Abstract
The H/ACA small nucleolar ribonucleoprotein (snoRNP) complexes guide the modification of uridine to pseudouridine at conserved sites in rRNA. The H/ACA snoRNPs each comprise a target-site-specific snoRNA and four core proteins, Nop10p, Nhp2p, Gar1p, and the pseudouridine synthase, Cbf5p, in yeast. The secondary structure of the H/ACA snoRNAs includes two hairpins that each contain a large internal loop (the pseudouridylation pocket), one or both of which are partially complementary to the target RNA(s). We have determined the solution structure of an RNA hairpin derived from the human U65 box H/ACA snoRNA including the pseudouridylation pocket and adjacent stems, providing the first three-dimensional structural information on these H/ACA snoRNAs. We have also determined the structure of Nop10p and investigated its interaction with RNA using NMR spectroscopy. Nop10p contains a structurally well-defined N-terminal region composed of a β-hairpin, and the rest of the protein lacks a globular structure. Chemical shift mapping of the interaction of RNA constructs of U65 box H/ACA 3′ hairpin with Nop10p shows that the β-hairpin binds weakly but specifically to RNA. The unstructured region of Nop10p likely interacts with Cbf5p.
Keywords: β-hairpin, NMR, pseudouridylation, RNA–protein
INTRODUCTION
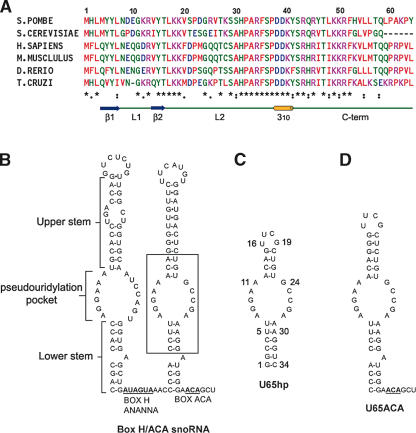
In eukaryotes and archaebacteria, pseudouridine (ψ) modification of ribosomal RNA is mediated by box H/ACA small nucleolar RNPs (snoRNP) (Ganot et al. 1997b; Ni et al. 1997; Tollervey and Kiss 1997). The H/ACA snoRNAs are composed of two hairpins of varying length separated and followed by short single-strand regions that include a conserved box H (5′-ANANNA-3′) sequence and ACA, respectively (Fig. 1B ▶). The 5′ and 3′ hairpins each contain a lower stem, an internal loop (pseudouridylation pocket), an upper stem, and a terminal loop (Ganot et al. 1997b; Ni et al. 1997; Tollervey and Kiss 1997). One or both of the internal loops in each H/ACA snoRNA are complementary to residues flanking a specific modification site, leading to the proposal that the substrate U is positioned for modification by base pairing of its flanking RNA to the internal loop, with the U and its 5′ neighboring residue unpaired at the top of the internal loop (Ganot et al. 1997a). Box H/ACA snoRNAs are therefore considered to be guides because they bind the substrate RNA and mediate conversion of U to ψ. Pseudouridine, which has been dubbed the fifth nucleotide, is the most abundant modified nucleotide in RNA (Ofengand et al. 1995). In eukaryotes, there are ~95 ψ modifications in 5.8S, 18S, and 28S rRNAs (Grosjean et al. 1995; Maden et al. 1995). Pseudouridines are typically clustered in functionally important regions in the ribosome, such as the peptidyl transferase region of 23S rRNA (Bakin and Ofengand 1993; Bakin et al. 1994). While the secondary structure of the box H/ACA snoRNPs has been established, their three-dimensional structures have not been determined.
FIGURE 1.
Sequence alignment of Nop10p and secondary structure of snoRNA. (A) Sequence alignments of Nop10. The residues are color coded as follows: red, hydrophobic; blue, negatively charged; pink, positively charged; and green, uncharged polar. The asterisks denote sequence identity, the colon denotes conserved substitutions, and the period denotes semiconserved substitutions. The sequence alignment was done using ClustalW (http://www.ebi.ac.uk/clustalw). A schematic of the secondary structure elements of Nop10p is indicated below the consensus. (B) Secondary structure of human U65snoRNA. (C,D) Sequence and secondary structure of U65hp (C) and U65ACA (D). Other RNAs used in this study include U65s (substrate RNA sequence 5′-UCGGCUCUUCCU), a 16-bp dsRNA capped by an AGAA tetraloop (Wu et al. 2004), and U65 3′ terminal hairpin (9 bp and UUCAUGU loop).
Box H/ACA snoRNAs are complexed with four different proteins to form the snoRNPs. In yeast (Saccharomyces cerevisiae), these are Nop10p (6 kDa), Nhp2p (22 kDa) (Henras et al. 1998), Gar1p (22 kDa) (Bousquet-Antonelli et al. 1997), and the putative pseudouridine synthase, Cbf5p (65 kDa) (Lafontaine et al. 1998). All four proteins of box H/ACA snoRNPs (Lafontaine et al. 1998; Dez et al. 2001) have been shown to be essential for viability. Nhp2p is a highly basic protein that is homologous to the 15.5-kDa protein (Nottrott et al. 1999) and to ribosomal protein L30, both of which have been shown to bind K-turn RNA (Vidovic et al. 2000; Klein et al. 2001; Chao and Williamson 2004). Nhp2p shows sequence alignment with the ribosomal proteins L32 and L7Ae over 53 amino acids (45% similarity between Nhp2 and the archaeal L7Ae from Methanocaldococcus jannaschii) (Henras et al. 1998). In archaea, Nhp2 is replaced by L7Ae, which is also common to their box C/D sRNPs involved in 2′O methylation (Rozhdestvensky et al. 2003). Structures of L7Ae–RNA complexes show how this protein binds to the K-turn found in box C/D sRNAs as well as K-turns and terminal loops of archaeal box H/ACA sRNAs (Hamma and Ferre-D’Amare 2004; Moore et al. 2004; Suryadi et al. 2005). Gar1p comprises a central core domain flanked by repetitive glycine–arginine repeat (GAR) domains (Bousquet-Antonelli et al. 1997; Bagni and Lapeyre 1998). Deletion studies have shown that the central core is required for interaction with the yeast box H/ACA snoRNAs snR10 and snR30 in vitro (Bagni and Lapeyre 1998). Cbf5p is highly homologous to other pseudouridine synthases and is thus the presumptive modifying enzyme (Lafontaine et al. 1998; Watkins et al. 1998). Nop10p is not homologous to any proteins outside the Nop10 family (Henras et al. 1998). The human homologs of these proteins also bind the box H/ACA snoRNA domain found at the 3′ end of human telomerase RNA (Dragon et al. 2000; Pogacic et al. 2000).
Recent studies have begun to shed light on the specific interactions of archaeal and eukaryotic H/ACA snoRNP proteins with RNA and with each other. In an investigation of H/ACA snoRNP assembly, Wang and Meier (2004) found that mouse Nop10 associates with Nap57 (mammalian pseudouridine synthase) and that this binding is a prerequisite for Nhp2 binding. Their results also showed that Gar1 independently associates with Nap57. UV cross-linking studies indicated that both Gar1 and Nap57 localize at the site of pseudouridylation on the RNA (Wang and Meier 2004). Henras et al. (2004) reported that Cbf5p can form a complex with Nop10p alone and with Nop10p and Gar1p in the absence of Nhp2p and snoRNA. These studies provide evidence that at least some of the H/ACA snoRNP proteins associate with each other in the absence of RNA, and this association may be relevant to snoRNP assembly in vivo. Consistent with these results, a study of archaeal box H/ACA sRNP components from Pyrococcus furiosus found that aCbf5 recruits binding of aGar1 and aNop10 independently (Baker et al. 2005). This study also found that L7Ae (the archaeal homolog of Nhp2p) does not interact with the other proteins in the absence of sRNA. In another recent study using the archaea Pyrococcus abyssi, it was found that a pseudouridylation-competent H/ACA sRNP could be reconstituted from the H/ACA RNA, aCBf5, and aNop10 alone (Charpentier et al. 2005). In all of the above studies, Nop10 was found to directly interact with the pseudouridine synthase (Cbf5p/Nap57 or aCbf5). The observed differential interaction of Nhp2 (L7Ae) or Gar1 with the pseudouridine synthase may reflect either a species-specific difference or may simply be attributed to differences in experimental conditions.
The secondary structure of human U65 snoRNA, used in the present study, is shown in Figure 1B ▶. Human U65 H/ACA snoRNA is homologous to yeast (S. cerevisiae) snR34 and trypanosome (Leptomonas collosoma) h1. The 5′ and 3′ pseudouridylation pockets of U65 guide the conversion of human 28S rRNA U4374 and U4428, respectively, to ψ. The equivalent Us in yeast 25S rRNA are targeted by snR34 (U2822/U2876) (Bortolin et al. 1999), and the single hairpin snoRNA h1 in trypanosome targets 25S rRNA at U3643 (U2876 in yeast) (Liang et al. 2001). Although eukaryotic H/ACA snoRNAs have two hairpins separated by a linker containing an H box motif, there is evidence that each hairpin assembles as a bipartite structure with a complete set of the core proteins (Watkins et al. 1998). In many of the H/ACA snoRNAs, only one of the two hairpins has been identified as a guide sequence. Furthermore, the L. collosoma h1 H/ACA snoRNA consists of a single hairpin with an AGA tail, as do some other H/ACA snoRNAs from archaea and early eukaryotes such as Euglena gracilis (Liang et al. 2001; Tang et al. 2002; Russell et al. 2004). The 3′ hairpin of human U17 and human telomerase RNA sequences have been shown to be sufficient for assembly to form a ribonucleoprotein complex when reconstituted with nuclear extracts containing the core proteins (Dragon et al. 2000). Thus, the 3′ hairpin of U65 provides a good model system for studying H/ACA snoRNA structure and core RNP protein assembly.
We have solved the three-dimensional structures of an RNA hairpin derived from the human U65 box H/ACA snoRNA including the 3′ pseudouridylation pocket and adjacent stems (U65hp) and of yeast Nop10p, and characterized their interaction with each other using nuclear magnetic resonance (NMR) spectroscopy. Eight of the 10 nt of the U65 3′ pseudouridylation pocket form mismatched base pairs in the absence of proteins and substrate RNA. Nop10p has a small structured region composed of two anti-parallel β-strands and a short 310 helix, and the rest of the protein is only partially structured. Chemical shift mapping studies show that the β-hairpin of Nop10p interacts with U65hp at the junction of the lower stem and internal pseudouridylation pocket and more weakly with the 3′ terminal hairpin loop. Nop10p does not become substantially more structured on RNA binding, suggesting that the less structured region of the protein is likely involved in interactions with Cbf5p.
RESULTS
Solution structure of the pseudouridylation pocket of the 3′ hairpin of U65 snoRNA
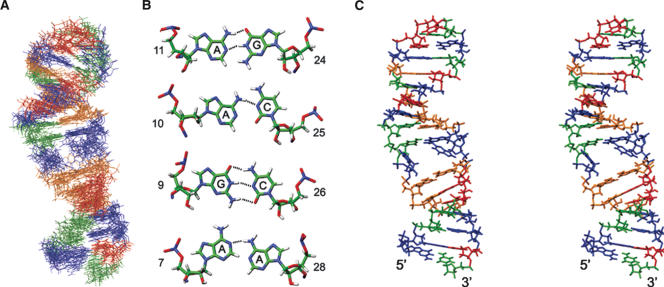
For structural studies, we focused on the region of the snoRNA that contains the pseudouridylation pocket (U65hp) (Fig. 1C ▶). The human U65 rather than yeast snR34 was chosen for structural studies because the stems and the pseudouridylation pocket were more clearly defined by the sequence. It has been demonstrated that human homologs hNop10 and hNhp2 can complement yeast cells depleted of Nop10p and Nhp2p (Pogacic et al. 2000), suggesting that the human and yeast snoRNP components are interchangeable. The U65hp RNA contains the sequence of the pseudouridylation pocket and adjacent base pairs of the 3′ hairpin of U65snoRNA (boxed region in Fig. 1B ▶). The top stem was shortened, and the terminal loop was replaced by a stable UUCG tetraloop to ensure formation of a hairpin rather than a duplex. NMR spectra of U65hp indicate that it folds into a hairpin structure with a UUCG tetraloop as designed. The imino proton spectra (data not shown) contain the characteristic high field shifted U and G imino resonances from the U-G base pair in the tetraloop, the expected iminos from the base-paired upper and lower stem, and some additional imino resonances from bases in the pseudouridylation pocket, providing an initial indication that the internal loop is at least partially base-paired. The 34-nt U65hp was assigned by analysis of 2D and 3D NMR spectra acquired on unlabeled and uniformly 13C,15N-labeled samples. Structure calculations were performed using 572 distance restraints derived from NOEs, 209 dihedral angle restraints, and 11 RDCs (Table 1 ▶). The 15 lowest-energy structures (Fig. 2A ▶) have an overall root mean squared deviation (rmsd) to the mean structure of 2.1 Å for all heavy atoms. The lower stem, the upper stem, and the tetraloop (C(UUCG)G) are well-defined (rmsd 0.78 Å, 0.67 Å, and 0.29 Å, respectively). Within the pseudouridylation pocket, the top six bases form well-determined base pairs (Fig. 2B,C ▶). The Watson–Crick faces of A11 and G24 hydrogen bond to form a symmetric A·G base pair. A10 amino is hydrogen bonded to C25 N3, and the N1 of A10 is partially protonated to hydrogen bond with the C25 carbonyl. G9 and C26 form a Watson–Crick base pair. The bottom of the pocket does not have a well-defined structure. A7 and A28 stack on the Watson–Crick U·A base pair at the top of the stem in the majority (9 of 15) of the structures to form a mismatch pair with a single A7N1-A28N6H hydrogen bond. The bases of G8 or G27 do not have any defined position in the structure, due to insufficient NOE restraints. The small number of NOE restraints may be due to conformational heterogeneity for G8, since resonances for this residue exhibit exchange broadening. The structural heterogeneity in this part of the pseudouridylation pocket leads to the relatively high rmsd for superpositions over the entire structure.
TABLE 1.
Restraint and structure statistics for U65hp and Nop10p
| U65hp | Nop10p | |
| Distance restraints | ||
| Total NOE restraints | 572 | 743 |
| Intraresidue | 186 | 354 |
| Sequential (i + 1) | 288 | 191 |
| Medium (i + 2 to i + 4) | 26 | 128 |
| Long range (>i + 4) | 72 | 70 |
| Hydrogen bond restraints | 64 | 10 |
| RDC restraints | 11 | |
| Dihedral angle restraints | 209 | |
| Structure statistics | ||
| No. of NOE violation >0.2 Å | 0.1 ± 0.0 | 4.6 ± 1.1 |
| No. of NOE violation >0.5 Å | 0 | 0 |
| Maximum violation, Å | 0.20 | 0.36 |
| Mean deviation of NOE violation, Å | 0.022 ± 0.007 | 0.032 ± 0.010 |
| Rms deviation from ideal covalent geometry | ||
| Bond lengths, Å | 0.0044 ± 0.0012 | 0.0026 ± 0.0008 |
| Angles, ° | 1.03 ± 0.27 | 0.47 ± 0.15 |
| Impropers, ° | 0.39 ± 0.10 | 0.30 ± 0.10 |
| Rms deviation (Å) from the mean structure | ||
| Backbone | Heavy atoms | |
| Nop10p (10 lowest energy structures) | ||
| Residues 4–44 | 1.97 | 2.95 |
| Residues 4–16 and 37–42 | 0.90 | 1.74 |
| U65hp (15 lowest energy structures) | ||
| Residues 1–34 | 2.38 ± 1.14 | 2.14 ± 1.03 |
| Residues 9–26 | 1.23 ± 0.69 | 1.08 ± 0.59 |
| Residues 1–6, 29–34 | 0.79 ± 0.22 | 0.78 ± 0.20 |
FIGURE 2.
Structure of U65hp. (A) Superpositions of the 15 lowest-energy structures. Nucleotides are colored blue (Gua), green (Cyt), orange (Ade), and red (Ura). (B) Base pairs in the pseudouridylation pocket. (C) Stereo view of the lowest-energy structure.
Structural characterization of Nop10p
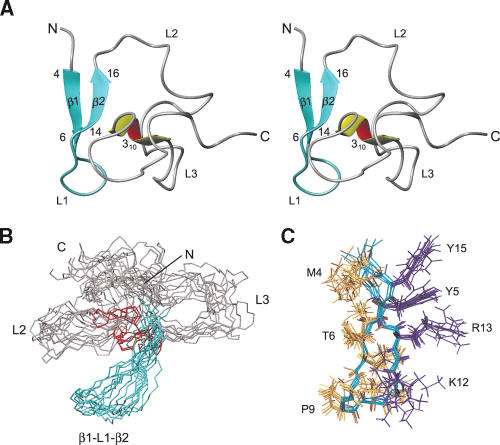
Nop10p is a 58–amino acid protein, with an isoelectric point of 10.5, which shows sequence homology with other Nop10 proteins (Fig. 1A ▶) but is not homologous to any other proteins. The protein amide resonances show limited chemical shift dispersion for most of the resonances, characteristic of a protein that lacks a high degree of secondary structure (see Fig. 5A ▶, below). A structural model of Nop10p (Fig. 3 ▶) was obtained using a total of 743 NOEs and 10 hydrogen bond restraints (Table 1 ▶). The NOE restraints show an uneven distribution along the protein sequence, with the majority of them between the first 25 residues. Structure calculations showed that Nop10p is only partly structured, and this was confirmed by dynamics studies discussed below. The backbone rmsd from the mean structure for residues 4–44 is 1.97 Å, while the rmsd for residues 4–16 and 37–42 is 0.9 Å. The N-terminal third of Nop10p contains a two-stranded anti-parallel β-sheet connected by a structured loop (L1) (β-hairpin; residues 4–16) (Fig. 3A ▶). The only other part of the protein with a well-defined secondary structure is a short 310 helix (residues 39–41) that interacts with the β-hairpin. The residues (L2) connecting β2 and the helix are only partially ordered, as is the C-terminal region (L3), consistent with the NOE distribution observed. Thus, Nop10p belongs to the growing family of proteins that are partially disordered or unfolded in the absence of their binding partners (Wright and Dyson 1999; Dunker et al. 2002; Dyson and Wright 2002).
FIGURE 5.
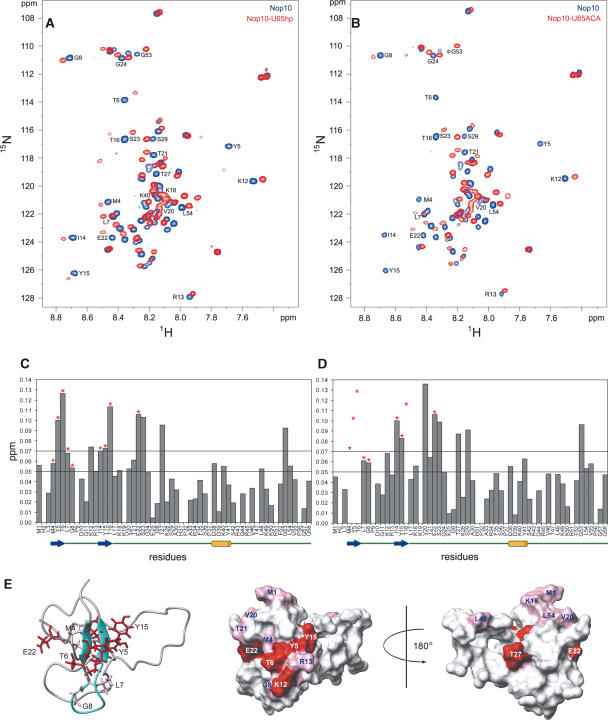
Chemical shift mapping of U65hp and U65ACA on Nop10p. 1H-15N HSQC of free Nop10p (blue) bound to U65hp (red) (A) and U65ACA (red) (B) at a 1:1 ratio of RNA:protein. (C,D) Plots of the chemical shift changes of Nop10p in the presence of U65hp (C) and U65ACA (D). Note that although Y5 is not visible in Figure 5A in the bound form, it is possible to observe the peak at a higher noise level. Asterisks indicate resonances that are exchange broadened in the presence of the RNA. In D some of these are broadened beyond detection and therefore their chemical shift change cannot be quantified. The dashed lines indicate the thresholds used in the surface mapping. A schematic of the secondary structure elements are shown underneath the sequence. (E) The chemical shift changes after the addition of U65hp mapped onto the structure of Nop10p: (Left) Ribbon representation of Nop10p with side chains of residues whose amides are significantly exchange broadened in the presence of U65hp shown; (middle) van der Waals surface representations of Nop10p with all residues that show significant chemical shift changes highlighted in pink (Δppm 0.05–0.07) and red (Δppm >0.07) with same view as ribbon representation; (right) surface representation rotated by 180°.
FIGURE 3.
Structure of Nop10p. (A) Stereo view of the lowest-energy structure. The β-hairpin is colored aqua, and the 310 helix is yellow-red. (B) Superposition of the eight lowest-energy structures. (C) Backbone superpositions of the β-hairpin, with side chains of aromatic and acidic residues shown in purple.
The residues on the solvent-exposed face of the β-hairpin are Tyr5 (β1 strand), Tyr15 (β2 strand), and Lys12, Arg13 (loop L1) (Fig. 3C ▶). The hydrophobic residues Met4, Thr6 (β1 strand), and Thr16, Ile14 (β2 strand) are on the inside of the protein and are in contact with other residues. The orientation of the 310 helix with respect to the β-hairpin is not well-defined. However, there are hydrophobic contacts between the side chains of Ile14 and Thr16 in the β2 strand and Asp39 and Lys40 in the 310 helix, respectively, in the majority of structures. These interactions indicate that the 310 helix is at least partially packed against the β-sheet.
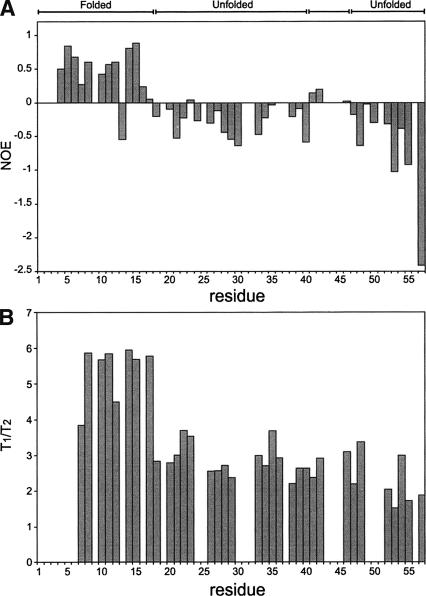
Nop10p dynamics
The lack of interresidue NOEs for parts of Nop10p suggested the presence of conformational heterogeneity. Regions of Nop10p flexibility were assessed by measuring steady-state heteronuclear 1H-15N NOEs (Farrow et al. 1994a). Residues 4–17, with the exception of Arg13, and residues Tyr41 and Ser42 have positive values, while the rest of the residues have near zero or negative heteronuclear 1H-15N NOEs (Fig. 4A ▶). All of the residues with positive heteronuclear NOEs are located in the β-hairpin or the 310 helix. The heteronuclear NOEs are consistent with the results of the structure calculations, indicating that the lack of a well-defined structure in the L2 and L3 regions of the protein is due to conformational dynamics. The T1/T2 ratio for residues in the β-hairpin region is very different from the rest of the protein (Fig. 4B ▶). The average value of T1/T2 for β1–L1–β2 (residues 7–17) gives a correlation time of 5.5 nsec, while the rest of the residues have an average correlation time of 2.6 nsec. The average T1/T2 value of the structured N-terminal region of Nop10p is in agreement with the average value obtained for structured proteins of comparable size (Huang et al. 1999). Based on the T1/T2 ratio, the 310 helix has the same correlation time as the L2 and L3 regions. This may explain why the orientation of the 310 helix relative to the β-sheet is not well-defined, in spite of the presence of NOEs between these two regions of the protein.
FIGURE 4.
Plots of Nop10p dynamics. Plots of 15N-1H heteronuclear NOE (A) and T1/T2 (B) as a function of the residue numbers.
Nop10p binds weakly to the 3′ hairpin of human U65snoRNA
Although Nop10 associates with H/ACA snoRNAs as part of the snoRNP (Henras et al. 1998; Pogacic et al. 2000), there is no direct experimental evidence showing that Nop10p interacts with the snoRNA. Studies using gel mobility shift assays did not detect any binding of Nop10 alone to H/ACA RNAs (Wang and Meier 2004; Baker et al. 2005). However, several lines of evidence indicate that Nop10 may interact with RNA in the RNP. Human Nop10 has been reported to UV cross-link to U17 RNA (Dragon et al. 2000). In archaea, Nop10 contains a putative zinc-finger motif consistent with a role in RNA binding (Baker et al. 2005). In the reconstitution experiments of Charpentier et al. (2005), the presence of a Nop10 was found to result in more efficient binding of the RNA substrate in RNP complex, implicating it in stabilization of the snoRNA:substrate interaction. Since gel shift experiments failed to detect any binding of Nop10p alone to H/ACA RNA up to 10 μM, this indicates that if Nop10p does bind to the RNA its Kd must be greater than micromolar in the absence of Cbf5p (Wang and Meier 2004; Baker et al. 2005; Charpentier et al. 2005). Since NMR can easily be used to detect millimolar binding, we tested for the interaction of Nop10p with RNA by monitoring the chemical shift changes of Nop10 upon addition of various RNA constructs using 1H-15N HSQC experiments. These constructs included the U65hp, U65ACA, the terminal stem-loop of U65, a double-stranded RNA (dsRNA) capped by a structured AGAA tetra-loop, and the single-stranded substrate RNA (U65s) (Fig. 1B ▶). No chemical shift changes in the 1H-15N HSQC of Nop10 were observed when the single-stranded substrate RNA or the dsRNA capped by an AGAA tetraloop were titrated into the protein. However, in the presence of the U65hp significant chemical shift changes occur for several resonances, indicating that Nop10p is binding to this RNA. For most of the affected resonances, incremental addition of RNA results in incremental shifts (up to an RNA:protein ratio of 1:1), indicating fast exchange on the NMR timescale between protein and RNA. However, several resonances showed significant line broadening as well as shifting, in some cases broadening to baseline, indicative of chemical exchange. Addition of RNA to Nop10p above a ratio of 1:1 (up to 2:1) did not result in any additional spectral changes. To confirm that the His6 tag on the protein was not contributing to binding affinity to the RNA, we repeated the titration with protein from which the his6 tag had been removed (data not shown). The same pattern of chemical shift changes was observed on the protein lacking the His6 tag. We also tested for binding of Nhp2p to U65hp and also observed no chemical shift changes in the 1H-15N HSQC spectrum of Nhp2p (N. Chim and J. Feigon, unpubl. data).
The residues in Nop10p that exhibit the largest amide chemical shift changes following addition of U65hp RNA are 4–7, 12–16, 18, (structured region) and 20–24, 27, 29, 40, 53, and 54. Among these, Met4, Tyr5, Thr6, Leu7, Gly8, Ile14, Tyr15, Thr16, and Glu22, all of which are on the β-hairpin except for Glu22, show significant exchange broadening (Fig. 5A,C ▶). The side chains of these residues are shown on the ribbon diagram of Nop10p (Fig. 5E ▶). The large chemical shift changes and exchange broadening of residues in the β-hairpin indicate that this region of the protein is interacting directly with the RNA. The chemical shift changes for residues in the adjacent L2 region of the protein as well as near the N terminus may be due to direct contacts to the RNA and/or some conformational rearrangement of the protein. However, the small magnitude of the chemical shift changes is not consistent with any major structural rearrangement in the L2–L3 regions of Nop10. The surface view of Nop10p with residues with significant chemical shift changes highlighted shows the potential interacting surface at and around the β-hairpin on one face of the protein (Fig. 5E ▶).
The H and ACA boxes are required for accumulation and stability of snoRNA in vivo (Balakin et al. 1996; Ganot et al. 1997b). Because it has been proposed that Nop10p binds to the H and ACA boxes (Henras et al. 1998), we also obtained chemical shift mapping results using a longer RNA that includes the ACA box and is closer in sequence to the full-length 3′ hairpin of U65snoRNA (U65ACA) (Fig. 1D ▶). This RNA contains two additional base pairs in the upper stem and the full-length lower stem including a bulged A residue, followed by the 3′ tail containing the ACA box found in the full-length U65snoRNA. The chemical shift changes observed upon addition of U65ACA to Nop10p are very similar to those for the shorter U65hp (Fig. 5B,D ▶). However, the resonances that exhibited the most exchange broadening during the titration with U65hp (Met4, Tyr5, Thr6, Ile14, Tyr15, Thr16) disappear at the first addition of U65ACA RNA (Fig. 5B ▶). Since this complex has a significantly larger molecular weight, the additional line broadening may be attributed to contributions from the larger line widths of the resonances in the complex. Alternatively, the exchange kinetics may be somewhat different between the two complexes, due to differences in RNA sequence and structure. However, overall the chemical shift changes on Nop10p observed in the presence of U65hp and U65ACA are so similar that it is clear that the interaction between Nop10p and H/ACA snoRNA does not require the ACA sequence. Finally, we checked for binding of the terminal stem-loop of U65 to Nop10p. In this case, a similar pattern of chemical shift changes was observed in Nop10p for this RNA (data not shown), indicating that Nop10 also binds this RNA. Like U65hp, the U65 terminal loop contains a lower stem and partially structured loop.
Taken together, the chemical shift mapping of the various RNA constructs on Nop10p indicates that Nop10 interacts weakly with the U65 H/ACA RNA (Kd ~ 0.1 mM based on NMR titration), and this interaction does not require either the H or ACA box for binding. Further, the binding is specific to a stem-loop junction. There is no binding to substrate RNA, double-stranded RNA, or a structured tetraloop. This interaction appears to involve the structured β-hairpin region of the protein. Although additional chemical shift changes in other regions of the protein are observed, they are not accompanied by the resonance line broadening that is observed for the shifted amide resonances on the two β strands. The chemical shift changes observed for amide resonances in other regions of the protein are likely due to secondary shifts rather than direct interaction with the RNA, due to some structural rearrangement of the protein. However, these changes are too small to be consistent with any major structural change in the rest of the protein.
Mapping of Nop10p binding to U65hp
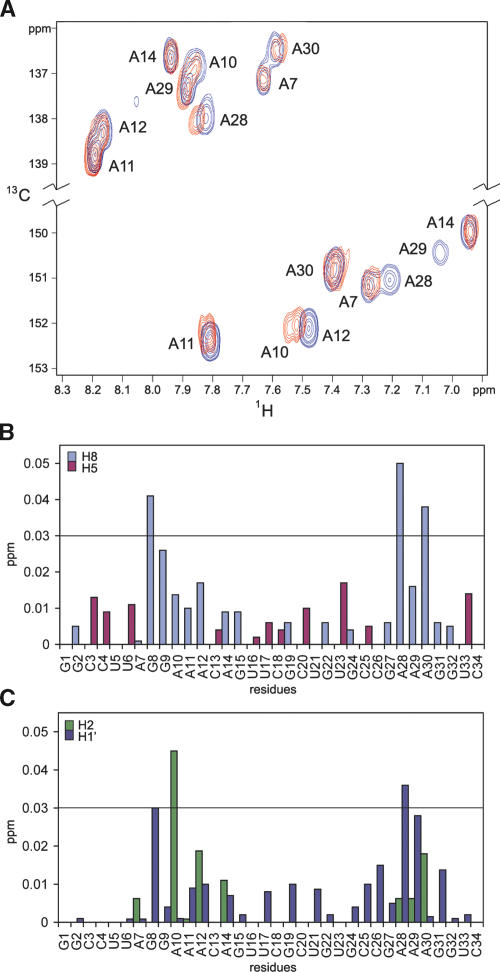
To further check for the specificity of the interaction between Nop10p and U65hp, we also obtained chemical shift data using 13C-1H HSQC experiments on the RNA as a function of added protein, up to a ratio of 1:1. As was the case for the reverse titration, some resonances shifted as a function of added protein, and in some cases this shifting was accompanied by resonance line broadening (Fig. 6A ▶). Nucleotides exhibiting 1H chemical shift changes of >0.02 ppm for one or more resonances are G8, G9, A10, A28, A29, and A30 (Figs. 6B,C ▶, 7A ▶). These results clearly indicate that Nop10p binds specifically to U65hp at the junction between the lower stem and the pseudouridylation pocket.
FIGURE 6.
Chemical shifts of Nop10p mapped on RNA. (A) 13C-HSQC of U65hp in the absence (blue) and the presence (red) of Nop10p. The resonances between 136 and 139 ppm in the 13C region are from H8-C8 residues, and between 150 and 153 ppm, from H2-C2 residues. (B,C) Graphs of the proton chemical shift differences of H8 (gray) and H5 (red) (B) and AH2 (green) and H1′ (blue) (C) resonances before and after the addition of Nop10p to U65hp.
FIGURE 7.
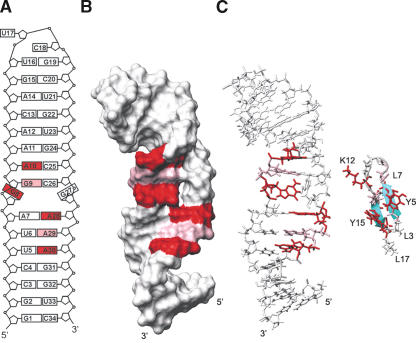
Mapping of Nop10p on U65hp structure. (A) Schematic of the structure of U65hp. Residues that show chemical shift changes upon addition of Nop10p are colored in red (Δppm >0.03) and pink (Δppm >0.02). (B) Surface view of U65hp is shown with the affected residues from Nop10p binding highlighted as in A. (C) Stick and ribbon representations of U65hp and the β-hairpin of Nop10p, shown to illustrate size complementarity of binding interface. Side chains of Y5, T6, K12, Y15, and T16 are red; L7, G8, and R13 are pink.
Figure 7B ▶ shows a surface view of U65hp with the nucleotides that the chemical shift mapping indicates interact with the protein highlighted. For protein interactions with double-stranded RNA or DNA that do not distort the structure of the duplex, chemical shift mapping can usually distinguish between minor groove and major groove interactions on the basis of which resonances (e.g., AH2 in minor groove, AH8 in major groove) shift. However, in this complex Nop10p is interacting with residues at or near the conformationally dynamic bottom half of the pseudouridylation pocket. In this case, binding of the protein likely results in some conformational changes in this region of the RNA. We observed chemical shift changes for protons located in both the major and minor grooves of the RNA. Thus, we cannot use the specific resonance shifts to distinguish between minor and major groove binding. However, the surface view shows that Nop10 likely binds on one face of the RNA and interacts with the phosphodiester backbone on opposing strands. The relatively small chemical shift changes as well as the requirement for Nop10p to be able to interact with many different H/ACA hairpins are consistent with a structure-specific rather than sequence-specific interaction. This may also explain the interaction with the U65 terminal hairpin, which also has a stem-loop junction. Similar small chemical shift changes were observed in an NMR study of the structure specific interaction of the dsRBD of S. cerevisiae RNase III (Rnt1p) with dsRNA capped by an AGNN tetraloop (Wu et al. 2004).
DISCUSSION
The H/ACA snoRNP is a unique particle required for conversion of U to ψ in eukaryotic rRNAs. Although the protein components of the mature H/ACA snoRNP have been identified and the secondary structure of the RNA has been determined, no structures of the RNA component have been previously reported and there is limited structural information on related protein components. As a first step in characterizing the structure of the H/ACA snoRNP complex and understanding how the four snoRNP proteins and the RNA assemble to form a structure for rRNA substrate binding and catalysis, we have determined the structures of Nop10p and the 3′ hairpin of U65 H/ACA snoRNA. We have further shown that these two components interact weakly but specifically with each other in the absence of other RNP components.
The pseudouridylation pocket is partially structured in the absence of proteins
The H/ACA snoRNAs form a bipartite secondary structure in which each half forms a hairpin that contains an internal loop of 9–15 nt, the pseudouridylation pocket (Fig. 1B ▶). The conserved H and ACA boxes are located 14–17 nt from the 5′ and 3′ pseudouridylation sites, respectively. Prior to base modification, the substrate rRNA is thought to bind the pseudouridylation pocket through base-pairing of complementary nucleotides flanking the U that is subsequently modified, leaving the U and its adjacent 3′ nucleotide unpaired at the top of the pocket. The pseudouridylation pockets of H/ACA snoRNA thus form the essential core of the snoRNP, since the substrate rRNA and the pseudouridylase, Cbf5p in yeast, must bind to this pocket for catalysis. We have determined the structure of the 3′ pseudouridylation pocket of U65 snoRNA, and find that in the absence of substrate RNA and snoRNP proteins, the internal loop is largely base-paired to form a “closed” conformation (Fig. 2C ▶). This is not surprising, as most internal loops exhibit at least some secondary structure from mismatch base pairs. The top half of the pseudouridylation pocket of the 3′ hairpin of human U65 snoRNA contains three well-defined base pairs, while the bases in the bottom half are more dynamic. Since the U65 3′ pseudouridylation pocket is partially ordered and base-paired in the free RNA, binding of the rRNA substrate will require opening of the internal loop. This is likely to be the general case since, as discussed above, most internal loops are partially structured. Binding of the snoRNP proteins to the snoRNA likely destabilizes the base pairing in the internal loop to prepare the snoRNP for substrate binding, as discussed below.
The pseudouridylation pocket contains successive A11·G24, A10·C25 mismatched, and G9-C26 Watson–Crick base pairs in the upper part of the pocket, next to the pseudouridylation site. For the bottom 2 nt on each site of the pocket, there is no defined structure for G8 and G27, and the A7·A28 bases interact via a single hydrogen bond and appear to be dynamic. It is interesting to note that the homologous internal loops from trypanosome h1 and yeast snR34 can form either the same 3 bp found at the top of the U65 3′ pseudouridylation pocket or an A·U in place of the A·C mismatch, respectively, while the sequence at the bottom of these pseudouridylation pockets differs and also contains additional nucleotides in the predicted secondary structures (Samarsky et al. 1995; Ganot et al. 1997b; Liang et al. 2001). In both cases, it appears that the bottom of the pocket will be less structured than the top of the pocket, as we observed for U65hp. Thus, for these three U65 homologs the bottom part of the pseudouridylation pocket is more open than the top. It remains to be seen whether this is the general case for H/ACA internal loops.
Role of Nop10p in H/ACA snoRNP assembly and catalysis
RNA-guided pseudouridylation of substrate RNAs requires a site-specific H/ACA RNA and four core proteins: Cbf5, Nop10, Gar1, and Nhp2. Nop10p is the smallest of the four proteins, and, like Gar1p and Nhp2p, its role is unknown. Although Cbf5 can bind snoRNA in the absence of other protein components, in archaea the minimal functional complex with the H/ACA RNA has been shown to require Nop10 in addition to Cbf5 (Charpentier et al. 2005). A direct interaction between Cbf5 and Nop10 appears to be universal among archaeal and eukaryotic H/ACA snoRNPs (Henras et al. 2004; Wang and Meier 2004; Baker et al. 2005; Charpentier et al. 2005). Here we have shown that in the absence of other RNP components, Nop10p is only partially structured, with a two-stranded anti-parallel β-sheet in the N-terminal third of the protein and a 310-helix loosely packed against the β-hairpin (Fig. 3 ▶). NMR relaxation measurements confirmed that the C-terminal two-thirds of Nop10p is less structured than the β-hairpin. Many proteins or protein domains are intrinsically unstructured or lack globular structure under physiological conditions, and only become structured upon binding to their target (Dyson and Wright 2002). These include a variety of nucleic acid binding proteins, where a disorder-to-order transition takes place on binding to the nucleic acid. However, we have found that Nop10p interacts with RNA via its structured β-hairpin, and the rest of the protein does not become significantly more structured in complex with RNA. This suggests that the flexible region of Nop10p rather plays a role in protein–protein interactions in the snoRNP, and therefore likely becomes structured on binding Cbf5p.
Although the binding of the free Nop10p to RNA is weak and probably occurs in vivo only in the presence of Cbf5p, this low affinity interaction is detectable by NMR and provides insight into probable interactions between Nop10 and H/ACA snoRNA in the intact snoRNP. The observed interaction is consistent with biochemical data that indicate that Nop10p binds to the RNA (Dragon et al. 2000; Pogacic et al. 2000). We found that Nop10p binds at the bottom of the U65 3′ pseudouridylation pocket under NMR conditions (protein and RNA concentrations of 0.1–1.0 mM). Nop10p does not bind the single-stranded substrate RNA or double-stranded RNA capped by an ordered tetraloop, but does interact with the U65 terminal loop, which also contains a stem-“unstructured” loop junction. Interestingly, Nop10p interacts via its structured β-hairpin. The β-hairpin was first identified as a DNA binding motif that interacts in the major groove (Somers and Phillips 1992) and has since been found in several rRNA–protein interactions, e.g., S6, S7, S11 (Wimberly et al. 2000) and L5, L14, L25 (Yusupov et al. 2001; Klein et al. 2004), as well as a BIV Tat peptide–TAR RNA complex (Puglisi et al. 1995). In these complexes, the mode of interaction of the β-hairpin with RNA varies; it has been found to bind in a widened major groove, the minor groove, and to loop regions. In the ribosome, both the isolated β-hairpin and β-hairpins that are part of β-barrels appear to play an important role in RNA–protein assembly. Thus it appears reasonable that the β-hairpin of Nop10p could recognize a stem-loop junction in H/ACA snoRNAs. Based on the chemical shift mapping, all of the residues on the β-strands of the β-hairpin interact with the RNA. These include two highly conserved aromatic residues (Tyr5 and Tyr15) and the positively charged Lys12 and Arg13 on L1 adjacent to β2 (Fig. 3C ▶), residues that are frequently found to interact with RNA (Jones et al. 2001; Treger and Westhof 2001). A comparison of the binding interfaces for Nop10p and U65hp shows that there is size complementarity between the interacting residues of the β-hairpin and the RNA (Fig. 7C ▶).
We propose that the intrinsically unstructured region of Nop10p functions to help assemble the H/ACA snoRNP complex by interacting with Cbf5p and that it becomes structured on binding Cbf5p. The weak interaction of Nop10p with the snoRNA is likely stabilized by the interaction of Cbf5p with the RNA in the snoRNP. Although NMR data showed that Nop10p can interact with both the bottom of the pseudouridylation pocket and the terminal loop of the 3′ U65 hairpin, which both contain a stem-loop junction, it appears most likely that binding of Nop10p to Cbf5p would position it at the bottom of the pseudouridylation pocket, allowing the catalytic core of Cbf5p to be in proximity to the upper part of the pocket where the modification occurs. Binding of Nop10p at the bottom of the pseudouridylation pocket would be consistent with the finding that the presence of Nop10p in the complex with Cbf5p increases the efficiency of substrate RNA binding to the H/ACA RNA (Charpentier et al. 2005). Once the snoRNP proteins assemble on the RNA, they form a stable complex and do not exchange their RNA components, suggesting that formation of new snoRNPs requires de novo synthesis (Wang and Meier 2004). Since each snoRNP should catalyze the conversion of U to ψ for many rRNA substrates, this implies that the rRNA binds subsequent to particle formation. Thus, the snoRNP proteins likely facilitate opening of the pseudouridylation pocket for rRNA binding. An interaction between Nop10p and the stem-loop junction at the bottom of the pseudouridylation pocket could play a role in opening of the pseudouridylation pocket for binding to the target RNA.
MATERIALS AND METHODS
RNA preparation and purification
Unlabeled, uniformly 15N,13C-labeled, and nucleotide-type selectively 15N,13C-labeled RNAs (Fig. 1C,D ▶) were synthesized by in vitro transcription using T7 RNA polymerase and purified as described (Batey et al. 1992; Dieckmann and Feigon 1997). The purified RNAs were exchanged into the NMR buffer and concentrated to 0.1 mM (for titrations) or 1.5 mM (for structure determination) using an AMICON filtering device. The single-strand substrate RNA 5′-UCGGCUCUUCCU was purchased from Dharmacon.
Protein expression and purification
Nop10p was cloned in a pET15b vector as described (Henras et al. 1998). Unlabeled and 13C,15N-labeled (media supplemented with 1 g/L of 15NH4Cl and 2 g/L [13C]-glucose) Nop10p were expressed in BL21(DE3) gold cells (Stratagene) and grown in M9 minimal media. Cells were lysed in the loading buffer used for the HiTrap chelating nickel affinity column (10 mM Tris-HCl, 0.5 M NaCl, 10 mM imidazole at pH 7.6) in the presence of protease inhibitor cocktails (Calbiochem). The protein was eluted using 10 mM Tris-HCl, 0.5 M NaCl, 0.5 M imidazole (pH 7.6). The sample was loaded on a HiTrap heparin sepharose column (Amersham) and eluted using a linear gradient from 0.05 to 1 M NaCl. Finally, the protein was loaded on a superdex 75 preparation grade size exclusion column (Amersham) and eluted in the NMR buffer (10 mM potassium phosphate, 100 mM KCl at pH 6.5). The sample was concentrated using an AMICON filtering device at 4°C.
NMR spectroscopy, assignment methodology, and structure calculations for U65hp
All spectra for assignments and structure determination were obtained on Bruker DRX 500 and 600 MHz spectrometers. Sample conditions were 1.5 mM RNA in 30 mM potassium phosphate (pH 6.5). Additional spectra were obtained on a sample with 100 mM KCl added for comparison since titrations with protein were done under the higher salt conditions. Spectra of samples in H2O (95% H2O/5% D2O) were recorded at 283 K, and spectra of samples in D2O were recorded at 298 K. Assignments of the nonexchangeable 1H and proton attached 13C resonances were obtained from analysis of 2D NOESY, CT-HSQC, MLEV-TOCSY, HCCH-COSY, and long-range 1H-15N HSQC and 3D HCCH-TOCSY experiments (Furtig et al. 2003). Filtered/edited NOESY spectra acquired on 13C,15N-A, -U, -G, and -C labeled samples were used to resolve some of the ambiguities in assignments (Peterson et al. 2004). Assignments of the exchangeable 1H and proton attached 15N were obtained from analysis of NOESY, 1H-15N HMQC, and 15N-correlated CPMG-NOESY spectra acquired on samples in H2O (Furtig et al. 2003). Residual dipolar couplings (RDCs) were measured for 1JHC in 5% C12E6/hexanol using CT-CE-HSQC.
Distance restraints were obtained from NOE cross peaks in 2D 1H-1H NOESY in H2O (τm = 75 msec) acquired on unlabeled RNA and filtered/edited (F2f, F2eF1f, and F2eF1e) NOESY experiments (τm = 200 msec) acquired on base specifically 15N,13C-A, -U, -C, and -G labeled RNAs (Peterson et al. 2004). Distance restraints obtained from spectra of D2O samples were classified as follows for structure calculations: 1.8–2.5 Å (strong), 1.8–3.5 Å (medium), 2.0–4.5 Å (weak), and 3.0–5.5 Å (very weak). Similar boundaries were used for cross-peaks in H2O spectra, except very weak was 3.0–6.5 Å. Hydrogen bond distance restraints for Watson–Crick base pairs, including G8·C26, were derived from JNN-COSY type experiments (Luy and Marino 2000). Additional hydrogen bond restraints for A11·G24 and A10·C25 mismatches were included near the end of the structure calculations based on the structure and NOE patterns. 3JH3′P and 3JCP were measured using 31P spin echo difference CT-HSQC to determine β and ɛ torsion angles. Ribose conformation and χ angles were analyzed as described (Wu et al. 2004). Ninety-six additional dihedral angle restraints (±30°) were included for stem nucleotides with NOE patterns consistent with A-form geometry.
Under low salt conditions, G8 is exchanging between two conformations with two different (N and S) sugar puckers, which give rise to two sets of NOE cross-peaks for this residue. A28 and A29 also have two sets of cross-peaks but with only small chemical shift differences, both with N-type sugar puckers. Thus, in low salt the bottom of the pseudouridylation pocket adopts at least two conformations that are in slow exchange on the NMR timescale. When 100 mM KCl was added to the RNA samples to obtain the salt conditions used for the titration experiment with Nop10p, cross-peaks for only one of the two conformations were seen, corresponding to an S-type sugar pucker for G8. Therefore, only the set of restraints for this conformation were used in the calculation of U65hp. The RDCs were obtained for a sample in 100 mM KCl.
Structures were calculated using the standard simulated annealing protocols with NIH-XPLOR (Schwieters et al. 2003) starting from an extended template with randomized angles. Sixty low-energy structures from 200 initial calculated structures were then subjected to refinement against 11 RDCs (Clore et al. 1998). The refinement used a simulated annealing procedure that consisted of 18 psec of high temperature dynamics at 1000 K followed by slow cooling to 100 K. The alignment tensor of the RDCs was determined by a grid-search protocol as described, which produced optimal values of Da = −41 Hz and R = 0.18. The force constants used in the refinement stage of the structure calculations were 50 kcal·mol−1·Å2 and 200 kcal·mol−1·Å2 for NOEs and dihedral angles, respectively. Structure statistics for the 15 lowest-energy structures are given in Table 1 ▶.
NMR spectroscopy, assignment methodology, and structure calculations for Nop10p
All spectra for assignments and structure determination were obtained on Bruker DRX 500 and 600 MHz spectrometers at 288 K and processed as above. Backbone assignments of Nop10p were obtained from 3D CBCANH (Grzesiek and Bax 1992b), 3D CBCA(CO)NH (Grzesiek and Bax 1992a,b) using a 15N, 13C-labeled proteins. Protein side-chain assignments were obtained using 3D HBHA(CO)NH (Grzesiek and Bax 1993), 3D HCCH-TOCSY (Bax et al. 1990), and 2D homonuclear TOCSY. The NOE cross-peaks were obtained from 3D 13C-NOESY-HMQC in D2O and 3D 15N-NOESY-HSQC in H2O (Nikonowicz and Pardi 1993). Eleven RDCs were also obtained at 288 K and the alignment medium used was C8E5/octanol.
Protein–protein NOE cross-peaks obtained from 3D 13C-NOESY-HMQC (τm = 150 msec) and 3D 15N-NOESY-HSQC (τm = 150 msec) were converted into distance restraints and categorized as follows: strong (1.8–2.5 Å), medium (1.8–4.5 Å), and weak (1.8–6.5 Å). In total, 743 NOE-derived restraints were used in the structure calculation, of which 354 NOEs are intraresidue and 191 are sequential NOEs (Table 1 ▶). The structure of the protein was calculated using XPLOR (NIH) with a simulated annealing protocol. We generated 50 templates from extended protein structures with random orientations. This was followed by several rounds of simulated annealing protocol employing the all-hydrogen force-field parallhgd.pro version 5.0. The simulated annealing procedure started from random coordinates and consisted of 30 psec of high temperature dynamics at 2000 K followed by slow cooling to 100 K in 30 psec. Following the simulation, the 10 lowest-energy structures were chosen for further refinement with a simulated annealing procedure that consisted of 30 psec of high temperature dynamics at 1000 K followed by slow cooling to 100 K. Due to the dynamic nature of the protein, incorporation of RDCs in the structure calculations did not improve the accuracy of the final structures.
To assess Nop10p dynamics, the modified T1 and T2 experiments with better water suppression and 1H-15N steady-state NOE experiments were acquired as described (Farrow et al. 1994a,b) at 288 K and 500 MHz. The T1/T2 ratio for each residue was plotted and used to estimate the correlation time of the molecule (Huang et al. 1999).
Chemical shift mapping
All spectra for chemical shift mapping were obtained on a Bruker DRX 800 MHz spectrometer at 298 K. Sample buffer conditions were the same as those used for the free Nop10p. In order to monitor the chemical shift changes of U65snoRNA hairpin upon addition of Nop10p, Nop10p was added in 0.05 mM increments to 0.1 mM base specifically 13C,15N-A, -G, -U, and -G,C labeled RNA samples, up to a protein:RNA ratio of 1:1. At higher protein:RNA ratios the sample began to precipitate. We were also unable to make soluble 1:1 complexes at concentrations above ~0.2 mM. The 1H chemical shift changes of the RNA were monitored using 1H-13C HSQC experiments. The 1H and 15N amide chemical shift changes on Nop10p were monitored by titration of RNA in 0.1 mM increments to a 0.5 mM 15N-labeled protein sample up to an RNA:protein ratio of 2:1 using 1H-15N HSQC experiments. The chemical shift changes found in the 1H and 15N resonances of the protein after addition were plotted according to the following equation:
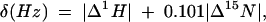 |
where |Δ1H| is the absolute value of the proton chemical shift difference and 0.101Δ15N is the absolute value of the nitrogen chemical shift difference weighted based on the relative proton and nitrogen gyromagnetic ratio. There were no significant changes in the 13C chemical shifts of the RNA in the titrations with Nop10p; consequently we only plotted Δ1H for the RNA.
Coordinate deposition
Coordinates for the 15 lowest-energy structures of U65hp and 10 lowest-energy structures of Nop10p have been deposited in the Protein Data Bank under accession codes 2EUY and 1Y2Y, respectively.
Acknowledgments
We thank Dr. Robert Peterson for assistance with NMR spectroscopy, Evan Feinstein for manuscript preparation, Craig Blois for sample preparation, and Dr. Anthony Henras for helpful discussion. This work was supported by NIH grants GM37254 and GM48123 to J.F.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2221606.
REFERENCES
- Bagni, C. and Lapeyre, B. 1998. Gar1p binds to the small nucleolar RNAs SNR10 and SNR30 in vitro through a nontypical RNA binding element. J. Biol. Chem. 273: 10868–10873. [DOI] [PubMed] [Google Scholar]
- Baker, D.L., Youssef, O.A., Chastkofsky, M.I.R., Dy, D.A., Terns, R.M., and Terns, M.P. 2005. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes & Dev. 19: 1238– 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin, A. and Ofengand, J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center—Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. [DOI] [PubMed] [Google Scholar]
- Bakin, A., Lane, B.G., and Ofengand, J. 1994. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry 33: 13475–13483. [DOI] [PubMed] [Google Scholar]
- Balakin, A.G., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Inada, M., Kujawinski, E., Puglisi, J.D., and Williamson, J.R. 1992. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 20: 4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax, A., Clore, G.M., and Gronenborn, A.M. 1990. 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88: 425–431. [Google Scholar]
- Bortolin, M.L., Ganot, P., and Kiss, T. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Henry, Y., Gelugne, J.P., Caizerguesferrer, M., and Kiss, T. 1997. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 16: 4770–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, J.A. and Williamson, J.R. 2004. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure 12: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Charpentier, B., Muller, S., and Branlant, C. 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33: 3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., and Tjandra, N. 1998. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 131: 159–162. [DOI] [PubMed] [Google Scholar]
- Dez, C., Henras, A., Faucon, B., Lafontaine, D.L.J., Caizergues-Ferrer, M., and Henry, Y. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann, T. and Feigon, J. 1997. Assignment methodology for larger RNA oligonucleotides—Application to an ATP-binding RNA aptamer. J. Biomol. NMR 9: 259–272. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Pogacic, V., and Filipowicz, W. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20: 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker, A.K., Brown, C.J., Lawson, J.D., Iakoucheva, L.M., and Obradovic, Z. 2002. Intrinsic disorder and protein function. Biochemistry 41: 6573–6582. [DOI] [PubMed] [Google Scholar]
- Dyson, H.J. and Wright, P.E. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12: 54– 60. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Formankay, J.D., and Kay, L.E. 1994a. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by N-15 NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Zhang, O.W., Formankay, J.D., and Kay, L.E. 1994b. A heteronuclear correlation experiment for simultaneous determination of N-15 longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4: 727–734. [DOI] [PubMed] [Google Scholar]
- Furtig, B., Richter, C., Wohnert, J., and Schwalbe, H. 2003. NMR spectroscopy of RNA. Chembiochem 4: 936–962. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M.L., and Kiss, T. 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Caizergues-Ferrer, M., and Kiss, T. 1997b. The family of Box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 11: 941– 956. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Bjork, G., and Maden, B.E.H. 1995. Nucleotide modification and base conversion of RNA—Summary and outlook. Biochimie 77: 3–6.7541253 [Google Scholar]
- Grzesiek, S. and Bax, A. 1992a. Correlating backbone amide and side chain resonances in layer proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114: 6291–6293. [Google Scholar]
- ———. 1992b. An efficient experiment for sequential backbone assignment of medium sized isotopically enriched proteins. J. Magn. Reson. 99: 201–207. [Google Scholar]
- ———. 1993. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 3: 185–204. [DOI] [PubMed] [Google Scholar]
- Hamma, T. and Ferre-D’Amare, A.R. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903. [DOI] [PubMed] [Google Scholar]
- Henras, A., Henry, Y., Bousquet-Antonelli, C., Noaillac-Depeyre, J., Gelugne, J.P., and Caizergues-Ferrer, M. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17: 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras, A., Capeyrou, R., Henry, Y., and Caizergues-Ferrer, M. 2004. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 10: 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., Ghose, R., Flanagan, J.M., and Prestegard, J.H. 1999. Backbone dynamics of the N-terminal domain in E-coli DnaJ determined by N-15- and (CO)-C-13-relaxation measurements. Biochemistry 38: 10567–10577. [DOI] [PubMed] [Google Scholar]
- Jones, S., Daley, D.T.A., Luscombe, N.M., Berman, H.M., and Thornton, J.M. 2001. Protein–RNA interactions: A structural analysis. Nucleic Acids Res. 29: 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., and Steitz, T.A. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340: 141–177. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.L.J., Bousquet-Antonelli, C., Henry, Y., Caizerguesferrer, M., and Tollervey, D. 1998. The Box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & Dev. 12: 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X.H., Liu, L., and Michaeli, S. 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 276: 40313–40318. [DOI] [PubMed] [Google Scholar]
- Luy, B. and Marino, J.P. 2000. Direct evidence for Watson–Crick base pairs in a dynamic region of RNA structure. J. Am. Chem. Soc. 122: 8095–8096. [Google Scholar]
- Maden, B.E.H., Corbett, M.E., Heeney, P.A., Pugh, K., and Ajuh, P.M. 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77: 22–29. [DOI] [PubMed] [Google Scholar]
- Moore, T., Zhang, Y.M., Fenley, M.O., and Li, H. 2004. Molecular basis of box C/D RNA–protein interactions: Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807– 818. [DOI] [PubMed] [Google Scholar]
- Ni, J., Tien, A.L., and Fournier, M.J. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Nikonowicz, E.P. and Pardi, A. 1993. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in C-13/N-15 labeled nucleic acids. J. Mol. Biol. 232: 1141–1156. [DOI] [PubMed] [Google Scholar]
- Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., and Luhrmann, R. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18: 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand, J., Bakin, A., Wrzesinski, J., Nurse, K., and Lane, B.G. 1995. The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol. 73: 915–924. [DOI] [PubMed] [Google Scholar]
- Peterson, R.D., Theimer, C.A., Wu, H.H., and Feigon, J. 2004. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA–protein complexes. J. Biomol. NMR 28: 59–67. [DOI] [PubMed] [Google Scholar]
- Pogacic, V., Dragon, F., and Filipowicz, W. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20: 9028– 9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi, J.D., Chen, L., Blanchard, S., and Frankel, A.D. 1995. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide–RNA complex. Science 270: 1200–1203. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., Tang, T.H., Tchirkova, I.V., Brosius, J., Bachellerie, J.P., and Huttenhofer, A. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 31: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.G., Schnare, M.N., and Gray, M.W. 2004. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA 10: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A., Balakin, A.G., and Fournier, M.J. 1995. Characterization of three new snRNAs from Saccharomyces cerevisiae: snR34, snR35 and snR36. Nucleic Acids Res. 23: 2548–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160: 65–73. [DOI] [PubMed] [Google Scholar]
- Somers, W.S. and Phillips, S.E. 1992. Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature 359: 387–393. [DOI] [PubMed] [Google Scholar]
- Suryadi, J., Tran, E.J., Maxwell, E.S., and Brown, B.A. 2005. The crystal structure of the Methanocaldococcus jannaschii multi-functional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry 44: 9657– 9672. [DOI] [PubMed] [Google Scholar]
- Tang, T.H., Bachellerie, J.P., Rozhdestvensky, T., Bortolin, M.L., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Huttenhofer, A. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. 99: 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. and Kiss, T. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Struct. Biol. 9: 337–342. [DOI] [PubMed] [Google Scholar]
- Treger, M. and Westhof, E. 2001. Statistical analysis of atomic contacts at RNA–protein interfaces. J. Mol. Recog. 14: 199–214. [DOI] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Luhrmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Wang, C. and Meier, U.T. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N.J., Gottschalk, A., Neubauer, G., Kastner, B., Fabrizio, P., Mann, M., and Luhrmann, R. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Wright, P.E. and Dyson, H.J. 1999. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 293: 321–331. [DOI] [PubMed] [Google Scholar]
- Wu, H.H., Henras, A., Chanfreau, G., and Feigon, J. 2004. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. 101: 8307–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]