) in 0.5× Tris Borate EDTA (TBE) using a Panther Semidry Electroblotter (Owl) at 600 mA for 1 h. Probes for htRNA3 Lys, JA141 (5′-CTGGACCCTCAGA), tRNAi Met, JA11 (5′-TCGGTTTCGATCCGAGGACATCAGGGTTATGA), or tRNALeu(CAA), JA151 (5′-TGGTTGCTAAGAGATTCGAAC), were made by incubating 10 pmol of the oligonucleotide with 50 μCi (γ-32P) ATP (GE Healthcare) and T4 polynucleotide kinase (New England Biolabs) at 37°C for 1 h. Probes were separated from unincorporated (γ-32P) ATP by one pass through a 1-mL G-25 column equilibrated with water. Hybridization was carried out overnight in 0.25 M Na2HPO4 (pH 7.5), 1 mM EDTA, 1% BSA, and 7% SDS, and results were visualized by autoradiography.
Mtase activity assay
Mtase activity assays of purified human enzyme were conducted as described (Anderson et al. 2000) by using an optimized 1× Mtase buffer (100 mM Tris [pH 7.6], 0.1 mM EDTA, 10 mM MgCl2, 100 mM NH4Cl, 1 mM DTT). A plasmid containing yeast tRNAi Met (HG300, described in Senger et al. 1992) was obtained as a gift from Dr. Henri Grosjean (CNRS, Gif-sur-Yvette, France). The A58U tRNAi Met mutant plasmid (p324) was created in HG300 using the QuikChange XL site-directed mutagenesis kit (Stratagene) and oligos JA318 (5′-GTCCTCGGATCGTAACCGAGCGGCGC) and JA319 (5′-CGCCGCTCGGTTACGATCCGAGGAC). The A58U mutation was confirmed by DNA sequencing. These plasmids were digested with BstNI and transcribed in vitro using T7 RNA polymerase; as a control, the pTRI-RNA 18S antisense control template (Ambion) was also transcribed. Transcribed RNAs were phenol extracted, ethanol precipitated, and quantitiated using ethidium bromide staining after agarose gel electrophoresis by comparison to known amounts of tRNAPhe from Escherichia coli (Sigma-Aldrich). Before use in Mtase assays, RNAs were renatured by first heating to 90°C for 1 min and 60°C for 15 min, and then cooling to 30°C. Once at 30°C, samples were brought to 2.5 mM MgCl2 and incubated at 30°C for 15 additional minutes, and then placed on ice until being used in an Mtase assay. Activity assays were carried out at 30°C for 10 min using 30 μM 3H-AdoMet, 150 nM RNA substrate, and 15 nM purified enzyme. Radioactivity incorporated into the substrate RNAs was measured by liquid scintillation as previously described (Anderson et al. 1998) using BetaMax scintillation cocktail (ICN Biomedical).
Acknowledgments
We thank Dr. Glen R. Björk and Kerstin Jacobsson for conducting HPLC analysis of modified nucleosides. G.R.B. and K.J. are supported by grants (BU-2930) and (proj 680) from the Swedish Science Research Council and Swedish Cancer Society. We appreciate help from S.A.K. and R.E.S. with Northern blots. We acknowledge the Department of Biological Sciences and the Office of Research and Sponsored Programs at Marquette University for their support of J.A. as a summer research intern and S.O. through an intramural regular research grant.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5040605.
REFERENCES
- Alexandrov, A., Martzen, M.R., and Phizicky, E.M. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, A., Grayhack, E., and Phizicky, E.M. 2005. tRNA m7G methyltransferase Trm8p/Trm82p: Evidence linking activity to growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, J.T., Paddy, M.R., and Swanson, M.S. 1993. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 6102–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., and Hinnebusch, A.G. 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 12: 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., and Hinnebusch, A.G. 2000. The Gcd10p/ Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansmant, I., Motorin, Y., Massenet, S., Grosjean, H., and Branlant, C. 2001. Identification and characterization of the tRNA:Psi 31- synthase (Pus6p) of Saccharomyces cerevisiae. J. Biol. Chem. 276: 34934–34940. [DOI] [PubMed] [Google Scholar]
- Björk, G.R. 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure biosynthesis and function (eds. D. Söll and U.L. RajBhandary), pp. 165–206. ASM Press, Washington, DC.
- Brule, H., Elliott, M., Redlak, M., Zehner, Z., and Holmes, W.M. 2004. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43: 9243–9255. [DOI] [PubMed] [Google Scholar]
- Bujnicki, J.M. 2001. In silico analysis of the tRNA:m1A58 methyltransferase family: Homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett. 507: 123–127. [DOI] [PubMed] [Google Scholar]
- Burnett, B.P. and McHenry, C.S. 1997. Posttranscriptional modification of retroviral primers is required for late stages of DNA replication. Proc. Natl. Acad. Sci. 94: 7210–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, O., Cuesta, R., Anderson, J., Gutierrez, N., Garcia-Barrio, M.T., Hinnebusch, A.G., and Tamame, M. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta, R., Hinnebusch, A.G., and Tamame, M. 1998. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 148: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A., Klaver, B., and Berkhout, B. 1995. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA3 Lys. J. Virol. 69: 3090–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans, L., Roovers, M., Bujnicki, J.M., Tricot, C., Hartsch, T., Stalon, V., and Grosjean, H. 2003. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 31: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauman, E.B., Cheng, X., and Blumenthal, R.M. 1999. Structure and evolution of AdoMet-dependent methyltransferases. In S-adenosylmethionine- dependent methyltransferases: Structures and functions (eds. X. Cheng and R.M. Blumenthal), pp. 1–38. World Scientific, River Edge, NJ.
- Gietz, R.D. and Sugino, A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gilboa, E., Mitra, S.W., Goff, S., and Baltimore, D. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Sprinzl, M., and Steinberg, S. 1995. Posttranscriptionally modified nucleosides in transfer RNA: Their locations and frequencies. Biochimie 77: 139–141. [DOI] [PubMed] [Google Scholar]
- Gupta, A., Kumar, P.H., Dineshkumar, T.K., Varshney, U., and Subramanya, H.S. 2001. Crystal structure of Rv2118c: An Ado- Met-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 312: 381–391. [DOI] [PubMed] [Google Scholar]
- Hiley, S., Jackman, J.E., Babak, T., Trochesset, M., Morris, Q., Phizicky, E.M., and Hughes, T.R. 2005. Detection and discovery of RNA modification using microarrays. Nucleic Acids Res. 33: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukada, Y., Murata, K., and Kimura, A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H.Q., Motorin, Y., Jin, Y.X., and Grosjean, H. 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: An in vitro study. Nucleic Acids Res. 25: 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., and Anderson, J. 2004. Nuclear surveillance and degradation of hypo-modified initiator tRNAMet in S. cerevisiae. Genes & Dev. 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan, R.M. and Clarke, S. 1994. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine–dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310: 417–427. [DOI] [PubMed] [Google Scholar]
- Liu, J. and Straby, K.B. 2000. The human tRNA(m2 2G26)dimethyltransferase: Functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res. 28: 3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, K., Kosloff, B., Kelly, N., Kirkman, R., Dupuy, L., McPherson, S., and Morrow, C. 2004. HIV type 1 that select tRNAHis or tRNALys1,2 as primers for reverse transcription exhibit different infectivities in peripheral blood mononuclear cells. AIDS Res. Hum. Retroviruses 20: 373–381. [DOI] [PubMed] [Google Scholar]
- Nordlund, M.E., Johansson, J.O., von Pawel-Rammingen, U., and Bystrom, A.S. 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda, M.J., Rosenblatt, J.D., Klimatcheva, E., Demeter, L.M., Bambara, R.A., and Planelles, V. 2001. Mutation of the methylated tRNA3 Lys residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J. Virol. 75: 9671–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda, M., Bradel-Tretheway, B., Planelles, V., Bambara, R.A., and Dewhurst, S. 2004. Inhibition of HIV type 1 replication using lenti-viral- mediated delivery of mutant tRNALys3 A58U. AIDS Res. Hum. Retroviruses 20: 1324–1334. [DOI] [PubMed] [Google Scholar]
- Roovers, M., Wouters, J., Bujnicki, J.M., Tricot, C., Stalon, V., Grosjean, H., and Droogmans, L. 2004. A primordial RNA modification enzyme: The case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas, C.E., Uschmann, B.D., and Leboy, P.S. 1982. Methyl-accepting RNA in 13762 mammary adenocarcinoma correlated with low adenine methyltransferase levels. Cancer Res. 42: 5004–5009. [PubMed] [Google Scholar]
- Senger, B., Despons, L., Walter, P., and Fasiolo, F. 1992. The anticodon triplet is not sufficient to confer methionine acceptance to a transfer RNA. Proc. Natl. Acad. Sci. 89: 10768–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M. and Vassilenko, K. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33: D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, U., Lee, C.P., and RajBhandary, U.L. 1991. Direct analysis of aminoacylation levels of tRNA as in vivo. J. Biol. Chem. 266: 24712–24718. [PubMed] [Google Scholar]
- Varshney, U., Ramesh, V., Madabushi, A., Gaur, R., Subramanya, H.S., and RajBhandary, U.L. 2004. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res. 32: 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield, J., Wolf, A., and Morrow, C. 1995. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA3 Lys. J. Virol. 69: 6021–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield, J., Kang, S.-M., and Morrow, C. 1996. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol. 70: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P., Brockenbrough, J.S., Paddy, M.R., and Aris, J.P. 1998. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene 220: 109–117. [DOI] [PubMed] [Google Scholar]
- Yoo, C.J. and Wolin, S.L. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. [DOI] [PubMed] [Google Scholar]