Abstract
Much of our knowledge about the mechanisms of vertebrate early development comes from studies using Xenopus laevis. The recent development of a remarkably efficient method for generating transgenic embryos is now allowing study of late development and organogenesis in Xenopus embryos. Possibilities are also emerging for genomic studies using the closely related diploid frog Xenopus tropicalis.
Xenopus as a model organism
Amphibians have played a key role in the elucidation of the mechanisms of early development over the last century. The eggs and embryos are relatively large and robust, and they develop externally in a simple salt solution. Because of their suitability for micromanipulation, amphibian embryos provided many of the core results of experimental embryology. These include the provision of fate maps (stage-specific diagrams indicating where cells will end up and which tissues they will form), states of developmental commitment of cells to different lineages, and inductive signals that allow one cell type to influence the development of another. Since its introduction as a model organism in the 1950s, most of this work has focused on the African clawed frog Xenopus laevis. The eggs and embryos of X. laevis can be produced in large numbers by means of a simple hormone injection and are, like other amphibian embryos, easily manipulated, injected, grafted or labeled. In the molecular era, the ability of the Xenopus embryo to translate injected, synthetic mRNA has led to many important contributions to the study of early developmental events. In particular, the unique advantages of Xenopus described above have allowed the identification of the major classes of inducing factor used by all animals (the transforming growth factor β (TGFβ)/bone morphogenetic protein, fibroblast growth factor (FGF) and Wnt classes). In fact, Xenopus would seem to be the ideal all-purpose research organism, except for the fact that it is not ideal for experimental genetics given its pseudotetraploid genome and long generation time. Recently, however, the emergence of a potential ideal model organism, the closely related diploid species Xenopus tropicalis, along with the development of transgenic methods and the initiation of a community-wide initiative for the production of genomic and genetic resources for the future, has secured the future of Xenopus as a genetic model as well.
Xenopus ESTs in expression screening and microarray analyses
The sequencing of expressed sequence tags (ESTs) from X. laevis has lagged behind efforts on many other model organisms and humans, in part because of the pseudotetraploid nature of the X. laevis genome. Acquiring large numbers of ESTs has recently been recognized as a priority for future Xenopus research, and, as a result of several parallel efforts, the number of deposited ESTs has now exceeded 100,000. According to the National Center for Biotechnology Information (NCBI [1]), this now places Xenopus eleventh in the ranking of species in terms of available EST numbers. Information on Xenopus ESTs can also be obtained from the XEST website [2].
Although there is still a long way to go, the Xenopus EST collection is already starting to have an impact on research. A large-scale gene-expression screen was performed by Gawantka and colleagues [3] and has formed the basis for the searchable Axeldb expression-pattern database [4,5]. In Gawantka's initial screen [3], 1,765 random Xenopus cDNAs were analyzed by whole-mount in situ hybridization to embryos. Of these, 273 unique, differentially expressed genes were identified, 204 of which had not been isolated previously from Xenopus. The relatively large size of the screen enabled Gawantka and colleagues [3] to identify four putative 'synexpression' groups, or sets of genes with identical expression patterns, two of which, named 'delta1' and 'BMP4' after well-known genes in the groups, were predicted to correspond to molecular pathways involved in patterning and differentiation. When you consider that the number of marker genes that have been submitted by the entire research community to the Xenopus Molecular Marker Resource (XMMR) [6] is less than 50, this is an impressive contribution from a single lab.
One of the forces driving the accumulation of ESTs is the wish to produce Xenopus microarrays, which can be used for genome-wide gene-expression analysis. Xenopus is ideal for this kind of screening, as it is relatively easy to isolate specific regions of the embryo in order to make tissue-specific RNA probes. Traditionally, researchers attempting to characterize a phenotype arising from their experiment would analyze changes in the expression levels of a small panel of genes that have well-documented expression patterns, known as markers, by reverse-transcriptase-coupled (RT) PCR, RNase protection assay, or Northern blot. Although this gives a good idea of the molecular changes that have occurred, it might be possible to miss key events if markers are not chosen carefully or if appropriate markers are not available. In a recent paper, Altmann and colleagues [7] have demonstrated the power of using Xenopus microarrays to address several key aspects of early development. Their prototype microarray contained 864 sequenced cDNAs prepared from embryos at the gastrula stage of development, 107 of which were not represented in the EST database and 96 of which were previously characterized markers (see the Xenopus microarray site [8]). Three test experiments were performed using the array. First, the authors searched for temporally regulated genes using either maternal or zygotic mRNA to generate probes; second, they compared mesoderm from dorsal and ventral regions of early gastrula embryos; and third, they compared untreated embryonic explants with those treated with the known mesoderm inducer activin. Novel temporally regulated, spatially restricted and activin-induced genes were isolated using this screen, and the results were confirmed by independent analysis using RT-PCR or in situ hybridization. Importantly, probes could be generated from a quantity of RNA equivalent to the amount in a single embryo, although this was thought to be the absolute minimum requirement.
Another use for the growing EST dataset is in the construction of UniGene sets for Xenopus. These are created automatically from the NCBI database, which includes both ESTs and well-characterized genes, by forming clusters of sequences, each representing a unique gene (see NCBI's UniGene [9]). The usefulness of this type of information is twofold. Firstly, the ideal microarray would contain a single, representative copy of each expressed gene - in other words, a non-redundant gene set. Secondly, a program has been developed by the NCBI that makes use of the UniGene data to enable 'digital differential display' (DDD) [10]. Each of the libraries that have been used to generate ESTs can be compared using DDD, to show the genes that are significantly differentially represented between them. For example, a liver library should have generated more EST hits for the albumin gene than would a heart library, and this will be displayed as a darker spot on the DDD (see Figure 1). As the number of ESTs increases and more libraries are used for sequencing, this should become a very powerful tool.
Figure 1.
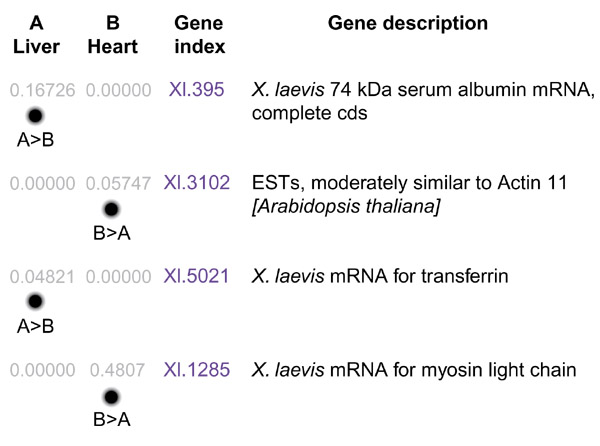
Example of digital differential display (DDD [10]), comparing gene expression between the liver (pool A) and heart (pool B) by comparing the available EST data from two Xenopus libraries. The top four results are shown. The numbers in grey represent the proportion of sequences within each pool that map to the UniGene cluster indicated in blue, for which a description is given on the right;cds, coding sequence. Dots are a visual representation of the numerical values. Statistically significant results are shown by indication of the relationship between two pools for a particular gene cluster, for example A > B for albumin and transferrin in the liver, B > A for myosin light chain and a probable actin homolog in the heart library.
Transgenic technology for studying later development and organogenesis
Many of the experiments using Xenopus in the study of early development have made use of injected mRNA, antibodies, or ordinary or morpholino antisense oligonucleotides. These methods are all essentially transient, however, as the genome is not altered and the injected substance decays with time. Arguably the biggest leap forward in the establishment of Xenopus as a model organism beyond the limits of early development has been the development of methods for generating transgenic embryos, by Enrique Amaya and Kristen Kroll [11,12]. Known as restriction enzyme mediated insertion (REMI) transgenesis, this method has been adopted across the Xenopus community with great enthusiasm. The key to REMI transgenesis involves mixing transgene DNA with purified and permeabilized sperm, along with a small quantity of restriction enzyme and an extract of egg cytoplasm generated by high-speed centrifugation (the key ingredient of which is thought to be nucleoplasmin, which serves to partially decondense the sperm chromatin, facilitating transgene insertion); this mixture is then injected into dejellied eggs at the rate equivalent to about one sperm per egg. Around a third of the recipient eggs usually cleave normally and a proportion (around 50%) of these integrate the transgene into the genome during the first cell cycle. This method produces large quantities of non-mosaic transgenics in the parent (F0) generation, allowing experiments to be performed without the generation of stable transgenic lines. Importantly, several labs have shown that the transgene is reliably transferred through subsequent generations [13,14,15].
A key advantage of Xenopus transgenesis is the ability to study transgene expression in living embryos using green fluorescent protein (GFP) as the reporter; this allows quick and easy promoter analysis [15,16,17,18,19]. Furthermore, the advantages of Xenopus embryos allowing the later roles of genes involved in early patterning to be elucidated (these were previously accessible only using the far more difficult procedure of making conditional knockouts in mice). Already the technique has been used to observe the onset of differentiation of the lens [15], to study the role of bone morphogenetic proteins in heart looping [20], and to investigate several aspects of metamorphosis [21,22,23,24]. In our lab, we have been making use of tissue-specific promoters to study the formation of the gut organs [25] (Figure 2). With the adaptation of existing methods to Xenopus transgenics - for example, the Cre and FLP conditional mutagenesis systems [26] and tetracycline-inducible constructs [27] - it is becoming clear that the golden age for studying late development is fast approaching.
Figure 2.
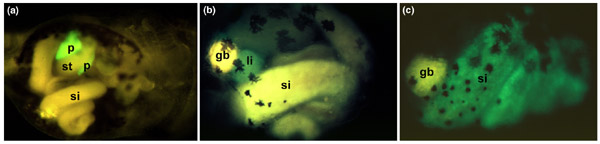
Analysis of gut-specific promoters in live X. laevis tadpoles. Green fluorescence is seen where the promoter is active, and yellow is due to autofluorescence of the tadpole gut. Black patches are pigment cells. (a) The Elastase enhancer (rat) driving the expression of GFP in the pancreas of a 7-day-old tadpole; ventral view, anterior to the right. (b) The Transthyretin promoter (mouse) driving expression of GFP in the liver of a 6-day-old tadpole; ventral/left view, anterior to the left. (c) The Intestinal fatty acid binding protein promoter (rat) driving expression of GFP in the small intestine of a 7-day-old tadpole, ventral view, anterior to the left. Abbreviations: gb, gall bladder; li, liver; p, pancreas; si, small intestine; st, stomach. Reproduced with permission from [25].
A genomic basis for Xenopus research
Although the establishment of a genomic base for Xenopus research is still in the planning stages, there are pilot muta-genesis studies already available. Amaya and colleagues [14] have shown that it is possible to adapt the transgenesis technique in order to trap genes with interesting, tissue-specific expression. Promoterless constructs containing GFP as the reporter, with or without splice-site acceptor sequences, were used to visualize 'trapped' genes (close to enhancers) in living F0 transgenics. Rapid PCR amplification of cDNA ends (5' RACE PCR) was then used to amplify the disrupted genes from reverse-transcribed RNA extracted from the embryos, and the genes were identified by sequencing. Although no novel genes were identified in this pilot screen, a variety of tissue-specific expression patterns were generated, showing the potential power of the technique. In the future, it should be possible to use transposons to increase the rate of gene trapping (P. Mead, personal communication). It is also possible to isolate developmental mutants using inbreeding techniques, in which siblings are interbred for several generations to create 'isogenic' lines, or 'gynogenesis', in which haploid individuals are made homozygous by suppression of the first cleavage. In one study, 12 heritable developmental mutations were isolated from just eight wild-caught X. laevis females [28].
Despite the many advantages of Xenopus as a model organism, it does have one or two major drawbacks for genetic studies. Firstly, X. laevis is a pseudotetraploid, as a result of an additional genome duplication (relative to other vertebrates) about 30 million years ago [29,30]. This is also the case for zebrafish, another favorite model organism, as genome duplication also occurred in the teleost fish around 420 million years ago [31,32]. Aside from the larger genome that results from pseudotetraploidy, mutagenesis screens are less likely to be successful given the functional redundancy between closely related paralogous genes. The second disadvantage of X. laevis is the relatively long generation time, usually around 1-2 years, making the generation of stable transgenic lines a slow process. Fortunately, a closely related frog, Xenopus (formerly Silurana) tropicalis, has neither of these disadvantages, while retaining the many advantages of its larger relative. X. tropicalis has a diploid genome and a generation time of around 4-5 months [30,33]. X. tropicalis adults are also smaller, making them more practical for large-scale laboratory use, and they have a genome size of around half the size of that of the mouse (1.7 × 109 base pairs per haploid nucleus [34]). Like X. laevis, they can be induced by hormone injection to lay around 1,000-3,000 eggs at a time. Although their eggs are around half the size of those of X. laevis, they are still big enough to be used for transgenesis and other manipulations [15]. Preliminary studies in Robert Grainger's lab have shown that it is possible to use existing X. laevis probes for in situ hybridization studies on X. tropicalis (see the Grainger laboratory's X. tropicalis site [35]), thus removing the need to re-clone existing genes in X. tropicalis.
Driven by the recent advances in genetics, the Xenopus community is now working together to secure the large amounts of funding needed for development of genomic tools, resource centers and the X. tropicalis system. Details of the current proposals can be found on the NIH Xenopus Initiative website [36] and include both an expansion on the work discussed here and descriptions of new projects. For example, the planned creation of a genetic map for Xenopus will make use of the same methods as were used for zebrafish, such as radiation hybrid panels [37] and random amplified polymorphic DNAs (RAPDs) [38]. Peter Vize's Xenbase [39], the main website resource for Xenopus researchers, is expanding to include sections on genetics and genomics, and further expansion of this facility is planned. As it becomes available, genomic technology, combined with the advantages of Xenopus as a model system, will allow Xenopus researchers to continue to make an extensive contribution to functional studies in the future.
References
- dbEST: Database of "Expressed Sequence Tags". Summary by Organism http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html
- A Xenopus laevis database (XEST) http://www.dkfz-heidelberg.de/tbi/services/axeldb_images/nicolas/xest.html
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–141. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- Axeldb http://www.dkfz-heidelberg.de/abt0135/axeldb.htm
- Pollet N, Schmidt HA, Gawantka V, Vingron M, Niehrs C. AxelDB: a Xenopus laevis database focusing on gene expression. Nucleic Acids Res. 2000;28:139–140. doi: 10.1093/nar/28.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenopus Molecular Marker Resource http://cbrmed.ucalgary.ca/pvize/html/WWW/Welcome.html
- Altmann CR, Bell E, Sczyrba A, Pun J, Bekiranov S, Gaasterland T, Brivanlou AH. Microarray-based analysis of early development in Xenopus laevis. Dev Biol. 2001;236:64–75. doi: 10.1006/dbio.2001.0298. [DOI] [PubMed] [Google Scholar]
- Xenopus microarray site http://arrays.rockefeller.edu/xenopus
- UniGene frog sequences collection at NCBI http://www.ncbi.nlm.nih.gov/UniGene/Xl.Home.html
- Xenopus laevis UniGene digital differential display (DDD) http://www.ncbi.nlm.nih.gov/UniGene/ddd.cgi?ORG=Xl
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Amaya E, Kroll KL. A method for generating transgenic frog embryos. Methods Mol Biol. 1999;97:393–414. doi: 10.1385/1-59259-270-8:393. [DOI] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, Berry DL, Brown DD. Germ-line transmission of transgenes in Xenopus laevis. Proc Natl Acad Sci USA. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronchain OJ, Hartley KO, Amaya E. A gene trap approach in Xenopus. Curr Biol. 1999;9:1195–1198. doi: 10.1016/S0960-9822(00)80025-1. [DOI] [PubMed] [Google Scholar]
- Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–1797. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- Hyde CE, Old RW. Regulation of the early expression of the Xenopus nodal-related 1 gene, Xnr1. Development. 2000;127:1221–1229. doi: 10.1242/dev.127.6.1221. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Casey ES, Harland RM, Smith JC, Beddington RS. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev Biol. 2001;234:304–316. doi: 10.1006/dbio.2001.0265. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Latinkic BV, Remacle JE, Huylebroeck D, Smith JC. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development. 2000;127:2729–2739. doi: 10.1242/dev.127.12.2729. [DOI] [PubMed] [Google Scholar]
- Ryffel GU, Lingott A. Distinct promoter elements mediate endodermal and mesodermal expression of the HNF1alpha promoter in transgenic Xenopus. Mech Dev. 2000;90:65–75. doi: 10.1016/s0925-4773(99)00230-0. [DOI] [PubMed] [Google Scholar]
- Breckenridge RA, Mohun TJ, Amaya E. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev Biol. 2001;232:191–203. doi: 10.1006/dbio.2001.0164. [DOI] [PubMed] [Google Scholar]
- Huang H, Marsh-Armstrong N, Brown DD. Metamorphosis is inhibited in transgenic Xenopus laevis tadpoles that overexpress type III deiodinase. Proc Natl Acad Sci USA. 1999;96:962–967. doi: 10.1073/pnas.96.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Brown DD. Prolactin is not a juvenile hormone in Xenopus laevis metamorphosis. Proc Natl Acad Sci USA. 97:195–199. doi: 10.1073/pnas.97.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, Remo BF, Liu TT, Brown DD. Asymmetric growth and development of the Xenopus laevis retina during metamorphosis is controlled by type III deiodinase. Neuron. 1999;24:871–878. doi: 10.1016/s0896-6273(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Coen L, du Pasquier D, Le Mevel S, Brown S, Tata J, Mazabraud A, Demeneix BA. Xenopus Bcl-X(L) selectively protects Rohon-Beard neurons from metamorphic degeneration. Proc Natl Acad Sci USA. 2001;98:7869–7874. doi: 10.1073/pnas.141226798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CW, Slack JM. Gut specific expression using mammalian promoters in transgenic Xenopus laevis. Mech Dev. 1999;88:221–227. doi: 10.1016/s0925-4773(99)00217-8. [DOI] [PubMed] [Google Scholar]
- Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 2001;29:E53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway P, Quivy JP, Almouzni G. Tetracycline-regulated gene expression switch in Xenopus laevis . Exp Cell Res. 2000;256:392–399. doi: 10.1006/excr.2000.4853. [DOI] [PubMed] [Google Scholar]
- Krotoski DM, Reinschmidt DC, Tompkins R. Developmental mutants isolated from wild-caught Xenopus laevis by gynogenesis and inbreeding. J Exp Zool. 1985;233:443–449. doi: 10.1002/jez.1402330313. [DOI] [PubMed] [Google Scholar]
- Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus). Science. 1977;195:785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Amaya E, Offield MF, Grainger RM. Frog genetics: Xenopus tropicalis jumps into the future. Trends Genet. 1998;14:253–255. doi: 10.1016/s0168-9525(98)01506-6. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Meyer A, Malaga-Trillo E. Vertebrate genomics: More fishy tales about Hox genes. Curr Biol. 1999;9:R210–R213. doi: 10.1016/s0960-9822(99)80131-6. [DOI] [PubMed] [Google Scholar]
- de Sa RO, Hillis DM. Phylogenetic relationships of the pipid frogs Xenopus and Silurana: an integration of ribosomal DNA and morphology. Mol Biol Evol. 1990;7:365–376. doi: 10.1093/oxfordjournals.molbev.a040612. [DOI] [PubMed] [Google Scholar]
- Tymowska J. Karyotype analysis of Xenopus tropicalis Gray, Pipidae. Cytogenet Cell Genet. 1973;12:297–304. doi: 10.1159/000130468. [DOI] [PubMed] [Google Scholar]
- X.tropicalis: amphibian model for vertebrate developmental genetics http://minerva.acc.virginia.edu/~develbio/trop/
- Trans-NIH Xenopus Initiative http://www.nih.gov/science/models/xenopus/
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci USA. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA. Using random amplified polymorphic DNAs in zebrafish genomic analysis. Methods Cell Biol. 1999;60:165–179. doi: 10.1016/s0091-679x(08)61899-3. [DOI] [PubMed] [Google Scholar]
- Xenbase: a Xenopus resource http://cbrmed.ucalgary.ca/pvize/html/Xenbase.html


