Abstract
Dysregulated production of IL-6 and its receptor (IL-6R) are implicated in the pathogenesis of multiple myeloma, autoimmune diseases and prostate cancer. The IL-6R complex comprises two molecules each of IL-6, IL-6R, and the signaling molecule, gp130. Here, we report the x-ray structure (2.4 Å) of the IL-6R ectodomains. The N-terminal strand of the Ig-like domain (D1) is disulfide-bonded to domain D2, and domains D2 and D3, the cytokine-binding domain, are structurally similar to known cytokine-binding domains. The head-to-tail packing of two closely associated IL-6R molecules observed in the crystal may be representative of the configuration of the physiological dimer of IL-6R and provides new insight into the architecture of the IL-6R complex.
Interleukin-6 (IL-6) is a polyfunctional cytokine that plays a central role in host defense because of its wide range of immune and hemopoietic activities and because of its potent ability to induce the acute-phase responses of the liver (1–5). IL-6 binds first to its cognate α-chain receptor (IL-6R) with low affinity, and then the complex binds to the signal-transducing molecule gp130 to form a high-affinity, functional hexameric receptor complex of two IL-6, IL-6R, and gp130 hetero-trimers (6). The aberrant production of both IL-6 and its receptor has been implicated in the pathogenesis of multiple myeloma (7), postmenopausal osteoporosis (8), autoimmune diseases (9), and prostate cancer (10).
The cDNA of the human IL-6R encodes a protein of 468 amino acids, which includes a signal peptide, an extracellular region, a transmembrane domain, and a short cytoplasmic domain of 19, 339, 28, and 82 amino acids, respectively (11). The mature 80-kDa IL-6R is a glycosylated form of the predicted 50-kDa precursor (12). Motif and homology analysis indicate that the extracellular region is highly modular, consisting of three domains (D1, D2, and D3, each ≈100 residues; ref. 13). The N-terminal domain D1 is characteristic of the Ig superfamily (14), and domains D2 and D3 are homologous to cytokine-binding domains (CBD) consisting of two fibronectin type III (FnIII) domains (an Ig-like-fold; ref. 15). In common with other members of this class I cytokine receptor family, the N-terminal domain of the CBD (D2) has four conserved cysteines, there is a proline-rich “hinge” region, and the C-terminal domain (D3) has a conserved “WSXWS” sequence motif (13). Although the Ig domain D1 has been shown to be dispensable for ligand recognition and signal initiation (16, 17), it is implicated in receptor internalization and protein stability (18).
The transmembrane and cytoplasmic domains of the IL-6R are not required for IL-6 signal transduction (17). Biologically active forms of the extracellular region of the IL-6R or “soluble IL-6R” (sIL-6R; ref. 19) are found naturally in low concentrations in human urine and serum of healthy individuals (20). Increased sIL-6R levels have been reported in a number of clinical conditions (e.g., HIV infection, multiple myeloma, rheumatoid arthritis, etc.), indicating that its production is correlated with a disease response (21). It is still unclear as to whether the sIL-6R in physiological fluids comes from proteolytic shedding of membrane-bound IL-6R (22) or from an alternatively spliced mRNA species (23). In contrast to other soluble cytokine receptors that act as inhibitors by competing for ligand binding with cellular receptors (19), the sIL-6R facilitates IL-6 activity (17). Inhibition of the formation of the IL-6/IL-6R complex is reported to be a primary target for antagonizing the action in vivo of IL-6 (24).
As a contribution to improving an understanding of the biochemistry and biology of the binary IL-6/IL-6R complex, we have purified the extracellular portion of human IL-6R from medium conditioned by Chinese hamster ovary (CHO) cell expression system (25) secreting recombinant IL-6R α-chain (26, 27). Previously, we have shown that this form of IL-6R binds IL-6, and then this binary complex binds to the soluble form of the extracellular portion of gp130 (sgp130) to form a 2:2:2 stoichiometry of IL-6, IL-6R, and sgp130 (6). sIL-6R is bioactive: in the presence of IL-6, the binary complex prevents the differentiation of embryonic stem cells (6). We now report the three-dimensional structure of the extracellular region of the human IL-6R.
Materials and Methods
Expression, Purification, and Crystallization of sIL-6R.
Monomeric sIL-6R was purified from the conditioned medium of glycosylation-defective mutant CHO cell line Lec3.2.8.1 (25) and transfected with the construct pEE14sIL6RL325, which encodes sIL-6R (R. L. Moritz, unpublished work). Briefly, sIL-6R-transformed Lec3.2.8.1 cells were grown in fermentation apparatus with a working volume of 1.25 liters (Celligen Plus fermenter, New Brunswick Scientific). Conditioned media was concentrated 20-fold by ultrafiltration. sIL-6R was purified from concentrated Lec3.2.8.1 cell media by binding to a 5-ml column of human IL-6-Sepharose. sIL-6R was further purified by preparative size-exclusion chromatography and concentrated to 10 mg/ml by using a 10,000 MWCO centrifugal concentrator (Centricon, Millipore).
The protein (8 mg/ml in 5 mM Tris⋅HCl, pH 8.0) was crystallized in hanging drops by vapor diffusion against a reservoir containing 50 mM ammonium sulfate, 18% PEG 3350, and 2.5 mM sodium citrate, at pH 5.6. Crystals grew in the space group P43212 (a = 51.4 Å, c = 305.4 Å) with 51% solvent. To ensure isomorphism for heavy metal derivatives, 0.1% glutaraldehyde was added to the well solution for 16 h. Crystals then were transferred to a stabilizing solution of 0.1 M lithium sulfate with 2.5 mM Tris⋅HCl at pH 6.5, and finally to a freezing solution of 20% PEG 3350/16% glycerol/0.1 M lithium sulfate/2.5 mM sodium citrate/2.5 mM Tris⋅HCl, pH 6.5.
Diffraction Data.
X-ray data were collected from crystals flash-frozen to 100K in a nitrogen stream. Data sets were collected for each of the native and two platinum derivatives [PIP, di-u-iodo-bis(ethylene-diamine)-di-platinum(II)nitrate; PTN, K2Pt(NO2)4]. Three data sets were collected for each native and two derivatives. A data set with the c* axis parallel to the spindle, one perpendicular to the spindle axis, and one low-resolution data set using R-AXIS II and IV image-plate detectors fitted with both mirror and monocapillary optics (28), respectively, were collected. A 2.4-Å native data set was collected at the Advance Photon Source (Argonne, IL) on beam line BM14C by using an Area Detector Systems Corporation (Poway, CA) Quantum-4 charge-coupled device detector. All data sets were collected at 0.5° or 1.0° oscillations and processed and scaled by using hkl (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org; ref. 29).
Structure Solution.
The heavy atom sites of the PIP and PTN derivatives were determined by Patterson methods (30) and SOLVE (31). The space group and hand of the heavy atom sites were determined by using the heavy atom anomalous data from both derivatives. The positions of sites were refined to 4 Å by using SHARP (32), and phasing statistics are shown on Table 1. The 4 Å solvent-flattened (51% solvent) MIRAS-phased electron density map was skeletonized and used to trace the Cα chain of the protein. This map clearly showed the CBD module and the Ig domain. A partial atomic model of the FnIII domains and part of the Ig domain was built by using Lego fragments in o (33). N-linked carbohydrates at asparagines N36, N74, N202, and N226 as well as 80 solvent molecules were included. The disulfide bonds were consistent with the earlier mass-spectrometric data (34). The model then was refined by using the Crystallography and NMR Solution (CNS) program (35), extending the resolution to 2.4 Å by an iterative process involving model rebuilding and gradual identification and refinement of the residues on the longer loops linking the strands. The quality of the electron density and the thermal parameters of the Ig domain were poorer than the CBD indicating a higher degree of disorder. Residues 1–5 and 82–84 were set to zero occupancy in the refinement, because the density was poor and residues beyond 299 (C terminus) were not observable. The side chains of loops 49–52 and 136–138 appeared highly mobile or disordered. Additional solvent (including two sulfate ions) was added during the course of the refinement. A previously reported cysteinyl-cysteine at residue 192 (34) was observed and built into the model. The final R-factor was 0.22 (R-free of 0.29 for 5% of the data) with rmsd in bond lengths and angles of 0.015 Å and 1.96°, respectively, with overall anisotropic temperature factors of B11 = B22 = −6.4 Å2 and B33 = 12.7 Å2 added to the individual atomic isotropic B-values. The coordinates of the IL-6R are deposited in the PDB under ID code .
Modeling the Complex.
The model of the hexameric interleukin-6 signal transduction complex (see Fig. 6a and Fig. 7, which is published as supporting information on the PNAS web site) was assembled by using a parallel version of the protein–protein docking algorithm FTDOCK 2.0 (36). The protein chains were taken from the NMR structure (PDB code ) for IL-6 (37), the x-ray structure of gp130 from the viral IL-6 gp130 complex (PDB code ; ref. 38) and the IL-6R dimer. By using a grid spacing of 0.5 Å and incorporating electrostatic treatment, the following protein–protein docking calculations were undertaken. IL-6 was docked with IL-6R, IL-6 with IL-6R dimer, IL-6 with gp130, and gp130 to IL-6R. Complexes were rescored (39), and the top ranking complexes satisfying the available mutational data (Table 2, which is published as supporting information on the PNAS web site) were subject to rigid body refinement and side-chain rotamer optimization by using the program MULTIDOCK (40). There was no significant difference in the docking solutions produced for IL-6 to IL-6R and IL-6 to the IL-6R dimer, so the solutions of IL-6 to IL-6R dimer were used, with each α-chain in the dimer binding an IL-6 molecule for assembly of the complex. The final complex was built by superimposing the chains in common from the best solutions by using INSIGHT II (Accelrys, San Diego, CA). The resulting complex was then minimized by using CNS (35), with a harmonic restraint applied to the α-carbon backbone (K. Branson, unpublished work). The coordinates of the complex are deposited in the PDB under ID code .
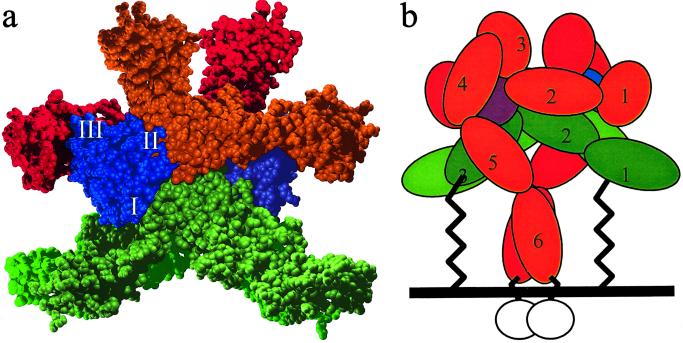
Fig 6.
(a) Space-filling model of the proposed hexameric receptor complex, consisting of two molecules each of IL-6, IL-6R, and gp130 chains (shown in purple and blue, light and dark green, orange and red, respectively). IL-6 is labeled with the binding sites I, II, and III. (b) A modular representation of the hexameric complex (D1–D6 are labeled 1–6) colored as in a, but with the additional three fibronectin domains (D4–D6) of gp130 that have not been modeled placed to indicate how the association of the two D5′ modules from both gp130 molecules, under the “tunnel” of the IL-6R dimer, can activate signaling by dimerization of the cytoplasmic domains (white circles). The membrane proximal extracellular region of IL-6R (49 residues) is shown as an extended chain connecting IL-6R to the membrane.
Results and Discussion
The IL-6R Extracellular Structure.
The complete extracellular region of human IL-6R was expressed and secreted from the CHO cell line, Lec3.2.8.1, which produces glycoprotein with reduced carbohydrate (25). This step was necessary because normal CHO cell lines produce heterogeneously glycosylated protein, which did not crystallize. The Lec3.2.8.1 cell secretes a form of the extracellular domain of IL-6R (residues 1–325) which was purified and crystallized to diffract to 2.4 Å. The IL-6R three-dimensional structure was determined by multiple isomorphous replacement with anomalous scattering from two heavy atom derivatives.
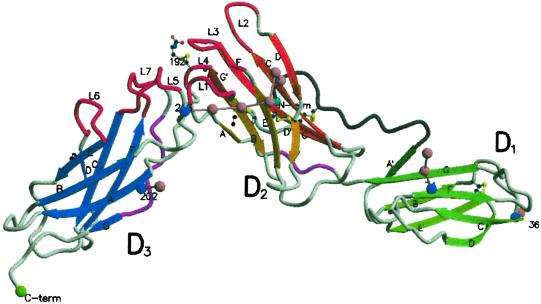
Our x-ray structure of the extracellular domain of the IL-6R (Fig. 1) consists of the N-terminal Ig domain (D1, residues 1–93) linked to a classical CBD (D2, residues 94–194; D3, residues 195–299). The first five residues of the N terminus are poorly defined, and amino acids past residue 299 are not visible in the x-ray structure. The three domains lie on a similar plane, making the receptor a long “flat” structure in which the domains D2 and D3 are connected at ≈90° to each other and D1 is connected at ≈135° to D2. Carbohydrate is attached at four sites on one of the faces of the molecule (see Fig. 8, which is published as supporting information on the PNAS web site), indicating that this face is unlikely to be involved in either the binding of IL-6 or the formation of the signaling complex with gp130. This conclusion is supported by the expression of functional sIL-6R in Escherichia coli, which lacks carbohydrate (41).
Fig 1.
A molscript (56, 57) diagram of sIL-6R indicating the β-sheet arrangement (shades of green, orange, and blue for domains D1, D2, and D3, respectively). The different β-strand shades represent the separate β-sheets in the structure. Four asparagine-linked sites (blue spheres) are shown with their associated carbohydrate moieties represented by pink spheres linked by pink bonds. The N-terminal residue 1–15 (gray tube) is tethered to strand F of D2 by a disulfide bond. The 310 helices (purple) and the loops (red) L1–L7 [L1–L7 consist of the polypeptides S106–N110 (L1), K133–P138 (L2), A160–F168 (L3), Q190–G193 (L4), S227–R233 (L5), M250–H256 (L6) and Q276–Q281 (L7)] that could interact with IL-6 around the juncture of D2 and D3 are shown. The single cysteine residue disulfide linked to C192 is also shown, as are the disulfide links.
The N-terminal cysteine residue C6 is disulfide bonded to residue C174 in D2, and there is an unusual β-sheet arrangement of D1 (Fig. 1). D1 has an S-type Ig topology (14) very similar to the ligand-binding domain of fibroblast growth factor (LBD-FGF; ref. 42), but the first “A” strand of the three-stranded β-sheet of the LBD-FGF Ig domain is not observed in IL-6R. It seems that these residues have peeled away from the sheet when compared with this strand in LBD-FGF (0.7 Å rmsd between the α-carbons of the two domain structures excluding the A strand atoms. The N terminus of this domain forms a fifth A′ strand, hydrogen-bonded to strand G, making a five-stranded β-sheet (A′GFCD) in an S-type Ig topology. This arrangement of the A′ strand of the β-sheet could be a result of crystal packing, and we suggest there is a degree of flexibility of this domain, which acts as a conformational switch, to allow D1 to rotate up to 20° by β-strand shifting. This flexibility could explain what seems to be a higher degree of disorder in this domain when compared with the other domains, as indicated by the higher temperature factors of residues in domain D1 compared with D2 or D3 (mean B-values for domains D1, D2, and D3 are 68 Å2, 49 Å2, and 44 Å2, respectively). The molecules of IL-6R in the crystal are packed head to tail in a double helix arrangement, with the helical axis along the fourfold crystal screw axis in the direction of the long cell edge (c-axis), and the pitch and diameter of the helix will vary if the angle between domains D1 and D2 changes. Such flexibility of domains would explain the lack of isomorphism between different crystals of IL-6R that were observed in this study.
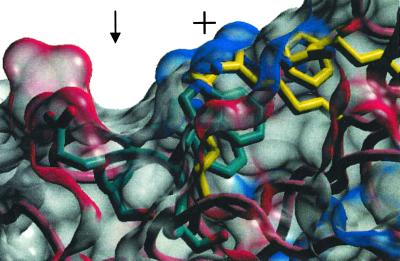
A characteristic of the CBD of IL-6R is the long tryptophane-arginine ladder (from the N terminus end of D3 is R239, F246, R237, W287, R274, W284, Q276) which is also found in other class 1 CBD structures (43). This ladder incorporates the conserved WSXWS motif (residues 284–287 in IL-6R) located in the carboxyl terminus region of domain D3 of the CBD (Fig. 2). The polypeptide backbone of the WSXWS sequence has an unusual motif: a left-handed 310 helix similar to a polyproline helix. This helix is stabilized by the stacking of the indole side chains of the two tryptophan residues of the WSXWS sequence with arginine side chains (R237 and R274), and an H-bonding of serine side-chain hydroxyls but not by main-chain H-bonds. The ladder produces a long surface stripe of positive charge (from the guanadinium and tryptophan indol nitrogens) running along D3 near the inside elbow of D2-D3 and a groove formed by the 310 helix running parallel to the stripe (Fig. 2). Although this structure is conserved in other known structures of CBDs (T. Locsei, unpublished work), its functional role has not been clarified as yet. It could be implicated in the folding of the CBD, complex formation, or it could be involved in a general receptor transport system (44).
Fig 2.
A view of the surface associated with the WSXWS (green bonds) sequence motif in IL-6R and its association with the other residues of the tryptophane/arginine ladder (yellow bonds) in D3. The backbone of the WSXWS motif is shown as part of the 310 helix found in cytokine-binding domains, and a groove (↓) on the surface lies above this helix. A stripe of positively charged (+) residues (arginines) lies parallel to this grove. Nitrogen and oxygen atoms of the side chains are colored blue and red spheres, respectively, as are the surfaces in proximity to these atoms.
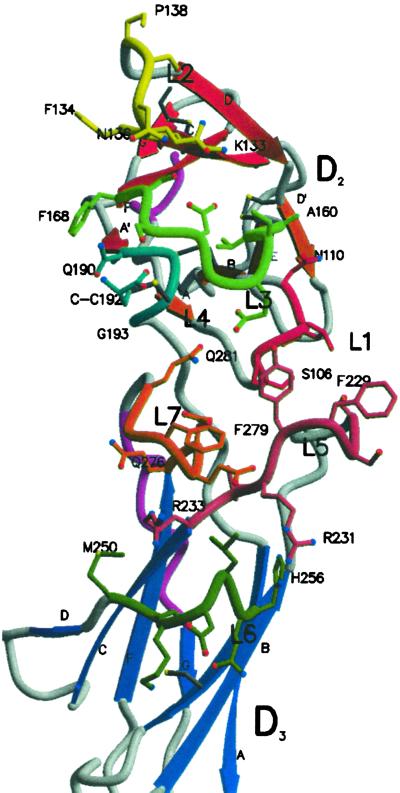
Following the paradigm of GH (45) and the available mutational data (46), it would be expected that IL-6 would bind in the region of the outer elbow formed at the junction of D2 and D3 (Fig. 3), characterized by four loops [S106–N110 (L1), K133–P138 (L2), A160–F168 (L3), Q190–G193 (L4)] from D2 and three loops [S227–R233 (L5), M250–H256 (L6) and Q276–Q281 (L7)] from D3 (Fig. 3). These loops present a long and narrow (43 Å × 9.5 Å) potential binding area held in place by the rigid D2 and D3 framework of the cytokine-binding domains. IL-6 would engage residues in some of these loops.
Fig 3.
A molscript (56, 57) diagram of the CBD of sIL-6R viewed from the top of Fig. 1 showing the IL-6 binding loops L1–L7. The side chains of the loops are shown as ball and stick representations, and the remaining structure is shown as in Fig. 1. The IL-6-binding loops are represented by a backbone worm representation colored red (L1), yellow (L2), green (L3), cyan (L4), pink (L5), dark green (L6), and orange (L7), respectively. The side-chain bonds are color matched to the respective loops. Oxygen, nitrogen, and sulfur atoms in the loop side chains are colored as red, blue, and yellow spheres, respectively (57). The cysteinyl-cysteine C192 is also shown on loop L4.
A Dimer in the Crystal.
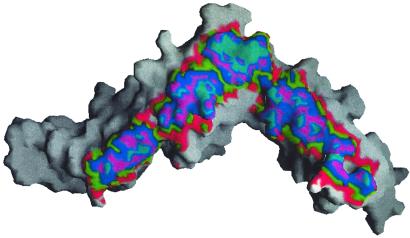
In the crystal lattice, two molecules of IL-6R are related by a crystallographic twofold axis and are closely associated along the length of each molecule (Fig. 4). These two molecules (with a total buried accessible surface area of ≈2,460 Å2) represent a potential physiological dimer of IL-6R. The association is primarily a hydrophobic contact around the twofold axis involving F134 and F168 of domain D2, the salt-link E97-R274, and hydrogen-bonding of the side-chain carboxylate of E283 with the main-chain amide of T186. The buried accessible surface area (Fig. 4) of each molecule [surface- (47) and electrostatic-complementarity (48) of 0.722 and 0.728, respectively] is much larger than that of a protein–protein interaction surface in solution (typically ≈800 Å2). Furthermore, the buried surface of membrane-associated receptor complexes do not have to be as extensive as their counterparts in solution, because their degrees of freedom are restricted to the two-dimensional surface of the membrane. Hence, the dimeric association observed here in the crystal may well represent a complex formed on the cell surface.
Fig 4.
The dimer interface of IL-6R showing the region of interaction by red, green, blue, purple, and cyan areas representing interdimeric distances (58) of less than 5, 4, 3, 2, and 1 Å, respectively. The view is rotated 180° relative to the view in Fig. 1; the crystal twofold axis is vertical, and the membrane is horizontal below the molecule.
The orientation of the monomers with the dimer is such that the interaction is along the carbohydrate-free flat face of the molecule (Fig. 5). In this crystal structure, if the twofold axis of the dimer is perpendicular to the membrane surface, then domain D3 projects away from the membrane at 45° and D2 points down toward the membrane at 30°. D1 would then run parallel to the membrane (Fig. 4). The dimer appears as a bridge structure (Fig. 6) similar to the common β-receptor of IL-3 (43), with a 50-Å long tunnel of triangular crosssection (base ≈40 Å; height ≈22 Å).
Fig 5.
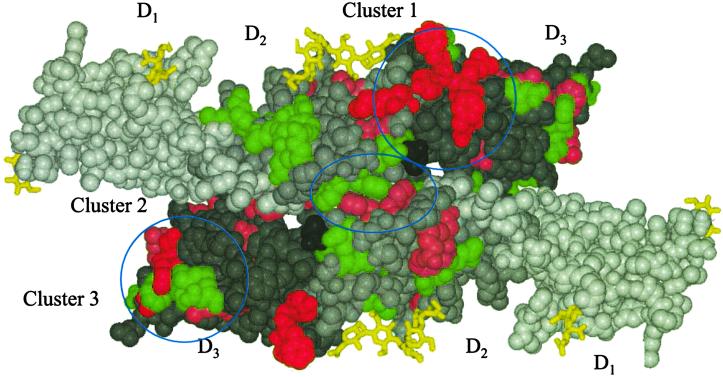
A molscript (56) CPK representation of a crystallographic dimer of IL-6R, viewed toward the membrane anchors, showing the three mutational clusters found on the IL-6R structure. The crystallographic twofold axis lies at the center of the figure and perpendicular to the paper; the dimer interface lies east to west through the twofold axis. Residues in the upper molecule in the figure are colored to represent mutations that affect IL-6 binding to IL-6R (red, pink, and green spheres representing the atoms of residues that have <25%, 25–75%, and >75% of the binding activity of the wild type, respectively). Residues in the lower molecule are colored to represent mutations that affect gp130 signaling (red, pink, and green spheres representing the atoms of residues that have <25%, 25–75%, and >75% of the signaling of wild type, respectively). Residues in domains D1, D2, and D3 that have no mutational data or alter the structure are colored white, light gray, and gray, respectively. The cysteinyl-cysteine C192 is colored dark gray, and carbohydrate is represented by yellow bonds.
Structural Implications of Mutations.
Mapping of the mutational data (17, 49, 50) onto the crystal structure of IL-6R provides some insights into regions on the surface of IL-6R that are critical to binding of IL-6 and are implicated in signaling through gp130. Analysis of these data indicate that most of the mutations, when mapped onto the crystal structure, cause altered binding because of compromised structural integrity of the molecule, in particular, mutations in the tryptophan/arginine ladder, cysteines forming disulfide bonds, and buried hydrophobic residues forming the hydrophobic core. The nonstructural mutations occur on three clusters on the surface of IL-6R (Fig. 5).
There is clearly a cluster of residues (see Fig. 5, Cluster 1) that form the binding site to IL-6. The mutations of residues having the greatest effect on IL-6 binding are those at P162 and E163 from L3, S228, F229, Y230, and R231 from L5, and E278 and F279 from L7. Two of these loops, L5 and L7, are both situated on D3, and this is consistent with the report that D3 by itself is able to bind IL-6 (18). This site at the juncture of D2 and D3 domains can be inferred to be primarily responsible for IL-6 binding.
Nonstructural mutations in IL-6R that affect gp130 signaling (Fig. 5) can be classified onto two main clusters. The first cluster occurs at a hydrophobic patch (residues F134 and F168) at the dimer interface (Fig. 5, Cluster 2) located around the crystallographic twofold axis relating the two molecules of the dimer. Mutations here reduce signaling but have no effect on binding. There are also other residues, not included in Cluster 2, on domain D3 in this interface that reduce signaling but not binding (see Fig. 5). These data indicate that mutations that would be expected to interfere with the formation of the IL-6R dimer have a significant effect on IL-6 signaling but not on IL-6 binding.
The other cluster of mutations affecting gp130 signaling occurs on a patch of residues (Fig. 5, Cluster 3) centered around residue H261 on domain D3. Mutational studies in this region of IL-6R were based on the GH paradigm, which demonstrated a contact region between the second CBD domains of the GH complex (45). This patch is clearly not involved in IL-6 binding or IL-6R dimerization, but when mutated, this region affects signaling. This region is most likely one that is involved in the formation of the IL-6/IL-6R complex with gp130.
Kalai et al. (51) predicted that two SSFY sequence repeats in IL-6R were involved in binding sites to IL-6. The first repeat S165-Y169 is located in L3 of D2. Mutations in S167 and Y169 reduce signaling, but this is likely because of the structural importance of these buried residues. F168 is solvent exposed and forms the central part of Cluster 2 that is involved in IL-6R dimer formation but not IL-6 binding. A second aromatic residue, F134, is the other major residue in Cluster 2. The second SSFY repeat (S227–Y230) is located in L5 on D3 of IL-6R (Fig. 3). These residues (apart from S227) are critical to IL-6 binding and are all surface-exposed residues.
Salvati et al. (50) reported that N211, H261, and D262 together form a group of residues that affect signaling but not binding of IL-6 to IL-6R. Surprisingly, these residues cluster with W214 and V259 and, when mutated (to N and Q, respectively), increase the signaling of IL-6 over wild type (17). The role of the cysteinyl-cysteine C192 remains unsolved. The residue is on the periphery the IL-6 binding site. When C192 is mutated to alanine, the binding of IL-6 to IL-6R and signaling are slightly increased. The human IL-6R sequence is the only vertebrate sequence in the database to date that has a cysteine in this position: pig and cow have a tyrosine in this position and mouse and rat have a leucine (34).
The Hexameric Signaling Complex.
A hexameric complex (Fig. 6a) incorporating the IL-6R dimer can be constructed in a manner that is compatible with the available mutational and functional data present in the literature except for the mutations in Cluster 3, which has no cognate binding partner in this model. Additionally, the placement of the three C-terminal fibronectin type III domains (D4, D5, and D6) from gp130 has not been considered. Several orientations of these domains are possible, but in the absence of any structural information, they are difficult to dock with sufficient surety. By using homology models of these three domains, one can orient in such a way so they contact Cluster 3 on D3 of IL-6R and form a disulfide link proposed (52) between D5 of gp130 underneath the complex, as shown schematically in Fig. 6b.
The recent structure of the Ig and CBD domain of gp130 complexed with viral IL-6 (38) reveals a possible dimeric relationship between the human IL-6 and the binding domains of gp130. The dimeric relationship of the viral IL-6/gp130 complex is a crystallographic twofold relationship, as in the IL-6R dimer. Incorporating the same dimeric relationship between all of the IL-6 binding receptor domains, it is possible to construct a model for the signaling complex (Fig. 6a). In this model, two IL-6 molecules bind to the IL-6R dimer by means of site I (Fig. 6), followed by two gp130 molecules each binding through sites II and III of different IL-6 molecules in a manner similar to the viral IL-6/gp130 fragment complex. The signaling molecule gp130 would bind the IL-6/IL-6R complex with D3 pointing away from the membrane. The remaining three FnIII domains of each gp130 would orient toward the membrane, and the signaling activated by dimerization and possible disulfide cross-linking of the gp130 membrane proximal FnIII domains (Fig. 6b) under the bridge of the IL-6R dimer. It is possible that domain D1 of IL-6R interacts with gp130 by means of the proposed conformational switch in D1 of IL-6R discussed earlier. Although this model is substantially different from current proposed models (2, 46), it is consistent with the available biological data (17, 49, 50). Other gp130 signaling complexes (e.g., IL-11) could act through a similar mechanism. A notable feature of this model is the binding orientation of IL-6 to IL-6R: it is rotated 180° compared with the analogous positions in GH (45), prolactin (53), erythropoietin (54) receptor structures, and the IL-12 structure (55). The validity of such a hexameric complex arrangement will be tested as more biophysical and biological data accumulates.
The crystal structure of IL-6R and the proposed model of the hexameric complex will provide the basis for the design of mutations of the proteins involved in this complex. It will also enable the design of small molecular weight antagonists and agonists to IL-6 signaling that can be developed into therapeutics targeted to the diseases modulated by IL-6 signaling.
Supplementary Material
Acknowledgments
We thank A. W. Burgess for critically reading the manuscript, G. Moont for the mpi version of ftdock v.2.0, and B. J. Smith for helpful discussions on the computational aspects of modeling. We also thank the Australian Synchrotron Research Program for funding and access to the Advance Photon Source and Cooperative Research Center–Cellular Growth factors for partial funding.
Abbreviations
CBD, cytokine-binding domain
CHO, Chinese hamster ovary
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Kishimoto T., Akira, S., Narazaki, M. & Taga, T. (1995) Blood 86, 1243-1254. [PubMed] [Google Scholar]
- 2.Simpson R. J., Hammacher, A., Smith, D. K., Matthews, J. M. & Ward, L. D. (1997) Protein Sci. 6, 929-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich P. C., Behrmann, I., Muller-Newen, G., Schaper, F. & Graeve, L. (1998) Biochem. J. 334, 297-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bravo J. & Heath, J. K. (2000) EMBO J. 19, 2399-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naka T., Nishimoto, N. & Kishimoto, T. (2002) Arthritis Res. 4, S233-S242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward L. D., Howlett, G. J., Discolo, G., Yasukawa, K., Hammacher, A., Moritz, R. L. & Simpson, R. J. (1994) J. Biol. Chem. 269, 23286-23289. [PubMed] [Google Scholar]
- 7.Klein B., Wijdenes, J., Zhang, X. G., Jourdan, M., Boiron, J. M., Brochier, J., Liautard, J., Merlin, M., Clement, C., Morel-Fournier, B., et al. (1991) Blood 78, 1198-1204. [PubMed] [Google Scholar]
- 8.Sugiyama T. (2001) J. Bone Miner. Metab. 19, 89-96. [DOI] [PubMed] [Google Scholar]
- 9.Nishimoto N., Kishimoto, T. & Yoshizaki, K. (2000) Ann. Rheum. Dis. 59, Suppl. 1, i21-i27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Twillie D. A., Eisenberger, M. A., Carducci, M. A., Hseih, W. S., Kim, W. Y. & Simons, J. W. (1995) Urology 45, 542-549. [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki K., Taga, T., Hirata, Y., Yawata, H., Kawanishi, Y., Seed, B., Taniguchi, T., Hirano, T. & Kishimoto, T. (1988) Science 241, 825-828. [DOI] [PubMed] [Google Scholar]
- 12.Hirata Y., Taga, T., Hibi, M., Nakano, N., Hirano, T. & Kishimoto, T. (1989) J. Immunol. 143, 2900-2906. [PubMed] [Google Scholar]
- 13.Bazan J. F. (1990) Proc. Natl. Acad. Sci. USA 87, 6934-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bork P., Holm, L. & Sander, C. (1994) J. Mol. Biol. 242, 309-320. [DOI] [PubMed] [Google Scholar]
- 15.Leahy D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992) Science 258, 987-991. [DOI] [PubMed] [Google Scholar]
- 16.Taga T., Hibi, M., Hirata, Y., Yawata, H., Natsuka, S., Yasukawa, K., Totsuka, T., Yamasaki, K., Hirano, T. & Kishimoto, T. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, Pt. 2, 713-722. [DOI] [PubMed] [Google Scholar]
- 17.Yawata H., Yasukawa, K., Natsuka, S., Murakami, M., Yamasaki, K., Hibi, M., Taga, T. & Kishimoto, T. (1993) EMBO J. 12, 1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozbek S., Grotzinger, J., Krebs, B., Fischer, M., Wollmer, A., Jostock, T., Mullberg, J. & Rose-John, S. (1998) J. Biol. Chem. 273, 21374-21379. [DOI] [PubMed] [Google Scholar]
- 19.Heaney M. L. & Golde, D. W. (1996) Blood 87, 847-857. [PubMed] [Google Scholar]
- 20.Narazaki M., Yasukawa, K., Saito, T., Ohsugi, Y., Fukui, H., Koishihara, Y., Yancopoulos, G. D., Taga, T. & Kishimoto, T. (1993) Blood 82, 1120-1126. [PubMed] [Google Scholar]
- 21.Jones S. A., Horiuchi, S., Topley, N., Yamamoto, N. & Fuller, G. M. (2001) FASEB J. 15, 43-58. [DOI] [PubMed] [Google Scholar]
- 22.Mullberg J., Schooltink, H., Stoyan, T., Gunther, M., Graeve, L., Buse, G., Mackiewicz, A., Heinrich, P. C. & Rose-John, S. (1993) Eur. J. Immunol. 23, 473-480. [DOI] [PubMed] [Google Scholar]
- 23.Horiuchi S., Koyanagi, Y., Zhou, Y., Miyamoto, H., Tanaka, Y., Waki, M., Matsumoto, A., Yamamoto, M. & Yamamoto, N. (1994) Eur. J. Immunol. 24, 1945-1948. [DOI] [PubMed] [Google Scholar]
- 24.Liautard J., Gaillard, J. P., Mani, J. C., Montero-Julian, F., Duperray, C., Lu, Z. Y., Jourdan, M., Klein, B., Brailly, H. & Brochier, J. (1994) Eur. Cytokine Network 5, 293-300. [PubMed] [Google Scholar]
- 25.Stanley P. (1989) Mol. Cell. Biol. 9, 377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasukawa K., Saito, T., Fukunaga, T., Sekimori, Y., Koishihara, Y., Fukui, H., Ohsugi, Y., Matsuda, T., Yawata, H., Hirano, T., et al. (1990) J. Biochem. (Tokyo) 108, 673-676. [DOI] [PubMed] [Google Scholar]
- 27.Ward L. D., Howlett, G. J., Hammacher, A., Weinstock, J., Yasukawa, K., Simpson, R. J. & Winzor, D. J. (1995) Biochemistry 34, 2901-2907. [DOI] [PubMed] [Google Scholar]
- 28.Balaic D. X., Barnea, Z., Nugent, K. A., Garrett, R. F., Varghese, J. N. & Wilkins, S. W. (1996) J. Synchrotron Rad. 3, 289-295. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z. & Minor, W. (1997) Macromol. Crystallogr. 276, Pt. A, 307-326. [Google Scholar]
- 30.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 31.Terwilliger T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, Pt. 4, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Fortelle E. & Bricogne, G. (1997) Macromol. Crystallogr. 276, Pt. A, 472-494. [DOI] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, Pt. 2, 110-119. [DOI] [PubMed] [Google Scholar]
- 34.Cole A. R., Hall, N. E., Treutlein, H. R., Eddes, J. S., Reid, G. E., Moritz, R. L. & Simpson, R. J. (1999) J. Biol. Chem. 274, 7207-7215. [DOI] [PubMed] [Google Scholar]
- 35.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, Pt. 5, 905-921. [DOI] [PubMed] [Google Scholar]
- 36.Gabb H. A., Jackson, R. M. & Sternberg, M. J. (1997) J. Mol. Biol. 272, 106-120. [DOI] [PubMed] [Google Scholar]
- 37.Xu G. Y., Yu, H. A., Hong, J., Stahl, M., McDonagh, T., Kay, L. E. & Cumming, D. A. (1997) J. Mol. Biol. 268, 468-481. [DOI] [PubMed] [Google Scholar]
- 38.Chow D., He, X., Snow, A. L., Rose-John, S. & Garcia, K. C. (2001) Science 291, 2150-2155. [DOI] [PubMed] [Google Scholar]
- 39.Moont G., Gabb, H. A. & Sternberg, M. J. (1999) Proteins 35, 364-373. [PubMed] [Google Scholar]
- 40.Jackson R. M., Gabb, H. A. & Sternberg, M. J. (1998) J. Mol. Biol. 276, 265-285. [DOI] [PubMed] [Google Scholar]
- 41.Stoyan T., Michaelis, U., Schooltink, H., Van Dam, M., Rudolph, R., Heinrich, P. C. & Rose-John, S. (1993) Eur. J. Biochem. 216, 239-245. [DOI] [PubMed] [Google Scholar]
- 42.Stauber D. J., DiGabriele, A. D. & Hendrickson, W. A. (2000) Proc. Natl. Acad. Sci. USA 97, 49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carr P. D., Gustin, S. E., Church, A. P., Murphy, J. M., Ford, S. C., Mann, D. A., Woltring, D. M., Walker, I., Ollis, D. L. & Young, I. G. (2001) Cell 104, 291-300. [DOI] [PubMed] [Google Scholar]
- 44.Hilton D. J., Watowich, S. S., Katz, L. & Lodish, H. F. (1996) J. Biol. Chem. 271, 4699-4708. [DOI] [PubMed] [Google Scholar]
- 45.de Vos A. M., Ultsch, M. & Kossiakoff, A. A. (1992) Science 255, 306-312. [DOI] [PubMed] [Google Scholar]
- 46.Grötzinger J., Kernebeck, T., Kallen, K.-J. & Rose-John, S. (1999) Biol. Chem. 380, 803-813. [DOI] [PubMed] [Google Scholar]
- 47.Lawrence M. C. & Colman, P. M. (1993) J. Mol. Biol. 234, 946-950. [DOI] [PubMed] [Google Scholar]
- 48.McCoy A. J., Epa, V. C. & Colman, P. M. (1997) J. Mol. Biol. 268, 570-584. [DOI] [PubMed] [Google Scholar]
- 49.Kalai M., Montero-Julian, F. A., Grotzinger, J., Fontaine, V., Vandenbussche, P., Deschuyteneer, R., Wollmer, A., Brailly, H. & Content, J. (1997) Blood 89, 1319-1333. [PubMed] [Google Scholar]
- 50.Salvati A. L., Lahm, A., Paonessa, G., Ciliberto, G. & Toniatti, C. (1995) J. Biol. Chem. 270, 12242-12249. [DOI] [PubMed] [Google Scholar]
- 51.Kalai M., Montero-Julian, F. A., Grotzinger, J., Wollmer, A., Morelle, D., Brochier, J., Rose-John, S., Heinrich, P. C., Brailly, H. & Content, J. (1996) Eur. J. Biochem. 238, 714-723. [DOI] [PubMed] [Google Scholar]
- 52.Moritz R. L., Hall, N. E., Connolly, L. M. & Simpson, R. J. (2001) J. Biol. Chem. 276, 8244-8253. [DOI] [PubMed] [Google Scholar]
- 53.Elkins P. A., Christinger, H. W., Sandowski, Y., Sakal, E., Gertler, A., de Vos, A. M. & Kossiakoff, A. A. (2000) Nat. Struct. Biol. 7, 808-815. [DOI] [PubMed] [Google Scholar]
- 54.Cheetham J. C., Smith, D. M., Aoki, K. H., Stevenson, J. L., Hoeffel, T. J., Syed, R. S., Egrie, J. & Harvey, T. S. (1998) Nat. Struct. Biol. 5, 861-866. [DOI] [PubMed] [Google Scholar]
- 55.Yoon C., Johnston, S. C., Tang, J., Stahl, M., Tobin, J. F. & Somers, W. S. (2000) EMBO J. 19, 3530-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 57.Merritt E. A. & Bacon, D. J. (1997) Macromol. Crystallogr. 277, Pt. B, 505-524. [Google Scholar]
- 58.Nicholls A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.