Abstract
Misincorporated ribonucleotides in DNA will cause DNA backbone distortion and may be targeted by DNA repair enzymes. Using double-stranded oligonucleotide probes containing a single ribose, we demonstrate a robust activity in human, yeast, and Escherichia coli cell-free extracts that nicks 5′ of the ribose. The human and yeast extracts also make a subsequent cut 3′ of the ribonucleotide releasing a ribonucleotide monophosphate. The resulting 1-nt gap is an ideal substrate for polymerase and ligase to complete a proposed repair sequence that effectively replaces the ribose with deoxyribose. Screening of yeast deletion mutant cells reveals that the initial nick is made by RNase H(35), a RNase H type 2 enzyme, and the second cut is made by Rad27p, the yeast homologue of human FEN-1 protein. RNase H type 2 enzymes are present in all kingdoms of life and are evolutionarily well conserved. We knocked out the corresponding rnhb gene in E. coli and show that extracts from this strain lack the nicking activity. Conversely, a highly purified archaeal RNase HII type 2 protein has a pronounced activity. To study substrate specificity, extracts were made from a yeast double mutant lacking the other main RNase H enzymes [RNase H1 and RNase H(70)], while maintaining RNase H(35). It was found that a single ribose is preferred as substrate over a stretch of riboses, further strengthening a proposed role of this enzyme in the repair of misincorporated ribonucleotides rather than (or in addition to) processing RNA/DNA hybrid molecules.
There is presently a nearly total lack of information about repair of deoxyribose modifications in DNA. Such modifications can be caused by external agents, such as oxidizing agents and ionizing radiation (1–3), and can also occur naturally by misincorporation of ribonucleotides into DNA during DNA replication (4). The presence of ribose in DNA is a hindrance to formation of normal B form DNA as evidenced by the structure of RNA/DNA hybrid molecules (5), and consequently a single ribose in DNA will result in a local DNA backbone distortion (6). Other bulky modified sugars are also likely to cause backbone distortions, and it can be hypothesized that they pose a hindrance for DNA polymerases and can be mutagenic.
Progressive DNA and RNA polymerases are similar in structure and belong to the same class of proteins (4), probably with a common evolutionary origin (7). The specificity toward deoxyribonucleoside triphosphates (dNTPs) or ribonucleoside triphosphates (rNTPs) has been found to be determined by subtle differences at the active site (4). Gao et al. (8) could largely eliminate the discrimination between the rNTPs and dNTPs by introducing a single amino acid change in a reverse transcriptase, and similar observations in mutant polymerases have recently been made by several investigators (7, 9–13). On the basis of such observations, it has been suggested that the discrimination against ribonucleotides by DNA polymerases is largely accomplished by a “steric gate” that will not give enough space for the 2′ hydroxyl group present in rNTPs (4). However, the discrimination against rNTPs is not 100%, and detectable incorporation of rNTPs has been found in vitro by using purified DNA polymerases with a wide variety of discrimination factors ranging from a few thousand-fold (7, 11) to several million-fold (13). However, it is presently not known to what extent ribonucleotides are misincorporated into DNA during normal in vivo DNA replication. The intranuclear milieu contains both ribonucleotides and deoxyribonucleotides, with the ribonucleotide concentration generally higher than the deoxyribonucleotide concentration (14). The deoxyribonucleotides are produced from the ribonucleotide pool by the enzyme ribonucleotide diphosphate reductase. This enzyme can be inhibited in eukaryotic cells by hydroxyurea (HU), which blocks DNA replication when given to cell cultures in sufficient concentration.
Gao and Goff (15) mutagenized a viral polymerase to increase ribose misincorporation during viral DNA replication in vivo. Although some minus-strand DNA was synthesized with significant ribose incorporation in virally infected cells, the synthesis was not completed, and the mutation was lethal. A possible explanation is that ribose in the template DNA strand acts as a replication block. Here we demonstrate in cell-free extracts an activity that excises a single ribonucleotide from DNA, and we identify the enzymes involved.
Materials and Methods
Probes with Misincorporated Ribose.
Two partly complementary 100-mer oligonucleotides (Fig. 1, probe 1) were extended with a modified T7 DNA polymerase (Sequenase Version 2, United States Biochemical) in a 50-μl reaction mixture containing 20 mM Tris⋅HCl, pH 7.5; 2 mM MnCl2; 5 mM dl-isocitrate; 5 mM DTT; 100 μM dGTP, dATP, and dTTP; 3 μM dCTP; 3 μM [α-32P]rCTP (800 Ci/mmol) (Amersham Pharmacia Biotech); 1 pmol annealed oligonucleotide; and 13 units of Sequenase. Reaction was for 30 min at 37°C followed by addition of 100 μM dCTP and continued incubation for 20 min. Nonincorporated 32P was then completely removed by three consecutive Sephadex G50 spin columns. The discrimination factor against rCTP compared with dCTP under these conditions was ≈300-fold. Incorporation of 32P-labeled rGTP, rATP, or rUTP was performed with the same strategy as for rCTP.
Fig 1.
Primers used for construction of probes containing a single ribose residue (probes 1–3) or a stretch of ribose residues (probe 4). Capital letters are used for deoxyribonucleotides and lowercase letters are used for ribonucleotides. A triangle (Δ) indicates the position of ribose misincorporation for probe 2 and the location of ribose residue in probe 3.
A 16-mer primer annealed to a 32-mer template (Fig. 1, probe 2) was extended in the reaction mixture specified above modified with the addition of 50 mM NaCl and excluding dCTP. Incubation was for 30 min at 37°C followed by two consecutive Sephadex G25 spin columns.
Primers 3 and 4 (18 mers) containing either a single ribose modification at the 3′ end (Fig. 1, probe 3) or consisting exclusively of ribonucleotides (Fig. 1, probe 4) were obtained from Integrated DNA Technologies (Coralville, IA). They were annealed to a 5′ phosphorylated 40-mer template and extended by using Klenow polymerase in a 100-μl reaction mixture containing 10 mM Tris⋅HCl, pH 7.9; 10 mM MgCl2; 50 mM NaCl; 1 mM DTT; 200 μM dATP, dTTP, dCTP, and dGTP; 37 pmol annealed template; and 5 units of Klenow polymerase. Incubation was for 30 min at 37°C. The extended oligonucleotides were then purified with a Quickstep2 purification kit (Edge Biosystems, Gaithersburg, MD) to remove protein, ssDNA, and low molecular-weight products. Small aliquots of the purified double-stranded probe were subsequently 5′ end-labeled with 32P by using T4 polynucleotide kinase. Nonincorporated 32P was removed by two consecutive Sephadex G25 spin columns.
Escherichia coli Strains.
An E. coli rnhB knockout strain was constructed by using the recombination system of Yu et al. (16). The host strain DY330 [W3110 ΔlacU169 gal490 λc1857 Δ(cro-bioA)] was obtained from Donald Court (National Cancer Institute, Frederick, MD). The method uses an inducible λ bacteriophage recombination system (RED), by which a targeted genomic sequence can be replaced by a drug-resistance cassette. We first used PCR to construct an ampicillin-resistance cassette with flanking sequences that targeted upstream and downstream regions of the E. coli rnhB gene. The primers were 5′ ATT TGT TTA TCC GCA CAC GCA GCT GGT TGC GGG TGT GGA TCA TTC AAA TAT GTA TCC GCT C and 5′ AAG TCC CAG TGC GCG TTT GAC AGG CCC AAA GCT GCG CCG AAG AGT TGG TAG CTC TTG ATC by using a previously amplified ampicillin-resistance cassette amp from pBluescript (Stratagene) as template. The underlined sequences are complementary to the amp template, whereas the nonunderlined sequences target the rnhB gene The gel-purified cassette was subsequently electroporated into heat-induced DY330 cells followed by selection of ampicillin-resistant colonies as described by Yu et al. (16). Three ampicillin-resistant colonies were picked and tested for RNase HII function.
An E. coli rnhA1 mutant strain FB2 (CGSC# 6585) and parental rnhA+ strain KS351 (CGSC# 6586) were obtained from E. coli Genetic Stock Center, Yale University (New Haven, CT). Additional markers in these strains were lacZ482(Am), relA1, spoT1, rha-9, and thi-1.
Yeast Strains and Procedures.
Construction of a complete set of yeast mutants deleted for each known nonessential gene or ORF has been described in detail elsewhere (17). In these strains, each deleted gene is replaced by a functional gene, KANMX4, that confers resistance to the antibiotic geneticin (G418). A haploid set of these deletions was obtained from Research Genetics, Huntsville, AL (now Invitrogen). Appropriate deletion strains were used for initial biochemical assays. Standard yeast genetic techniques (18) were used to demonstrate cosegregation of the rnh35 deletion with the absence of ribonucleotide excision activity in meiotic tetrads. The rnh35Δ and other deletion segregations were followed by scoring geneticin resistance, by using 100 μg/ml geneticin (obtained from Sigma) added in solution to cooled yeast extract/peptone/dextrose medium (18) before pouring plates. Genetic crosses with tetrad analysis were used to construct the rnh1Δ rnh70Δ double deletion mutant strain and the rnh1Δ rnh35Δ rnh70Δ triple deletion mutant. To construct the triple mutant, we replaced by transformation the geneticin-resistance gene in the MATα rnh35Δ mutant with a functional LEU2 gene [provided by Jim Brown (Department of Radiation Oncology, Stanford University School of Medicine, Stanford, CA)], so that we could use leucine prototrophy to monitor the segregation of the rnh35 deletion independently of the rnh1 and rnh70 deletions. We confirmed the genotypes of double and triple mutant strains by appropriate backcrosses to the single mutants.
Cell-Free Extracts.
A 10-liter HeLa cell pellet (obtained from National Institutes of Health, National Cell Culture Center) was washed three times in PBS, treated with a hypotonic buffer (10 mM Hepes⋅KOH, pH 7.9/10 mM KCl/1.5 mM MgCl2/0.5 mM DTT/protease inhibitors) and the cells broken with a Dounce homogenator (20 strokes with tight pestle). The homogenate was spun at 760 × g for 15 min followed by 22,500 × g for 20 min and the supernatants collected and pooled as a low-salt extract. The pellet was resuspended in a high-salt buffer (20 mM Hepes⋅KOH, pH 7.9/420 mM NaCl/1.5 mM MgCl2/0.2 mM EDTA/0.5 mM DTT/25% glycerol/protease inhibitors), stirred on ice for 30 min, and spun at 22,500 × g for 30 min. The supernatant was recovered as a high-salt extract and stored aliquoted at −70°C. A small-scale cell-free extract from human Jurkat cells was also prepared with the same protocol.
Strains of Saccharomyces cerevisiae were grown at 30°C in 40 ml of liquid yeast extract/peptone/dextrose growth medium (18) and collected in late log phase. After three washes in water, the pellets were resuspended in one volume of 20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/protease inhibitors. To the slurry was added an equal volume of glass beads (size 425–600 μm, acid-washed; Sigma), and the mixtures were vortexed in 1.5 ml of Eppendorf tubes at maximum speed with intermittent cooling on ice for a total of 2 min to break the cells. The tubes were then spun at 15,000 × g for 10 min at 4°C. The supernatants were collected, supplemented with glycerol to a final concentration of 20% vol/vol, and stored at −70°C. E. coli cells were grown in 100 ml of LB until late log phase, washed twice in 10 mM Tris⋅HCl, pH 7.5/100 mM NaCl/1 mM EDTA, and the pellets frozen to −70°C. Thawed pellets were suspended in 1 ml of 20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/2 mM EDTA/10 mM DTT/0.5% Nonidet P-40/10% glycerol/5 mg/ml lysozyme/protease inhibitors and incubated on ice for 15 min. The cells were then disrupted by sonication on ice for 1 min, spun at 15,000 × g for 10 min, and the supernatants collected and stored at −70°C.
In Vitro Assay.
In a standard 10-μl reaction, 1–10 fmol 32P-labeled probe was mixed with 1 μl of variously diluted cell-free extracts in a buffer containing 10 mM Tris⋅HCl, pH 7.5, 35 mM KCl, 1 mM DTT, 1 mM EDTA, 4 mM MgCl2, and 5% glycerol. The mixture was incubated for 15 min at 37°C and the reaction terminated by the addition of an equal volume of loading buffer (80% formamide/10 mM EDTA, pH 8/1 mg/ml xylene cyanol FF/1 mg/ml bromophenol blue). Separation of the products was achieved on denaturing polyacrylamide gels by using standard techniques, followed by exposure on phosphor screens and evaluation on a PhosphorImager (Molecular Dynamics).
Results
Probes.
We constructed a set of oligonucleotide probes that contain a single ribonucleotide (or in one case a stretch of ribonucleotides) (Fig. 1). Probes 1 and 2 are obtained by in vitro misincorporation of 32P-labeled ribonucleotides during primer extension, by using a modified T7 DNA polymerase in a Mn-containing buffer. Probe 1 is a 172-bp double-stranded oligomer obtained by extending two partly complementary 100-mers. Random misincorporation of any 32P-labeled rNTP present in the reaction results in labeled probes containing a ribose, whereas products formed without ribose misincorporation will be unlabeled. Because misincorporation is a rare event, most labeled molecules will have a single ribose. Probe 2 is obtained by extending a 16-mer primer on a 32-mer template containing a single location of G for misincorporation with 32P-rCTP. This probe will thus have a site-specific location for a single ribose with a 32P present immediately 5′ of the ribose sugar. Probe 3 is a 40-mer obtained by extending a commercially obtained primer (Integrated DNA Technologies) that is modified with a single ribose at the 3′ end, by using Klenow polymerase. This probe is 32P-labeled at the 5′ end with T4 polynucleotide kinase before use. Probe 4, which is used for comparative purposes, is obtained by extending an 18-mer RNA primer with deoxyribonucleotides on a 40-mer template. Probes 3 and 4 are identical in sequence, with probe 3 containing a single ribose, whereas probe 4 contains a stretch of 18 ribonucleotides on the 5′ end annealed to the complementary normal DNA strand. We verified that ribose was present in the probes by specific cleavage 3′ of the ribose by potassium hydroxide or piperidine (an example is seen in Fig. 3 below).
Fig 3.
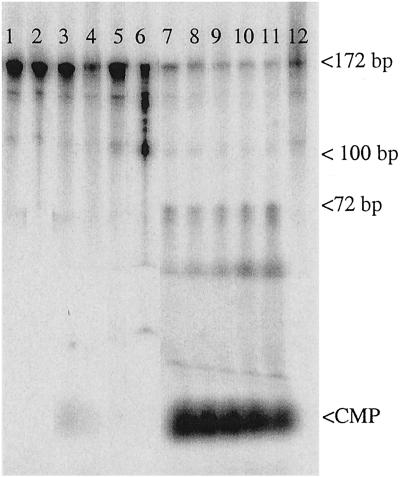
Nicking of probe 1. Lanes 1–4, control probe with no ribose incorporation; lanes 5–12, with ribo C incorporation; lanes 1 and 5, no extract; lanes 2 and 6, probe treated with KOH; lanes 3 and 7–11, incubation with various amounts of Jurkat crude extract; lanes 4 and 12, incubation with heat-inactivated extract.
Nicking Activity in Cell-Free Extracts.
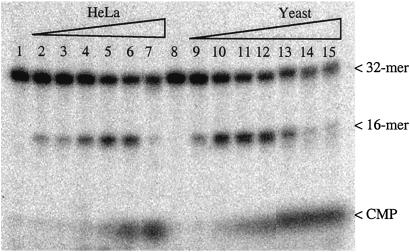
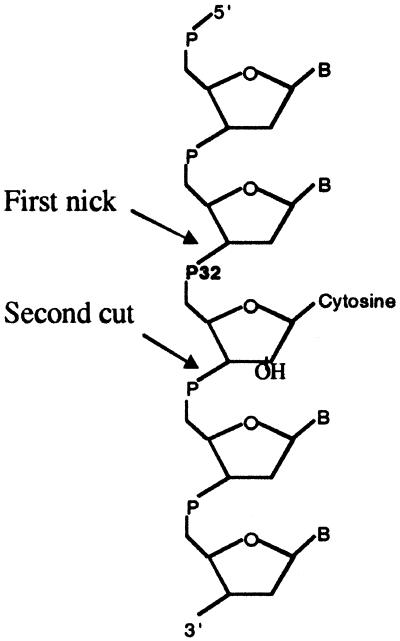
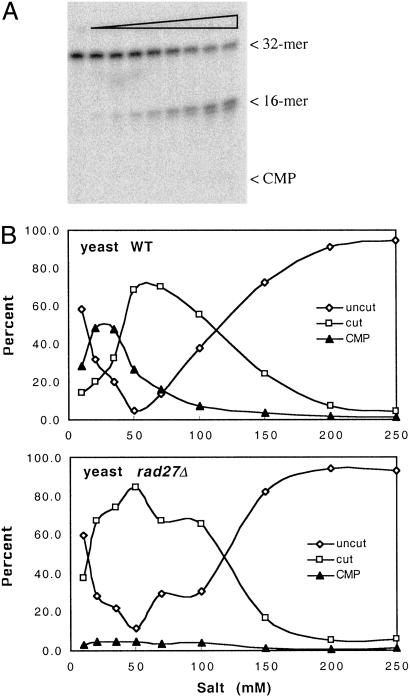
Cell-free extracts were prepared from human cells (Jurkat and HeLa cells), yeast cells (S. cerevisiae), and E. coli by using standard techniques (19) with Dounce homogenization (human cells), disruption with glass beads (yeast cells), or sonication (E. coli). Double-stranded probes 1 and 2 containing a single ribose with adjoining 32P were then incubated with the cell-free extracts and analyzed on denaturing PAGE gels. A robust nicking activity was detected in all cell-free extracts, with a nicked molecule seen as product using lower extract concentrations and with release of free cytidine monophosphate (CMP) at higher extract concentrations (Fig. 2). The activity depends on magnesium and is completely abolished by EDTA (data not shown). The release of CMP was not caused by general degradation of the oligomers, as could be determined by the use of end-labeled probes (results shown below). The initial nick occurred 5′ of the 32P label, as evidenced by the use of probe 1 (Fig. 3). As can be seen, KOH hydrolysis generated, as expected, a nick at the 3′ side of the ribose sugar, generating labeled products of 100–172 bp. On the other hand, the nicking activity in the extracts generated 32P-labeled products of 1–72 bp, as expected from nicking on the 5′ side of the 32P in this substrate. The 32P-CMP seen for higher extract concentrations comigrated with authentic CMP in several gel systems (data not shown). These results strongly suggest an activity that initiates the repair of misincorporated ribose in DNA. The 1-nt gap that results from excision of the ribonucleotide could probably be filled in by one of several pathways, for example by polymerase β and ligase III in association with XRCC1 in human cells (20). Fig. 4 shows the indicated locations of the nicks in relation to the ribose sugar. The position of the initial nick will classify the responsible enzyme as a class II endonuclease, belonging to the same family as, for example, the human AP endonuclease and most RNase H enzymes.
Fig 2.
Nicking of probe 2 by crude cell-free extracts. Lanes 1 and 8, no extract; lanes 2–7, increasing amounts of HeLa cell extract (0.001–0.1 μl/assay); lanes 9–15, increasing amounts of S. cerevisiae extract (0.004–0.5 μl/assay).
Fig 4.
Excision of ribonucleotide from oligomer probes by crude cell-free extracts.
Screen of Yeast Deletion Mutants.
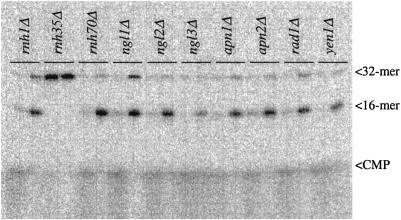
To identify the enzyme that causes nicking, we screened selected mutants from an annotated library of yeast deletion strains recently constructed by an international consortium (17). This strain collection contains >4,600 separate mutants involving almost every ORF in the genome that can be deleted without causing loss of viability. There are <100 known or putative endonucleases in S. cerevisiae, and only some of them are known as class II endonucleases. In the first test of 10 nuclease knockout strains, we included the deletion mutants for genes encoding each of the three yeast RNase H enzymes previously known to degrade the RNA strand in a DNA/RNA hybrid molecule, i.e., rnh1Δ, rnh35Δ, and rnh70Δ. As shown in Fig. 5, the rnh35Δ strain completely lacked the nicking activity at a single ribose, whereas all of the other strains in the panel had normal levels. We later confirmed this finding by backcrossing the rnh35Δ knockout strain with wild-type cells, finding that the lack-of-nicking phenotype cosegregated as expected with the drug resistance marker used to replace the rnh35 gene (data not shown). On the basis of primary sequence analysis (21, 22), RNase H(35) is a type 2 RNase H enzyme, whereas RNase H1 and RNase H(70) belong to type 1.
Fig 5.
Crude extracts from 10 S. cerevisiae knockout strains were assayed for nicking activity by using probe 2. Each strain was assayed with two extract concentrations (1 and 0.1 μl per assay). RNH1, RNH35, and RNH70 are the genes for the three known RNase H type enzymes present in S. cerevisiae. NGL1, NGL2, NGL3, and YEN1 are putative endonuclease genes, as determined by sequence homology. APN1 and APN2 are AP-endonuclease genes, and RAD1 is a gene for a 5′-endonuclease that functions in nucleotide excision repair. Note the absence of nicking in the extract generated from the rnh35Δ knockout strain. The amount of protein in the extracts was in the range of 1.4–2.1 mg/ml.
In previous studies related to the removal of Okazaki primers (22, 23), the human enzyme FEN-1 was found to be able to remove a single ribonucleotide at the 5′ end of DNA. This makes FEN-1 a candidate enzyme for the second cut that we observe in substrates with a single ribose. For this reason, we also studied yeast rad27 deletion mutant cells, because yeast Rad27p is the homologue of human FEN-1 (24). As seen in Fig. 6A, the second cut was indeed severely compromised in these knockout cells, suggesting that FEN-1 is the major enzyme involved in removal of the ribonucleotide after the initial nick has been made by the RNase H(35). Because the activities of the two enzymes depend on salt concentration, the activity was also measured in a wide range of salt concentrations in the assay. As seen in Fig. 6B, the initial nicking activity is similar in wild-type and rad27 knockout strains, whereas the release of free CMP is virtually absent in extracts of the rad27 mutant. The results support a novel DNA repair pathway, acting on misincorporated ribonucleotides in DNA, that consists of an initial nick by an RNase H type 2 enzyme followed by removal of the ribonucleotide by FEN-1 (Rad27p), as depicted in Fig. 4.
Fig 6.
(A) Increasing amounts of extract from a rad27Δ knockout cell line were incubated with probe 2. Compared with extracts with wild-type RAD27 (Fig. 5), only a very limited amount of CMP is seen, with the majority of product being the 16 mer even at a high concentration of extract per assay. (B) Products generated as a function of salt concentration in the assay for wild-type (Upper) and rad27Δ knockout (Lower) cell lines. Note the absence of CMP in the reaction using extract from the rad27Δ mutant.
Substrate Specificity.
Using probe 1, we tested nicking activity in probes containing misincorporated rC, rG, or rA and found that crude extracts from HeLa cells, yeast, and E. coli could nick the probes, with slight preference for rG and rA over rC. Interestingly, misincorporated rU was also found to be a good substrate in human and yeast extracts, including yeast extracts lacking Apn1p, the main AP endonuclease in yeast cells, whereas extracts from RNase H(35) knockout cells lacked the nicking activity (data not shown). This suggests that the ribose repair activity described here, rather than uracil DNA glycosylase and AP endonuclease, predominantly acts at such sites.
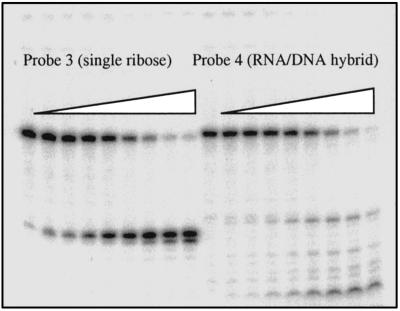
Using probes 3 and 4, we also compared the efficiencies of nicking by RNase H(35) at a single ribose with nicking of a RNA/DNA hybrid molecule as has been mainly studied previously (6, 22, 23). Using extracts from a rnh1Δ rnh70Δ double-knockout strain, lacking the other two main RNase H enzymes present in yeast cells, we find that the single ribose is cut 3-fold more efficiently than all cuts together within the stretch of 18 ribonucleotides present in probe 4 (Fig. 7). This further supports the notion that a main function for this enzyme is in repair of single misincorporated ribonucleotides. We also crossed the rnh1Δ rnh70Δ double-mutant strain with a rnh35Δ strain and observed viability and normal colony size in all 84 spore clones isolated from 21 meiotic tetrads from the triple heterozygote. This demonstrates viability for each of the double mutant genotypes and the triple mutant involving these three genes. We were readily able to identify a triple mutant spore clone and observed normal colony formation on plates in this strain. Hence, viability of the three single mutant rnh strains in yeast cannot be attributed to compensatory functions of either of the others in any essential process.
Fig 7.
Probes 3 and 4 were incubated with increasing amounts of crude extract from a double yeast mutant lacking RNaseH1 and RnaseH(70). The single ribose in probe 3 is the preferred substrate.
Nicking by Bacterial and Archaeal RNase HII.
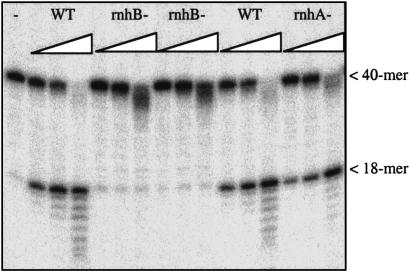
On the basis of sequence homology (23), the E. coli rnhB gene product RNase HII corresponds to the yeast RNase H(35) and human RNase H1, all type 2 enzymes. We therefore constructed an E. coli rnhB gene knockout strain by using the inducible λ bacteriophage recombination system (RED) of Yu et al. (16). As we expected, extracts made from rnhB knockout strains lack the nicking activity at a single ribose present in wild-type cells as well as in rnhA− cells lacking the RNase HI activity in E. coli (Fig. 8). The E. coli RNase HII enzyme is very weak in degrading RNA/DNA hybrid molecules, with <1% of the activity of the well known E. coli RNase HI enzyme (25). However, the nicking activity at a single ribose is very robust, pointing to a strong preference for a single ribose or deoxyribose/ribose link for this enzyme.
Fig 8.
Nicking of probe 3 (40 mer, leftmost lane) by increasing amounts of E. coli extracts from wild-type strain (WT), two independent rnhB knockout strains, and a rnhA− strain. Laddering is due to nonspecific nucleases in the extracts. Note the lack of specific nicking for the rnhB knockouts.
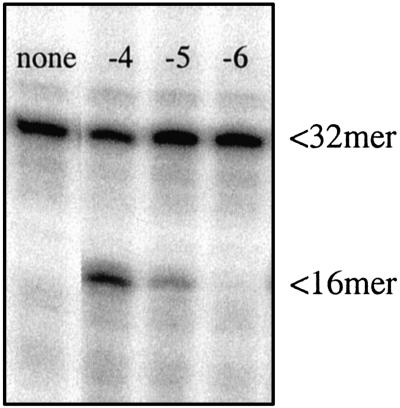
To further test the substrate specificity of this type of highly conserved RNase H type 2 enzymes, we used a cloned and purified RNase HII enzyme from the archaea Pyrococcus furiosus (a generous gift from John Tainer, The Scripps Research Institute, La Jolla, CA). This enzyme is very similar to the Archaeoglobus fulgidus RNase HII, studied by Chapados et al. (22). On SDS/PAGE, we found the purity of the enzyme to be ≈90%. We tested the activity of this preparation on our ribose-containing substrates and found a robust nicking activity down to a 10−5 dilution (original concentration, 50 mg/ml) (Fig. 9).
Fig 9.
Nicking of probe 2 by purified P. furiosus RNase HII. First lane: no enzyme. Next three lanes: 1 μl of enzyme added after dilution of the original stock solution 10−4 to 10−6, as indicated. The original stock solution had a concentration of 50 μg/μl.
Discussion
A Repair Pathway for Removal of Misincorporated Ribose in DNA.
Our results show biochemical evidence for a DNA repair pathway in bacteria, yeast, and human cells for removal of ribose residues in DNA. Such residues may be misincorporated during normal DNA replication, and it may be essential for cells to remove such residues. On the basis of the in vitro results, the pathway looks intriguingly simple. An endonuclease (RNase H, type 2) cuts 5′ to the lesion followed by FEN-1 (or homologous enzyme) cutting 3′ to the lesion leaving a 1-nt gap. The gap has the proper end groups to be a direct substrate for a DNA polymerase and DNA ligase without any further modifications. In human cells, DNA polymerase β and DNA ligase III in association with XRCC1 can complete repair at such sites (24). We believe the suggested simplicity of this repair pathway may hold true in prokaryotic systems. However, as has been generally demonstrated by other DNA repair pathways, a new level of complexity typically comes into play in higher eukaryotic systems. An indication of this is the presence of a second smaller subunit of the mammalian RNase H1 type 2 enzyme (26) with hitherto unknown function. It is the large subunit of the mammalian protein that has substantial sequence homology to yeast and prokaryotic RNase H type 2 enzymes (26) and the cloned large subunit functions as an RNase H enzyme on its own in vitro.
The suggested pathway for repair of sugar lesions has similarities to repair of AP sites, where an AP endonuclease makes a first cut at the same position relative to the damaged nucleotide as the RNase H type 2 enzyme. However. in this case, DNA polymerase β is generally believed to remove the abasic sugar fragment present on the 5′ end, and repair can be completed without participation of FEN-1 (27).
RNase H Enzymes and Their Proposed Functions.
The functional definition of RNase H enzymes is the ability to endonucleolytically degrade the RNA strand of a DNA/RNA hybrid molecule. RNase H enzymes are present in all kingdoms of life, often in multiple forms, but their cellular functions are still not well understood. On the basis of the primary amino acid sequence, two major unrelated families of RNase H enzymes are known, proposed to be called types 1 and 2 (21, 22). Type 1 enzymes have been proposed to be involved mainly in functions related to transcription, whereas the predominantly suggested function of type 2 enzymes is in removal of Okazaki primers during lagging strand DNA replication (22, 23). The main support for this function is given by in vitro studies of hybrid molecules that simulate the situation of an RNA primer extended as DNA on a DNA template. This substrate is preferred by the enzyme over a simple RNA/DNA hybrid molecule, with the strongest nicking occurring one base 5′ of the RNA-DNA link (6, 22, 23). The last remaining ribonucleotide still attached to the DNA can then be removed by FEN-1 (23). However, evidence for this function for RNase H type 2 enzymes in vivo is lacking and, as an alternative pathway, removal of Okazaki primers is believed to be efficiently accomplished by strand displacement followed by cutting by FEN-1, thus not requiring the activity of the RNase H enzyme in the process (23). An alternative view that these enzymes are involved in removal of misincorporated ribose in DNA has been expressed previously (6, 28, 29). In particular, early work by Eder and Walder (28) shows that purified human RNase H1 (a type 2 enzyme) is able to incise at the site of a single ribose, as opposed to E. coli RNase H1 (a type 1 enzyme), which requires a stretch of at least 4 ribose nt for activity. This ability, which is studied in more detail in the present paper, suggests a possible repair function of misincorporated ribose for type 2 enzymes. However, this possible function has not been emphasized before in literature about either RNase H enzymes or DNA repair. In recent years, it has become evident that type 2 enzymes are highly conserved and universally present in organisms of all kingdoms, with no known exceptions (26). A search of current databases identifies >100 RNase H type 2 enzymes with significant sequence homology from Rickettsia to Homo sapiens, indicating an important biological function.
The phenotypic properties of RNase H(35) type 2 knockout cells of the yeast S. cerevisiae have recently been studied (23, 29, 30). Such knockout cells have normal growth characteristics, arguing against an essential role for the enzyme in DNA replication. However, in a double mutant combination, rnh35Δ has been shown to further slow the reduced growth rate of rad27Δ mutants at 30°C, and this has been interpreted as implying an accessory role in replication (23). Also, rnh35Δ mutants are sensitive to HU (29), which is an inhibitor of ribonucleotide reductase, the enzyme that produces deoxyribonucleotides from ribonucleotides. High levels of HU may result in the collapse of replication forks, and sensitivity to HU can be argued to imply a function in replication, such as Okazaki primer processing. However, equally possible is the notion that depletion of deoxyribonucleotides promotes an increase in misincorporation of ribonucleotides into DNA, and lacking the ability to repair such misincorporation will result in sensitivity to HU. Knockout cells also have a moderate increase in spontaneous mutation rate (23), which also is consistent with a repair function. The spontaneous mutations are mainly frameshifts in the form of small deletions, reportedly with a preference for 4-bp deletions (30). Knockout cells are also moderately sensitive to ethyl methane sulfonate, an alkylating agent, suggesting a repair deficiency of lesions other than ribose (29).
The structure of the archaeal A. fulgidus RNase HII protein, which belongs to the family of type 2 enzymes, has recently been resolved at 1.95-Å resolution (22). Among the most interesting features is a DNA-binding cleft with evolutionarily conserved charged amino acids proposed to form a phosphate ruler motif that interacts in particular with the distorted backbone resulting from the inability of a RNA/DNA hybrid to form B form DNA. From such a mode of substrate discrimination based on backbone distortion, it can be speculated that substrate specificity would be quite wide and could include other sugar modifications in addition to ribose.
Acknowledgments
We thank Dr. John Tainer for generously providing purified P. furiosus RNase HII protein and Dr. Donald Court for providing the E. coli DY330 strain. J.G. acknowledges the technical assistance of Clelia Baccari. Work on this project was supported by National Institutes of Health Grant CA80207 to P. K. Cooper and by the Office of Biological and Environmental Research, Office of Energy Research, U.S. Department of Energy, under Contract No. DE-AC03-76SF00098. Work by J.G. was additionally supported by National Institutes of Health Grant GM599701 and Department of Energy Contract DEAC 76SF00098.
Abbreviations
HU, hydroxyurea
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.von Sonntag C. & Schulte-Frohlinde, D. (1978) Mol. Biol. Biochem. Biophys. 27, 204-226. [PubMed] [Google Scholar]
- 2.Isildar M., Schuchmann, M. N., Schulte-Frohlinde, D. & von Sonntag, C. (1981) Int. J. Radiat. Biol. 40, 347-354. [DOI] [PubMed] [Google Scholar]
- 3.von Sonntag C. (1984) Int. J. Radiat. Biol. 46, 507-519. [DOI] [PubMed] [Google Scholar]
- 4.Joyce C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 1619-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton N. C. & Finzel, B. C. (1996) J. Mol. Biol. 264, 521-533. [DOI] [PubMed] [Google Scholar]
- 6.Eder P. S., Walder, R. Y. & Walder, J. A. (1993) Biochimie 75, 123-126. [DOI] [PubMed] [Google Scholar]
- 7.Patel P. H. & Loeb, L. A. (2000) J. Biol. Chem. 275, 40266-40272. [DOI] [PubMed] [Google Scholar]
- 8.Gao G., Orlova, M., Georgiadis, M. M., Hendrickson, W. A. & Goff, S. P. (1997) Proc. Natl. Acad. Sci. USA 94, 407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sousa R. & Padilla, R. (1995) EMBO J. 14, 4609-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabor S. & Richardson, C. C. (1995) Proc. Natl. Acad. Sci. USA 92, 6339-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Astatke M., Ng, K., Grindley, N. D. & Joyce, C. M. (1998) Proc. Natl. Acad. Sci. USA 95, 3402-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner A. F. & Jack, W. E. (1999) Nucleic Acids Res. 27, 2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnin A., Lazaro, J. M., Blanco, L. & Salas, M. (1999) J. Mol. Biol. 290, 241-251. [DOI] [PubMed] [Google Scholar]
- 14.Kornberg A. & Baker, T. A., (1992) DNA Replication (Freeman, New York).
- 15.Gao G. & Goff, S. P. (1998) J. Virol. 72, 5905-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G. & Court, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C., Michaelis, S. & Mitchell, A., (1994) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 19.Deutscher M. P., (1990) Guide to Protein Purification (Academic, New York).
- 20.Kubota Y., Nash, R. A., Klungland, A., Schar, P., Barnes, D. E. & Lindahl, T. (1996) EMBO J. 15, 6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani N., Haruki, M., Morikawa, M., Crouch, R. J., Itaya, M. & Kanaya, S. (1999) Biochemistry 38, 605-618. [DOI] [PubMed] [Google Scholar]
- 22.Chapados B. R., Chai, Q., Hosfield, D. J., Qiu, J., Shen, B. & Tainer, J. A. (2001) J. Mol. Biol. 307, 541-556. [DOI] [PubMed] [Google Scholar]
- 23.Qiu J., Qian, Y., Frank, P., Wintersberger, U. & Shen, B. (1999) Mol. Cell. Biol. 19, 8361-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieber M. R. (1997) BioEssays 19, 233-240. [DOI] [PubMed] [Google Scholar]
- 25.Itaya M. (1990) Proc. Natl. Acad. Sci. USA 87, 8587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank P., Braunshofer-Reiter, C., Wintersberger, U., Grimm, R. & Busen, W. (1998) Proc. Natl. Acad. Sci. USA 95, 12872-12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett R. A., Wilson, D. M., III, Wong, D. & Demple, B. (1997) Proc. Natl. Acad. Sci. USA 94, 7166-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eder P. S. & Walder, J. A. (1991) J. Biol. Chem. 266, 6472-6479. [PubMed] [Google Scholar]
- 29.Arudchandran A., Cerritelli, S., Narimatsu, S., Itaya, M., Shin, D. Y., Shimada, Y. & Crouch, R. J. (2000) Genes Cells 5, 789-802. [DOI] [PubMed] [Google Scholar]
- 30.Chen J. Z., Qiu, J., Shen, B. & Holmquist, G. P. (2000) Nucleic Acids Res. 28, 3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]