Abstract
We have estimated phylogenies of fungus-growing termites and their associated mutualistic fungi of the genus Termitomyces using Bayesian analyses of DNA sequences. Our study shows that the symbiosis has a single African origin and that secondary domestication of other fungi or reversal of mutualistic fungi to a free-living state has not occurred. Host switching has been frequent, especially at the lower taxonomic levels, and nests of single termite species can have different symbionts. Data are consistent with horizontal transmission of fungal symbionts in both the ancestral state of the mutualism and most of the extant taxa. Clonal vertical transmission of fungi, previously shown to be common in the genus Microtermes (via females) and in the species Macrotermes bellicosus (via males) [Johnson, R. A., Thomas, R. J., Wood, T. G. & Swift, M. J. (1981) J. Nat. Hist. 15, 751–756], is derived with two independent origins. Despite repeated host switching, statistical tests taking phylogenetic uncertainty into account show a significant congruence between the termite and fungal phylogenies, because mutualistic interactions at higher taxonomic levels show considerable specificity. We identify common characteristics of fungus-farming evolution in termites and ants, which apply despite the major differences between these two insect agricultural systems. We hypothesize that biparental colony founding may have constrained the evolution of vertical symbiont transmission in termites but not in ants where males die after mating.
Colonies of fungus-growing macrotermitine termites and attine ants are among the most impressive animal phenomena in the world. They can have nest volumes of thousands of liters, may persist for decades, and contain millions of sterile helper individuals, which are normally the offspring of a single queen (1, 2). The agricultural symbiosis with fungi has allowed both the ants and the termites to occupy previously inaccessible niches that have abundant resources (3). The phylogenetically most-derived genera of the attine ants have become dominant herbivores of the New World tropics (3). Analogously, the fungus-growing termites have become major decomposers of the Old World tropics (4) and form perhaps the most complex colony and mound structures of any invertebrate group. The two main symbioses of social insects with fungi are similar in many respects, but they differ in others. The fungal symbionts of the attine ants rarely fruit and are normally propagated clonally and vertically by dispersing queens (5, 6). In contrast, the symbionts of the Macrotermitinae often produce sexual fruiting bodies such that horizontal acquisition of symbionts has been inferred to be the rule, although exceptions do occur (7–9). Recent studies have broadened our understanding of the evolution of the symbiosis between the attine ants and their fungi considerably (5, 6, 10–13), but similar large-scale studies of the Macrotermitinae and their Termitomyces symbionts have been lacking.
Symbiotic relationships have had an essential role in termite evolution and involve a range of intestinal microorganisms including protists, methanogenic Archaea, and bacteria (14). However, only a single Termitidae subfamily, the Macrotermitinae, has evolved a mutualistic ectosymbiosis with fungi of the genus Termitomyces [tribe Termitomyceteae (Jülich) Singer, family Tricholomataceae Roze, Basidiomycotina]. The fungus helps the termites to degrade the plant-derived material (e.g., wood, dry grass, and leaf litter) on which they live (8). It grows on a special structure in the nest, the fungus comb, maintained by the termites through continuous addition of predigested plant substrate while the older comb material is consumed (15).
The Macrotermitinae have been divided into 11 taxonomically well supported genera and ≈330 species (ref. 16; see the supporting information, which is published on the PNAS web site, www.pnas.org). Most of the diversity occurs in Africa, where 10 of the 11 genera are found. Five genera occur in Asia (one of these exclusively) and two genera in Madagascar (16). Approximately 40 species of the Termitomyces symbiont have been described (17). Although additional varieties have been recognized, the low number of fungal species suggests that many of these fungi are shared by different termite species unless morphospecies frequently consist of several sibling species.
In this paper we present phylogenies of both the fungus-growing termites and their associated fungal mutualists, and we use these phylogenies to address a number of key questions. First, we determine whether the fungus-growing termites are a monophyletic group, i.e., whether there is a unique origin of fungus farming within the termites. Second, we establish whether the cultivated fungi are monophyletic, i.e., whether there is a single origin of symbiosis with termites within the basidiomycetes or whether secondary domestications or reversals to a free-living state have occurred. Third, we use the phylogenies to infer whether the geographic origin of the symbiosis is Asian or African, and we analyze patterns of cospeciation and interaction specificity across the clades. Fourth, we use the termite phylogeny to reconstruct the evolution of symbiont transmission, and we determine whether alternative modes of symbiont transmission have affected cospeciation. Finally, we compare the overall pattern of coevolution of the fungus-growing termites and fungus-growing ants with their respective fungal mutualists.
Materials and Methods
Technical details and results of additional analyses can be found in the supporting information.
Taxon Sampling.
We sampled fungus-growing termites from three African (Cameroon, Senegal, and Gabon) and three Asian [Sumatra, Kalimantan (Indonesia), and Sri Lanka] locations spanning most of the distribution of the Macrotermitinae. Our total collection included 38 colonies belonging to 32 termite species, covering 9 of the 11 genera. Both termites and symbiotic fungi from the same nest were analyzed. In addition, for 10 species we analyzed the fungi of multiple nests to look for intraspecific variation in symbionts. For information on the taxonomic affiliation of the Termitomyces symbionts we also obtained DNA sequences from herbarium specimens of seven well described species. We developed specific primers for two fungal gene fragments so that we could amplify fungal sequences from termite guts (see below). The use of termite gut contents as a source of fungal sequences allowed us to definitively match fungi with their hosts. It also made sampling more effective, because nests did not necessarily need to be opened to get access to comb material.
The following species of non-fungus-growing Termitidae were included in an initial analysis to test the monophyly of the fungus-growing termites and to find the sister group: Amalotermes phaeocephalus, Astalotermes quietus (Apicotermitinae), Foraminitermes valens, Labritermes butelreepeni, Amitermes evuncifer, Microcerotermes parvus, Cubitermes sp. (Termitinae), and Nasutitermes latifrons (Nasutitermitinae). One species belonging to the sister group of the Termitidae (Coptotermes sjostedti, Rhinotermitidae) was included to root the trees. Sphaerotermes sphaerothorax was included as well, because this species has been placed in the Macrotermitinae by some authors, although it does not cultivate fungi (16).
The outgroups for the Termitomyces fungi were based on refs. 18 and 19 (published in GenBank before this study). We used Lyophyllum semitale as the first outgroup and included Lyophyllum atratum and Tephrocybe rancida to test the monophyly of Termitomyces.
General Information on Phylogenetic Analysis.
We used Bayesian techniques [with the program MRBAYES, versions 2.01 and 3.0* (20)] to estimate the phylogenetic histories of the two interacting mutualists. Posterior probabilities were calculated by using a Metropolis-coupled Markow chain Monte Carlo approach with sampling according to the Metropolis–Hastings algorithm (20). All our analyses used four chains, one cold and three incrementally heated, where the heat of the ith chain is B = 1/[1 + (i − 1)T] and T = 0.2. Starting trees for each chain were random and used default starting values of MRBAYES 2.01. A single run consisted of 1.5 million generations that were sampled every 50th tree. Likelihood values reached a stable value after 10,000–30,000 generations. To assure that we included only trees after the chain had reached a stable (“burnin”) value, we fixed the burnin for all analyses at 100,000 generations, which produced 28,000 sampled trees and corresponding posterior probability distributions in every analysis. We present the results of one such Bayesian analysis in the form of a majority-rule consensus tree of all the trees sampled in the analysis. We also performed maximum-likelihood and maximum-parsimony analyses to check for consistency with the Bayesian results.
Termite DNA Sequences and Phylogenetic Analyses.
For the termites, a 937-bp region of cytochrome oxidase subunit 1 corresponding to positions 1,890–2,826 of the Apis mellifera ligustica mitochondrial genome (21) was sequenced as two fragments with an overlapping region. For most species we used the primer pairs Bl1834 (5′-tcaacaaatcataaagatattgg-3′) and TH2472 (5′-aataggtgttggtataggat-3′) and TL2350 (5′-ccmctrttygtatgatcagt-3′) and TH2877 (5′-gtrtcrtgtartacratgtc-3′), but for some species we used the alternative primer pairs TL1862 (5′-tacttcgtattcggagcttga-3′) and TH2397 (5′-gttagtagtattgtgattgctcc-3′) and TL2341 (5′-cgaacgaatcccactatttgt-3′) and TH2928 (5′-aatactgctcctatagatag-3′). A PCR (using AmpliTaq gold, Applied Biosystems) consisted of an initial denaturing step of 10 min at 95°C followed by 35 cycles (30 s at 95°C, 30 s at 57°C, and 30 s 72°C) finished by a final extension step at 72°C of 5 min. Sequences were generated on an ABI 3700 automated sequencer (Applied Biosystems) by using BigDye terminator chemistry (Applied Biosystems) and the two PCR primers for each sequence. The two sequences generated were combined by using the overlapping sequence in the computer program SEQUENCHER 3.11 (Gene Codes, Ann Arbor, MI). The final alignment was straightforward, because no insertions/deletions had to be inferred.
Bayesian methods were used to estimate the phylogenies of the fungus-growing termites. An initial analysis including 10 species of non-fungus-growing termites (see above) was performed to test the monophyly of the fungus-growing termites (using the general time-reversible model with site-specific rates). Theoretical and empirical data have shown that including (members of) the sister group as the outgroup is to be preferred over (members of) more distant clades as outgroups (22). Therefore, the data set used in all subsequent analyses consisted of the fungus-growing termites and their sister group as outgroup. However, we did some additional analyses to check how inclusion of other outgroups affected the topology of the trees. We used MODELTEST (23) and parametric bootstrapping (24) to estimate the best-fit model of sequence evolution. The model selected was the general time-reversable model with site-specific rates. Additional maximum-likelihood and maximum-parsimony analyses were performed by using PAUP*4.0b10 (25). For the maximum-likelihood analysis, the model and model parameters found with MODELTEST were used. Maximum-likelihood bootstrap analyses were conducted by using 100 bootstrap replicates generated by CODONBOOTSTRAP (26). CODONBOOTSTRAP takes the protein-coding structure of the data set into account by bootstrapping codons instead of single-nucleotide positions.
Termitomyces DNA Sequences and Phylogenetic Analyses.
Fungal DNA was obtained from comb material, basidiocarps, and termite gut contents. To be able to use gut contents for obtaining fungal sequences, specific primers were developed (D.K.A. and J.J.B., unpublished data). Two sequences were determined for the fungal symbionts: (i) ≈530 bp from the 5′ side (alignment 553 nucleotides) of 25S nuclear RNA gene (nLSU-rDNA) by using the primers 25S4R (5′-acaagtgctgagttcctcag-3′, a specific primer) and ITS4R (5′-gcatatcaataagcggagga-3′, the reverse complement of the universal primer ITS4 (27) (a region of maximally 11 nucleotides was excluded from the analysis, because it could not be aligned unambiguously) and (ii) ≈320 bp of the 12S mitochondrial RNA gene (mtSSU-rDNA) by using the primers ssufw105 (specific for Termitomyces, 5′-tcgcgttagcatcgttactagt-3′) and ssurev475 (specific for some Lyophylleae including Termitomyces, 5′-gccagagacgcgaacgttagtcg-3′) (a region of maximally 28 nucleotides was excluded from the analysis, because it could not be aligned unambiguously). To test the method and look for intracolonial variation in symbionts, we compared fungal sequences of different sources (guts and combs or basidiocarps) for seven nests and obtained identical sequences in all cases.
The two fungal sequences were tested for combinability by using the partition homogeneity test (28) implemented in PAUP*4.0b10 (25), which showed that there was no significant incongruence between the two data sets (1,000 artificial data sets, P = 0.24). Bayesian analyses [using MRBAYES 3.0* (20)] therefore were performed on the combined fungal data set. The two partitions were defined, and for each partition a separate model was used [determined by using MODELTEST (23)]: the GTR + I + Γ (lset nst = 6 rates = invgamma) for the nuclear 25S and the GTR + Γ (lset nst = 6 rates = gamma) for the mitochondrial 12S. We also analyzed the two fungal sequences separately and did some additional analyses on the combined data set by varying the number of outgroups. Additional maximum-likelihood and maximum-parsimony analyses were performed by using PAUP*4.0b10 (25). Maximum-likelihood analyses used the best-fit model of sequence evolution as determined with MODELTEST, Tamura and Nei's model (29) with gamma-distributed rate variation and a proportion of invariable sites (TrN + I + Γ). Nonparametric maximum-likelihood bootstrap analyses were conducted by using 100 bootstrap replicates.
Analysis of Congruence Between Symbiont Trees.
We used COMPONENT (30) to test for congruence between the termite and fungal trees. The procedure used in COMPONENT is “tree reconciliation” (31). Tree reconciliation estimates the number of duplications and losses (extinctions, unsampled taxa) necessary to make the associate tree exactly fit the host tree. COMPONENT quantifies the fit of the host tree to the reconciled associate tree by three different measures: number of duplications, number of terminal nodes added, and number of independent losses (30). We determined these measures for our data set. To account for uncertainty in the estimation of both phylogenies, we analyzed a random sample of the trees visited during a Bayesian analysis. We did a separate Bayesian analysis by using only the termite taxa for which we had recovered both sequences of the fungal symbiont (30 of 38). We calculated the fit between 100 fungal trees and 100 termite trees (randomly chosen from all the trees saved in the Bayesian analysis) in all possible combinations (10,000 comparisons). To determine statistical significance, the observed fit was compared with the fit between 100 random fungal trees and 100 random termite trees.
Results and Discussion
Termite Phylogeny.
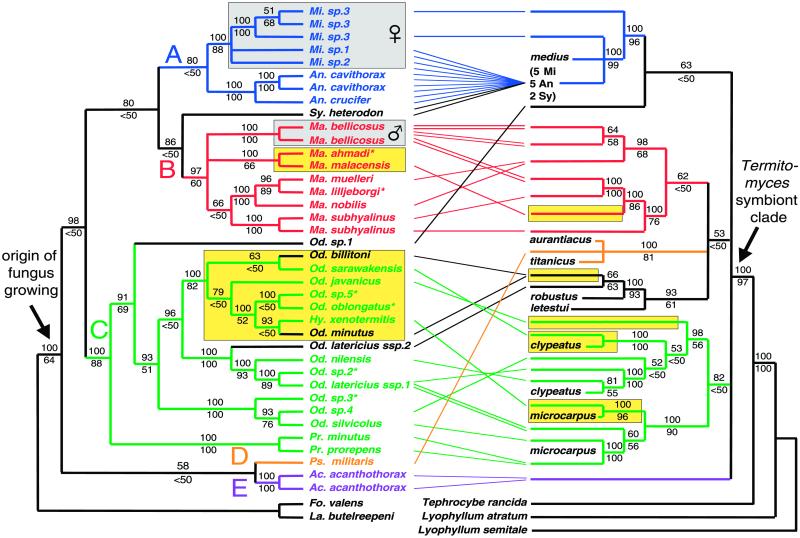
An initial analysis including 10 species of non-fungus-growing termites strongly supports the monophyly of the fungus-growing termites (posterior probability 0.98), which implies that fungus growing has a single origin within the termites. The non-fungus-growing species S. sphaerothorax, placed in the Macrotermitinae by some authors (16), falls outside the fungus-growing Macrotermitinae. It therefore is highly unlikely that S. sphaerothorax has secondarily lost its fungal symbiont as has been suggested previously (32). The sister group of the fungus-growing termites consists of L. butelreepeni and F. valens (posterior probability 0.99). We used this sister group as outgroup in subsequent analyses (Fig. 1). Using this outgroup we obtained ingroup relationships that only slightly differed from the initial analysis. Most noticeably, the genus Microtermes has a basal position in the first analysis, whereas it has a terminal position as the sister group of the genus Ancistrotermes in the second analysis. The latter result is consistent with morphological evidence (33). Three separate Bayesian analyses, using different random starting trees, and additional analyses that varied the number of outgroups resulted in identical trees and the posterior probabilities for individual nodes differed only slightly. Maximum-likelihood analysis produced identical and maximum-parsimony analysis almost identical results, although nonparametric bootstrap support was sometimes lower than Bayesian posterior probabilities (see supporting information for further details on the analyses). This has been found previously (e.g., ref. 34) and is consistent with the suggestion of Hillis and Bull (35) that nonparametric estimates of bootstrap support are too conservative. All polytypic genera form well supported monophyletic groups with the exception of Odontotermes, because the Asiatic genus Hypotermes is derived from it. All basal groups in the termite cladogram are African, whereas the Asiatic species form two terminal clades, one within Macrotermes and one within Odontotermes. Our analysis thus documents at least two colonizations of Asia from Africa, but the true figure of independent colonizations is likely to be at least four, because species of Ancistrotermes and Microtermes also occur in Asia (16) but were not sampled from those locations for the present study.
Fig 1.
Majority-rule consensus trees of fungus-growing termites (Left) and their fungal symbionts (Right) of trees sampled in a Bayesian analysis. The numbers above the branches refer to the Bayesian posterior probability of the nodes (>50%) and were derived from 28,000 Markow chain Monte Carlo-sampled trees. Three independent Markow chain Monte Carlo runs, each starting with random trees for each of four simultaneous chains, resulted in concordant joint posterior probability distributions for the topologies. Alternative estimations based on maximum-likelihood were almost identical. The values below branches represent percent support in maximum-likelihood nonparametric bootstrapping. Termites and their corresponding fungal symbionts are connected by thin lines. The five major termite clades, labeled A–E, and their corresponding fungal symbionts are indicated with different colors to illustrate the degree of specificity. Exceptions to this pattern of higher-level specificity are indicated in black. The two termite clades with vertical uniparental symbiont transmission [by different sexes as indicated (7, 8)] are contained in gray boxes, whereas yellow boxes indicate the two termite clades and five fungal clades from Asia. Termites for which we were not able to obtain one or both fungal DNA sequences are indicated with an asterisk. Sequences derived from herbarium basidiocarps of seven fungal morphospecies were included to link a number of termite symbionts with known strains of Termitomyces. Mi, Microtermes; An, Ancistrotermes; Sy, Synacanthotermes; Ma, Macrotermes; Od, Odontotermes; Hy, Hypotermes; Pr, Protermes; Ps, Pseudacanthotermes; Ac, Acanthotermes; Fo, Foraminitermes; and La, Labritermes.
Termitomyces Phylogeny.
The fungi cultivated by the fungus-growing termites also form a monophyletic group together with seven described morphospecies of the genus Termitomyces. The fruiting bodies of Termitomyces are always associated with termite nests (8, 15, 32, 33), which implies that within the basidiomycetes there is a single evolutionary origin of mutualistic symbiosis with termites and that there are no known reversals to a nonsymbiotic lifestyle. These results are consistent with earlier findings (18, 19, 33) based on much less-complete data sets. Additional analyses varying the number of outgroups and using maximum likelihood and maximum parsimony resulted in similar topologies, although the basal relationships between the main clades of fungal symbionts were never strongly supported. The position of the Asiatic Termitomyces symbionts in the fungal phylogeny indicates at least five migrations between Africa and Asia [three equally most parsimonious reconstructions: (i) three times Africa → Asia and twice Asia → Africa, (ii) four times Africa → Asia and once Asia → Africa, and (iii) five times Africa → Asia]. This higher figure, compared with the two inferred intercontinental migrations of the termites, suggests that fungal symbionts can disperse independently of their hosts. This is confirmed by the phylogenetic position of the Asiatic and African samples of Termitomyces microcarpus and Termitomyces clypeatus, which belong to monophyletic groups of exclusively Asiatic and African termite symbionts, respectively.
Patterns of Cospeciation and Specificity.
All three measures of fit between the termite and fungal phylogeny estimates were significant [number of duplications, number of terminal nodes added, and number of independent losses; all P < 0.0001 (30, 31)]. Therefore, the termite and fungal trees are more similar than would be expected by chance under a Markov branching model. Specificity of fungal symbionts occurs mainly at the higher taxonomic levels. We can recognize five main clades of termites (called here groups A–E). These are broadly associated with particular clades of fungal symbionts, although there are some exceptions (Fig. 1). For example, the basal species of clade B (Synacanthotermes heterodon) rears fungi that are otherwise associated with clade A, and four termite species of clade C have symbionts that do not belong to the main clade of fungi associated with clade C.
Within the five recognized clades of termites there is no strong association between the evolutionary history of the termites and that of their symbionts. Therefore, termites within the recognized clades must have repeatedly exchanged their fungal symbionts. Single termite species have a variety of symbionts, and the symbionts of the species Macrotermes bellicosus, Macrotermes subhyalinus, and Odontotermes latericius do not form monophyletic groups. This finding is in line with recently found intraspecific variation in the Asiatic species Odontotermes formosanus (7). The different samples of the species in our study were collected from one (M. bellicosus) or two (the other two species) localities in Cameroon, and thus the variation of symbionts within these species is unlikely to be due to geographical variation. Switching to a fungal perspective, we found that several group A fungi with identical sequences are found across a range of termite species (Fig. 1). In addition, Odontotermes bilitoni and Odontotermes minutus are not sister groups but have symbionts that are 100% identical.
Evolution of Transmission Modes.
The patterns of cospeciation and specificity are consistent with transmission of fungal symbionts from host to host outside the vertical host lineage [horizontal transmission (36)]. Horizontal symbiont transmission has also been shown experimentally to be the norm in several species of Macrotermitinae and is consistent with the frequent formation of sexual fruiting bodies in many fungal symbionts (8, 9). However, clonal uniparental transmission (vertical transmission) of fungal symbionts has been experimentally documented in the genus Microtermes (all five species studied) and the species M. bellicosus. In line with this observation, sexual fruiting bodies have never been found in these species (8, 9). For Microtermes it is the female that transmits the fungus, whereas it is the male in M. bellicosus (8, 9). This difference in sex specificity is consistent with our present analysis, showing that the termites with vertical transmission of fungal symbionts do not form a monophyletic group (see Fig. 1; posterior probability that they form a monophyletic group, <0.00004). This pattern indicates that horizontal transmission is the ancestral transmission mode, and uniparental clonal transmission is a derived trait with two independent origins.
Interestingly, neither the fungi of the genus Microtermes nor the fungi of M. bellicosus form a monophyletic group (Fig. 1), suggesting that some horizontal transmission also occurs for those groups. The Microtermes species included in this study all share a fungus (identical in two DNA sequences) with species of Ancistrotermes and Synacanthotermes, which indicates that symbionts have been exchanged recently between these divergent termite lineages. The six nests of M. bellicosus included in this study have fungi that fall into two strongly supported clades, but these clades together do not form a monophyletic group (see Fig. 1; posterior probability that the fungi of M. bellicosus form a monophyletic group, <0.00004). Two individuals of M. bellicosus from nests with fungi belonging to the two different clades were included in the termite phylogeny and shown to form a monophyletic group, in contrast to the fungi that they cultivate (Fig. 1).
Comparison Between the Fungus-Growing Termites and Fungus-Growing Ants.
The results presented allow a comparison between the convergent evolution of obligate fungus growing in two different groups of social insects. The double monophyly of the partner taxa implies a single origin of the fungus-farming mutualism in termites without secondary reversals to a free-living state. For the fungal symbionts, this contrasts markedly with the polyphyletic origin of the extant fungi grown by the fungus-growing ants, where support for a minimum of four independent domestications has been found (10, 11). At least some of the fungi reared by the lower attine ants have close free-living relatives, indicating that the ants continue to recruit symbionts from nonsymbiotic, sexually reproducing fungi (5, 11). Only the higher attine ants live in symbiosis with a monophyletic group of highly specialized fungi of which free-living relatives have not been found, similar to the fungal symbionts of termites (5, 10). A second major difference is the predominant mode of symbiont transmission as discussed above. Vertical transmission of Termitomyces fungi is restricted to two apical termite clades (Fig. 1) so that horizontal symbiont transmission is the ancestral state within this symbiosis. In contrast, vertical transmission of fungal symbionts occurs in all attine lineages and may even have preceded the origin of fungicultural behavior in the ants (5).
Despite the seemingly fundamentally different ways of controlling fungus transmission in the two types of insect societies, the degree of specificity of fungal symbionts found in this study is similar to the specificity observed in the attine ants (10, 11). In both symbioses, groups of fungi can be defined that are generally associated with specific host clades, but within these clades the specificity is low. Given the different modes of symbiont transmission, this convergence must have been achieved through different mechanisms. In the attine ants a moderate level of host symbiont specificity can be explained by vertical transmission of symbionts with occasional cultivar exchanges between closely and sometimes distantly related ant species (5, 6). In most of the macrotermitine termites, on the other hand, associations between fungi and termites arise de novo in every generation such that active or passive selection of mutualistic partners must be the primary force inducing specificity. Once irreversibly committed to symbiosis with termites, Termitomyces clades apparently have evolved specific adaptations to the comb substrates built by the specific clades of termites (15), and the termites may have evolved active selection of specific symbiont clades. However, both niche specialization by the fungi and symbiont selection by the termites will be constrained by availability of comb substrates and fungal strains in the environment, and thus less-specific associations are likely to arise frequently.
The niche-specialization scenario for the Macrotermitinae–Termitomyces symbiosis implies that many of the evolutionary modifications in this mutualism may be as much “fungus-driven” as “insect-driven.” The essentially “symmetrical” symbiosis with Termitomyces fungi allowed termites to occupy new food niches, and these diverging niches selected for fungal adaptations to combs built from different plant-derived materials. The roles of the fungal symbionts for termite nutrition therefore may differ across genera, as present data indicate (15). In contrast, the symbiosis between attine ants and fungi remained “asymmetrical” in most of the basal lineages, i.e., ants were obligatory dependent on fungal symbionts that were not necessarily obligatory dependent on them. The nutritious role of the fungal symbionts therefore changed rather little until the symbiosis became symmetrical in the higher attine ants, a clade characterized by specifically adapted fungi producing unique nutritious structures (gongylidia) for the ant brood (10). In other words, the attine ants primarily evolved specific adaptations to be farmers of rather unspecified fungal crops, and their fungi realized crucial adaptations only in the apical clade of the higher attine ants. The obligate symmetrical interactions that followed allowed the symbiosis to become highly specialized and ultimately produced the leafcutter ants. The Macrotermitinae, on the other hand, specialized on a single group of fungi, which quickly became genetically isolated from its free-living sister group and cospeciated and cospecialized in response to the increasing diversity of fungus–comb substrates across termite species and habitats.
Transmission Modes and the Evolution of Mutualism.
Our results imply that strict vertical transmission is not a necessary condition for highly interdependent obligate mutualism despite obvious advantages for hosts in preventing competition between unrelated coexisting strains of symbionts in a single nest (36, 37). A crucial and as-yet-unanswered question is to what extent the Macrotermitinae manage to reduce the genetic diversity of horizontally acquired symbionts to a single strain, preventing the evolution and expression of noncooperative symbiont traits and achieving a symbiosis that is structurally similar to what the attine ants realize through vertical transmission. We obtained identical fungal sequences in multiple samples from four nests of species that have horizontal symbiont transmission (see also ref. 7), which strongly suggests that genetic screening of Termitomyces strain diversity happens in at least some of the genera either directly through active selection of symbionts or indirectly through interstrain competition for comb space. However, the fact that vertical transmission evolved twice in terminal clades of the Macrotermitinae indicates that this transmission mode incurs advantages for maintaining specialized agricultural symbioses even when the symbionts have close relatives that regularly fruit and are propagated horizontally when associated with other Macrotermitinae. We hypothesize that the evolution of vertical symbiont transmission in termites has been constrained, because it produces single-strain fungus gardens only when restricted to a single termite sex. This is realized by default in ants where males do not survive beyond mating but not in the termites where colonies are founded by a female and male together.
Supplementary Material
Acknowledgments
Bo Vest-Pedersen, Sylvia Mathiasen, Pernille Selmer-Olsen, Jakob Damgaard, Pia Friis, Peter Arctander, and David Nash gave valuable technical advice, and Stefan Hauser, Karen Machielsen, Neree Unguene, Michel Diouf, Mahfouss Sarr, Luc Dibog, Thom Kuyper, Birgith Pedersen, and Philippe Delabarre assisted in the field. David Bignell's coordinating suggestions were crucial for getting this project started. Ulrich Mueller, Hans Siegismund, an anonymous referee, and Ted Mes commented on the manuscript, Thomas Læssøe gave expert advise on Termitomyces taxonomy, and Bart Buyck and Tiina Saarimäki provided herbarium material of Termitomyces. This research has been supported by an European Union Marie Curie Fellowship HPMF-CT-2000-00642 (to D.K.A.).
References
- 1.Hölldobler B. & Wilson, E. O., (1990) The Ants (Belknap, Cambridge, MA).
- 2.Shellman-Reeve J. S. (1997) in Social Competition and Cooperation in Insects and Arachnids, eds. Choe, J. C. & Crespi, B. J. (Cambridge Univ. Press, Cambridge, U.K.), pp. 52–93.
- 3.Waller D. A. (1988) in Advances in Myrmecology, ed. Trager, J. C. (E. J. Brill, Leiden, U.K.), pp. 337–345.
- 4.Bignell D. & Eggleton, P. (2000) in Termites: Evolution, Sociality, Symbioses, Ecology, eds. Abe, T., Bignell, D. E. & Higashi, M. (Kluwer Academic, Dordrecht, The Netherlands), pp. 363–388.
- 5.Mueller U. G., Schultz, T. R., Currie, C. R., Adams, R. M. M. & Malloch, D. (2001) Q. Rev. Biol. 76, 169-197. [DOI] [PubMed] [Google Scholar]
- 6.Green A. M., Mueller, U. G. & Adams, R. M. M. (2002) Mol. Ecol. 11, 191-195. [DOI] [PubMed] [Google Scholar]
- 7.Katoh H., Miura, T., Maekawi, K., Shinzato, N. & Matsumoto, T. (2002) Mol. Ecol. 11, 1565-1572. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R. A., Thomas, R. J., Wood, T. G. & Swift, M. J. (1981) J. Nat. Hist. 15, 751-756. [Google Scholar]
- 9.Darlington J. E. C. P. (1994) in Nourishment and Evolution in Insect Societies, eds. Hunt, J. H. & Nalepa, C. A. (Westview, Boulder, CO), pp. 105–130.
- 10.Chapela I. H., Rehner, S. A., Schultz, T. R. & Mueller, U. G. (1994) Science 266, 1691-1694. [DOI] [PubMed] [Google Scholar]
- 11.Mueller U. G., Rehner, S. A. & Schultz, T. R. (1998) Science 281, 2034-2038. [DOI] [PubMed] [Google Scholar]
- 12.Currie C. R., Scott, J. A., Summerbell, R. C. & Malloch, D. (1999) Nature 398, 701-704. [Google Scholar]
- 13.Mueller, U. G. (2002) Am. Nat. 160, in press. [DOI] [PubMed]
- 14.Bignell D. E. (2000) in Termites: Evolution, Sociality, Symbioses, Ecology, eds. Abe, T., Bignell, D. E. & Higashi, M. (Kluwer Academic, Dordrecht, The Netherlands), pp. 189–208.
- 15.Rouland-Lefèvre C. (2000) in Termites: Evolution, Sociality, Symbioses, Ecology, eds. Abe, T., Bignell, D. E. & Higashi, M. (Kluwer Academic, Dordrecht, The Netherlands), pp. 289–306.
- 16.Kambhampati S. & Eggleton, P. (2000) in Termites: Evolution, Sociality, Symbioses, Ecology, eds. Abe, T., Bignell, D. E. & Higashi, M. (Kluwer Academic, Dordrecht, The Netherlands), pp. 1–24.
- 17.Kirk P. M., Cannon, P. F., David, J. C. & Stalpers, J. A., (2001) Ainsworth & Bigby's Dictionary of the Fungi (CAB Int., Wallingford, U.K.).
- 18.Moncalvo J.-M., Lutzoni, F. M., Rehner, S. A., Johnson, J. & Vilgalys, R. (2000) Syst. Biol. 49, 278-305. [DOI] [PubMed] [Google Scholar]
- 19.Moncalvo J. M., Vilgabys, R., Redhead, S. A., Johnson, J. E., James, T. Y., Aime, M. C., Hofstetter, V., Verduin, S. J. W., Larsson, E., Baroni, T. J., et al. (2002) Mol. Phylogenet. Evol. 23, 357-400. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 21.Crozier H. & Crozier, Y. C. (1993) Genetics 13, 97-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith A. B. (1994) Biol. J. Linn. Soc. 51, 279-292. [Google Scholar]
- 23.Possada D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck J. P. & Crandall, K. A. (1997) Annu. Rev. Ecol. Syst. 28, 437-466. [Google Scholar]
- 25.Swofford D. L., (2001) PAUP* (Sinauer, Sunderland, MA), Version 4.0b10.
- 26.Bollback J. P., (2001) CODONBOOTSTRAP (Dept. of Biology, Univ. of Rochester, Rochester, NY), Version 3.0b4.
- 27.White T. J., Bruns, T., Lee, S. & Taylor, J. (1990) in PCR Protocols: A Guide to Methods and Applications, eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J. (Academic, San Diego), pp. 315–322.
- 28.Farris J. S., Kallersjo, M., Kluge, A. G. & Bult, C. (1994) Cladistics 10, 315-319. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K. & Nei, M. (1993) Mol. Biol. Evol. 10, 512-526. [DOI] [PubMed] [Google Scholar]
- 30.Page R. D. M., (1993) COMPONENT (Natural History Museum, London), Version 2.0.
- 31.Page R. D. M. (1990) Cladistics 6, 119-136. [DOI] [PubMed] [Google Scholar]
- 32.Heim R., (1977) Termites et Champignons (Boubée, Paris).
- 33.Rouland-Lefèvre C., Diouf, M. N., Brauman, A. & Neyra, M. (2002) Mol. Phylogenet. Evol. 22, 423-429. [DOI] [PubMed] [Google Scholar]
- 34.Murphy W. J., Eizirik, E., O'Brien, S. J., Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Science 294, 2348-2351. [DOI] [PubMed] [Google Scholar]
- 35.Hillis D. M. & Bull, J. J. (1993) Syst. Biol. 42, 182. [Google Scholar]
- 36.Frank S. A. (1996) Proc. R. Soc. London Ser. B 263, 339-344. [Google Scholar]
- 37.Bot A. N. M., Rehner, S. A. & Boomsma, J. J. (2001) Evolution (Lawrence, Kans.) 55, 1980-1991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.