Abstract
The enzyme POR [Pchlide (protochlorophyllide) oxidoreductase] catalyses the reduction of Pchlide to chlorophyllide, which is a key step in the chlorophyll biosynthesis pathway. This light-dependent reaction has previously been studied in great detail but recent reports suggest that a mixture of MV (monovinyl) and DV (divinyl) Pchlides may have influenced some of these properties of the reaction. Low-temperature absorbance and fluorescence spectroscopy have revealed several spectral differences between MV and DV Pchlides, which were purified from a Rhodobacter capsulatus strain that was shown to contain a mixture of the two pigments. A thorough steady-state kinetic characterization using both Pchlide forms demonstrates that neither pigment appears to affect the kinetic properties of the enzyme. The reaction has also been monitored following illumination at low temperatures and was shown to consist of an initial photochemical step followed by four ‘dark’ steps for both pigments. However, minor differences were observed in the spectral properties of some of the intermediates, although the temperature dependency of each step was nearly identical for the two pigments. This work provides the first detailed kinetic and spectroscopic study of this unique enzyme using biologically important MV and DV substrate analogues. It also has significant implications for the DV reductase enzyme, which is responsible for converting DV pigments into their MV counterparts, and its position in the sequence of reactions that comprise the chlorophyll biosynthesis pathway.
Keywords: chlorophyll biosynthesis, chlorophyllide, divinyl, low-temperature absorbance, monovinyl, protochlorophyllide oxidoreductase
Abbreviations: Chlide, chlorophyllide; DV, divinyl; MV, monovinyl; Pchlide, protochlorophyllide; POR, Pchlide oxidoreductase
INTRODUCTION
The chlorophyll biosynthesis enzyme POR [Pchlide (protochlorophyllide) oxidoreductase; NADPH:protochlorophyllide oxidoreductase; EC 1.3.1.33] catalyses the reduction of the C-17–C-18 double bond of the D-ring of Pchlide to form Chlide (chlorophyllide; Scheme 1) [1,2]. The reaction is driven by light and consequently it is an important regulatory step in the chlorophyll biosynthetic pathway and in the subsequent assembly of the photosynthetic apparatus [2]. It is one of the only two enzymes known to require light for catalysis; the other is DNA photolyase [3]. In addition to POR, non-flowering land plants, algae and cyanobacteria possess a light-independent Pchlide reductase, consisting of three separate subunits, which is able to operate in the dark [4].
Scheme 1. Scheme of the chlorophyll biosynthesis pathway, illustrating the possible locations of the DV reductase enzyme.
POR catalyses the reduction of the C-17–C-18 double bond of Pchlide and DV reductase catalyses the reduction of the vinyl group at the C-8 position of the tetrapyrrole molecule to an ethyl group. As MV and DV Pchlides are accepted equally as substrates by POR, we have shown that DV reductase can operate before or after the reduction of Pchlide. Therefore DV Chlide must act as a substrate for DV reductase (solid arrow), whereas the dashed arrows indicate DV reductase reactions that are still to be established. The dashed boxes indicate the groups that are reduced during each reaction and the IUPAC numbering system of the tetrapyrrole ring is shown.
POR is a member of the ‘RED’ (Reductases, Epimerases, Dehydrogenases) superfamily of enzymes [5,6], which generally catalyse NADP(H)- or NAD(H)-dependent reactions involving hydride and proton transfers [7]. Indeed, a homology model of POR has been constructed using other family members as a structural template [8]. All members of this large protein family contain highly conserved Tyr and Lys residues, which have been shown to be essential for activity. In the reaction catalysed by POR, it has been proposed that the conserved Tyr residue donates a proton to the C-18 position with the Lys residue thought to be important for lowering the apparent pKa of the Tyr, allowing deprotonation to occur [5]. The hydride is transferred from the pro-S face of NADPH to the C-17 position of the Pchlide molecule [9,10].
The fact that POR is light-activated means the enzyme–substrate complex can be formed in the dark, removing the diffusive components out of the reaction. This has recently been exploited by studying Pchlide reduction at low temperatures to trap intermediates in the reaction pathway [11–13]. As a result, the reaction has been shown to consist of at least three distinct steps: an initial light-driven step, followed by a series of ‘dark’ reactions. An initial photochemical step can occur below 200 K [11], whereas two ‘dark’ steps were identified for Synechocystis POR, which can only occur close to or above the ‘glass transition’ temperature of proteins [12]. This implies a role for protein motions during these stages of the catalytic mechanism. A thermophilic form of the enzyme has been used to identify two additional ‘dark’ steps, which were shown to represent a series of ordered product release and cofactor binding events. First, NADP+ is released from the enzyme and then replaced by NADPH, before release of the Chlide product and subsequent binding of Pchlide have taken place [13]. Additionally, the reaction has been shown to proceed on an ultrafast timescale after catalysis was initiated with a 50 fs laser pulse. This suggests that the dynamic motions that accompany catalysis must therefore occur on similar short timescales [14].
By using a range of Pchlide analogues, it has been possible to gain information about the structural regions of the pigment that are essential for catalysis [15]. Pchlide analogues with different side chains at the C-17 position could not be used as substrates by POR from oat etioplasts, which suggests that the free carboxylic acid group (C-17) plays an important role in the binding of the pigment to the enzyme. Similarly, the central Mg atom and the structure of the isocyclic ring of the Pchlide molecule have also been shown to be crucial for activity. Conversely, the enzyme does not differentiate between vinyl and ethyl groups at the C-8 position of ring B as both MV (monovinyl) and DV (divinyl) forms of Pchlide can be converted into the respective Chlides [15]. However, there are currently no quantitative kinetic or spectroscopic data to establish the relative merits of the MV and DV forms of Pchlide as substrates for POR.
An additional reason for requiring such data arises from recent reports suggesting that impurities in the pigment preparations or the use of a mixed pool of MV and DV Pchlides may influence the kinetics and spectral properties of the POR-catalysed reaction [16,17]. Therefore it is important to analyse the reaction by using highly purified preparations of MV and DV Pchlide. In addition, a preference for MV or DV Pchlide may be an important factor in determining the exact location in the pathway where the DV reductase enzyme [18,19] is able to operate. This enzyme has recently been identified in Arabidopsis thaliana and is proposed to catalyse the conversion of either DV Pchlide or DV Chlide into the respective MV pigments (Scheme 1) [18]. In the present study, MV and DV forms of Pchlide have been purified, allowing the first detailed kinetic and spectroscopic characterization of the reaction with both pigments.
MATERIALS AND METHODS
Preparation and separation of MV and DV Pchlides
Pchlide was isolated from Rhodobacter capsulatus ZY5 cultures as described previously [12]. MV and DV Pchlides were purified on a preparative scale by HPLC using C30 reverse-phase column (3 μm, 250 mm×4.6 mm; YMC Europe GmbH) as described previously [17]. For analytical purposes, MV and DV Pchlides were separated using the same column in methanol/acetonitrile/hexane (7:2:1, by vol.) at 0.8 or 1 ml·min−1 flow rate and absorption detection at 437 nm. The equipment used was a Jasco PU-980 Pump, a Jasco UV-970 UV/VIS detector and Borwin v. 1.5 software.
Steady-state measurements
POR from the cyanobacterium Thermosynecoccus elongatus BP-1 was overproduced and purified from Escherichia coli as described previously [13]. Activity assays were carried out as described previously [20] in the presence of 5 μM NADPH. The Km for Pchlide and Vmax values were obtained by fitting the initial rates of Chlide synthesis against the concentration of Pchlide using the following equation:
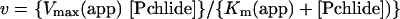 |
(1) |
where v is the initial rate, Vmax (app) is the initial rate achieved as [Pchlide] approaches infinity and Km (app) is the apparent value of [Pchlide] giving Vmax/2. The data were fitted and standard errors calculated by nonlinear regression analysis using the Sigma Plot program (SPSS).
The binding of Pchlide to POR was measured by performing 77 K fluorescence emission spectra as described previously [11]. The apparent Kd values were obtained by fitting the fluorescence changes against the concentration of POR using the following equation:
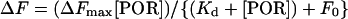 |
(2) |
where ΔF is the change in the ratio of fluorescence emission at 631 nm/644 nm upon binding to Pchlide, ΔFmax is the apparent maximum change in the fluorescence ratio, Kd is the apparent dissociation constant for Pchlide binding to POR and F0 is the initial ratio of fluorescence emission at 631 nm/644 nm. The data were fitted and standard errors were calculated by nonlinear regression analysis using the Sigma Plot program (SPSS).
Low-temperature absorbance and fluorescence spectroscopy measurements
All spectra were measured in 44% (v/v) glycerol, 20% (w/v) sucrose, 50 mM Tris/HCl (pH 7.5), 0.1% (v/v) Genapol X-080 and 0.1% (v/v) 2-mercaptoethanol and samples were maintained at the required temperature using an OpstistatDN nitrogen bath cryostat (Oxford Instruments). The temperature of the sample was monitored directly with a thermocouple sensor (Comark) and intermediates in the reaction pathway were isolated as described previously [12]. Fluorescence spectra were recorded using a SPEX FluoroLog spectrofluorimeter (Jobin Yvon) at 77 K. The exciting light was provided from a xenon light source, using excitation monochromator slit widths of 4.5 nm and emission monochromator slit widths of 3.6 nm. Absorbance spectra were recorded with a Cary 500 Scan UV–visible-NIR (Varian) spectrophotometer at 77 K. Normalization of the spectra, spectral deconvolutions and difference spectra were calculated using the Galactics software.
RESULTS
Analysis of Pchlide from R. capsulatus
Analysis of the Pchlide obtained from R. capsulatus cells showed that the pigment was a mixture of DV and MV Pchlides in a 5:1 molar ratio (results not shown). The MV and DV Pchlides were purified on a C30 reverse-phase column and the pigments that were used for all further experiments showed no cross-contamination.
Spectroscopic differences between MV and DV Pchlides
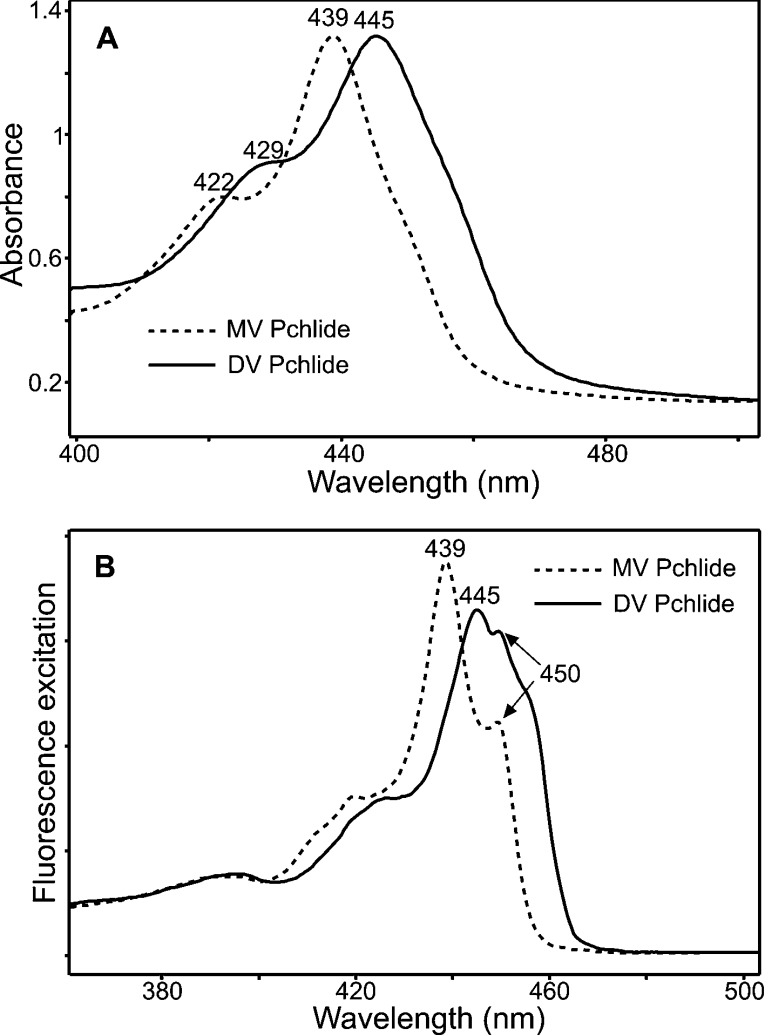
The MV and DV Pchlides were characterized by using 77 K absorbance and fluorescence spectroscopy. Low-temperature absorbance measurements revealed that MV Pchlide has an absorbance maximum at 629 nm and that this peak is slightly red-shifted to 630 nm for DV Pchlide (results not shown). However, in the Soret region, there are more considerable differences in the spectra of the two pigments. MV Pchlide has two main absorbance bands at 422 and 439 nm, whereas DV Pchlide has two major absorbance peaks at 429 and 445 nm (Figure 1A). A similar phenomenon is also observed for the low-temperature fluorescence spectra. MV Pchlide has a single fluorescence band at 631 nm, whereas DV Pchlide has a fluorescence maximum at 632 nm (results not shown). Figure 1(B) shows the excitation spectra of these emission bands, which further emphasizes the distinctive properties of the two pigments. The fluorescence band at 631 nm, arising from MV Pchlide, has two main excitation peaks at 439 and 450 nm, whereas the excitation spectrum of the DV Pchlide band at 632 nm has two maxima at 445 and 450 nm and a shoulder at 457 nm.
Figure 1. Low-temperature absorbance and fluorescence excitation spectra of MV and DV Pchlides.
(A) The 77 K absorbance spectra of MV and DV Pchlides (6.5 μM). (B) The 77 K fluorescence excitation spectra of the 632 and 631 nm emission bands of MV and DV Pchlides (1.1 μM). The spectra are normalized for ease of clarification.
Steady-state kinetic and binding measurements
The kinetic parameters for the reaction were measured over a range of MV and DV Pchlide concentrations with NADPH in excess. For both pigments, the dependence of the initial rate on Pchlide concentration followed Michaelis–Menten kinetics. The Vmax was determined to be 0.53±0.05 μM·min−1 for MV Pchlide and 0.61±0.05 μM·min−1 for DV Pchlide. The Km for MV Pchlide was calculated to be 1.36±0.34 μM and the Km for DV Pchlide was 0.92±0.33 μM.
The binding of both pigments to the enzyme has been monitored by measuring Pchlide fluorescence emission spectra at 77 K. The binding constant was determined by measuring the ratio of the bound (644 nm) and unbound (631 nm) Pchlide peaks at a range of enzyme concentrations. Under these conditions, the apparent dissociation constant was calculated to be 1.37±0.28 μM for the binding of MV Pchlide and 0.83±0.24 μM for the binding of DV Pchlide.
Initial light-driven step
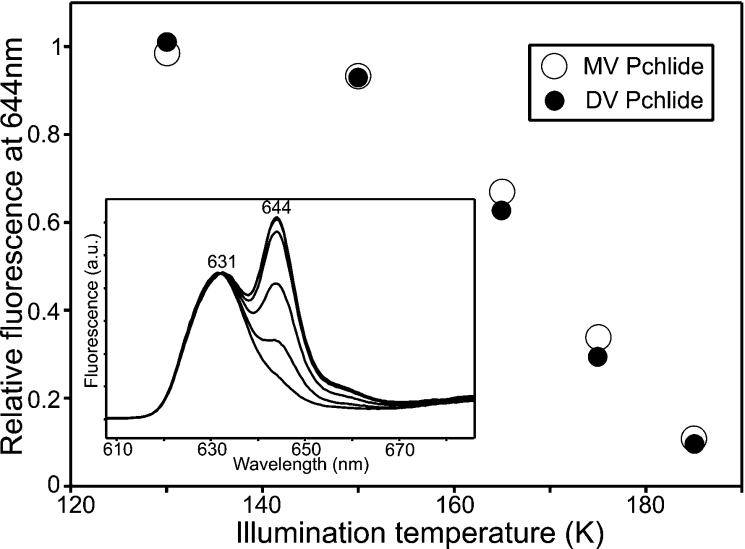
Five distinct steps in the reaction pathway have previously been identified for the T. elongatus enzyme, an initial light-driven one followed by four ‘dark reactions’ [13]. In the present study, all of these steps have been analysed for both the MV and DV Pchlides by using low-temperature fluorescence measurements. The initial photochemistry, which involves the conversion of the POR–Pchlide–NADPH fluorescence band at 644 nm into a non-fluorescent intermediate (Figure 2, inset), was almost identical for the two pigments. A plot of the temperature dependence revealed that the light-driven step occurred over the same temperature range for both the MV and DV Pchlides (Figure 2).
Figure 2. Characterization of the initial light-dependent step of the reaction for MV and DV Pchlides.
The inset shows the 77 K fluorescence emission spectra of samples containing 1.1 μM DV Pchlide, 30 μM POR and 200 μM NADPH after illumination for 10 min at 130, 150, 165, 175 and 185 K. The intensity of the fluorescence band at 644 nm was measured at 77 K using an excitation wavelength of 450 nm. The temperature dependence of the initial photochemical step for MV and DV Pchlides, derived from these spectra, is shown. The decrease in fluorescence after illumination for 10 min at the respective temperatures was calculated relative to the largest decrease.
The ‘dark’ steps
Any differences that may exist between the two pigments during the subsequent ‘dark’ reactions have also been explored. All samples were illuminated at 185 K for 10 min to trigger the initial photochemical step and were then warmed to progressively higher temperatures between 185 and 330 K in the dark. During the first ‘dark’ step, the non-fluorescent intermediate is converted into a new state that has previously been shown to represent a POR–Chlide–NADP+ complex. For the MV pigment, this new species fluoresces at 685 nm, but in the DV sample the fluorescent maximum is slightly blue-shifted at 684 nm (results not shown). The temperature dependence of this first ‘dark’ step, which is calculated by measuring the increase in fluorescence at 684 nm/685 nm, is very similar for both pigments (results not shown).
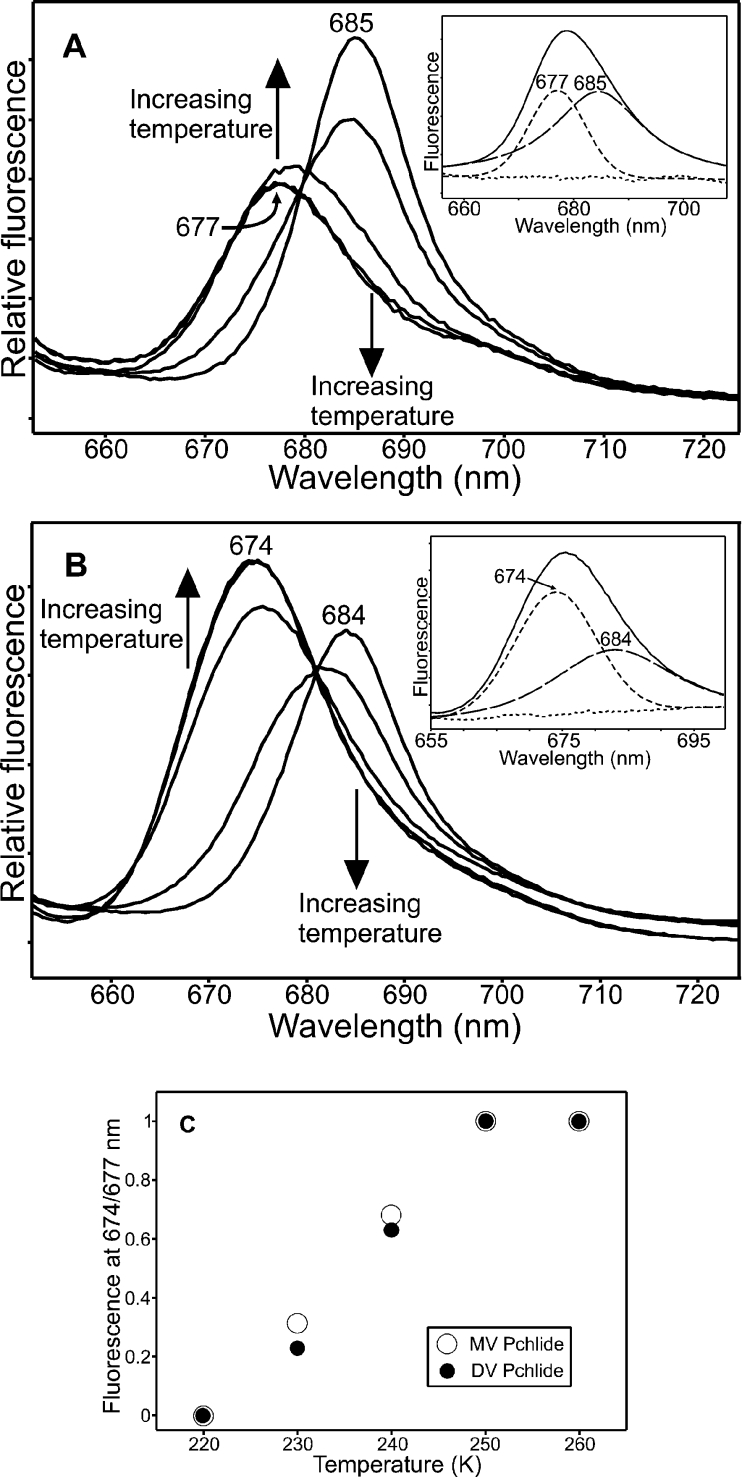
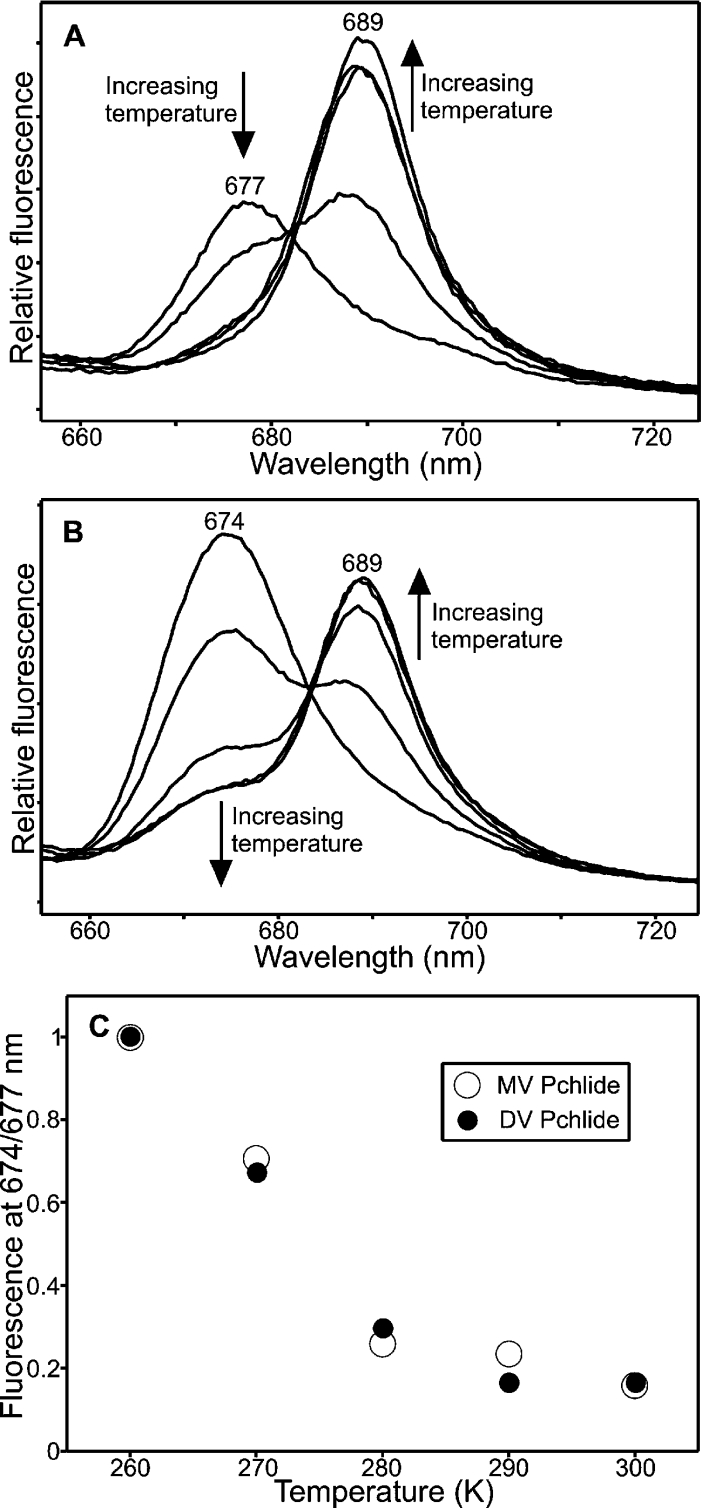
Conversely, significant differences between the two pigments are observed during the remaining ‘dark’ steps of the reaction. For both pigments, the second ‘dark’ step involves a decrease in the POR–Chlide–NADP+ fluorescence band at either 684 or 685 nm, but the states that are formed as a result exhibit differing spectral properties. In the MV samples, the new fluorescence band is centred at 677 nm with a half-bandwidth of 12 nm (Figure 3A), whereas in the DV samples the new species has a fluorescence peak at 674 nm with a half-bandwidth of 15 nm (Figure 3B). In addition, the fluorescence yield from the DV species at 674 nm is considerably greater than that from the MV species at 677 nm. The temperature dependence of this step, which is obtained by measuring the increase in fluorescence intensity at either 674 or 677 nm, shows that it can occur between 220 and 260 K and is almost identical between the two pigments (Figure 3C).
Figure 3. Spectroscopic characterization of the second ‘dark’ step of the reaction for MV and DV Pchlides.
The 77 K fluorescence emission spectra of samples containing 1.1 μM MV Pchlide (A) or DV Pchlide (B), 30 μM POR and 200 μM NADPH after illumination for 10 min at 185 K and incubation in the dark for 10 min at 220, 230, 240, 250 and 260 K are shown. The arrows indicate the formation of a fluorescent band at either 677 nm (MV Pchlide) or 674 nm (DV Pchlide) and the disappearance of the fluorescent band at either 685 nm (MV Pchlide) or 684 nm (DV Pchlide) at increasing temperatures. Spectra were recorded with an excitation wavelength of 450 nm. The respective insets show deconvolutions of the 240 K spectra with the residuals represented by the small dotted lines. (C) The temperature dependence of the second ‘dark’ step was calculated by measuring the relative increase in fluorescence at either 677 nm (MV Pchlide) or 674 nm (DV Pchlide) over the temperature range 220–260 K.
For both pigment types, the third ‘dark’ step involves the disappearance of the fluorescence band at either 674 or 677 nm together with the formation of a new state, which fluoresces at 689 nm (Figures 4A and 4B). The temperature dependence of the third ‘dark’ step, which can be measured by plotting out the relative fluorescence at either 674 or 677 nm, revealed that it could occur over the same temperature range for both the MV and DV pigments (Figure 4C).
Figure 4. Spectroscopic characterization of the third ‘dark’ step of the reaction for MV and DV Pchlides.
The 77 K fluorescence emission spectra of samples containing 1.1 μM MV Pchlide (A) or DV Pchlide (B), 30 μM POR and 200 μM NADPH after illumination for 10 min at 185 K and incubation in the dark for 10 min at increasing temperatures. Samples were incubated at 260, 270, 280, 290 and 300 K are shown. The formation of the fluorescence band at 689 nm and the simultaneous disappearance of the fluorescence band at either 677 nm (MV Pchlide) or 674 nm (DV Pchlide) at higher temperatures are indicated by the arrows. Spectra were recorded with an excitation wavelength of 450 nm. (C) The temperature dependence of the third ‘dark’ step was calculated by measuring the relative decrease in fluorescence at either 677 nm (MV Pchlide) or 674 nm (DV Pchlide) over the temperature range 260–300 K.
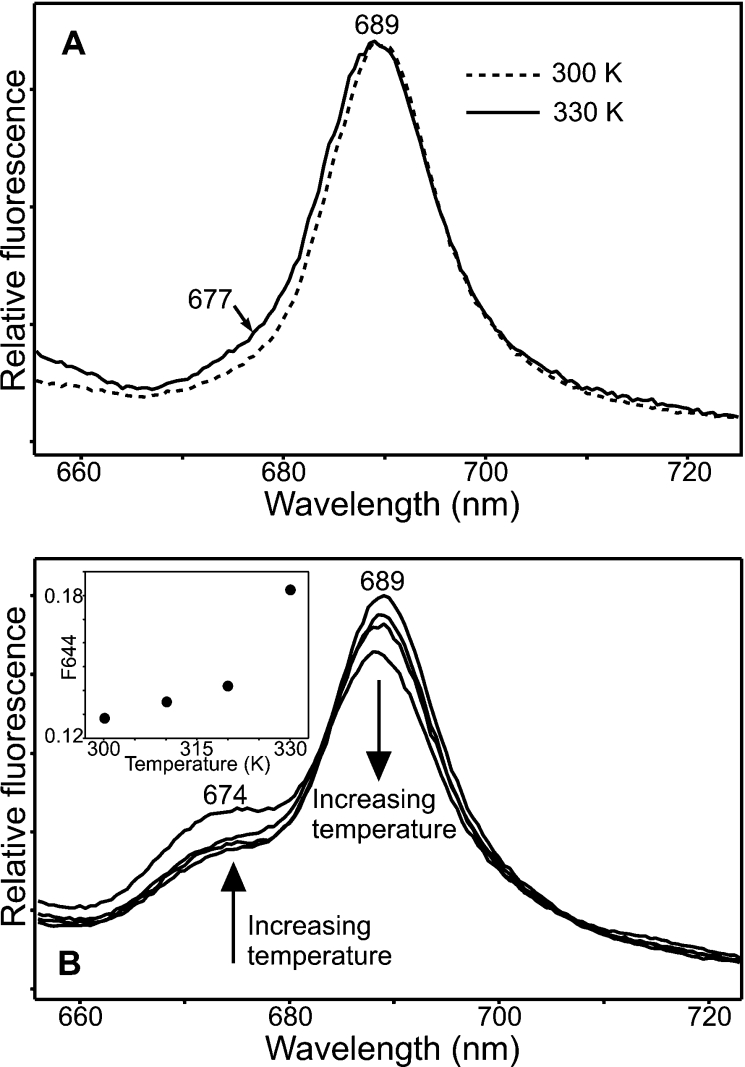
During the fourth ‘dark’ step, the state that fluoresces at 689 nm appears to be converted back into a species that fluoresces at either 674 or 677 nm (Figure 5). In the MV samples, this spectral change is rather subtle with only a small increase in fluorescence at 677 nm as the temperature is increased (Figure 5A). However, the conversion is much more apparent for the DV samples, allowing a temperature dependence of this fourth ‘dark’ step to be measured by plotting out the relative fluorescence at 674 nm over a range of temperatures (Figure 5B, inset).
Figure 5. Spectroscopic characterization of the fourth ‘dark’ step of the reaction for MV and DV Pchlides.
(A) The 77 K fluorescence emission spectra of samples containing 1.1 μM MV Pchlide, 30 μM POR and 200 μM NADPH after illumination for 10 min at 185 K and incubation in the dark for 10 min at either 300 or 330 K are shown. Spectra were recorded with an excitation wavelength of 450 nm. (B) The 77 K fluorescence emission spectra of samples containing 1.1 μM DV Pchlide, 30 μM POR and 200 μM NADPH after illumination for 10 min at 185 K and incubation in the dark for 10 min at 300, 310, 320 and 330 K are shown. The arrows indicate the formation of a fluorescence band at 674 nm and the disappearance of the fluorescence band at 689 nm at increasing temperatures. Spectra were recorded with an excitation wavelength of 450 nm. The inset shows the temperature dependence of the fourth ‘dark’ step for DV Pchlide, which was calculated by measuring the relative decrease in fluorescence at 674 nm over the temperature range 300–330 K.
DISCUSSION
The reduction of Pchlide to Chlide, catalysed by the light-dependent enzyme POR, is an important reaction in the chlorophyll biosynthetic pathway [2]. The steady-state kinetic properties of the enzyme from a range of organisms have previously been measured [11,15,20–25] and the reaction has also been analysed at low temperatures in order to trap intermediates in the reaction pathway [11–13]. Consequently, it was shown to consist of an initial light-dependent reaction [11] followed by a series of ‘dark’ reactions that involve a series of ordered product release and substrate rebinding events [12,13]. However, it has been suggested that some of these properties of the reaction may be affected by using a mixture of MV and DV Pchlides [16,17]. Therefore in the present study, both MV and DV forms of Pchlide have been purified and all aspects of the reaction have been studied using both pigment types.
Many previous studies on POR have used Pchlide preparations from a strain of R. capsulatus that has a mutation in one of the subunits of the light-independent Pchlide reductase and is unable to reduce Pchlide to Chlide [12]. The pigment content from this R. capsulatus strain has now been analysed and shown to consist of a mixture of MV and DV Pchlides with DV Pchlide present at approx. 5-fold higher levels than MV Pchlide. Therefore this implies that the DV reductase enzyme [18,19], which is responsible for converting the vinyl group at the C-8 position of the tetrapyrrole macrocycle into an ethyl group, can operate either before or after the reduction of Pchlide to Chlide (Scheme 1). This is in agreement with previous hypotheses suggesting that the reduction of the 8-vinyl group may occur at various steps of the chlorophyll biosynthesis pathway [19].
In order to explore the effects that the two pigment types may have on the POR-catalysed reaction, purified MV and DV forms of Pchlide were successfully obtained. The spectral properties of both pigments have been analysed using low-temperature absorbance and fluorescence spectroscopy. The spectra exhibit similar features to those previously reported at room temperature [16,26] and reveal several differences between the MV and DV Pchlides. Consequently, this may help us to understand the complex spectral diversity of Pchlide forms occurring in vivo [27–30].
Most previous steady-state kinetic measurements on POR have been carried out using pigment preparations that are a mixture of MV and DV Pchlides. These measurements have now been repeated for POR from the thermophilic cyanobacterium T. elongatus using either purified MV or DV Pchlide. Both Pchlide types appear to be accepted equally as substrates by POR with only minor discrepancies between the Vmax and Km values obtained for each pigment. These kinetic parameters are also in very close agreement with those that were recently calculated for the same enzyme [20]. In addition, the Kd values for the binding of each pigment are also very similar although the DV Pchlide appears to bind slightly more tightly than the MV Pchlide. All of these measurements suggest that it is not necessary for the vinyl group at the C-8 position to be reduced for POR activity and provides further evidence that the DV reductase enzyme can operate before or after POR (Scheme 1). This is significant because if POR had only been capable of reducing MV Pchlide, then vinyl reduction would have to precede Pchlide reduction, whereas if DV Pchlide had been the only accepted substrate, then vinyl reduction must follow Pchlide reduction.
As POR is a light-driven enzyme, it is possible to trap intermediates in the reaction pathway by illumination at low temperatures. As a result, it has previously been shown that the reaction for the T. elongatus enzyme consists of the initial photochemistry followed by four ‘dark’ steps [13]. Each of these steps has now been characterized and the temperature dependence measured using either MV or DV Pchlide. The reaction for both pigments was shown to proceed via the same five steps with only subtle differences in the spectral properties of some of the intermediates that are formed. The POR–Chlide–NADP+ complex, which is formed after the first ‘dark’ step, is very slightly red-shifted for the MV samples (685 nm) compared with the DV samples (684 nm). Similarly, the MV Chlide–POR state (677 nm), formed after the second ‘dark’ step, and the ‘free’ MV Chlide (677 nm) are both red-shifted with respect to their DV counterparts (674 nm). Hence, these discrepancies may help to explain some of the different spectral intermediates and Chlide forms that are formed in vivo [27–30]. However, apart from these minor differences, the reaction pathway was remarkably similar for both pigments with virtually identical temperature dependencies.
In conclusion, we have shown that previous studies, which have used Pchlide preparations from R. capsulatus, have indeed used a mixture of MV and DV pigments. However, neither pigment appears to influence the enzyme kinetics or the steps in the reaction pathway as has been previously suggested. These results also have implications for the DV reductase enzyme, which we have shown can operate before or after the reduction of Pchlide.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council, U.K.
References
- 1.Griffiths W. T. Reconstruction of chlorophyllide formation by isolated etioplast membranes. Biochem. J. 1978;174:681–692. doi: 10.1042/bj1740681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heyes D. J., Hunter C. N. Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem. Sci. 2005;30:642–649. doi: 10.1016/j.tibs.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Aubert C., Vos M. H., Mathis P., Eker A. P., Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature (London) 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 4.Fujita Y., Bauer C. E. Reconstitution of light-independent protochlorophyllide reductase from purified BchL and BchN-BchB subunits – in vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 2000;275:23583–23588. doi: 10.1074/jbc.M002904200. [DOI] [PubMed] [Google Scholar]
- 5.Wilks H. M., Timko M. P. A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase. Identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl. Acad. Sci. U.S.A. 1995;92:724–728. doi: 10.1073/pnas.92.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M. E. Protochlorophyllide reductase is homologous to human carbonyl reductase and pig 20-β-hydroxysteroid dehydrogenase. Biochem. J. 1994;300:605–607. doi: 10.1042/bj3000605b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jörnvall H., Persson B., Krook M., Atrian S., Gonzalez-Duarte R., Jeffery J., Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 8.Townley H. E., Sessions R. B., Clarke A. R., Dafforn T. R., Griffiths W. T. Protochlorophyllide oxidoreductase: a homology model examined by site-directed mutagenesis. Proteins. 2001;44:329–335. doi: 10.1002/prot.1098. [DOI] [PubMed] [Google Scholar]
- 9.Valera V., Fung M., Wessler A. N., Richards W. R. Synthesis of 4R- and 4S-tritium labeled NADPH for the determination of the coenzyme stereospecificity of NADPH-protochlorophyllide oxidoreductase. Biochem. Biophys. Res. Commun. 1987;148:515–520. doi: 10.1016/0006-291x(87)91141-7. [DOI] [PubMed] [Google Scholar]
- 10.Begley T. P., Young H. Protochlorophyllide reductase. 1. Determination of the regiochemistry and the stereochemistry of the reduction of protochlorophyllide to chlorophyllide. J. Am. Chem. Soc. 1989;111:3095–3096. [Google Scholar]
- 11.Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N. Enzymology below 200 K: the kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11145–11150. doi: 10.1073/pnas.182274199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyes D. J., Ruban A. V., Hunter C. N. Protochlorophyllide oxidoreductase: spectroscopic characterization of the ‘dark’ reactions. Biochemistry. 2003;42:523–528. doi: 10.1021/bi0268448. [DOI] [PubMed] [Google Scholar]
- 13.Heyes D. J., Hunter C. N. Identification and characterization of the product release steps within the catalytic cycle of protochlorophyllide oxidoreductase. Biochemistry. 2004;43:8265–8271. doi: 10.1021/bi049576h. [DOI] [PubMed] [Google Scholar]
- 14.Heyes D. J., Hunter C. N., van Stokkum I. H. M., van Grondelle R., Groot M. L. Ultrafast enzymatic reaction dynamics in protochlorophyllide oxidoreductase. Nat. Struct. Biol. 2003;10:491–492. doi: 10.1038/nsb929. [DOI] [PubMed] [Google Scholar]
- 15.Klement H., Helfrich M., Oster U., Schoch S., Rudiger W. Pigment-free NADPH:protochlorophyllide oxidoreductase from Avena sativa L. Eur. J. Biochem. 1999;265:862–874. doi: 10.1046/j.1432-1327.1999.00627.x. [DOI] [PubMed] [Google Scholar]
- 16.Lebedev N., Karginova O., McIvor W., Timko M. P. Tyr275 and Lys279 stabilize NADPH within the catalytic site of NADPH:protochlorophyllide oxidoreductase and are involved in the formation of the enzyme photoactive state. Biochemistry. 2001;40:12562–12574. doi: 10.1021/bi0105025. [DOI] [PubMed] [Google Scholar]
- 17.Kruk J., Mysliwa-Kurdziel B. M. Separation of monovinyl and divinyl protochlorophyllides using C-30 reverse phase high performance liquid chromatography column: Analytical and preparative applications. Chromatographia. 2004;60:117–123. [Google Scholar]
- 18.Parham R., Rebeiz C. A. Chloroplast biogenesis 72: a [4-vinyl]chlorophoryllide a as an exogenous substrate. Anal. Biochem. 1995;231:164–169. doi: 10.1006/abio.1995.1516. [DOI] [PubMed] [Google Scholar]
- 19.Nagata N., Tanaka R., Satoh S., Tanaka A. Identification of a vinyl reductase gene for chlorophyll biosynthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell. 2005;17:233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFarlane M. J., Hunter C. N., Heyes D. J. Kinetic characterisation of the light-driven protochlorophyllide oxidoreductase (POR) from Thermosynechococcus elongatus. Photochem. Photobiol. Sci. 2005;12:1055–1059. doi: 10.1039/b506035d. [DOI] [PubMed] [Google Scholar]
- 21.Martin G. E. M., Timko M. P., Wilks H. M. Purification and kinetic analysis of pea (Pisum sativum L.) NADPH:protochlorophyllide oxidoreductase expressed as a fusion with maltose-binding protein in Escherichia coli. Biochem. J. 1997;325:139–145. doi: 10.1042/bj3250139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townley H. E., Griffiths W. T., Nugent J. P. A reappraisal of the mechanism of the photoenzyme protochlorophyllide reductase based on studies with the heterologously expressed protein. FEBS Lett. 1998;422:19–22. doi: 10.1016/s0014-5793(97)01589-5. [DOI] [PubMed] [Google Scholar]
- 23.Lebedev N., Timko M. P. Protochlorophyllide oxidoreductase B-catalyzed protochlorophyllide photoreduction in vitro: insight into the mechanism of chlorophyll formation in light-adapted plants. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9954–9959. doi: 10.1073/pnas.96.17.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyes D. J., Martin G. E. M., Reid R. J., Hunter C. N., Wilks H. M. NADPH:protochlorophyllide oxidoreductase from Synechocystis: overexpression, purification and preliminary characterization. FEBS Lett. 2000;483:47–51. doi: 10.1016/s0014-5793(00)02081-0. [DOI] [PubMed] [Google Scholar]
- 25.Urbig T., Knaust R. K. C., Schiller H., Senger H. Solubilization and hydrophobicity test by Triton X-114-partitioning of NADPH-protochlorophyllide oxidoreductase from the unicellular alga Scenedesmus obliquus, mutant C-2A′. Z. Naturforsch. C Biosci. 1995;50:775–780. [Google Scholar]
- 26.Mysliwa-Kurdziel B., Kruk J., Strzalka K. Fluorescence lifetimes and spectral properties of protochlorophyllide in organic solvents in relation to the respective parameters in vivo. Photochem. Photobiol. 2004;79:62–67. [PubMed] [Google Scholar]
- 27.Boddi B., Ryberg M., Sundqvist C. Identification of 4 universal protochlorophyllide forms in dark-grown leaves by analyses of the 77K fluorescence emission-spectra. J. Photochem. Photobiol. B Biol. 1992;12:389–401. [Google Scholar]
- 28.Boddi B., Ryberg M., Sundqvist C. Analysis of the 77K fluorescence emission and excitation-spectra of isolated etioplast inner membranes. J. Photochem. Photobiol. B Biol. 1993;21:125–133. [Google Scholar]
- 29.Boddi B., Franck F. Room temperature fluorescence spectra of protochlorophyllide and chlorophyllide forms in etiolated been leaves. J. Photochem. Photobiol. B Biol. 1997;41:73–82. [Google Scholar]
- 30.Mysliwa-Kurdziel B., Amirjani M. R., Strzalka K., Sundqvist C. Fluorescence lifetimes of protochlorophyllide in plants with different proportions of short-wavelength and long-wavelength protochlorophyllide spectral forms. Photochem. Photobiol. 2003;78:205–212. doi: 10.1562/0031-8655(2003)078<0205:flopip>2.0.co;2. [DOI] [PubMed] [Google Scholar]