Abstract
Colony-stimulating factor-1 receptor (CSF-1R) is the major regulator of macrophage development and is associated with epithelial cancers of the breast and ovary. Immunohistochemistry analysis of murine prostate development demonstrated epithelial expression of CSF-1R during the protrusion of prostatic buds from the urogenital sinus, during the prepubertal and androgen-driven proliferative expansion and branching of the gland, with a decline in older animals. Models of murine prostate cancer showed CSF-1R expression in areas of carcinoma- and tumor-associated macrophages. Several human prostate cancer cell lines and primary cultures of human prostate epithelial cells had low but detectable levels of CSF-1R. Human prostatectomy samples showed low or undetectable levels of receptor in normal glands or benign prostatic hypertrophy specimens. Staining was strongest in areas of prostatic intraepithelial neoplasia or carcinoma of Gleason histological grade 3 or 4. The activated form of the receptor reactive with antibodies specific for phosphotyrosine modified peptide sequences was observed in samples of metastatic prostate cancer. Immunohistochemistry showed strong expression of CSF-1R by macrophage lineage cells, including villous macrophages and the syncytiotrophoblast layer of placenta, Kupper cells in the liver, and histiocytes infiltrating near prostate cancers. These observations correlate CSF-1R expression with changes in the growth and development of the normal and neoplastic prostate.
Perturbation of protein tyrosine kinase signaling is frequently associated with malignant transformation (1). Tyrosine kinase receptors and their ligands have been implicated in prostate development and cancer, including transforming growth factor α, epidermal growth factor, insulin-like growth factor 1 (IGF-1), fibroblast growth factors, hepatocyte growth factor (HGF), platelet-derived growth factor (PDGF), and nerve growth factors (reviewed in refs. 2 and 3). Strategies attacking EGF and PDGF receptors are currently being evaluated in prostate cancer (4–7).
We defined the tyrosine kinase expression profile of normal prostatic epithelial cells during a phase of rapid growth in a relatively low androgen environment from day 10 murine prostate. CD44 was used as a marker of early progenitor cells within prostatic epithelium and is expressed by actively proliferating epithelia at sites of epithelial–mesenchymal interaction (8, 9). Embryonic formation of the prostate occurs through epithelial budding from the urogenital sinus. Elongation and branching of the ducts begin prenatally and are extensive during the first 21 days after birth. Although ductal morphogenesis of the prostate is androgen-dependent, the early postnatal period is marked by low levels of circulating androgen (10, 11). Mice with loss-of-function mutations in the homeobox genes NKX 3.1 or Hox D13 show mild defects in prostate development (10). A more severe block in prostate development is seen in P63−/− mice, which do not develop a recognizable prostate (12).
We prepared cDNA libraries from highly enriched CD44+ prostate cells from day 10 mice. A PCR-based strategy targeting highly conserved tyrosine kinase catalytic domain sequences was used (13, 14). One of the most frequently recovered tyrosine kinases was the colony-stimulating factor-1 receptor (CSF-1R). CSF-1R is encoded by the cellular homolog of the retroviral oncogene v-fms (15) and is the major regulator of development and response for all cells belonging to the mononuclear phagocyte lineage (16–18).
In osteoclastogenesis, one of the critical factors produced by bone stromal cells is CSF-1 (19, 20). The Csf1op/Csf1op mouse has inactivated the CSF-1 gene. These mice are osteopetrotic, toothless, and have low fecundity and reduced macrophage numbers (21, 22). CSF-1R null mutation (Csf1r−/Csf1r−) mice have a very similar but more severe phenotype (23). Prostate histology and function have not been well characterized in either Csf1op/Csf1op or Csf1r−/Csf1r− mice.
CSF-1R is expressed in testis, uterus, ovary, placenta, and mammary glands (21, 24). Elevated expression of CSF-1R has been seen in breast, ovarian, and uterine cancers, and the extent of expression in these tumors correlates with high grade and poor prognosis (24, 25). High circulating levels of CSF-1 correlate with active disease in ovarian and endometrial cancers and with metastatic breast and prostate cancer (24–26). In this article, we show that CSF-1R is expressed during the early phases of murine prostate development and prostate cancer progression in mouse and human.
Materials and Methods
Animal and Cell Lines.
Prostate cancer cell lines LNCaP (27), PC-3 (28) and DU145 (29), and breast cancer line BT-20 (30) were obtained from American Type Culture Collection (Rockville, MD). BT-20 cells were incubated in medium with 1 μΜ dexamethasone (synthetic glucocorticoid, Sigma) (31); LAPC-4 was provided by R. Reiter [University of California, Los Angeles (UCLA); ref. 32]; and basaloid PrEC prostate cells were from Clonetics (Walkersville, MD). Mouse prostate (C57BL/6) was fixed with 10% buffered formalin and embedded in paraffin wax. Prostate tumors were from transgenic adenocarcinoma mouse prostate (TRAMP) (33) and phosphatase and tensin homolog deleted from chromosome 10 (PTEN) +/− mice (34, 35). Hong Wu (UCLA) and Norman Greenberg (Baylor College of Medicine) kindly provided PTEN+/− and TRAMP mice, respectively. E. Richard Stanley and Xu-Ming Dai (Albert Einstein College of Medicine) kindly provided tissue sections of Csfr−/Csf1r− mice. The M-NFS-60 murine macrophage line was used as a control in some experiments (36). Csf1rop/Csf1rop mice were from The Jackson Laboratory.
Prostate tissues were minced and digested with collagenase I (Sigma) at a concentration of 1,500 units/ml for 1 hr at 37°C. Single-cell suspensions were analyzed with FACS Vantage (Becton Dickinson). For depletion of macrophages from mouse prostate tissues, cells were incubated with mouse monoclonal antibody CD11b for 30 min at 4°C, washed three times, and incubated with sheep anti-mouse IgG Dynabeads M-450 (Dynal, Oslo) for 30 min at 4°C, then separated by magnetic column. The retained CD11b-positive and CD11b-negative cells that flow through the column were tested.
RNA Expression Analysis.
Total RNAs were extracted by the RNeasy Mini kit (Qiagen, Chatsworth, CA). Each portion of total RNAs from the cells was reverse-transcribed by oligo(dT) primer and SuperScript reverse transcriptase (Life Technologies, Gaithersburg, MD) in a volume of 25 μl after DNase I treatment. The resulting cDNA was subjected to PCR. DNA sequences of the primer pairs used are as follows: human CSF-1R, 5′-ACACTAAGCTCGCAATCCC-3′ and 5′-GTATCGAAGGGTGAGCTCAAA-3′; mouse CSF-1R, 5′-GACCTGCTCCACTTCTCCAG-3′ and 5′-GGGTTCAGACCAAGCGAGAAG-3′; human CSF-1, 5′-GGAGTGGACACCTGCAGTCT-3′ and 5′-TGTGCAGGGGCTGCTCACCA-3′; prostate-specific antigen, 5′-GGTCGGCACAGCCTGTTTCA-3′ and 5′-CCACGATGGTGTCCTTGATC-3′; β-actin, 5′-GACTACCTCATGAAGATCCT-3′ and 5′-GCGGATGTCCACGTCACACT-3′.
Immunoblot Analysis.
For Western blot analyses, the cells were resuspended in boiling sample buffer, and each sample was separated on a 4–12% gradient SDS Tris Glycine Gel (NOVEX, San Diego). Proteins were transferred onto a nitrocellulose membrane (Micron Separations, Westboro, MA) and visualized with a chemiluminescence kit (Amersham Pharmacia). Rabbit anti-human CSF-1R antibody (Santa Cruz Biotechnology, lot K130, 1:200 dilution) was used for Western blot analysis. Anti-ABL Western blots were probed with 21–24 mouse monoclonal antibodies as an internal loading control, as described (37). Goat anti-mouse IgG and goat anti-rabbit antibody conjugated by horseradish peroxidase (Bio-Rad) were used as secondary antibodies.
Tissue Microarray Analysis (TMA).
Archival formalin-fixed paraffin-embedded tissue samples were provided through the Department of Pathology at the UCLA Medical Center under Institutional Review Board approval. Primary radical prostatectomy cases from 1984 to 1995 were randomly selected from the pathology database to represent a wide spectrum of tumor grades and primary disease stages. Three TMAs were constructed by the method of Kononen et al. (38), encompassing a total of 246 individual patients. Patients treated preoperatively with neoadjuvant hormones were then excluded from the analysis (n = 20). An additional 11 cases were uninformative due to missing spots or lack of target tissues. Overall, 215 prostatectomy cases encompassing 1,109 informative tissue spots on three TMAs were used for analysis. Included were 214 radical prostatectomies and one cystoprostatectomy. The median age at diagnosis of this cohort was 65 years. Semiquantitative assessment of antibody staining on the TMAs was performed by one pathologist (D.B.S.). Target tissue for scoring included only the glandular elements of the prostate tissue. The maximal intensity of diaminobenzidine brown chromogen staining was graded on a 0–2 scale (0 = negative, 1 = weak, and 2 = strong staining).
Immunohistochemistry (IHC).
Serial 4-μm-thick sections were deparaffinized in three changes of xylene and rehydrated through a 100–70% descending series of ethanol, immersed in citrate buffer (pH 6.0) in a 95°C water bath for 25 min, and then placed in 3% H2O2 in methanol for 10–20 min at room temperature to block endogenous peroxidase activity. After the blocking of nonspecific protein binding by incubation for 30 min to 1 h with 5% goat serum, the whole-tissue sections were incubated with each primary antibody against either monoclonal mouse anti-human CD68 (DAKO), polyclonal rabbit anti-human CSF-1R (Santa Cruz Biotechnology, lot K130, 1:200 dilution), or polyclonal rabbit anti-human phosphorylated tyrosine 723 peptide (PY723) of CSF-1R (1:50 dilution) overnight at 4°C (39). Subsequently, sections were processed for IHC by using the EnVision+ System (DAKO), StreptABComplex/HRP kit (DAKO), or Vector ABC Elite (Vector Laboratories), as described (34). For PY723 of CSF-1R staining, we used the EnVision+ (DAKO) and tyramide signal amplification (NEN) systems, according to the manufacturers' guidelines. For double staining of CD68 and CSF-1R, we used the EnVision Doublestain system (DAKO), according to the protocol recommended by the manufacturer. Specificity of staining was confirmed by replacing the primary antibody with mouse IgG for CD68 or rabbit IgG for other antibodies at the same concentration and by blocking of positive staining using excess purified peptides.
Results
CSF-1R Expression in Mouse Prostate Development and Cancer.
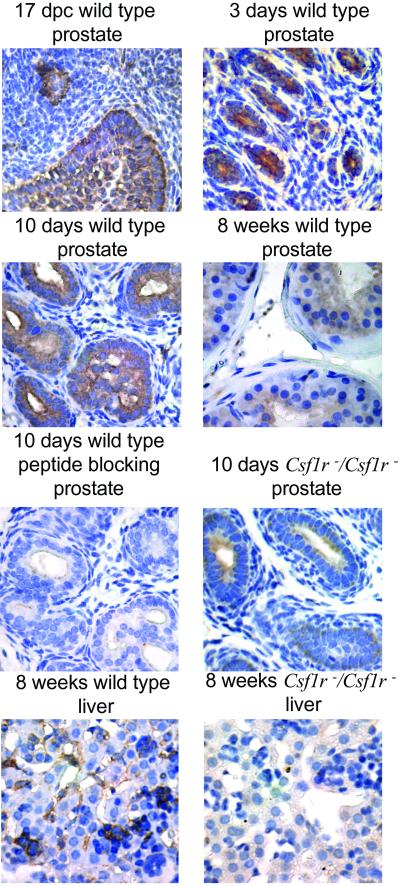
Limited data are available on the expression of CSF-1R in rodent prostate. One group has reported that a rat prostate cell line is positive for CSF-1R by microarray analysis, and the level of expression was found to increase with added androgen (40). RT-PCR analysis for CSF-1R on a panel of mouse prostate-derived cell lines showed a low frequency of positive lines (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). The midgestational budding of epithelium from the urogenital sinus first defines the mouse embryonic prostate. IHC for CSF-1R with a commercial rabbit anti-CSF-1R peptide antiserum (Santa Cruz Biotechnology) strongly and uniformly stains these structures (Fig. 1). Because the gland actively proliferates after birth, the level of CSF-1R remains high at 3 and 10 days (Fig. 1). Postpuberty at 8 weeks of age, limited cell division is occurring, and the expression of CSF-1R is reduced to background levels (Fig. 1). The staining on day 10 from a wild-type mouse is blocked with competing peptide. Sections of prostate from a day 10 Csf1r−/Csf1r− animal show a much lower level of staining. We also examined liver sections from 8-week-old wild-type and CSF-1R −/− mice by IHC. Positive strong staining was detected in Kupper cells of wild-type mice but not in CSF-1R−/− mice (Fig. 1). Strong staining for CSF-1R was seen in day 10 sections of prostate harvested from Csf1rop/Csf1rop mice (Fig. 8, which is published as supporting information on the PNAS web site).
Fig 1.
Immunohistochemical analysis of CSF-1R in mouse developing prostate. Tissue sections from 17 days past conception as well as 3 or 10 days and 8 weeks of age after birth of wild-type mice (C57BL/6) stained with polyclonal rabbit anti-CSF-1R antibody (Santa Cruz Biotechnology, lot K130, 1:200 dilution) based on the ABC method [DAKO or EnVison System (DAKO)]. The specificity of this antibody was confirmed by serial sections stained with the antibody after preincubation with immunizing peptide. Ten-day prostate and liver sections from Csf1r−/Csf1r− mice (23) were also stained with rabbit anti-CSF-1R antibody at the same dilution.
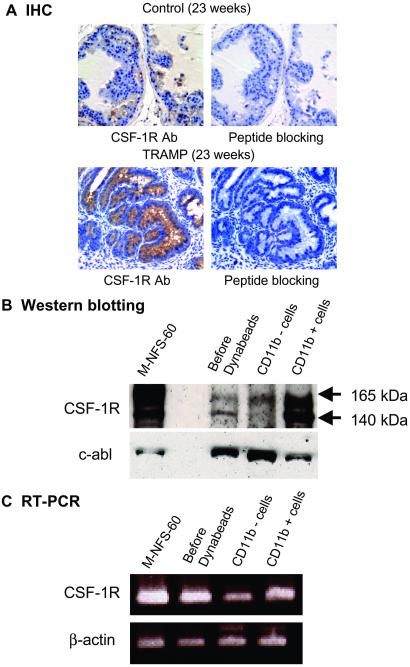
TRAMP is a transgenic mouse model in which the promoter of the probasin gene controls simian virus 40 T antigen expression. These mice develop prostatic intraepithelial neoplasia by 12–18 and cancer by 19–25 weeks of age (33). Immunohistochemical analysis of tissue sections from TRAMP tumors harvested between 23 and 30 weeks of age showed moderate to high expression of CSF-1R in 20–25% of the cancerous glands (Fig. 2A). The staining is largely intracellular, in concert with previous analyses of the membrane versus internal pools of CSF-1R (41). Background staining was detected in normal prostate glands from age-matched control mice. After preincubation with the immunizing peptide, staining for CSF-1R is negative (Fig. 2A).
Fig 2.
CSF-1R expression analyses in the prostate tumors of TRAMP mice. (A) CSF-1R expression in age-matched control mouse prostate and in the prostate tumor of a TRAMP mouse was examined by IHC with polyclonal rabbit anti-CSF-1R antibody (Santa Cruz Biotechnology, lot K130, 1:200 dilution) or after preincubation with immunizing peptide developed with the EnVison System (DAKO). (B) Immunoblot and (C) RT-PCR analyses of CSF-1R expression in single-cell suspensions prepared from a TRAMP tumor (23 weeks) before and after depletion of CD11b-positive macrophages by Dynabead magnetic bead technology as described in Materials and Methods. The M-NFS-60 cell line was derived from a myelogenous leukemia (36).
To verify the specificity of CSF-1R staining in prostate cancer epithelial cells in TRAMP tumors, single-cell suspensions were prepared, and CD11b-positive tissue macrophages were depleted by adherence to antibody-conjugated magnetic beads (Dynabeads; see Materials and Methods). The per-cell level of CSF-1R was, as expected, much higher in the CD11b-positive macrophage cell fraction by immunoblotting, as normalized to the c-abl protein levels (Fig. 2B) or PCR analysis normalized to β-actin levels (Fig. 2C). Interestingly, the CD11b-negative cell fraction is enriched for expression of the slower migrating mature glycosylated form (165 kDa) of CSF-1R compared with the macrophage-enriched CD11b-positive fraction, which has roughly equal levels of the 165-kDa form and the immature mannose-rich 140-kDa form of the receptor (42). A macrophage cell line, NFS60, predominately shows the mature form.
CSF-1R expression was analyzed in prostate tumors from PTEN+/− mice from 12 to 18 months of age. Moderate diffuse expression of CSF-1R was detected in the prostate tumors of these mice compared with age-matched controls (Fig. 9, which is published as supporting information on the PNAS web site).
Expression of CSF-1R in Human Prostate Cancer Cell Lines and in Human Prostate Cancer Tissue.
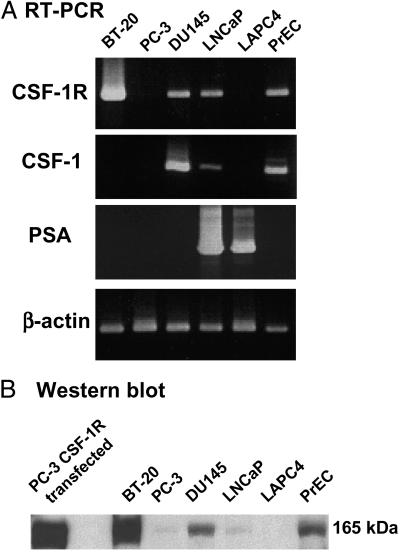
Several papers have noted increased levels of CSF-1R in human breast, ovarian, and uterine cancers and representative cell lines like BT-20 (24, 25). There are reports of the expression of CSF-1R in human prostate cancer cell lines PC-3, DU-145, and LNCaP by RT-PCR (43). Gene expression databases using microarray data (Unigene, www.ncbi.nlm.nih.gov/UniGene/Hs.Home.html), EST analyses (GeneCards, Weizmann Institute of Science, http://bioinformatics.weizmann.ac.il/cards/), or serial analysis of gene expression (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/SAGE/) show moderate to high levels of expression of CSF-1R sequences in human prostate cancer-derived samples. It is difficult to evaluate the contribution of expression from the prostate epithelial component of the tumor versus the stromal component, which can include tissue macrophage-derived cells by such techniques. We evaluated prostate cancer-derived clonal cell lines and commercially available prostate-derived basaloid epithelial cell populations (PrEC). We could detect low levels of CSF-1R mRNA expression in LNCaP, DU145, and PrEC cells by RT-PCR analysis but not in the PC3 or LAPC4 cell lines (Fig. 3A). BT-20 cells are derived from a human breast cancer and are used as a positive control of CSF-1R expression (30). All three of the prostate cell preparations positive for CSF-1R expression were also positive for the CSF-1 ligand RNA, raising the possibility of an autocrine or paracrine type of stimulation in some situations. Western blot analysis of extracts from this panel of cell lines (Fig. 3B) demonstrates the CSF-1R mature-sized protein (165 kDa) in BT-20, DU145, and PrEC cells, with lower levels in LNCaP and borderline to undetectable levels in the PC3 and LAPC4.
Fig 3.
Expression analyses of CSF-1R in human prostate cancer cell lines. (A) RT-PCR analysis. The PCR products at representative cycles were shown: CSF-1R, CSF-1, and prostate-specific antigen, cycle 40 and β-actin, cycle 25. The primers sequences are listed in Materials and Methods. (B) Immunoblotting analysis. Protein lysates of each 2 × 105 cells were subjected to SDS gel electrophoresis and polyclonal rabbit anti-human CSF-1R antibodies (1:250 dilution) were used for this analysis, as described in Materials and Methods. Human breast cancer cell line BT-20; human prostate cancer cell lines PC3, DU-145, LNCap, and LAPC4, and normal basaloid PrEC prostate cells were used in these analyses. PC-3 infected with human CSF-1R expressing retrovirus was used as a control.
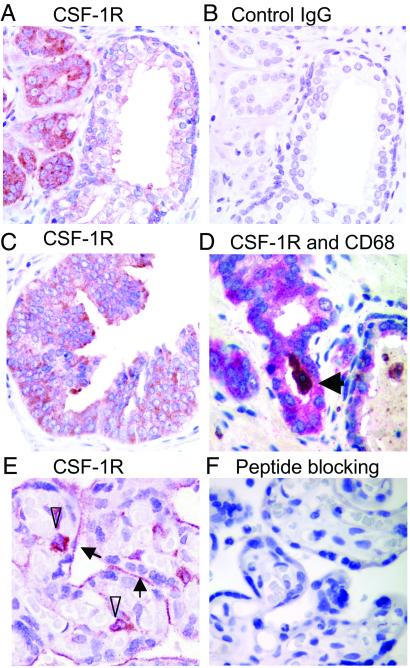
Human prostate cancer tissue specimens were examined by IHC for CSF-1R expression with rabbit anti-CSF-1R peptide antiserum (Santa Cruz Biotechnology). Reactivity, shown as red in cancerous glands, was much higher than in noncancerous glands in the same field (Fig. 4A). Control rabbit IgG (Fig. 4B) did not stain serial sections, and preincubation with the immunizing peptide blocked reactivity of the antiserum (Fig. 10, which is published as supporting information on the PNAS web site). CSF-1R staining was heterogeneous and predominately cytoplasmic, with some membranous staining in prostate cancer (Fig. 4A) and prostatic intraepithelial neoplasia (PIN) (Fig. 4C), which is consistent with prior reports of breast cancer staining (44). The presence of CSF-1R was also detected in macrophages near prostate cancers and within the lumens of some glands. The level of expression in macrophages was much higher than prostate cancer cells on a per-cell basis (Fig. 10). We confirmed the identity of macrophages stained with anti-CSF-1R by double staining with CD68 (Fig. 4D) (45). The CSF-1R antibody stained placental syncytiotrophoblasts and villous macrophages at the same dilution (Figs. 4E and 10) and was inhibited with the immunizing peptide (Fig. 4F). This antibody stained tumors derived from xenographs of PC-3 cells engineered to express CSF-1R (Fig. 11, which is published as supporting information on the PNAS web site). A series of rat anti-human CSF-1R monoclonal antibodies reactive with the ectodomain of the native receptor (46) were evaluated for IHC but did not recognize the antigen after tissue fixation.
Fig 4.
CSF-1R immunostaining in human primary prostate cancer with polyclonal rabbit anti-human CSF-1R antibodies (Santa Cruz Biotechnology) based on the ABC method (DAKO) or the EnVision+ System (DAKO). (A) Stronger staining in tumor cells was seen compared with adjacent normal glands. (B) Control IgG staining on a serial section. (C) Specific immunostaining was observed in the cytoplasm of PIN as well as malignant epithelial tumor cells. (D) Macrophage origin defined by CD68 staining (arrow: luminal macrophage in prostatic gland) using the double-staining method as described in Materials and Methods. CD68 staining used DAB (brown) and CSF-1R staining used Fast red (red purple) as substrates. (E) CSF-1R staining was detected in brush border of placental syncytiotrophoblasts (arrows) and villous macrophages (arrowheads). (F) After preincubation with the immunizing peptide, the staining for CSF-1R was blocked.
IHC Analysis of CSF-1R Expression in Primary Prostate Cancers Using Tissue Microarrays.
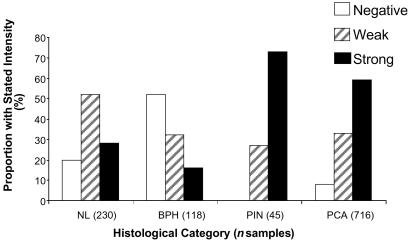
We examined samples from 215 informative cases, which had not received neoadjuvant therapy, by IHC analysis using tissue microarrays. Specimen cores with normal histology (230), benign prostatic hypertrophy (118), PIN (45), and tumor (716) were evaluated. Sections were scored as undetectable, weak, or strong-staining. The majority of all specimens (whether normal or malignant) showed some staining, but a clear and important trend is the higher percentage of cases of PIN and prostate cancer with strong staining (Fig. 5). Further analysis will be needed to accurately correlate such staining results with clinical and pathological variables. We have noted that the most intense and uniform staining for lesions within the prostate occurred in areas of PIN or carcinoma of histological Gleason grade 3 or 4. Examples of representative tissue microarray IHC results are shown in Fig. 12, which is published as supporting information on the PNAS web site.
Fig 5.
Immunohistochemical staining distribution of prostate by anti-CSF1R antibody on tissue microarrays. Two hundred fifteen prostatectomy cases encompassing 1,109 informative tissue spots on three TMAs were used for analysis grading on a 0–2 scale (0 = negative, 1 = weak, and 2 = strong staining). Immunostaining conditions were as described in Materials and Methods. NL, normal glands; BPH, benign prostatic hyperplasia; PIN, prostatic intraepithelial neoplasia; PCA, prostate cancer. Tissue spot histology and grading were confirmed on hematoxylin/eosin slides.
IHC Staining of Activated CSF-1R in Metastatic Sites by Tyrosine 723-Phosphospecific Peptide Antibody.
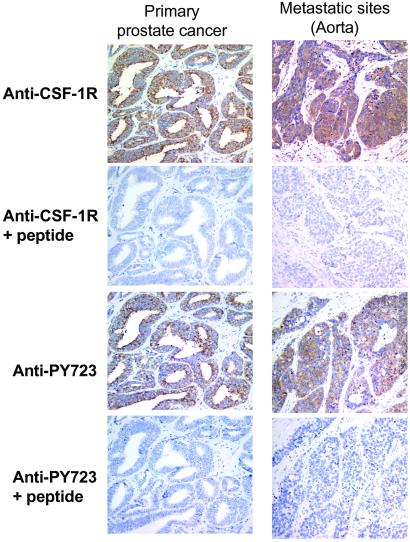
Prostate cancer can spread to distant sites, including lymph nodes, soft tissues, and, most commonly, bone and bone marrow (47). We used antibodies recognizing CSF-1R and specialized activation-specific reagents raised against the phosphorylated tyrosine 723 peptide (PY723) of CSF-1R (39). Phosphorylation of this tyrosine is important for the CSF-1R-driven phenotypic traits of anchorage-independent growth and metastasis (42, 48). Specimens of metastases were obtained that stained positive for prostate-specific antigen, confirming that they were of prostate cancer origin (data not shown). In each of the specimens, carcinoma cell areas showed staining for the CSF-1R and the PY723 modification of CSF-1R, which was blocked by the cognate peptide (Fig. 6; also see Fig. 13, which is published as supporting information on the PNAS web site).
Fig 6.
Immunohistochemical analysis of CSF-1R and activated CSF-1R expression in metastatic prostate cancer. Tissue sections from primary and metastatic prostate cancer (aorta) were stained with polyclonal rabbit anti-CSF-1R (Santa Cruz Biotechnology) antibody using the EnVision+ System (DAKO) and polyclonal rabbit anti-PY723 CSF-1R antibody using both the EnVision+ system and the tyramide signal amplification system (NEN). Coincidence of CSF-1R with activated CSF-1R in some sections was confirmed by serial sections. The specificity of anti-PY723 CSF-1R antibody was confirmed after preincubation with immunizing PY723 peptide (10 μM) blocked reactivity (39).
Discussion
Our results correlate the expression of the CSF-1 receptor with murine prostate development, proliferation, and cancer progression. Murine prostate carcinoma, initiated by distinct oncogenic signals, demonstrated enhanced expression of this receptor. Several cultured human prostate cancer-derived cell lines, primary cultures of prostate epithelium, and a large proportion of human prostate cancer specimens, most prominent in PIN lesions and carcinomas of histological grades 3 and 4, were positive for CSF-1R expression. The mechanisms regulating this pattern remain unknown.
Role of the CSF-1 Receptor in Steroid-Regulated Epithelial Development.
Mammary gland development in Csf1op/Csf1op mice revealed a lactational defect secondary to incomplete ductal growth, with precocious development of the lobuloalveolar system (49). Crossing Csf1op/Csf1op mice with a mammary cancer-susceptible strain of mice did not affect the incidence or growth of primary tumors but delayed their development to invasive metastatic carcinomas associated with lessened infiltration and function of tumor-associated macrophages (50). CSF-1 signaling provides a critical function for mammary gland development during pregnancy, lactation, and cancer progression (24).
Prostate development depends on steroid sex hormones, including androgen. Androgen may not directly stimulate the proliferation of normal prostate epithelial cells (10, 11). Paracrine factors, which are produced by the mesenchyme and regulated by steroid hormones, play a critical role. Several growth factors that act as paracrine mediators have been identified, including IGF-I, fibroblast growth factor (FGF)-7, FGF-10, HGF, and transforming growth factor-β (11, 51). Our observations suggest that CSF-1 is a candidate for such a paracrine factor.
CSF-1/1R and Bone Metastasis.
The most favored site of metastasis of prostate cancer is bone (47). The ability of prostate cancer to incite an osteoblastic reaction suggests there are bidirectional pathways between prostate cancer cells and osteoblasts and between osteoclasts and bone stromal cells. Prostate cancer metastases in bone have an extensive bone resorption ability, and osteoclastogenesis may play a role in the establishment of bone metastasis (52).
CSF-1 is an important regulator of hematopoiesis and bone resorption (19, 20). Studies performed with Csf1op/Csf1op mice established that CSF-1 function is essential for proliferation and differentiation of osteoclasts and is locally synthesized by bone marrow stromal fibroblasts and osteoblasts. In addition to effects on cell proliferation, differentiation, and invasion, the membrane-bound CSF-1 may mediate adhesion (53).
Other Receptor Tyrosine Kinases Expressed in Prostate Cancer.
Several protein tyrosine kinases, such as PDGF receptor (PDGF-R), c-met, HER-2/neu, and IGF-1R, are expressed in prostate cancer, including metastatic sites (2). CSF-1R is more closely related structurally to the receptors for PDGF than to other PTK receptor subfamilies (54). Immunohistochemical analyses show moderate to strong PDGF-Rα expression in PIN and primary prostate cancers but weak or absent expression in nonneoplastic prostate epithelium and stroma (13). The expression of PDGF-Rα protein by metastatic prostate cancer was also confirmed by IHC in a series of bone marrow metastases (13). HER-2/neu overexpression in primary prostate cancer and in metastatic sites of prostate tumors was noted before and after androgen-deprivation therapy (55). HGF is overexpressed in prostate cancer, and its receptor c-met expression in prostate cancer has been linked to higher histologic-grade and -stage disease (56). The finding of activated CSF-1R in metastatic tumors supports a role for CSF-1/CSF-1R signaling in prostate cancer progression.
Supplementary Material
Acknowledgments
We thank Norman Greenberg (Baylor College of Medicine) for providing TRAMP mice; Dr. Hong Wu (UCLA) for providing PTEN+/− mice; Drs. E. Richard Stanley and Xu-Ming Dai (Albert Einstein College of Medicine) for Csf1r−/Csf1r− tissues; Martine Roussel (Department of Tumor Cell Biology, St. Jude Children's Research Hospital) and Charles Sherr (Howard Hughes Medical Institute at St. Jude Children's Research Hospital) for reagents and valuable advice; J. C. White for preparation of the manuscript and figures; Yoon Kim, Shirley Quan, Benjamin Rafii, Adam Gottesfeld, Gregory Ferl, Sheila Tze, and Gregg Kanter for excellent technical assistance; and Drs. Tetsuro Watabe, Robert E. Reiter, and members of the Witte laboratory for helpful discussions. We thank the members of the Human Tissue Research Center at UCLA for processing and sectioning of tissues. We thank Dr. Arie Belldegrun (Department of Urology, UCLA) for help in providing specimens used in the construction of the tissue microarray used in these studies. Portions of this study were supported by a CaP CURE grant (to O.N.W.), National Institutes of Health Grant GM08243-13 (to D.B.S.), and funds from the Tina and Richard Carolan Prostate Research Fund (to A.P.). O.N.W. is an Investigator and H.I. an Associate of the Howard Hughes Medical Institute.
Abbreviations
IGF-1, insulin-like growth factor 1
HGF, hepatocyte growth factor
PDGF, platelet-derived growth factor
CSF-1R, colony stimulating factor-1 receptor
TRAMP, transgenic adenocarcinoma mouse prostate
PTEN, phosphatase and tensin homolog deleted from chromosome 10
TMA, tissue microarray analysis
IHC, immunohistochemistry
PrEC, prostate-derived basaloid epithelial cell populations
PIN, prostatic intraepithelial neoplasia
UCLA, University of California, Los Angeles
References
- 1.Blume-Jensen P. & Hunter, T. (2001) Nature 411, 355-365. [DOI] [PubMed] [Google Scholar]
- 2.Djakiew D. (2000) Prostate 42, 150-160. [DOI] [PubMed] [Google Scholar]
- 3.Ware J. L. (1999) Cancer Metastasis Rev. 17, 443-447. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs J. B. (2000) J. Clin. Invest. 105, 9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwaab T., Lewis, L. D., Cole, B. F., Deo, Y., Fanger, M. W., Wallace, P., Guyre, P. M., Kaufman, P. A., Heaney, J. A., Schned, A. R., et al. (2001) J. Immunother. 24, 79-87. [DOI] [PubMed] [Google Scholar]
- 6.Ko Y. J., Small, E. J., Kabbinavar, F., Chachoua, A., Taneja, S., Reese, D., DePaoli, A., Hannah, A., Balk, S. P. & Bubley, G. J. (2001) Clin. Cancer Res. 7, 800-805. [PubMed] [Google Scholar]
- 7.Bergan R. C., Waggle, D. H., Carter, S. K., Horak, I., Slichenmyer, W. & Meyers, M. (2001) Urology 57, 77-80. [DOI] [PubMed] [Google Scholar]
- 8.Yu Q. & Toole, B. P. (1997) Dev. Dyn. 208, 1-10. [DOI] [PubMed] [Google Scholar]
- 9.Gakunga P., Frost, G., Shuster, S., Cunha, G., Formby, B. & Stern, R. (1997) Development (Cambridge, U.K.) 124, 3987-3997. [DOI] [PubMed] [Google Scholar]
- 10.Abate-Shen C. & Shen, M. M. (2000) Genes Dev. 14, 2410-2434. [DOI] [PubMed] [Google Scholar]
- 11.Hayward S. W. & Cunha, G. R. (2000) Radiol. Clin. North Am. 38, 1-14. [DOI] [PubMed] [Google Scholar]
- 12.Signoretti S., Waltregny, D., Dilks, J., Isaac, B., Lin, D., Garraway, L., Yang, A., Montironi, R., McKeon, F. & Loda, M. (2000) Am. J. Pathol. 157, 1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chott A., Sun, Z., Morganstern, D., Pan, J., Li, T., Susani, M., Mosberger, I., Upton, M. P., Bubley, G. J. & Balk, S. P. (1999) Am. J. Pathol. 155, 1271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson D. R., Wu, Y. M. & Lin, S. F. (2000) Oncogene 19, 5548-5557. [DOI] [PubMed] [Google Scholar]
- 15.Sherr C. J., Rettenmier, C. W., Sacca, R., Roussel, M. F., Look, A. T. & Stanley, E. R. (1985) Cell 41, 665-676. [DOI] [PubMed] [Google Scholar]
- 16.Roth P. & Stanley, E. R. (1992) Curr. Top. Microbiol. Immunol. 181, 141-167. [DOI] [PubMed] [Google Scholar]
- 17.Cecchini M. G., Dominguez, M. G., Mocci, S., Wetterwald, A., Felix, R., Fleisch, H., Chisholm, O., Hofstetter, W., Pollard, J. W. & Stanley, E. R. (1994) Development (Cambridge, U.K.) 120, 1357-1372. [DOI] [PubMed] [Google Scholar]
- 18.Pollard J. W. & Stanley, E. R. (1996) Adv. Dev. Biochem. 4, 153-193. [Google Scholar]
- 19.Cecchini M. G., Hofstetter, W., Halasy, J., Wetterwald, A. & Felix, R. (1997) Mol. Reprod. Dev. 46, 75-83. [DOI] [PubMed] [Google Scholar]
- 20.Ryan G. R., Dai, X. M., Dominguez, M. G., Tong, W., Chuan, F., Chisholm, O., Russell, R. G., Pollard, J. W. & Stanley, E. R. (2001) Blood 98, 74-84. [DOI] [PubMed] [Google Scholar]
- 21.Pollard J. W. (1997) Mol. Reprod. Dev. 46, 54-61. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H., Hayashi, S. I., Kunisada, T., Ogawa, M., Nishikawa, S., Okamura, H., Sudo, T., Shultz, L. D. & Nishikawa, S. I. (1990) Nature 345, 442-444. [DOI] [PubMed] [Google Scholar]
- 23.Dai X. M., Ryan, G. R., Hapel, A. J., Dominguez, M. G., Russell, R. G., Kapp, S., Sylvestre, V. & Stanley, E. R. (2002) Blood 99, 111-120. [DOI] [PubMed] [Google Scholar]
- 24.Sapi E. & Kacinski, B. M. (1999) Proc. Soc. Exp. Biol. Med. 220, 1-8. [DOI] [PubMed] [Google Scholar]
- 25.Kacinski B. M. (1997) Mol. Reprod. Dev. 46, 71-74. [DOI] [PubMed] [Google Scholar]
- 26.McDermott R. S., Deneux, L., Mosseri, V., Vedrenne, J., Clough, K., Fourquet, A., Rodriguez, J., Cosset, J. M., Sastre, X., Beuzeboc, P., et al. (2002) Eur. Cytokine Network 13, 121-137. [PubMed] [Google Scholar]
- 27.Horoszewicz J. S., Leong, S. S., Chu, T. M., Wajsman, Z. L., Friedman, M., Papsidero, L., Kim, U., Chai, L. S., Kakati, S., Arya, S. K. & Sandberg, A. A. (1980) Prog. Clin. Biol. Res. 37, 115-132. [PubMed] [Google Scholar]
- 28.Kaighn M. E., Narayan, K. S., Ohnuki, Y., Lechner, J. F. & Jones, L. W. (1979) Invest. Urol. 17, 16-23. [PubMed] [Google Scholar]
- 29.Stone K. R., Mickey, D. D., Wunderli, H., Mickey, G. H. & Paulson, D. F. (1978) Int. J. Cancer 21, 274-281. [DOI] [PubMed] [Google Scholar]
- 30.Filderman A. E., Bruckner, A., Kacinski, B. M., Deng, N. & Remold, H. G. (1992) Cancer Res. 52, 3661-3666. [PubMed] [Google Scholar]
- 31.Sapi E., Flick, M. B., Gilmore-Hebert, M., Rodov, S. & Kacinski, B. M. (1995) Oncogene 10, 529-542. [PubMed] [Google Scholar]
- 32.Klein K. A., Reiter, R. E., Redula, J., Moradi, H., Zhu, X. L., Brothman, A. R., Lamb, D. J., Marcelli, M., Belldegrun, A., Witte, O. N., et al. (1997) Nat. Med. 3, 402-408. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg N. M., DeMayo, F., Finegold, M. J., Medina, D., Tilley, W. D., Aspinall, J. O., Cunha, G. R., Donjacour, A. A., Matusik, R. J. & Rosen, J. M. (1995) Proc. Natl. Acad. Sci. USA 92, 3439-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubey P., Wu, H., Reiter, R. E. & Witte, O. N. (2001) Cancer Res. 61, 3256-3261. [PubMed] [Google Scholar]
- 35.Stiles B., Gilman, V., Khanzenzon, N., Lesche, R., Li, A., Qiao, R., Liu, X. & Wu, H. (2002) Mol. Cell. Biol. 22, 3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakoinz I., Lee, M. T., Weaver, J. F. & Ralph, P. (1990) J. Immunol. 145, 860-864. [PubMed] [Google Scholar]
- 37.Era T. & Witte, O. N. (2000) Proc. Natl. Acad. Sci. USA 97, 1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kononen J., Bubendorf, L., Kallioniemi, A., Barlund, M., Schraml, P., Leighton, S., Torhorst, J., Mihatsch, M. J., Sauter, G. & Kallioniemi, O. P. (1998) Nat. Med. 4, 767-768. [DOI] [PubMed] [Google Scholar]
- 39.Flick M. B., Sapi, E., Perrotta, P. L., Maher, M. G., Halaban, R., Carter, D. & Kacinski, B. M. (1997) Oncogene 14, 2553-2561. [DOI] [PubMed] [Google Scholar]
- 40.Whitacre D. C., Chauhan, S., Davis, T., Gordon, D., Cress, A. E. & Miesfeld, R. L. (2002) Cell Growth Differ. 13, 1-11. [PubMed] [Google Scholar]
- 41.Guilbert L. J. & Stanley, E. R. (1986) J. Biol. Chem. 261, 4024-4032. [PubMed] [Google Scholar]
- 42.Roussel M. F., Downing, J. R. & Sherr, C. J. (1990) Oncogene 5, 25-30. [PubMed] [Google Scholar]
- 43.Savarese D. M., Valinski, H., Quesenberry, P. & Savarese, T. (1998) Prostate 34, 80-91. [DOI] [PubMed] [Google Scholar]
- 44.Maher M. G., Sapi, E., Turner, B., Gumbs, A., Perrotta, P. L., Carter, D., Kacinski, B. M. & Haffty, B. G. (1998) Clin. Cancer Res. 4, 1851-1856. [PubMed] [Google Scholar]
- 45.Pulford K. A., Sipos, A., Cordell, J. L., Stross, W. P. & Mason, D. Y. (1990) Int. Immunol. 2, 973-980. [DOI] [PubMed] [Google Scholar]
- 46.Ashmun R. A., Look, A. T., Roberts, W. M., Roussel, M. F., Seremetis, S., Ohtsuka, M. & Sherr, C. J. (1989) Blood 73, 827-837. [PubMed] [Google Scholar]
- 47.Scher H. I. & Yagoda, A. (1987) Am. J. Med. 82, 6-28. [DOI] [PubMed] [Google Scholar]
- 48.Roussel M. F., Dull, T. J., Rettenmier, C. W., Ralph, P., Ullrich, A. & Sherr, C. J. (1987) Nature 325, 549-552. [DOI] [PubMed] [Google Scholar]
- 49.Pollard J. W. & Hennighausen, L. (1994) Proc. Natl. Acad. Sci. USA 91, 9312-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin E. Y., Nguyen, A. V., Russell, R. G. & Pollard, J. W. (2001) J. Exp. Med. 193, 727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein J., Borzillo, G. V. & Rettenmier, C. W. (1990) Blood 76, 1308-1314. [PubMed] [Google Scholar]
- 52.Zhang J., Dai, J., Qi, Y., Lin, D. L., Smith, P., Strayhorn, C., Mizokami, A., Fu, Z., Westman, J. & Keller, E. T. (2001) J. Clin. Invest. 107, 1219-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uemura N., Ozawa, K., Takahashi, K., Tojo, A., Tani, K., Harigaya, K., Suzu, S., Motoyoshi, K., Matsuda, H. & Yagita, H. (1993) Blood 82, 2634-2640. [PubMed] [Google Scholar]
- 54.Hamilton J. A. (1997) J. Leukocyte Biol. 62, 145-155. [DOI] [PubMed] [Google Scholar]
- 55.Osman I., Scher, H. I., Drobnjak, M., Verbel, D., Morris, M., Agus, D., Ross, J. S. & Cordon-Cardo, C. (2001) Clin. Cancer Res. 7, 2643-2647. [PubMed] [Google Scholar]
- 56.Humphrey P. A., Zhu, X., Zarnegar, R., Swanson, P. E., Ratliff, T. L., Vollmer, R. T. & Day, M. L. (1995) Am. J. Pathol. 147, 386-396. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.