Abstract
We found that the well-studied nematode Caenorhabditis elegans can use various yeasts, including Cryptococcus laurentii and Cryptococcus kuetzingii, as a sole source of food, producing similar brood sizes compared with growth on its usual laboratory food source Escherichia coli OP50. C. elegans grown on these yeasts had a life span similar to (C. laurentii) or longer than (C. kuetzingii) those fed on E. coli. However, the human pathogenic yeast Cryptococcus neoformans killed C. elegans, and the C. neoformans polysaccharide capsule as well as several C. neoformans genes previously shown to be involved in mammalian virulence were also shown to play a role in C. elegans killing. These included genes associated with signal transduction pathways (GPA1, PKA1, PKR1, and RAS1), laccase production (LAC1), and the α mating type. C. neoformans adenine auxotrophs, which are less virulent in mammals, were also less virulent in C. elegans. These results support the model that mammalian pathogenesis of C. neoformans may be a consequence of adaptations that have evolved during the interaction of C. neoformans with environmental predators such as free-living nematodes and amoebae and suggest that C. elegans can be used as a simple model host in which C. neoformans pathogenesis can be readily studied.
Studies of microbial pathogenesis in nonvertebrate hosts during the past decade have resulted in important insights into the molecular mechanisms of microbial pathogenesis and host defense. It is now apparent that many of the same bacterial virulence factors are involved in pathogenesis in evolutionarily disparate hosts, including plants, nematodes, and mammals (1–15). The nematode worm Caenorhabditis elegans has proven to be a particularly facile host for studying bacterial pathogenesis. For example, both Gram-negative and Gram-positive human bacterial pathogens kill C. elegans, bacterial virulence factors important for mammalian pathogenesis have been shown to enhance the rate at which C. elegans are killed by these pathogens, and bacterial mutant libraries can be readily screened by using a C. elegans killing assay for avirulent clones. C. elegans can also be readily used to study the host innate immune response to microbial infection (16, 17). C. elegans feed on bacteria and the C. elegans bacterial killing assay simply involves transferring worms from their normal laboratory food, Escherichia coli strain OP50, to a lawn of the pathogen of interest growing on agar medium.
In contrast to bacterial pathogens, C. elegans has not been reported as a host for studying human fungal pathogens. Recently Steenbergen et al. (18) reported the use of the free-living amoebae Acanthamoeba castellanii as a model for the study of survival strategies used by the human opportunistic fungal pathogen Cryptococcus neoformans after ingestion by macrophages. Steenbergen et al. found that C. neoformans was phagocytosed by A. castellanii, and that once inside, C. neoformans replicated, eventually killing the amoebae. The process was remarkably similar to that seen in mammalian macrophages (19). An acapsular strain of C. neoformans did not survive when incubated with A. castellanii and a phospholipase mutant exhibited a decreased replication rate in amoebae, similar to results obtained in macrophages (18). These observations suggested that cryptococcal characteristics that contribute to mammalian virulence also promote fungal survival in free-living amoebae.
Given the successful use of C. elegans and A. castellanii to study bacterial and fungal pathogenesis, respectively, and because fungal infections are significant causes of human morbidity and mortality, especially among immunocompromised patients (20–26), we sought to develop a fungal–C. elegans model pathogenesis system. The increasing number of serious fungal infections, the paucity of new antifungal agents, and the likelihood of the emergence of drug resistance in fungi all contribute to a pressing need for new model systems to study the mechanisms of fungal virulence. However, the ability of C. elegans to use yeasts as a food source, and the susceptibility of C. elegans to killing by yeasts that are pathogenic to animals and humans, has not been previously reported. Indeed, because of the small size of C. elegans and the relatively large size of fungal cells compared with bacterial cells, it was not apparent that a fungal–C. elegans model for the study of pathogenesis could be developed.
To determine whether C. elegans could consume fungi as a food source and whether human fungal pathogens could kill C. elegans, we initially chose to test free-living basidiomycetous yeasts belonging to the genus Cryptococcus (27) that presumably interact with nematodes in natural habitats. One reason for selecting cryptococcal species for these experiments was that they form transparent lawns on various growth media, thereby allowing us to easily monitor the survival of C. elegans by using a dissecting microscope. C. laurentii and C. kuetzingii are two generally nonpathogenic cryptococcal species that were used as controls for varieties of the pathogenic species C. neoformans. C. neoformans is heterothallic with two mating types, MATa and MATα and a variety of serotypes (28), and it has been used as a model for the study of fungal pathogenesis. Despite frequent environmental exposure to C. neoformans, immunologically intact individuals uncommonly develop clinical disease. However, the incidence of infection rises dramatically in patients with impaired T-cell immunity (26, 29). Study of the pathogenic mechanisms of C. neoformans has been enhanced substantially by the development of transformation protocols, homologous recombination for genetic manipulations, and reproducible animal models (30–34). The most important C. neoformans virulence factors identified so far include the polysaccharide capsule (30, 35), laccase (an enzyme essential for melanin production) (18, 36–40), the α allele of the mating type locus (41, 42), and at least two signal transduction cascades (42–47).
In this paper, we show that C. elegans can feed on cryptococci, that C. neoformans but not C. laurentii or C. kuetzingii infects and kills C. elegans, and that killing by C. neoformans depends on a number of virulence genes that are also important in mammalian infection. These data suggest that important aspects of C. neoformans pathogenesis in humans (and possibly that of other fungal pathogens) can be modeled by using the simple invertebrate nematode C. elegans as an experimental host.
Materials and Methods
Strains and Media.
The C. neoformans strains used in these experiments are summarized in Table 1 or described in the text. C. laurentii strains ATCC#18803, ATCC#66036, and ATCC#76483 and C. kuetzingi strain ATCC#42276 were obtained from the American Type Culture Collection (ATCC). Yeast cultures were maintained on yeast extract/peptone/dextrose (YPD; Difco) agar. The standard C. elegans strain N2 Bristol (55) was maintained at 15°C and propagated on E. coli strain OP50 by using established procedures (6, 55, 56).
Table 1.
Yeast strains used and their interaction with C. elegans
| C. neoformans strains (ref.) | Relevant characteristics or phenotype | LT50 for C. elegans killing (P value compared to parent strain, when relevant) |
|---|---|---|
| H99 ATCC #208821 (48) | Serotype A; clinical isolate; produces laccase; genome sequence being determined | 5.5–7.0 days |
| H99 gpa1 (42) | GPA1 encodes a G-protein alpha subunit homolog; attenuated in mammalian models | >10 days (P < 0.0001) |
| H99 gpa1 + GPA1 (42) | Complementation of the gpa1 mutant with wild-type GPA1 restored virulence in rabbits | 5.0 days (no difference) |
| H99 pka1 (44) | PKA1 encodes the major cAMP-dependent protein kinase catalytic subunit; attenuated in mammalian models | 9.0 days (P = 0.0007) |
| H99 pka1 + PKA1 (44) | Complementation of the pka1 mutant with wild-type PKA1 restored virulence in rabbits and mice | 5.0 days (no difference) |
| H99 ras1 (46) | ras1 mutant is avirulent in an animal model of cryptococcal meningitis | >10 days (P < 0.0001) |
| H99 ras1 + RAS1 (46) | Complementation of the ras1 mutant with wild-type RAS1 restored virulence in rabbits | 5.0 days (no difference) |
| H99 pkr1 (44) | PKR1 encodes the protein kinase A (PKA) regulatory subunit; in mice, a pkr1 mutant overproduces capsule and is hypervirulent | 3.5 days (P = 0.0001) |
| H99 M001 (49, 50) | Adenine auxotroph lacking phosphoribosyl-aminoimidazole; avirulent when inoculated intrathecally into immunosuppressed rabbits | >10 days (P < 0.0001) |
| H99 M001.1c (49) | Prototrophic transformant of M001 that has received a cloned ADE2 cDNA; virulence was completely restored in rabbit model | 6.0 days (no difference) |
| H99 pMFα1:GFP (51) | Promoter from the C. neoformans α pheromone gene fused to GFP | 5.5 days (no difference compared to H99) |
| H99 cap59 (52) | CAP59 is essential for capsule formation | 6.0 days in rich media (no difference); in minimal media, LT50 = 5.0 days compared to 3.5 days for H99 (P < 0.0001) |
| JEC20/JEC21 (48, 53, 54) | A pair of congenic MATa (JEC20) and MATα (JEC21) strains | LT50 = 9.5 days for JEC20; LT50 = 5.5 for JEC21 (P = 0.0043; JEC20 compared to JEC21) |
| ATCC #208820 (2e-tuc4) (40) | Serotype D; MATα CNLAC1/laccase positive | 6.0 days (LT50 = 2.0 days in the presence of dopamine) |
| ATCC #208819 (2e-tu4) (40) | Serotype D; MATα cnlac1/laccase negative | 6.0 days (no difference from 2e-tuc4 in the absence of dopamine; LT50 = 4.0 days and P < 0.0001 compared to 2e-tuc4 in the presence of dopamine) |
C. elegans Killing Assays.
Yeast lawns were grown for C. elegans killing assays as follows: C. neoformans strains were inoculated into 2 ml of YPD and grown at 28°C for 48 h; 10 μl of the culture was spread on 35-mm tissue-culture plates (Falcon) containing brain heart infusion (BHI) agar (Difco). The plates were incubated at 28°C overnight. C. laurentii and C. kuetzingii lawns for killing assays were prepared similarly except that growth was at 25°C. Ampicillin (100 μg/ml) or gentamicin (25–50 μg/ml) was added to the medium to selectively prevent growth of E. coli OP50 carried over on transfer of worms to the yeast-containing plates. For melanization studies, dopamine (100 μg/ml) was added to the BHI medium. For some killing studies, standard nematode growth medium [NGM, minimal medium containing NaCl, agar, peptone, cholesterol, CaCl2, MgSO4, and potassium phosphate (55)] supplemented with Synthetic Complete Supplement Mixture (SC Amino Acid Mixture, Qbiogene, Carlsbad, CA) was used instead of BHI.
Between 40 and 50 C. elegans animals at the young adult developmental stage were transferred from a lawn of E. coli OP50 on NGM (55) to a lawn of the yeast to be tested on BHI medium, incubated at 25°C, and examined for viability at 12- to 24-h intervals with a dissecting microscope. Worms were considered dead when they did not respond to touch with a platinum wire pick. Each experimental condition was tested in duplicate or triplicate. Nematodes exposed to C. laurentii or C. kuetzingii were able to grow and produce progeny similarly to those growing on E. coli OP50. For these latter two yeasts, we transferred nematodes to new plates every 48 h to allow determination of survival of the original worms without the presence of progeny. Worms growing on C. neoformans did not produce any progeny.
Plotting of killing curves, calculation of LT50 (time for half of the worms to die) and estimation of differences in survival (log-rank and Wilcoxon tests) with the Kaplan–Meier method were performed by using STATA 6 statistical software (Stata, College Station, TX). P values < 0.05 were considered statistically significant.
Evaluation of the Life Span of C. elegans.
Evaluation of the life span of C. elegans when feeding on E. coli OP50, C. kuetzingii, or C. laurentii was carried out at 25°C, as described (56). In brief, 3-day-old nematodes at the L4 developmental stage maintained on E. coli OP50 on NGM were transferred to fresh NGM plates containing 100 μg/ml 5-fluoro-2′-deoxyuridine (FdUrd; Sigma), which had been seeded with grown cultures of the bacterium or yeast under study. The animals remained on the FdUrd NGM plates for the remainder of their life spans; surviving nematodes were counted every day and dead ones were removed. Alternatively, nematode survival was evaluated on BHI plates. For this, nematodes were transferred to new plates every 24–48 h to allow determination of survival of the original worms without the presence of progeny.
Microscopic Studies.
Nematodes were exposed to yeast on BHI plates for various times and then placed on a pad of 2% agarose in a 5-μl drop of 30 mM NaN3 in M9 medium. The worms were examined with an Axioplan2 microscope (Zeiss) fitted with Nomarski optics. For experiments involving cryptococcal strains containing the pYGFP3 plasmid (57) expressing the synthetic GFP, the same methodology was used except that worms were viewed with a Leica TCS NT confocal spectrophotometer.
Results
Survival and Killing of C. elegans by Cryptococci.
To determine whether cryptococci can serve as a food source for C. elegans, and to determine whether pathogenic cryptococci can kill C. elegans, we provided C. neoformans, C. laurentii, and C. kuetzingii to wild-type N2 C. elegans as the sole source of food.
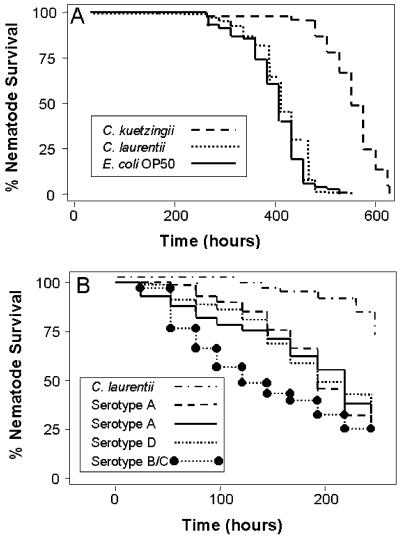
When L4 worms were transferred from lawns of E. coli OP50 to lawns of three different C. laurentii strains or a strain of C. kuetzingii on NGM or BHI medium containing ampicillin or gentamicin to prevent the growth of E. coli OP50, the nematodes appeared healthy and produced numbers of progeny similar to those of E. coli-fed worms. The C. elegans progeny produced when feeding exclusively on C. laurentii and C. kuetzingii for several generations were morphologically indistinguishable from E. coli-fed worms, and it appeared likely that C. elegans could be propagated indefinitely on C. laurentii or C. kuetzingii. In addition, C. elegans lived at least as long when feeding on C. laurentii or C. kuetzingii as when feeding on E. coli OP50. As shown in Fig. 1A, on NGM medium containing 5-fluoro-2′-deoxyuridine (FdUrd) to prevent the production of progeny (the standard assay for determining C. elegans longevity), the nematodes had the longest life span when feeding on C. kuetzingii [LT50 (time for half of the worms to die) = 23 days; P < 0.0001 compared with worms feeding on E. coli OP50], whereas longevity on C. laurentii was similar to that on E. coli OP50 (LT50 = 17 days). As reported previously (6), we found that C. elegans died prematurely when fed E. coli OP50 that had been grown on BHI medium compared with E. coli grown on NGM medium (ref. 6 and data not shown), suggesting that E. coli OP50 can be pathogenic to C. elegans when grown on the appropriate medium.
Fig 1.
(A) Life span of wild-type C. elegans N2 on NGM plates containing 5-fluoro-2′-deoxyuridine (FdUrd) feeding on lawns of C. kuetzingii ATCC#42276, C. laurentii ATCC#18803, and E. coli OP50. P < 0.0001 for E. coli OP50 or C. laurentii compared with C. kuetzingii. Similar results were obtained with C. laurentii strains ATCC#66036 and ATCC#76483. (B) Survival of C. elegans N2 feeding on lawns of C. neoformans serotype A (ATCC#62067, ATCC#62068), serotype B/C (ATCC#34877), or serotype D (ATCC#36556), or on a lawn of C. laurentii ATCC#18803. P < 0.001 for each of the C. neoformans strains compared with C. laurentii.
In contrast to C. laurentii and C. kuetzingii, all strains of C. neoformans that we tested killed C. elegans, and the nematodes were unable to make progeny. C. elegans killing was seen with human isolates of all known serotypes, including C. neoformans serotype A (ATCC#208821, which is also referred to as H99, ATCC#62067, and ATCC#62070), serotype B/C (ATCC#56695), and serotype D (ATCC#36556 and ATCC#24067) (Fig. 1B, Table 1, and data not shown). Environmental isolates of C. neoformans (ATCC#14116, ATCC#34870, ATCC#32308, ATCC#32045, and ATCC#62068) were also pathogenic to C. elegans (data not shown). Significant killing started at day 2 or 3 and most wild-type C. neoformans strains demonstrated an LT50 of 5–7 days with a range of 4–8.5 days (Fig. 1B, Table 1 and data not shown). C. elegans also died when exposed to mixed lawns of C. neoformans and C. laurentii (data not shown), indicating that C. neoformans actively kills C. elegans and excluding the trivial possibility that C. neoformans is simply not a nutritious food source for C. elegans.
In previous studies using C. elegans as a host to study bacterial pathogenesis, we observed that some bacterial pathogens induce the internal hatching of eggs in hermaphroditic worms, leading to matricide (1, 2, 6, 15). To exclude the possibility that killing of C. elegans by C. neoformans is caused solely by this matricidal effect (and not associated with an infectious-like process), we tested killing of C. elegans glp-4 mutant worms. C. elegans glp-4 animals have normal morphology and brood sizes at 15°C but do not make gonads and are unable to produce eggs at 25°C (58, 59). The LT50 of glp-4 worms feeding on C. neoformans H99 at 25°C was ≈2 days longer compared with the LT50 for wild-type worms, but still substantially shorter than on C. laurentii or C. kuetzingii.
Infection of C. elegans by Cryptococci.
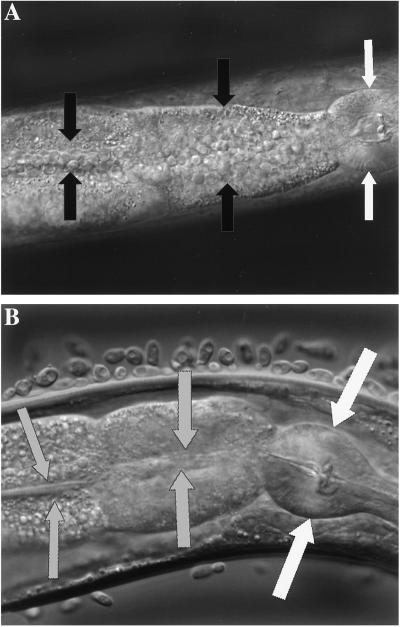
Microscopic evaluation of nematodes exposed to C. neoformans H99 showed that the yeast cells accumulated inside an abnormally distended gastrointestinal tract (Fig. 2A). Similar results have been observed previously with C. elegans feeding on a variety of bacterial pathogens (2, 6, 13). Yeast cells were mostly concentrated at the area directly distal to the pharyngeal grinder (the organ that functions to disrupt ingested organisms). However, yeast cells could be seen throughout the nematode gastrointestinal tract. It is unlikely that the accumulation is solely due to proliferation of the yeast in the worm intestine, as the gastrointestinal tract was filled with C. neoformans within 6 h of the start of feeding. In contrast to C. neoformans, neither C. laurentii (Fig. 2B) nor C. kuetzingii cells (not shown) were seen inside nematodes feeding on these yeasts, and there was no dilation of the intestinal tract for at least 7 days. Of note, when we allowed nematodes to feed on a lawn of C. neoformans H99 and then, at different times, transferred them to lawns of C. laurentii, they were rescued from C. neoformans killing for each of the time points tested (up to 48 h). Moreover, the intestines of the rescued worms regained a normal-nondistended anatomy, no yeast cells could be observed in the intestines of the rescued worms, and no H99 cells could be cultured following disruption of the rescued worms (data not shown). These latter data suggest that C. neoformans strain H99 is not capable of permanently colonizing the intestine.
Fig 2.
C. neoformans but not C. laurentii accumulates in the gastrointestinal tract of C. elegans. (A) Intact yeast cells present in the distended gastrointestinal tract after feeding 24 h on C. neoformans strain H99. (B) No C. laurentii cells can be detected in the gastrointestinal tract after 24 h of feeding. The round structure (white arrows) is the pharyngeal grinder organ, which functions to disrupt ingested organisms. Black and gray arrows point to the intestinal lumen.
Genes Associated with Pathogenesis in Mammals Cause Enhanced Killing of C. elegans.
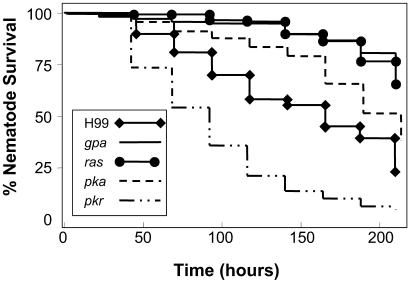
Capsule growth and melanin production in C. neoformans are regulated by a G-α protein-cAMP-PKA (cAMP-dependent protein kinase A) signaling pathway. PKA is composed of catalytic and regulatory subunits encoded by the PKA1 and PKR1 genes, respectively (44). Strains in which GPA1 (which encodes the G-α protein) or PKA1 are disrupted display attenuated virulence in experimental murine cryptococcosis. On the other hand, a C. neoformans strain with a disruption in the PKR1 gene overproduces capsule and is hypervirulent in animal models (43, 44). As shown in Fig. 3 and Table 1, the gpa1 and pka1 mutants were significantly less virulent than the wild type, whereas the pkr1 mutant was hypervirulent in the C. elegans model, similar to the results in mice. Reconstitution of a Pka+ phenotype (by integrating the wild-type PKA1 gene linked to the hygromycin B resistance gene into the mutant strain) that restored virulence in mammalian models (42–44), also restored virulence in the C. elegans model (Table 1).
Fig 3.
C. neoformans virulence factors for mammalian infection also enhance killing of C. elegans. Survival of C. elegans N2 animals feeding on C. neoformans mutants with disruptions in the genes encoding the G protein-cAMP-PKA and the RAS1-controlled signal transduction cascades demonstrated hypovirulence (gpa1, ras1, and pka1) or hypervirulence (pkr1), similar to results in mammalian models. P < 0.001 for each of the mutants compared with the parental strain H99 (see Table 1).
The results described in the preceding paragraph suggest that capsule and/or melanization may play a role in C. neoformans-mediated C. elegans killing. In mammalian hosts, the C. neoformans polysaccharide capsule, which distinguishes C. neoformans from many other pathogenic fungi, protects against phagocytosis and killing by immune effector cells and blocks the production of cytokines and antigen presentation to T cells (20, 60, 61). To determine the role of capsule in C. elegans killing, we tested two different C. neoformans acapsular strains, 602 (ATCC#36555) and cap59, a derivative of H99 (52). Both 602 and cap59 caused killing of C. elegans without accumulation in the nematode gastrointestinal tract (LT50 = 5 and 6 days, respectively; Table 1 and data not shown), suggesting that neither capsule nor accumulation is necessary for nematode killing. In rich media (BHI), killing by cap59 was similar to that caused by the wild-type parent H99 (LT50 = 6 days). However, under conditions that promote capsule formation, such as growth in minimal media (NGM) (20, 60), killing by cap59 was significantly slower compared with H99 (LT50 = 3.5 and 5, respectively; P < 0.0001; Table 1). Moreover, lawns of heat killed H99 grown on rich media led to C. elegans killing with an LT50 of 9 days, whereas nematodes survived at least 14 days when exposed to lawns of heat-killed 602 or cap59 (data not shown). In summary, these results suggest that capsule plays a significant, but not essential, role in C. neoformans-mediated killing of C. elegans.
Melanization is also regulated by the G-α protein-cAMP-PKA signaling pathway. C. neoformans is often melanized in the environment (37), and melanization has been associated with protection against harsh environmental conditions such as UV radiation and extremes of temperature (62). Melanin is also a potent free radical scavenger and melanization has been shown to protect C. neoformans from macrophages and has been associated with virulence in murine studies (36–38, 62). Melanin is produced when laccase (encoded by CNLAC1) catalyzes the oxidation of dopamine to quinones, which then polymerize to form melanin (36). Two otherwise isogenic CNLAC1 and cnlac1 strains (2e-tuc4 and 2e-tu4, respectively) have been extensively studied (40). In the C. elegans model, in the presence of dopamine (100 μg/ml), the laccase-positive strain that was able to synthesize melanin was significantly more virulent compared with the laccase-deficient strain. As expected, the two strains exhibited no difference in virulence when the experiment was carried out by using standard media in the absence of L-dopa (LT50 = 6 days-no difference compared with the parent strain in the absence of dopamine). In the presence of dopamine, the LT50 of the laccase-negative strain decreased to 4 days, whereas the LT50 of the laccase-positive strain decreased to 2 days (P < 0.0001; Table 1).
In addition to the G-α protein-cAMP-PKA signaling pathway, another regulatory pathway associated with C. neoformans maintenance of infection in mammalian hosts involves RAS1-specific signaling cascades that are required for growth at high temperature (37°C) (46). In the C. elegans model, a ras1 mutant that was previously found to be attenuated in mammalian models was significantly attenuated at 25°C compared with wild type; reconstitution of the Ras1+ phenotype (by integrating the wild-type RAS1 gene linked to the hygromycin B resistance gene into the ras1 mutant strain) restored virulence (Table 1). These findings indicate that RAS1 plays a role in C. neoformans beyond its role in promoting growth at 37°C.
The phosphoribosylaminoimidazole gene (ADE2) has been demonstrated to be essential for growth of C. neoformans in the cerebrospinal fluid, and an ade2 auxotroph of H99 (M001) was avirulent in a cryptococcal meningitis model in corticosteroid-treated rabbits (49). In the C. elegans model, the LT50 for killing by the M001 ade2 mutant was >10 days, compared with 6 days for the H99 parents; virulence was completely restored by the reintroduction of the wild-type ADE2 gene in the M001.1c strain (Table 1).
Activation of Mating Type α Gene During C. elegans Infection.
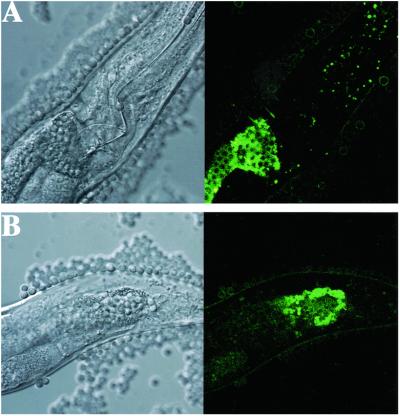
C. neoformans can reproduce asexually by budding or can form spores when cells of opposite mating types (MATα and MATa) undergo conjugation. The MATα mating type has been associated with increased virulence in animal models (41). Similarly, MATα mutants lacking the MFα pheromone were subtly or modestly attenuated compared with wild type in a murine tail vein injection model (63). We found that the MATα mating type was also more virulent than the MATa mating type with respect to C. elegans killing (LT50 = 5.5 days compared with 9.5 days; P = 0.0043; Table 1). The GFP from the jellyfish Aequorea victoriae has been used as a reporter of gene expression in a variety of heterologous systems. Cormack et al. (57) developed a modified GFP protein (yEGFP3) that generates significantly more fluorescence than wild-type GFP in the fungi Saccharomyces cerevisiae and Candida albicans. Using this modified GFP, del Poeta and coworkers (51) showed that a C. neoformans mating type α-specific reporter gene fusion pMFα1:GFP (pMFα1 regulates production of MFα pheromone) was activated during the proliferative stage of infection in the rabbit CNS but not during earlier phases of infection. Similar to the results in the rabbit CNS, as shown in Fig. 4, we found that there was no pMFα1:GFP expression in C. neoformans cells that were not ingested by nematodes, but that there was significant GFP expression in yeast cells within the nematode intestine by three days after ingestion. Although the C. elegans model has been used in the study of virulence by a number of pathogenic bacteria, this is the first direct evidence demonstrating that expression of a gene associated with virulence is temporally related to C. elegans killing.
Fig 4.
The C. neoformans MFα1 promoter is specifically expressed in the C. elegans intestine (magnification ×40). Fluorescent microscopy of wild-type C. elegans N2 after feeding for 3 days on C. neoformans H99 expressing GFP fused to the MFα1 promoter. Green fluorescence is seen in yeast cells inside the proximal (A) and the distal (B) end of the C. elegans intestine. There was no fluorescence associated with yeast cells outside the nematodes and no fluorescence was observed on day 1 or 2 of this experiment.
Discussion
In this paper, we report the observations that C. elegans can feed on yeasts, that a human pathogenic yeast can kill C. elegans, and that fungal virulence factors important for mammalian pathogenesis also play a role in C. elegans killing, findings that validate the use of C. elegans as a model host to study fungal pathogenesis. Similar to the hypothesis proposed by Steenbergen et al. (18) as a consequence of their studies examining phagocytosis and replication of C. neoformans in A. castellanii (18), mammalian pathogenesis of C. neoformans may be a consequence of adaptations that have evolved during the interaction of C. neoformans with environmental predators such as free-living nematodes and amoebae.
C. elegans Can Feed on Yeasts.
Despite the relatively large size of yeast cells compared with bacterial cells, it appears that C. elegans can use at least two different yeasts, C. laurentii and C. kuetzingii, as a sole source of food, producing the same number of healthy progeny as when feeding on E. coli strain OP50, the standard laboratory food source. Indeed, C. kuetzingii may be even more healthful than E. coli because the longevity of the worms is significantly longer when feeding on C. kuetzingii than on E. coli. This latter observation and other unpublished data from our laboratory (D. Garsin, J. Villanueva, J. Begun, C. Sifri, D. Kim, G. Ruvkun, and F.M.A., in preparation) indicates that E. coli OP50 may be mildly pathogenic to C. elegans.
C. neoformans Kills C. elegans.
In contrast to C. laurentii and C. kuetzingii, the human pathogenic yeast, C. neoformans, infects and kills C. elegans. Ecological studies have examined the interactions between fungi and free-living rhabditid nematodes (64), but only in the case of the endoparasitic fungus Drechmeria coniospora has a model of infection with C. elegans been established (65). Infection of C. elegans by D. coniospora starts by the adhesion of fungal spores to the head and is followed by the extension of hyphal processes into the nematodes (65). In contrast to D. coniospora, the mechanism by which C. neoformans accumulates within the C. elegans intestine and kills the worms is not immediately apparent from the studies reported here.
In previous studies, our laboratory and others have shown that the Gram-negative human pathogens Pseudomonas aeruginosa, Salmonella enterica, and Burkholderia pseudomallei, and Gram-positive bacteria such as Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus also kill adult C. elegans (1–15). The C. neoformans/C. elegans model shares a variety of features with these C. elegans models involving bacterial pathogens. For example, as in the case of C. neoformans, accumulation within the gastrointestinal tract is associated with killing of C. elegans in most of the bacterial models. It is very unlikely that C. neoformans-mediated “killing” is a consequence of C. neoformans being a nonnutritious food source. Mixed lawns of C. neoformans and C. laurentii also killed and >50% of the nematodes exposed to the C. neoformans adenine auxotroph M001 survived for more than 10 days. Although we cannot formally rule out the possibility that accumulation per se of C. neoformans in the C. elegans intestine and the resulting distention of the intestine is sufficient for killing, we have observed previously that Enterococcus faecium accumulates to very high levels causing intestinal distention but does not kill C. elegans (6). In any case, C. neoformans acapsular strains (such as cap59) also kill C. elegans without accumulation inside the nematode intestine. A similar phenomenon was observed in the case of the P. aeruginosa/C. elegans model, where P. aeruginosa also killed without accumulation by a toxin-mediated process when grown on particular media (13). Killing by the acapsular C. neoformans strains could also be attributable to toxins produced by the yeast or by components of the yeast cell (such as parts of the cell wall) that are generated as the nematode grinds the yeast cells. It is not clear why heat-killed C. neoformans strains also killed, but this could also be attributable to the production of toxins or cell-wall fragments.
C. neoformans Virulence Factors.
An important feature of the C. elegans/bacterial models of pathogenesis developed previously is that a variety of bacterial virulence factors involved in mammalian pathogenesis were also shown to be involved in nematode killing. We provide several examples in this paper that demonstrate that this is also the case for C. neoformans-mediated killing of C. elegans.
One of the most extensively studied C. neoformans virulence factors is its polysaccharide capsule (30, 35). In the C. elegans model, capsule may be sufficient for nematode killing because heat-killed capsular strains killed much more efficiently than heat-killed acapsular strains. On the other hand, capsule was not absolutely necessary for virulence as the acapsular strains cap59 and 602 also killed. Moreover, in rich media where relatively little capsule is made, there was no observable difference in nematode killing by cap59 and its isogenic wild-type parent. This differentiates the C. elegans and the A. castellanii pathogenesis systems, because capsule was necessary for C. neoformans virulence in the free-living amoebae (18). On the other hand, although the C. neoformans cap59 mutant is acapsular, it still may synthesize certain components of the capsule that are toxic. Of note is that C. laurentii also forms capsule, but it is rarely identified from clinical specimens and was not pathogenic to nematodes. The lack of pathogenicity of C. laurentii has been attributed to the inability of most strains to grow at 37°C. However, in the C. elegans model, C. laurentii was nonpathogenic at 25°C, suggesting that it may lack additional virulence traits unique to C. neoformans.
In addition to the C. neoformans capsule, we found extensive correlation between C. neoformans factors involved in both C. elegans and mammalian pathogenesis. C. neoformans mutants with disruptions in the genes associated with two signal transduction systems demonstrated hypo- (GPA1, PKA1, and RAS1) or hypervirulence (PKR1), as in established mammalian models. In the presence of the melanin substrate L-dopamine, a laccase-positive strain was significantly more virulent than an isogenic laccase-deficient strain. Also, the C. neoformans ADE2 gene is important for C. elegans killing, as in established mammalian models. A final example of the similarity between mammalian models and the C. elegans model of C. neoformans pathogenesis is the fact that the C. neoformans MFα1 gene, which is specifically activated during pathogenesis in a mammalian host (51), is similarly activated during infection of the C. elegans intestine. It is likely that the conditions inside the nematode gastrointestinal tract stimulate this pathway and it would be interesting to identify both C. neoformans and C. elegans mutants that failed to induce the MFα1-GFP reporter gene.
Previously, our laboratory and others have shown that C. elegans can be used to identify and study bacterial virulence factors relevant for mammalian pathogenesis by screening individual clones from bacterial mutant libraries for clones that exhibited impaired C. elegans killing (2, 6, 10–12). The results reported here suggest that a similar strategy could be used to identify C. neoformans virulence factors important for mammalian pathogenesis, irrespective of the fact that there may be many unique features of the C. elegans/C. neoformans model that are not directly relevant to mammalian pathogenesis or that particular virulence factors may play a more significant role in one of the models compared with the other.
In conclusion, the interaction of C. elegans with the yeast C. neoformans involves a number of genes that are also important during the host/pathogen interaction during mammalian infection. Identification of new C. neoformans virulence factors using this model may lead to new targets for antifungal therapies as well as a deeper understanding of the host–fungus interaction.
Acknowledgments
We thank J. A. Alspaugh, G. M. Cox, and J. K. Lodge for generous gifts of strains and advice and J. Plotnikova for technical assistance in using the confocal spectrophotometer. This work was supported by a grant from Aventis, SA (to F.M.A. and S.B.C.) and by a postdoctoral fellowship from the Howard Hughes Medical Institute (to E.M.).
Abbreviations
ATCC, American Type Culture Collection
BHI, brain heart infusion
NGM, nematode growth medium
PKA, protein kinase A
References
- 1.Aballay A. & Ausubel, F. M. (2002) Curr. Opin. Microbiol. 5 97-101. [DOI] [PubMed] [Google Scholar]
- 2.Aballay A., Yorgey, P. & Ausubel, F. M. (2000) Curr. Biol. 10 1539-1542. [DOI] [PubMed] [Google Scholar]
- 3.Aballay A. & Ausubel, F. M. (2001) Proc. Natl. Acad. Sci. USA 98 2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J. Y., Sifri, C. D., Goumnerov, B. C., Rahme, L. G., Ausubel, F. M. & Calderwood, S. B. (2002) J. Bacteriol. 184 952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darby C., Cosma, C. L., Thomas, J. H. & Manoil, C. (1999) Proc. Natl. Acad. Sci. USA 96 15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garsin D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., Calderwood, S. B. & Ausubel, F. M. (2001) Proc. Natl. Acad. Sci. USA 98 10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurz C. L. & Ewbank, J. J. (2000) Trends Microbiol. 8 142-144. [DOI] [PubMed] [Google Scholar]
- 8.Labrousse A., Chauvet, S., Couillault, C., Kurz, C. L. & Ewbank, J. J. (2000) Curr. Biol. 10 1543-1545. [DOI] [PubMed] [Google Scholar]
- 9.Plotnikova J. M., Rahme, L. G. & Ausubel, F. M. (2000) Plant Physiol. 124 1766-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahme L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995) Science 268 1899-1902. [DOI] [PubMed] [Google Scholar]
- 11.Rahme L. G., Tan, M.-W., Le, L., Wong, S. M., Tompkins, R. G., Calderwood, S. B. & Ausubel, F. M. (1997) Proc. Natl. Acad. Sci. USA 94 13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahme L. G., Ausubel, F. M., Cao, H., Drenkard, E., Goumnerov, B. C., Lau, G. W., Mahajan-Miklos, S., Plotnikova, J., Tan, M.-W., Tsongalis, J., et al. (2000) Proc. Natl. Acad. Sci. USA 97 8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan M.-W., Mahajan-Miklos, S. & Ausubel, F. M. (1999) Proc. Natl. Acad. Sci. USA 96 715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan M.-W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G. & Ausubel, F. M. (1999) Proc. Natl. Acad. Sci. USA 96 2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan M.-W. & Ausubel, F. M. (2000) Curr. Opin. Microbiol. 3 29-34. [DOI] [PubMed] [Google Scholar]
- 16.Kim D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M.-W. & Ausubel, F. M. (2002) Science 297 623-626. [DOI] [PubMed] [Google Scholar]
- 17.Mallo G. V., Kurz, C. L., Couillault, C., Pujol, N., Granjeaud, S., Kohara, Y. & Ewbank, J. (2002) Curr. Biol. 12 1209-1214. [DOI] [PubMed] [Google Scholar]
- 18.Steenbergen J. N., Shuman, H. A. & Casadevall, A. (2001) Proc. Natl. Acad. Sci. USA 98 15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitz S. M. (2001) Proc. Natl. Acad. Sci. USA 98 14760-14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casadevall A. & Perfect, J. R., (1998) Cryptococcus neoformans (Am. Soc. Microbiol., Washington, DC).
- 21.Marty F. & Mylonakis, E. (2002) Exp. Opin. Pharmacother. 3 91-102. [DOI] [PubMed] [Google Scholar]
- 22.Mylonakis E., Rich, J., Skolnik, P. R., De Orchis, D. F. & Flanigan, T. (1997) Medicine (Baltimore) 76 249-255. [DOI] [PubMed] [Google Scholar]
- 23.Mylonakis E., Barlam, T. F., Flanigan, T. & Rich, J. D. (1998) Chest 114 251-262. [DOI] [PubMed] [Google Scholar]
- 24.Mylonakis E., Paliou, M., Sax, P. E., Skolnik, P. R., Baron, M. J. & Rich, J. D. (2000) Medicine (Baltimore) 79 269-280. [DOI] [PubMed] [Google Scholar]
- 25.Pappas P. G., Perfect, J. R., Cloud, G. A., Larsen, R. A., Pankey, G. A., Lancaster, D. J., Henderson, H., Kauffman, C. A., Haas, D. W., Saccente, M., et al. (2001) Clin. Infect. Dis. 33 690-699. [DOI] [PubMed] [Google Scholar]
- 26.Thomas C. J., Lee, J. Y., Conn, L. A., Bradley, M. E., Gillespie, R. W., Dill, S. R., Pinner, R. W. & Pappas, P. G. (1998) Ann. Epidemiol. 8 212-216. [DOI] [PubMed] [Google Scholar]
- 27.Vandenkoornhuysem P., Baldauf, S. L., Leyval, C., Straczek, J. & Young, J. P. (2002) Science 295 2051. [DOI] [PubMed] [Google Scholar]
- 28.Kwon-Chung K. J. (1976) Mycologia 68 821-833. [PubMed] [Google Scholar]
- 29.Gumbo T., Kadzirange, G., Mielke, J., Gangaidzo, I. T. & Hakim, J. G. (2002) Pediatr. Infect. Dis. J. 21 54-56. [DOI] [PubMed] [Google Scholar]
- 30.Chang Y. C. & Kwon-Chung, K. J. (1994) Mol. Cell. Biol. 14 4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edman J. C. & Kwon-Chung, K. J. (1990) Mol. Cell. Biol. 10 4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua J., Meyer, J. D. & Lodge, J. K. (2000) Clin. Diagn. Lab. Immunol. 7 125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lodge J. K., Jackson-Machelski, E., Toffaletti, D. L., Perfect, J. R. & Gordon, J. I. (1994) Proc. Natl. Acad. Sci. USA 91 12008-12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toffaletti D. L., Rude, T. H., Johnston, S. A., Durack, D. T. & Perfect, J. R. (1993) J. Bacteriol. 175 1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perfect J. R., Wong, B., Chang, Y. C., Kwon-Chung, K. J. & Williamson, P. R. (1998) Med. Mycol. 36, Suppl. 1 79-86. [PubMed] [Google Scholar]
- 36.Salas S. D., Bennett, J. E., Kwon-Chung, K. J., Perfect, J. R. & Williamson, P. R. (1996) J. Exp. Med. 184 377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casadevall A., Rosas, A. L. & Nosanchuk, J. D. (2000) Curr. Opin. Microbiol. 3 354-358. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Aisen, P. & Casadevall, A. (1995) Infect. Immun. 63 3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson E. S. (2000) Clin. Microbiol. Rev. 13 708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson P. R., Wakamatsu, K. & Ito, S. (1998) J. Bacteriol. 180 1570-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon-Chung K. J., Edman, J. C. & Wickes, B. L. (1992) Infect. Immun. 60 602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alspaugh J. A., Perfect, J. R. & Heitman, J. (1997) Genes Dev. 11 3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Souza C. A. & Heitman, J. (2001) FEMS Microbiol. Rev. 25 349-364. [DOI] [PubMed] [Google Scholar]
- 44.D'Souza C. A., Alspaugh, J. A., Yue, C., Harashima, T., Cox, G. M., Perfect, J. R. & Heitman, J. (2001) Mol. Cell. Biol. 21 3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lengeler K. B., Davidson, R. C., D'Souza, C. A., Harashima, T., Shen, W. C., Wang, P., Pan, X., Waugh, M. & Heitman, J. (2000) Microbiol. Mol. Biol. Rev. 64 746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alspaugh J. A., Cavallo, L. M., Perfect, J. R. & Heitman, J. (2000) Mol. Microbiol. 36 352-365. [DOI] [PubMed] [Google Scholar]
- 47.Waugh M. S., Nichols, C. B., DeCesare, C. M., Cox, G. M., Heitman, J. & Alspaugh, J. A. (2002) Microbiology 148 191-201. [DOI] [PubMed] [Google Scholar]
- 48.Heitman J., Casadevall, A., Lodge, J. K. & Perfect, J. R. (1999) Mycopathologia 148 1-7. [DOI] [PubMed] [Google Scholar]
- 49.Perfect J. R., Toffaletti, D. L. & Rude, T. H. (1993) Infect. Immun. 61 4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudarshan S., Davidson, R. C., Heitman, J. & Alspaugh, J. A. (1999) Fungal Genet. Biol. 27 36-48. [DOI] [PubMed] [Google Scholar]
- 51.del Poeta M., Toffaletti, D. L., Rude, T. H., Sparks, S. D., Heitman, J. & Perfect, J. R. (1999) Infect. Immun. 67 1812-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson R. T., Hua, J., Pryor, B. & Lodge, J. K. (2001) Genetics 157 935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore T. D. & Edman, J. C. (1993) Mol. Cell. Biol. 13 1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karos M., Chang, Y. C., McClelland, C. M., Clarke, D. L., Fu, J., Wickes, B. L. & Kwon-Chung, K. J. (2000) J. Bacteriol. 182 6222-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brenner S. (1974) Genetics 77 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honda S., Ishii, N., Suzuki, K. & Matsuo, M. (1993) J. Gerontol. 48 B57-B61. [DOI] [PubMed] [Google Scholar]
- 57.Cormack B. P., Bertram, G., Egerton, M., Gow, N. A., Falkow, S. & Brown, A. J. (1997) Microbiology 143 303-311. [DOI] [PubMed] [Google Scholar]
- 58.Beanan M. J. & Strome, S. (1992) Development (Cambridge, U.K.) 116 755-766. [DOI] [PubMed] [Google Scholar]
- 59.Roussell D. L. & Bennett, K. L. (1993) Proc. Natl. Acad. Sci. USA 90 9300-9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doering T. L. (2000) Trends Microbiol. 8 547-553. [DOI] [PubMed] [Google Scholar]
- 61.Feldmesser M., Rivera, J., Kress, Y., Kozel, T. R. & Casadevall, A. (2000) Infect. Immun. 68 3642-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosas A. L. & Casadevall, A. (2001) Mycopathologia 151 53-56. [DOI] [PubMed] [Google Scholar]
- 63.Shen W. C., Davidson, R. C., Cox, G. M. & Heitman, J. (2002) Eukaryot. Cell 1 366-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mendoza De Gives P. M., Davies, K. G., Clark, S. J. & Behnke, J. M. (1999) Parasitology 119 95-104. [DOI] [PubMed] [Google Scholar]
- 65.Jansson H. B., Jeyaprakash, A. & Zuckerman, B. M. (1985) Appl. Environ. Microbiol. 49 552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]