Abstract
Erv41p and Erv46p form an integral membrane protein complex that cycles between the endoplasmic reticulum (ER) and Golgi. Both proteins contain a large lumenal domain and short N- and C-terminal tail sequences exposed to the cytosol. The coat protein complex II (COPII) packages the Erv41p–Erv46p complex into ER-derived vesicles for delivery to the Golgi. We determined signals in the Erv41p–Erv46p complex that are required for COPII-dependent export from the ER. Mutants lacking the Erv41p or Erv46p C-terminus accumulated in the ER and were not packaged efficiently into vesicles. We identified an isoleucine–leucine sequence in the Erv41p tail that was required for COPII binding and inclusion of the complex into vesicles. This signal was sufficient for COPII binding but not for ER export. The Erv46p tail contains a phenylalanine–tyrosine sequence required together with the isoleucine–leucine signal in Erv41p for export of the complex. Surprisingly, Erv41p– Erv46p tail-swapped chimeras were not exported from the ER, indicating that signals in both the Erv41p and the Erv46p tail sequences are required in a specific orientation for efficient packaging of the Erv41p–Erv46p complex.
Keywords: coat proteins/ER/sorting/trafficking/vesicles
Introduction
Proteins destined for the secretory pathway are synthesized at the rough endoplasmic reticulum (ER) and co-translationally translocated into its lumen, where they are folded and may undergo post-translational modification. At ER exit sites, soluble and transmembrane cargo proteins are sorted into transport vesicles, which are formed by the coat protein complex II (COPII) coat (reviewed by Antonny and Schekman, 2001). It is thought that the COPII coat is required for anterograde transport of proteins and lipids from the ER to the Golgi, whereas the COPI coat is responsible for retrograde transport from the Golgi to the ER as well as for intra-Golgi transport (reviewed by Mellman and Warren, 2000).
It has been shown that the COPII subunits Sar1p and Sec23/24p form pre-budding complexes with cargo molecules (Aridor et al., 1998; Kuehn et al., 1998; Springer and Schekman, 1998), thus providing a mechanistic explanation for selective capture of cargo molecules into forming vesicles. For some cargo molecules, ER export motifs that act in COPII-dependent export have been defined. For example, vesicular stomatitis virus (VSV) G protein (Nishimura and Balch, 1997) and potassium channels (Ma et al., 2001) contain a di-acidic (DXE) motif near their C-termini that is required for efficient ER export. In the case of VSV-G, however, it appears that additional elements mediate COPII binding (Nishimura et al., 1999), and an overlapping tyrosine signal has been identified that is required for optimal ER export rates (Sevier et al., 2000). The di-acidic motif also functions in yeast Sys1p (Votsmeier and Gallwitz, 2001).
A second class of ER export motifs has been described for integral membrane proteins cycling between the ER and Golgi, such as the p24 proteins and ERGIC-53. Nakamura et al. (1998) showed that transport of a chimeric invertase–Wbp1p reporter protein was accelerated by the addition of the Emp24p tail, an effect that was dependent on a pair of hydrophobic C-terminal residues (LV). Dominguez et al. (1998) reported that mutation of a di-phenylalanine (FF) motif in mammalian p24 tail sequences interfered with binding to the Sec23 subunit of COPII and caused accumulation in the ER. In yeast, export of the p24 complex also requires the presence of a pair of aromatic residues: FF on the Emp24p tail and YF on the Erv25p sequence. Curiously, the tails were not fully interchangeable between Erv25p and Emp24p, as chimeric Erv25p–Emp24p proteins were not packaged as efficiently as the wild-type proteins (Belden and Barlowe, 2001a).
Efficient ER export of ERGIC-53 depends on an FF sequence at the extreme C-terminus (Kappeler et al., 1997). A recent study (Nufer et al., 2002) showed that this motif can be functionally substituted by a single phenylalanine in position –2 from the C-terminus, by two isoleucines or leucines in position –2 and –1 or by a single valine in position –1. Consistent with the concept of selective capture of cargo molecules into vesicles by interactions with COPII, export efficiency and COPII binding ability of these hydrophobic motifs have been shown to correlate (Dominguez et al., 1998; Belden and Barlowe, 2001a; Nufer et al., 2002). However, the mechanisms by which these diverse COPII signals act in export from the ER remain unclear in comparison with the other highly conserved coat-binding motifs (Cosson and Letourneur, 1997; Robinson and Bonifacino, 2001).
We previously have described Erv41p and Erv46p, two abundant ER vesicle (Erv) proteins of 41 and 46 kDa, which are contained on COPII vesicles (Otte et al., 2001). Both are packaged efficiently by COPII and cycle between the ER and Golgi complex. Expression levels of Erv41p and Erv46p are interdependent, and both proteins associate physically as they can be co-immunoprecipitated. Erv46p contains a conserved C-terminal KKXX COPI-binding motif (Cosson and Letourneur, 1994) that is predicted to act in retrograde transport of the Erv41p–Erv46p complex from the Golgi to the ER.
The exact biological functions of Erv41p and Erv46p are unknown; however, both genetic and biochemical evidence point to a role in vesicular traffic between the ER and Golgi. The erv41Δ and/or erv46Δ deletion mutants are viable and display no apparent general transport defects. Mutations in these genes do produce synthetic slow growth phenotypes when combined with other mutations in the early secretory pathway, including erv14Δ, a gene required for packaging of a transmembrane cargo protein into COPII vesicles (Otte et al., 2001; Powers and Barlowe, 2002). Possible roles for the Erv41p–Erv46p complex have been proposed as part of the glycoprotein processing machinery in the early secretory pathway, as adaptors in sorting secretory cargo into COPII vesicles, as a Golgi to ER retrieval system, or in lipid transport.
Here we report a detailed analysis of the cytoplasmic C-terminal tail sequences of Erv41p and Erv46p in cycling of the complex between the ER and Golgi. Immuno precipitation data (Otte et al., 2001) suggested that the Erv41p–Erv46p complex probably consists of single Erv41p and Erv46p subunits with little functional redundancy encoded by the tails, especially since only Erv46p contains a KKXX motif. Neither the Erv41p nor the Erv46p tails carry a DXE motif, but the Erv41p tail includes two hydrophobic residues, Ile349 and Leu350 (IL), in positions –4 and –3 from the C-terminus. The Erv46p tail contains a phenylalanine–tyrosine (FY) motif in positions –14 and –13 (Figure 1A). We therefore chose the Erv41p–Erv46p complex as a model substrate to study the general requirements for the sorting of multisubunit transmembrane proteins into COPII vesicles. We find that the IL residues in the Erv41p tail were indeed necessary and sufficient for COPII binding. However, this signal was not sufficient for packaging into vesicles. A second signal comprised of the FY residues in Erv46p was required in combination with the Erv41p IL sequence for COPII-dependent export. These findings suggest that efficient ER export of transmembrane complexes may not rely on simple linear exit motifs but on more complex combinatorial signals.
Fig. 1. Schematic diagram of Erv41p and Erv46p tail mutants. (A) Wild-type sequences and membrane topology. The C-terminal tail sequences are given in one-letter code, and black boxes represent predicted transmembrane helices. Potential sorting signals are in bold. Underlined residues of Erv41p were mutated individually to alanines for the alanine scan experiment. (B) Summary of C-terminal tail constructs. Exchanged tail sequences are in italics.
Results
Membrane topology of Erv41p and Erv46p
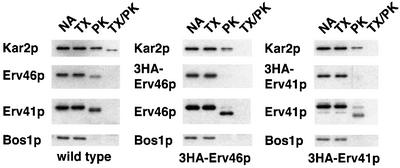
Erv41p, Erv46p and their respective homologs in other species have been predicted to contain two segments of sufficient length and hydrophobicity to form transmembrane domains, and their short N- and C-terminal tails were predicted to reside on the cytosolic side (Otte et al., 2001). To test this topology (Figure 1A), we used a protease protection assay to examine the accessibility of both proteins to proteinase K in the presence or absence of a detergent (Figure 2). In wild-type membranes, proteinase K treatment without detergent results in a slightly increased electrophoretic mobility, indicating that the tails were degraded but the large domain between the membrane spanners, which is recognized by the Erv41p and Erv46p antisera, was protected. In strains expressing N-terminally hemagglutinin (HA)-tagged Erv46p and Erv41p, an anti-HA antibody did not detect the epitope after proteinase K treatment alone, and the shift in mobility was greater in the tagged version, consistent with the loss of longer N-terminal tails. Kar2p, a protein of the ER lumen, was degraded only in the presence of detergent, while Bos1p, a largely cytoplasmically exposed SNARE protein, was degraded without detergent. Taken together, these data indicate that the N-termini of both Erv41p and Erv46p are exposed to the cytosol. Combined with the presence of the conserved C-terminal KKXX COPI-binding motif (Cosson and Letourneur, 1994) on Erv46p, this shows that both N- and C-terminal tails are located on the cytosolic side of the ER and Golgi membrane and potentially could interact with cytosolic factors such as coat proteins.
Fig. 2. The N-terminal tails of both Erv41p and Erv46p are exposed to the cytosol. Microsomes from wild-type (FY834), 3HA-Erv46p- (CBY770) and 3HA-Erv41p-expressing (CBY783) cells were treated with buffer alone (NA), 1% Triton X-100 (TX), proteinase K (PK) or both (TX/PK). Samples were resolved on a polyacrylamide gel and blotted for Kar2p (a lumenal ER protein) and Bos1p (a cytoplasmically exposed membrane protein). Wild-type as well as N-terminally 3HA-tagged Erv41p and Erv46p were detected with polyclonal antisera and with 3HA-specific monoclonal antibody.
ERV41 and ERV46 C-terminal tail mutants accumulate in the ER
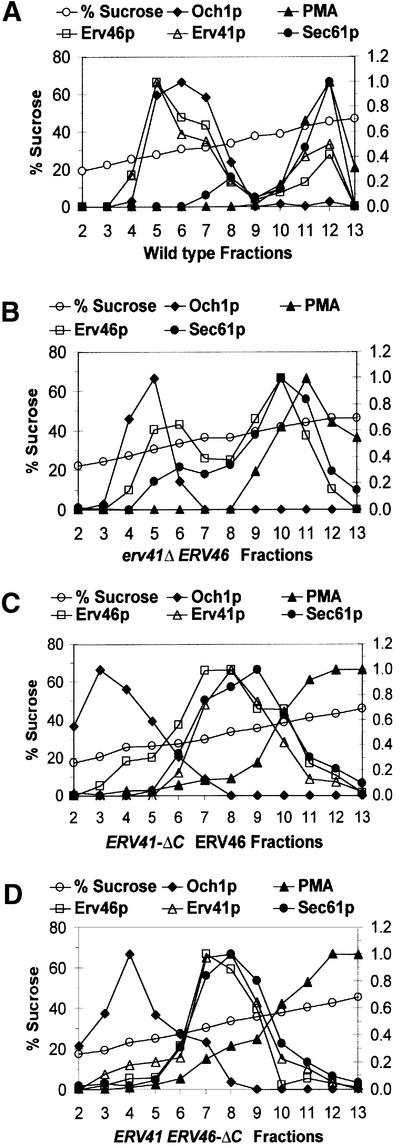
We had observed earlier that expression of Erv41p and Erv46p is interdependent. Specifically, Erv41p was not detected by western blot in an erv46Δ strain and Erv46p expression was reduced in an erv41Δ strain (Otte et al., 2001). At the beginning of the present study, we therefore asked whether the reduced level of Erv46p expression also reflected a mislocalization of this protein when the Erv41p subunit is absent. Cell lysates were prepared from wild-type and erv41Δ strains, and organelles were resolved on sucrose gradients. Fractions were collected and blotted for the early Golgi marker Och1p, the ER marker Sec61p, the plasma membrane marker PMA1, Erv41p and Erv46p. In wild-type cells (Figure 3A), the major fraction of Erv46p (∼70%) and Erv41p is located in the Golgi and a minor fraction in the ER (∼30% of Erv46p). The erv41Δ deletion (Figure 3B) reverses this distribution, with the majority of Erv46p (∼70%) accumulating in the ER and a second minor peak in the Golgi (∼30%). This redistribution of Erv46p to the ER may be caused by loss of ER export information contained on the Erv41p subunit or by retention of uncomplexed Erv46p by the ER quality control system (Ellgaard et al., 1999).
Fig. 3. Subcellular localization of Erv41p and Erv46p depends on the presence of both C-terminal tail sequences. Cell lysates were resolved on 18–60% (w/v) sucrose gradients and fractions were collected from the top. Relative amounts of the ER marker Sec61p, the Golgi marker Och1p, the plasma membrane marker PMA1, Erv41p and Erv46p were quantified by densitometry of immunoblots. Sucrose percentages were determined by refractometry. In the first experiment, (A) wild-type (FY834) and (B) erv41Δ (CBY797) strains were compared. In the second experiment, (C) ERV41-ΔC (CBY1199) and (D) ERV46-ΔC (CBY1200) were analyzed.
We next investigated the requirements for subcellular localization of the Erv41p–Erv46p complex. The cytoplasmically exposed N-terminal tails of both proteins are relatively short, 23 residues in Erv41p and 24 residues in Erv46p, and we had found earlier that Erv41p and Erv46p tagged at their N-termini with the 3HA epitope are packaged efficiently into COPII vesicles (Otte et al., 2001). This suggested that the N-terminal tails do not play a role in ER export, and we thus focused on the C-terminal tails. However, a contribution of the N-terminal tails currently cannot be ruled out.
To examine whether the C-terminal tails of Erv46p and/or Erv41p are required for subcellular localization, we generated a set of truncations and tail-swapped chimeras. Individual constructs were expressed under the ERV46 and ERV41 promoters from CEN-based plasmids covering deletions (Figure 1B). Stop codons were introduced by site-directed mutagenesis, resulting in an Erv46p truncation mutant, Erv46p-ΔC, that lacked 14 C-terminal residues, and an Erv41p truncation mutant, Erv41p-ΔC, lacking 13 C-terminal residues. To test for transplantability of potential tail functions, we also generated strains expressing two sets of chimeric proteins. In the first set, Erv46p-41Tail and Erv41p-46Tail, 14 C-terminal residues of Erv46p and 24 residues of Erv41p were exchanged, preserving the sequence contexts of both the KKXX and IL motifs. In the second set, Erv46p-ILDR and Erv41p-KKSQ, only the four C-terminal residues containing the actual KKXX and IL motifs were swapped. In this second set of constructs, both motifs are out of their native sequence context, but effects of these mutations on the structure and folding of the proteins should be minimal.
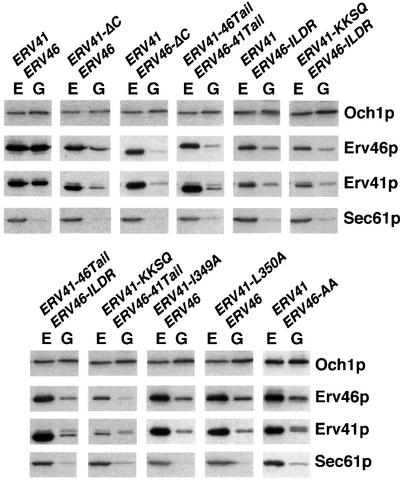
All of the mutants were expressed stably as verified by western blot analysis of membrane fractions (Figure 4), although in all cases where the KKXX-containing Erv46p tail was deleted or out of its immediate sequence context, overall expression of both proteins was lower. We also confirmed by immunoprecipitation that Erv41p-ΔC and Erv46p still form a complex, indicating that the tails are dispensable for this interaction and that Erv41p and Erv46p most probably interact through their lumenal and/or transmembrane domains. Even in strains expressing the truncated versions and more extensive tail swaps of Erv46p and Erv41p, Kar2p expression levels were not elevated (data not shown), indicating that the unfolded protein response pathway was not activated (reviewed by Patil and Walter, 2001).
Fig. 4. Expression levels and subcellular distribution of the indicated Erv41p and Erv46p constructs. Membrane fractions enriched in ER (E, 13 000 g pellets) and Golgi (G, 100 000 g pellets) were analyzed by immunoblotting using Och1p as a Golgi marker and Sec61p as an ER marker, as well as polyclonal antisera against Erv41p and Erv46p.
In order to assess the subcellular localization of the mutant proteins, we prepared membrane fractions enriched for ER and Golgi and monitored the distribution of Erv41p and Erv46p between these compartments (Figure 4). As expected, wild-type Erv41p and Erv46p co-fractionated with both the Sec61p-containing ER fraction and the fraction enriched in the early Golgi marker Och1p. In contrast, all mutants showed an accumulation of the Erv41p–Erv46p complex in the ER at steady state, although this effect was less pronounced in the strain expressing Erv41p–Erv46p-ILDR.
Figure 4 also documents the reduced levels of mutant Erv41p–Erv46p complexes in which the KKXX motif is either missing or in the context of the Erv41p tail, i.e. Erv41p–Erv46p-ILDR, Erv41p-KKSQ–Erv46p-ILDR or Erv41p-KKSQ–Erv46p-41Tail. Therefore, the KKXX motif appeared to be functional in the chimeric constructs only if the complete Erv46p tail was transplanted but not if it was in the context of the Erv41p tail. We presume that the mutants without a functional KKXX motif slowly leak from the ER, are not efficiently retrieved by COPI and are degraded in post-Golgi compartments.
To confirm our crude fractionation method shown in Figure 4, we also examined the subcellular distribution of truncated Erv41p-ΔC and truncated Erv46p-ΔC through resolution of organelles on sucrose gradients (Figure 3C and D). As expected, Erv41p-ΔC caused a strong accumulation of the Erv41p–Erv46p complex in the ER (∼86% of Erv46p), and Erv46p-ΔC resulted in a similar mislocalization with ∼94% of Erv46p-ΔC in the ER fractions. These results are in good agreement with the fractionation data shown in Figure 4 and validate this method.
Based on the collective fractionation data, we conclude that both C-terminal tails of the Erv41p–Erv46p complex are required for a wild-type subcellular localization pattern and that mutations cause the complex to accumulate in the ER. Furthermore, sorting information contained in the C-terminal sequences was not exchangeable, regardless of whether long or short swaps were employed.
ERV41 and ERV46 C-terminal tail mutants are not packaged by COPII
We hypothesized that the observed ER accumulation was due to a failure of COPII to package these tail mutants into ER-derived vesicles. To test this idea, we performed in vitro vesicle budding reactions from microsomes with isolated COPII subunits (Figure 5). Wild-type Erv41p and Erv46p are sorted efficiently into these vesicles (Otte et al., 2001), as is Erv25p (Belden and Barlowe, 1996), whereas Sec61p, an ER resident protein, is excluded. As anticipated from the mislocalization results, both tail truncation mutants failed to incorporate into ER-derived vesicles. Similarly, the chimeric Erv41p-46Tail–Erv46p-41Tail complex also failed to sort into vesicles (Figure 5A). The same results were observed when the shorter tail mutants Erv41p-KKSQ–Erv46p-ILDR or combinations of these with Erv41p-46Tail and Erv46p-41Tail were examined. The only mutant strain that exhibited efficient packaging expressed the Erv41p–Erv46p-ILDR complex, in which the KKXX motif had been inactivated but the Erv41p tail was intact (Figure 5B). This result suggests that the KKXX motif is not required for ER export, but other parts of the Erv46p tail seem to be necessary. To summarize, these findings lead to two conclusions. First, both the Erv41p tail and the Erv46p tail but not the KKXX motif are required for sorting into COPII vesicles. Secondly, the tails are not functionally interchangeable among subunits of the same complex.
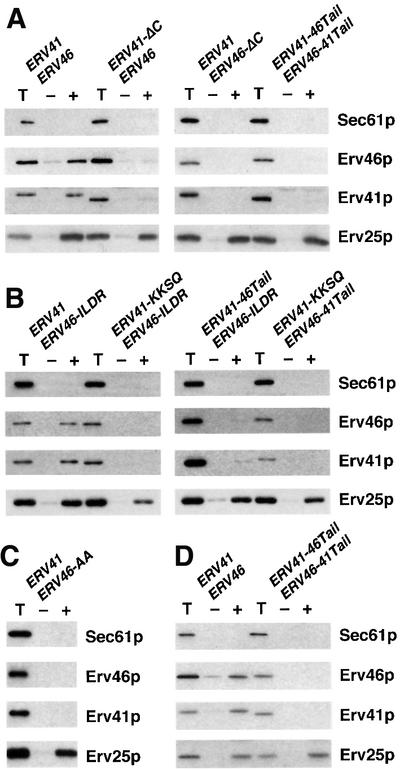
Fig. 5. Packaging of Erv41p and Erv46p into COPII vesicles requires both C-terminal tail sequences in their wild-type orientation and is not influenced by GTP hydrolysis. (A) In vitro budding reactions were performed in the presence of GTP with microsomes prepared from strains with wild-type ERV41 and ERV46 (CBY1198), ERV41-ΔC and wild-type ERV46 (CBY1199), wild-type ERV41 and ERV46-ΔC (CBY1200), or chimeric ERV41-46Tail and ERV46-41Tail (CBY1204). (B) Budding reactions in the presence of GTP were performed with microsomes from strains with wild-type ERV41 and the ERV46-ILDR mutation (CBY 1305), ERV41-KKSQ and ERV46-ILDR (CBY1306), ERV41-46Tail and ERV46-ILDR (CBY1307), or ERV41-KKSQ and ERV46-41Tail (CBY1308). (C) Budding reactions with microsomes from a strain with wild-type ERV41 and the ERV46-AA mutation (CBY1334). (D) Budding reactions were carried out as in (A) except that GMP-PNP was added instead of GTP. One-tenth of total reactions (T), budded vesicles formed in the presence of COPII proteins (+) and mock reactions without COPII proteins (–) were analyzed on 12.5% polyacryl amide gels and immunoblotted.
It has been reported that sorting of Golgi resident proteins into COPI vesicles requires GTP hydrolysis by Arf-1, a GTPase related to Sar1p (Lanoix et al., 1999, 2001). To determine whether GTP hydrolysis influenced the incorporation of the chimeric protein complex into COPII vesicles, we performed budding assays in the presence of the non-hydrolyzable GTP analog, GMP-PNP. Results identical to those obtained with GTP were observed (Figure 5D). Therefore, the selection of Erv41p–Erv46p by COPII is not influenced by GTP hydrolysis, as has been observed for the bulk of vesicle cargo proteins (Barlowe et al., 1994).
Two hydrophobic residues on the Erv41p tail are required for packaging by COPII
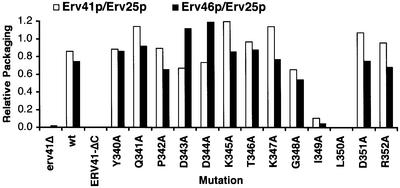
The Erv41p tail contains two acidic residues and two hydrophobic residues near the C-terminus that are similar to previously described ER exit signals (Nakamura et al., 1998; Nufer et al., 2002). To characterize the roles of the individual residues on the Erv41p tail, we performed an alanine scan experiment, individually mutating all 13 residues to alanines in the part of the tail that had been shown to be required for packaging. In this screen, mutations were introduced into the ERV41 gene on a single-copy plasmid, covering a chromosomal erv41Δ deletion. We confirmed by immunoblot of membrane fractions that all mutants were expressed at wild-type levels (data not shown). In vitro budding reactions were performed by adding COPII subunits to washed membranes from these strains, and packaging efficiencies for Erv25p, Erv41p and Erv46p were determined individually by densitometry of western blots. Since Erv25p and the Erv41p–Erv46p complex are sorted into vesicles independently of each other (data not shown), it was possible to use Erv25p packaging as a standard to insure comparable levels of overall budding in the individual membrane preparations (Figure 6). In two of these point mutants, Erv41p-I349A and Erv41p-L350A, packaging of the Erv41p–Erv46p complex was completely abolished, while none of the others showed a significantly lower packaging efficiency. As seen in Figure 4, these two mutants are stably expressed at near wild-type levels but accumulate in ER membranes. As a positive control, a strain in which the erv41Δ deletion was covered by wild-type Erv41p on a CEN-based plasmid (wt) and, as negative controls, erv41Δ strains that carried a blank vector (41Δ) and an Erv41p-ΔC-expressing plasmid (ERV41-ΔC) were included in Figure 6. These data demonstrate that Ile349 and Leu350 in positions –4 and –3 from the C-terminus of Erv41p are required for sorting of the Erv41p–Erv46p complex into COPII vesicles in vitro and form an anterograde IL transport signal for ER export.
Fig. 6. Ile349 and Leu350 of the C-terminal tail of Erv41p are required for efficient packaging of Erv41p and Erv46p by COPII in vitro. Amino acid residues 340–352 of Erv41p were mutated to alanine, and erv41Δ cells were transformed with the mutant plasmids as indicated. erv41Δ strains carrying an empty pRS314 vector (CBY1036), an ERV41 wild-type plasmid (CBY1037) or an ERV41-ΔC plasmid (CBY1038) are shown as controls. COPII vesicles were synthesized in vitro from washed semi-intact cells, and individual packaging efficiencies for Erv41p, Erv46p and Erv25p were calculated by densito metry of western blots. Packaging efficiencies of Erv41p and Erv46p were plotted relative to Erv25p inclusion into vesicles.
The FY sequence in Erv46p is required for COPII packaging
The C-terminal tail sequence of Erv41p and specifically the IL residues are necessary for ER export of the Erv41p–Erv46p complex. However, the Erv46p-ΔC mutant also fails to exit the ER in COPII vesicles (Figure 5A), indicating that additional sorting determinants are present in the Erv46p C-terminal tail sequence. In this instance, the FY residues in position –14 and –13 of the Erv46p tail formed a probable signal (Dominguez et al., 1998). To test this possibility, the FY residues were converted to alanines to generate a strain with wild-type Erv41p and mutant Erv46p-AA. Subcellular fractionation (Figure 4) and in vitro budding assays (Figure 5C) with this mutant indicated that the FY residues were required for COPII-dependent export from the ER. Based on this finding, we conclude that both the IL signal in Erv41p and the FY signal in Erv46p are required in concert for export of this complex.
The IL motif is sufficient for COPII binding but is not sufficient for packaging into vesicles
Next, we explored the role of the Erv41p and Erv46p sorting signals in binding to COPII subunits in vitro. We also asked whether the tail mutants failed to be packaged in the previous experiments because they could not bind to COPII subunits, or whether they bound but were not packaged into COPII vesicles. To this end, we used an assay (Kuehn et al., 1998) in which pre-budding COPII–cargo complexes are formed on microsomal membranes when incubated with a Sar1p–GST fusion protein and Sec23/24p in the presence of GMP-PNP. These membranes are then solubilized and Sar1p–GST pre-budding complexes are collected on glutathione– agarose beads. First, we established that wild-type Erv41p–Erv46p complex binds specifically to COPII subunits (Figure 7). In binding reactions that contained Sar1p–GST and GMP-PNP, a basal level of Erv41p– Erv46p binding was detected; however, this interaction was stimulated by the addition of Sec23/24p and not by Sec13/31p. This interaction was specific, because the ER resident protein Sec61p was not bound. Erv25p also forms pre-budding complexes (Powers and Barlowe, 2002) and was shown here as a positive control.
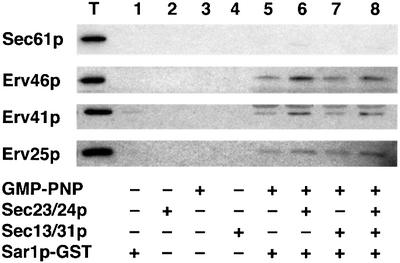
Fig. 7. Erv41p and Erv46p form pre-budding complexes with COPII subunits. Microsomes from a wild-type strain (FY834) were incubated with GMP-PNP, Sar1p–GST, Sec23/24p and Sec13/31p as indicated. The total lane (T) represents 0.6% of binding reactions. Proteins recovered from reactions 1–8 on glutathione–agarose were monitored by immunoblot. Note that binding to Sar1p–GST required GMP-PNP and was increased by addition of Sec23/24p but not by Sec13/31p.
We then tested the mutants described above for their ability to form pre-budding complexes with COPII (Figure 8). As expected from the in vitro packaging experiments, in strains expressing Erv41p-ΔC–Erv46p or the point mutants Erv41p-I349A and Erv41p-L350A, neither Erv46p nor the Erv41p mutants bound COPII. Surprisingly, in strains expressing Erv41p–Erv46p-ΔC or chimeric Erv41p-46Tail–Erv46p-41Tail, both proteins displayed some COPII-binding activity. This result contrasted with our observation that they were not packaged into budded vesicles (Figure 5). This interaction was also detected in the presence of Sar1p–GST and GMP-PNP alone and was not stimulated by Sec23/24p (Figure 9, left panel), although the present assay does not allow for a precise quantification of this effect. In contrast, mutation of Ile349 of the IL motif on Erv41p abolished all binding (Figure 9, right panel), even the basal interaction observed with Sar1p–GST and GMP-PNP alone in wild-type membranes (Figure 7). Low levels of binding to Sar1p–GST were also detected in the presence of GMP-PNP when the short swaps or the Erv41p–Erv46p-AA complex were examined (data not shown).
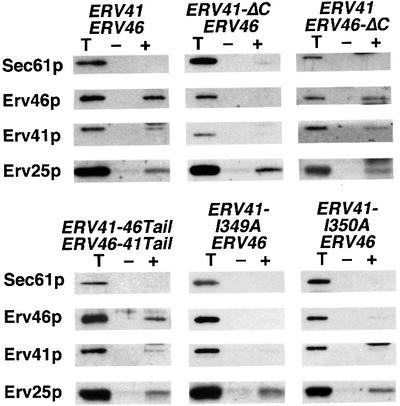
Fig. 8. Erv41p and Erv46p bind to COPII subunits if the Erv41p tail is intact or present on either subunit. Binding reactions with tail mutants were performed as in Figure 7. Totals (T) represent 0.6% of binding reactions, and proteins recovered from reactions containing Sar1p–GST alone (–) or Sar1p–GST, GMP-PNP and Sec23/24p (+) were analyzed by immunoblot. Microsomes were from cells with wild-type ERV41 and ERV46 (CBY1198), truncated ERV41-ΔC and wild-type ERV46 (CBY1199), wild-type ERV41 and truncated ERV46-ΔC (CBY1200), chimeric ERV41-46Tail and ERV46-41Tail (CBY1204), point mutant ERV41-I349A (CBY1159) or point mutant ERV41-L350A (CBY1090).
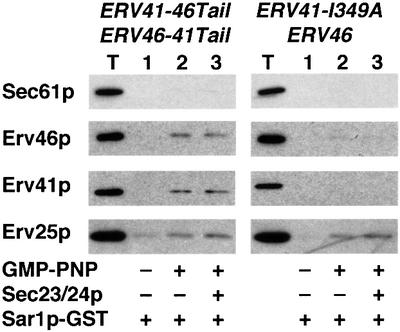
Fig. 9. The Erv41p tail sequence is necessary and sufficient for binding to the COPII subunit Sar1p. Binding reactions were performed as in Figure 7 with GMP-PNP, Sar1p–GST and Sec23/24p added as indicated. Microsomes were from the tail-swapped chimera (CBY1204) or the point mutant ERV41-I349A strain CBY1090. Note that addition of Sec23/24p did not increase Erv41p–Erv46p binding as observed for Erv25p.
These results suggest that multiple sorting motifs in the Erv41p–Erv46p complex are required for efficient formation of pre-budding complexes and export from the ER. The IL motif on the Erv41p tail is indeed a COPII-binding motif, and this motif may be sufficient for binding to Sar1p-GTP. In contrast, the Erv46p tail sequence appears to be dispensable for Sar1p-GTP binding but required for Sec23/24p-stimulated pre-budding complex formation and for packaging into COPII vesicles. Taken together, these results indicate that the tail mutants fail to be packaged into ER-derived vesicles for different reasons. First, if the IL motif is mutated, the complex cannot bind to Sar1p-GTP. Secondly, sequence information in the Erv46p tail in a particular spatial arrangement is necessary for formation of pre-budding complexes with Sec23/24p.
Discussion
In this study, we have characterized sorting determinants in the Erv41p–Erv46p complex that are necessary for COPII-dependent export from the ER. Unexpectedly, sequence information contained in both the Erv41p and the Erv46p subunits is required in its proper context for efficient packaging of the Erv41p–Erv46p complex into COPII vesicles. We reach this conclusion after determining the membrane topology of the Erv41p–Erv46p complex and analyzing the influence of mutations in the cytoplasmically exposed C-terminal tails of Erv41p and Erv46p. Other characterized coat-dependent sorting events appear to employ single linear polypeptide signals that are necessary and sufficient for selection into vesicles (Cosson and Letourneur, 1997; Robinson and Bonifacino, 2001). Therefore, our findings may have broad implications for COPII-dependent export of multisubunit protein complexes from the ER.
We demonstrated that wild-type Erv41p–Erv46p complex binds to subunits of the COPII coat. Partial binding of Erv41p–Erv46p complex to activated Sar1p was detected; however, optimal binding required both activated Sar1p and the Sec23/24p subunit. We propose that the sequential assembly of Erv41p–Erv46p into these pre-budding complexes is a prerequisite for incorporation into COPII vesicles, in accord with current models for selective export of proteins from the ER (Antonny and Schekman, 2001). Using an alanine scan approach, we identified two hydrophobic residues in the Erv41p tail, isoleucine in position –4 and leucine in position –3, that were required for sorting of the complex into COPII vesicles. Furthermore, this pair of hydrophobic residues was required for a normal cellular distribution in vivo and for COPII binding activity in vitro. These results indicate that the hydrophobic residues in position –4 or –3 of Erv41p form an anterograde transport signal that is required for COPII binding and for sorting into ER-derived vesicles.
Though the Erv41p tail sequence was necessary for incorporation of the Erv41p–Erv46p complex into COPII vesicles, it was not sufficient. Deletion of the Erv46p tail sequence or mutation of the FY residues in position –14 and –13 caused an ER accumulation of the Erv41p–Erv46p complex in vivo that correlated with a failure to incorporate into COPII vesicles in vitro. Thus, the Erv41p tail sequence in the absence of the Erv46p FY signal was not sufficient for ER export. However, Erv41p–Erv46p mutants lacking the Erv46p tail retained some COPII-binding activity. This result contrasts with the COPII-binding properties of complex lacking the Erv41p tail or the IL sequence in which all COPII-binding activity was abolished. Finally, exchange of the tail sequences produced stable Erv41p-46Tail–Erv46-41Tail complexes that bound to activated Sar1p but were not packaged into COPII vesicles. A failure to incorporate the tail-swapped chimeras into COPII vesicles suggests that additional spatial constraints must also be satisfied for export of the Erv41p–Erv46p complex from the ER.
Based on the collective results, we propose that efficient packaging of the Erv41p–Erv46p complex into ER-derived vesicles relies on multiple sorting determinants in a specific spatial arrangement. One possibility is that the Erv41p tail contacts activated Sar1p-GTP while the Erv46p tail interacts with the Sec23/24p complex. These associations in concert could lead to formation of stabilized pre-budding complexes that then are gathered by Sec13/31p into COPII vesicles. If the Erv41p and Erv46p tail sequences contain motifs that interact with distinct domains of Sar1p and Sec23/24p, a specific geometry may be required to conform with pre-budding complexes. This geometric constraint may explain why tail-swapped chimeras fail to be packaged into COPII-derived vesicles.
Alternatively, the Erv41p/46p tail sequences may contain COPII-independent sorting determinants that are responsible for localization of the complex to specific regions of the ER. For example, the FY signal within the Erv46p tail could direct the Erv41p–Erv46p complex to transitional zones of the ER, perhaps mediated through association with auxiliary budding factors such as Sec16p (Campbell and Schekman, 1997). It has also been proposed that COPI association with cargo may be necessary for ER export (Stephens and Pepperkok, 2002); however, this seems unlikely for the Erv41p– Erv46p complex because the Erv41p–Erv46-ILDR chimera that lacks a KKXX motif was packaged efficiently into COPII vesicles (Figure 5B).
We cannot exclude the possibility that these mutations influence the overall structure of the Erv41p–Erv46p complex and cause ER retention, as has been observed for misfolded VSV-G protein (de Silva et al., 1990). However, several of our results argue against this possibility. First, expression of Erv41p depends on its interaction with Erv46p (Otte et al., 2001) and we detect stable expression of Erv41p in all of our strains with the exception of mutants that lack a functional di-lysine retrieval signal. Secondly, single point mutations and tail deletions displayed identical properties with respect to formation of pre-budding complexes and incorporation into COPII vesicles. Thirdly, we constructed both long (13 amino acids) and short (four amino acids) tail-swap chimeras and observed qualitatively identical results.
Other linear amino acid sequence motifs have been identified that act in ER export. The DXE motif on VSV-G is required for efficient ER export (Nishimura and Balch, 1997) but, surprisingly, not for COPII binding, leading to the proposal that more than one step is involved in cargo sorting (Nishimura et al., 1999). More recently, a second tyrosine-based signal (YXXØ) that overlaps with the DXE motif was identified in the VSV-G tail sequence and found to be required for optimal ER export (Sevier et al., 2000). It remains to be determined whether the tyrosine-based signal is necessary for VSV-G incorporation into pre-budding complexes. Interestingly, it is known that VSV-G forms homotrimers before export from the ER (Doms et al., 1988). Therefore, multiple sorting determinants may be exposed on a VSV-G trimer that couple this complex to distinct components of the ER budding machinery as we suggest for the Erv41p–Erv46p complex.
The IL motif we identified in the Erv41p tail is similar to hydrophobic motifs described by Nufer et al. (2002). These authors reported that the C-terminal diphenylalanine motif of ERGIC-53 could be functionally replaced by two leucines or isoleucines in positions –2 and –1, a single valine at position –1 or single phenylalanine or tyrosine residues in position –2. Our binding studies with purified COPII subunits corroborate their data obtained by incubation of immobilized tail peptides with cell lysates. However, our results show that COPII-binding motifs consisting of pairs of leucine or isoleucine residues are not strictly position dependent and can function not only at the extreme C-terminus but at least also in the –4, –3 positions. More recently, studies on Emp46p, an ERGIC-53 homolog in yeast, have identified two ER export signals contained within the C-terminal tail of Emp46p: a tyrosine-containing motif centered on position –15 and a pair of leucine residues in the –1, –2 positions. Both motifs are required for incorporation of Emp46p into pre-budding complexes and into COPII vesicles (Sato and Nakano, 2002). The presence of multiple ER export signals on Emp46p resembles our findings reported here although in the Erv41p–Erv46p complex these signals reside on different polypeptide chains. The Emp46p export signals could also act on separate subunits because the oligomeric status of Emp46p, or ERGIC-53, has not been determined. Lastly, studies on the multimeric Emp24p–Erv25p complex also suggest that multiple COPII-binding motifs operate in transport of this complex (Marzioch et al., 1999; Belden and Barlowe, 2001a).
It is not clear how signals as disparate as the di-acidic and the di-aromatic/hydrophobic motifs direct cargo into COPII vesicles. It is possible that distinct mechanistic pathways are responsible or that multiple binding sites in the COPII coat recognize different sorting signals. Evidence for the latter is provided by genetic and biochemical studies in yeast where diversity in the Sec24p subunit of the Sec23/24p complex expands the variety of cargo that are packaged into ER-derived vesicles (Shimoni et al., 2000). It is also possible that a single Sec24p subunit could have more than one cargo recognition site.
Recent structural studies on individual COPII subunits and on the pre-budding Sec23/24p–Sar1p complex provide a molecular level view of this GTPase-directed sorting mechanism (Lederkremer et al., 2001; Matsuoka et al., 2001; Bi et al., 2002). Notably, the Sec23/24p–Sar1p complex possesses a number of potential cargo-binding sites that appear to face the vesicle surface (Bi et al., 2002). Our data indicate that multiple sorting determinants are contained on oligomeric cargo, and further structural studies can test whether these signals are required to contact distinct regions of the Sec23/24–Sar1p complex.
Materials and methods
Plasmid construction
To generate plasmid pRS314-ERV41, the ERV41 coding sequence and ∼300 bp of flanking upstream and downstream sequences were excised from pRS424-ERV41 (Otte et al., 2001) with NotI and BamHI, and ligated into the single-copy vector pRS314 (Sikorski and Hieter, 1989). Similarly, the ERV46 sequence was excised from pRS426-ERV46 (Otte et al., 2001) with the same enzymes and inserted into the single- copy vector pRS316 (Sikorski and Hieter, 1989) to yield plasmid pRS316-ERV46.
Point mutations were introduced into these two plasmids using the Transformer site-directed mutagenesis kit (Clontech, Palo Alto, CA) and universal selection primer JP28 (Powers and Barlowe, 2002) as recommended by the manufacturer, and verified by DNA sequencing. Primer sequences are listed in Table I. To obtain plasmid pRS314-ERV41-ΔC, a stop codon was introduced after Arg339, using mutagenic primer ERV41R. Plasmid pRS316-ERV46-ΔC was generated by introducing a stop codon after Leu401 with primer ERV46L. Erv41p alanine scan mutants were produced by the same method, replacing individual residues between Tyr340 and Arg352 with alanine, using mutagenic primers Y340A–R352A.
Table I. Primer sequences.
| Primer | Sequence (5′→3′) |
|---|---|
| JP28 | GGGGGGGCCCAGTACTCAGCTTTTGTTCCC |
| ERV41R | GGGACCTAAGTGGTCCTTGCGTTAGTAAGCTTATGATAAAACTAAGGGTATTTTAG |
| ERV46L | GGGCACTGTCATGGACAAGCTATAGTAAGCTTCACAGAGATCGATCTGGGGCAAG |
| ERV41-SpeI | CGTGGATTTTCACACTACTGGATATGGCACTAGTCACCATCATGGGACCTAAG |
| ERV46-SpeI | GGGCACTGTCATGGACAAACTAGTCTACAAAGCACAGAGATCGATCTGGGGCAAG |
| ERV46-ILDR | CAGAGATCGATCTGGGGCATTTTAGATAGGTAGAGGAAGAGACTGTCATAG |
| ERV41-KKSQ | GCCAGATGATAAAACTAAGGGTAAGAAGAGCCAGTAGATATGTAAAAGAAA |
| ERV46-AA | GGGCACTGTCATGGACAAGCTAGCAGCCAAAGCACAGAGATCGATCTGGGGC |
| Y340A | GGGACCTAAGTGGTCCTTGCGTGCTCAGCCAGATGATAAAACTAAGGGTATTTTAG |
| Q341A | GGGACCTAAGTGGTCCTTGCGTTATGCGCCAGATGATAAAACTAAGGGTATTTTAG |
| P342A | CCTAAGTGGTCCTTGCGTTATCAGGCAGATGATAAAACTAAGGGTATTTTAGATAGG |
| D343A | CCTAAGTGGTCCTTGCGTTATCAGCCAGCTGATAAAACTAAGGGTATTTTAGATAGG |
| D344A | GGTCCTTGCGTTATCAGCCAGATGCTAAAACTAAGGGTATTTTAGATAGGTAG |
| K345A | GGTCCTTGCGTTATCAGCCAGATGATGCAACTAAGGGTATTTTAGATAGGTAG |
| T346A | GGTCCTTGCGTTATCAGCCAGATGATAAAGCTAAGGGTATTTTAGATAGGTAG |
| K347A | GCGTTATCAGCCAGATGATAAAACTGCGGGTATTTTAGATAGGTAGATATGTAAAAG |
| G348A | GCGTTATCAGCCAGATGATAAAACTAAGGCTATTTTAGATAGGTAGATATGTAAAAG |
| I349A | CAGCCAGATGATAAAACTAAGGGTGCTTTAGATAGGTAGATATGTAAAAGAAAAGAG |
| L350A | CAGCCAGATGATAAAACTAAGGGTATTGCAGATAGGTAGATATGTAAAAGAAAAGAG |
| D351A | GATGATAAAACTAAGGGTATTTTAGCTAGGTAGATATGTAAAAGAAAAGAGAATGCAG |
| R352A | GATGATAAAACTAAGGGTATTTTAGATGCGTAGATATGTAAAAGAAAAGAGAATGCAG |
To construct the plasmids with exchanged tail sequences, singular SpeI sites were introduced into pRS314-ERV41 with primer ERV41-SpeI and into pRS316-ERV46 with primer ERV46-SpeI, changing Ile329 of Erv41p to valine and Phe402 of Erv46p to valine. The plasmids were then digested with SpeI and BamHI, and the ERV41 tail fragment was ligated in-frame into the ERV46 plasmid and vice versa to yield plasmids pRS314-ERV41-46Tail and pRS316-ERV46-41Tail. Plasmids pRS314-ERV41-KKSQ, pRS316-ERV46-ILDR and pRS316-ERV46-AA were generated by site-directed mutagenesis using mutagenic primers ERV41-KKSQ, ERV46-ILDR and ERV46-AA, respectively.
Yeast strains and media
Strains used in this study are listed in Table II. Cultures were grown at 30°C in rich medium (YPD, 1% Bacto yeast extract, 2% Bacto peptone, 2% dextrose) or in minimal medium (YMD, 0.67% yeast nitrogen base without amino acids, 2% dextrose, appropriate amino acid supplements) to maintain selective pressure on strains carrying plasmids. Standard yeast (Sherman, 1991) and cloning methods (Ausubel et al., 1987) were used.
Table II. Strain list.
| Strain | Genotype | Reference |
|---|---|---|
| FY833 | MATa his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63 | Winston et al. (1995) |
| FY834 | MATα his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63 | Winston et al. (1995) |
| CBY770 | FY834 with ERV46::HIS3MX6-PGAL1-3HA | Otte et al. (2001) |
| CBY783 | FY834 with ERV41::TRP1-PGAL1-3HA | Otte et al. (2001) |
| CBY797 | FY834 with erv41::HIS3 | Otte et al. (2001) |
| CBY795 | FY834 with erv41::HIS3 erv46::KAN | Otte et al. (2001) |
| CBY1036 | CBY797 with pRS314 | This study |
| CBY1037 | CBY797 with pRS314-ERV41 | This study |
| CBY1038 | CBY797 with pRS314-ERV41-ΔC | This study |
| CBY1074 | CBY797 with pRS314-ERV41-Y340A | This study |
| CBY1075 | CBY797 with pRS314-ERV41-P342A | This study |
| CBY1076 | CBY797 with pRS314-ERV41-D343A | This study |
| CBY1077 | CBY797 with pRS314-ERV41-T346A | This study |
| CBY1078 | CBY797 with pRS314-ERV41-K347A | This study |
| CBY1080 | CBY797 with pRS314-ERV41-G348A | This study |
| CBY1089 | CBY797 with pRS314-ERV41-D344A | This study |
| CBY1090 | CBY797 with pRS314-ERV41-L350A | This study |
| CBY1091 | CBY797 with pRS314-ERV41-D351A | This study |
| CBY1092 | CBY797 with pRS314-ERV41-R352A | This study |
| CBY1096 | CBY797 with pRS314-ERV41-Q341A | This study |
| CBY1158 | CBY797 with pRS314-ERV41-K345A | This study |
| CBY1159 | CBY797 with pRS314-ERV41-I349A | This study |
| CBY1198 | CBY795 with pRS314-ERV41, pRS316-ERV46 | This study |
| CBY1199 | CBY795 with pRS314-ERV41-ΔC, pRS316-ERV46 | This study |
| CBY1200 | CBY795 with pRS314-ERV41, pRS316-ERV46-ΔC | This study |
| CBY1204 | CBY795 with pRS314-ERV41-46Tail, pRS316-ERV46-41Tail | This study |
| CBY1305 | CBY795 with pRS314-ERV41, pRS316-ERV46-ILDR | This study |
| CBY1306 | CBY795 with pRS314-ERV41-KKSQ, pRS316-ERV46-ILDR | This study |
| CBY1307 | CBY795 with pRS314-ERV41-46Tail, pRS316-ERV46-ILDR | This study |
| CBY1308 | CBY795 with pRS314-ERV41-KKSQ, pRS316-ERV46-41Tail | This study |
| CBY1334 | CBY795 with pRS314-ERV41, pRS316-ERV46-AA | This study |
Subcellular fractionation
Membrane fractions enriched in ER (13 000 g pellets) and Golgi (100 000 g pellets) were prepared as described (Belden and Barlowe, 2001b), except that pellets were resuspended in 200 µl of 2× sample buffer. Microsomes (Wuestehube and Schekman, 1992) and semi-intact cells (Baker et al., 1988) were prepared as described. Organelles were resolved on sucrose gradients according to Powers and Barlowe (1998).
Protease protection assay
Microsomes (∼45 µg) were mixed with either buffer 88 (20 mM HEPES pH 7.0, 250 mM sorbitol, 150 mM potassium acetate, 5 mM magnesium acetate), buffer 88 with proteinase K (final concentration 0.3 mg/ml), buffer 88 with Triton X-100 (final concentration 1%) or buffer 88 with both protease and detergent. After 30 min on ice, reactions were stopped by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), mixed with sample buffer and resolved on a polyacrylamide gel.
In vitro vesicle budding and COPII binding assays
Analytical scale in vitro budding reactions with recombinant COPII subunits were performed as described previously (Barlowe et al., 1994). In the alanine scan experiment, packaging efficiencies for Erv41p and Erv46p were divided by the Erv25p packaging efficiency to give relative packaging levels for Erv41p and Erv46p in all strains. COPII binding assays were performed as reported by Kuehn et al. (1998) with the following modification: if assays included conditions with Sec13/31p, all binding reactions were centrifuged for 15 min at 100 000 g through 0.5 ml of 0.1 M sucrose to sediment both membranes and vesicles.
Antibodies and immunoblotting
Polyclonal antibodies raised against fragments of the lumenal domains of Erv41p and Erv46p as well as against Och1p have been described (Otte et al., 2001). Antibodies directed against Erv25p, Kar2p, plasma membrane ATPase (PMA1) and Sec61p (Powers and Barlowe, 1998) as well as Bos1p (Cao and Barlowe, 2000) were published earlier. A monoclonal anti-HA antibody was obtained from Berkeley Antibody Co. (Richmond, CA). Western blots were developed using the enhanced chemiluminescence (ECL) method (Amersham, Arlington Heights, IL). For densitometric analysis, films were scanned and plotted using NIH Image 1.52.
Acknowledgments
Acknowledgements
This work was supported by grants from the National Institute of General Medical Sciences. S.O. was supported by a fellowship from Deutsche Forschungsgemeinschaft.
References
- Antonny B. and Schekman,R. (2001) ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol., 13, 438–443. [DOI] [PubMed] [Google Scholar]
- Aridor M., Weissmann,J., Bannykh,S., Nuoffer,C. and Balch,W.E. (1998) Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol., 141, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel R.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, NY, pp. 3.0.1–3.14.3.
- Baker D., Hicke,L., Rexach,M., Schleyer,M. and Schekman,R. (1988) Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell, 54, 335–344. [DOI] [PubMed] [Google Scholar]
- Barlowe C. et al. (1994) COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the ER. Cell, 77, 895–907. [DOI] [PubMed] [Google Scholar]
- Belden W.J. and Barlowe,C. (1996) Erv25p, a component of COPII-coated vessicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem., 271, 26939–26946. [DOI] [PubMed] [Google Scholar]
- Belden W.J. and Barlowe,C. (2001a) Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J. Biol. Chem., 276, 43040–43048. [DOI] [PubMed] [Google Scholar]
- Belden W.J. and Barlowe,C. (2001b) Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell, 12, 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Corpina,R.A. and Goldberg,J. (2002) Structure of the Sec23/24–Sar1 pre-budding complex of the COPII vesicle coat. Nature, 419, 271–277. [DOI] [PubMed] [Google Scholar]
- Campbell J. and Schekman,R. (1997) Selective packaging of cargo molecules into endoplasmic reticulum-derived COPII vesicles. Proc. Natl Acad. Sci. USA, 94, 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. and Barlowe,C. (2000) Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with acceptor membranes. J. Cell Biol., 149, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P. and Letourneur,F. (1994) Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science, 263, 1629–1631. [DOI] [PubMed] [Google Scholar]
- Cosson P. and Letourneur,F. (1997) Coatomer (COPI)-coated vesicles: role in intracellular transport and protein sorting. Curr. Opin. Cell Biol., 9, 484–487. [DOI] [PubMed] [Google Scholar]
- de Silva A.M., Balch,W.E. and Helenius,A. (1990) Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J. Cell Biol., 111, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M., Dejgaard,K., Fullekrug,J., Dahan,S., Fazel,A., Paccaud,J.P., Thomas,D.Y., Bergeron,J.J. and Nilsson,T. (1998) gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COPI and II coatomer. J. Cell Biol., 140, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R.W., Ruusala,A., Machamer,C., Helenius,J., Helenius,A. and Rose,J.K. (1988) Differential effects of mutations in three domains on folding, quaternary structure and intracellular transport of vesicular stomatitis virus G protein. J. Cell Biol., 107, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Molinari,M. and Helenius,A. (1999) Setting the standards: quality control in the secretory pathway. Science, 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Kappeler F., Klopfenstein,D.R.C., Foguet,M., Paccaud,J.-P. and Hauri,H.-P. (1997) The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J. Biol. Chem., 272, 31801–31808. [DOI] [PubMed] [Google Scholar]
- Kuehn M., Herrmann,J.M. and Schekman,R. (1998) COPII–cargo interactions direct protein sorting into ER-derived transport vesicles. Nature, 391, 187–190. [DOI] [PubMed] [Google Scholar]
- Lanoix J., Ouwendijk,J., Lin,C.C., Stark,A., Love,H.D., Ostermann,J. and Nilsson,T. (1999) GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COPI vesicles. EMBO J., 18, 4935–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J., Ouwendijk,J., Stark,A., Szafer,E., Cassel,D., Dejgaard,K., Weiss,M. and Nilsson,T. (2001) Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J. Cell Biol., 155, 1199–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer G.Z., Cheng,Y., Petre,B.M., Vogan,E., Springer,S., Schekman,R., Walz,T. and Kirchhausen,T. (2001) Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl Acad. Sci. USA, 98, 10704–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Zerange,N., Lin,Y.F., Collins,A., Yu,M., Jan,Y.M. and Jan,L.Y. (2001) Role of ER export signals in controlling surface potassium channel numbers. Science, 291, 316–319. [DOI] [PubMed] [Google Scholar]
- Marzioch M., Henthorn,D.C., Herrmann,J.M., Wilson,R., Thomas,D.Y., Bergeron,J.J., Solari,R.C. and Rowley,A. (1999) Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell, 10, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Schekman,R., Orci,L. and Heuser,J.E. (2001) Surface structure of the COPII-coated vesicle. Proc. Natl Acad. Sci. USA, 98, 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I. and Warren,G. (2000) The road taken: past and future foundations of membrane traffic. Cell, 100, 99–112. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Yamazaki,S., Sato,K., Nakano,A., Sakaguchi,M. and Mihara,K. (1998) Identification of potential regulatory elements for the transport of Emp24p. Mol. Biol. Cell, 9, 3493–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N. and Balch,W.E. (1997) A di-acidic signal required for selective export from the endoplasmic reticulum. Science, 277, 556–558. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Bannykh,S., Slabough,S., Matteson,J., Altschuler,Y., Hahn,K. and Balch,W.E. (1999) A di-acidic (DXE) code directs concentration of cargo during export from the endoplasmic reticulum. J. Biol. Chem., 274, 15937–15946. [DOI] [PubMed] [Google Scholar]
- Nufer O., Guldbrandsen,S., Degen,M., Kappeler,F., Paccaud,J.-P., Tani,K. and Hauri,H.-P. (2002) Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J. Cell Sci., 115, 619–628. [DOI] [PubMed] [Google Scholar]
- Otte S., Belden,W.J., Heidtman,M., Liu,J., Jensen,O.N. and Barlowe,C. (2001) Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J. Cell Biol., 152, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C. and Walter,P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol., 13, 349–355. [DOI] [PubMed] [Google Scholar]
- Powers J. and Barlowe,C. (1998) Transport of Axl2p depends on Erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J. Cell Biol., 142, 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J. and Barlowe,C. (2002) Erv14p directs a transmembrane secretory protein into COPII coated vesicles. Mol. Biol. Cell, 13, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.S. and Bonifacino,J.S. (2001) Adaptor related proteins. Curr. Opin. Cell Biol., 13, 444–453. [DOI] [PubMed] [Google Scholar]
- Sato K. and Nakano,A. (2002) Emp47p and its close homolog Emp46p have a tyrosine-containing endoplasmic reticulum exit signal and function in glycoprotein secretion in Saccharomyces cerevisiae.Mol. Biol. Cell, 13, 2518–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier C.S., Weisz,O.A., Davis,M. and Machamer,C.E. (2000) Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell, 11, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–20. [DOI] [PubMed] [Google Scholar]
- Shimoni Y., Kurihara,T., Ravazzola,M., Amherdt,M., Orci,L. and Schekman,R. (2000) Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J. Cell Biol., 151, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae.Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S. and Schekman,R. (1998) Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science, 281, 698–700. [DOI] [PubMed] [Google Scholar]
- Stephens D.J. and Pepperkok,R. (2002) Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci., 115, 1149–1160. [DOI] [PubMed] [Google Scholar]
- Votsmeier C. and Gallwitz,D. (2001) An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J., 20, 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard,C. and Ricupero-Hovasse,L.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Wuestehube L.J. and Schekman,R. (1992) Reconstitution of transport from the endoplasmic reticulum to the Golgi complex using an ER-enriched membrane fraction from yeast. Methods Enzymol., 219, 124–136. [DOI] [PubMed] [Google Scholar]