Abstract
The Bin1/amphiphysin/Rvs167 (BAR) domain proteins are a ubiquitous protein family. Genes encoding members of this family have not yet been found in the genomes of prokaryotes, but within eukaryotes, BAR domain proteins are found universally from unicellular eukaryotes such as yeast through to plants, insects, and vertebrates. BAR domain proteins share an N-terminal BAR domain with a high propensity to adopt α-helical structure and engage in coiled-coil interactions with other proteins. BAR domain proteins are implicated in processes as fundamental and diverse as fission of synaptic vesicles, cell polarity, endocytosis, regulation of the actin cytoskeleton, transcriptional repression, cell-cell fusion, signal transduction, apoptosis, secretory vesicle fusion, excitation-contraction coupling, learning and memory, tissue differentiation, ion flux across membranes, and tumor suppression. What has been lacking is a molecular understanding of the role of the BAR domain protein in each process. The three-dimensional structure of the BAR domain has now been determined and valuable insight has been gained in understanding the interactions of BAR domains with membranes. The cellular roles of BAR domain proteins, characterized over the past decade in cells as distinct as yeasts, neurons, and myocytes, can now be understood in terms of a fundamental molecular function of all BAR domain proteins: to sense membrane curvature, to bind GTPases, and to mold a diversity of cellular membranes.
INTRODUCTION
The BAR domain proteins form a rapidly expanding protein family defined by the presence of a homologous α-helical domain of 250 to 280 amino acids named after the founding members of this family: Bin1, Amphiphysin, and Rvs167 (BAR) (278, 298). BAR domain proteins have been implicated in an extraordinary diversity of cellular processes, including fission of synaptic vesicles, cell polarity, endocytosis, regulation of the actin cytoskeleton, transcriptional repression, cell-cell fusion, signal transduction, apoptosis, secretory vesicle fusion, excitation-contraction coupling, learning/memory, tissue differentiation, ion flux across membranes, and tumor suppression. The conserved features of the BAR domain suggest there may exist an underlying common molecular mechanism that is provided by the BAR domain and that has been adapted for use in these different physiological processes.
Insight into what this common molecular mechanism may be has come from recent key discoveries. The first discovery was that BAR domains bind liposomes in vitro and convert low-curvature spheres to high-curvature tubules (316). The second discovery was that the BAR domain is itself curved (banana-shaped) and is therefore exquisitely designed both to sense membrane curvature and to actively influence membrane curvature (237). Furthermore, subsequent structure comparisons revealed that many proteins that bind GTPases do so through a similar protein fold (109). These discoveries have created great excitement, since it may now be possible to explain the diverse cellular roles of BAR domain proteins in terms of sensing membrane curvature, binding GTPases, and actively molding cellular membranes.
BAR domain proteins are encoded by most, if not all, eukaryotic genomes. They are found in organisms from lower unicellular eukaryotes such as budding yeast and fission yeast to insects, plants, and vertebrates. The phylogeny of BAR domain proteins has been the subject of an excellent recent review (109). Database homology searches with known BAR domains have not yet identified an obvious BAR domain encoded by the genome of any prokaryote. Sequence homology between known BAR domains is, however, relatively modest. The sequence features that confer folding of a polypeptide into a structure that can bind and bend membranes are still not fully understood. It is possible that proteins with similar properties exist also in prokaryotes but that low sequence homology has made them difficult to recognize. The apparent origin of BAR domain proteins in eukaryotes suggests a function(s) unique to eukaryotes. Some of the roles of BAR domain proteins (e.g., in membrane traffic) would be relevant only in eukaryotic cells. Hence, the evolution of BAR domain proteins may parallel the evolution of cellular compartmentation and complexity.
This article will first review the physiological roles and interactions of the BAR domain proteins, focusing primarily on the amphiphysins (including the Bin1-3 proteins) and the endophilins. Other BAR domain proteins, such as the sorting nexins (SNXs), will be discussed only briefly. In particular, this article focuses on the roles of BAR domain proteins in yeasts (budding yeast and fission yeast) and in mammals (with occasional reference to their roles in flies). This article then reviews the three-dimensional structure of the BAR domain. The article concludes with a somewhat speculative attempt to explain the known physiological roles of BAR domain proteins in yeasts and in mammals in terms of binding membranes and generating membrane tubules in vivo.
BUDDING YEAST Rvs PROTEINS
Nutrient Availability and the Control of Cell Proliferation
Yeasts as free-living unicellular organisms are at the mercy of their environment and experience a variety of stresses, including changes in osmolarity, temperature, pH, redox potential, and nutrient deprivation. Therefore, yeast cells must continuously monitor their extracellular environment and respond to changes in ways that ensure survival. A common response of yeast cells when confronted by stress is to arrest progression through the cell division cycle and become quiescent. For example, when starved of essential nutrients, dividing yeast cells arrest uniformly in late G1 of the cell division cycle prior to initiation of chromosome replication and formation of a daughter cell (bud) (185, 186). This arrest point in yeast is analogous to the “restriction point” in mammalian cells. Following cell division cycle arrest the starved yeast cells enter a quiescent phase (G0) (135). Yeast cells quiescent in G0 survive for long periods and are resistant to stress (247). However, if yeast cells cannot enter this quiescent state, their futile attempts to transit the cell division cycle without an adequate nutrient supply have dire consequences and the cells perish. Hence, the ability to stop dividing and become quiescent is vital for yeast cells to survive under adverse environmental conditions.
To identify genes important for cell cycle arrest in response to starvation, Michel Aigle and colleagues screened a random collection of UV-induced mutants for those specifically defective in their ability to adapt to starvation conditions. Mutants that were fully viable in the presence of nutrients but that lost viability more rapidly than wild-type cells when starved of glucose (carbon), ammonium (nitrogen), or sulfate (sulfur) were retained. Two mutants found to exhibit a Reduced Viability upon Starvation (Rvs−) phenotype and to carry mutations in distinct genes were named rvs161 and rvs167 the corresponding wild-type genes are RVS161 and RVS167, respectively). In cell cycle terminology, the proteins Rvs161p and Rvs167p are negative cell cycle regulators that link nutrient availability to cell cycle progression (12, 42).
Rvs161p and Rvs167p Proteins and Their Common BAR Domain
Comparison of Saccharomyces cerevisiae Rvs161p and Rvs167p revealed strong amino acid sequence homology between the two proteins (Fig. 1). The homology extended over the total length (265 residues) of Rvs161p and over the N-terminal 281 residues of Rvs167p. Over this region the two proteins exhibit 27% amino acid sequence identity and 52% amino acid sequence similarity. The homologous domain was predicted to contain two regions with predominantly α-helical structure (12, 47, 220, 298). The N-terminal homologous domain was initially named the Rvs domain (299). Because vertebrate amphiphysin 1 and Bin1 (see below) also feature a homologous domain, Sakamuro et al. renamed the Rvs domain the Bin1/Amphiphysin/Rvs167 (BAR) domain (Fig. 1) (278). The designation BAR domain is now commonly used to describe this homologous domain in both yeast and nonyeast proteins. More recently, this domain has been subdivided into a short N-terminal amphipathic α-helix (∼40 to 45 residues) and the BAR domain itself, as some BAR domain proteins lack the N-terminal amphipathic α-helix (Fig. 1).
FIG. 1.
Domain structure of yeast and human amphiphysin family proteins. Schematic representation of the domain organization of the budding yeast amphiphysin family proteins Rvs161p and Rvs167p, the fission yeast amphiphysin family proteins Hob1 and Hob3, and the human amphiphysin family proteins amphiphysin 1, Bin1/amphiphysin 2 (several tissue-specific and ubiquitous splice variants), Bin2, and Bin3, and endophilins A1, A2, A3, B1, and B2.
Rvs167p has two additional C-terminal domains that are not present in Rvs161p. Following the BAR domain is the glycine-, proline-, and alanine-rich (GPA-rich) region which contains no charged amino acids, includes a hydrophobic sequence, and is not predicted to adopt a defined secondary structure (Fig. 1). The GPA-rich region is followed by a Src Homology 3 (SH3) domain (Fig. 1) (12). SH3 domains are short, 50- to 70-amino-acid modules found in a diverse range of signal transduction and actin cytoskeletal proteins (234). They mediate protein-protein interactions by binding primarily (but not exclusively) to short linear proline-rich target motifs (261, 382).
BAR Domains of Rvs161p and Rvs167p Assemble into Heterodimers
Secondary-structure prediction identified two regions of high α-helical potential in both the Rvs161p BAR domain (residues 22 to 65 and 127 to 183) and the Rvs167p BAR domain (residues 30 to 57 and 144 to 191) (47, 220). Moreover, the COILS algorithm revealed that the BAR domain α-helices are amphipathic and have a propensity to form coiled-coil structures that may mediate interaction of the BAR domain with other coiled-coil proteins. Two-hybrid analysis revealed that Rvs167p and Rvs161p interact and that the interaction is mediated by their BAR domains. In this study, neither Rvs161p nor Rvs167p interacted with itself, suggesting that the Rvs proteins form obligate heterodimers. The in vivo association of Rvs161p with Rvs167p was confirmed by coimmunoprecipitation of Rvs161p with Rvs167p from yeast lysates in vitro (220).
Subsequently, no fewer than eight other two-hybrid studies have analyzed interactions between Rvs161p and Rvs167p and each protein with itself (18, 38, 59, 84a, 97, 136, 180, 339). All these studies confirmed interaction between Rvs161p and Rvs167p via the respective BAR domains. In contrast to the initial study (220), two of these later studies found that Rvs167p also forms homodimers via the BAR domain (38, 180). Furthermore, two-hybrid interaction of Rvs167p with itself persists when RVS161 is deleted, indicating that Rvs167p-Rvs167p interaction is either direct or mediated by a protein other than Rvs161p (180). Two later studies, however, concluded that Rvs167p does not form homodimers in vegetatively growing cells (i.e., it only forms heterodimers) (84a, 97). Interestingly, in cells lacking Rvs161p the steady-state level of Rvs167p is considerably reduced due to accelerated proteolysis and vice versa (180). These results represent strong evidence that in vivo Rvs161p and Rvs167p form heterodimers and function in concert.
Loss of Rvs161p and Loss of Rvs167p Cause Similar and Diverse Spectra of Phenotypes
The yeast rvs161 and rvs167 mutations are highly pleiotropic and the reported phenotypes are summarized below. For many of the phenotypes tested, rvs161 and rvs167 mutants have identical phenotypes and the rvs161 rvs167 double mutant is not more severely affected than either single mutant. A recent genome-wide analysis of genetic interactions of Rvs161p and Rvs167p revealed that loss of each protein is lethal in pairwise combination with loss of the same set of 49 other yeast proteins. This indicates that the roles of Rvs161p and Rvs167p in vegetative growth are identical. The rvs161-1 and rvs167-1 mutations identified in the initial mutant screen appear to be complete loss-of-function alleles because deletion or disruption of the RVS161 gene (rvs161Δ) or the RVS167 gene (rvs167Δ) in almost every case reveals a phenotype identical to that with the original mutant allele (12, 42, 53, 84a).
Reduced viability upon starvation (Rvs−).
As introduced above, rvs161 and rvs167 mutants were originally isolated based on their inability to maintain viability when starved of a source of nitrogen, carbon, or sulfur. For example, rvs161 cells grown to stationary phase in glucose-limited or nitrogen-limited medium and then left in the depleted medium for 60 h exhibit only 20 and 65% viability, respectively. In contrast, wild-type cells subjected in the same way to either glucose or nitrogen starvation maintain close to 100% viability. A similar loss of viability under starvation conditions has been shown for rvs167 cells. In nitrogen- or glucose-limited minimal medium, 90 to 100% of wild-type cells arrest without buds after 50 h. In contrast, under each condition ∼20% of rvs161 cells still exhibit a bud. Again, a similar defect in starvation-induced cell cycle arrest was found for rvs167 cells. Hence, under starvation conditions rvs mutants still attempt to transit the cell cycle and likely die as a consequence of their failure to arrest (12, 42).
Small GTPases of the Ras family regulate response to starvation in yeast. They do this by regulating adenylyl cyclase activity and production of the second messenger cyclic AMP (282). Yeast mutants with constitutively activated Ras exhibit reduced viability upon starvation and an inability to mount a normal physiological response to nutrient deprivation similar to rvs mutants. Wild-type yeast cells accumulate the storage carbohydrate glycogen when starved (177). One characteristic of the constitutively activated ras phenotype is the failure to accumulate glycogen upon glucose starvation (185, 325, 329). Can rvs mutant cells accumulate glycogen when nutrients become limiting, e.g., when cultures reach stationary phase and the cells cease growth? When they reach stationary phase both rvs161 and rvs167 mutant cells do indeed accumulate glycogen normally (12, 42, 53). This suggests the defect in rvs mutant cells is distinct from that in hyperactivated ras mutants or mutants in which adenylyl cyclase activity is constitutive. The molecular basis of the defect in response to starvation in rvs mutants is not yet known.
Growth sensitivity to salt (especially Na+).
rvs mutants are not only sensitive to starvation. They are also more sensitive to a range of other stresses that include the presence of high concentrations of various salts (e.g., NaCl, KCl, MgCl2, and Na2SO4) in the medium. This sensitivity is to salt rather than to osmotic strength, because rvs mutants are not sensitive to sorbitol at high osmotic strength. Moreover, there is selectivity for certain cations. For example, rvs161 mutants exhibit morphological defects at fivefold lower concentrations of NaCl (0.25 M) than of KCl (1.2 M) while wild-type cells are unaffected by either salt. Furthermore, rvs161 mutants are no more sensitive to LiCl than wild-type cells (although both are sensitive) (42). In this review the rvs salt sensitivity will be referred to as Na+ sensitivity to reflect this. The molecular basis of the enhanced sensitivity of rvs mutants to Na+ is not known.
Growth sensitivity to cytotoxic compounds.
The growth of both rvs161 and rvs167 mutants is hypersensitive to the presence of various cytotoxic compounds, including 3-amino-1,2,4-triazole (3-AT) and canavanine (12, 42). 3-AT is a histidine biosynthesis inhibitor that induces an artificial starvation response when added to cells. Canavanine is an arginine analog that can be incorporated into proteins in place of arginine and produce nonfunctional proteins. Yeast mutants defective in ubiquitin-mediated proteolysis are especially sensitive to canavanine and overexpression of ubiquitin confers canavanine resistance (32, 115). In addition, rvs161 cells have been reported to be hypersensitive to the effects of sinefungin, which completely blocks growth of rvs161 cells at 0.1 μM (rvs167 cells were not tested) (42). Sinefungin is a toxic analog of S-adenosylmethionine that inhibits methylation reactions. In yeast, sinefungin has recently been shown to inhibit methylation of guanosine to form m7GpppN, which is used to cap the 5′ end of mRNAs. Capping of mRNAs is in turn important for mRNA stability and efficient translation (36). Whether hypersensitivity to these compounds is specific or reflects a general hypersensitivity of rvs161 and rvs167 cells to all cytotoxic compounds is not known.
Growth sensitivity to elevated temperature.
The standard growth temperature for budding yeast is 30°C. Elevated temperature presents a stress to yeast cells and while wild-type cells continue to grow well at 37°C they stop dividing at 42°C. Yeast mutants with constitutively activated ras mutations exhibit not only reduced viability upon starvation, but also an inability to grow at normal temperature after a short heat shock at 55°C. In contrast, rvs161 and rvs167 mutant cells both survive heat shock at 55°C. In two reports, rvs161 cells did not exhibit defects in growth at low (e.g., 15°C) or normal (28°C) temperature and neither rvs161 nor rvs167 cells exhibit defects in growth at elevated (e.g., 36°C) temperature (12, 42).
The ability of rvs161 and rvs167 cells to grow at elevated temperature may be dependent on genetic background or the accumulation of second-site suppressor mutations. In a somewhat different genetic background rvs161 and rvs167 cells did not grow at 37°C (215). Exposure to elevated temperature causes induction of heat shock proteins (HSP) which aid in the refolding of damaged proteins. However, HSP induction upon exposure to elevated temperature was found to be normal in rvs167 mutant cells (12). rvs mutants are not sensitive to all environmental stresses, e.g., rvs161 and wild-type yeast cells are equally resistant to extremes of pH (12, 42).
Growth on nonfermentable carbon sources.
Both rvs161 and rvs167 mutants grow well on a range of fermentable carbon sources (e.g., glucose, galactose, mannose, and sucrose), but are unable to grow on nonfermentable carbon sources (e.g., glycerol, lactate, or acetate) (12, 42). An inability to utilize nonfermentable carbon sources suggests a possible mitochondrial defect. These carbon sources are metabolized in mitochondria (370). rvs161 mutant cells possess a normal spectrum of mitochondrial cytochromes, but respire threefold more slowly than wild-type cells as measured by oxygen consumption. It is not clear, however, whether this level of respiratory defect is sufficient to fully account for the observed failure to utilize nonfermentable carbon sources. Defects in glycerol utilization are a common phenotype of mutations that affect the actin cytoskeleton, but the molecular basis for the defect is still unclear. Mitochondrial defects alone cannot account for the Rvs− phenotype since deletions in the mitochondrial genome ([rho−]) that abolish growth on nonfermentable carbon sources do not give rise to the other Rvs− phenotypes such as reduced viability upon starvation (12, 42).
Meiosis and sporulation.
In the laboratory, budding yeast cells can be maintained as either stable diploids or stable haploids. Diploid yeast cells can be induced to undergo sporulation (meiosis) by nitrogen starvation on nonfermentable carbon sources. Sporulation has been well studied in yeast because it represents a cellular differentiation process that can be studied in budding yeast and that may have molecular mechanisms in common with more complex cellular differentiation pathways used in vertebrate development. Sporulation involves switching off the expression of blocks of genes required for vegetative growth and initiating a developmental program in which spore-specific genes are expressed in a highly regulated pattern. In addition to meiosis, sporulation also involves de novo bilayer membrane biogenesis to form the plasma membrane of the resultant haploid cell and also cell wall biogenesis to form the spore wall.
Desfarge et al. reported that homozygous rvs161/rvs161 diploid cells do not sporulate upon nitrogen starvation (53). Subsequently, Colwill et al. showed that rvs167/rvs167 homozygous diploid cells, although not entirely compromised for sporulation, form spores with only 10% the frequency of wild-type diploids (38). rvs/rvs homozygous diploids may not sporulate because they are unable to utilize the nonfermentable carbon source provided or because they lose viability upon nitrogen starvation. The sporulation defect in these mutants could be indirect. However, genetic interaction studies have suggested that the Rvs proteins play a direct role in sporulation (53) (discussed below). The molecular mechanisms that underlie this defect have yet to be elucidated.
Heterogeneous cell size and morphology upon starvation or exposure to Na+.
Although rvs161 and rvs167 mutant cells exhibit the relatively uniform size and ellipsoid shape of wild-type cells under optimal growth conditions, when starved or exposed to Na+ (or the cytotoxic compounds referred to above) cultures of both rvs161 and rvs167 cells accumulate a high proportion of cells that are either grossly enlarged with swollen vacuoles or abnormally tiny. Some mother cells also have multiple buds. Each bud has a nucleus but is apparently unable to complete cytokinesis and separate from the mother cell (12, 42). In wild-type yeast, there is a minimum cell size that is required at Start for commitment to a new cell division cycle (137). The rvs mutations appear to compromise this cell cycle regulation with under-sized mother cells continuing to divide.
Loss of actin cytoskeleton polarization to sites of polarized growth.
Yeast cells possess an actin cytoskeleton comprising filamentous actin (F-actin) and a diverse set of actin-associated proteins (71, 213, 371). Yeasts express orthologs of many, although not all, vertebrate actin-associated proteins, including the Arp2/3 complex and its activators, both conventional (filament forming) and unconventional myosins, profilin, tropomyosin, fimbrin, capping protein, and cofilin. When yeast cells are stained with fluorophore-conjugated phalloidin (an F-actin-specific reagent) several distinct structures are visible. Actin patches are small highly motile spots located at or near the cortex. Actin cables are long thick fibers comprising bundled actin filaments that extend through the cortical cytoplasm. Although individual actin patches and actin cables turn over, patches and cables are visible throughout the cell cycle.
Dividing yeast cells display a third F-actin structure known as the contractile actomyosin ring (16, 178). This ring structure localizes at the neck between the mother cell and bud. Contraction of the actomyosin ring occurs during cytokinesis and is accompanied by deposition of new cell wall material (septum). Septum deposition eventually separates the cytoplasm of the mother cell and bud and cleavage of the septum allows cell separation.
During the cell division cycle, the distribution of actin patches and actin cables changes. Immediately prior to bud emergence, cables align with their tips focused at the nascent bud site and patches concentrate at this site. When the bud starts to emerge, the cables align with their tips inside the growing bud and the patches localize at the bud tip. In G2 the actin patches remain polarized to the bud but switch from a polarized to an isotropic distribution within the bud. After the switch the bud expands laterally as well as at the tip. During mitosis actin is recruited to the actomyosin ring, cables become randomly oriented, and patches distribute randomly throughout the mother cell and bud. Finally, upon exit from mitosis the patches in the mother cell and bud repolarize to either side of the bud neck and cables in the mother cell and bud realign with their tips focused to the bud neck. At this stage of the cell cycle the actomyosin ring contracts to a dot, septum is deposited and cleaved, and the cells divide. The newly divided mother and daughter cells transiently retain polarized actin patches and cables, and then polarity is lost until a new bud site is selected (16, 178, 371).
When rvs161 and rvs167 mutant cells were first stained to visualize F-actin it was apparent that the actin cytoskeleton in these cells was abnormal. Actin patches were not as polarized at nascent bud sites and growing buds. Actin cables were more difficult to visualize and appeared less well aligned than those in wild-type cells. When rvs161 or rvs167 mutant cells were exposed to high levels of salt (e.g., NaCl) or starved, the loss of actin cytoskeleton polarity became complete. Under both stress conditions actin cables disappeared completely and actin patches depolarized fully (12, 298).
Delocalized cell wall chitin deposition.
In wild-type yeast cells the cell wall polysaccharide chitin is specifically deposited at the site of bud formation during both bud emergence and bud growth, and to seal the scar after separation of the mother cell and the bud. In rvs167 mutant cells, however, staining of chitin with Calcofluor and microscopic examination reveal that chitin is not restricted to sites of active bud formation and at scars from previous budding events. There appears to be an accumulation of chitin distributed evenly throughout the cell wall and this defect is exacerbated by the presence of a sublethal concentration of Na+ in the growth medium (12, 84a).
Loss of bipolar bud site selection.
Yeast cells do not bud randomly. After a bud has emerged from a mother cell, a scar made of chitin is left on the surface of both mother (bud scar) and daughter (birth scar). These scars are permanent and can be visualized by use of the fluoresent stain Calcofluor. Bud scars on mother cells accurately record all sites where previous buds have emerged. In wild-type haploid cells, the bud sites are visualized as a cluster at one pole of the cell in an “axial” pattern, which is generated when budding occurs in new mothers at a site adjacent to the birth scar and in old mothers at a site adjacent to the previous bud scar. In diploid cells the bud sites form clusters at both poles of the cell in a “bipolar” pattern, which is generated when budding occurs in new mothers at a site on the opposite pole to the birth scar and in old mothers at a site that alternates between opposite cell poles.
Mutant studies have shown that the requirements for axial and bipolar bud site selection are distinct. Mutations in the genes BUD1, BUD2, and BUD5 cause haploid cells to bud at random sites, while mutations in BUD3 and BUD4 cause haploid cells to bud in a bipolar pattern (29, 30). So some genes are required for axial budding and some for all patterns of budding. Interestingly, RVS161 and RVS167 were among the first genes discovered that are specifically required for bipolar bud site selection. Haploid rvs161 and rvs167 mutant cells bud in an axial pattern like wild-type cells. However, rvs161/rvs161 and rvs167/rvs167 homozygous mutant diploids bud at random sites (12, 53, 65, 298). The defect is not specific to diploid cells, but to the process of bipolar bud site selection. When haploid cells are induced to undergo bipolar budding (e.g., by mutation of BUD3 or BUD4) additional mutation of RVS161 or RVS167 results in random budding (65).
It is possible that spatial landmarks at the cell poles are not formed properly in rvs161 or rvs167 mutant cells. This would mean that when the next division ensues the cell can no longer “remember” where the previous bud formed to place the next bud site appropriately. Alternatively, spatial landmarks may be formed in rvs mutant cells, but not recognized or interpreted correctly. Subsequently, a close correlation has been established between those mutations that affect actin patch polarization and those that abolish bipolar bud site selection (375).
Defective fluid-phase and receptor-mediated endocytosis.
Endocytosis is the process by which cells internalize plasma membrane material as well as ligands, particles, and fluid from the extracellular environment via invagination of the plasma membrane and formation of endocytic vesicles. To identify genes important for endocytosis, Riezman and colleagues conducted a screen for yeast mutants unable to internalize plasma membrane receptor-ligand complexes. A bank of random yeast mutants was screened using an assay for receptor-mediated internalization of α-factor, the peptide ligand secreted by haploid yeasts of the α mating type. One of several mutants recovered from this screen carried a mutation in ENDocytosis-defective 6 (END6). Isolation and characterization of the END6 gene showed that END6 is identical to RVS161 and that the end6 mutation affects polarization of the actin cytoskeleton as observed for rvs161. A previously isolated rvs167Δ mutant was also blocked in internalization of α-factor. Hence, efficient receptor-mediated endocytosis in budding yeast requires both Rvs161p and Rvs167p. Uptake and accumulation in the vacuole of the membrane-impermeant fluid-phase endocytic dye lucifer yellow are also blocked in both rvs161 and rvs167 mutant cells (215).
Inefficient cell-cell fusion during mating.
Haploid budding yeast cells exist in two mating types, a (MATa) and α (MATα), that have an identical physical appearance. MATa cells secrete a peptide pheromone known as a-factor that binds to the a-factor receptor (a G-protein-coupled receptor called Ste3p) expressed only by MATα cells. Conversely, MATα cells secrete a peptide pheromone known as α-factor that binds to the α-factor receptor (a G-protein-coupled receptor called Ste2p) expressed only by MATa cells. Binding of each pheromone to its receptor activates a mitogen-activated protein kinase signal transduction cascade that has two main readouts. First, cell cycle progression is arrested in G1 to ensure that each mating cell has a normal 1C DNA content in preparation for nuclear fusion. Second, expression of various proteins specifically required for mating is induced, e.g., proteins that promote cell-cell adhesion and fusion (41, 365).
During mating, each haploid cell chooses a mate from the surrounding cells based on the level of pheromone each cell secretes. Yeast cells are able to detect gradients of pheromone with extraordinary sensitivity and form a tube-like projection at their surface known as a mating projection or shmoo. This projection grows up the gradient to the source of pheromone, i.e., towards the cell that produces the highest level. When mating yeast cells are treated with synthetic mating pheromone they mate at random with cells of the opposite mating type even if those cells produce no pheromone. This behavior, known as “default” mating, is approximately 10-fold less efficient than normal pheromone gradient mating. Supersensitive 2 (sst2) mutants are hypersensitive to endogenous pheromone and are consequently unable to accurately detect subtle pheromone gradients. They mate by default with random partners even in the presence of endogenous pheromone gradients (56).
In a search for mutants defective in default mating, candidate mutants were tested for their ability to mate with a “pheromoneless” partner in the presence of synthetic pheromone. One of the mutants tested was rvs161Δ. Earlier work had shown rvs161Δ sst2 double mutants (which mate by default due to sst2) mate extremely inefficiently. Indeed, the rvs161Δ mutant was specifically defective in default mating since rvs161Δ and wild-type cells mated with efficiency similar to that of wild-type cells of the opposite mating type (56).
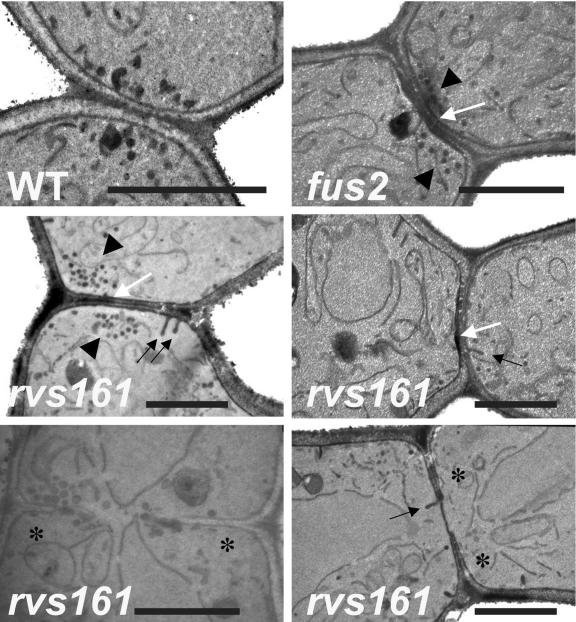
In an independent study, a screen was performed for mutants defective in mating (using endogenous pheromone) but only when both parents carry the mutation (i.e., bilateral mating defect) (158). One such mutation blocked mating at the stage of cell-cell fusion and was named fusion 7 (fus7) (91). Further work showed fus7 is a mutation in RVS161 (21). The mating defect of rvs161 cells was not apparent in the study of Dorer et al. (56) because only mating of rvs161Δ cells to wild-type cells was tested (which only detects unilateral mating defects). Interestingly, rvs167 cells do not exhibit a mating defect in either test, one case where rvs161 and rvs167 cells differ in phenotype.
Rvs161p and Rvs167p Structure-Function Relationships
The diverse range of phenotypes displayed by rvs161 and rvs167 mutants suggested that Rvs161p and Rvs167p are multifunctional proteins. In the case of Rvs167p, which comprises three distinct domains, each domain may confer different biological activities. Two studies investigated structure-function relationships in Rvs167p (38, 299). In the first study the ability of various truncated Rvs167p constructs, each lacking one or more Rvs167p domains, to complement the phenotypes of rvs167 was examined, i.e., viability upon glucose starvation, growth in the presence of Na+ or 3-AT, and utilization of a nonfermentable carbon source. The BAR domain alone was sufficient to rescue each phenotype tested, although less efficiently than full-length Rvs167p. In contrast, a fragment comprising only the GPA-rich and SH3 domains did not rescue any of the phenotypes (299).
The ability of the various truncated Rvs167p fragments to rescue the loss of actin cables and depolarization of actin patches in rvs167 mutant cells exposed to a sublethal concentration of Na+ was also examined. In general, the same Rvs167p fragments that were functional in growth assays were also able to rescue the actin cytoskeleton defects, i.e., the BAR domain alone was able to correct the actin cytoskeleton defects, but rescue by the BAR domain was not as complete as rescue by full-lengthRvs167p. The fragment comprising the GPA-rich and SH3 domains was not able to correct the actin cytoskeleton defects. Expression of the BAR domain alone restored bipolar bud site selection in 75% of rvs167/rvs167 homozygous diploid cells but full rescue of bipolar budding required full-lengthRvs167p. The GPA-rich and SH3 domain fragment did not rescue bipolar bud site selection (299).
In a later study, various Rvs167p constructs were tested for their ability to restore growth in the presence of Na+, bipolar bud site selection, fluid-phase endocytosis, and sporulation to cells lacking Rvs167p (rvs167Δ). In addition to a truncated Rvs167p construct lacking an SH3 domain (BAR-GPA), this study also employed a full-length Rvs167p construct featuring a P473L substitution in the SH3 domain that abolishes binding to proline-rich motifs. Unlike the BAR-GPA fragment, the full-length P473L mutant construct was expressed at the same steady-state level as full-length Rvs167p and fully rescued all rvs167Δ defects. This study concluded that Rvs167p is fully functional without a functional SH3 domain if expressed at normal levels.
Do the GPA-rich and SH3 domains contribute to Rvs167p function? The GPA-rich and SH3 fragment (GPA-SH3) did not rescue growth in the presence of Na+, bipolar bud site selection, or endocytosis, but unexpectedly was able to fully rescue the sporulation defect of rvs167Δ. The SH3 domain alone was also able to rescue the sporulation defect, showing that the SH3 domain was sufficient for this function (38).
The BAR domains of Rvs161p and Rvs167p are highly homologous. Sivadon et al. tested the effect of swapping the BAR domains. The Rvs167p GPA-rich and SH3 domains were fused to Rvs161p to create an artificial Rvs167p-like protein (Rvs161p-GPA-SH3). The fragment of Rvs167p comprising the BAR domain only was used as the corresponding Rvs161p-like protein (Rvs167p-BAR). Rvs161p-GPA-SH3 retained full ability to rescue the growth, actin cytoskeleton, and bud site selection defects of rvs161Δ, but did not rescue these defects in rvs167 cells. Rvs167p-BAR was partially functional in rescuing the phenotypes of rvs167 cells, but it lacked the ability to rescue the defects of rvs161 cells. Nor could Rvs167p-BAR and Rvs161p-GPA-SH3 when coexpressed rescue the growth, actin cytoskeleton, or bud site selection defects in rvs161 rvs167 double mutant cells (299). Clearly, the two BAR domains have important differences.
Enforced overexpression of Rvs167p from a strong promoter induces lethality at normal growth temperature. In contrast, overexpression of the GPA-SH3 construct does not affect growth. However, the SH3 domain is important for the overexpression phenotype of full-length Rvs167p. Overexpression of the full-length P473L mutant has milder deleterious effects than wild-type Rvs167p and these only become apparent at elevated temperature (38). The mechanism by which Rvs167p overexpression inhibits growth is not known. Consistent with an important function for the Rvs167p SH3 domain, a recent study identified conditions under which the Rvs167p SH3 domain becomes important for growth (84a).
Rvs Proteins Localize to the Cortical Actin Cytoskeleton
Subcellular localization of Rvs161p and Rvs167p.
Where do Rvs161p and Rvs167p localize in the cell? Rvs161p and Rvs167p interact with each other and there is compelling evidence that these two proteins function as a heterodimer in most, if not all, of their various cellular functions (18, 38, 97, 180, 220). Indeed, as discussed above, in the absence of either Rvs protein the other is unstable and is degraded (180). Paradoxically, Rvs161p and Rvs167p appear to exhibit somewhat distinct subcellular localizations in live cells when fused to the green fluorescent protein (GFP) for visualization by fluorescent imaging. It should be noted, however, that fusion to GFP alters the subcellular distribution of some proteins, so differences may reflect differences in the ability of Rvs161p and Rvs167p to tolerate fusion to GFP without perturbing their subcellular localization rather than differences between Rvs161p and Rvs167p in subcellular localization per se.
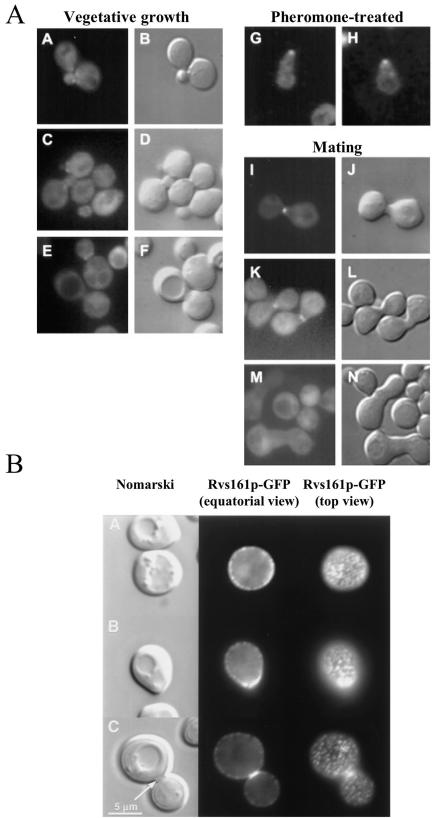
In growing cells a fusion protein comprising Rvs161p and GFP (Rvs161p-GFP) was reported to exhibit a predominantly diffuse cytoplasmic distribution in cells without buds (Fig. 2A). In cells with small buds Rvs161p-GFP localized to patches at the bud neck, but these patches were no longer apparent in large buds. Expression of Rvs161p-GFP fully rescued the cell fusion defect of an rvs161Δ mutant (see below), suggesting this fusion protein is functional (at least in cell fusion). This study focused on the role of Rvs161p in cell fusion during mating, so the ability of this fusion protein to rescue other rvs161Δ defects was not tested. Both N- and C-terminal Rvs161p fusions to GFP exhibited the same subcellular distribution (21). A more recent report demonstrated localization of a similar Rvs161p-GFP fusion protein to numerous small cortical patches (Fig. 2B). These patches exhibited polarization to nascent bud sites and small buds during bud emergence and to the bud neck in dividing cells. However, this particular Rvs161p-GFP fusion protein was not able to rescue the defects of an rvs161Δ mutant, so it is nonfunctional (10).
FIG. 2.
Subcellular localization of Rvs161p in growing and mating yeast cells. A. Shown are budding yeast cells that express fusion proteins that comprise full-length Rvs161p fused to the fluorescent reporter Aequoria victoria green fluorescent protein (GFP). The same fields of cells were viewed by fluorescence optics (left) to visualize GFP and by differential interference contrast optics (right) to visualize the cell profiles. In panels A to J, M, and N the cells express Rvs161p-GFP with the reporter fused at the Rvs161pC terminus. In panels K and L the cells express GFP-Rvs161p with the reporter fused at the Rvs161pN terminus. Panels A to F show vegetatively growing cells, panels G and H show cells arrested in G1 and forming mating projections after pheromone treatment, and panels I to N show mating cells. In pheromone-treated cells Rvs161p-GFP concentrates at the tip of the mating projection (shmoo) and in mating cells Rvs161p-GFP and GFP-Rvs161p concentrate at the site of cell-cell fusion. In vegetatively growing cells Rvs161p-GFP exhibits a diffuse cytoplasmic distribution and is excluded from the vacuole (large indentation apparent in the cell profiles). (Reproduced from reference 21 by copyright permission of The Rockefeller University Press.) B. Shown are vegetatively growing budding yeast cells that express a fusion protein (Rvs161p-GFP) that comprises full-length Rvs161p with GFP fused at the Rvs161pC terminus. The same fields of cells were viewed by fluorescence optics (center and right columns) to visualize GFP and by differential interference contrast (left column) to visualize the cell profiles. For each field of cells two focal planes were viewed by fluorescence optics: an equatorial view (center column) and a top view (right column). A, an unbudded cell; B, a cell at an early stage of bud emergence; C, cells undergoing cell division (cytokinesis). This Rvs161p-GFP fusion protein localizes to numerous small cortical patches. (Reprinted with permission from reference 10.)
A very recent report described the subcellular localization of an apparently fully functional Rvs161p-GFP fusion protein (144). This Rvs161p-GFP fusion localized to cortical actin patches but also exhibited a strong diffuse cytoplasmic distribution. Cortical actin patches are larger and less numerous than the patches to which the nonfunctional Rvs161p-GFP fusion used by Balguerie et al. localizes (10). Localization of Rvs161p-GFP to cortical actin patches is F-actin dependent, since localization to patches is abolished by depolymerization of all F-actin by treatment with latrunculin A (144). Confirmation of the subcellular distribution of native (untagged) Rvs161p by immunofluorescence staining with Rvs161p-specific antisera has not yet been reported. This may be due to the extreme sensitivity of Rvs161p antigenicity and/or subcellular localization to chemical fixation (our unpublished data).
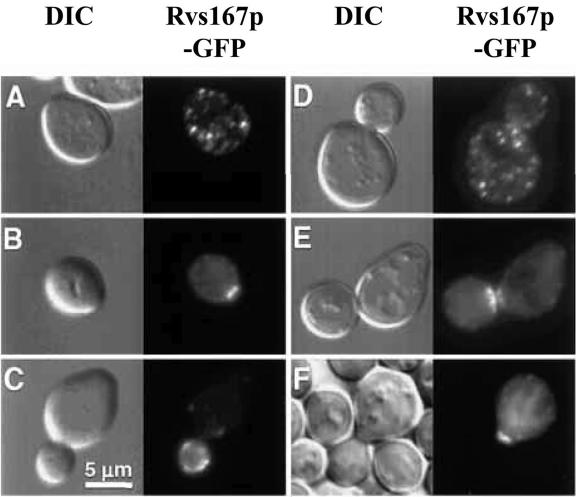
An Rvs167p-GFP fusion protein localizes in vegetatively growing cells to cortical actin patches (Fig. 3). The Rvs167p-GFP fusion protein was able to fully rescue the various defects of rvs167Δ cells and hence appears to be functional (11). This gives one confidence that this fusion protein displays an authentic Rvs167p subcellular localization. Double labeling of F-actin and Rvs167p-GFP shows that most patches that contain F-actin also contain Rvs167p-GFP and vice versa, although the relative signal intensity of the two proteins varies from one patch to another (11). This difference may reflect the age of the patch, since a number of studies have shown that the protein composition of an individual patch can vary during its lifetime (132, 139, 143, 144).
FIG. 3.
Subcellular localization of Rvs167p in growing and mating yeast cells. Shown are budding yeast cells that express a fusion protein (Rvs167p-GFP) that comprises full-length Rvs167p with GFP inserted between the BAR and GPA-rich domains of Rvs167p. The same fields of cells were viewed by fluorescence optics (right) to visualize GFP and by differential interference contrast (left) to visualize the cell profiles. A, unbudded cell; B, cell at an early stage of bud emergence; C, cell with small bud; D, cell with large bud; E, cells undergoing division (cytokinesis); F, cells arrested in G1 and forming a mating projection (shmoo) after pheromone treatment (only one cell expressing Rvs167p-GFP is depicted in this panel). In vegetatively growing cells Rvs167p-GFP localizes to large cortical patches that polarize to nascent bud sites, small buds, and the bud neck during cell division (cytokinesis). In mating cells Rvs167p-GFP concentrates at the tip of the mating projection (shmoo). (Reprinted from reference 11 with permission of the Company of Biologists Ltd.)
The distribution of Rvs167p-GFP patches changes as cells progress through the cell cycle exactly as for actin patches. Rvs167p-GFP specifically associates with actin patches and has not been observed to localize to other F-actin structures such as actin cables. Localization of Rvs167p is predominantly independent of F-actin, as disassembly of all F-actin in cells by treatment with the actin polymerization inhibitor latrunculin A does not abolish Rvs167p localization to cortical patches (11). A recent study reported, however, that loss of F-actin appears to cause a partial redistribution of Rvs167p from cortical patches, as there is an apparent increase in diffuse cytoplasmic Rvs167p after latrunculin A treatment (144).
Interactions between Rvs167p and actin patch proteins.
How does Rvs167p localize to actin patches? As loss of Rvs161p does not perturb actin patch localization of Rvs167p, it seems unlikely that interaction with Rvs161p mediates actin patch localization of Rvs167p (11).
Does Rvs167p associate with actin? In one study a two-hybrid screen was performed to identify actin-interacting proteins and a fragment encoding the Rvs167p SH3 domain was recovered. Hence, there is some evidence Rvs167p associates with actin. The Rvs167p-actin interaction requires the Rvs167p SH3 domain since deletion of the SH3 domain-encoding sequence abolishes the two-hybrid interaction (4). SH3 domains interact with short proline-rich motifs (e.g., PXXP) (331). The actin sequence contains only a single motif (PMNP) that might mediate SH3 domain interaction. Subsequently, it was proposed that the Rvs167p-actin interaction is indirect (38, 176, 180). A third protein might bind actin directly and contain proline-rich motifs that then bind the Rvs167p SH3 domain.
Systematic “charged-to-alanine” scanning mutagenesis of the yeast ACTin gene (ACT1) has been performed (363). Charged residues predicted to be surface exposed and potentially able to engage in interactions with other proteins were replaced singly and in clusters with uncharged alanine residues and a collection of 35 act1 mutations was generated. These act1 mutants vary in phenotype (e.g., some are inviable and others are without obvious phenotype). Two-hybrid analysis using these 35 mutant act1 genes and a panel of actin-interacting proteins revealed that different interactions are affected by different act1 mutations. Unexpectedly, all those act1 mutations that abolish interaction with Rvs167p also abolish interaction with another actin-binding protein, profilin. Conversely, the act1 mutations that do not affect interaction with profilin do not affect interaction with Rvs167p (4). So either Rvs167p binds to actin in the same way as profilin (i.e., makes the same contacts) or Rvs167p binds to profilin and the Rvs167p-actin interaction is indirect and mediated by profilin. To date, interaction between Rvs167p and profilin has not been demonstrated.
One actin patch component that has been proposed to mediate actin-Rvs167p interaction is Abp1p, encoded by the Actin Binding Protein 1 (ABP1) gene. Abp1p was first identified biochemically as a protein that binds with high affinity to F-actin in vitro (60). The Rvs167p SH3 domain recognizes Abp1p in a Far-Western blot and in two-hybrid screens. Consistent with a model that Abp1p contributes to Rvs167p function in vivo, loss of Abp1p results in defects in sporulation, growth in the presence of Na+, and growth on nonfermentable carbon sources, similar to but weaker than defects associated with loss of Rvs167p. Abp1p and Rvs167p also exhibit functional redundancy with the same set of proteins (i.e., with Sla1p, Sla2p, and Sac6p). Moreover, loss of Rvs167p reduces the deleterious effect of Abp1p overexpression on growth and cell morphology, which suggests that the deleterious effect of Abp1p overexpression requires an intact Rvs167p-Abp1p complex (38, 176). A recent study identified conditions under which Rvs167p-Abp1p interaction becomes important for vegetative growth (84a).
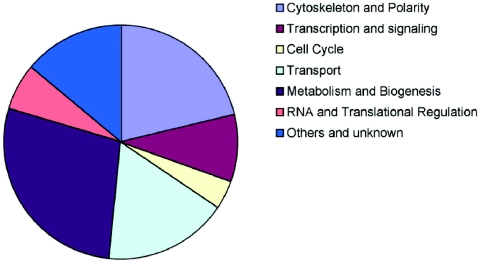
Despite the appeal of models in which Abp1p recruits Rvs167p into actin patches via interaction with the Rvs167p SH3 domain, experimental evidence suggests other mechanisms must also exist: deletion of ABP1 (abp1Δ) does not affect Rvs167p localization to actin patches (11), two-hybrid interaction between the Rvs167p SH3 domain and actin does not require Abp1p (180), the act1 two-hybrid data suggest that Rvs167p interaction with actin is mediated by a protein that binds monomeric actin (e.g., profilin) while Abp1p is an F-actin binding protein, localization of Rvs167p to cortical patches does not require F-actin, the SH3 domain of Rvs167p alone does not associate with actin patches in vivo, and the SH3 domain of Rvs167p is not essential for localization to actin patches (11, 38). Abp1p may function in Rvs167p localization to actin patches, but its role may be redundant with that of other proteins (see Table 1 for a list of other cortical actin patch proteins known to interact with Rvs167p).
TABLE 1.
Comprehensive list of Rvs161p and Rvs167p interactors in budding yeasta
| Group | Gene | Encoded protein | Type(s) of interaction (reference[s])b | Experimental evidence |
|---|---|---|---|---|
| Cytoskeleton and polarity | ABP1c | Actin Binding Protein involved in actin cytoskeleton assembly and cell polarity establishmentc | 7 (38, 59, 160, 176) | Two-hybrid, peptide scanning |
| ACF2/PCA1 | Actin Assembly Complementing Factor, intracellular β-1,3-endoglucanase | 7 (59, 97, 160, 331) | Two-hybrid, peptide scanning | |
| ACF4 | Actin Assembly Complementing Factor, molecular function unknown | 7 (59, 160, 331) | Two-hybrid, peptide scanning | |
| ACT1 | Yeast ACTin | 1 (19, 215), 7 (4, 19, 59, 180) | Two-hybrid, synthetic lethal, nonallelic noncomplementation | |
| APP1 | Actin Patch Protein, molecular function unknown | 1 (18), 7 (18, 59, 160, 331) | Two-hybrid, peptide scanning | |
| ARP2 | Actin-Related Protein, essential component of the Arp2/3 complex | 1 (332), 7 (160, 332) | Two-hybrid, synthetic lethal (rvs161), synthetic sick (rvs167) | |
| BBC1 | Protein possibly involved in assembly of actin patches; interacts with actin assembly factor Las17p and with the SH3 domains of type 1 myosins Myo3p and Myo5p | 1 (332), 7 (332) | Synthetic sick | |
| BSP1 | Binding protein of Synaptojanin Polyphosphoinositide phosphatase domain, adaptor that links synaptojanins Inp52p (Sjl2p) and Inp53p (Sjl3p) to the cortical actin cytoskeleton | 7 (59, 160, 331) | Two-hybrid, peptide scanning | |
| CAP1 | CAPping protein, binds to barbed ends of actin filaments, preventing further polymerization | 1 (332), 7 (332) | Synthetic lethal | |
| CAP2 | CAPping protein, binds to barbed ends of actin filaments, preventing further polymerization | 1 (332), 7 (332) | Synthetic lethal | |
| CDC24 | Cell Division Cycle, guanine nucleotide exchange factor for Cdc42p | 1 (136) | Two-hybrid | |
| CLA4 | Protein serine/threonine kinase, homologous to Ste20p | 1 (332), 7 (332) | Synthetic sick | |
| END3 | ENDocytosis defect, EH domain-containing protein involved in endocytosis, actin cytoskeletal organization, and cell wall morphogenesis | 1 (332), 7 (85, 332) | Synthetic sick | |
| EXO70 | EXOcyst, 70-kDa subunit of exocyst complex | 7 (18) | Two-hybrid | |
| GIM3 | Gene Involved in Microtubule biogenesis, subunit of the heterohexameric cochaperone prefoldin complex | 1 (332), 7 (332) | Synthetic sick | |
| GIM4 | Gene Involved in Microtubule biogenesis, subunit of the heterohexameric cochaperone prefoldin complex | 1 (332), 7 (332) | Synthetic sick | |
| GIM5 | Gene Involved in Microtubule biogenesis, subunit of the heterohexameric cochaperone prefoldin complex | 1 (332), 7 (332) | Synthetic lethal | |
| LAS17 | Yeast mutant that is Local Anesthetic Sensitive, yeast WASp | 1 (123), 7 (18, 38, 59, 97, 123, 136, 160, 181, 331) | Two-hybrid, peptide scanning, ELISA, complex isolation, and mass spectrometry | |
| MYO1 | Yeast type II MYOsin | 1 (19), 7 (19) | Synthetic lethal | |
| MYO2 | Yeast type V MYOsin | 1 (19), 7 (19) | Synthetic sick | |
| MYO3 | Yeast type I MYOsin | 7 (331) | Two-hybrid | |
| MYO5 | Yeast type I MYOsin | 1 (332), 7 (331, 332) | Two-hybrid, synthetic sick | |
| PAC10 | Protein required in the Absence of Cin8p, part of heteromeric cochaperone GimC/prefoldin complex, also called GIM2 | 1 (332), 7 (332) | Synthetic lethal | |
| RVS161 | Reduced Viability upon Starvation | 7 (18, 38, 59, 97, 123, 136, 180, 220, 339) | Two-hybrid, affinity purification, complex isolation, and mass spectrometry | |
| RVS167 | Reduced Viability upon Starvation | 1 (18, 38, 59, 97, 123, 136, 180, 220, 339), 7 (18, 180) | Two-hybrid, affinity purification, complex isolation, and mass spectrometry | |
| SAC6 | Suppressor of ACtin mutations, yeast fimbrin | 1 (97), 7 (97, 176) | Two-hybrid, synthetic lethal | |
| SEC8 | SECretory, essential 121-kDa subunit of the exocyst complex | 7 (18) | Two-hybrid | |
| SLA1 | Synthetic Lethal with ABP1, protein required for assembly of the cortical actin cytoskeleton | 1 (123, 332), 7 (59, 85, 97, 123, 176, 307, 332) | Two-hybrid, Co-IP, affinity purification, synthetic lethal, complex isolation, and mass spectrometry | |
| SLA2 | Synthetic Lethal with ABP1, protein required for assembly of the cortical actin cytoskeleton | 1 (97), 7 (59, 97, 176, 364) | Two-hybrid, synthetic lethal | |
| SUR7 | SUppressor of Rvs167 | 1 (300, 381), 7 (300, 381) | Genetic suppression | |
| VRP1 | Very Rich in Proline, yeast WASp-interacting protein | 1 (123), 7 (123, 275, 331) | Two-hybrid, synthetic sick, complex isolation, and mass spectrometry | |
| YKE2 | Yeast nuclear gene encoding a protein showing homology to mouse KE2 and containing a putative leucine zipper motif | 1 (332), 7 (332) | Synthetic sick | |
| YSC84 | SH3 domain-containing protein with function unknown, also called LSB4 (LaS17p-Binding) | 7 (59) | Two-hybrid | |
| Transcription and signaling | BCK1 | Bypass of C Kinase, mitogen-activated protein kinase kinase kinase | 1 (332), 7 (332) | Synthetic sick |
| CTI6 | Cyc8-Tup1-Interacting protein, transcription factor binding protein | 7 (18) | Two-hybrid | |
| EAP1 | EIF4E-Associated Protein, implicated in TOR signaling | 1 (332), 7 (332) | Synthetic lethal (rvs161), synthetic sick (rvs167) | |
| GTS1 | Glycine Threonine Serine repeat protein, transcription activator activity | 7 (160) | Peptide scanning | |
| RIM101 | Regulator of IME2, transcriptional repressor involved in the response to pH | 7 (160) | Peptide scanning | |
| RVB2 | RuVB-like, involved in transcription regulation | 7 (123) | Affinity purification | |
| SAP30 | SIT4 protein phosphatase-Associated Protein, subunit of a histone deacetylase complex | 1 (332), 7 (332) | Synthetic sick | |
| SDS3 | Suppressor of Defective Silencing, involved in transcriptional silencing and required for sporulation | 1 (332), 7 (332) | Synthetic lethal | |
| SIN3 | Switch INdependent, DNA binding subunit of Sin3p-Rpd3p histone deacetylase complex | 1 (332), 7 (332) | Synthetic sick | |
| SLN1 | Synthetic Lethal of N-end rule, histidine kinase osmosensor that regulates a MAP kinase cascade | 7 (18) | Two-hybrid | |
| SLT2 | Suppression at Low Temperature, serine/threonine MAP kinase | 1 (20, 332), 7 (20, 332) | Synthetic lethal | |
| SUM1 | SUppresor of Mar1-1, nuclear protein involved in silencing | 1 (332), 7 (332) | Synthetic sick | |
| SWI4 | SWItching deficient, involved in cell cycle-dependent gene expression | 1 (332), 7 (332) | Synthetic lethal | |
| TFC6 | Transcription Factor C, subunit of RNA polymerase III transcription initiation factor | 7 (18) | Two-hybrid | |
| TY1B | Transposon Ty1 protein B | 7 (18) | Two-hybrid | |
| Cell cycle | BBP1 | Bfr1p Binding Protein, required for the spindle pole body (SPB) duplication | 7 (160) | Peptide scanning |
| CYK3 | CYtoKinesis, SH3 domain protein located in the mother-bud neck, molecular function unknown | 1 (332), 7 (332) | Synthetic sick | |
| ESP1 | Extra Spindle Pole bodies, sister chromatid separase | 7 (18) | Two-hybrid | |
| PCL2 | PHO85 CycLin, forms a functional kinase complex with Pho85p, activated by Swi5p | 7 (163) | Two-hybrid, direct binding, Co-IP | |
| PCL9 | PHO85 CycLin, forms a functional kinase complex with Pho85p, activated by Swi5p | 7 (163) | Two-hybrid | |
| RED1 | REDuctional division, involved in chromosome segregation during the first meiotic division | 1 (114) | Two-hybrid | |
| Transport | FEN1/ELO2, ISUR5/VBM2 | FENpropimorph resistance, fatty acid elongase, involved in sphingolipid biosynthesis | 1 (262) | Genetic suppression |
| GRD19 | Golgi Retention Deficient, sorting nexin required to maintain late-Golgi-resident enzymes | 7 (350) | Two-hybrid | |
| GUP1 | Glycerol Uptake, plasma membrane protein with a possible role in proton symport of glycerol | 1 (332), 7 (332) | Synthetic lethal | |
| GYL1 | GYp-Like, putative GTPase-activating protein, stimulates Gyp5p GAP activity on Ypt1p | 1 (18), 7 (18, 84, 317, 331) | Two-hybrid, affinity purification | |
| GYP5 | GTpase-activating protein for Ypt Proteins, GAP for yeast rab1 | 1 (18), 7 (18, 84, 160, 317) | Two-hybrid, peptide scanning, affinity purification | |
| HSE1 | Has Symptoms of class E mutants, subunit of the endosomal Vps27p-Hse1p complex | 7 (18) | Two-hybrid | |
| IVY1 | Phospholipid-binding protein that Interacts with both Ypt7p and Vps33p | 7 (18) | Two-hybrid | |
| KAP122 | KAryoPherin, responsible for import of the Toa1p-Toa2p complex into the nucleus | 7 (18) | Two-hybrid | |
| MGE1 | Mitochondrial GrpE, involved in protein import into mitochondria | 1 (123) | Affinity purification | |
| MNN2 | MaNNosyltransferase | 1 (332), 7 (332) | Synthetic sick | |
| MNN9 | MaNNosyltransferase | 1 (332), 7 (332) | Synthetic lethal | |
| MNN10 | MaNNosyltransferase | 1 (332), 7 (332) | Synthetic lethal | |
| MRS3 | Mitochondrial RNA Splicing, mitochondrial iron transporter | 7 (160) | Peptide scanning | |
| PEX14 | PEroXisome related, peroxisomal membrane protein, a central component of the peroxisomal protein import machinery | 7 (385) | Peptide scanning | |
| PHO84 | PHOsphate metabolism, phosphate transporter and low-affinity manganese transporter | 7 (123) | Affinity purification | |
| POR1 | PORin, mitochondrial porin (voltage-dependent anion channel) | 1 (123) | Affinity purification | |
| PSE1 | Protein Secretion Enhancer, karyopherin/importin that interacts with the nuclear pore complex | 7 (18) | Two-hybrid | |
| RSP5 | Reversal of Spt−Phenotype 5, yeast NEDD4 ubiquitin ligase | 7 (123, 307) | Two-hybrid, co-immunoprecipitation, affinity purification, complex isolation, and mass spectrometry | |
| RUD3 | Relieves Uso1-1 transport Defect, Golgi matrix protein involved in the structural organization of the cis-Golgi | 1 (332), 7 (332) | Synthetic sick | |
| SEC21 | SECretory, COP1 complex component | 7 (18) | Two-hybrid | |
| SEC22 | SECretory, R-SNARE protein, cycles between ER and Golgi | 1 (332), 7 (332) | Synthetic sick | |
| SEC27 | SECretory, essential beta-coat protein of the COPI coatomer | 1 (123) | Affinity purification | |
| SRP54 | Signal Recognition Particle subunit | 7 (18) | Two-hybrid | |
| SPF1 | Sensitivity to Pichia farinosa killer toxin, P-type ATPase, ion transporter of the ER membrane involved in ER function and Ca2+ homeostasis | 1 (332), 7 (332) | Synthetic lethal | |
| SUR4/ELO3, /VBM1 | SUppressor of rvs161 and rvs167 mutations, elongase III synthesizes 20-26-carbon fatty acids from C18-CoA primers | 1 (53) | Genetic suppression | |
| SXM1 | Suppressor of mRNA eXport Mutant, nuclear transport factor (karyopherin) | 1 (18) | Two-hybrid | |
| VPS21/YPT51 | Vacuolar Protein Sorting, Rab5-like GTPase | 1 (97, 332), 7 (97, 296, 332) | Synthetic sick | |
| Metabolism and biogenesis | ARG1 | ARGinine requiring, acetylglutamate synthase | 7 (123) | Affinity purification |
| BNI4 | Bud Neck Involved, required for localization of chitin synthase III to the bud neck | 1 (332), 7 (332) | Synthetic sick (rvs161), synthetic lethal (rvs167) | |
| CCW12 | Covalently linked Cell Wall protein, expression down-regulated by alpha factor | 1 (332), 7 (332) | Synthetic lethal | |
| CHS3 | CHitin Synthase-related, chitin synthase III | 1 (332), 7 (332) | Synthetic sick | |
| CHS5 | CHitin Synthase-related, molecular function unknown | 1 (332), 7 (332) | Synthetic sick | |
| CHS6 | CHitin Synthase-related, molecular function unknown | 1 (332), 7 (332) | Synthetic sick | |
| CHS7 | CHitin Synthase-related, molecular function unknown | 1 (332), 7 (332) | Synthetic sick | |
| COR1 | CORe protein of QH2 cytochrome c reductase | 7 (123) | Affinity purification | |
| CSF1 | Cold Sensitive for Fermentation | 1 (332), 7 (332) | Synthetic lethal | |
| DEP1 | Disability in regulation of Expression of genes involved in Phospholipid biosynthesis | 1 (332), 7 (332) | Synthetic sick | |
| DOA1 | Degradation Of Alpha2, regulatory component of the proteasome pathway | 1 (332), 7 (332) | Synthetic sick | |
| ECM29 | ExtraCellular Mutant, major component of the proteasome | 7 (123) | Affinity purification | |
| FKS1 | FK506 Sensitivity, catalytic subunit of 1,3-β-d-glucan synthase | 7 (332) | Synthetic lethal | |
| FUS2 | cell FUSion, required for the alignment of parental nuclei before nuclear fusion during mating | 1 (18, 21, 97, 136, 221) | Two-hybrid, Co-IP | |
| GDH3 | Glutamate DeHydrogenase 3 | 7 (97) | Two-hybrid | |
| HOC1 | Homologous to OCh1, α-1,6-mannosyltransferase involved in cell wall mannan biosynthesis | 1 (332), 7 (332) | Synthetic sick (rvs161) synthetic lethal (rvs167) | |
| HOM6 | HOMoserine requiring, homoserine dehydrogenase | 7 (123) | Affinity purification | |
| HSP90 | Heat Shock Protein, cytoplasmic chaperone | 1 (389), 7 (332) | Synthetic lethal | |
| IDH1 | Isocitrate DeHydrogenase | 7 (123) | Affinity purification | |
| ILV5 | IsoLeucine-plus-Valine requiring, acetohydroxy acid reductoisomerase | 7 (123) | Affinity purification | |
| IPT1 | Inositol PhosphoTransferase | 1 (10) | Genetic suppression | |
| LPD1 | LiPoamide Dehydrogenase | 7 (123) | Affinity purification | |
| KGD2 | α-KetoGlutarate Dehydrogenase | 7 (123) | Affinity purification | |
| KRE1 | Killer toxin REsistant, cell wall glycoprotein involved in β-glucan assembly | 1 (332), 7 (332) | Synthetic sick | |
| KRE6 | Killer toxin REsistant, cell wall glycoprotein involved in β-glucan assembly | 7 (20) | Synthetic sick | |
| LYS12 | LYSine requiring | 7 (123) | Affinity purification | |
| MET18 | METhionine requiring | 7 (123) | Affinity purification | |
| MUS81 | MMS and UV Sensitive, Helix-hairpin-helix protein | 7 (18) | Two-hybrid | |
| PDI1 | Protein Disulfide Isomerase | 1 (114) | Two-hybrid | |
| PHO23 | PHOsphate metabolism, component of the Rpd3 histone deacetylase complex | 1 (332), 7 (332) | Synthetic sick | |
| PMI40 | PhosphoMannose Isomerase | 7 (123) | Affinity purification | |
| PRE10 | PRoteinase yscE, 20S proteasome α-type subunit | 7 (123) | Affinity purification | |
| PSK1 | Pas domain-containing Serine/threonine protein Kinase | 7 (160) | Peptide scanning | |
| SKT5 | Activator of Chs3p (chitin synthase III) | 1 (332), 7 (332) | Synthetic sick (rvs161), synthetic lethal (rvs167) | |
| SMI1 | Suppressor of MAR Inhibitor, involved in (1,3)-β-glucan synthesis | 7 (332) | Synthetic sick | |
| SML1 | Suppressor of Mec Lethality, ribonucleotide reductase inhibitor | 7 (160) | Peptide scanning | |
| SRV2 | Suppressor of RasVal19, adenylyl cyclase-associated protein | 7 (59, 176) | Two-hybrid, synthetic sick | |
| SUR1 | SUppressor of rvs161 and rvs167 mutations, catalytic subunit of a mannosylinositol phosphorylceramide synthase | 1 (53) | Genetic suppression | |
| SUR2 | SUppressor of rvs161 and rvs167 mutations, sphingosine hydroxylase | 1 (53) | Genetic suppression | |
| TPS1 | Trehalose-6-Phosphate Synthase, regulator of glucose influx into the cell and into glycolytic pathway | 1 (332), 7 (332) | Synthetic sick | |
| TRP5 | TRyPtophan requiring | 7 (123) | Affinity purification | |
| UBI4 | UBIquitin | 7 (123) | Affinity purification | |
| UBP7 | UBIquitin-specific Protease | 7 (123) | Affinity purification | |
| URA7 | URAcil requiring | 7 (18, 123) | Affinity purification | |
| RNA and translational regulation | CDC33 | Cell Division Cycle, cytoplasmic mRNA cap binding protein | 7 (123) | Affinity purification |
| DED1 | DEaD-box protein, ATP-dependent DEAD (Asp-Glu-Ala-Asp)-box RNA helicase | 7 (123) | Affinity purification | |
| KRS1 | Lysyl (K) tRNA Synthetase | 7 (123) | Affinity purification | |
| LCP5 | Lethal with Conditional Pap1, essential protein involved in maturation of 18S rRNA | 7 (18) | Two-hybrid | |
| MSU1 | Degradosome associates with the ribosome and mediates turnover of RNAs | 7 (18, 331) | Two-hybrid | |
| PRP40 | Pre-mRNA Processing, U1 snRNP protein involved in splicing | 7 (114) | Two-hybrid | |
| RPC40 | RNA Polymerase C subunit | 7 (123) | Affinity purification | |
| SES1 | SEryl-tRNA Synthetase | 7 (123) | Affinity purification | |
| SLF1 | Associates with translating ribosomes | 7 (18) | Two-hybrid | |
| SNP1 | U1snRNP 70K protein homolog | 7 (160) | Peptide scanning | |
| Others and unknown | CUE5 | Coupling of Ubiquitin conjugation to ER degradation | 7 (160) | Peptide scanning |
| DEM1 | Differentially Expressed in Malignancies, molecular function unknown | 7 (160) | Peptide scanning | |
| ECM30 | ExtraCellular Mutant, function unknown | 7 (18) | Two-hybrid | |
| GFD2 | Great for Full DEAD box protein activity, molecular function unknown | 7 (18) | Two-hybrid | |
| HUA1 | Unknown function | 7 (59, 136) | Two-hybrid | |
| HUA2 | Unknown function | 7 (59) | Two-hybrid | |
| RXT2 | Molecular function unknown | 1 (332), 7 (332) | Synthetic sick | |
| SDS23 | Homolog of S. pombe SDS23, implicated in APC/cyclosome regulation | 1 (114) | Two-hybrid | |
| YBP2 | Yap1 Bing Protein, protein with a role in resistance to oxidative stress | 7 (18, 136) | Two-hybrid, affinity purification | |
| YBR108W | Hypothetical ORF | 1 (339), 7 (59, 97, 136, 160) | Two-hybrid, peptide scanning | |
| YBR239C | Hypothetical ORF | 7 (160) | Peptide scanning | |
| YBR255W | Hypothetical ORF | 1 (332), 7(332) | Synthetic sick | |
| YDL156W | Hypothetical ORF | 7 (160) | Two-hybrid, peptide scanning | |
| YDR154C | Hypothetical ORF | 1 (136) | Two-hybrid | |
| YDR239C | Hypothetical ORF | 7 (160) | Peptide scanning | |
| YGL085W | Hypothetical ORF | 7 (160) | Peptide scanning | |
| YLR111W | Hypothetical ORF | 1 (332), 7 (332) | Synthetic sick | |
| YLR243W | Hypothetical ORF | 7 (123) | Affinity purification | |
| YNL086W | Hypothetical ORF | 7 (59) | Two-hybrid | |
| YNL152W | Hypothetical ORF | 7 (160) | Peptide scanning | |
| YPL009C | Hypothetical ORF | 7 (160) | Peptide scanning | |
| YPR091C | Hypothetical ORF | 7 (160) | Peptide scanning |
This table contains a comprehensive listing of all reported genetic and physical interactions. Many interactions require further confirmation. Note that some interactions of Rvs proteins are with proteins whose predominant subcellular localization seems incompatible with interaction (e.g., Pdi1p is a protein of the ER lumen). The existence of additional (perhaps minor) cytoplasmic pools of these proteins cannot be formally excluded and so these putative interactions are included here. Abbreviations: ELISA, enzyme-linked immunosorbent assay; Co-Ip, coimmunoprecipitation; MAP, mitogen-activated protein; SPB, spindle pole body; R-SNARE, R-type soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor; CoA, coenzyme A; ORF, open reading frame; APC, anaphase promoting complex; MMS, methyl methanesulfonate. This table does not include all of the synthetic lethal interactions recently reported in reference 84a.
1, interaction with Rvs161; 7, interaction with Rvs167.
Information on gene/gene product nomenclature and function was obtained from the Saccharomyces Genome Database (R. Balakrishnan, K. R. Christie, M. C. Costanzo, K. Dolinski, S. S. Dwight, S. R. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. Nash, R. Oughtred, M. Skrzypek, C. L. Theesfeld, G. Binkley, C. Lane, M. Schroeder, A. Sethuraman, S. Dong, S. Weng, S. Miyasato, R. Andrada, D. Botstein, and J. M. Cherry [http://www.yeastgenome.org/ {November 2005}]).
There is evidence that the BAR and SH3 domains of Rvs167p both contribute to its actin patch localization. For example, a fragment comprising only the Rvs167p BAR domain localizes poorly to cortical actin patches, while the Rvs167p SH3 domain shows no localization to actin patches. In contrast, the full-length protein containing the BAR and SH3 domains localizes efficiently to actin patches (11). Perhaps other actin patch components mediate recruitment of Rvs167p to actin patches via interactions with the BAR domain. Possible candidates are the products of three genes, synthetic lethal with abp1 1 (SLA1) and SLA2 and suppressor of ras Val19 2 (SRV2), which localize to actin patches and have functions redundant with that of Abp1p (126, 176).
Sla1p associates with Rvs167p in vivo and directly binds via multiple domains to recombinant Rvs167p in vitro, but the Rvs167p domain that mediates these interactions is not yet known (59, 123, 307). Interestingly, Sla2p possesses a coiled-coil domain (coil1) that has been reported to interact with the Rvs167p BAR domain. Furthermore, the Sla2p coil1 domain has a function important for growth, actin patch polarization, and endocytosis. Intriguingly, however, this function of coil1 is fully redundant with a function mediated by Abp1p (or Srv2p), i.e., the Sla2p coil1 mutant phenotypes only become apparent in cells that lack Abp1p (or Srv2p) (364). Interactions of the Rvs167p BAR domain with Sla2p coil1 and the Rvs167p SH3 domain with Abp1p may work in concert to facilitate Rvs167p localization to actin patches.
The Rvs167p SH3 domain may also recruit actin monomers to patches via interaction with other actin patch components such as Local Anaesthetic Sensitive 17 (Las17p, also known as Bee1p) and Very Rich in Proline 1 (Vrp1p, also known as End5p or verprolin) (Table 1). Las17p is the yeast ortholog of human Wiskott-Aldrich syndrome protein (WASp) that is mutated in patients with the inherited immunodeficiency (8). Both WASp in mammals and Las17p in yeast are implicated in de novo assembly of actin monomers into actin filaments by a highly conserved seven-subunit complex known as the Arp2/3p complex (119, 372). Vrp1p is the yeast ortholog of human WASp-interacting protein (WIP), a protein identified by two-hybrid screens with WASp. Vrp1p and Las17p in yeast interact analogous to WASp and WIP in mammals (181, 219).
Consistent with a possible role for Vrp1p in recruitment of Rvs167p to actin patches, loss of VRP1 (vrp1Δ) displays exactly the same pattern of negative genetic interactions with the yeast myosins (Myo1p-Myo5p) as loss of Rvs167p (see below) (19, 275). However, Rvs167p must retain significant function in cells lacking Vrp1p even if its localization to actin patches is affected, since additional loss of Rvs167p (rvs167Δ) dramatically affects the growth of vrp1Δ mutants (las17Δ was not tested). Interestingly, additional loss of Rvs161p (rvs161Δ) did not have any obvious effect on the growth of vrp1Δ cells (las17Δ was not tested) (275). Hence, Vrp1p may be required for all cellular functions of Rvs161p, which would be consistent with a possible role in Rvs161p localization to actin patches. Interestingly, in the fission yeast Schizosaccharomyces pombe there is recent evidence that the Las17p ortholog (Wsp1p) is required for actin patch localization of the Rvs167p ortholog (Hob1p) (131).
An interesting possibility that has not yet been explored is that Vrp1p and/or Las17p mediates the two-hybrid interaction of Rvs167p with actin (4). Rvs167p interacts with both Las17p and Vrp1p via its C-terminal SH3 domain (38, 181, 331; our unpublished data). Both Vrp1p and Las17p possess actin monomer binding WASP Homology 2 (WH2) domains. Although the contacts that WH2 domains make with actin are not yet known, they may resemble the contacts that profilin makes with actin, since both WH2 domains and profilin bind specifically to actin monomers. If correct, this would account for the observation that actin mutations that perturb profilin binding also perturb Rvs167p binding (4).
Roles of Rvs161p and Rvs167p in Endocytosis
Rvs161p and Rvs167p in receptor-mediated internalization of α-factor.
In vertebrates, receptor endocytosis occurs at specific sites on the plasma membrane that bear a cytoplasmic protein coat. This coat comprises the coat protein clathrin (comprising heavy and light chains) and the clathrin coat assembly factor known as the clathrin Associated (or Assembly) Protein 2 (AP-2) adaptor. The assembly of clathrin coats on the cytoplasmic face of the plasma membrane by the AP-2 adaptor generates clathrin-coated endocytic pits. Binding of the AP-2 adaptor to the cytoplasmic tails of plasma membrane receptors to be endocytosed concentrates the receptors in the forming clathrin-coated pits. The conversion of an invaginated clathrin-coated pit into a clathrin-coated vesicle is mediated by the GTPase dynamin. Dynamin assembles into rings at the neck of invaginated clathrin-coated pits, and hydrolysis of GTP is accompanied by fission of the neck of the pit and the release of a clathrin-coated vesicle into the cytoplasm. In vertebrates, the Rvs161p and Rvs167p homolog amphiphysin has been shown to function in endocytosis via recruitment of dynamin from the cytoplasm to plasma membrane clathrin-coated pits. Could rvs161 and rvs167 mutants perturb the recruitment of a yeast dynamin to clathrin-coated pits?
In yeast cells, in contrast to vertebrates, receptor endocytosis is predominantly independent of clathrin. Yeast cells in which the single genes encoding clathrin heavy chain or clathrin light chain are deleted continue to internalize α-factor, although at a reduced rate (37, 235). A temperature-sensitive clathrin mutant (chc1-ts) was constructed that can be inactivated by a shift to elevated temperature. This mutant was used to examine the acute effects of clathrin inactivation on α-factor endocytosis. Even in this study, only a slower kinetics of α-factor internalization and not a complete block was observed (318). Furthermore, single and combined deletions of the various genes encoding the subunits of the putative yeast equivalent of the AP-2 adaptor and related proteins failed to block the formation of clathrin-coated vesicles and had less impact on α-factor endocytosis even than deletion of clathrin heavy or light chain. Clathrin does not even appear to associate with the AP-2 adaptor in yeast (130, 377).
Other adaptors are known to bind clathrin heavy chain and to function in clathrin-mediated endocytosis in vertebrates. Mammalian epsin (57, 58) and AP180 (2, 216) are both known to bind clathrin heavy chain, promote assembly of clathrin coats, and function in clathrin-dependent endocytosis. In yeast, clathrin heavy chain does associate with the yeast epsins (Ent1p and Ent2p) (362) and AP180 adaptors (Yap1801 and Yap1802) (361). The Ent1p and Ent2p adaptors are required for endocytosis in yeast (362), however, Yap1801 and Yap1802 are not required individually or collectively (361). Yap1801 and Yap1802 are also not required for clathrin coat assembly or clathrin-coated vesicle formation in yeast (130).
For some time it was not clear whether clathrin-coated pits exist on the yeast plasma membrane as they do in vertebrates. Two very recent studies, discussed below, were the first to demonstrate that a pool of clathrin exists on (or near) the plasma membrane in yeast cells and colocalizes with cortical actin patches (144, 223).
Is dynamin required for receptor endocytosis in yeast? Yeast cells possess three dynamin-like proteins (Vps1p, Dnm1p, and Mgm1p). Vps1p localizes to the Golgi apparatus and/or peroxisomes (124, 274). Deletion of the VPS1 gene that encodes Vps1p (vps1Δ) does not abolish endocytic internalization (226). A recent study showed that deletion or mutation of Vps1p perturbs actin patch polarization and abolishes endocytic internalization, but only at elevated temperature (383). Dnm1p localizes to the surface of mitochondria (86, 166). Deletion of the DNM1 gene that encodes Dnm1p did not affect the kinetics of α-factor internalization, although subsequent trafficking of α-factor to the vacuole was delayed (92). Mgm1p localizes to the intramembrane space of mitochondria and therefore a role in endocytosis seems unlikely (although it has not been directly tested) (373). Dnm1p and Mgm1p regulate mitochondrial morphology (17, 86, 229, 289). Interestingly, none of the yeast dynamin-like proteins have been shown to interact physically with clathrin, AP-2 adaptor, or either Rvs protein.
In yeast, internalization of receptors by endocytosis is dependent on prior covalent attachment of the protein ubiquitin to the receptor cytoplasmic tail (117). Ubiquitin is attached to specific lysine residues in the receptor tail by the ubiquitin protein ligase Rsp5p (63, 64). Ubiquitin-dependent endocytosis requires the attachment of only a single ubiquitin molecule (monoubiquitination) (327). This is in contrast to targeting of cytoplasmic proteins for degradation by the 26S proteasome, which requires attachment of chains containing multiple ubiquitin molecules (polyubiquitination). The requirement for monoubiquitination for receptor endocytosis can be bypassed by the use of recombinant receptors in which the C terminus of the receptor tail is fused to ubiquitin (327). Are rvs161 and rvs167 mutants defective in receptor-mediated endocytosis of α-factor because they cannot ubiquitinate the α-factor receptor cytoplasmic tail? This has not yet been thoroughly investigated, but a recent study indicated that the α-factor receptor tail is still ubiquitinated in rvs167Δ cells (307).
The polymerization of actin monomers into actin filaments is a process that can produce mechanical force. It has been proposed that the requirement for actin cytoskeletal proteins for endocytic internalization in yeast may reflect a role for actin polymerization in producing the force necessary to sever endocytic pits and release endocytic vesicles (71, 213, 214). Live cell imaging has been used to visualize endocytosis in yeast and to look for colocalization of endocytosed plasma membrane receptors with proteins important for actin polymerization during internalization (143). Transient colocalization of endocytosed receptors with proteins implicated in de novo actin filament assembly occurs at sites of endocytic internalization. Moreover, correlations have been observed between the movement of actin filament assembly proteins and endocytic vesicles (132, 139, 143, 144, 223).
During endocytic internalization clathrin first assembles to form a cortical patch. Clathrin is soon joined in the patch by Las17p and a second Arp2/3p activator protein, Pan1p. Other actin filament assembly proteins are then recruited, including End3p, Sla1p, and Sla2p. These proteins assemble sequentially to form cortical patches that are initially stationary and do not yet contain F-actin. Next, Abp1p, the Arp2/3p complex itself, and actin are recruited to the patch. The Arp2/3p complex is responsible for de novo nucleation of actin filaments. Las17p, Pan1p, and Abp1p are believed to promote actin filament assembly by interacting with and stimulating the Arp2/3p complex. Following recruitment of actin and the Arp2/3p complex an actin cloud is formed via Arp2/3p-dependent de novo actin filament assembly. Finally, type I myosins are transiently recruited to the stationary patch. Type I myosins are a class of actin-dependent motor that also have the potential to interact with and activate the Arp2/3p complex and have been proposed to function in endocytic vesicle fission. Las17p, Pan1p, and type I myosins remain in a stationary patch on the membrane, while the cortical patch initiates slow movement for a distance of ∼200 nm perpendicular to the plane of the membrane. At first End3p, Pan1p, Sla1p, and Sla2p move with the actin patch but then they gradually dissociate. In contrast, Abp1p, Arp2/3p, and actin move slowly into the cell with the actin patch and remain associated as the actin patch undergoes a subsequent fast long-range movement (132, 139, 143, 144).
Important questions remain concerning the role of Arp2/3p-activating proteins such as Las17p, Pan1p, Abp1p, and type I myosins in endocytosis. In vitro evidence has suggested that the yeast Arp2/3p complex is inactive unless activated by these proteins (372). This is consistent with the requirement of WASp (mammalian ortholog of Las17p) for mammalian Arp2/3 activity in actin filament nucleation. A recent study, however, showed that the dependence of Arp2/3p activity on activating proteins in vitro is affected by the source of actin. The original assays that showed the Las17p dependence of Arp2/3p activity used mammalian muscle actin. When yeast actin was used instead there was strong Arp2/3p-dependent actin polymerization even in the absence of Las17p (although the addition of Las17p further enhanced Arp2/3p activity). This study concluded that Arp2/3p-dependent polymerization of yeast actin does not absolutely require an activating protein (at least in vitro) (360). However, other studies found a strict requirement for Arp2/3p-activating proteins for Arp2/3p-dependent actin assembly even when the assay was performed using only yeast actin (101). Whether activating proteins are required for Arp2/3p activity in yeast is an important question. If they are indeed dispensable for Arp2/3p activation in vitro and this is also true in vivo, then the role of Arp2/3p complex “activators” in actin-dependent movement and endocytosis may need to be revisited.
Interestingly, two recent studies found that there is a cortical pool of clathrin in yeast in the form of small motile patches (144, 223). Inhibition of actin patch assembly by treatment of wild-type cells with the actin polymerization inhibitor latrunculin A or in cells lacking the actin assembly protein Sla2p (sla2Δ) or End3p (end3Δ) arrests cortical clathrin patch movement, extends clathrin patch lifetime, and results in a dramatic accumulation of clathrin at the cortex (144, 223). Interestingly, in cells lacking Rvs161p or Rvs167p clathrin patch lifetime is not extended and clathrin does not accumulate at the cortex. Hence, not all mutations that block endocytic internalization have this effect on clathrin. It is not yet clear why loss of Sla2p or End3p has such different effects from loss of Rvs161p or Rvs167p.
Several lines of evidence suggest that cortical clathrin in yeast, while not essential, is involved in endocytic internalization. First, after treatment of cells with latrunculin A to induce clathrin accumulation at the cortex, some cortical clathrin patches colocalize with endocytic cargo (e.g., α-factor receptors). In untreated yeast cells some cortical clathrin patches colocalize with Arp2/3p-activating proteins and are recruited to patches at the cortex just prior to recruitment of Arp2/3p-activating proteins to the patch. Moreover, accumulation of clathrin at the cortex upon latrunculin A treatment requires either the yeast AP180 adaptor Yap1801p or Yap1802p or a different type of yeast clathrin adaptor related to vertebrate epsin (Ent1p or Ent2p). Finally, loss of clathrin heavy chain or clathrin light chain reduces the number and the lifetime of cortical patches containing Las17p and Sla1p, although the movement of the remaining Las17p and Sla1p patches and disassembly of these proteins during movement appears unaffected. This evidence directly implicates yeast clathrin in actin-dependent receptor-mediated endocytosis (144, 223).
Although Rvs161p and Rvs167p have not been shown to activate the Arp2/3p complex for actin filament assembly in vitro, they do interact with several other proteins that are known Arp2/3p activators (see below). This, together with their ability to directly bind membranes (see below), makes the Rvs proteins ideal candidates for linking actin filament assembly to membrane dynamics during endocytic internalization. Is actin patch movement off the plasma membrane or the coupling of this actin patch movement to internalization of endocytic cargo such as plasma membrane receptors dependent on Rvs161p and/or Rvs167p? Rvs161p and Rvs167p have recently been shown to assemble into cortical patches at sites of endocytosis. Rvs161p and Rvs167p are recruited after Abp1p to patches during the initial actin cloud formation but while the patches are still stationary on the plasma membrane. When the patches exhibit the initial slow movement over a distance of ∼200 nm perpendicular to the plane of the membrane Rvs161p and Rvs167p undergo a very rapid movement over a distance of ∼100 nm and then immediately dissociate. While treatment of cells with latrunculin A to disassemble all F-actin does not prevent Rvs167p localization to cortical patches it does block movement of Rvs167p patches off the membrane (144).
The effect of rvs161Δ and rvs167Δ mutations on actin patch dynamics during endocytosis has been examined recently and is particularly interesting (144). In the absence of Rvs161p and Rvs167p the actin patch (as defined by Sla1p) still assembles on the plasma membrane. Upon recruitment of Abp1p the patch undergoes the initial slow movement perpendicular to the plane of the plasma membrane and Sla1p gradually dissociates as in wild-type cells. However, in cells lacking Rvs161p or Rvs167p these patches do not undergo the second phase of rapid long-range movement. Instead, many of the actin patches appear to stop and then exhibit an abberant retrograde movement back to the plasma membrane. Interestingly, Abp1p, which normally is retained on the patch during rapid long-range movement in wild-type cells, dissociates prior to the retrograde movement in rvs161Δ and rvs167Δ mutants. It has been proposed that invagination proceeds in the absence of Rvs161p and Rvs167p but membrane scission fails. As a consequence the coat complex defined by Sla1p is then retracted back to the plasma membrane driven by membrane tension. The actin assembly complex defined by Abp1p that normally drives the subsequent rapid long-range patch movement in wild-type cells falls off the invaginated membrane as the invagination retracts in rvs161Δ and rvs167Δ mutant cells (144).
Rvs161p and Rvs167p function in endocytosis as a heterodimer. The ability of Rvs protein overexpression to perturb receptor-mediated endocytosis of α-factor in wild-type cells requires simultaneous overexpression of both proteins. The level of overexpression achieved in these experiments (which used elevated gene dosage rather than a strong promoter) did not affect the growth of wild-type cells or exacerbate the growth defect of rvs167Δ mutant cells, but overexpression of Rvs167p in rvs161Δ cells caused mild inhibition of growth. Rvs161p overexpression in rvs167Δ cells or, conversely, Rvs167p overexpression in rvs161Δ cells enhances the defect in receptor-mediated endocytosis of α-factor. Interestingly, this exacerbation of the endocytic defect occurs without obvious exacerbation of the actin patch polarization defect. Clearly, endocytosis is very sensitive to the total amount and ratio of Rvs161p and Rvs167p, but actin patch polarization and cell growth are less sensitive (180).
Role for Rvs161p and Rvs167p in postinternalization trafficking through endosomes?
The strong defect in receptor internalization in rvs161 and rvs167 mutants has made it difficult to test possible defects in postinternalization traffic to the vacuole (213, 215). Defects in postinternalization traffic are often (although not always) associated with defects in biosynthetic traffic through endosomes to the vacuole. This is because the endocytic pathway and the vacuole biosynthetic pathway meet in the prevacuolar compartment, which is a type of endosome (214). Vacuole biogenesis appears normal in rvs161 and rvs167 mutant cells. Neither end6-1 (Rvs161p-R59K mutation) nor rvs167Δ affects the appearance of the vacuole. Traffic of newly synthesized soluble vacuolar proteins from the late Golgi apparatus via endosomes to the vacuole can be monitored based on the kinetics of endoplasmic reticulum (ER)-dependent and Golgi apparatus-dependent glycosylation and the proteolytic processing event that occurs upon arrival in the vacuole. These events appear normal in rvs161 and rvs167 mutant cells even at elevated temperature (37°C). Neither rvs161 nor rvs167 cells are defective in vacuolar protein sorting because they do not missort soluble vacuolar proteins into the extracellular medium (215).
It is possible that Rvs161p and Rvs167p function in the late endocytic pathway but redundantly with other proteins. In support of this possibility, combining mutations in rvs167 with mutations in two genes encoding proteins known to function in the late endocytic pathway, YPT51 (296) and VPS20 (97), results in double mutant cells that either have severely reduced viability compared to either single mutant or are totally inviable, respectively (Table 1). YPT51 (also known as VPS21) encodes Ypt51p, which is one of three yeast orthologs of human early endosomal Rab5 and is important for vacuolar protein sorting and traffic of internalized α-factor from early endosomes to late endosomes. The interaction with Ypt51p is specific because Rvs167p does not genetically interact with Ypt7p, the ortholog of human late endosomal Rab7, which functions in trafficking of internalized α-factor from late endosomes to the vacuole (297). VPS20 encodes a small coiled-coil protein (Vps20p) also required for postinternalization endocytic trafficking and vacuolar protein sorting (376). Interestingly, vps20 mutants exhibit severely reduced viability upon entry into stationary phase (i.e., nutrient starvation) (7). This further supports a functional link between Vps20p and the Rvs161p and Rvs167p proteins.
Roles of Rvs161p and Rvs167p in the Secretory Pathway
Rvs161p and Rvs167p are not essential for all secretory membrane traffic.
A functional secretory pathway is essential for viability of all cells, including budding yeast. In the absence of a functional secretory pathway cells are unable to deliver newly synthesized plasma membrane components to the cell surface and the cell can neither grow in size nor divide. The viability of both rvs161 and rvs167 single mutants and the rvs161 rvs167 double mutant shows that the secretory pathway is not blocked (at least under normal growth conditions) in the absence of Rvs proteins (12, 42, 53). Trafficking of newly synthesized soluble vacuolar proteins from the ER to the Golgi apparatus was also unaffected in rvs161 or rvs167 mutants even at elevated temperature (37°C) (215). Transport of the soluble secreted enzyme invertase from the ER via the Golgi apparatus to the cell surface was not affected in either rvs161 or rvs167 mutant cells (298). Hence, Rvs proteins are not strictly essential for a functional secretory pathway.
Rvs proteins may be required for polarized secretion during cell division.
Several lines of evidence suggest the rvs mutants have a secretory pathway that is not fully functional. rvs167 mutant cells exhibit delocalization of cell wall chitin (12). This suggests a defect in polarized secretion of chitin synthases to the bud site. Defects in cell wall biogenesis are further supported by an electron microscopy study of cell wall (septum) deposition at the neck of dividing rvs161 and rvs167 mutant cells (20). Wild-type yeast cells deposit septum at the bud neck and then cleave the septum and divide so rapidly that electron micrographs reveal few cells in the process of forming a septum. In contrast, a much greater proportion of rvs161 and rvs167 mutant cells are seen in electron micrographs in the process of depositing septa, strongly, suggesting that deposition of septum is considerably slowed (20). Defects in cell wall deposition may arise because of inefficient delivery of cell wall biosynthetic enzymes to the division site.
Rvs proteins and post-Golgi apparatus traffic.
Further evidence for a role of Rvs161p and Rvs167p in the secretory pathway comes from genetic studies. rvs161 and rvs167 mutations exhibit negative genetic interactions with mutations affecting the unconventional type V myosin Myo2p (myo2). rvs161 myo2 and rvs167 myo2 double mutants exhibit a lethal phenotype (19). Myo2p plays a role in motor-driven polarized transport of Golgi apparatus-derived vesicles along actin cables to the bud (104, 138, 150, 253, 273, 286). myo2 mutations are also lethal in combination with a subset of secretion (sec) mutations that affect delivery of Golgi apparatus-derived transport vesicles to the plasma membrane (104). This suggests rvs161 and rvs167 may also have defects in this late step of the secretory pathway. Moreover, as discussed further below, mutations in two genes, SUR4 and SUR5/FEN1, that suppress the defects of rvs161 and rvs167 mutants also suppress the defects in snc1 snc2 double mutants, which are known to be defective in post-Golgi apparatus traffic (48).
Further hints for a role in vesicle traffic for Rvs161p and Rvs167p comes from large-scale two-hybrid screens that identified interactions between Rvs167p and components of the exocyst complex, notably Sec8p and Exo70p (Table 1) (18). The exocyst complex is required for fusion of Golgi apparatus-derived transport vesicles with the plasma membrane. Moreover, a recent genome-wide genetic interaction study of Rvs161p and Rvs167p revealed that loss of either protein is lethal in pairwise combination with deletions affecting the same set of (nonessential) secretory proteins. This independent evidence is consistent with the results of the earlier two-hybrid study and further supports the view that the Rvs proteins function in secretion (84a).
Rvs161p and Rvs167p also interact with two proteins that regulate Rab GTPases: Gyp5p (GYP5/YPL249c gene product) and Gyl1p (GYL1/YMR192w gene product) (Table 1). The SH3 domain of Rvs167p binds Gyp5p, whereas full-length Rvs161p appears to be required for its interaction with Gyp5p. The SH3 domain of Rvs167p binds Gyl1p and phosphorylation of Rvs167p inhibits this interaction (18, 84, 85, 123, 317, 331). Rab GTPases cycle between an active GTP-bound form and an inactive GDP-bound form. Rab GTPase-activating proteins (Rab-GAPs) stimulate GTP hydrolysis and promote inactivation, while Rab GDP/GTP exchange proteins (Rab-GEFs) stimulate GDP/GTP exchange and promote activation (287, 309). Gyp5p has strong Rab-GAP activity in vitro and is most active on the Rab GTPase Ypt1p, which functions in ER-to-Golgi apparatus traffic (see below), although it also has significant activity on the Rab GTPase Sec4p, which functions in post-Golgi apparatus traffic (49). During subcellular fractionation Gyp5p and Gyl1p copurified with post-Golgi apparatus vesicles and plasma membrane. Gyp5p was shown to physically interact with Sec4p. gyp5Δ interacts genetically with sec2 mutations affecting the Sec4p GEF. At low temperature, gyp5Δ gyl1Δ double mutant cells exhibit a slight defect in growth and polarized secretion and accumulate secretory vesicles at sites of polarized growth (33).
In wild-type cells, transport vesicles bud from donor compartments and efficiently dock and fuse with acceptor compartments. Hence, transport vesicles are transient and rarely visible in electron micrographs. In contrast, both rvs161 and rvs167 mutant cells accumulate vesicles at sites of polarized growth. rvs161 mutants accumulate vesicles mainly at the bud neck during cell division, however, some vesicle accumulation at the nascent bud site and in the small growing buds is also observed. In contrast, rvs167 mutants accumulate vesicles mainly at the nascent bud site and in small buds, however, some vesicle accumulation at the bud neck during cell division is also apparent. The vesicle accumulation in both rvs mutants is apparent even under optimal growth conditions, but becomes dramatic under conditions of stress, e.g., in the presence of sublethal levels of Na+. In both mutants, exposure to Na+ initially results in vesicle accumulate at sites of growth, but later vesicles distribute throughout the mother cell and bud (20). myo2 mutants (see above) accumulate what appear to be similar vesicles (104, 138).
If Rvs proteins transport vesicles to the plasma membrane, what cargo is transported in these vesicles? The content of the vesicles that accumulate in rvs mutants has not been examined. Several other actin cytoskeletal mutations also cause accumulation of vesicles and in some cases the contents have been analyzed. For example, the vesicles that accumulate in act1 and sla2 mutant cells contain Ypt1p. Ypt1p is a Rab family GTPase equivalent to mammalian Rab1 that localizes to the Golgi apparatus and functions in ER-Golgi apparatus traffic in yeast (206). It has been proposed that the Ypt1p vesicles are not transport vesicles but rather Golgi apparatus fragments.
Interestingly, there are emerging links between Rvs167p and regulators of Ypt1p GTPase activity (see below) and it is possible that altered activity of Ypt1p may lead either to accumulation of transport vesicles or perhaps to organelle fragmentation. Insight into the nature of the cargo transported in the vesicles that accumulate in rvs mutant cells came from a recent genomewide mutant screen (250). RVS161 was one of 137 genes (including sterol and sphingolipid biosynthesis genes) that, when deleted, resulted in reduced cell surface delivery and intracellular accumulation of a lipid raft-associated integral membrane protein marker but are not required for secretion of the soluble protein invertase into the extracellular medium.
Rvs proteins and ER-to-Golgi apparatus traffic.
Recent evidence suggests that the Rvs proteins may also function earlier in the secretory pathway. Rvs167p exhibits two-hybrid interaction with the COP-I component Sec21p (γ-COP) (18). COP-I is a coat complex that functions early in the secretory pathway and mediates the budding of transport vesicles that shuttle cargo between the ER and Golgi apparatus. A genomewide mass spectrometry analysis of yeast protein complexes identified Sec27p in a complex with Rvs161p and Rvs167p (123). Sec27p is the β-COP subunit of the COP-I coat complex. These interactions are thought-provoking but await more detailed analysis.
Negative genetic interactions have been observed with two mutations that affect ER-to-Golgi apparatus membrane traffic, sec22 and rud3 (84). Sec22p is a vesicle-associated soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor that functions specifically in ER-to-Golgi apparatus transport. Rud3p (also known as Grp1p) is a matrix protein of the early Golgi apparatus. Genetic interactions suggest that Sec22p, present on ER-derived transport vesicles, and Rud3p, present on an early Golgi apparatus compartment, function in concert to promote fusion of ER-derived transport vesicles with the early Golgi apparatus. sec22 and rud3 exhibit negative genetic interactions and overexpression of Rud3p rescues the phenotype of some sec22 mutations (153).
Further evidence for a role of Rvs proteins in ER-to-Golgi apparatus traffic came from the discovery that both Rvs161p and Rvs167p interact with regulators of the ER-to-Golgi apparatus Rab GTPase Ypt1p. Ypt1p, like other Rab GTPases, cycles between an active GTP-bound form and an inactive GDP-bound form. Rvs161p and Rvs167p both directly bind Gyp5p (which as mentioned above has strong Rab-GAP activity in vitro on Ypt1p) and Gyl1p (Table 1) (18, 84, 85, 123, 317, 331). Consistent with Gyp5p's being a Ypt1p Rab-GAP, genetic interaction data are consistent with an in vivo role for Gyp5p in negatively regulating Ypt1p (49). Gyl1p shares extensive amino acid sequence homology with Gyp5p and was initially proposed to also be a Rab-GAP for Ypt1p (317). Unexpectedly, Gyl1p lacks detectable Rab-GAP activity on Ypt1p in vitro. However, Gyl1p binds to Gyp5p, possibly via coiled-coil interactions, and stimulates its GAP activity on Ypt1p (84).
There is some evidence that Gyp5p and Gyl1p function in ER-to-Golgi apparatus traffic with Rvs167p, despite the lack of strong mutant phenotypes. Gyl1p overexpression is slightly inhibitory for growth of wild-type cells. Interestingly, simultaneous overexpression of both Gyl1p and Gyp5p is lethal specifically in sec22Δ and rud3Δ cells, which are partially compromised in ER-to-Golgi apparatus transport (although the effect of overexpression of Gyl1p and Gyp5p on ER-to-Golgi apparatus transport has not yet been directly tested). In this way, overexpression of Gyp5p and Gyl1p mimics loss of Rvs167p (84). Another study, however, found that Gyp5p and Gyl1p function in post-Golgi apparatus traffic (see above) (33). Perhaps Gyp5p and Gyl1p function in early steps of the secretory pathway with Ypt1p and in late steps of the secretory pathway with Sec4p.
Neither Gyp5p nor Gyl1p localizes to cortical actin patches; instead they exhibit diffuse cytoplasmic staining (49). Some reports also find cortical staining at nascent bud sites, the tips of small growing buds, and the bud neck during cytokinesis. These are also areas where Rvs167p patches localize, but the distributions only partially overlap. Interestingly, Gyp5p-GFP localization to the bud tip and bud neck is lost in both rvs161Δ/rvs161Δ and rvs167Δ/rvs167Δ homozygous diploid cells, suggesting both Rvs proteins may be important for Gyp5p localization to these sites. Loss of Gyp1p and Gyl1p, either singly or in combination, did not result in Rvs phenotypes with the possible exception of bipolar bud site selection. Furthermore, overexpression of Gyp5p and Gyl1p did not perturb actin patch polarization or endocytosis, so the inhibitory effect seems to be restricted to ER-to-Golgi apparatus transport (33, 84, 317).
A model has been proposed in which Gyp5p, Gyl1p, and Rvs167p function together in a complex whose role in ER-to-Golgi apparatus transport overlaps that of Sec22p and Rud3p. In this model Gyp5p and Gyl1p negatively regulate Rvs167p function (84). Another possibility to consider is that Rvs167p negatively regulates Gyp5p and Gyl1p. Loss of Rvs167p would lead to increased Gyp5p Rab-GAP activity and a reduction in the pool of active GTP-bound Ypt1p. As GTP-bound Ypt1p is required for vesicle fusion, it may be interesting to test if the vesicles that accumulate in rvs161 and rvs167 mutants bear Ypt1p and furthermore if hyperactivation of Gyp5p Rab-GAP activity in rvs161 and rvs167 mutant cells plays an important role in the observed vesicle accumulation.
Rvs proteins and mating.
Unlike animal cells, yeast cells are surrounded by a thick polysaccharide cell wall. When the tips of each mating projection come into contact the two mating cells are still separated by intervening cell wall material. Examination of wild-type cells in the process of mating reveals that after mating projections come into contact a seal is made at the outer perimeter of the initial contact zone. This outer perimeter seal remains intact, but intervening cell wall material within the initial zone of contact is rapidly removed. This process of cell wall removal continues until the plasma membrane of each mating cell is exposed. The two plasma membranes then undergo a fusion event and cytoplasmic continuity is established. The nucleus of each mating cell migrates to the site of cell-cell fusion, and these haploid nuclei then also fuse, resulting in one diploid nucleus.
An inability to mate can result from defective cell cycle arrest, mating projection formation, cell fusion, or nuclear fusion. As mentioned above, rvs161 (fus7) was identified as a bilateral cell fusion mutation. In matings between rvs161 mutants (rvs161 × rvs161) mating projections are formed and cell-cell contact is established, but cell-cell fusion is compromised (Fig. 4) (20, 21, 91, 158).
FIG. 4.
Rvs161p functions in cell-cell fusion during mating. Shown are matings between two wild-type haploids (panels A and B) or between two rvs161Δ mutant haploids (panels C to F) visualized by fluorescence/differential interference contrast optics. Cell nuclei are stained with the fluorescent dye 4′,6′-diamidino-2-phenylindole. Cell-cell fusion allows cytoplasmic mixing and the nuclei of each mating partner meet at the site of cell fusion to fuse and create a single diploid nucleus. In the case of rvs161Δ × rvs161Δ matings, delays in cell-cell fusion (apparent by differential interference contrast) result in delays in nuclear fusion. This leads to the persistence of two nuclei. In panels C and D nuclear fusion is taking place despite incomplete removal of the intervening cell wall (partial Fus− phenotype) and in panels E and F nuclear fusion has been completely blocked by the inability to remove intervening cell wall (full Fus− phenotype). (Reproduced from reference 21 by copyright permission of the Rockefeller University Press.)
Consistent with a novel function in mating cells, an Rvs161p-GFP fusion was observed to redistribute to the tip of the mating projection upon treatment of haploid cells with α-factor mating pheromone. Interestingly, Rvs167p-GFP has also been reported to redistribute to the tip of the mating projection upon pheromone treatment. Upon contact between the tips of the two mating projections, Rvs161p-GFP remains concentrated at the site of cell contact throughout the processes of cell wall removal and plasma membrane fusion. Electron microscopy showed that rvs161 mutant cells form cell-cell contacts that are abnormally large (Fig. 5). In mating wild-type cells, the intervening cell wall at sites of contact appears to be rapidly removed. Electron micrographs that show wild-type cells in the process of removing the intervening cell wall are less common. In contrast, rvs161 mutant cells are often seen with wide contacts and at intermediate stages of cell wall removal (11, 20, 21, 91).
FIG. 5.
Ultrastructure of wild-type, fus2, rvs161, and rvs161 fus2 cells during cell-cell fusion. Shown are electron micrographs of wild-type (WT), fus2Δ, rvs161Δ, and rvs161Δ fus2Δ cells undergoing cell-cell fusion during mating. Note that wild-type cells exhibit restricted zones of cell-cell contact during fusion while fus2Δ, rvs161Δ, and rvs161Δ fus2Δ mutants exhibit very wide zones of cell-cell contact during fusion. Wild-type and mutant cells all exhibit numerous vesicles that cluster at the zone of cell-cell fusion. Note that plasma membrane invaginations/tubules accumulate at the zone of fusion in the mutant cells, but are less common in wild-type cells. Labels indicate plasma membrane invaginations (black arrows), vesicle clusters (black arrowheads), electron-dense plaques (white arrows), and nonremoved cell wall remnants (asterisks). Bars = 1 μm. (These previously unpublished images are shown here with the kind permission of Alison Gammie.)
Although the frequency of successful matings of rvs161 cells with wild-type cells is normal, electron micrographs reveal apparent delays and abnormalities even in these matings (21). Matings of rvs167 cells (rvs167 × rvs167) also show apparent delays in cell wall removal at contact sites similar to that of rvs161 × rvs161 matings despite the fact that the frequency of successful rvs167 × rvs167 matings is similar to that of matings between wild-type cells (Fig. 6) (20). Evidence of a role for both Rvs161p and Rvs167p in mating was also found in a recent genome-wide genetic interaction study (84a). The apparent delay in localized removal of cell wall at sites of cell-cell contact in rvs161 and rvs167 cells may be due to the larger area of wall that has to be removed. Alternatively, polarized delivery of transport vesicles carrying cell wall-degrading enzymes to the site of contact may be affected.
FIG. 6.
Ultrastructure of wild-type, rvs161Δ, and rvs167Δ cells during cell-cell fusion. Shown are electron micrographs of wild-type, rvs161Δ, and rvs167Δ cells undergoing cell-cell fusion during mating. Panels a to c, matings between two wild-type cells; panels d to f, matings between two rvs161Δ cells; panels g and h, matings between two rvs167Δ cells. Bar, 1 μm. Labeled organelles: n, nucleus; v, vacuole; vs, vesicles; g, Golgi apparatus. (Reproduced from reference 20 with permission of the publisher. Copyright John Wiley and Sons Ltd.)
Electron micrographs show that in wild-type cells vesicles appear at sites where mating projections achieve cell-cell contact. These vesicles do not spread out evenly across the entire area of the contact, but instead form a tight cluster with a diameter of 0.6 μm, which is considerably less than the diameter of the zone of cell-cell contact (1 μm in wild-type and 2 to 3 μm in rvs161 cells). Cell wall is degraded not throughout the region of cell-cell contact, but rather at a small zone within this region known as the fusion pore. Plasma membrane contact and fusion likely occur at the fusion pore. Intriguingly, the site where vesicles cluster is precisely the site of the fusion pore. The diameter of the fusion pore is ∼0.5 μm, which is similar to the diameter of the vesicle cluster. The vesicles that cluster at the fusion pore may carry enzymes that degrade cell wall carbohydrates. Interestingly, some cell fusion mutants do not accumulate any vesicles at the site of cell contact (e.g., fus1 cells). Other cell fusion mutants accumulate vesicles but they do not form tight clusters at the fusion pore (e.g., spa2 mutants). In contrast, both vesicle accumulation and clustering at the fusion pore appear normal in rvs161 mutants (91).
Electron micrographs reveal that mating rvs161 cells exhibit an electron-dense plaque at the site of cell-cell contact (Fig. 5). This plaque may represent a fusion intermediate comprising newly deposited membrane material. This plaque is not observed in cell fusion mutants that do not accumulate or cluster vesicles, suggesting that this plaque material is transported by the vesicles observed in clusters at the fusion pore. Interestingly, the electron-dense plaques observed in rvs161 cells are major sites of plasma membrane invagination (Fig. 5) (91). Perhaps rvs161 cells have defects in fission of the necks of these plasma membrane invaginations resulting in accumulation of plaque material at the fusion pore.
In crosses to wild-type cells, why do rvs161 cells exhibit a defect in default mating but not in mating in response to a pheromone gradient? Comparison of several mutants defective in default mating revealed a close correlation between defects in cell fusion and defects in default mating (56). It was proposed that default mating requires a much higher level of cell fusion activity than mating in response to pheromone gradients. Hence, cell fusion defects so mild that they barely affect mating using pheromone gradients still cause strong defects in default mating. When mating in response to a pheromone gradient, yeast cells may use the gradient to more accurately localize the cell fusion machinery at the projection tip. Yeast cells mating in response to a pheromone gradient are also more persistent in their attempts to mate. Either of these factors may compensate for a lowered cell fusion activity during normal pheromone gradient mating but not during default mating.
Could the cell fusion defect in rvs161 cells be attributable to the defect in actin patch polarization or endocytosis? To address this question a collection of rvs161 mutant alleles was generated. rvs161 mutations confer defects in growth on media containing 1 M Na+ or nonfermentable carbon sources such as glycerol (12, 42). Mutations in RVS161 that specifically affect sensitivity to Na+ and utilization of glycerol (these mutations affect actin cytoskeletal organization and endocytosis [ACE]) or that specifically affect cell-cell fusion during mating (CF) were isolated. Intriguingly, rvs161 CF mutations do not affect actin cytoskeletal organization or endocytosis. Conversely, rvs161 ACE mutations do not affect cell-cell fusion during mating. For example, the rvs161 mutant originally identified in the screen for endocytosis mutants (end6) (Rvs161p-R59K) is not defective in cell-cell fusion during mating. Hence, Rvs161p has at least two independent cellular roles, endocytosis/actin cytoskeleton (ACE) and cell-cell fusion during mating (CF) (21).
Rvs161p consists only of a BAR domain, and hence both ACE and CF functions lie within this domain (Fig. 1). Strikingly, all ACE mutations map to the N-terminal 65% of Rvs161p, while all CF mutations map to the C-terminal 35% of Rvs161p. Thus, the Rvs161p BAR domain can be subdivided into an N-terminal ACE domain regulating actin cytoskeleton and endocytosis and a C-terminal CF domain regulating cell fusion. Rvs161p transcript levels increase fourfold and the level of Rvs161p protein also increases following exposure of haploid cells to mating pheromone. This strongly supports the view that Rvs161p provides some function specific for CF despite the fact that Rvs161p clearly has functions other than CF and is still expressed in both haploid cells not treated with mating pheromone and in diploid cells which cannot mate (21).
Vesicles and plasma membrane invaginations accumulate at the site of cell-cell fusion in cells lacking Rvs161p (rvs161Δ). Are these defects linked to the CF defect or the ACE defect? The rvs161 CF mutants were used to address this important question. Interestingly, rvs161 CF mutants still accumulate both vesicles and plasma membrane invaginations at the site of cell-cell fusion, although the total number of vesicles and invaginations per cell was somewhat lower than in cells lacking Rvs161p. Therefore, while some of the vesicles and plasma membrane invaginations are probably attributable to the ACE defect, a subset certainly appear to be specific for the CF defect.
Electron micrographs reveal that rvs161Δ cells accumulate two types of vesicle. One type of vesicle has a diameter of 86 ± 5 nm and is found in vegetative cells predominantly at sites of polarized growth (Fig. 7). These are presumably the vesicles that accumulate when the ACE function is perturbed. A second type of vesicle with a diameter of 54 ± nm was observed in mating cells at the tip of the mating projection and at the site of cell-cell fusion (Fig. 7). These vesicles may be a novel type of secretory vesicle that delivers cell wall-degrading enzymes to the tips of mating projection and that accumulates when the CF function of Rvs161p is perturbed. The diameters of the vesicles that accumulate and form clusters at sites of cell-cell contact in the rvs161 CF mutant fus7 are also small (∼50 nm) (Fig. 5) (20, 21, 91).
FIG.7.
Defects in vesicle fusion in rvs161 mutant cells result in vesicle accumulation. Shown are electron micrographs of wild-type (A), rvs161Δ (B), and rvs167Δ (C) yeast cells. The panels on the left depict cells grown under standard conditions,while those on the right depict cells grown in medium containing sublethal levels (3.4%, wt/vol) of Na+. Bar, 1 μm. Labeled organelles: n, nucleus; v, vacuole; vs, vesicles; g, Golgi apparatus apparatus; ps, primary septum; s, septum. The arrow indicates cell wall abnormalities. (Reproduced from reference 20 with permission of the publisher. Copyright John Wiley and Sons Ltd.)
What is the role of Rvs161p in cell-cell fusion during mating? Two genetically defined pathways important for fusion during mating have been identified. One pathway is dependent on the Fus1p protein and the other on the Fus2p protein. Both Fus1p and Fus2p are important for cell-cell fusion during both default mating and mating in response to pheromone gradients, and the expression of both proteins is induced by treatment with mating pheromone. Fus1p is an O-glycosylated integral plasma membrane protein, while Fus2p is a cytoplasmic coiled-coil protein that localizes to patches at the cortex of the mating projection. The functions of Fus1p and Fus2p are partially redundant, since loss of both proteins has more severe effects on cell-cell fusion than loss of each individual protein (56, 67, 335, 336).
Analysis of fus1Δ rvs161Δ and fus2Δ rvs161Δ double mutants places Rvs161p in the Fus2p pathway of cell-cell fusion. Indeed, Rvs161p and Fus2p coimmunoprecipitate from yeast extracts. Moreover, CF mutations in Rvs161p abolish interaction with Fus2p. The Rvs161p BAR domain and Fus2p are both predicted to form coiled-coil structures so it is likely that Fus2p interacts via coiled-coil interactions with Rvs161p. Why is Rvs161p-Fus2p interaction important for mating? Further work revealed that when Fus2p is bound to Rvs161p it is stable, but when Rvs161p-Fus2p interaction is abrogated by CF mutations Fus2p becomes unstable and is degraded. Overexpression of Fus2p compensates for an inability of a CF mutant form of Rvs161p to bind Fus2p and restores efficient mating. However, the converse is not true, i.e., overexpression of Rvs161p cannot compensate for loss of Fus2p. Hence, a major role of Rvs161p is to stabilize Fus2p (21).
An understanding of Fus2p function in cell-cell fusion is starting to emerge. A key finding was that mating yeast cells exhibit a transient flux of Ca2+ ions across the plasma membrane immediately prior to cell-cell fusion. This results in a transient increase in the level of free intracellular Ca2+ and activation of various downstream signaling proteins. There are two Ca2+ influx systems, one of high affinity and one of low affinity. The latter has been named the low-affinity Ca2+ influx system. Induction of the low-affinity Ca2+ influx system (but not of the high-affinity system) is dependent on an integral plasma membrane protein known as Fig1p and on several known cell-cell fusion proteins, including Rvs161p, Fus1p, and Fus2p. Loss of Fig1p (fig1Δ) results in a mating defect that can be rescued by elevated Ca2+ in the growth medium. Rvs161p and Fus2p may be essential for the low-affinity Ca2+ influx system because they play an important role in delivery of Fig1p to the plasma membrane or in the activation of Fig1p Ca2+ channel activity at the plasma membrane (210).
Rvs161p and Fus2p have at least one additional role in mating. Unlike Fig1p (or the low-affinity Ca2+ influx syste), Rvs161p and Fus2p are required for mating pheromone-induced activation of the cell wall integrity Mpk1p/Slt2p mitogen-activated protein kinase cascade important for remodeling of the cell wall during mating (210). In fus2Δ cells, cell wall material, in particular β(1,3)-glucan, is improperly deposited at the tip of the mating projection, where removal of cell wall is required for cell-cell fusion. The mating defects caused by rvs161Δ or fus2Δ are partially suppressed by overexpression of the protein Lrg1p, which is a Rho-GAP specific for Rho1p. Rho1p in turn is an activator of the cell wall biosynthetic enzyme β(1,3)-glucan synthase (52). Loss of Lrg1p (lrg1Δ) appears to increase the activity of β(1,3)-glucan synthase and enhance cell wall deposition. This in turn inhibits cell-cell fusion. Lrg1p localizes in wild-type cells to the tip of the mating projection, where it may act to locally inhibit Rho1p and β(1,3)-glucan synthase to restrict cell wall deposition. Overexpression of Lrg1p in fus2Δ mutants reduces the improper β(1,3)-glucan deposition at the tip of the mating projection and corrects the mating defect (76).
rvs Mutations Interact with Actin and Myosin Mutations
Rvs− phenotypes are associated with mutations affecting other actin cytoskeleton proteins.
In the original screens for rvs mutants, only a very few of the many mutations screened affected survival upon starvation without also having nonspecific effects on cell viability and thereby satisfied the criteria for a true Rvs− phenotype (42). However, subsequent screens for mutations that affect arrest in G0 when cells are starved of nutrients identified not only the Rvs proteins, but also other actin cytoskeletal and endocytosis proteins. Moreover, proteins such as Whi2p that were originally thought to be specific regulators of the cell cycle in response to nutrients are now known to also be required for actin patch polarization and endocytosis. Hence, cell cycle arrest in response to starvation and long-term viability upon starvation appear to require not only proteins such as Whi2p, Rvs161p, and Rvs167p, but a functional actin cytoskeleton and/or endosomal system in general (24).
Essentially all the phenotypes associated with loss of Rvs161p or Rvs167p are also phenotypes associated with mutations in actin (10, 15, 19, 206, 215, 363). Among the collection of act1 charged-to-alanine mutant alleles described above (363), there is a strong correlation between act1 alleles that affect growth in the presence of Na+ and those that affect utilization of nonfermentable carbon sources. Interestingly, a number of act1 alleles confer reduced viability upon starvation. These alleles tend to be those that also affect growth in the presence of Na+ (19). These phenotypic similarities between mutations affecting actin, Rvs161p, and Rvs167p suggest a close connection between Rvs161p and Rvs167p function and actin cytoskeleton function.
Genetic interactions between Rvs proteins and actin.
Further support for a functional connection between the Rvs proteins and the actin cytoskeleton comes from genetic interaction data. The first genetic interaction was discovered by chance. Both the act1-1 mutation affecting actin and the end6 (later renamed rvs161-E6) R59K substitution in Rvs161p are recessive (i.e., act1-1/ACT1 and rvs161-E6/RVS161 heterozygous diploids are without obvious phenotype). Remarkably, however, a diploid doubly heterozygous for act1-1 and rvs161-E6 (act1-1/ACT1 rvs161-E6/RVS161) displays the mutant phenotype (i.e., is unable to grow at elevated temperature). This type of genetic interaction is known as nonallelic noncomplementation because rvs161-E6 and act1-1 affect different genes (i.e., are nonallelic), but nevertheless do not exhibit genetic complementation. This genetic interaction is specific for the rvs161-E6 allele, because rvs161Δ and act1-1 fully complement, as one would expect. Furthermore, rvs167Δ and act1-1 also fully complement (215). A subsequent study showed rvs161-E6 exhibits nonallelic noncomplementation with act1-1 for several other phenotypes, including bipolar bud site selection, growth in the presence of Na+, and utilization of nonfermentable carbon sources (19).
What does nonallelic noncomplementation tell us about Rvs161p and actin? There are two ways in which nonallelic noncomplementation can arise. One is based on gene dosage. A doubly heterozygous diploid will have only half the normal level of each protein and this may be insufficient. This does not account for the case of rvs161-E6 and act1-1 because nonallelic noncomplementation was not observed with rvs161Δ and act1-1 even though this diploid would have only half the normal level of each protein. The other way in which nonallelic noncomplementation can arise is by the formation of a toxic complex between the two mutant proteins. This better accounts for the nonallelic noncomplementation of rvs161-E6 and act1-1 because it explains why rvs161Δ, which does not encode a protein, would not show the same genetic interaction. Nonallelic noncomplementation suggests the mutant form of Rvs161p (Rvs161p-R59K) binds the mutant form of actin in vivo and this complex has deleterious effects.
A more extensive study of genetic interactions between actin and Rvs161p and Rvs167p was subsequently reported. Actin and Rvs161p or actin and Rvs167p mutations were combined in haploid cells and the double mutants created were examined for combinations that resulted in more severe defects than found in the original single mutants. A striking result was that genetic interaction is allele specific. Some actin mutations (e.g., act1-1) in combination with either rvs161Δ or rvs167Δ resulted in lethality. In contrast, other actin mutations (e.g., act1-4) in combination with rvs161Δ or rvs167Δ yielded double mutants that were no more defective than the single mutants. The act1 alleles that are lethal in combination with rvs mutations may affect the interaction of actin with a protein that is functionally redundant with Rvs proteins in some essential cellular process (19).
The three-dimensional structure of rabbit muscle actin has been determined and the very high amino acid sequence homology of rabbit muscle actin to yeast actin has led to a plausible predicted structure for yeast actin. The residues altered in each act1 mutant have been mapped onto the predicted yeast actin structure (125, 363). Several mutations that are lethal when combined with rvs167Δ map to a patch on the surface of the actin molecule where myosins are known to bind. Although only a few act1 mutations were tested for lethality with rvs161Δ, those that exhibit lethality also map to the same patch on the surface of actin (19).
Genetic interactions between Rvs proteins and yeast myosins.
The yeast genome encodes five MYOsin genes (MYO1 to MYO5). Myosins all contain a globular domain known as the S1 fragment or head domain. This domain has ATPase activity and binds actin. The energy of ATP hydrolysis is coupled to association and dissociation of the myosin head domain with actin filaments and directed movement of the myosin along actin filaments. Myosins in muscle have long coiled-coil tail domains that associate in a tail-to-tail orientation to form dimers known as thick filaments. The myosin thick filaments interdigitate actin thin filaments, and hydrolysis of ATP by myosin is coupled to sliding of the myosin thick filaments over the actin thin filaments during muscle fiber contraction.
Not all myosins have coiled-coil tail domains, nor do all myosins form dimers. The myosins that do have coiled-coil tail domains and form dimers are known as type II myosins or conventional myosins because they resemble muscle myosin. Other myosins may have short tail domains that bind cargo, organelles, vesicles, mRNA, etc., that is then transported along actin filaments by the activity of the ATPase head domain (70, 71, 95, 213, 214, 252, 358).
Genetic interactions between each yeast myosin and Rvs161p and Rvs167p have been examined. The strongest genetic interaction resulted when loss of the only type II conventional myosin in yeast, Myo1p (myo1Δ), was combined with rvs161Δ or rvs167Δ; these double mutants were inviable (19). Myo1p is the myosin that forms the actomyosin contractile ring during cytokinesis (16, 178, 354). Hence, this genetic interaction suggests a role for Rvs161p and Rvs167p in cytokinesis. Neither rvs161 nor rvs167 mutants have been tested for defects in cytokinesis, however, loss of the Rvs167p-associated proteins Las17p and Vrp1p causes defects in cytokinesis. In the case of Vrp1p the cytokinesis and actin patch polarization defects are genetically separable and probably represent distinct activities (173, 218, 260, 328). Loss of the Rvs161p ortholog in the fission yeast Schizosaccharomyces pombe (Hob3) affects cytokinesis (276). Like rvs161Δ and rvs167Δ, both las17Δ and vrp1Δ mutations are lethal in combination with myo1Δ (275). A tight block in cytokinesis may explain why some rvs161 and rvs167 mutant cells continue to display buds with nuclei even under conditions of nutrient starvation (12, 42).
The myo2-66 mutation is not lethal in combination with rvs161Δ or rvs167Δ, but the double mutants grow much more poorly than either single mutant (19). MYO2 encodes Myo2p, which is a type V unconventional myosin and the only essential myosin in yeast. Myo2p localizes to zones of polarized growth where actin patches are also found, but does not strictly colocalize with actin patches. A conditional allele, myo2-66, causes temperature-sensitive defects in polarized secretion to the bud and accumulation of what appear to be secretory vesicles, similar to rvs161 and rvs167 mutants. Moreover, myo2-66 is lethal in combination with mutations that affect fusion of Golgi apparatus-derived secretory vesicles with the plasma membrane (104, 138). Myo2p has been proposed to move secretory vesicles along actin cables to sites of surface growth, where they dock and fuse with the plasma membrane (273). The negative genetic interaction between myo2-66 suggests a role for Rvs161p and Rvs167p in polarized secretion.
No genetic interactions between Rvs161p or Rvs167p and the other yeast myosins, Myo4p (type V myosin), Myo3p (type I myosin), or Myo5p (type I myosin), were observed (19). Myo3p and Myo5p colocalize with Rvs167p in actin patches and have redundant functions in actin patch polarization and endocytosis (5, 96, 102). The type I myosins interact via an SH3 domain with Vrp1p and Las17p and via a C-terminal acidic tail with the Arp2/3p complex and play a role in de novo actin filament assembly (72, 161). Given the redundancy of Myo3p and Myo5p, it would be interesting to know the phenotype of myo3Δ myo5Δ rvs161Δ and myo3Δ myo5Δ rvs167Δ triple mutants.
Actin mutations that map to the surface patch and affect binding of myosin head domains may genetically interact with rvs mutations because the myosin(s) whose binding is affected has redundant function with the Rvs proteins (19). If this explanation is correct, the myosins whose binding is affected may be Myo1p and Myo2p. The Rvs proteins may help to stabilize actin-myosin interactions and become essential when actin-myosin interaction is affected by mutation of either protein. Alternatively, the Rvs proteins may act downstream to tether actin filaments to sites of vesicle fusion or play a role in vesicle fusion itself.
Rvs Proteins and Membranes
Association of cortical actin cytoskeleton with membranes.
The ability of yeast Rvs proteins to associate with lipid membranes has been examined only recently. The diffuse cytoplasmic distribution initially reported for Rvs161p did not suggest an association with membranes (21). In the case of Rvs167p, which localizes to cortical actin patches, a membrane association seemed easier to envisage (11). Cortical actin patches have been visualized by electron microscopy and at least some actin patches are found at sites where the plasma membrane forms “finger-like” invaginations. The invaginations exhibit immunoreactivity for Abp1p and actin. Antibodies against actin decorate transverse striations along the invagination that were proposed to be actin filaments. While some actin patches were observed in contact with the plasma membrane invaginations, other actin patches did not appear to be in contact with the plasma membrane. This may be due to an ability of actin patches to reversibly associate and dissociate from plasma membranes. Alternatively, due to technical limitations, electron microscopy may not reveal all the plasma membrane contacts.
Fractionation of Rvs proteins with membranes.
Subcellular fractionation approaches initially revealed that the majority of Rvs167p and a significant fraction of Rvs161p are in tight association with membranes in vivo (10, 98). Recently, purified Rvs161p and Rvs167p were shown to directly bind liposomes in vitro (84a).
Rvs proteins and lipid rafts.
Membranes both in mammalian cells and in yeast have been shown to consist of a number of distinct “microdomains” characterized by distinct protein and lipid compositions. The best characterized of these microdomains are lipid rafts. Lipid rafts are formed by the lateral association of sphingolipids and sterols (cholesterol in mammalian cells and ergosterol in yeast) and have the biochemical property that, when maintained on ice, they resist solubilization by nonionic detergents such as Triton X-100. This property has led to lipid rafts' being also referred to as detergent-resistant membranes or detergent-insoluble glycosphingolipid-enriched membranes (22, 113, 159, 233). In wild-type cells, both Rvs167p and Rvs161p are associated with detergent-insoluble membranes. In mutant cells deficient in sphingolipid biosynthesis Rvs161p becomes detergent soluble, suggesting the Rvs proteins may partition into glycosphingolipid-enriched lipid raft microdomains (10, 97). Further evidence that Rvs proteins are associated with glycosphingolipid-enriched lipid raft membranes in vivo has come from genetic approaches. A key discovery was that mutations that affect glycosphingolipid biosynthesis suppress the full range of rvs phenotypes (10, 53, 97, 300, 381) (see below).
In yeast, the glycosylphosphatidylinositol (GPI)-anchored protein Gas1p and the integral plasma membrane H+-ATPase Pma1p are markers of lipid rafts (9). Lipid rafts form in the ER (9). Export of lipid raft proteins such as GPI-anchored proteins from the ER has requirements distinct from those for nonraft proteins, in particular a requirement for continuing glycosphingolipid biosynthesis (127, 311).
A distinct class of transport vesicle that is lipid raft enriched mediates the transport of lipid rafts from the ER. The formation of this type of transport vesicle has been reconstituted in vitro and requires the Rab GTPase Ypt1p. In reactions deficient in Ypt1p lipid rafts are still exported from the ER but instead of being transported in lipid raft-enriched vesicles they are transported by default in vesicles containing nonraft proteins (202). Mutations that affect sphingolipid biosynthesis perturb delivery of Pma1p to the cell surface (9). This suggests that sorting of Pma1p to the cell surface involves partitioning into lipid rafts. Interestingly, mutations affecting the actin patch protein Sla2p reduce Pma1p levels at the plasma membrane, suggesting that sorting of lipid raft proteins to the cell surface requires a functional actin cytoskeleton (217).
One of the important functions of lipid rafts is in providing a platform for integration of signal transduction pathways. One of the best examples is the activation of the mitogen-activated protein kinase Raf by the small GTPase H-Ras, which only occurs when H-Ras partitions into lipid raft microdomains (27, 249). In yeast there exist five integral plasma membrane proteins, Wsc1p to Wsc4p and Mid2p, that act as sensors of different stress conditions (152, 179, 241, 346). Wsc1p is the sensor responsible for signaling actin patch depolarization in response to elevated temperature or exposure to Na+ (10, 52). Wsc1p localizes in part to lipid rafts (179). Protein kinase C (PKC) and mitogen-activated protein kinases regulate polarization and depolarization of actin patches upon exposure of cells to stress and downstream of Wsc1p. Exposure of wild-type yeast cells to high temperature or to high Na+ induces depolarization of actin patches over a period of about 30 min. In the continual presence of the stress, cells gradually adapt, and after 90 min the actin patches have returned to a fully polarized distribution (35).
Protein kinase C (Pkc1p) is implicated both in depolarization of actin patches and in actin patch repolarization. In contrast, the PKC1-dependent mitogen-activated protein kinase Slt2p/Mpk1p is not implicated in depolarization of actin patches in response to heat stress, but is specifically required for adaptation to heat stress and subsequent actin patch repolarization. The PKC effectors that mediate actin patch depolarization are yet to be identified (52). Loss of PKC results in loss of cell wall integrity and cell lysis during bud growth. In comparison, loss of the downstream mitogen-activated protein kinase Slt2p has less severe phenotypes and these mutants are viable but exhibit depolarized actin patches, mild cell wall defects, and temperature sensitivity as do cells deficient in Rvs161p or Rvs167p. Intriguingly, rvs mutations exhibit strong genetic interactions with slt2 mutations: while both rvs and slt2 single mutants are viable, an rvs slt2 double mutation is lethal (10). The terminal phenotype of rvs slt2 mutants is reminiscent of that of pkc1 mutants (arrested cells exhibit cell lysis during bud growth).
Are rvs mutants defective in actin patch repolarization? Indeed, loss of either Rvs protein does not affect stress-induced actin patch depolarization, but blocks repolarization. Depolarization of actin patches is a physiological response to stress. Actin patch depolarization observed in untreated rvs mutant cells may be due to stresses encountered normally during growth to which wild-type cells but not rvs mutant cells adapt easily. Restoration of actin patch polarity may require repair of cell wall damage induced by elevated temperature or Na+. Slt2p and Rvs proteins may function in distinct pathways downstream of PKC that are important for repair of cell wall damage and actin patch repolarization. Slt2p may be required for induction of cell wall repair enzymes via phosphorylation of specific transcription factors. Rvs proteins may be required for polarized delivery of newly synthesized cell wall repair enzymes to the cell surface. Consistent with this, mutation of the cell wall repair enzyme β-glucan synthase (kre6) causes only mild growth defects, but kre6 rvs167 double mutants are barely viable (10). Mapping of in vivo phosphorylation sites in Rvs167p has not revealed phosphorylation of any site that matches the PKC consensus, and it is not yet clear if Rvs167p is a PKC substrate.
Suppression of Rvs− phenotypes by mutations affecting glycosphingolipid biosynthesis.
Loss of Rvs161p or Rvs167p results in diverse phenotypes. Several studies have sought to determine if all these phenotypes result from one basic underlying deficiency. In one study, extragenic suppressor mutations that rescue the various rvs defects were isolated and characterized. The rationale behind this approach is that by characterizing the mutations that rescue the rvs mutant defects we may gain insight into what caused those rvs defects. Extragenic mutations were isolated that suppress rvs161Δ (sur). Four separate screens were performed to isolate sur mutations that restore different rvs phenotypes, i.e., viability upon glucose starvation, utilization of nonfermentable carbon sources (glycerol/ethanol), growth in the presence of 3-AT, and growth in the presence of Na+. Since the starting strain had a deletion of the RVS161 gene, all suppressor mutations isolated affect other genes (i.e., they are extragenic) (53).
Of >100 extragenic suppressor mutations isolated, some suppressed only the specific Rvs− phenotype they were selected on (e.g., restored growth in the presence of Na+, but not survival upon glucose starvation). Ten suppressor mutations, however, that suppressed all Rvs− phenotypes were recovered. These sur mutations suppressed the phenotypes not only of rvs161 but also rvs167 and rvs161 rvs167 double mutants. All sur mutations were recessive and genetic complementation analysis showed they affect four separate genes called SUR1 to SUR4 (53). Interestingly, sur1-4 mutations suppress vesicle accumulation in rvs161 and rvs167 mutant cells (20). sur4 (vbm1) and fen1/sur5 (vbm2) were also isolated in an independent study as mutations that suppress the growth and vesicle accumulation defects of snc1 snc2 double mutants (48). Snc1p and Snc2p are vesicle-associated soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors that localize to post-Golgi apparatus secretory vesicles and mediate docking and fusion of these vesicles with the plasma membrane (251). Snc1p interacts genetically with both Ras GTPases and Srv2p, suggesting a role in response to nutrient availability (98).
The sur single mutants do not exhibit reduced viability upon starvation and they grow well on nonfermentable carbon sources. sur1-3 mutants sporulate well and exhibit a bipolar budding pattern in diploids, but sur4 mutants have severe defects in sporulation and bipolar bud site selection, like rvs mutants. The sporulation defect exhibited by sur4 arises in spite of the ability of sur4 mutants to utilize nonfermentable carbon sources and survive nitrogen starvation. Moreover, sur4 rvs161 and sur4 rvs167 double mutant diploids are able to sporulate efficiently even though each single mutant diploid is not. These data suggest Rvs and Sur4p proteins have specific roles in sporulation. Where tested, deletion of each SUR gene suppresses rvs161 and rvs167 mutant phenotypes, similar to the original sur mutation. This suggests the sur mutations isolated in the mutant screen are loss-of-function mutations (53).
The genes affected by the sur1, sur2, and sur4 mutations have been identified (the gene affected by the sur3 mutation is still unknown). SUR1, SUR2, and SUR4 encode ER-localized integral membrane proteins that function in biosynthesis of glycosphingolipids (Fig. 8). Unlike glycerol-based phospholipids that comprise a glycerol base to which one phosphate and two long-chain fatty acids (C16 to C18) are esterified, glycosphingolipids contain a phytosphingosine base with one very long chain fatty acid (C26) attached via an amide linkage (54). The phytosphingosine base is synthesized by the transfer of serine to palmitic acid (as palmitoyl-coenzyme A) to form 3-ketodihydrosphingosine followed by two reduction steps, first yielding dihydrosphingosine and then phytosphingosine (Fig. 8). The C26 fatty acid is then attached to phytosphingosine to yield ceramide. Ceramide is converted into glycosphingolipids, which in yeast are inositolphosphoceramide, mannosylinositolphosphoceramide, and mannosyldiinositylphosphoceramide (54).
FIG. 8.
Sphingolipid biosynthesis in yeast. The yeast sphingolipid biosynthetic pathway indicating the steps blocked by sur1, sur2, sur4, fen1/sur5, and ipt1 mutations that suppress the phenotypes of rvs161 and rvs167 mutants. CoA, coenzyme A; LCB, long-chain base; PI, phosphatidylinositol; Inos-P, inositol-1-P; Cer, ceramide; IPC, inositolphosphorylceramide; MIPC, mannosyl-inositolphoshorylceramide; M(IP)2C, mannosyl-diinositolphoshorylceramide; Man-P-Inos, mannose(α-1,2)inositol-1-P. (Reprinted from reference 54 with permission from Elsevier.)
The product of the SUR2 gene (Sur2p) is the reductase that converts dihydrosphingosine to phytosphingosine during synthesis of the ceramide base. The product of SUR4 (Sur4p) is required for elongation of fatty acid chains by fatty acid synthase, e.g., conversion of palmitate (C16) to very long chain fatty acids (C26). As very long chain fatty acids are incorporated into sphingolipids, sur4 mutants are also defective in sphingolipid biosynthesis. SUR4 is also known as ELOngation 3 (ELO3). The SUR1 gene product (Sur1p) acts downstream of SUR2 and SUR4 and is required for conversion of ceramide to glycosphingolipids. The mannosylation of inositolphosphoceramide to yield mannosylinositolphosphoceramide is dependent on Sur1p (54). IPT1 is the gene required for conversion of mannosylinositolphosphoceramide to mannosyldiinositylphosphoceramide (the step after the Sur1p step). Deletion of IPT1 (ipt1Δ) suppresses the phenotypes of rvs161Δ mutants (rvs167Δ was not tested), and hence IPT1 is also a SUR gene (10).
When the amino acid sequence of SUR4/ELO3 was determined it was noticed that two other yeast genes encode homologous proteins. One, FEN1, was originally identified as the gene affected in a mutant resistant to the toxic sterol biosynthesis inhibitor fenpropimorph. Indeed, mutation of SUR4/ELO3 also results in resistance to fenpropimorph. The mechanism by which these mutations confer resistance to fenpropimorph has not been elucidated, however, this result does suggest that Sur4p has a function closely related to that of Fen1p. Further characterization of Fen1p revealed that it, like Sur4p, is involved in the elongation of fatty acids to give the C26 fatty acid utilized in sphingolipid biosynthesis and FEN1 is also known as ELO2. fen1 mutations, like sur4 mutations, affect bipolar budding in homozygous diploids. Also like sur4 mutations, fen1 mutations suppress the phenotypes of rvs161 and rvs167, and FEN1/ELO2 is also known as SUR5. sur4Δ fen1Δ double mutants are inviable. This shows Sur4p and Fen1p have redundant functions in an essential process, presumably synthesis of very long chain fatty acids (262, 300).
The other gene that encodes a protein homologous to Sur4p is also required for fatty acid elongation and has been named ELO1. Unlike FEN1/ELO2 and SUR4/ELO3, which are required for fatty acid elongation to yield very long chain fatty acids with a chain length of C26, ELO1 is involved in elongation of medium-chain C12 to C16 fatty acids to long-chain C16 to C18 fatty acids. Deletion of ELO1 does not affect doubling time or bipolar bud site selection. Moreover, elo1 mutations do not suppress the phenotypes of rvs mutants (so ELO1 is not a SUR gene). This finding shows that suppression of rvs is specific to mutations that affect glycosphingolipid synthesis, which requires C26 but not C16-18 fatty acids (262, 285).
The phospholipid composition of rvs and sur single and double mutants has been compared. The total amount of phospholipid in rvs161Δ was similar to that of the wild type, but the phospholipid content was reduced twofold in sur1, rvs161Δ sur1, sur4, and rvs161Δ sur4 mutant cells. The rvs161Δ mutant does not have an altered phospholipid composition. In all sur1 and sur4 mutants the percentage of phosphatidylcholine and phosphatidylethanolamine increased, while the percentage of phosphatidylglycerol plus cardiolipin and phosphatidylinositol decreased. The decrease in phosphatidylinositol was slight in sur1 mutants but dramatic in sur4 mutants (approximately 70% reduction). The abundance of phosphatidylserine was relatively normal in all mutants (53). It is more likely the suppression of rvs phenotypes is associated with the altered glycosphingolipid composition rather than the altered phospholipid composition.
SUppressor of Rvs167 7 (SUR7) was identified in a screen to identify wild-type genes that, when overexpressed, suppress the Rvs− phenotype of rvs167-1. Overexpression of SUR7 partially rescued all the defects of rvs167 as well as rvs161 and rvs161 rvs167 double mutants. Deletion of SUR7 (sur7Δ) did not confer any of the phenotypes of rvs mutants except for a somewhat reduced efficiency of sporulation. sur7Δ did not suppress any phenotypes when combined with the rvs161 or rvs167 mutation. The mechanism of SUR7 suppression therefore appears to differ from that of sur1-5. SUR7 encodes an integral membrane protein (Sur7p) with three or four predicted transmembrane domains whose expression is regulated and peaks in late G2-M of the cell cycle. Sur7p localizes to numerous small cortical patches, which are distinct from actin patches (300, 381).
Sur7p is implicated in glycosphingolipid biosynthesis, but at a different step than Sur1-5p. The phytosphingosine base of sphingolipids can be either C18 or C20 in length and is also variably hydroxylated. Glycosphingolipids are classified as class A, B, C, or D based on whether the phytosphingosine base has 1, 2, 3, or 4 hydroxyl groups, respectively. The plasma membranes of sur7Δ cells contain the normal ratio of sphingolipids with C18 and C20 phytosphingosine bases, but they exhibit a reduced amount of the more heavily hydroxylated class C and D glycosphingolipids and a corresponding accumulation of class B glycosphingolipids with only two hydroxyl groups. The phospholipid composition was unaffected in plasma membranes from sur7Δ cells (381). Given that SUR7 overexpression, but not deletion of SUR7, suppresses rvs161 and rvs167 mutant phenotypes, it will be interesting to see the effect of SUR7 overexpression on glycosphingolipid and phospholipid composition.
How do sur mutations suppress Rvs− phenotypes? Does altered glycosphingolipid composition in sur mutant cells affect lipid raft-based signaling pathways that control depolarization and/or repolarization of actin patches? In support of this idea, sur4 and fen1 cells depolarize their actin patches somewhat faster than wild-type cells upon exposure to Na+. In contrast, sur1, sur2, and ipt1 cells are not able to fully depolarize their actin patches upon exposure to Na+. Adaptation to the Na+ stress and repolarization of actin patches were normal in sur4, fen1, sur1, and sur2 cells, but slower in ipt1 cells. Importantly, each rvs161 sur double mutant cell was able to adapt to stress and repolarize actin patches after Na+-induced depolarization, whereas rvs161 single mutant cells were not. The suppressing effect of sur mutations on Rvs− phenotypes is not restricted to rvs mutants: sur1, sur2, sur4, fen1, and ipt1 also restore growth of act1 mutant cells in the presence of Na+ although effects on actin patches in act1 mutant cells were not investigated (10).
How do sphingolipids influence actin patch depolarization and repolarization? In mammalian cells, sphingolipids such as ceramide act as important second messengers in a wide variety of signaling pathways that regulate differentiation, proliferation, apoptosis, and malignancy (111, 112, 156, 228). Sphingoid bases (e.g., phytosphingosine) are important second messengers in yeast and exogenous phytosphingosine is sufficient to restore endocytosis in yeast mutants unable to synthesize endogenous sphingolipids. Restoration of endocytosis in sphingolipid-deficient mutants by exogenous phytosphingosine does not require incorporation into ceramide or glycosphingolipids, so the phytosphingosine does not have to be incorporated into lipid raft components. Instead, restoration of endocytosis by exogenous phytosphingosine depends on its ability as a second messenger to activate protein kinases Pkh1p/Pkh2p, which in turn activate PKC, which results in activation of the downstream mitogen-activated protein kinase Mpk1p/Slt2p. These signaling pathways regulate many cellular responses, including actin patch repolarization (82, 83, 156, 384).
There are two views about the importance of actin patch repolarization for suppression. According to one view, actin patch repolarization is critical and suppression of Rvs− phenotypes is interpreted according to the effects of both phytosphingosine base and lipid raft-dependent Wsc1p on actin patch distribution. The levels of endogenous phytosphingosine and lipid rafts in sur mutants are proposed to influence the phenotype of rvs mutants. According to the second view, suppression still may involve phytosphingosine but can occur without actin patch repolarization.
Balguerie et al. reported data in support of the first view. fen1 mutants, which repolarize more rapidly than the wild type, are blocked at an early step of glycosphingolipid synthesis and accumulate phytosphingosine. Excess endogenous phytosphingosine in fen1 mutants (and possibly also sur4 mutants, which are also blocked at an early step) is proposed to induce repolarization of actin patches to such an extent that Rvs161p is no longer necessary. sur1, sur2, and ipt1 mutant cells are blocked later in the glycosphingolipid biosynthetic pathway and do not accumulate phytosphingosine but may also have reduced levels of lipid rafts due to lowered glycosphingolipid levels. When exposed to Na+ these mutants are not able to fully depolarize their actin patches, so repolarization may not require Rvs161p. As mentioned above, one pool of Wsc1p localizes to lipid rafts (179). If lipid rafts are important for signaling actin patch depolarization, this may explain why mutants such as sur1, sur2, and ipt1 mutants cannot fully depolarize their actin patches (10).
Germann et al. reported data in support of the second view. This study confirmed that sur4Δ suppresses the Na+ sensitivity of rvs161 and rvs167 mutants. It did so, however, without significantly restoring actin patch polarization. Furthermore, in sur4Δ cells Rvs167p does not localize to actin patches but redistributes to the cytoplasm, where it is proteolytically degraded. This reinforces the view that, despite improved growth, the actin patch defect is not rescued by sur4Δ. This study identified several novel Rvs167p-interacting proteins and showed that some do not localize to actin patches. Intriguingly, the most critical Rvs167p-interacting proteins for sur4-mediated suppression were two proteins, Acf2p and Gdh3p (Table 1), that do not localize to actin patches. In contrast, the Rvs167p-interacting proteins Abp1p and Sla1p, which do localize to actin patches, were not required for sur4-dependent suppression of rvs161Δ and Abp1p was not required for sur4-dependent suppression of rvs167Δ (note that Sla1p could not be tested because sla1Δ rvs167Δ is lethal [176]). Interestingly, Acf2p and Gdh3p fractionate with Rvs167p in detergent-insoluble membranes, suggesting all three proteins localize to lipid rafts such as Rvs161p (10, 97).
These findings are in agreement with recent evidence from several other studies that found actin patch polarization is not strictly required for growth even at elevated temperatures. The idea that actin patch polarization may be important for growth came from two observations: no viable mutant that lacks actin patches has yet been reported (hence the existence of actin patches appears to be essential) (149), and many mutants that exhibit depolarized actin patches also exhibit slow growth at normal temperature and are unable to grow at elevated temperature. Examples include loss of Rvs167p, Rvs161p, Sla1p, Sla2p/End4p, Sac6p, Srv2p, Vrp1p, Las17p, and point mutations in actin, Pan1p, and Arp2/3p complex subunits (15, 60, 126, 173, 176, 181, 197, 215, 219, 321, 340, 363, 364). There are, however, strains in which actin patch polarization is lost without abolishing growth at elevated temperature, for example, in the wild type or in vrp1Δ mutants overexpressing Las17p (219), vrp1Δ mutants expressing a fragment of Vrp1p (328), or mutants deficient in capping protein Cap2p (cap2Δ) (3) or the actin-regulatory kinase Prk1p (prk1Δ) (387). Some mutations in Ysl2p (ysl2-1) (296) and in actin (act1) (15) also severely perturb actin patch polarization without resulting in temperature-sensitive growth.
It is possible that alterations in cellular lipid composition induce growth defects in rvs mutants and suppression of growth defects in sur mutants via mechanisms unrelated to their effects on actin patch distribution. It is interesting that the sphingoid base component of sphingolipids can cause cell damage and induce apoptosis in mammalian cells (111, 112, 228). In S. cerevisiae the sphingoid base phytosphingosine regulates growth via ceramide-activated protein phosphatase (CAPP) (75, 224). Lipids other than sphingolipids can also affect growth and viability. A recent study found that endogenous free fatty acids and diacylglycerol induce apoptosis upon entry into stationary phase in S. pombe. This apoptosis pathway is caspase independent, i.e., independent of the yeast caspase-like protease (388). Finally, protein N-myristoylation is known to be essential for survival under starvation conditions in S. cerevisiae and requires the medium-chain fatty acid myristate (7). Perhaps levels of myristate vary depending on the level of very long chain fatty acids. The contribution of lipid-mediated signaling to the various defects in rvs mutant cells is an area that has not yet been fully explored.
Regulation of Rvs167p by Phosphorylation
Insight into how Rvs proteins function to link nutrient availability to G1-S cell cycle progression came from the discovery that Rvs167p is regulated by the kinase Pho85p (163). Pho85p is a yeast cyclin-dependent kinase (Cdk) like the major cell cycle-regulatory Cdk known as Cdc28p in budding yeast (Cdc2/Cdk2 in fission yeast and mammalian cells) (200). The mammalian ortholog of budding yeast Pho85p is Cdk5, and its activity is critical for neuronal function (303). Mammalian Cdk5 has been shown to functionally substitute for Pho85p when expressed in budding yeast, so Cdk5 and Pho85p are functional orthologs (129, 225).
As for all Cdks, the protein kinase activity of both Cdc28p and Pho85p is dependent on their association with regulatory subunits known as cyclins whose expression level often varies through the cell cycle. Cdc28p is activated by one set of cyclins known as G1 cyclins (Cln1p to Cln3p) in late G1 for entry into the cell cycle and by two other sets of cyclins known as S-phase and mitotic cyclins (Clb1p to Clb6p) at subsequent stages of the cell cycle. Pho85p has 10 known cyclins known as Pho85 cyclins (Pcls). The Pcls have been classified into two subfamilies based on homology, the Pcl1p/2p subfamily (comprising Pcl1p, Pcl2p, Clg1p, Pcl5p, and Pcl9p) and the Pho80p subfamily (comprising Pho80p, Pcl6p-Pcl8p, and Pcl10p) (6, 170, 190).
In conjunction with different Pcls, Pho85p plays a role in phosphate and glycogen metabolism as well as a role in G1-S cell cycle progression. Pcl1p and Pcl2p in association with Pho85p are functionally redundant with Cdc28p and the G1 cyclins Cln1p and Cln2p in G1-S cell cycle progression. This late G1 role is specific to Pcl1p and Pcl2p, as other Pcls are not able to perform this function (in the case of the highly Pcl2p-homologous Pcl9p, this may simply be due to insufficient expression in late G1). Two-hybrid screens identified Rvs167p as a Pcl2p- and Pcl9p-associated protein and the interaction was confirmed by coimmunoprecipitation of Rvs167p with Pcl2p from lysates and binding of recombinant Pcl2p and Rvs167p in vitro. All Pcl1p/2p subfamily Pcls bind strongly to Rvs167p, but Pho80p subfamily Pcls bind Rvs167p only weakly (163).
Do pho85, pcl2, or pcl9 mutants exhibit Rvs− phenotypes? As the Pcl1p/2p subfamily Pcls exhibit functional redundancy, a strain with deletions of all five genes encoding Pcl1p/2p subfamily Pcls (referred to in reference 163 as quintΔ) was constructed. Both quintΔ and loss of Pho85p (pho85Δ) resulted in essentially the full range of Rvs− phenotypes. Consistent with a role for Pho85p/Pcl2p in regulation of Rvs167p, Pho85p/Pcl2p phosphorylates Rvs167p in vitro and phosphorylation of Rvs167p in vivo is partially dependent on Pcl1p/Pcl2p subfamily Pcls. Rvs167p is phosphorylated only in G1 phase, consistent with in vivo phosphorylation by Pho85p in association with Pcl1p and Pcl2p (which are expressed specifically in G1). There are three major phosphorylation sites in vivo and all are located in the GPA-rich domain, S299, S379, and S321. When S321 is mutated T323 is phosphorylated instead, so T323 is a potential phosphorylation site but is not normally phosphorylated. Phosphorylation of S299 and S379 in vivo is partially dependent on Pcl1p/Pcl2p, but phosphorylation of S321 is not (85, 163).
Rvs167p is hyperphosphorylated upon exposure of cells to mating pheromone, suggesting a role for Rvs167p phosphorylation also in the mating response. Hyperphosphorylation requires Pho85p and specifically Pcl2p (whose expression is induced by mating pheromone). Hyperphosphorylation also requires the mitogen-activated protein kinase Fus3p, which is induced by mating pheromone and functions in the mating pheromone response pathway. In vitro, Fus3p phosphorylates the same sites in the Rvs167p GPA-rich domain as Pho85p/Pcl2p (i.e., S299, S321, S370, and, when S321 is missing, T323) (85). Hyperphosphorylation of Rvs167p by Fus3p supports a role for Rvs167p in mating, as proposed previously (11, 20).
Is phosphorylation of Rvs167p important for its function? An Rvs167p mutant protein in which S299, S321, T323, and S379 were all replaced with alanine (Rvs167-4A) did not result in Rvs− phenotypes. This was perhaps not unexpected since these phosphorylation sites all lie within the GPA-rich domain, which has been shown previously to be nonessential for known Rvs167p functions (38, 299). Under normal conditions, rvs167-4A was also not lethal in combination with any mutations that are known to be lethal in combination with rvs167Δ (176). However, under conditions of high temperature and in the presence of Na+ rvs167-4A was lethal in combination with sla1Δ. Moreover, testing of the set of systematic deletions covering each nonessential S. cerevisiae open reading frame revealed that a mutant carrying the novel combination end3Δ rvs167Δ is inviable. Furthermore, end3Δ is also lethal in combination with rvs167-4A. Rvs167p phosphorylation may therefore be important in the absence of Sla1p and essential in the absence of End3p (85).
End3p and Sla1p form a complex with the Arp2/3p complex-activating protein Pan1p (62, 322). Activation of the Arp2/3p complex by Pan1p may be essential in the absence of Las17p, as loss of both Pan1p and Las17p results in lethality (62). Given the deleterious interactions between rvs167-4A and mutations affecting the Sla1p-End3p-Pan1p complex, loss of phosphorylation of Rvs167p may perturb Las17p function. This would result in dependency on the Sla1p-End3p-Pan1p complex for Arp2/3p activation. Indeed, the Rvs167p SH3 domain binds Las17p in vitro (38, 181). Binding of GPA-SH3 to Las17p (and also Ymr192p/Gyl1p) in vitro is considerably reduced by phosphorylation of the GPA-rich domain by Pho85p/Pcl2p. Introduction of a charge via phosphorylation into the uncharged GPA-rich domain may induce major conformational changes in the GPA-rich domain that affect the neighboring SH3 domain and its ability to bind Las17p and other proteins. In this case Rvs167-4A (which cannot be phosphorylated) may bind Las17p too strongly and perhaps inhibit Las17p-dependent Arp2/3p activation, causing dependency on Sla1p-End3p-Pan1p (85).
Regulation of Rvs167p by Ubiquitination
The role of ubiquitination in endocytosis extends beyond ubiquitination of the receptor tail by the ubiquitin protein ligase Rsp5p. Ubiquitination of the endocytic machinery may also be essential for endocytosis (64). Genetic interactions between Rsp5p and a number of actin patch proteins required for endocytosis support an additional role for Rsp5p in regulation of actin patches and the endocytic machinery (146). Substrates ubiquitinated by Rsp5p often associate with Rsp5p and can be identified by protein interaction screens.
In a high-throughput mass spectrometry analysis of the yeast proteome, a protein complex containing Rsp5p and Rvs167p was identified (123). Rvs167p also associates with Sla1p (59, 123) and both Rvs167p and Sla1p immunoprecipitate with Rsp5p from yeast lysate, showing Rvs167p, Sla1p, and Rsp5p form a complex in vivo (307). Consistent with the possibility that Rvs167p is ubiquitinated by Rsp5p, mass spectrometry analysis of the yeast proteome detected ubiquitin attached to K481 in the Rvs167p SH3 domain (122, 236).
The Rvs167p-Rsp5p association is direct and mediated by the Rsp5p WW domains and the Rvs167p GPA-rich domain. WW domains bind motifs of the consensus PPXY and the Rvs167p GPA-rich domain includes one motif (PPAY) that matches exactly and two motifs (PSY and PQY) that match less closely. All three motifs contribute to the interaction with Rsp5p. The Sla1p-Rsp5p association is also direct and mediated by residues 420 to 720 of Sla1p, which contains Sla1p Homology Domain 1 (SHD1) and a coiled-coil domain. It has been proposed that the Sla1p-Rvs167p complex may be the yeast equivalent of the vertebrate CIN8-endophilin complex. While Rvs167p is often compared to vertebrate amphiphysin, it is also related to vertebrate endophilin, which has a domain structure similar to that of amphiphysin but was discovered more recently (Fig. 1). Sla1p exhibits sequence homology to CIN8 and the two proteins have a similar domain structure. Moreover, like the yeast Sla1p-Rvs167p complex, the vertebrate CIN8-endophilin complex interacts with the ubiquitin ligase Rsp5p (known as Nedd4 in vertebrates) and functions in endocytosis (240, 305, 307, 313).
The ubiquitin ligase Rsp5p is responsible for Rvs167p monoubiquitination, since an rsp5 mutation results in loss of the modified form of Rvs167p. Moreover, Rsp5p binding to Rvs167p is also important for monoubiquitination, since combined mutation of the PPXY, PSY, and PQY motifs in the Rvs167p GPA-rich domain or combined mutation of the three Rsp5p WW domains not only reduce Rvs167p-Rsp5p association but also reduce the level of monoubiquitinated Rvs167p in vivo. Interestingly, although the GPA-rich domain mediates binding of Rvs167p to Rsp5p, it is not the site at which ubiquitin is attached (indeed, the GPA-rich domain contains no lysine residues and hence cannot be ubiquitinated). K481 in the SH3 domain is the only site ubiquitinated in Rvs167p. Hence, binding of Rsp5p to the Rvs167p GPA-rich domain induces monoubiquitination of K481 in the Rvs167p SH3 domain (307).
Is monoubiquitination of Rvs167p important for in vivo function? As discussed above, neither the GPA-rich nor SH3 domain is required for any of the known biological roles of Rvs167p (38). Consistent with this, mutated forms of Rvs167p lacking PPXY and PXY motifs in the GPA-rich domain, with a K481R substitution in the SH3 domain (to prevent ubiquitin attachment), or lacking the SH3 domain entirely were fully functional for receptor-mediated endocytosis (307). An Rvs167p mutant construct lacking the SH3 domain was previously found to be only partially functional for growth in the presence of Na+, possibly due to lowered expression (38, 299). Similarly, the mutant Rvs167p construct with a K481R substitution in the SH3 domain was also only partially functional for growth in the presence of Na+. However, this requirement is for K481 only, not ubiquitination of K481. An Rvs167p mutant construct lacking a PPXY motif does not bind Rsp5p and is not ubiquitinated on K481, but nevertheless is fully functional for growth in the presence of Na+. Hence, K481, but not ubiquitination of K481, is important for some functions of Rvs167p (307).
Genomic and Proteomic Approaches and the Diverse Network of Rvs Interactions
Large-scale two-hybrid screens for Rvs161p- and Rvs167p-interacting proteins.
The first large-scale screen to identify all Rvs161p- and Rvs167p-interacting proteins was reported by Bon et al. Full-length RVS161 and RVS167 genes in both centromeric (low copy) and multicopy plasmids were used as bait. Each prey sequence isolated by either Rvs161p or Rvs167p was then tested against the other bait. Finally, selected preys were tested against each individual domain of both proteins. Numerous potential interacting proteins were identified (Table 1). Interestingly, five preys interacted with Rvs161p and Rvs167p, specifically Las17p, Rvs167p itself, App1p, Gyp5p, and Gyl1p (18).
Subsequently, more global analyses of protein-protein interactions among all proteins encoded by the S. cerevisiae genome were performed by Uetz et al. and by Ito et al. Uetz et al. used a genomewide set (6,000) of full-length genes as baits and identified two interactors for Rvs161p and 10 for Rvs167p. Ito et al. also used a genomewide set of 6,000 full-length genes as baits and preys and tested each combination systematically. The interacting proteins identified in this screen overlapped only slightly with those obtained by Uetz et al., however, only three interactions for Rvs161p and five for Rvs167p were identified in this screen (Table 1) (136, 339).
A third study by Drees et al. aimed to produce a protein interaction map for proteins implicated in cell polarity development; 68 selected baits relevant to cell polarity were chosen and used to screen an array of preys representing about 90% of the S. cerevisiae genome. A network of interactions was revealed that links signaling proteins, the actin cytoskeleton, and organelles to polarity cues. This network included Rvs161p and Rvs167p homo- and heterointeractions as well as Rvs167p interactions with 14 other proteins (59) (Table 1).
Identification of Rvs167p SH3 domain-interacting proteins by phage display.
Phage display is a technique in which libraries of ligand peptides are displayed on the surface of bacteriophages and screened for the ability to interact with an immobilized receptor the bait). In phage display, sequencing the bacteriophage DNA is used to identify those proteins that confer interaction with the bait. This technique has the drawback of low accuracy (false-positives). One study combined computational prediction of interactions using phage display ligand consensus sequences with large-scale two-hybrid interaction analysis to identify interactions involving yeast SH3 domains, including the Rvs167p SH3 domain. Phage display was used to define the binding consensus motifs of each SH3 domain (which were analyzed computationally by a position-specific scoring matrix) to identify potential binding partners for each SH3 domain. Then the SH3 domain baits were screened against yeast two-hybrid libraries and an ordered array of interacting open reading frames was established. A phage display network containing 394 interactions among 206 proteins and a two-hybrid network containing 233 interactions among 145 proteins were established. Graph theoretical analysis identified 59 highly likely interactions common to both networks. This revealed nine potential partners for the Rvs167p SH3 domain (331).
Another study combined phage display and synthesis of peptides on cellulose membranes to search for all the peptides in the yeast proteome with the potential to bind to certain SH3 domains, including the Rvs167p SH3 domain. Peptides that matched a relaxed consensus as deduced by phage display experiments were identified. Then all these peptides were synthesized at high density on a cellulose membrane and probed with the SH3 domains. The SH3 domains were grouped by this approach into five classes with partially overlapping specificity. This approach, named the whole interactome scanning experiment, permits identification of the partners of any peptide recognition module by peptide scanning of a proteome. The whole interactome scanning experiment found 24 potential partners for the Rvs167p SH3 domain (160) (Table 1).
Identification of Rvs161p- and Rvs167p-interacting proteins by high-throughput proteomics.
In the quest for comprehensive analysis of protein-protein interactions, high-throughput mass spectrometric analyses leading to protein complex identification have been developed as a powerful proteomic approach. Selected yeast proteins were chosen as baits, epitope tagged, transiently overexpressed, and used for immunoprecipitation of protein complexes from cell extracts. Proteins in each purified complex were resolved by polyacrylamide gel electrophoresis, stained, excised from gels, and trypsin digested before mass spectrometric analysis. With 725 bait proteins, 3,617 associated proteins covering 25% of the yeast proteome were detected, including four Rvs161p and 27 Rvs167p interactors (Table 1) (123).
Mapping the genetic interactions of Rvs161p and Rvs167p by synthetic genetic array.
To identify functional relationships between genes, a method for systematic construction of double mutants, termed synthetic genetic array analysis, was developed for S. cerevisiae. A haploid carrying a query mutation (conditional or deletion) linked to a dominant selectable marker and expressed from a mating type-specific promoter is crossed to an array of approximately 4,700 haploid open reading frame deletion mutants (nonessential genes) of the opposite mating type, where each open reading frame deletion is marked with a drug resistance gene. The diploid progeny are then sporulated to yield recombinant meiotic haploid progeny and those recombinant progeny expressing both deletion markers (double mutants) are selected. The inviability or sickness of these meiotic double mutant progeny represents negative genetic interactions that may suggest the affected genes are functionally related. Synthetic genetic array analysis of 132 queries allowed the construction of a network of approximately 1,000 genes and 4,000 functional interactions. Of these, 47 involved Rvs161p and 51 involved Rvs167p (Table 1) (332). A more recent SGA analysis focusing specifically on Rvs161p and Rvs167p found 49 lethal combinations for each gene. Strikingly, the genetic interactors identified for each Rvs protein were identical (84a).
Rvs161p and Rvs167p physical and genetic interactions suggest multiple functions in vivo.
Interactions found by high-throughput approaches should be regarded as putative until further characterized, however, these studies have provided the first global view of the cellular processes in which Rvs161p and Rvs167p are (or may be) implicated. What is interesting is that the Rvs161p and Rvs167p interactors represent a diversity of both organelles and structures as well as cellular processes (Fig. 9). Some Rvs161p and Rvs167p physical interactors are cytoplasmic, while others are nuclear. Some interactors are components of cortical actin patches, while others are not.
FIG. 9.
Functional classification of Rvs161p and Rvs167p-interacting proteins. Pie chart depicting the proportions of the total set of known Rvs161p- and Rvs167p-interacting proteins (including both physical and genetic) which belong to each functional category.
One interesting finding from the large-scale two-hybrid and peptide-scanning screens was the potential interaction of Rvs167p with Bsp1p (59, 160, 331). Bsp1p is a protein that interacts with two of the three yeast orthologs of vertebrate synaptojanin (Inp52p/Sjl2p and Inp53p/Sjl3p). As will be discussed further below, in vertebrates the function of BAR domain proteins is closely linked to the function of synaptojanin. In yeast, Bsp1p has been implicated in the recruitment of Inp52p/Sjl2p and Inp53p/Sjl3p to cortical actin patches (366).
The diversity of potential Rvs161p and Rvs167p interactors is striking. Even among those interactors known to function in membrane traffic, some function in endocytosis and others in exocytosis. Many Rvs161p- and Rvs167p-interacting proteins function in nucleocytoplasmic transport, some in translocation of secreted proteins into the ER, and others in mitochondrial import. Some interactors are involved in transcription, others in translation, and still others in degradation of proteins and mRNAs. Several are cell cycle regulators. Many are components of the cytoskeleton. However, the largest number of Rvs161p and Rvs167p interactors are metabolic enzymes. Hence, Rvs161p and Rvs167p should not be thought of as strictly endocytic proteins or as strictly actin patch components. They perhaps should be thought of as proteins that perform a basic molecular function that is so important and fundamental that it has been utilized again and again by multiple cellular structures for the provision of multiple cellular functions.
FISSION YEAST BAR DOMAIN PROTEINS Hob1p AND Hob3p
Hob3p, Fission Yeast Ortholog of Rvs161p
Hob3p has a domain structure similar to that of Rvs161p.
A fission yeast (Schizosaccharomyces pombe) gene encoding a protein that comprises a BAR domain only and is homologous to S. cerevisiae Rvs161p was identified by homology search with the Rvs161p sequence and named homologue of bin3 (hob3+). Hob3p exhibits 56 and 29% sequence identity to S. cerevisiae Rvs161p and human Bin3 (see below), respectively. Hob3p is not essential, but Hob3p-deficient cells (hob3Δ) are more elongated than wild-type cells and tend to form clumps, suggesting a defect in cytokinesis or cell separation. Wild-type fission yeast cells, unlike budding yeast cells, are cylindrical. A small percentage of hob3Δ cells are spherical rather than cylindrical, indicating that in some Hob3p-deficient cells there is a loss of cell polarity. These morphological defects do not worsen upon a shift in temperature, osmotic strength, or nutrient availability, and loss of Hob3p does not affect the rate of cell proliferation at high (37°C) or low (21°C) temperature or upon nitrogen starvation. Furthermore, overexpression of Hob3p does not inhibit the growth of wild-type S. pombe cells (276).
Hob3p functions in polarization of the cortical actin cytoskeleton but not endocytosis.
Loss of Hob3p perturbs the actin cytoskeleton. Actin patches are not polarized to the growing cell tips in interphase hob3Δ cells as is the case in wild-type cells but are often randomly distributed along the length of the cell. During cytokinesis, actin patches are not efficiently polarized to the medial site of division in hob3Δ. Actin patches are often distributed randomly in the daughter cell, mother cell, or both. Loss of actin cytoskeleton polarization often causes defects in cytokinesis and accumulation of multinucleate cells. Indeed, hob3Δ cells tend to become multinucleate, consistent with an impairment of cytokinesis. Unlike S. cerevisiae rvs161 mutants, S. pombe hob3Δ cells endocytose the membrane-soluble fluorescent dye FM4-64 with the same kinetics as wild-type cells, and the appearance of the endocytic compartments is normal. Moreover, hob3Δ mutants mate with the same efficiency as wild-type cells and thus lack the cell-cell fusion defect of S. cerevisiae rvs161 mutants (276).
Functional conservation between Hob3p, Rvs161p, and human Bin3.
Hob3p function is fully conserved in Bin3 (see below), but only partially conserved in Rvs161p. The cell morphology and actin patch polarization defects of hob3Δ mutants are fully rescued by ectopic expression of the human Bin3 protein. In contrast, ectopic expression of S. cerevisiae Rvs161p restored normal cell morphology to hob3Δ mutants but only partially restored the actin cytoskeleton. Rvs161p restored F-actin localization to the cell middle during cell division, but there was no significant improvement in actin patch polarization to cell tips in interphase cells. In contrast, expression of S. cerevisiae Rvs167p did not rescue any of the defects observed in hob3Δ mutants. Interestingly, ectopic expression of Hob3p in S. cerevisiae rvs161Δ mutants rescued growth in the presence of Na+, but ectopic expression of human Bin3 (or Bin1) in rvs161Δ cells did not (276). This shows that Rvs161p function is still conserved in Hob3p, but that human Bin3 has diverged during evolution.
Hob1p, Fission Yeast Ortholog of Rvs167p
Hob1p has a domain structure similar to that of Rvs167p.
An S. pombe gene encoding a protein with homology to S. cerevisiae Rvs167p and mammalian Bin1 was identified by homology search. This gene was named homolog of Bin1 (hob1+). Hob1p has a domain structure similar to Rvs167p with an N-terminal BAR domain, a central domain, and a C-terminal SH3 domain. The BAR domain of Hob1p exhibits 51% homology to the BAR domain of Rvs167p and only 27 to 28% homology to the BAR domains of mammalian amphiphysin 1 and Bin1 (277).
Hob1p is not required for polarization of the cortical actin cytoskeleton or endocytosis.
Hob1p is not an essential protein and Hob1p-deficient cells (hob1Δ) exhibit relatively normal morphology, but are slightly more elongated than wild-type S. pombe cells. Hob1p-deficient cells exhibit no defect in cell proliferation or in mating and retain a fully polarized actin cytoskeleton. Endocytosis of the membrane-soluble fluorescent dye FM4-64 is also unaffected by loss of Hob1p (or even by loss of both Hob1p and Hob3p). Hob1p-deficient cells also remain fully viable upon nitrogen starvation, at a range of temperatures, and in the presence of 250 mM Na+. The expression of S. pombe Hob1p in S. cerevisiae rvs167Δ cells did not restore growth on medium containing Na+. This suggests that Hob1p function has diverged from that of Rvs167p during evolution. Overexpression of Hob1p weakly inhibits growth but does not affect cell morphology. In contrast, overexpression of the Hob1p BAR domain alone does not inhibit growth, while overexpression of the central and SH3 domains is inhibitory for growth only when combined with exposure to high Na+ (131, 277).
Hob1p interacts with actin assembly proteins and protein kinases that regulate cell polarity.
Hob1p localizes to cortical actin patches whose distribution is polarized to sites of surface growth at cell tips (equivalent to buds in budding yeast) and the medial region in cells undergoing division (equivalent to the bud neck in budding yeast). This localization is similar to that of Rvs167p in budding yeast. Unlike Rvs167p, however, localization of Hob1p to patches is abolished by disassembly of F-actin by treatment with the actin polymerization inhibitor latrunculin A (131).
Hob1p interacts with proteins important for actin filament assembly, such as the fission yeast ortholog of budding yeast Las17p, Wsp1p. Interaction requires the Hob1p SH3 domain, as in the case of Rvs167p-Las17p interaction in budding yeast, but differs in that it also requires the BAR domain. Wsp1p is essential for Hob1p localization to cortical patches since Hob1p adopts a diffuse cytoplasmic distribution when Wsp1p expression is repressed. Overexpression of Hob1p causes Wsp1p to redistribute from patches to the cytoplasm, and this effect requires the Hob1p SH3 domain that interacts with Wsp1p. Overexpression of Hob1p also reduces the ability of cell extracts to support Wsp1p-dependent actin filament assembly in vitro, suggesting that Hob1p may inhibit Wsp1p. Repression or complete loss of Wsp1p alone has mild effects on cell polarity and growth, but repression of Wsp1p expression in hob1Δ mutants severely inhibits growth and induces aberrant round-cell morphology. Overexpression of Wsp1p also inhibits growth and causes cells to become somewhat roundish, and these defects are corrected by additional overexpression of Hob1p. Hence, Hob1p and Wsp1p appear to have multiple functions in growth and polarity, and some of these are probably synergistic, while others are antagonistic (131).
Hob1p also interacts with the protein kinase Nak1p. Nak1p plays an essential role in polarized growth in fission yeast and is a member of the germinal center kinase-like kinases that form a subgroup of the p21-activated kinase family of kinases that have a conserved role in cell polarity. The Hob1p BAR domain mediates interaction of Hob1p with Nak1p. Repression of Nak1p expression causes S. pombe cells to become round. Loss of Nak1p does not affect Hob1p localization to patches, but causes depolarization of Hob1p patches. Repression of Nak1p expression has mild effects on growth in the presence of Na+, but combination with loss of Hob1p causes severe growth sensitivity to Na+, suggesting that Nak1p and Hob1p have some redundant functions. Consistent with this, overexpression of Hob1p or of the Hob1p BAR domain alone partially restores growth and normal elongated cell morphology to cells in which Nak1p expression is repressed. In contrast, overexpression of the Hob1p central and SH3 domains did not restore normal cell morphology to Nak1p-deficient cells, indeed, it induced even more severe growth defects, perhaps by interfering with endogenous Hob1p function (131).
These results strongly implicate Hob1p in organization of the actin cytoskeleton, in growth under stress conditions, and in cell polarity. It seems that the lack of effect of hob1Δ on cell polarity, growth in the presence of Na+, or actin cytoskeleton organization is due to functional redundancy with both Wsp1p and Nak1p. Hence, Hob1p plays a role in cell polarity, actin cytoskeleton organization, and growth under stress conditions much like Rvs167p in budding yeast, but its role is more redundant with Wsp1p/Las17p family proteins in fission yeast than in budding yeast, and this obscures the hob1 phenotypes in fission yeast.
Hob1p functions in regulating cell cycle progression in response to nutrient availability.
hob1Δ cells have a defect in cell cycle progression under nitrogen-limiting conditions. Unlike S. cerevisiae cells, which spend most of their cell cycle in G1 phase prior to DNA replication, S. pombe cells spend most of their cell cycle in G2 phase after DNA replication. Hence, S. pombe cells normally exhibit a 2C DNA content. Upon nitrogen starvation, however, S. pombe cells continue to divide and at each division exhibit a smaller cell size than during the previous division. Eventually, the starved cells reach a minimum size at which they are unable to satisfy the cell size requirement to enter the next S phase. The starved cells therefore accumulate in G1 with a 1C DNA content and eventually enter G0. In contrast to wild-type cells, hob1Δ cells starved of nitrogen maintain a 2C DNA content. The hob1Δ cells do arrest and presumably enter G0, but do so directly from G2. Despite their 2C DNA content, hob1Δ cells remain viable even after prolonged periods of nitrogen starvation (277).
Functional Conservation of Hob1p and Bin1 and Divergence of Rvs167p
Ectopic expression in hob1Δ cells of the ubiquitous human Bin1 isoform that lacks exon 10, Bin1−10−12+13 (SH3p9), but not human amphiphysin 1 or Bin2, restored the ability to arrest in G1 and enter G0 with a 1C DNA content upon nitrogen starvation. This suggests that Hob1p function is most conserved with human Bin1−10−12+13/SH3p9. Ectopic expression of human Bin1−10−12+13/SH3p9 in rvs167Δ S. cerevisiae cells did not rescue growth in the presence of Na+, again suggesting that Rvs167p function has diverged from that of Bin1−10−12+13/SH3p9 and Hob1p. Intriguingly, overexpression of hob3+ in hob1Δ cells efficiently rescued the cell cycle arrest defect. Hence, it has been proposed that Hob3p functions in the same signaling pathway as Hob1p, but downstream of Hob1p (277).
Hob1p functions in regulation of cell cycle progression in response to DNA damage.
Loss of Hob1p also causes hypersensitivity to both chemical (phleomycin)- and UV-induced DNA damage, but not to inhibitors of DNA replication (e.g., hydroxyurea). hob1Δ cells arrest cell cycle progression normally in response to DNA damage (so the checkpoint is still functional), but presumably the ability of the mutant cells to repair the DNA damage and then reenter the cell cycle is impaired. Combination of hob1Δ with rad3Δ and chk1Δ mutations, which are also hypersensitive to DNA damage, revealed that hob1Δ rad3Δ double mutants are considerably more sensitive to phleomycin than either single mutant, i.e., Hob1p and Rad3p genetically interact. In contrast, hob1Δ chk1Δ double mutants are only as sensitive to phleomycin as the more sensitive single mutant (hob1Δ). It has been proposed that Rad3p and Hob1p act in distinct pathways that each contribute to repair of DNA damage (277).
Hob1p functions in a cell stress signal transduction pathway upstream of Hob3p.
Unexpectedly, the lethality caused by DNA damage in hob1Δ cells is dependent on hob3+. hob1Δ hob3Δ double mutants, unlike hob1Δ single mutants, are not hypersensitive to DNA damage. The mechanisms involved are still unclear, but the data seem consistent with a function for Hob3p in cell cycle arrest downstream of Hob1p. Interestingly, the steady-state levels of both Hob1p and Hob3p increase dramatically upon exposure of cells to DNA damage. The level of Hob1p increases within 15 min of phleomycin treatment, while the increase in Hob3p levels is slightly delayed and becomes apparent only after 30 min and the levels of both proteins remain elevated for at least 24 h. The increase in Hob3p protein levels is absolutely dependent on Hob1p, as it is absent in hob1Δ cells (277). Whether Rvs161p and Rvs167p levels increase upon exposure of S. cerevisiae cells to DNA damage and if loss of Rvs proteins causes sensitivity to DNA damage has not been examined.
VERTEBRATE BAR DOMAIN PROTEIN AMPHIPHYSIN 1
Endocytic Recycling of Synaptic Vesicles
Nerve terminals are known to be sites highly active in endocytosis. When synaptic vesicles fuse with the plasma membrane (plasmalemma) and release their cargo of neurotransmitters into the presynaptic cleft, synaptic vesicle membrane material is rapidly recovered back into the cell via endocytosis. These empty synaptic vesicles are then used for storage of newly synthesized neurotransmitters. Endocytosis of the synaptic vesicle membrane occurs at clathrin-coated pits. Clathrin assembles to form a coat on the cytoplasmic face of the plasma membrane at subdomains where endocytosis occurs and this assembly is driven by the plasma membrane-clathrin assembly complex AP-2. Clathrin assembly promotes curvature of the plasma membrane, leading to the formation of an invagination known as a clathrin-coated pit. Progressive invagination results in the formation of a deeply invaginated clathrin-coated pit connected to the plasma membrane by a narrow neck of membrane (120, 121, 155, 283, 290, 293, 295, 315, 368).
Role of dynamin 1 in synaptic vesicle recycling.
The GTPase dynamin 1 is required for the scission of the neck of deeply invaginated clathrin-coated pits to release free clathrin-coated vesicles into the cytoplasm. Dominant negative mutant forms of dynamin 1 block receptor-mediated endocytosis of transferrin and epidermal growth factor, but do not affect fluid-phase endocytosis (45, 116, 121, 344). Upon a shift to the restrictive temperature, Drosophila melanogaster cells carrying a temperature-sensitive mutation of dynamin (shibire) become paralyzed due to failure of nerve transmission. Examination of neurons from shibire mutant flies reveals the accumulation of deeply invaginated pits at the synaptic plasmalemma that appear to be arrested prior to fission (155, 184). In vitro treatment of permeabilized nerve terminals (synaptosomes) with nonhydrolyzable GTP analogs to inhibit dynamin 1 results in accumulation of deeply invaginated clathrin-coated pits blocked prior to fission. Moreover, electron-dense rings that contain dynamin 1 are visible at the necks of these deeply invaginated pits (121, 315).
Discovery of Amphiphysin 1 in Chicken Brain
The function of amphiphysin 1 in the brain has been the topic of an earlier review (368).
Chicken amphiphysin 1 has a domain structure similar to that of budding yeast Rvs167p.
Amphiphysin 1 was originally identified by expression cloning in a large-scale immunoscreen for novel proteins of the chicken synaptic plasma membrane. Amphiphysin 1 is a hydrophilic protein with the only hydrophobic segment at residues 478 to 499. Chicken amphiphysin 1 has a predicted size of 75 kDa, but its electrophoretic mobility suggests a protein of 115 kDa (175). Aberrant gel mobility is not specific to chicken amphiphysin and has been reported also for rat, mouse, and human amphiphysin 1 (47, 50), Bin1+10+13 (278), and Bin1+6a+12+13 (amphiphysin 2) (23, 168, 256, 367) (Fig. 1).
The N terminus of amphiphysin 1 features a short amphipathic α-helix. This is followed by the BAR domain, which, as in the case of Rvs161p and Rvs167p, is also predicted to have a strongly α-helical secondary structure and has a high density of charged residues (Fig. 1) (47, 175, 220). The BAR domain is followed by a highly basic region with a high content of glycine, proline, alanine, serine, and threonine residues, similar to the Rvs167p GPA-rich region (Fig. 1). This region of amphiphysin 1 is predicted to adopt extensive β-turn secondary structure. The C-terminal half of amphiphysin 1 is highly acidic and terminates, as in the case of Rvs167p, with a single SH3 domain (Fig. 1) (12, 175).
Chicken amphiphysin 1 is expressed in neurons.
A single major chicken amphiphysin 1 transcript was shown to be most abundant in chicken brain (both forebrain and cerebellum) and also present in the adrenal gland. Initial studies reported an absence of amphiphysin 1 transcripts in other tissues of chicken, including skeletal muscle, lung, testis, liver, spleen, pancreas, and heart. Amphiphysin 1 protein is highly expressed in the chicken central nervous system, including forebrain, cerebellum, hippocampus, olfactory bulb, and spinal cord. A cross-reacting protein in the rat is expressed in the brain and adrenal gland but also present in the anterior and posterior pituitary (175). A subsequent study showed a rat protein recognized by antiserum to human amphiphysin 1 is also expressed in the testis (256). In the chicken forebrain, amphiphysin 1 is present throughout the cortex and is highly enriched at punctate structures that represent presynaptic terminals. Electron microscopy reveals that amphiphysin 1 immunoreactivity is concentrated around clusters of synaptic vesicles in the presynaptic cytoplasm, but is absent from postsynaptic neurons (175).
Minor pool of chicken amphiphysin 1 is tightly associated with synaptic vesicles.
During subcellular fractionation a small pool of amphiphysin 1 is present in purified chicken synaptic vesicles, but it is also present in purified chicken synaptosomal plasma membrane. The rat brain contains an antigen that cross-reacts with both chicken and human amphiphysin 1 antisera (see below). In subcellular fractionation of the rat brain, amphiphysin 1 distributes partly in the soluble fraction and partly in the sedimentable fraction. Highly purified synaptic vesicles from rat and chicken also contain amphiphysin 1, but it is only a minor component. The cross-reacting rat protein copurifies with rat synaptophysin-positive synaptic vesicles and, although peripheral, behaves almost like an integral membrane component. As for the integral membrane protein synaptophysin, neither physiological salt (0.15 M KCl), high salt (1 M KCl), nor pH 3 buffer was able to efficiently strip the rat amphiphysin 1-cross-reacting protein from the synaptic vesicles, however, both proteins were solubilized from synaptic vesicles by detergent (50, 80, 175, 256).
Because only a small pool of the rat amphiphysin 1-cross-reacting protein is associated with synaptic vesicles and there is a large cytoplasmic pool, it is not enriched in synaptic vesicles during purification. Fractionation of crude rat synaptosome lysates reveals that approximately 75% of the amphiphysin 1-cross-reacting protein in the initial lysate remains in a high speed (260,000 × g) supernatant, while only 25% pellets with membranes. When rat synaptosomal membranes are subjected to Triton X-114 extraction to separate hydrophilic peripheral membrane proteins (recovered in the aqueous phase) from hydrophobic integral membrane proteins (recovered in the detergent phase) the rat amphiphysin 1-cross-reacting protein is present exclusively in the aqueous phase. There is no apparent difference in electrophoretic migration of the membrane-associated or soluble form of the rat amphiphysin 1-cross-reacting protein, and the biochemical feature that distinguishes these two forms is unknown. The name amphiphysin was chosen to reflect the existence of both soluble and membrane-associated pools of this protein (175).
Discovery of Human Amphiphysin 1
Human amphiphysin 1 is the autoantigen in stiff-man syndrome with breast cancer.
Subsequently, human amphiphysin 1 was discovered as the 128-kDa autoantigen in breast cancer patients with the rare neurological condition stiff-man syndrome (SMS). SMS is characterized by severe muscle rigidity throughout the body and intermittent spasms and is caused by an autoimmune response against antigens in the central nervous system. How breast cancer leads to an autoimmune response against amphiphysin 1 is not clear, but it has been proposed that elevated expression of amphiphysin 1 or a related protein in breast tumors may trigger an autoimmune reaction against amphiphysin 1.
Cloning of the human amphiphysin 1 gene based on homology to chicken amphiphysin 1 revealed that human amphiphysin 1 has a predicted size of 76 kDa, which is smaller than the apparent size of 128 kDa based on gel electrophoresis. Aberrant electrophoretic behavior was also observed for chicken amphiphysin 1. Human amphiphysin 1 has the same domain structure as chicken amphiphysin 1, with an N-terminal BAR domain, a central alanine- and glutamate-rich domain containing a proline-rich motif and hydrophobic region, and a C-terminal SH3 domain (Fig. 1). The autoimmune sera from SMS patients were predominantly directed against the amphiphysin 1 SH3 domain (47, 50). Amphiphysin 1 does have some tissue-specific splice variants, e.g., amphiphysin Ir, which is retina specific (128), but few compared to Bin1 (Table 2) (see below).
TABLE 2.
Nomenclature of various BAR domain proteins and their splice variantsa
| Type | Nameb (reference) | Other name(s) | Exon(s) missing | Remarks |
|---|---|---|---|---|
| Different amphiphysin 1 isoforms | Amphiphysin 1 (or I) isoform 1 (47) | Longer amphiphysin isoform expressed in brain. | ||
| Amphiphysin 1 (or I) isoform 2 (77) | Shorter amphiphysin isoform expressed in brain. Also detected in breast carcinoma. | |||
| Amphiphysin 1 (or I) (326) | Retina-specific amphiphysin isoform, contains two novel insertions. | |||
| Different BIN1/amphiphysin 2/amphiphyin II/Amph2/BRAMP2/SH3p9/ALP1/ amphiphysin IIm isoforms | BIN1-iso1 (245)1 | Amphiphysin IIa (255); S11/R3-a; BRAMP2 (168) | 10 | Expressed in brain and concentrated in nerve terminals. Localizes to the cytoplasm. |
| BIN1-iso2 (245)1 | Amphiphysin IId (255); S11/R3-b | 10; 12a | Expressed in brain. Localizes to the cytoplasm and nucleus. | |
| BIN1-iso3 (245)1 | Amphiphysin IIc1 (255) | 10; 12 (a, b, c) | Expressed in brain. Localizes to the cytoplasm and nucleus. | |
| BIN1-iso4 (245)2 | 6a; 12 (b, c, d) | Expressed in brain and possibly in muscle. Localizes to the cytoplasm and nucleus. | ||
| BIN1-iso5 (245)3 | Amphiphysin IIb (255) | 6a; 10; 12 (b, c) | Expressed in brain. Localizes to the cytoplasm and nucleus. | |
| BIN1-iso6 (245)3 | AP II2 (338) | 6a; 10; 12 (b, c, d) | Expressed in brain. Localizes to the cytoplasm and nucleus. Aberrantly expressed in melanoma (93). | |
| BIN1-iso7 (245)3 | Amphiphysin IIc2 (255) | 6a; 10; 12 (a, b, c) | Expressed in brain. Localizes to the cytoplasm and nucleus. | |
| BIN1-iso8 (245)2 | Amph 2-7 (369) | 6a; 12 (a, b, c, d) | Expressed in muscle and localizes to the nucleus. | |
| BIN1-iso9 (245)4 | BIN1-10 (356); SH3P9 (306); AP II3 (338) | 6a; 10; 12 (a, b, c, d) | Ubiquitously expressed. | |
| BIN1-iso10 (245)5 | BIN1-10-13 (356); ALP1 (141); amphiphysin IIm (100); Amph 2-6 (369) | 6a; 10; 12 (a, b, c, d); 13 | Ubiquitously expressed, the only Amph II/Bin1 isoform in macrophages. | |
| BIN1-iso11 | Amph 2-1 (369) | 10; 13 | Expressed in brain. | |
| BIN1-iso12 | Amph 2-2 (369) | 10; 12d; 13 | Expressed in brain. | |
| BIN1-iso13 | Amph 2-3 (369) | 10; 12 (a, b, c, d); 13; 14; 15; 16 | Expressed in brain, lacks SH3 domain. | |
| BIN1-iso14 | Amph 2-4 (369) | 6a; 10; 11; 12 (a, b, c, d); 13; 14; 15; 16 | Expressed in brain, lacks SH3 domain. | |
| BIN1-iso15 | Amph 2-5 (369) | 10; 12 (a, b, c, d); 13 | Expressed in brain. | |
| Endophilin family proteins | Endophilin A1 (134) | Endophilin 1 (323); SH3P4 (323); SH3GL2 (99); EEN-B1 (304) | Expressed mainly in brain. | |
| Endophilin A2 (134) | Endophilin 2 (323); SH3P8 (323); SH3GL1 (99); EEN (304) | Ubiquitously transcribed (270), but only translated in brain and testis (269). | ||
| Endophilin A3 (134) | Endophilin 3 (323); SH3P13 (323); SH3GL3 (99); EEN-B2 (304) | Expressed mainly in brain and testis. | ||
| Endophilin B1 (134) | SH3GLB1 (244); Bif-1 (43) | Expressed mainly in heart, placenta, and skeletal muscle, cytoplasmic protein. | ||
| Endophilin B2 (134) | SH3GLB2 (244) | Expressed mainly in heart, placenta, and skeletal muscle, cytoplasmic protein. |
This table adopts the Bin1 exon nomenclature proposed by Wechsler-Reya et al. (356); however, one additional missing exon located between exons 6 and 7 is referred to here as exon 6a. In the recently introduced systematic nomenclature, exon 6a is renamed exon 7 and the following exons have been assigned new numbers (n + 1) with respect to the exon numbers referred to here and in the previous literature reports. The new systematic Bin1 splice isoform nomenclature and some of the exon information shown in this table was obtained from the National Center for Biotechnology Information and the U.S. National Library of Medicine, Bethesda, Md., via the following internet URL: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=274.
Superscript numbers are as follows: 1, Bin1+6a+12+13 in the text; 2, Bin1+10+13 in the text; 3, Bin1−6a+12+13 in the text; 4, Bin1−10−12+13 in the text; 5, Bin1−10−12−13 in the text. Note: Bin1+12+13 in the text refers collectively to Bin1−6a+12+13 and Bin1+6a+12+13; Bin1+12 in the text refers collectively to Bin1−6a+12+13, Bin1+6a+12+13, Bin1−6a+12−13, and Bin1+6a+12−13; Bin1+10 in the text refers collectively to Bin1+10−13 and Bin1+10+13.
Human amphiphysin 1 has a domain structure similar to that of yeast Rvs167p.
Human amphiphysin 1 displays strong sequence similarity to Rvs161p and Rvs167p within the N-terminal amphipathic α-helix and BAR domain (domain A in reference 47) (Fig. 1). Within this region, a stretch of 209 residues in amphiphysin 1 shares 42% amino acid sequence similarity and 25% identity with the corresponding 208 residues in yeast Rvs161p and 39% similarity and 22% identity with the corresponding 223 residues in yeast Rvs167p. Amphiphysin 1 also shares homology with Rvs167p within the C-terminal SH3 domain (domain D in reference 47). Although the central insert domain of amphiphysin 1 (domains B and C in reference 47) is not as conserved in Rvs167p, the Rvs167p GPA-rich region and amphiphysin 1 central insert domain both feature proline-rich motifs and a single hydrophobic stretch (12, 47, 298). The latter are also conserved in chicken amphiphysin 1 (175).
Mammalian amphiphysin 1 is highly expressed in the brain but is also expressed in other tissues.
Like chicken and rat amphiphysin 1, human amphiphysin 1 is highly expressed in the brain, where it localizes to punctate presynaptic termini (46, 175, 256). Human amphiphysin 1 and dynamin 1 colocalize extensively in the brain to presynaptic termini and both also exhibit a diffuse cytoplasmic localization (46). Although highly expressed in the brain, amphiphysin 1 transcripts and antigen are also expressed in certain other tissues (e.g., testis, ovary, pituitary, pancreas, and adrenal gland) (50, 80, 256, 367). Amphiphysin 1 has also been reported to be overexpressed in some breast tumors and breast cancer cell lines (77).
Amphiphysin 1 interacts with the endocytic proteins dynamin 1 and synaptojanin 1.
Consistent with the possibility that amphiphysin 1 functions with dynamin 1 in brain, gel overlay experiments using recombinant fragments of amphiphysin 1 as probes revealed interaction between an SH3 domain-containing fragment of amphiphysin 1 and a brain protein with a size (100 kDa) characteristic of dynamin 1. Affinity purification approaches using the amphiphysin 1 SH3 domain as affinity ligand confirmed that dynamin 1 is a major amphiphysin 1 SH3 binding protein in the brain. Dynamin 1 and amphiphysin 1 also coimmunoprecipitate from brain extracts, suggesting they form complex in vivo (46). Binding of a recombinant glutathione S-transferase (GST) fusion protein containing only the human amphiphysin 1 SH3 domain to endogenous dynamin 1 in rat brain extracts has also been demonstrated (256).
Dynamin 1 is not the only major binding partner for the amphiphysin 1 SH3 domain in the brain. A major 145-kDa protein was also affinity purified from brain extracts using the amphiphysin 1 SH3 domain (46). This protein was subsequently identified as a polyphosphoinositide 5-phosphatase and named synaptojanin 1. Amphiphysin 1 and synaptojanin 1 extensively colocalize in brain sections to presynaptic terminals. Like dynamin 1, synaptojanin 1 is implicated in synaptic vesicle endocytosis via its regulation of phosphoinositide levels (188). Synaptojanin 1 also is a major binding partner of the SH3 domain of another BAR domain protein, endophilin A1 (also called SH3p4 and SH3GL2) (51, 192) (see below).
Intriguingly, the amphiphysin 1 SH3 domain is also able to mediate an intramolecular interaction. This is predicted to involve the SH3 domain binding to highly conserved proline-rich sequences located within the central insert domain. Intramolecular interactions of the amphiphysin 1 SH3 domain may be in competition with intermolecular interactions and regulate amphiphysin 1 SH3 domain interactions with other proteins such as dynamin 1 and synaptojanin 1 (74). The three-dimensional structure of the amphiphysin 1 SH3 domain has not yet been solved, but is predicted to be similar to that of the Bin1 SH3 domain. The Bin1 SH3 domain also interacts with dynamin 1 and synaptojanin 1 (see below) and its three-dimensional structure has been solved (232).
The amphiphysin 1 SH3 domain interacts with a single PSRPNR motif (residues 833 to 838) in the C-terminal proline-rich domain (PRD) of dynamin 1. Deletion of this motif or R835D/R838D/P836A substitutions within this motif abolish association of a recombinant dynamin 1 PRD fragment with both endogenous amphiphysin 1 in brain extracts and purified recombinant amphiphysin 1 in overlay blots. The amphiphysin 1 SH3 domain is critical for interaction with dynamin 1. The double amino acid substitution G684R/P687L within the target binding cleft of the amphiphysin 1 SH3 domain abolishes interaction with both dynamin 1 and synaptojanin 1. The dissociation constant for amphiphysin 1 SH3 domain interaction with the dynamin 1 PRD is approximately 190 nM, which is relatively high affinity for an interaction mediated by an SH3 domain. A similar motif, PXRPXR in the C-terminal PRD of synaptojanin 1, may mediate interaction of synaptojanin 1 with the amphiphysin 1 SH3 domain (46, 105, 302).
Amphiphysin 1 Interacts with Clathrin and the AP-2 Adaptor
Other domains of amphiphysin 1 also interact with proteins implicated in synaptic vesicle endocytosis. The αc (and to a lesser extent αa) subunit of the plasma membrane clathrin assembly complex AP-2 has been shown to associate with amphiphysin 1 (46, 352). Moreover, αc adaptin and amphiphysin 1 colocalize in the brain. However, although the αa and αc subunits of AP-2 bind SH3 domains of other proteins (e.g., the SH2-SH3 adaptor protein Grb2) neither protein binds the amphiphysin 1 SH3 domain. The interaction of amphiphysin 1 with αa and αc is mediated by non-SH3 domains of amphiphysin 1 (see below) (46). Interestingly, amphiphysin 1 (and Bin1, see below) also binds clathrin heavy chain. This interaction is also mediated by sequences in amphiphysin 1 that are distinct from the SH3 domain (see below) (187, 301, 302).
Regulation of Amphiphysin 1 Interactions by Phosphorylation
Amphiphysin 1, like dynamin 1 and synaptojanin 1, is constitutively phosphorylated in resting neurons, but is rapidly dephosphorylated upon nerve terminal depolarization and synaptic vesicle exocytosis. Stimulated dephosphorylation affects only certain residues in amphiphysin 1, because some basal phosphorylation remains even after nerve depolarization. Stimulated dephosphorylation is not dependent on exocytosis of synaptic vesicles because it is not affected by prior treatment of neurons with tetanus toxin to block synaptic vesicle exocytosis. Dephosphorylation is, however, dependent on extracellular Ca2+. This suggests transmembrane Ca2+ ion flux may be required (13, 189, 193, 222, 271, 302).
Stimulated dephosphorylation of amphiphysin 1 in nerve terminals is also blocked by inhibitors of Ca2+/calmodulin-dependent protein phosphatase 2B (calcineurin) (e.g., FK506 and cyclosporine A), but not by inhibitors of protein phosphatase 1 or 2A (e.g., okadeic acid). This suggests a role for calcineurin in stimulated dephosphorylation of amphiphysin 1. Moreover, purified recombinant calcineurin (rendered constitutively active by mutation) dephosphorylates amphiphysin 1 directly in vitro. Amphiphysin 1 can be induced to undergo multiple rounds of phosphorylation and dephosphorylation in synaptosomes in response to a series of induced nerve depolarizations separated by rest periods to restore polarization. This suggests that cycles of constitutive phosphorylation and stimulated dephosphorylation are physiologically important for amphiphysin 1 function in synaptic vesicle recycling in vivo (13).
Subcellular fractionation shows that while the sedimentable membrane-bound pool of amphiphysin 1 contains equal amounts of the fully phosphorylated and partially dephosphorylated forms, the cytoplasmic pool is predominantly dephosphorylated. Interestingly, this does not reflect the localization of the kinase that phosphorylates amphiphysin 1. Addition of ATP and incubation at 37°C resulted in phosphorylation of amphiphysin 1 present in the cytosolic fraction but not amphiphysin 1 present in the membrane fraction. Interestingly, nonhydrolyzable GTP analogs strongly stimulate amphiphysin 1 phosphorylation in the presence of brain cytosol in vitro. Hence, amphiphysin 1 may be phosphorylated by a kinase whose activity is regulated by a GTP-binding protein (13).
Amphiphysin 1 Subcellular Localization in Presynaptic Terminals
Amphiphysin 1 localizes to endocytic intermediates.
There is a large cytoplasmic pool of amphiphysin 1 in rat brain (13, 50, 80, 175, 256). Permeabilization of rat brain synaptosomes followed by treatment with a nonhydrolyzable GTP analog is known to induce the formation of deeply invaginated clathrin-coated pits. Immunoelectron microscopy on synaptosomes treated in this way shows that amphiphysin 1 localizes to the accumulated clathrin-coated buds and also partially colocalizes with dynamin 1 at the necks of deeply invaginated clathrin-coated pits. Amphiphysin 1 also localized to the less abundant clathrin-coated pits observed in untreated synaptosomes, showing that localization of amphiphysin 1 to endocytic intermediates is physiologically relevant and not simply an artifact induced by nonhydrolyzable GTP analogs. Amphiphysin 1 is not present on all clathrin-coated pits and dynamin 1-coated tubules, however, and its distribution on those clathrin-coated pits where it is found is irregular and patchy. This suggests that, unlike clathrin or AP-2, amphiphysin 1 is not a major coat component (13).
Amphiphysin 1 also localizes to the actin-rich cytomatrix.
Amphiphysin 1 is also found in the actin-rich cortical cytomatrix, which is distinct from endocytic intermediates but surrounds clusters of synaptic vesicles (13). This localization suggests amphiphysin 1 may have a role in the cortical actin cytoskeleton, perhaps analogous to the role of Rvs167p in cortical actin patches in yeast (11). It is interesting that in Drosophila melanogaster, amphiphysin is expressed in a range of polarized cells (including epithelial cells), where it localizes to the actin-rich apical plasma membrane domain (386). This suggests a conserved role for amphiphysin family proteins in organization of the cortical actin cytoskeleton.
Amphiphysin 1 functions in polarized growth.
The strongest evidence that amphiphysin 1 plays a role in organization of the cortical actin cytoskeleton and cell polarity comes from a study looking at neurite outgrowth in cultured hippocampal neurons. Hippocampal neurons plated at low cell density in culture form axonal and dendritic processes that exhibit dramatic polarized growth. As the cells approach confluence the axonal processes eventually contact other cell bodies and form synapses in vitro. When hippocampal neurons are initially plated, the amphiphysin 1 expression level is detectable but low, however, a dramatic increase in amphiphysin 1 expression accompanies neurite outgrowth and synapse formation. Moreover, amphiphysin 1 and dynamin 1 become highly concentrated at the tips of actively growing axons and dendrites and at high magnification can be seen to localize to numerous patches containing F-actin distributed throughout the cortex of the growth cone. As growth cones come into contact with other neurons in culture and mature into synapses, amphiphysin 1 and dynamin 1 concentrate at the synaptic termini, analogous to amphiphysin 1 and dynamin 1 localization in mature brain (212).
Neurons derived from temperature-sensitive Drosophila dynamin mutants (shibire mutants) and cultured in vitro fail to form neurites upon a shift to the restrictive temperature (184). Also, down-regulation of dynamin 1 in cultured hippocampal neurons prevents neurite outgrowth and causes growth cone collapse (333). Does amphiphysin 1 also play a role in neurite outgrowth and synaptogenesis? Consistent with a role for amphiphysin 1 in neurite outgrowth, down-regulation of amphiphysin 1 expression using antisense oligonucleotides leads to a severe block in neurite outgrowth such that many cells fail to form neurites and the cells that do form neurites exhibit only a few short neurites. Hence, loss of amphiphysin 1 causes collapse of growth cones on both dendrites and axons. Interestingly, this effect is reversible, as removal of the antisense oligonucleotides leads to reestablishment of growth cones and recommencement of neurite outgrowth (212).
The defect in neurite outgrowth caused by amphiphysin 1 down-regulation is not due to effects on endocytosis, because the level of down-regulation achieved was insufficient to block receptor-mediated endocytosis. The cortical actin cytoskeleton is grossly perturbed by amphiphysin 1 down-regulation, however, with loss of F-actin polarization to growing neurite tips. In contrast, there is no apparent effect of this treatment on the microtubule cytoskeleton. It has been proposed that disorganization of the actin cytoskeleton may be responsible for the block in neurite outgrowth (212). This loss of cell polarity in neurons is perhaps analogous to what is seen in yeast cells lacking Rvs161p or Rvs167p (12, 215, 298). No interaction of amphiphysin 1 with G-actin or F-actin has been observed, however, despite their colocalization in patches. In contrast, yeast Rvs167p not only colocalizes with actin in patches, but also interacts genetically and in two-hybrid screens with actin (4, 11, 180, 212, 215).
Amphiphysin 1 Functions in Endocytosis
Amphiphysin 1 interaction with dynamin 1 is essential for endocytosis in neurons.
The essential role of Rvs proteins in endocytosis in yeast together with the observed interactions of amphiphysin 1 with dynamin 1, clathrin, and AP-2 in neurons suggested a possible role for amphiphysin 1 in synaptic vesicle endocytosis. The first evidence for a role of amphiphysin 1 in endocytosis came from experiments in which a recombinant fusion protein containing the amphiphysin 1 SH3 domain was microinjected into the giant reticulospinal synapse of the lamprey eel. Injection of the SH3 domain had no effect on the number of synaptic vesicles or plasmalemmal clathrin-coated pits in resting neurons. However, electrical excitation of the injected neurons to induce synaptic vesicle exocytosis resulted in persistent depletion of synaptic vesicle numbers and the dramatic accumulation of deeply invaginated clathrin-coated pits at the plasmalemma (Fig. 10). Injection of a synthetic peptide corresponding to the motif in the dynamin 1 PRD that binds the amphiphysin 1 SH3 domain had similar effects, while a mutant amphiphysin 1 SH3 domain unable to bind dynamin 1 and an unrelated SH3 domain were without effect (293). The results obtained from such domain microinjection experiments should be taken as suggestive of a cellular role rather than proof, however, as the injected domain can have off-target effects, especially at these concentrations.
FIG. 10.
Amphiphysin 1 SH3 domain interactions are required for fission of endocytic vesicles. Lamprey giant reticulospinal synapses were injected with a fusion protein comprising glutathione S-transferase and the human amphiphysin 1 SH3 domain (GST-amphSH3) and effects on synaptic vesicle recycling from the plasmalemma (plasma membrane) to regenerate the cytoplasmic vesicle pool were examined. Panel A, electron micrograph of a synapse in an axon injected with GST-amphSH3 and then maintained in low Ca2+ (0.1 mM Ca2+, 4 mM Mg2+), without stimulation. Panel B, electron micrograph of a synapse in an axon injected with GST-amphSH3 and then electrically stimulated at 0.2 Hz for 30 min in high Ca2+ (2.6 mM Ca2+ and 1.8 mM Mg2+) to induce exocytosis of synaptic vesicles. Panel C, electron micrograph of a synapse in an axon treated as in panel B showing the accumulation of invaginated endocytic pits at the plasmalemma. Panel D, electron micrograph of a synapse in an axon injected with GST-amphSH3 and then electrically stimulated at 5 Hz for 30 min in high Ca2+ (2.6 mM Ca2+ and 1.8 mM Mg2+) to induce exocytosis of synaptic vesicles. Bars, 0.2 μm. The bar in panel A applies to panels A to C. (Reprinted from reference 293 with permission of the publisher. Copyright 1997 American Association for the Advancement of Science.)
Amphiphysin 1 does not seem to play an essential role in exocytosis in neurons. Injection of the amphiphysin 1 SH3 domain did not affect synaptic vesicle exocytosis and release of neurotransmitters at low levels of nerve terminal stimulation. However, at higher levels of stimulation, where the ability to rapidly recycle synaptic membranes becomes more important, neurotransmitter release was affected (293). These data are consistent with the view that the amphiphysin 1 SH3 domain-mediated interaction with a proline-rich motif in dynamin 1 is essential for endocytosis but not for exocytosis of synaptic vesicles. This is consistent with the finding that Rvs161p and Rvs167p are not essential for exocytosis in yeast but loss of these proteins does nevertheless perturb exocytosis, leading to vesicle accumulation (20, 298).
As dynamin 1 performs the scission event at the necks of invaginated clathrin-coated pits, it is possible that amphiphysin 1-dynamin 1 interaction is important for dynamin 1 recruitment to the necks of clathrin-coated pits. Consistent with this, the necks of the deeply invaginated clathrin-coated pits that accumulate in neurons injected with the amphiphysin 1 SH3 domain appear to lack the electron-dense rings characteristic of dynamin 1 (121, 293). What was not clear was if this role of amphiphysin 1 in recruitment of dynamin 1 is specific to neurons. A potential SH3 domain binding motif in the dynamin 1 PRD was shown to be essential for localization to clathrin-coated pits when dynamin 1 was expressed ectopically in COS-7 cells. However, this motif is distinct from the PSRPNR motif that has been shown to interact specifically with the amphiphysin 1 SH3 domain and likely mediates interaction with another SH3 domain protein (292).
Amphiphysin interaction with dynamin is also essential for endocytosis in nonneuronal cells.
Amphiphysin 1 may have a more general function in recruiting dynamin family GTPases to clathrin-coated pits during endocytosis in nonneuronal cells. In support of this is the known role of Rvs proteins in endocytosis in yeast. Further support came from the finding that a 125-kDa isoform of amphiphysin is also expressed at lower but still significant levels in nonneuronal cells, including muscle, testis, lung, and fibroblast. Moreover, the amphiphysin 1 SH3 domain interacts with a nonneuronal isoform of dynamin expressed endogenously in the fibroblast cell line COS-7 (369).
To investigate whether amphiphysin 1 functions in endocytosis in nonneuronal cells, the amphiphysin 1 SH3 domain was transiently expressed in COS-7 cells and the effect on receptor-mediated endocytosis was examined. Expression of the amphiphysin 1 SH3 domain abrogated endocytic uptake of both transferrin and epidermal growth factor in COS-7 fibroblasts, but had no effect on fluid-phase uptake of fluorescein isothiocyanate-labeled dextran. Overexpression of other SH3 domains that bind to dynamin, but via motifs distinct from the amphiphysin 1 SH3, did not inhibit transferrin endocytosis. The effect of the amphiphysin 1 SH3 domain on endocytosis is due to perturbation of dynamin, since transient overexpression of dynamin neutralized the effect of the amphiphysin 1 SH3 domain and restored endocytosis (369). Similarly, microinjection of the amphiphysin 1 SH3 domain into cultured 3T3-L1 adipocytes inhibited receptor-mediated endocytosis of transferrin and caused accumulation of the Glut4 glucose transporter at the plasma membrane (349). These results are suggestive of a role for amphiphysin 1 in endocytosis, but the overexpressed amphiphysin 1 SH3 domain may engage in off-target interactions leading to nonspecific effects.
As discussed above, there appears to be a role for amphiphyin 1 in regulation of the cortical actin cytoskeleton during polarized growth. Intriguing, links between dynamin and the actin cytoskeleton are also emerging. Dynamin 2 in nonneuronal cells modulates the activation of the Arp2/3 actin filament nucleation complex and thereby de novo actin filament assembly at the cortex (151, 157, 280, 281). Mechanical force generated by de novo actin filament assembly may be important for dynamin-mediated scission of the neck of clathrin-coated vesicles during endocytosis as well as in cortical actin cytoskeletal dynamics during polarized growth.
Amphiphysin 1 interacts with clathrin and AP-2.
Amphiphysin 1 also interacts with other components of clathrin coats. Amphiphysin 1 binds directly to clathrin heavy chain (187). Furthermore, amphiphysin 1 binds the α-adaptin subunit of the AP-2 adaptor in vitro (352). Subsequently, it was found that the appendage domain of α-adaptin binds a non-SH3 domain of amphiphysin 1 (46). Binding of a recombinant central insert domain of human amphiphysin 1 to endogenous clathrin heavy chain and α-adaptin (αa and αc) in rat brain extract was subsequently confirmed (256).
Does amphiphysin 1 act as an adaptor to link dynamin to clathrin-coated pit components such as clathrin heavy chain and AP-2? Although an early report suggested that dynamin 1 binds directly to the ear domain of α-adaptin (352), subsequent studies found that the direct dynamin 1-α-adaptin interaction is very weak. When amphiphysin 1 and dynamin 1 are both overexpressed in COS-7 cells α-adaptin forms complexes containing both proteins but the α-adaptin appendage domain binds poorly to dynamin 1 compared to amphiphysin 1. The ability of amphiphysin 1 to form heterotrimeric complexes with α-adaptin and dynamin 1 is consistent with the possibility that amphiphysin 1 associates with clathrin-coated pits via interaction with α-adaptin and then recruits dynamin 1 to these pits via its SH3 domain, a process that is likely to be perturbed by overexpression of the isolated amphiphysin 1 SH3 domain. The effects of amphiphysin 1 SH3 domain overexpression on dynamin subcellular localization were not investigated directly, however, subcellular fractionation revealed that dynamin levels in the membrane fraction are reduced by overexpression of the amphiphysin 1 SH3 domain (367, 369).
The clathrin heavy chain and α-adaptin binding sites on amphiphysin 1 have been defined. Residues 262 to 405 of human amphiphysin 1 mediate binding of both clathrin heavy chain and α-adaptin. The sites that interact with clathrin heavy chain and α-adaptin are distinct, but adjacent. Minimal α-adaptin binding requires residues 322 to 340, however, residues 340 to 363 are required for full-strength interaction. The latter overlap with the clathrin heavy-chain binding site and contain a DPF motif found in multiple copies in other α-adaptin binding proteins. Mutation of residues 323 to 326 (FFED) abolishes α-adaptin binding without affecting clathrin heavy-chain binding. Binding to clathrin heavy chain is conferred by residues 347 to 386 (256, 301, 302). The three-dimensional structure of the α-adaptin appendage domain has been determined and a model has been proposed to account for its remarkable ability to mediate binding not only to amphiphysin 1 but also to several other proteins in clathrin coats via a single interaction interface (231, 334).
Mutational analysis identified two motifs that are important for binding to clathrin heavy chain, LLDLD and WDLW. LLDLD fits the consensus for clathrin heavy chain binding, L(L/I)(D/E/N)(L/F)(D/E), proposed based on analysis of clathrin binding sites in other clathrin coat components such as the β-adaptin subunit of AP-2 and AP-180. The affinity of the recombinant clathrin-binding domain of amphiphysin 1 for native clathrin triskelia was estimated at 1 nM (255). Mutation of either LLDLD or WDLW motifs in amphiphysin 1 weakens clathrin heavy-chain binding and the double mutation abolishes clathrin heavy-chain binding, but has no effect on α-adaptin binding. Combination of the mutations in α-adaptin and clathrin heavy-chain binding sites, as expected, abolish amphiphysin 1 interaction with both α-adaptin and clathrin heavy chain.
Amphiphysin 1 binds an N-terminal fragment of clathrin heavy chain (residues 1 to 579 in rat clathrin heavy chain) in vitro and this fragment contains the β-propeller “foot” or “terminal” domain of clathrin heavy chain. The foot domain is also the site in clathrin heavy chain that binds a range of other clathrin-associated proteins, including the β-adaptin subunit of AP-2, the adaptor AP-180, epsin, and β-arrestin. These proteins all have motifs similar to the LLDLD and WDLW motifs in amphiphysin 1 that mediate interaction with the foot domain of clathrin heavy chain. In vitro, amphiphysin 1 competes with AP-2 and AP-180 for binding to clathrin heavy chain, suggesting all three proteins recognize the same site within the foot domain of clathrin heavy chain. As AP-2 plays a key role in clathrin assembly into plasma membrane clathrin-coated pits, competition with AP-2 for binding clathrin heavy chain may explain the loss of clathrin from plasma membrane-coated pits observed upon overexpression of clathrin-binding fragments of amphiphysin 1 (57, 58, 187, 194, 255, 256, 301).
Appendage domain of α-adaptin mediates interaction with amphiphysin 1.
Amphiphysin 1 is required for efficient association of dynamin 1 in brain lysates with α-adaptin appendage domain or clathrin heavy chain in vitro. Similarly, the addition of recombinant amphiphysin 1 to extracts increases the association of both clathrin heavy chain and AP-2 with the dynamin 1 PRD in vitro. Hence, amphiphysin 1 can bind clathrin heavy chain, AP-2, and dynamin 1 simultaneously and thus has the potential to recruit dynamin 1 to plasma membrane clathrin-coated pits (46, 231, 256, 301, 302, 334, 352).
Amphiphysin 1 interaction with clathrin and AP-2 is essential for endocytosis.
Transient overexpression of amphiphysin 1 fragments that contain both the clathrin heavy chain- and AP-2-binding domains in CHO cells potently blocks receptor-mediated endocytosis of transferrin. Expression of these amphiphysin 1 fragments causes loss of clathrin heavy chain from plasma membrane-coated pits and results in a diffuse cytoplasmic clathrin heavy-chain distribution. It also causes relocalization of AP-2 adaptors from clathrin-coated pits into larger plasma membrane-associated aggregates. Transient overexpression in CHO cells of a mutated amphiphysin 1 fragment specifically unable to bind AP-2 has similar effects on clathrin heavy chain and AP-2 subcellular distribution and also blocks transferrin endocytosis. In contrast, overexpression of a mutated amphiphysin 1 fragment specifically unable to bind clathrin heavy chain does not cause redistribution of clathrin heavy chain but still induces aggregation of AP-2 adaptors and blocks receptor-mediated endocytosis of transferrin. Hence, interaction of amphiphysin 1 with both clathrin heavy chain and AP-2 may be important for clathrin-dependent endocytosis (301, 302). Again, however, these results from overexpression experiments should be interpreted with caution due to the possibility of off-target interactions and nonspecific effects.
Amphiphysin 1 interacts with endophilin A1/SH3p4/SH3GL2.
As well as clathrin heavy chain and AP-2, the amphiphysin 1 central insert domain also binds the other synapse-enriched BAR domain protein endophilin A1 (also called SH3p4 and SH3GL2) (Fig. 1). Recombinant endophilin A1 binds endogenous amphiphysin 1 in rat brain extracts via its SH3 domain (the BAR domain does not interact). The recombinant central insert domain of amphiphysin 1 binds endogenous endophilin A1 in brain extracts and recombinant endophilin A1. These results show that the endophilin A1-amphiphysin 1 interaction is direct and mediated by the endophilin A1 SH3 domain and the amphiphysin 1 central insert domain. A similar interaction has been reported between endophilin A1 and a neuronal splice variant of Bin1 (Bin1+6a+12+13) (see below). The central insert domains of both amphiphysin 1 and Bin1+6a+12+13 include a proline-rich SH3 domain binding consensus motif that may mediate the interaction (193, 255).
Amphiphysin 1 forms homodimers.
The recombinant amphiphysin 1 BAR domain associates in overlay assays with full-length endogenous amphiphysin 1 in brain extract, suggesting that amphiphysin 1 forms homodimers (302). Amphiphysin 1 also forms heterodimers with Bin1+6a+12+13 in the brain (see below).
Regulation of amphiphysin 1 complex formation by phosphorylation and dephosphorylation.
Interestingly, amphiphysin 1, dynamin 1, and synaptojanin 1 are phosphorylated in resting synapses but are rapidly dephosphorylated upon synapse stimulation and exocytosis of synaptic vesicle neurotransmitters (they are referred to collectively as dephosphins) (13, 78, 193, 271, 272, 302, 330). Are associations of amphiphysin 1 with clathrin heavy chain, AP-2, dynamin 1, and synaptojanin 1 regulated during the cycle of synaptic vesicle exocytosis and endocytosis at nerve terminals? Dynamin 1, synaptojanin 1, clathrin heavy chain, and AP-2 coimmunoprecipitate with amphiphysin 1 from brain extracts. However, when brain extract is preincubated with ATP and protein phosphatase inhibitors to induce phosphorylation, amphiphysin 1, dynamin 1, and synaptojanin 1 undergo a shift to slower electrophoretic mobilities consistent with phosphorylation. Under these conditions dynamin 1, synaptojanin 1, clathrin heavy chain, and AP-2 no longer coimmunoprecipitate with amphiphysin 1. The presence of a protein kinase inhibitor (K252a) prevents both mobility shift and loss of coimmunoprecipitation with amphiphysin 1, consistent with regulation by phosphorylation (302).
Is phosphorylation of amphiphysin 1 or its binding partners responsible for loss of complex formation? To address this, studies were performed in which brain extracts were preincubated with ATP and protein phosphatase inhibitors to induce phosphorylation and then tested for binding to recombinant amphiphysin 1 SH3 domain. This treatment reduced binding of both dynamin 1 and synaptojanin 1 to the amphiphysin 1 SH3 domain in vitro. This shows that it is phosphorylation of dynamin 1 and synaptojanin 1 (not amphiphysin 1) that regulates these protein interactions. In contrast, this treatment had little effect on the binding of clathrin heavy chain or AP-2 to a recombinant fragment of amphiphysin 1 comprising the clathrin heavy chain and AP-2 binding domains in vitro. This suggests that phosphorylation of clathrin heavy chain and AP-2 may not regulate association with amphiphysin 1. However, the treatment did reduce amphiphysin 1 binding to a recombinant fragment comprising the appendage domain of α-adaptin and the unphosphorylated form of amphiphysin 1 bound more efficiently than the phosphorylated form (302). Hence, phosphorylation of amphiphysin 1 regulates association with α-adaptin.
Protein phosphatase 2B/calcineurin is responsible for dephosphorylation of amphiphysin 1, dynamin 1, and synaptojanin 1 upon nerve stimulation.
The protein phosphatase responsible for stimulation-dependent dephosphorylation of amphiphysin 1, dynamin 1, and synaptojanin 1 in synapses is the Ca2+-dependent protein phosphatase calcineurin (13, 189, 271, 302, 367). The addition of phosphatase inhibitors is necessary to observe a shift in electrophoretic mobility of dynamin 1 and synaptojanin 1 upon treatment of brain extract with ATP. The specific calcineurin inhibitor cyclosporine A alone is sufficient to observe this shift in electrophoretic mobility upon ATP treatment. However, addition of okadaic acid and vanadate as well as cyclosporine A increases the shift in electrophoretic mobility compared to cyclosporine A alone. Protein phosphatases other than calcineurin are therefore also likely to play a role in dephosphorylation of endocytic proteins in synapses (302).
Proline-rich central insert domain of amphiphysin 1 is phosphorylated by Cdk5/p35 in vivo.
Protein kinase C and cyclin-dependent kinase 5 (Cdk5) have both been reported to mediate constitutive phosphorylation of dynamin 1 and synaptojanin 1 in resting synapses (40, 165, 271, 319, 330). Different results have been obtained with respect to phosphorylation of amphiphysin 1 by PKC. One study found that, unlike dynamin 1 and synaptojanin 1, an inhibitor of PKC (Ro 31-8220) did not block amphiphysin 1 rephosphorylation after stimulation-induced dephosphorylation in brain synaptosomes (40). A subsequent study found that Ro 31-8220 does block phosphorylation of amphiphysin 1 in brain synaptosomes and that activators of PKC such as phorbol 12-myristate 13-acetate (PMA) stimulate phosphorylation of amphiphysin 1 (367). However, PMA nonspecifically activates many kinases. Hence, despite evidence for PKC phosphorylation of amphiphysin 1 in vitro, there is no consensus on whether PKC phosphorylates amphiphysin 1 in vivo.
More recently, attention has focused on the potential role of Cdk5 in regulation of amphiphysin 1. Amphiphysin 1 binds the Cdk5 cyclin p35 in vitro and a recombinant fragment comprising residues 1 to 306 of amphiphysin 1 is sufficient. This shows that binding is mediated by either the N-terminal BAR domain or the central proline-rich insert domain, but not by the C-terminal SH3 domain. This may also be analogous to the situation in yeast, where the central GPA-rich domain of Rvs167p mediates Pcl2p binding (163). The amphiphysin 1-p35 interaction is likely to occur in vivo, as p35 coimmunoprecipitates with amphiphysin 1 (but not Cdk5) from rat brain extract (78).
Interestingly, p35 and the related Cdk5 cyclin p39 are expressed only in neurons and developing muscle cells (171, 242, 320, 337). Immunostaining of rat cortical neurons in culture shows that amphiphysin 1 and p35 colocalize in growth cones. Consistent with a role for Cdk5 and p35 in regulation of amphiphysin 1 in vivo, both Cdk5 and p35 have been implicated in neurite outgrowth, synapse formation, and neuronal migration such as amphiphysin 1. Amphiphysin 1 can be phosphorylated by immunoprecipitates of the Cdk5/p35 complex from rat brain or by recombinant Cdk5/p35 in vitro. Amphiphysin 1 Cdk5/p35 in vitro phosphorylation sites have been mapped to S261, S272, S276, S285, and T310 (78, 272, 330). These Cdk5/p35 phosphorylation sites are all within the central proline-rich domain of amphiphysin 1.
Two approaches have been used to determine whether Cdk5/p35 phosphorylates amphiphysin 1 in vivo. One study compared the phosphorylation status of amphiphysin 1 in resting synaptosomes from wild-type and p35-deficient mice with and without treatment with the calcineurin inhibitor FK506. In this study FK506 induced a very slight decrease in electrophoretic mobility, corresponding to phosphorylation in wild-type mouse synaptosomes. This slight mobility shift was not induced by FK506 in p35-deficient mice synaptosomes (330). This suggests that Cdk5/p35 contributes to amphiphysin 1 phosphorylation in presynaptic terminals, although other kinases may be required for full phosphorylation.
The second study examined rephosphorylation of amphiphysin 1 following dephosphorylation induced by nerve terminal stimulation. Purified synaptosomes were radiolabeled with 32P and then stimulated with K+ to induce dephosphorylation of amphiphysin 1. Then the labeled synaptosomes were washed to remove K+ and allow rephosphorylation. Finally, the synaptosomes were stimulated again with K+. Complexes containing amphiphysin 1 and its associated proteins were isolated from the labeled synaptosomes and analyzed for phosphorylation status after each treatment. Amphiphysin 1 was rephosphorylated following the removal of K+ and this was unaffected by the Cdk5-specific inhibitor roscovitine. In contrast, rephosphorylation of dynamin 1 and synaptojanin 1 was blocked by roscovitine. This suggests that rephosphorylation of dynamin 1 and synaptojanin 1 is dependent on Cdk5/p35, but rephosphorylation of amphiphysin 1 is not (319). Hence, if Cdk5 does play a role in amphiphysin 1 phosphorylation in synaptosomes, this role is redundant with other kinases.
This is reminiscent of yeast Rvs167p, which binds Pcl2p, a Pho85p cyclin, and is phosphorylated by Pho85p kinase in vitro. Yeast Pho85p and Pcl2p are functional orthologs of mammalian Cdk5 and p35/p39, respectively, as shown by the ability of mammalian Cdk5/p35 to functionally replace Pho85p/Pcl2p in yeast (129, 225). The central GPA-rich domain of yeast Rvs167p, which contains the major in vivo phosphorylation sites for Pho85p/Pcl2p, is equivalent to the central proline-rich domain of amphiphysin 1 that is phosphorylated by Cdk5/p35 in vitro. As in the case of amphiphysin 1, Rvs167p phosphorylation in vivo is only partially dependent on Pho85p/Pcl2p. Deletion of all five Pcl2p-like cyclins somewhat reduces, but does not abolish, Rvs167p phosphorylation in vivo (85, 163). This suggests that in yeast as well as in mammalian cells other kinases act redundantly with Cdk5 family kinases to phosphorylate amphiphysin family proteins.
Does Cdk5-dependent phosphorylation of amphiphysin 1 affect interaction with its partner proteins dynamin 1, clathrin heavy chain, and AP-2? In one study in vitro binding assays showed that phosphorylation of amphiphysin 1 in vitro by Cdk5/p35 had no effect on binding to dynamin 1, however, it inhibited binding to AP-2. In the same study phosphorylation of dynamin 1 in vitro by Cdk5 inhibited its interaction with the isolated SH3 domain of amphiphysin 1 (330). In contrast, another report showed that Cdk5/p25 (p25 is a truncated form of p35) phosphorylation of dynamin 1 in vitro does not affect dynamin 1 binding to full-length amphiphysin 1 (319). This study did find, however, that preincubation of dynamin 1 with the amphiphysin 1 SH3 domain blocked subsequent phosphorylation of dynamin 1 by Cdk5. This suggests that phosphorylation of dynamin 1, instead of regulating binding to amphiphysin 1, may in fact be regulated by binding to amphiphysin 1. Maybe phosphorylation of dynamin 1 in vivo only occurs at the termination of synaptic vesicle endocytosis after dynamin 1 and amphiphysin 1 dissociate.
These studies showing effects of phosphorylation on the interactions of dynamin 1 and amphiphysin 1 have made use of proteins that are phosphorylated in vitro by Cdk5. As it is not yet clear to what extent Cdk5 phosphorylates amphiphysin 1 in vivo and as other kinases that phosphorylate amphiphysin 1 in vivo may exist, the physiological relevance of these findings remains uncertain. In some cases effects on binding were investigated using protein fragments that represent individual domains (e.g., SH3 domain). In these cases phosphorylation may affect the interactions of these domains but not necessarily affect the interactions of the full-length proteins (which may interact via multiple domains). Whether interaction per se is the only aspect that is important (e.g., for holding proteins together in a complex) or whether the precise way in which the proteins interact is also important (e.g., in terms of conformation and activity of the complex) is an interesting area that has barely been explored. There are examples in which interactions involving SH3 domains appear to be transient and more important for regulation than for formation of stable multiprotein complexes (260).
As mentioned above, dephosphorylation of amphiphysin 1 and its associated proteins following nerve terminal depolarization induces synaptic vesicle recycling via endocytosis (40, 183). To determine if Cdk5/p35-dependent phosphorylation is also important for synaptic vesicle recycling, the effect of Cdk5-specific kinase inhibitors was examined. In the study that found some Cdk5-dependent phosphorylation of amphiphysin 1 (see above), Cdk5 inhibitors were also found to increase the number of synaptic vesicles formed during that round of synaptic vesicle endocytosis in cultured hippocampal neurons (330). This suggests that dephosphorylation of amphiphysin 1 favors synaptic vesicle endocytosis. In contrast, the study that found Cdk5-dependent phosphorylation of dynamin 1 but not amphiphysin 1 also found that addition of Cdk5 inhibitors after nerve depolarization (to prevent rephosphorylation) actually inhibited the subsequent rounds of synaptic vesicle endocytosis (319). This study concluded that rephosphorylation of endocytic proteins (although not amphiphysin 1) by Cdk5/p35 is required for the maintenance of the synaptic vesicle pool. The difference between the results of the two studies may be due to one study's observing acute and the other study's observing long-term effects of Cdk5/p35 inhibition.
When amphiphysin 1 is transiently expressed in CHO cells that lack p35 or p39 amphiphysin 1 is still phosphorylated, indicating the existence of other kinases that can potentially phosphorylate amphiphysin 1 in vivo. Cell cycle synchronization reveals that amphiphysin 1 in CHO cells specifically undergoes phosphorylation during mitosis. Moreover, amphiphysin 1 can be phosphorylated in vitro by the major cell cycle regulatory Cdk Cdc2/cyclin B. The Cdc2/cyclin B kinase sites mapped to date correspond to the known Cdk5/p35 phosphorylation sites, S272, S276, and S285 (78). While amphiphysin 1 is predominantly expressed in neurons, it is also expressed at lower levels in some other cell types (see above). In these nonneuronal cells phosphorylation by Cdc2/cyclin B may have a physiological role.
Amphiphysin 1 Knockout Mice
The initial evidence for a role of amphiphysin 1 in synaptic vesicle recycling and endocytosis came from cell-based assays employing overexpression of dominant negative constructs. Is amphiphysin 1 essential for synaptic vesicle recycling and endocytosis in the context of a whole animal? The amphiphysin 1 (AMPH1) gene has been knocked out in mice and the consequence of amphiphysin 1 deficiency on endocytosis at the synapse examined. Mice homozygous for the amphiphysin 1 knockout are devoid of amphiphysin 1 protein but do not exhibit developmental or anatomical abnormalities, are physically robust, and reproduce to yield viable progeny. However, the amphiphysin 1-deficient mice are more susceptible to seizures that resemble epilepsy upon reaching adulthood than wild-type mice. The amphiphysin 1 knockout mice also exhibit severe learning deficiencies. Hence, amphiphysin 1 is important for several aspects of brain function (55).
Various endocytic proteins associate in vitro with liposomes prepared from purified brain lipids. These associations occur via interaction of endocytic proteins with amphiphysin 1, which directly binds to membranes. When brain extracts from amphiphysin 1-deficient mice and wild-type mice were compared, all endocytic proteins tested except amphiphysin 1 and Bin1+6a+12+13 (see below) were present at similar levels, showing that endocytic proteins are stable in the absence of amphiphysin 1. However, the ability of clathrin heavy chain, AP-2, and synaptojanin 1 present in brain extracts from amphiphysin 1-deficient mice to associate in vitro with brain lipid liposomes was severely reduced. In contrast, association of dynamin 1 with brain lipid liposomes was unaffected by loss of amphiphysin 1, presumably because dynamin 1 can bind membranes directly via its pleckstrin homology domain, which is known to bind phosphoinositides (55).
Is recruitment of dynamin 1 to clathrin coats affected by loss of amphiphysin 1? The recombinant amphiphysin 1-binding C-terminal PRD of dynamin 1 forms complexes in vitro with amphiphysin 1 and also with clathrin heavy chain and AP-2 in wild-type brain extracts. When amphiphysin 1 knockout brain extracts were analyzed, the absence of amphiphysin 1 resulted in reduced association of clathrin heavy chain and AP-2 with the dynamin 1 PRD in vitro. However, binding of clathrin heavy chain and AP-2 to the dynamin 1 PRD was restored when the brain extract was supplemented with physiological amounts of recombinant amphiphysin 1 (55). Hence, although dynamin 1 binds liposomes directly in the absence of amphiphysin 1, in vivo amphiphysin 1 is important for linking dynamin 1 to components of clathrin coats on membranes.
Amphiphysin 1 is important but not essential for synaptic vesicle recycling in vivo.
Are there defects in synaptic vesicle exocytosis or endocytic recycling in the synapses of amphiphysin 1 knockout mice? Electron micrographs showed that the synapses of amphiphysin 1 knockout mice, either in situ or after culturing in vitro, exhibit normal domain organization and contain the same number of synaptic vesicles as wild-type synapses. Significantly, there was no significant accumulation of coated invaginations at the plasmalemma in amphiphysin 1 knockout synapses. This result was surprising since it contrasted with the situation in neurons that had been microinjected with the recombinant amphiphysin 1 SH3 domain (293). It also contrasted with what had been observed in neurons of dynamin mutant (shibire) flies (155). Hence, thorough examination did not reveal any apparent defect in resting nerve terminals of amphiphysin 1 knockout mice (55).
Could subtler defects in synaptic vesicle exocytosis or endocytic recycling be revealed under conditions of nerve stimulation? Primary synaptosomes were isolated from the cerebral cortex of wild-type and amphiphysin 1 knockout mice and tested for high-K+-stimulated release of the neurotransmitter glutamate. The kinetics of K+-stimulated exocytosis and the quantity of glutamate release are both unaffected in amphiphysin 1 knockout compared to wild-type synaptosomes. Quantitative assays of high-K+-stimulation-dependent endocytic internalization of fluorescent lipophilic dyes were performed and here it was observed that following stimulation, endocytosis was 40% less efficient in amphiphysin 1 knockout compared to wild-type synaptosomes. Moreover, the dye that was internalized after a brief high-K+ stimulation was less efficiently delivered back to the plasmalemma after a second high-K+ stimulation in amphiphysin 1 knockout compared to wild-type synaptosomes (55). This indicates that the efficiency of both endocytic recycling and the subsequent return of endocytosed synaptic vesicles to an “active exocytic pool” are reduced by loss of amphiphysin 1.
Cerebral cortex neurons cultured from amphiphysin 1 knockout mice also exhibit a reduced percentage of labeled synaptic vesicles and endosomes after high-K+ stimulation in the presence of the fluid-phase endocytic marker horseradish peroxidase compared to cultured neurons from wild-type mice. This difference in the percentage of horseradish peroxidase-labeled synaptic vesicles and endosomes is dependent on K+ stimulation, however. In resting synapses the fraction of synaptic vesicles and endosomes labeled with horseradish peroxidase is equivalent in amphiphysin 1 knockout and wild-type neurons. Hence, there are mild defects in synaptic vesicle recycling and exocytosis in amphiphysin 1 knockout mice that only become apparent under conditions of strong nerve stimulation (55).
Interestingly, while neurons cultured from amphiphysin 1 knockout and wild-type mice have the same total number of synaptic vesicles, the pool that participates in action potential stimulated exocytosis differs. Measurements of fluorescent lipophilic dye uptake showed that the percentage of the total pool of synaptic vesicles that undergo exocytosis (and become labeled with fluorescent dye at the plasma membrane) increases in proportion to the number and duration of stimulatory action potentials applied. However, the percentage of the total pool of vesicles that participate in exocytosis and can be labeled is 25 to 30% lower for the amphiphysin 1 knockout neurons than wild-type neurons at each level of stimulation. Labeled wild-type and amphiphysin 1 knockout neurons lose their fluorescent dye with comparable efficiency during initial action potentials, however, after more action potentials the ability of the mutant neurons to recycle fluorescent dye into the medium declines compared to that of wild-type neurons (55). Hence, amphiphysin 1 knockout neurons exhibit a reduced active vesicle pool.
Synaptic vesicles that have been reinternalized must be reprimed before they can undergo another round of stimulated exocytosis. The reduction in fluorescent dye recycling into the medium observed in the amphiphysin 1 knockout neurons is consistent with a possible defect in synaptic vesicle repriming. Repriming is assayed by labeling neurons with a fluorescent lipophilic dye using an action potential and then examining the effect of extending the duration of the action potential beyond the period of dye exposure. The prolonged action potential can result in immediate recycling of the internalized dye into the medium if repriming of endocytosed synaptic vesicles is rapid. If repriming is delayed, the recycling of the internalized dye into the medium is slower and only apparent if the duration of the action potential after dye removal is extended. Consistent with a longer synaptic vesicle repriming time in the amphiphysin 1 knockout synapses, the action potential has to be extended for longer times after removal of dye before the internalized dye is reexocytosed compared to wild-type neurons (55). Defects in repriming may account for the reduced pool of active synaptic vesicles in amphiphysin 1 knockout neurons.
Essential role for amphiphysin 1 in learning.
Particularly exciting is the finding that amphiphysin 1 knockout mice have severe learning deficiencies as revealed by standardized behavioral tests (55). Knockout of other genes in mice that encode synaptic vesicle proteins, e.g., synapsin 1, causes epileptic seizures like those caused by knockout of amphiphysin 1, but in contrast to amphiphysin 1 knockout these do not affect learning (294). It may be relevant that a human gene mutated in a mental retardation disorder encodes a neuronal Rab GDP dissociation inhibitor (44). In yeast, Rvs167p interacts with a catalytically active Rab-GAP (Gyp5p) as well as a catalytically inactive Rab-GAP family member (Gyl1p) (33, 84, 317), rvs167 genetically interacts with a gene encoding an endosomal Rab protein (Ypt51p/Vps21p) (296), and loss of Rvs161p or Rvs167p cause accumulation of what appear to be exocytic vesicles that likely carry the Rab protein Ypt1p (20).
Could amphiphysin 1 in brain function in connection with Rab-GTPase regulators? Altered Rab-GTPase activity in amphiphysin 1 knockout mice, if it indeed occurs, might explain the delayed repriming of endocytosed synaptic vesicles, since Rab-GTPases are key regulators of vesicle exocytosis and their association with vesicles is regulated by their GTP/GDP status. Indeed, the apparent endocytic defects in amphiphysin 1 knockout synapses may be a consequence of a failure of vesicles internalized in one round of endocytosis to undergo exocytosis and thus contribute membranes for the next round of endocytosis. Although there is no direct evidence for amphiphysin 1-Rab-GAP interactions in mammals, such interactions may be worth exploring.
Amphiphysin 1 and Molding of Membranes
Dynamin 1 binds membranes and evaginates tubules in vitro.
Binding of clathrin coat components to protein-free liposomes results in deformation of the initially spherical liposomes to create clathrin-coated buds. Furthermore, binding of dynamin 1 to protein-free liposomes results in evagination of the liposomes to form narrow membrane tubules that resemble the necks of deeply invaginated clathrin-coated pits. These tubules have a striated appearance and can be immunolabeled with antiserum to dynamin 1, suggesting that assembly of dynamin 1 rings on the liposome drives membrane deformation to form tubules (314). Dynamin 1 spontaneously assembles into closed rings and open spirals in vitro even in the absence of liposomes, and these structures have dimensions similar to the membrane tubules (121). This suggests dynamin 1 ring and spiral assembly at the membrane surface may drive membrane evagination to form tubules (121, 315). Addition of GTP to dynamin 1-coated tubules causes them to undergo fission to yield small vesicles (312). This reaction in vitro may mimic dynamin 1 function in cleavage of the necks of deeply invaginated clathrin-coated pits during clathrin-mediated endocytosis.
Amphiphysin 1 binds membranes and evaginates tubules in vitro.
Amphiphysin 1 also binds directly to liposomes and has membrane-tubulating activity in vitro (Fig. 11A). The liposome-tubulating activity of amphiphysin 1 is even greater than that of dynamin 1. The tubules formed by amphiphysin 1 display a striated appearance under electron microscopy, suggesting amphiphysin 1 assembles into rings that surround membrane tubules such as dynamin 1 (Fig. 11B). When clathrin coat components are also present, the membrane tubules formed by amphiphysin 1 are almost always associated with clathrin-coated buds (Fig. 11C). Other clathrin coat components (e.g., clathrin and AP-180) are unable to evaginate liposomes into tubules in vitro (316). Similarly, other known lipid-binding domains such as the pleckstrin homology (PH) domain of phospholipase Cδ are also unable to evaginate liposomes into tubules in vitro (73). This suggests the liposome-tubulating activity of amphiphysin 1 in vitro is specific and requires more than the ability to bind membranes. Amphiphysin 1 activity in liposome tubulation in vitro likely reflects an important in vivo function of amphiphysin 1 in clathrin-dependent endocytosis.
FIG. 11.
Amphiphysin 1 evaginates liposomes in vitro to generate coated membrane tubules. A, electron micrographs of negatively stained membrane tubules generated in vitro from spherical liposomes composed of brain lipids a) after incubation with recombinant full-length amphiphysin 1 (cleaved from a GST fusion protein) or b) after incubation with a recombinant fusion protein comprising GST and the amphiphysin 1 BAR domain only (residues 1 to 286). Bar, 500 nm. B, electron micrographs of negatively stained membrane tubules generated in vitro from spherical liposomes composed of brain lipids after incubation with clathrin coat components plus: a, recombinant dynamin 1; b, both recombinant dynamin 1 and recombinant amphiphysin 1; or c, recombinant amphiphysin 1. The insets at the top right of each panel show the ends of tubules at high magnification (a, negatively stained sample; b and c, positively stained embedded and thin-sectioned samples). For comparison, the bottom inset in panel a shows clathrin-coated buds at high magnification. Note that the presence of amphiphysin 1 in panels b and c increases the frequency of clathrin-coated bud formation at tubule ends compared to dynamin 1 alone in panel a. C, electron micrographs showing the appearance of the tubular coat that forms when spherical liposomes are incubated in vitro with recombinant amphiphysin 1 alone (a and b); recombinant dynamin 1 alone (c and d); both recombinant amphiphysin 1 and recombinant dynamin 1 (e and f), or total brain cytosol with ATP and GTPγS (g). The images in panels a, c, and e represent negatively stained samples, while those in panels b, d, f, and g represent samples embedded, thin sectioned, and then positively stained. Note that recombinant amphiphysin 1 or dynamin 1 alone forms thin tightly packed rings on membrane tubules while recombinant amphiphysin 1 and dynamin 1 together or endogenous amphiphysin 1 and dynamin 1 in total brain extract form thick rings with regular spacing on membrane tubules. (Reprinted from reference 316 with permission of the publisher.)
Amphiphysin 1 binding to membranes is dependent on lipid composition.
The membrane tubulation activity of amphiphysin 1 is affected by the lipid composition of the membrane. Liposomes composed of crude mixtures of brain lipids or of pure phosphatidylserine and phosphatidylcholine are efficiently tubulated by amphiphysin 1, but tubulation activity is proportional to the percentage of phosphatidylserine (316). Hence, similar to dynamin 1, amphiphysin 1 preferentially binds and tubulates membranes composed of acidic phospholipids.
Amphiphysin 1 and dynamin 1 coassemble into rings on membrane tubules.
Incubation of liposomes with total brain extract results in very obvious thick striations with regular spacing along the evaginated membrane tubules. When either purified dynamin 1 or amphiphysin 1 is used instead, the evaginated tubules display a continuous and tightly spaced arrangement of thin rings. Interestingly, incubation of liposomes with purified dynamin 1 and amphiphysin 1 in combination results in thick striations along the evaginated tubules, as seen when total brain extract is used. This suggests the rings assembled on evaginated tubules in the presence of total brain extract contain both dynamin 1 and amphiphysin 1. The formation of these thicker rings requires not only the membrane binding BAR domain of amphiphysin 1, but also the dynamin 1-interacting amphiphysin 1 SH3 domain. The presence of the isolated amphiphysin 1 SH3 domain does not alter the appearance of coats on tubules evaginated by dynamin 1. Similarly, the isolated amphiphysin 1 BAR domain did not alter the appearance of coats on tubules evaginated by dynamin 1. Hence, the formation of thick rings on evaginated tubules by dynamin 1 requires full-length amphiphysin 1 (314, 316).
Amphiphysin 1 stimulates assembly of dynamin 1 into thick rings in solution.
Purified amphiphysin 1, unlike purified dynamin 1, is unable to assemble into rings in the absence of liposomes. However, amphiphysin 1 does stimulate the assembly of dynamin 1 into rings in solution. Moreover, the rings formed by dynamin 1 in the presence of amphiphysin 1 in solution contain both proteins and resemble the thick rings formed when liposomes are also present (316). Ring assembly by amphiphysin 1 and dynamin 1 is influenced by their phosphorylation state. Dephosphorylated forms of amphiphysin 1 and dynamin 1 are highly active for assembly and their mixture results in highly efficient thick-ring assembly in vitro. In contrast, phosphorylated amphiphysin 1 and dynamin 1 are less active for ring assembly and form few thick rings (330).
Amphiphysin 1 coordinates clathrin bud and dynamin 1 tubule formation.
It is important that budding and fission be coordinated during clathrin-mediated endocytosis. However, when both clathrin coat components and dynamin 1 are incubated with liposomes in vitro, although clathrin-coated buds and dynamin 1-coated tubules both form on the liposomes, there is a lack of coordination between the two structures. Few clathrin-coated buds have tubular dynamin 1-coated necks and few dynamin 1-coated tubules have clathrin-coated buds. Because amphiphysin 1 interacts via the central insert domain with clathrin coat components and via its C-terminal SH3 domain with dynamin 1, a key question is whether amphiphysin 1 links clathrin coat components to dynamin 1 and thereby coordinates vesicle budding with vesicle fission. Indeed, if amphiphysin 1 is added to liposomes together with clathrin coat components and dynamin 1, there is an increase in the percentage of dynamin 1-coated tubules that are associated with clathrin-coated buds and vice versa (316).
Amphiphysin 1 stimulates the activity of dynamin 1 in membrane tubule fission.
Although amphiphysin 1 alone is unable to mediate membrane fission in vitro, the addition of purified amphiphysin 1 substantially increases the extent of liposome tubulation and subsequent GTP-induced fission mediated by purified dynamin 1. Under the conditions used in this study the addition of amphiphysin 1 did not increase dynamin 1 association with membranes or stimulate dynamin 1 GTPase activity (316). Interestingly, the activity of amphiphysin 1 and dynamin 1 in liposome tubulation and fission is regulated by their phosphorylation state. Dephosphorylated forms of both dynamin 1 and amphiphysin 1 were highly active in tubulation and fission, and incubation with liposomes resulted in dramatic generation of small vesicles. In contrast, phosphorylation by Cdk5/p35 of only dynamin 1 or amphiphysin 1 slightly reduced vesicle formation, while phosphorylation of both proteins almost completely inhibited the generation of small vesicles (330).
In vivo role for amphiphysin 1 in membrane tubule formation.
Is the ability of amphiphysin 1 to bind and tubulate membranes important in vivo? The first evidence that amphiphysin 1 generates membrane tubules in vivo came from experiments in which a fragment comprising the amphiphysin 1 N-terminal amphipathic α-helix and BAR domain (N-BAR) was overexpressed in COS-7 cells. Overexpression of the N-BAR fragment induced massive tubulation of the plasma membrane in vivo (237). The importance of the N-terminal amphipathic helix for high-affinity liposome binding and tubulation in vitro was first demonstrated for the BAR domain protein endophilin A1 (see below), but the amphiphysin 1 N-terminal amphipathic helix appears to play a similar role (73). There is also evidence that amphiphysin 1 plays a physiologically important role in dynamin 1-mediated membrane fission. Brain cytosol prepared from amphiphysin 1 knockout mice (and also deficient in Bin1+6a+12+13, see below) has considerably reduced ability to support dynamin 1-dependent fission of large unilamellar liposomes into small vesicles in vitro. This deficiency can be rescued by addition of purified recombinant amphiphysin 1 (380). This shows that the membrane fission defect in these extracts is specifically due to a lack of amphiphysin 1 (although Bin1+6a+12+13 is also deficient, it appears that amphiphysin 1 alone can stimulate dynamin 1-dependent fission).
Membrane curvature and amphiphysin 1 binding.
The N-terminal amphipathic α-helix is critical for high-affinity binding to liposomes and tubulation, however, the BAR domain alone retains lower affinity binding to liposomes. A key finding was that binding of the amphiphysin 1 BAR domain alone to liposomes in vitro is critically dependent on liposome size and therefore membrane curvature. While the amphiphysin 1 BAR domain binds poorly to large liposomes (0.8-μm diameter) that have low curvature, it binds strongly to small liposomes (0.05-μm diameter) that have high curvature. This result is important because it shows the BAR domain not only generates membrane curvature (in conjunction with the N-terminal amphipathic α-helix), but alone can also act as a sensor of membrane curvature. An exciting possibility is that amphiphysin 1 may recruit dynamin 1 specifically to high-curvature membranes such as those at the tubular neck of clathrin-coated pits. Hence, amphiphysin 1 may potentiate dynamin 1-dependent membrane fission by localizing dynamin 1 within the membrane to sites where fission is most efficient (237, 316, 380).
The effects of membrane curvature on amphiphysin 1- and dynamin 1-dependent membrane fission were investigated recently. In the presence of large liposomes that can be evaginated into high-curvature tubules, addition of amphiphysin 1 results in a dose-dependent stimulation of both dynamin 1 GTPase activity and recruitment to liposomes. In the presence of small liposomes that, because of their size, cannot support evagination of high-curvature tubules, the basal GTPase activity of dynamin 1 was higher than in the presence of large liposomes. However, in contrast to what was seen with large liposomes, addition of amphiphysin 1 in this case led to a dose-dependent inhibition of dynamin 1 GTPase activity. Small liposomes contain insufficient membrane material to support the formation of dynamin 1- and amphiphysin 1-coated high-curvature tubules, and amphiphysin 1 under these conditions may engage in inhibitory interactions with dynamin 1. However, in the case of large liposomes the conversion of low-curvature membranes into evaginated tubules with high curvature allows extensive coassembly of dynamin 1 and amphiphysin 1 into rings. This promotes a different type of interaction between amphiphysin 1 and dynamin 1 that in turn stimulates dynamin 1 GTPase activity (380).
Amphiphysin 1 tubulation of membranes is dependent on lipid composition.
The influence of membrane lipid composition on stimulation of dynamin 1 GTPase activity by amphiphysin 1 was also investigated. In the absence of liposomes dynamin 1 GTPase activity was low and only very weakly stimulated by amphiphysin 1. Large liposomes containing brain lipids and supplemented with cholesterol and phosphatidylinositol-4,5-phosphatase [PtdIns(4,5)P2]-stimulated dynamin 1 GTPase activity, even in the absence of amphiphysin 1. Addition of amphiphysin 1 in the presence of these liposomes resulted in further strong stimulation of dynamin 1 GTPase activity (380). This is consistent with the view that stimulation of dynamin 1 GTPase activity by amphiphysin 1 is not due to direct amphiphysin 1 interaction with dynamin 1 in solution, but instead related to the ability of these two proteins to coassemble into rings on high curvature membranes.
PtdIns(4,5)P2 binds the PH domain of dynamin 1 and is known to not only recruit dynamin 1 to membranes but also activate dynamin 1 GTPase activity in vitro. If PtdIns(4,5)P2 was substituted with phosphatidylcholine in the large liposomes, dynamin 1 GTPase activity was not as strongly stimulated by liposomes alone, but addition of amphiphysin 1 still resulted in strong stimulation (380). Perhaps amphiphysin 1 is especially important for dynamin 1 recruitment to membranes low in PtdIns(4,5)P2. Dynamin 1 and amphiphysin 1 have been shown to tubulate liposomes containing high levels of phosphatidylserine (314, 316). Furthermore, liposomes containing phosphatidylserine are subject to GTP-dependent fission by dynamin 1 in vitro (312, 316). When PtdIns(4,5)P2 was replaced with phosphatidylserine, dynamin 1 GTPase activity was strongly stimulated by liposomes alone and activity was directly proportional to liposome phosphatidylserine content. Moreover, the addition of amphiphysin 1 led to a very dramatic further stimulation of dynamin 1 GTPase activity that exhibited positive cooperativity with liposome phosphatidylserine content (380).
Domains of amphiphysin 1 required for membrane tubulation.
Testing of various recombinant fragments of amphiphysin 1 revealed that the N-terminal BAR domain (residues 1 to 286) is sufficient for full liposome tubulation activity and that an N-terminal piece of the BAR domain (residues 1 to 161) retains low tubulation activity. In contrast, the C-terminal SH3 domain has no tubulation activity (316). Tubulation activity correlates with stimulation of dynamin 1 GTPase activity. In the presence of large liposomes containing cholesterol and PtdIns(4,5)P2, the isolated amphiphysin 1 BAR domain stimulates dynamin 1 GTPase activity as efficiently as full-length amphiphysin 1. This is probably due to the BAR domain's promoting high-curvature tubule formation, which in turn facilitates dynamin 1 binding and assembly into rings. In contrast, the isolated amphiphysin 1 SH3 domain was not able to stimulate dynamin 1 GTPase activity in the presence of large liposomes (380).
The BAR domain of amphiphysin 1 is predicted to be highly acidic and hence to bear a strongly negative net charge. That the BAR domain is the part of amphiphysin 1 that mediates binding to acidic lipids, including phosphatidylserine and PtdIns(4,5)P2, initially seemed somewhat puzzling. However, the three-dimensional structure of the amphiphysin BAR domain provides a simple explanation for this apparent paradox because it reveals that the membrane binding surface of BAR domains bears a net positive charge (see below).
Interestingly, a construct containing both the BAR and SH3 domains but missing the central insert domain is considerably more active in stimulation of dynamin 1 GTPase activity than full-length amphiphysin 1. Hence, full stimulation of dynamin 1 GTPase activity requires both the BAR and SH3 domains, but not the central insert domain. The central insert domain of amphiphysin 1 is known to bind the SH3 domain via an intramolecular interaction and this inhibits interaction of the SH3 domain with dynamin 1 (74). Deletion of the proline-rich motif in the central insert domain that binds the SH3 domain enhances the stimulatory effect of amphiphysin 1 on dynamin 1 GTPase activity. The isolated amphiphysin 1 SH3 domain does not further enhance the ability of the isolated amphiphysin 1 BAR domain to stimulate dynamin 1 GTPase activity in the presence of large liposomes (380). This shows that the function of the SH3 domain is to link dynamin 1 to the amphiphysin 1 BAR domain and that the SH3 domain does not independently stimulate dynamin 1 GTPase activity.
The stimulatory effect of these amphiphysin 1 constructs on dynamin 1 GTPase activity correlates with their ability to recruit dynamin 1 to membranes in vitro, e.g., the amphiphysin 1 construct lacking the central insert domain was more active than full-length amphiphysin 1 both in recruiting dynamin 1 to liposomes and in stimulating dynamin 1 GTPase activity (380). This suggests that coassembly on membranes and stimulation of dynamin 1 GTPase activity are linked. This is consistent with results from other studies that show dynamin 1 assembly stimulates its GTPase activity (353). Stimulation is due to intermolecular interactions between the GTPase effector domain of one dynamin 1 monomer and the GTPase catalytic domain of the adjacent dynamin 1 monomer that can take place only within a multimeric assembly (203).
Domains of amphiphysin 1 required for coassembly with dynamin 1 into rings in solution.
The domains of amphiphysin 1 required for coassembly with dynamin 1 into rings in solution have also been investigated. Visualization of negatively stained protein samples by electron microscopy reveals that full-length amphiphysin 1 and amphiphysin 1 constructs lacking either the proline-rich motif or the entire central insert domain are able to coassemble with dynamin 1 into rings in solution. In contrast, the isolated BAR and SH3 domains lack the ability to coassemble with dynamin 1 into rings in solution (380). This requirement for the BAR domain even in the absence of membranes suggests that coassembly of amphiphysin 1 and dynamin 1 in solution may require that amphiphysin 1 make contacts with dynamin 1 via the BAR domain as well as the SH3 domain. However, direct evidence of amphiphysin 1 BAR domain-dynamin 1 interaction is lacking.
SECOND ISOFORM OF AMPHIPHYSIN, Bin1
Discovery of Bin1
A number of findings suggested the existence of a second, more ubiquitously expressed amphiphysin isoform in vertebrates. These included the ability of the amphiphysin 1 SH3 domain to powerfully inhibit endocytosis in nonneuronal cells that express only very low levels of amphiphysin 1 (369), the existence of amphiphysin-related proteins in unicellular eukaryotes such as yeast (12, 42, 47, 298), and the finding that amphiphysin 1-reactive sera from stiff-man syndrome patients also recognize an additional protein distinct in size from amphiphysin 1 (338). Various approaches led to the isolation of this second isoform of amphiphysin, which has resulted in its having several names, e.g., amphiphysin 2 (168), amphiphysin II (23, 256, 338), Amph2 (367), BRAMP2 (168), ALP1 (141), SH3P9 (306), and Bin1 (278). The Human Genome Organization-approved designation for this family of amphiphysin-related proteins is Bin1, so Bin1 is the name that will be used in this review.
Sequence comparison revealed that Bin1 is the product of a novel gene and that Bin1 transcripts are subject to extensive differential splicing, leading to a diversity of Bin1 forms. Table 2 lists all the known splice variants of Bin1 and the various names that have been assigned to them. Bin1 has a domain structure similar to that of amphiphysin 1, featuring an N-terminal BAR domain with predicted coiled-coil structure and a C-terminal SH3 domain (Fig. 1) (23, 141, 168, 256, 278, 306, 367).
The central insert domain of Bin1 is more divergent than that of amphiphysin 1. Some splice variants include a central insert domain that is highly homologous to the central insert domain of amphiphysin 1 and that interacts with clathrin and AP-2/α-adaptin (sometimes referred to as the endocytosis domain) (Fig. 1). Other splice variants lack this central insert domain or have alternative central insert domains that do not interact with clathrin or AP-2/α-adaptin (Fig. 1) (168, 255, 278, 338, 356). The central insert domain contains two proline-rich motifs (residues 297 to 306 and 340 to 346 in mouse Bin1) that may bind SH3 domains (168). Although the hydrophobic region in the central insert domain of human and chicken amphiphysin 1 is missing in Bin1, there is a region of hydrophobicity near the Bin1 SH3 domain (338). In some brain splice variants a 31-residue insert (N-terminal insert domain [NTID]) occurs within what was originally predicted to be a coiled-coil motif in the BAR domain (338). Comparison with the recently elucidated three-dimensional structure of Drosophila amphiphysin now suggests this site of insertion is a disordered loop between α-helices (237).
Four studies identified Bin1 partial sequences simply by searching expressed sequence tag databases and these were then used to obtain full-length cDNAs by hybridization screens (23, 256, 338, 367). In one study, a mouse cDNA expression library was screened with a synthetic peptide known to be a ligand of the Src SH3 domain in an attempt to identify novel SH3 domain proteins. One of the novel murine SH3 domain proteins identified was named SH3p9 and is a splice variant of Bin1 (Table 2) (306). Bin1 was also isolated in two-hybrid screens for proteins that interact with the oncoproteins c-Myc (278) and c-Abl (141) (Table 2). Bin1 was independently identified in a two-hybrid screen for mouse brain proteins that interact with the proline-rich C-terminal domain of the human Ras GDP/GTP exchange factor son of sevenless 1 (Sos-1) and named BRain form of AMPhiphysin 2 (BRAMP2) (Table 2). Purified recombinant BRAMP2 bound in vitro-translated human SOS-1 and -2, confirming the yeast two-hybrid interaction and showing that BRAMP2 binding to SOS-1 and -2 is direct (168).
Characterization of independent Bin1 cDNAs from different tissues and even from the same tissue revealed complete sequence identity over included domains but a high degree of heterogeneity in domain structure, consistent with the possibility that the Bin1 transcript is subject to differential splicing (Fig. 1 and Table 2) (23, 168, 256, 278, 338, 356, 367). In two reports at least 10 different transcripts could be amplified by PCR from human or rat brain tissue using primers that recognize sequences encoding the N- and C-terminal domains of Bin1 (338, 367). The longest Bin1 splice variant in human brain exhibits 71% amino acid sequence similarity and 55% amino acid sequence identity with human amphiphysin 1 (23). Interestingly, in one study two of the Bin1 cDNAs appeared to encode splice variants that lacked a C-terminal SH3 domain, similar to yeast Rvs161p (Table 2) (367).
The intron-exon boundaries of the 54-kb human BIN1 gene have been mapped by Wechsler-Reya et al. BIN1 encodes at least 20 exons, of which at least seven are differentially spliced, giving rise to a vast array of Bin1 splice variants (Fig. 1 and Table 2). This review will adopt the splice variant nomenclature proposed by Wechsler-Reya et al., but with the addition of one originally overlooked exon located between exons 6 and 7, which will be referred to in this review as exon 6a (356). It should be noted that in the new systematic exon nomenclature for Bin1, exon 6a is renamed exon 7 and all following exons have numbers that are n + 1 with respect to those assigned by Wechsler-Reya. The majority of published reports have used the exon nomenclature of Wechsler-Reya and not the new systematic exon nomenclature. Our decision to retain the older nomenclature in this review ensures the exon numbers referred to here are consistent with those in most of the published literature.
Although there exist many splice variants, the major forms vary in four exons: 6a, 10, 12, and 13 (Fig. 1 and Table 2). Exon 6a encodes the 31-residue brain-specific NTID (257). There is no sequence homologous to the NTID in the reported clones of amphiphysin 1 (23, 367). Exon 10 is muscle specific and encodes a 15-residue sequence that contains a putative nuclear localization sequence and lipid binding sequence (162, 278). Exon 12 includes a series of alternative brain-specific exons (12A to -D) that encode the central insert domain. Depending on the particular exon 12 variant this central insert domain can interact with α-adaptin/AP-2 and/or clathrin heavy chain and/or endophilin A1 (23, 168, 187, 255, 256, 338, 356, 367). For simplicity, except in Table 2 this review will not distinguish which of the exon 12 variants is present, only that an exon 12 is present.
Finally, exon 13 includes part of the Myc binding domain and is differentially spliced in a tissue-independent manner (100, 141, 338, 356, 357, 367). Bin1+6a+12+13 represents the neuronal isoforms of Bin1 (amphiphysin 2, amphiphysin II, Amph2, and BRAMP2), and Bin1+10+13 represents the muscle isoform of Bin1 (commonly referred to as Bin1). Bin1−10−12+13 represents the widely expressed Bin1 isoform known as SH3p9. Finally, Bin1−10−12−13 is the most ubiquitously expressed Bin1 isoform, known as ALP1 or amphiphysin IIm.
Different Bin1 splice variants exhibit different tissue distributions. In the rat, every tissue tested expressed transcripts lacking exons 6a and 10 except the retina (only +6a) and skeletal muscle (only +10). Exon 6a was present in at least some transcripts from all nerve tissues, but was not present in transcripts from any other tissues. In contrast, all skeletal muscle transcripts included exon 10, but exon 10 was not present in transcripts from any other tissue (Fig. 1 and Table 2). In the human, rat, and mouse, transcripts containing exon 12 that encodes the 127-residue central insert domain are expressed only in the brain (in particular the cerebral cortex and pons), spinal cord, other neuronal tissue, and neuron-like pheochromocytoma (PC12) cells, similar to amphiphysin 1 (23, 168, 256, 278, 338, 356, 367). Some nonneuronal cells misexpress transcripts containing exon 12 following tumorigenesis (Table 2) (93, 356). In rat brain Bin1 represents 0.1% of the total protein, similar to rat brain amphiphysin 1 (367).
In the rat, forms of Bin1 protein lacking the large brain-specific central insert domain encoded by exon 12 were expressed at high levels in skeletal muscle and could be detected (at approximately 10-fold lower levels) in many tissues, including heart, brain, lung, liver, skeletal muscle, kidney, ovary, and thymus (the skeletal muscle forms include the 15-residue muscle-specific domain encoded by exon 10 but the presence of this domain is difficult to distinguish by electrophoretic mobility) (367). In the mouse, transcripts lacking exon 12 were expressed in multiple tissues, including brain, liver, lung, kidney, heart, ovary, testis, spleen, and skeletal muscle (168, 278). In the human, transcripts lacking exon 12 were present in the brain, heart, placenta, lung, liver, kidney, pancreas, spleen, ovary, testis, and skeletal muscle (23, 141). They were expressed at equal levels in cultured normal human fibroblasts and in some cultured human tumor cell lines (e.g., HeLa cervical carcinoma cells), however, they were absent in other tumor cell lines (e.g., HepG2 hepatocarcinoma cells and MCF7 breast carcinoma cells) (278).
The various Bin1 splice variants exhibit anomolous electrophoretic mobilities. In humans the brain isoform of Bin1 migrates at 85 kDa, while in rats it migrates at 92 kDa. Ectopic expression of the equivalent mouse cDNA gave rise to a doublet at 88 and 96 kDa. In contrast, the longest Bin1 transcript in human, rat, or mouse brain is predicted to encode a protein of only 65 kDa (23, 168, 256, 367). The aberrant gel mobility of brain Bin1 isoforms is similar to what has been observed for amphiphysin 1. The amino acid sequence encoded by exon 12 corresponds to a sequence that in amphiphysin 1 has been shown to confer aberrant electrophoretic mobility (47, 175). Indeed, in the rat the presence of exon 12 in Bin1 has been shown to confer aberrant electrophoretic mobility (367). Interestingly, the muscle isoform containing the 15-residue domain encoded by exon 10 migrates on gels at 60 to 70 kDa although its predicted size is 50 kDa, so it also exhibits aberrant electrophoretic mobility (278).
Bin1+6a+12+13/Amphiphysin 2/Amphiphysin II/BRAMP2 in the Brain
The function of Bin1+6a+12+13 in the brain has been the topic of an excellent earlier review (368). There are several brain-specific isoforms of Bin1 that vary in exon 12 subtype (12A to 12D) (encoding the central insert domain) and the presence of exon 6a (encoding the NTID) (Fig. 1 and Table 2).
Distribution of Bin1+12+13 in the brain.
Different localizations have been reported for Bin1+12 in the brain (the antisera used for these studies could not distinguish between isoforms containing and lacking exons 6a and 13, so they will collectively be referred to here as Bin1+12). In one study immunocytochemistry revealed a distribution of Bin1+12 in the brain similar to that reported for amphiphysin 1. Bin1+12 was strongly enriched in a subset of nerve terminals within all three layers of the cerebellum as well as the hippocampus and pontine nucleus (367). Similarly, in a second study immunofluorescence staining of brain sections showed that Bin1+12 staining is punctate and colocalizes with known synaptic vesicle markers and a similar localization to punctate synapses was seen in spinal cord. The subcellular distribution of Bin1+12 was cytoplasmic and never nuclear, in contrast to what has been reported for some other splice variants of Bin1 (see below) (256). Subcellular fractionation showed that Bin1+12 cofractionates with isolated nerve terminals. Immunoelectron microscopy of rat brain sections shows that Bin1+12 is present on the surface of synaptic vesicles and also in isolated patches on the plasma membrane (367).
In contrast, another study found Bin1+12 distribution in the brain differs from that of amphiphysin 1. While amphiphysin 1 is enriched at synapses in the cerebral cortex, hippocampus, and cerebellum, this study found Bin1+12 enriched at other sites in the brain. In gray matter, which contains perikarya (neuronal cell bodies), dendrites, and axons, Bin1+12 localized to axon initial segments. Axon initial segments are short unmyelinated tubular segments innervated by basket cells where the axon is attached to the perikaryon. Bin1+12 also localized to the nodes of Ranvier. The nodes of Ranvier are fine ring-like structures distributed at intervals along the length of axons. In white matter, which contains mainly axons, the densely clustered nodes of Ranvier were found to be the main sites of Bin1+12 localization (23).
Both axon initial segments and nodes of Ranvier exhibit a dense submembranous cytoskeletal matrix. This matrix contains actin and a neuron-specific isoform of ankyrin 3 as major components. In high-magnification images of both axon initial segments and nodes of Ranvier, Bin1+12 localizes within the submembranous cytomatrix (23). This subcellular localization suggests a role for Bin1+12 in organization or function of the cortical actin cytoskeleton and/or in defining these specialized membrane domains within neurons. In this context it is interesting that in fruit flies (Drosophila melanogaster) amphiphysin also localizes to actin-rich membrane domains in a variety of polarized cell types, including the actin-rich apical domain in intestinal epithelial cells and the actin-rich sensory microvilli of photoreceptor neurons (386).
Interestingly, axon initial segments and nodes of Ranvier are active in endocytosis. Within neurons, axon initial segments and nodes of Ranvier have a higher concentration of clathrin-coated pits and vesicles than any other site except the presynaptic terminal. Moreover, they are also enriched in a variety of integral membrane proteins that mediate ion flux, including Na+ channels, Na+/K+-ATPase, and Na+/Ca2+ exchangers (23). This suggests axon initial segments and nodes of Ranvier are important sites within neurons for the generation and propagation of action potentials. This is interesting, as yeast rvs161 and rvs167 mutants exhibit growth that is severely sensitive to Na+. Regulation of ion channel activity may turn out to be a conserved role of amphiphysin family proteins.
While the reason for the different subcellular localizations of Bin1+12 observed in different studies is not yet clear, it appears to be related to the antibody used for detection. Independent monoclonal antibodies raised to the SH3 domain of Bin1+12 preferentially label axon initial segments and nodes of Ranvier. In contrast, polyclonal antibodies raised against Bin1+12 preferentially label presynaptic termini. One possibility is that the SH3 domain epitopes recognized by the monoclonal antibodies are masked at synapses due to conformational changes or posttranslational modification.
Localization of Bin1+6a+12 on purified plasma membranes.
Those isoforms of Bin1 in the brain that include the NTID encoded by exon 6a specifically localize to the plasma membrane upon transient expression in COS-7 cells. The distribution of Bin1+6a+12 (this includes isoforms with or lacking sequences encoded by exon 13) on plasma membrane is punctate, and the puncta partially colocalize with clathrin-coated pits. Bin1+6a+12 is detected at 62% of plasma membrane clathrin-coated pits. This degree of colocalization with clathrin-coated pits is similar to that reported for dynamin 1, but less than that reported for other clathrin-coated pit components, e.g., intersectin (257). There were puncta of Bin1+6a+12 on the plasma membrane that did not colocalize with clathrin-coated pits, but the identity of these structures is not known. Membranes other than plasma membrane were not examined in this study, so Bin1+6a+12 may also associate with other intracellular membranes.
Association of Bin1 with membranes.
Subcellular fractionation of purified nerve terminals revealed that Bin1+6a+12 (including isoforms with or lacking sequences encoded by exon 13) is present at approximately equal levels in a soluble pool and in a membrane-associated sedimentable pool (256, 367). When Bin1+6a+12 is expressed ectopically in COS-7 fibroblasts it is also found in association with a sedimentable fraction. Hence, association of Bin1+6a+12 with membranes does not require brain-specific integral membrane proteins. As mentioned above, the membrane association of Bin1+6a+12 is very strong and resists washes with 0.5 M Tris-HCl, 1 M KCl, and 1 M NaCl that strip all clathrin heavy chain and most AP-2 from membranes (257, 367). Tight and salt-resistant association with membranes is also a property of one pool of amphiphysin 1 (175).
Recruitment of Bin1+6a+12 to the plasma membrane.
Bin1+6a+12 contains the central insert domain encoded by exon 12 which, depending on the subtype of exon 12 (A to D), may bind both clathrin heavy chain and/or AP-2 (Fig. 1). Could this domain mediate plasma membrane clathrin-coated pit association? The discovery that Bin1+6a+12 also localizes to regions of plasma membrane other than clathrin-coated pits together with the finding that Bin1+6a+12 remains associated with isolated plasma membranes after clathrin heavy chain and AP-2 are both removed by salt washes suggests that other interactions may play an important role. Consistent with this view, ectopic expression in COS-7 fibroblasts of four different splice variants of Bin1 that differ in the presence or absence of sequences encoded by exons 6a and 12 revealed that some isoforms that lack the central insert domain encoded by exon 12 (and that therefore lack the ability to bind clathrin heavy chain or AP-2) are still efficiently targeted to the plasma membrane. Intriguingly, plasma membrane targeting of Bin1 in neurons correlates with the presence of the NTID. Furthermore, deletion of the NTID from splice variants that include this sequence completely abolishes plasma membrane localization of all Bin1 isoforms examined, including those that also contain clathrin heavy chain- and AP-2-binding domains (257).
The NTID also plays an important role in general association of Bin1 with membranes. When expressed in COS-7 fibroblasts the BAR domain alone of each plasma membrane isoform of Bin1 associates with membranes such as the full-length protein during subcellular fractionation. Hence, the SH3 domain is not essential for association of Bin1 with membranes. In contrast, deletion of the BAR domain from plasma membrane isoforms of Bin1 completely abolishes association with membranes (257). These data indicate that the BAR domain of plasma membrane isoforms of Bin1 is both necessary and sufficient for association with membranes. This finding is consistent with the observation that the isolated BAR domain of yeast Rvs167p retains some ability to localize to cortical actin patches and to functionally substitute for full-length Rvs167p (11, 299).
Deletion of the NTID from either full-length Bin1+6a+12 or its isolated BAR domain significantly reduces, but does not abolish, association with the sedimentable fraction (257). Hence, the NTID contributes to membrane association but other sequences in the BAR domain are sufficient to preserve membrane association of full-length Bin1+6a+12 or its isolated BAR domain in the absence of the NTID. Although yeast Rvs161p and Rvs167p also associate with membranes they lack a BAR domain sequence that resembles the Bin1+6a+12+13 NTID.
Interaction of Bin1+12 with Other Brain Proteins
Bin1+12 (with or lacking exons 6a and 13) interacts with dynamin 1 and synaptojanin 1 in the brain.
The first indication that the brain isoforms of Bin1 may function in endocytosis was their ability to bind the same set of endocytic proteins as amphiphysin 1. Bin1+6a+12+13 binds several dynamin isoforms, including brain dynamin 1 and also brain synaptojanin 1 (23, 168, 187, 256). The interaction of Bin1+6a+12+13 with dynamin 1 is direct and the Kd is 240 nM (168, 187). The Bin1 SH3 domain and the proline-rich motif GPPPQVPSRPNR in the dynamin 1 PRD appear to mediate the interaction (23, 168). Although both amphiphysin 1 and Bin1+6a+12+13 bind dynamin 1 and synaptojanin 1, peptide competition experiments suggest the two SH3 domains may differ in their affinity for these proteins (e.g., the Bin1+6a+12+13 SH3 domain binds synaptojanin 1 with much lower affinity than the amphiphysin 1 SH3 domain) (23, 302, 367). Interestingly, one study found that interaction of endogenous Bin1+6a+12+13 with dynamin 1 in PC12 cells is regulated by nerve growth factor (168).
Bin1+12 (with or lacking exons 6a and 13) interacts with the α-adaptin subunit of AP-2 in the brain.
Recombinant Bin1+6a+12+13 binds the α-adaptin subunit of AP-2 in extracts of NIH-3T3 fibroblasts or rat brain (168, 187). The BAR domain and central insert domain (residues 1-422) of Bin1+6a+12+13 and the appendage domain of α-adaptin mediated this interaction (187). In contrast, another study found that α-adaptin does not bind to the central insert domain of Bin1+6a+12+13 but rather to the SH3 domain (256). The difference in α-adaptin binding is not due to different exon 12 splice variants because a sequence motif (LFED) similar to the FFED α-adaptin binding motif of amphiphysin 1 is present in the Bin1+6a+12+13 constructs used in all three studies.
Bin1+12 (with or lacking exons 6a and 13) interacts with clathrin heavy chain in the brain.
Clathrin heavy chain also binds some splice variants of Bin1+12 depending on which of the four exons 12 (A to D) are present in the spliced transcript. Binding to clathrin heavy chain is direct and mediated by the Bin1+12 central insert domain (187, 256). In one study it was observed that full-length Bin1+12 is less efficient at binding clathrin heavy chain than a fragment comprising the BAR and central insert domains only and that the addition of dynamin 1 inhibits binding of full-length Bin1+12 to clathrin heavy chain (187). While the mechanism by which dynamin 1 binding to the Bin1+12 SH3 domain inhibits clathrin heavy chain binding to the central insert domain is not understood, one possibility is that coassembly of Bin1+12 and dynamin 1 into rings in solution reduces the ability of Bin1+12 to bind clathrin heavy chain.
Only those central insert domains that contain a 44-residue domain (residues 378 to 422 in human Bin1+12) exhibit high-affinity clathrin heavy-chain binding in vitro. This domain is located at the C terminus of the central insert domain. Within this domain there are two motifs highly conserved in the sequence of amphiphysin 1 (that also binds clathrin heavy chain), LLDLDFDP and PWDLW (255). Fragments of Bin1+12 lacking either LLDLDFDP or PWDLW bind clathrin heavy chain with approximately equal affinity, but deletion of both motifs abrogates clathrin heavy-chain binding. Clathrin triskelia bind to a human Bin1+12 fragment containing residues 378 to 422 with an affinity of 3 nM, but deletion of either LLDLDFDP or PWDLW reduces the affinity to 9 nM (255).
Interaction of Bin1+12 with endophilin A1/SH3p4/SH3GL2 in the brain.
In addition to clathrin heavy chain and α-adaptin/AP-2, the central insert domain of Bin1+12 also binds the other BAR domain protein endophilin A1 (also called SH3p4 and SH3GL2) (Fig. 1). This interaction is direct and mediated by the SH3 domain of endophilin A1 and residues 335-378 of the central insert domain of Bin1+12. Residues 335 to 378 of the central insert domain contain one proline-rich motif that fits the consensus for SH3 domain binding and may bind the endophilin A1 SH3 domain (193, 255).
Bin1+6a+12 forms homodimers and heterodimers in the brain.
Bin1+12 isoforms that contain exon 6a (Bin1+6a+12) bind amphiphysin 1 and fractionate on density gradients with amphiphysin 1 during subcellular fractionation. Chemical cross-linking experiments confirmed the existence of 1:1 heterodimers of amphiphysin 1 and Bin1+6a+12 in the brain. Heterodimer formation is mediated by residues 1 to 150 of the amphiphysin 1 BAR domain and residues 1 to 328 of the Bin1+6a+12 BAR domain and the latter also mediate Bin1+6a+12 homodimer formation. The formation of homodimers or heterodimers by the Bin1+6a+12 BAR domain was considerably reduced, but not abolished, by deletion of the NTID. Hence, the NTID contributes to dimer formation but other sequences in the BAR domain are sufficient to maintain dimer formation when the NTID is absent (257, 302, 367).
Interaction of amphiphysin 1/Bin1+6a+12 heterodimers with dynamin 1 in the brain.
Amphiphysin 1/Bin1+6a+12 heterodimers associate with dynamin 1 in a 1:2 stoichiometry in the brain, suggesting that each SH3 domain in the heterodimer binds one dynamin 1 molecule (367). The assembly of dynamin 1 monomers into oligomers in vitro stimulates dynamin 1 GTPase activity (353). Amphiphysin 1/Bin1+6a+12 heterodimers stimulate dynamin 1 GTPase activity in vitro, suggesting that their ability to bind two dynamin 1 molecules via their SH3 domains may promote dynamin 1 assembly into rings in solution (367). Stimulation of dynamin 1 GTPase activity by amphiphysin 1/Bin1+6a+12 heterodimers is likely to be important in vivo during dynamin 1-mediated severing of the necks of deeply invaginated clathrin-coated pits.
The three-dimensional crystal structure of the Bin1+6a+12 SH3 domain has been solved to 2.2 Å resolution (232). Unlike most other SH3 domains, the Bin1+6a+12 SH3 domain has a highly acidic patch positioned close to the binding cleft. The existence of this acidic patch explains the curious requirement for two basic (arginine) residues in the proline-rich motif within the dynamin PRD recognized by this SH3 domain (168). The Bin1+6a+12 SH3 domain also has a long insert sequence not found in other SH3 domains that is predicted to form an extended loop near the dynamin-binding site. The Bin1+6a+12 SH3 domain not only binds dynamin, but also interferes with dynamin assembly into rings in vitro. Deletion of the insert loop abolishes the ability of the Bin1+6a+12 SH3 domain to inhibit dynamin assembly in vitro and receptor-mediated endocytosis of transferrin in vivo. Moreover, transfer of this insert loop to an unrelated dynamin-binding SH3 domain (Grb2) confers the ability to inhibit dynamin assembly in vitro (232).
Bin1+6a+12 is regulated by phosphorylation and dephosphorylation.
One study did not observe significant phosphorylation of Bin1+6a+12 in resting synapses, in contrast to amphiphysin 1 (193). Subsequent studies, however, showed that when brain extract is incubated with ATP and protein phosphatase inhibitors, Bin1+6a+12, like dynamin 1, synaptojanin 1, clathrin heavy chain, and AP-2, undergoes a shift to lower electrophoretic mobility suggestive of phosphorylation. As in the case of the other endocytic proteins, Bin1+6a+12 is rapidly dephosphorylated upon nerve terminal depolarization. Dephosphorylation occurs concomitant with synaptic vesicle exocytosis, suggesting that Bin1+6a+12 may have to be dephosphorylated to become active and that Bin1+6a+12 may play a role in synaptic vesicle recycling via endocytosis like amphiphysin 1 and dynamin 1.
The interaction between amphiphysin 1 and Bin1+6a+12 is not inhibited by phosphorylation of either protein, however, as observed for amphiphysin 1, association of Bin1+6a+12 with dynamin 1, synaptojanin 1, clathrin heavy chain, and AP-2 is inhibited by phosphorylation (of either Bin1+6a+12 or its partner) (302). Hyperphosphorylation of Bin1+6a+12 occurs in response to treatment of neurons with the protein kinase C activator phorbol 12-myristate 13-acetate. This hyperphosphorylation is inhibited by pretreatment with the protein kinase C inhibitor Ro 31-8220, suggesting that protein kinase C is responsible for Bin1+6a+12 phosphorylation in synapses (367).
Bin1+6a+12 functions in synaptic vesicle recycling/endocytosis.
Does Bin1+6a+12 function in endocytosis? A hint that it may came from studies that tested the effect of transient overexpression of amphiphysin 1, Bin1+6a+12, or both proteins on receptor-mediated endocytosis of transferrin in COS-7 cells. Overexpression of either amphiphysin 1 or Bin1+6a+12 had a similar deleterious effect on transferrin endocytosis, but in contrast coexpression of amphiphysin 1 and Bin1+6a+12 did not. Since amphiphysin 1 and Bin1+6a+12 both bind dynamin 1 via their SH3 domains, this result suggests that overexpression of the individual proteins may lead to sequestration of dynamin 1 into amphiphysin 1-dynamin 1 or Bin1+6a+12-dynamin 1 complexes that are inactive for endocytosis. Perhaps only amphiphysin 1/Bin1+6a+12 heterodimers are able to functionally interact with dynamin 1 and stimulate endocytosis. A subsequent study showed an association of other clathrin coat components, including dynamin 1, synaptojanin 1, Eps15, and AP-180, with the dynamin 1 PRD in vitro that is dependent on binding of amphiphysin 1/Bin1+6a+12 heterodimers to the dynamin 1 PRD (302).
Further evidence for a role of Bin1+6a+12 in clathrin-dependent endocytosis came from studies similar to those of Shupliakov et al. (293), Wigge et al. (369), and Volchuk et al. (349) that first showed amphiphysin 1 has a role in endocytosis. Transient overexpression of the Bin1+6a+12 SH3 domain inhibited receptor-mediated endocytosis of transferrin in intact cells. Moreover, the Bin1+6a+12 SH3 domain caused potent inhibition of in vitro assays for clathrin-dependent endocytosis (half-maximal inhibition at 6 μM). The Bin1+6a+12 SH3 domain inhibited a target present in the membrane fraction and most potently inhibited the late step of membrane fission and clathrin-coated vesicle release. The inhibition by the Bin1+6a+12 SH3 domain is specific, because control SH3 domains did not inhibit endocytosis in this in vitro assay even at much higher concentrations (295).
The role of Bin1+12 in endocytosis and synaptic vesicle recycling has also been examined using knockout mice. Bin1+12 expression in the brain is critically dependent on amphiphysin 1 expression. Brain extracts of amphiphysin 1-deficient mice contained normal levels of Bin1+12 (with or lacking exons 6a and 13) transcript, but were almost devoid of Bin1+12 protein. This suggests that in the brain only amphiphysin 1/Bin1+12 heterodimers are stable. It is likely that Bin1+12 is subject to proteolysis when amphiphysin 1 is absent. The loss of Bin1+12 expression in the brain of amphiphysin 1 knockout mice is specific. No reduction in expression level in the brain was observed for other synaptic proteins that interact with amphiphysin 1. Moreover, the loss of Bin1 expression is brain specific because Bin1+10 was expressed normally in the muscle of amphiphysin 1 knockout mice (amphiphysin 1 is not expressed in muscle). Despite the loss of both amphiphysin 1 and Bin1+12, amphiphysin 1 knockout mice exhibit relatively mild defects in synaptic vesicle recycling that become apparent only under conditions of continuous nerve stimulation (55).
Mice homozygous for a deletion of the BIN1 gene that encodes Bin1 exhibit postnatal inviability. Although Bin1+6a+12 is highly expressed in the brain, the brains of homozygous Bin1 knockout mice appear to possess normal structures. Interestingly, amphiphysin 1 expression in the brain is not dependent on Bin1. Hence, while in yeast each Rvs protein is unstable in the absence of the other, in mouse brain only Bin1+6a+12+13 is unstable in the absence of its partner. As expected from analysis of homozygous amphiphysin 1 knockout mice that also lack Bin1 in the brain, homozygous Bin1 knockout mice also show no severe endocytic defects in the brain. Cultured Bin1-deficient neurons exhibit normal neurite outgrowth and form synapses in vitro. There is no obvious accumulation of clathrin-coated pits or vesicles at these synapses. Bin1-deficient embryonic fibroblasts express no detectable Bin1 isoforms but grow and divide as rapidly in culture as wild-type embryonic fibroblasts, endocytose fluorescein isothiocyanate-conjugated transferrin with kinetics even more rapid than that of wild-type embryonic fibroblasts, and exhibit a normal actin cytoskeleton that includes normal actin stress fibers (207).
The reason for the apparent discrepancy between the results obtained by transient overexpression or microinjection experiments in cultured cells and the results obtained by gene knockout in mice is not yet clear. It is possible that loss of endocytic function is only observed transiently upon acute perturbation of Bin1+6a+12 function. In knockout cells that have been without Bin1 expression for some time, long-term adaptation processes that compensate for loss of Bin1may have been activated. Alternatively, in the transient overexpression or microinjection experiments there may have been effects that were not limited to perturbation of Bin1+12 alone. These nonspecific effects, rather than loss of Bin1+12 interactions, may have been the cause of the observed endocytic defect.
Bin1+12 may function in postinternalization transport through endosomes.
While attention has mainly focused on the possible role of Bin1+12 isoforms in endocytosis at clathrin-coated pits on the plasma membrane, one study found that Bin1+6a+12+13 binds sorting nexin 4 (SNX4), a protein that localizes predominantly to intracellular compartments. Interaction is mediated by the C-terminal part of the Bin1+6a+12+13 BAR domain (BAR-C) and a short sequence at the extreme C terminusof SNX4 (167). Interestingly, the amino acid sequence of SNX4 suggests that SNX4 may possess a C-terminal BAR domain such as SNX1 (25). If so, the Bin1+6a+12+13-interacting sequence would lie within the SNX4 BAR domain. A pool of Bin1+6a+12+13 localizes to SNX4-containing compartments that contain internalized transferrin and are therefore a type of early endosome. Hence, Bin1+12 isoforms may not only function in the internalization step, but may also direct postinternalization traffic through endosomes (167).
Bin1+10+13/Bin1
Identification of Bin1+10+13/Bin1.
c-Myc is an important regulator of cell proliferation versus terminal differentiation decisions and is mutated in many human cancers. Nondividing cells do not express c-Myc, but after mitogenic stimulation c-Myc is one of the first proteins to be induced before cells enter the cell cycle. The role of c-Myc in cell proliferation is highlighted by recent studies using Drosophila c-Myc. Overexpression of Drosophila c-Myc in select cells in a tissue induces these cells to proliferate at the expense of surrounding low-expressor cells in the same tissue, i.e., to become “supercompetitors.” Indeed, in these competition experiments the low-expressor cells are not only overgrown by the high-expressors but undergo apoptosis (199). However, high c-Myc expression is not always advantageous. When mammalian cells in culture are serum starved c-Myc expression is down-regulated and cells stop dividing and become quiescent. Ectopic c-Myc expression in serum-starved cells prevents the cells from becoming quiescent. c-Myc-overexpressing cells that continue to divide under serum-starved conditions eventually undergo c-Myc-dependent apoptosis (278).
c-Myc has been shown to shuttle between the cytoplasm and the nucleus and can act as a transcriptional activator or repressor. The N-terminal domain of c-Myc is a transcription activation/repression domain and the C-terminal domain is a DNA binding domain. The activity of c-Myc both in oncogenic transformation and in apoptosis requires two conserved N-terminal domains known as Myc boxes 1 (MB1) and 2 (MB2). MB1 is positioned within the transcriptional activation domain of c-Myc and is subject to cell cycle-dependent phosphorylation on residues T58 and S62. MB1 is critically important for control of cell proliferation since most c-Myc mutations in primary human tumors and cultured tumor cell lines map to the MB1 domain. The observation that MB1 is mutated in tumors suggests a negative regulator of c-Myc may interact via the MB1 domain and mutation of MB1 may result in loss of this interaction and deregulation of c-Myc activity (278).
A Bin1 splice variant (referred to here as Bin1+10+13) was isolated in a two-hybrid screen with the c-Myc MB1 domain as the bait and named Box-dependent Myc-interacting protein (or subsequently bridging integrator) 1 (Bin1) (Fig. 1 and Table 2) (276, 278). As predicted for a negative regulator of c-Myc, expression of Bin1+10+13 inhibited the ability of c-Myc to induce oncogenic transformation in culture (69, 278).
Domain structure of Bin1+10+13.
In comparison to Bin1+6a+12 (which can either have or lack exon 13), Bin1+10+13 lacks the NTID domain encoded by exon 6a that mediates the formation of heterodimers with amphiphysin 1 and plasma membrane targeting (Fig. 1 and Table 2). It also lacks the central insert domain encoded by exon 12 that in some isoforms of Bin1+12 (with or lacking exons 6a and 13), depending on the choice of exon 12, binds clathrin heavy chain, α-adaptin, and/or endophilin A1 (Fig. 1 and Table 2). Bin1+10+13 contains, however, an additional 15-residue sequence encoded by exon 10 that includes a nuclear localization sequence and lipid binding sequence that is absent in all variants of Bin1+12 (with or lacking exons 6a and 13). Bin1+10+13 and some splice variants of Bin1+12 (with or lacking exon 6a) contain a complete c-Myc binding sequence, a third of which is encoded by exon 13 (Fig. 1 and Table 2). In this way Bin1+10+13 and Bin1+6a+12+13 differ from ALP1/amphiphysin IIm, which lack the sequences encoded by exon 13 (see below) (Fig. 1 and Table 2) (23, 168, 256, 257, 278, 338, 356, 357, 367). Hence, according to the nomenclature of Wechsler-Reya et al., this original Bin1 isoform is named Bin1+10+13 (356).
Interaction of Bin1+10+13 with c-Myc.
The interaction of Bin1+10+13 with c-Myc was originally identified by two-hybrid screens but has been confirmed by protein binding in vitro and is direct (278). c-Myc interacts with a central domain of Bin1+10+13 located between the N-terminal BAR domain and the C-terminal SH3 domain and this domain has been named the Myc-binding domain (MBD, residues 270 to 377). A recent study, however, found that the SH3 domain of Bin1+10+13 mediates binding to c-Myc (245). Bin1+10+13 binding to c-Myc in vitro requires both MB1 and MB2 on c-Myc, although in the two-hybrid assay MB1 is sufficient (278).
An initial study did not observe coimmunoprecipitation of c-Myc and Bin1+10+13 from cell extracts, suggesting these proteins may not exist in a stable complex in vivo (278). Prior to differentiation into myotubes, proliferating C2C12 cells express both c-Myc and Bin1+10+13, however, after differentiation c-Myc expression is lost and Bin1+10+13 expression is dramatically elevated (357). A subsequent study using proliferating C2C12 cells found endogenous Bin1+10+13 and c-Myc do coimmunoprecipitate from cell extracts and hence do form stable complexes in vivo, at least in this cell type. Bin1+10+13 also coimmunoprecipitates with c-Myc from extracts prepared from insect Sf9 cells ectopically expressing both proteins (69).
Bin1+10+13 is a tumor suppressor.
Bin1+10+13 is a negative regulator of cell cycle progression and plays a role in exit from the cell cycle in response to serum deprivation. The ability of Bin1+10+13 to suppress oncogenic transformation by c-Myc demonstrates that Bin1+10+13 has properties characteristic of a tumor suppressor. Moreover, Bin1+10+13 may be part of an important mechanism preventing oncogenic transformation and tumorigenesis in vivo, since the domain of c-Myc that mediates Bin1+10+13 binding (MB1) is frequently mutated in tumor cells. Moreover, the human and murine BIN1 genes map to chromosomal loci that are hotspots for deletion in tumors and Bin1+10+13 expression is absent in a high percentage of tumor cell lines and primary breast and other tumors. Furthermore, ectopic expression of Bin1+10+13 in tumor cells that have lost endogenous Bin1+10+13 expression causes arrest of cell proliferation. This strongly supports the idea that loss of Bin1+10+13 expression contributes significantly to tumorigenesis and is not simply a consequence of genetic instability in tumor cells (278).
This role of Bin1+10+13 as a negative regulator of cell cycle progression is reminiscent of the role of the yeast Rvs proteins (12, 42). While Bin1+10+13 plays a role in exit of serum-starved mammalian cells from the cell cycle (278), the Rvs proteins play a role in exit of nutrient-starved yeast cells from the cell cycle (12). Exit from the cell cycle and arrest in G0 are in turn important for prevention of c-Myc-induced apoptosis in mammalian cells and for survival under starvation conditions in yeast cells. Bin1+10+13 and Rvs167p are not functionally interchangeable, however, as Bin1+10+13 expression in rvs167Δ yeast mutants does not rescue the phenotype (278). Rvs167p also lacks the MBD found in Bin1+10+13 (Rvs167p has the GPA-rich region in place of the MBD) (Fig. 1). In addition, yeast does not have an ortholog of mammalian c-Myc.
Is interaction of Bin1+10+13 with c-Myc important for suppression of oncogenic transformation? Deletion of the MBD abolishes the ability of Bin1+10+13 to suppress oncogenic transformation by c-Myc. Moreover, expression of a fragment of Bin1+10+13 comprising only the MBD interferes in a dominant negative manner with the ability of full-length Bin1+10+13 to suppress oncogenic transformation by c-Myc. This suggests binding of Bin1+10+13 to c-Myc is physiologically important. Interestingly, mutations in MB1 that do not appear to affect Bin1+10+13 interaction but that prevent MB1 phosphorylation (e.g., T58M) also abolish suppression activity. This shows that Bin1+10+13 binding to MB1 is necessary but not sufficient for c-Myc suppression (69, 278).
Another Bin1+10+13 domain essential for suppression of oncogenic transformation by c-Myc is the C-terminal half of the BAR domain (BAR-C) (Fig. 1). Deletion of the BAR-C domain causes partial redistribution of Bin1+10+13 from the nucleus to the cytoplasm. Moreover, the BAR-C domain contains a nuclear localization sequence. The effect of the BAR-C deletion shows that this nuclear localization sequence, rather than that encoded by exon 10, is the critical sequence for Bin1+10+13 localization to the nucleus. Deletion of Bin1+10+13 domains other than the MBD and BAR-C domains had little or no effect on suppression of c-Myc transformation. The ability of Bin1+10+13 to inhibit proliferation of tumor cells that lack endogenous Bin1+10+13 (e.g., HepG2) is critically dependent on the presence of BAR-C. In contrast, despite the importance of the MBD in suppression of oncogenic transformation by c-Myc, Bin1+10+13 lacking the MBD retains significant ability to inhibit tumor cell proliferation, suggesting this activity may be partially independent of c-Myc binding (69).
Further insight into the role of Bin1+10+13 in suppression of oncogenic transformation by c-Myc came from a study of a Bin1 splice variant that lacks exon 10 but contains neuron-specific exon 12 (Bin1+12+13) and whose expression in nonneuronal cells correlates with oncogenesis (93, 245, 356). A highly sensitive protein interaction assay revealed that many Bin1 isoforms, including those that lack the MBD encoded in part by exon 13, do in fact interact with c-Myc. In this study the isolated MBD was not sufficient to bind c-Myc in vitro. The novel c-Myc interaction involves the C-terminal SH3 domain of various Bin1 isoforms interacting with a proline-rich motif positioned within the MB1 domain of c-Myc. This interaction is regulated by phosphorylation of S62 within MB1.
Intriguingly, Bin1+12+13 differs from all other Bin1 isoforms tested in that its SH3 domain is unable to bind c-Myc. The explanation comes from the finding that exon 12 contains a proline-rich motif. When exon 12 is present the SH3 domain engages in an intramolecular interaction with exon 12 that prevents its engagement with c-Myc (245).
The effect of Bin1+10+13 may be on the transcriptional activity of c-Myc. c-Myc stimulates transcription of various genes, including those encoding ornithine decarboxylase and α-prothymosin and also stimulates transcription of minimal viral promoters that have been engineered to include upstream c-Myc binding elements. The ability of c-Myc to activate transcription from each promoter was inhibited by a construct expressing Bin1+10+13. Deletion of the Bin1+10+13 MBD abolished its ability to inhibit c-Myc-dependent activation of some promoters (e.g., ornithine decarboxylase) but not other promoters (e.g., α-prothymosin). Hence, Bin1+10+13 inhibits c-Myc-dependent transcriptional activation but does so by distinct mechanisms, only one of which requires binding of c-Myc via the MBD.
The effect of Bin1+10+13 on transcriptional activation is specific to c-Myc. Bin1+10+13 does not inhibit transcriptional activation by other proteins, e.g., herpesvirus VP16. The N-terminal domain of c-Myc that contains MB1 and MB2 confers inhibition of transcriptional activation by Bin1+10+13, as fusion of this domain to the yeast Gal4p transcriptional activator results in the ability of Bin1+10+13 to inhibit transcriptional activation of Gal4p-specific promoters (69).
How does Bin1+10+13 inhibit transcriptional activation by c-Myc? Initially, it was proposed that Bin1+10+13 binding to MB1 and MB2 acts sterically to inhibit the ability of MB1 and MB2 to activate transcription at c-Myc-dependent promoters (278). Subsequently, it was suggested that Bin1+10+13 recruits a transcriptional repressor to c-Myc. Fusion of Bin1+10+13 to Gal4p is sufficient to repress transcription at a Gal4p-dependent promoter and this repression can be relieved by coexpression of unfused Bin1+10+13, perhaps via titration of a transcriptional repressor. Interestingly, the Bin1+10+13 MBD is not required either for repression of Gal4p-dependent transcription or for relief of this repression. Hence, the putative repressor must bind a domain of Bin1+10+13 distinct from the MBD. The nature of the repressor recruited by Bin1+10+13 is not known (69).
Bin1+10+13 activity in tumor suppression is not restricted to effects on c-Myc. Bin1+10+13 also suppresses oncogenic transformation by the unrelated adenovirus E1A oncoprotein and by a dominant negative mutated form of the tumor suppressor p53. The mechanism by which Bin1+10+13 suppresses transformation by these oncoproteins appears distinct, however, as neither the MBD nor the BAR-C domain is essential. Moreover, the Bin1+10+13 MBD does not bind E1A in vitro. A short sequence (U1) encoded by exon 9 is essential for suppression of both adenovirus E1A and mutant p53, while the SH3 domain is also essential for suppression of mutant p53. Adenovirus E1A and p53 both function in the nucleus, and hence the partial loss of nuclear localization of Bin1+10+13 caused by deletion of the BAR-C domain may not fully account for loss of c-Myc suppression. Bin1+10+13 does not inhibit oncogenic transformation by all oncoproteins, however. For example, Bin1+10+13 has no effect on oncogenic transformation by the simian virus 40 large T antigen (69, 278). Hence, the mechanisms by which Bin1+10+13 inhibits oncogenic transformation and tumorigenesis are specific.
The frequency with which human tumors either lack Bin1 expression or misexpress Bin1+12+13 together with the ability of ectopic Bin1+10+13 expression in tumor cells lacking endogenous Bin1+10+13 to inhibit proliferation suggests that Bin1 plays a critical role in tumor suppression in vivo (69, 245, 278). Further support for an important role for Bin1+10+13 in tumor suppression in vivo comes from analysis of the neoplastic potential of murine embryonic fibroblasts (MEFs) from homozygous Bin1 knockout mice. Bin1 knockout MEFs, but not wild-type MEFs, cotransfected with c-Myc and a mutant form of Ras exhibited morphological features of oncogenically transformed cells. Coexpression of c-Myc and mutant Ras in Bin1 knockout MEFs, but not wild-type MEFs, also led to an increased ability to form tumors in mice. This effect was specific to c-Myc, as Bin1 knockout MEFs transfected with viral oncogenes did not exhibit increased tumor formation (209).
Bin1+10+13 also prevents tumor cells from being recognized and killed by anti-tumor T cells. Bin1+10+13 has recently been shown to be a key regulator of the immunoregulatory enzyme indoleamine 2,3-dioxygenase (IDO). Overexpression of IDO in Bin1 knockout cells may be a major factor in immune escape and accelerated progression of tumor cells, as treatment of tumor-bearing mice with IDO inhibitors helps induce tumor regression (208).
Bin1+10+13 induces apoptosis specifically in tumor cells.
One mechanism by which Bin1+10+13 may suppress tumors is by inducing apoptosis (programmed cell death). Ectopic expression of Bin1+10+13 in the cultured tumor cell line HepG2 (which has lost endogenous Bin1+10+13 expression) induces cell death via activation of an apoptotic pathway. This apoptotic pathway involves reduction of cell volume, loss of adherence, accumulation of cytoplasmic vacuoles, and DNA degradation. This apoptotic role of Bin1+10+13 in tumor cells requires the BAR-C domain. Apoptosis and DNA degradation are induced independently of cell cycle stage. This apoptotic mechanism does not require the tumor suppressor p53 or retinoblastoma protein, caspases, Bcl-2, or Fas-dependent apoptotic signaling. Inhibition of the apoptotic pathway induced by c-Myc upon serum withdrawal also inhibits DNA degradation induced by Bin1+10+13, suggesting some apoptotic effects of Bin1+10+13 are mediated via mechanisms common to c-Myc-dependent apoptosis (68).
The role of Bin1+10+13 in c-Myc-dependent apoptosis has been tested in vivo using homozygous Bin1 knockout mice. Ectopic expression of c-Myc in primary homozygous Bin1 knockout MEFs still induced apoptosis upon serum withdrawal (207). This is perhaps not unexpected, since Bin1+10+13 has only been shown to be important for induction of apoptosis in oncogenically transformed cells (e.g., HepG2) (68). Ectopic expression of c-Myc in homozygous Bin1 knockout MEFs is insufficient to induce oncogenic transformation (207).
Subcellular localization of Bin1+10+13.
The subcellular localization of Bin1+10+13 has been an area in which different findings have been reported. Bin1+10+13 was initially reported to exhibit a nuclear localization when ectopically expressed in HepG2 hepatocarcinoma cells (278). This localization is consistent with the ability of Bin1+10+13 to interact both physically and functionally with the nuclear transcriptional activator c-Myc and the presence of a nuclear localization sequence encoded by exon 10 (69, 278). Examination of endogenous Bin1+10+13 in a collection of human and rodent Bin1+10+13-expressing tumor cell lines showed again that Bin1+10+13 localizes to the nucleus and in part to a subnuclear compartment. In some tumor cells Bin1+10+13 localizes entirely to the subnuclear compartment (355). In another study the expression level and subcellular localization of Bin1+10+13 in various normal human tissues were compared and differences were observed between tissues (e.g., bone marrow, breast, and intestine). Within intestinal epithelia, an expression gradient of Bin1+10+13 was observed, with strongest expression in those cells located at the tips of intestinal projections (villi) that are committed to undergo apoptosis (61).
Subsequently, two studies found Bin1+10+13 is predominantly nonnuclear in muscle cells, where this isoform is endogenously expressed at high levels (23, 162). When transiently expressed in COS-7 cells, both brain Bin1+6a+12+13 and muscle Bin1+10+13 were found to localize exclusively to the cytoplasm. Even when ectopically expressed in HepG2 cells the localization of Bin1+10+13 was predominantly cytoplasmic. The reason for the different subcellular localizations reported for Bin1+10+13 is not known. It has been suggested that Bin1+10+13 may shuttle between the cytoplasm and the nucleus (23). This process may be regulated in response to some yet-to-be-identified environmental signal.
During differentiation of C2C12 myoblasts into myotubes in vitro there is a switch of splice variant from Bin1−10 to Bin1+10 (each with or lacking exon 13) (357). Both the former (including SH3p9 and ALP1/amphiphysin IIm) and the latter have been reported to be present strictly in the cytoplasm, strictly in the nucleus, or distributed between the cytoplasm and nucleus in different studies (23, 69, 141, 162, 278, 355). Hence, while exon 10 encodes a predicted nuclear localization sequence, this exon is not required for nuclear localization of Bin1. As mentioned above, a second nuclear localization sequence that is critical for nuclear localization is present in the BAR-C domain (69). It is worth noting that the antibodies used for immunofluorescence localization of Bin1+10 may not have distinguished between those splice variants that include exon 10 sequences and those that lack exon 10 sequences. The domain encoded by exon 10 is required for the plasma membrane binding and tubulating activity of Bin1+10+13 (see below) (162). Perhaps this property is responsible for a shift from a nuclear to a cytoplasmic distribution when this exon is incorporated.
Stability of Bin1+10+13 in vivo.
The in vivo stability of Bin1+10+13 has been examined by pulse-chase radiolabeling and Bin1+10+13 is rather unstable in cells, with a half-life of only about 2 h (355). This is interesting in view of recent evidence that Rvs167p in yeast interacts with a ubiquitin protein ligase (Rsp5p, the yeast ortholog of Nedd4) and is subject to ubiquitination (307), and that Rvs167p is rapidly degraded in the absence of Rvs161p (180).
Phosphorylation of Bin1+10+13.
Like amphiphysin 1 and other splice variants of Bin1, Bin1+10+13 is phosphorylated in vivo (355). The role of this phosphorylation, the sites phosphorylated, and the kinase(s) responsible are not yet known. Interestingly, Bin1+10+13 lacks a region of homology to the proline-rich sequences in the central insert domain of amphiphysin 1 and GPA-rich domain of Rvs167p that are subject to phosphorylation by Cdk5/Pho85p (78, 85, 330). Therefore, phosphorylation of Bin1+10+13 may have some unique features compared to that of amphiphysin 1 or yeast Rvs167p.
Role for Bin1+10+13 in muscle differentiation.
Several findings suggested that Bin1+10+13 may play a role in muscle differentiation: Bin1+10+13 is a negative regulator of cell cycle progression, it is encoded by a transcript that undergoes differential splicing during differentiation of proliferating C2C12 myoblasts into terminally differentiated myotubes, and it is highly expressed in adult skeletal muscle. Moreover, Bin1+10+13 was identified in a large-scale screen of muscle-specific proteins as a protein required for muscle cell differentiation. The murine skeletal muscle-derived cell line C2C12 can be induced to differentiate into muscle in culture. Confluent C2C12 myoblasts subjected to serum withdrawal in culture undergo dramatic elongation and then lateral cell-cell fusion to form huge multinucleate myotubes similar to those found in muscle. When Bin1+10+13 is down-regulated differentiation of C2C12 myoblasts into myotubes upon serum withdrawal is blocked (172).
To investigate the role of Bin1+10+13 in muscle cell differentiation its expression during C2C12 differentiation in culture was examined. Bin1+10+13 transcript and protein levels were observed to increase upon serum withdrawal concomitant with terminal differentiation and cell-cell fusion into myotubes. c-Myc is a marker of proliferating cells. As expected, c-Myc expression levels declined after serum withdrawal as cells exited the cell cycle and became committed to terminal differentiation. Interestingly, higher-molecular-weight Bin1 protein species appear during differentiation. These forms arise by altered transcript splicing such that exon 10 is included. Bin1 in undifferentiated C2C12 cells lacks sequences encoded by exon 10 and localizes predominantly to the nucleus. In contrast, the form of Bin1 found in terminally differentiated C2C12 myotubes contains sequences encoded by exon 10 and forms a filamentous network in the cytoplasm (357).
Expression of Bin1+10+13 has also been examined during embryonic development in mice (182). By embryonic day 10.5 expression of Bin1+10+13 has already been induced in myotomes, which are the precursors of skeletal muscle cells. Bin1+10+13 appears to regulate cell cycle progression via induction of the Cdk inhibitor p21WAF1. Down-regulation of Bin1+10+13 in C2C12 cells prevents induction of p21WAF1 and hence interferes with cell cycle arrest upon serum withdrawal. Hence, Bin1+10+13 functions at a very early stage in differentiation of C2C12 myoblasts into myotubes (182).
Overexpression of human Bin1+10+13 slows the proliferation of C2C12 cells in culture and promotes differentiation into myotubes. A decrease in the rate of cell proliferation is observed even in the presence of serum, however, differentiation still requires serum withdrawal. Lowering Bin1+10+13 expression increases the rate of cell proliferation in the presence of serum and also abolishes differentiation upon serum withdrawal. C2C12 cells in which expression of Bin1+10+13 is down-regulated do not elongate, nor do they undergo cell-cell fusion to form multinucleate myotubes. Induction of muscle myosin II is normally observed in C2C12 cells prior to cell-cell fusion, however, after Bin1+10+13 down-regulation serum withdrawal no longer induces myosin II expression. The ability of Bin1+10+13 overexpression to slow cell proliferation is dependent on the presence of exon 10. In contrast, exon 10 is not essential for differentiation. Accelerated differentiation of C2C12 myoblasts occurs upon serum withdrawal even in cells expressing a form of Bin1+10+13 in which exon 10 is deleted. Hence, increased expression of Bin1+10+13 (although not exon 10) is essential for differentiation of C2C12 myoblasts into myotubes in culture (162, 357).
A role for Bin1+10+13 in cell differentiation is consistent with aspects of the function of the Rvs161p and Rvs167p proteins in yeast. In budding yeast two cell differentiation pathways have been described, sporulation and pseudohyphal growth. The sporulation of vegetative diploid cells to produce haploid spores is dependent on the function of both Rvs161p and Rvs167p (10, 12, 38, 53). This suggests a possible evolutionarily conserved role in cell differentiation. Furthermore, the importance of Bin1+10+13 expression for the cell-cell fusion of myoblasts to form multinucleate myotubes is reminiscent of the role of Rvs161p/Fus7p and Rvs167p in fusion of haploid yeast cells during mating (20, 21, 91). Mating leads initially to the formation of cells with two haploid nuclei (multinucleate), however, subsequent nuclear fusion results in a single diploid nucleus. Fusion of myoblasts requires elevated expression of type II conventional muscle myosin and this is dependent on Bin1+10+13 (357). It is interesting that in yeast, of all the myosins, the strongest genetic interactions of RVS161 and RVS167 are with MYO2, which encodes the sole conventional type II myosin (Myo1p) (19).
Bin1+10+13 localization in adult skeletal muscle.
In skeletal muscle, Bin1+10+13 localizes to distinctive transverse bands that appear with regular periodicity along the muscle fiber. The Bin1+10+13-specific bands were compared with known bands of muscle fiber and found to be within the broad actin filament-containing I-band and positioned on either side of the thinner desmin-containing Z-line. The submembranous cytoskeletal protein ankyrin 3 localizes to what appear to be the same bands as Bin1+10+13 within the muscle fiber (23). Ankyrin 3 is a specific marker for invaginations of the muscle cell plasmalemma known as transverse (T)-tubules (79). That Bin1+10+13 localizes to T-tubules in muscle was confirmed by colocalization with another T-tubule-specific marker, triadin (108), and immunoelectron microscopy of ultrathin skeletal muscle frozen sections (23). Interestingly, like axon initial segments and nodes of Ranvier in the brain, T-tubules in muscle bear on their cytoplasmic face a dense cytomatrix and it is to this cytomatrix to which ankyrin 3 localizes. Bin1+10+13 may function within this cortical cytomatrix to regulate traffic to and from T-tubules and may play a role in defining these specialized membrane subdomains (23, 79, 162).
T-tubules are membrane tubules that extend from the plasmalemma to form an extensive intracellular membrane reticulum. This T-tubule membrane reticulum is closely associated with the bundles of muscle fibers within muscle cells. T-tubules have been proposed to play a key role in passing the Na+/K+ action potential at the cell surface to each of the muscle fibers located in the cytoplasm. This propagation of the action potential ensures that the response in muscle fibers is coordinated and synchronous and results in efficient depolarization-contraction coupling. Moreover, T-tubules are enriched in integral membrane proteins that function in ion transport, which suggests a possible role for Bin1+10+13 in regulation of ion fluxes across membranes. T-tubules may also be sites of endocytosis in skeletal muscle. Clathrin heavy chain, like Bin1+10+13, localizes to the I-band. The localizations are not identical, however, as clathrin heavy chain localizes to a region between Bin1+10+13 and the Z-line. The insulin-regulated plasma membrane glucose transporter Glut4 is a protein that undergoes clathrin-dependent endocytosis. Glut4 localization in skeletal muscle extensively overlaps that of both clathrin heavy chain and Bin1+10+13 (23).
Interestingly, in the fruit fly Drosophila melanogaster the single amphiphysin ortholog is enriched in postsynaptic neuromuscular junctions in larvae. In contrast, Drosophila amphiphysin expression is low or absent in the central nervous system. Flies in which the amphiphysin gene is disrupted are viable and exhibit normal synaptic vesicle recycling and neurotransmission, but display severe locomotory defects in both larvae and adult flies (e.g., the adults cannot fly). The function of the neuromuscular junction appears normal. Amphiphysin localizes to T-tubules in muscle and amphiphysin-deficient flies have a disorganized T-tubule system. This suggests the locomotory defects in amphiphysin-deficient flies result from inefficient excitation-contraction coupling in which the T-tubule plays a critical role. Moreover, although Drosophila amphiphysin has both an N-terminal BAR domain and C-terminal SH3 domain highly conserved with mammalian amphiphysin 1 and Bin1 isoforms, the central insert domain of mammalian amphiphysin 1 and Bin1+12 that interacts with clathrin heavy chain and AP-2 is not well conserved in Drosophila amphiphysin. Indeed, Drosophila amphiphysin does not bind clathrin heavy chain (169, 258, 259, 386). Hence, Drosophila amphiphysin resembles mammalian Bin1+10+13.
Bin1+10+13 and T-tubule formation in muscle.
Although endogenous amphiphysin 1 and both neuronal Bin1+6a+12 (with or lacking exon 13) and muscle Bin1+10+13 localize to the cortical cytomatrix, they differ in their ability to localize to the cortex when ectopically expressed in CHO cells. Amphiphysin 1 and neuronal Bin1+6a+12+13 both exhibit a predominantly diffuse localization throughout the cytoplasm. In contrast, muscle Bin1+10+13 concentrates at the cytoplasmic face of the plasma membrane (162).
Ectopic expression of muscle Bin1+10+13, but not amphiphysin 1 or neuronal Bin1+6a+12+13, in CHO cells that normally lack T-tubules leads to the dramatic appearance of tubules that resemble T-tubules. These tubules are contiguous with the plasma membrane and are enriched in PtdIns(4,5)P2. The ability of Bin1+10+13 to induce membrane tubules in nonmuscle cells requires the BAR domain and the 15-residue muscle-specific domain encoded by exon 10 (Fig. 1) (162). This domain is highly basic with the sequence RKKSKLFSRLRRKKN and contains a putative nuclear localization sequence originally thought to confer nuclear localization on Bin1+10+13 (278). Full-length Bin1+10+13 is able to recruit a muscle isoform of dynamin (dynamin 2) to the induced tubules in CHO cells. Although the Bin1+10+13 BAR domain and exon 10-encoded domain are sufficient to induce tubules in CHO cells, the SH3 domain must also be present for recruitment of dynamin 2 to the tubules. The tubules induced by the BAR domain and exon 10-encoded domain also appear to lack surrounding cytomatrix, in contrast to the tubules induced by full-length Bin1+10+13 (162).
The role of endogenous Bin1+10+13 in T-tubule formation in muscle cells has been investigated using the myoblast cell line C2C12. During serum starvation-induced differentiation of C2C12 cells, expression of both Bin1+10+13 and caveolin 3 increases and both proteins localize to tubules that are contiguous with the plasma membrane and enriched in PtdIns(4,5)P2. Mature T-tubules are narrow tubules containing Bin1+10+13 but not caveolin 3. However, newly formed T-tubules resemble a series of vesicular structures connected by narrow tubules. Caveolin-3 is concentrated at the vesicular elements and Bin1+10+13 at the interconnecting tubular elements. Hence, Bin1+10+13 appears to play an important in vivo role in deforming membranes to generate narrow tubules, consistent with its ability to bind and deform membranes in vitro (see below) (162).
Further support for a role of Bin1+10+13 in muscle differentiation has come from studies of homozygous Bin1 knockout mice that lack Bin1+10+13. Homozygous Bin1 knockout mice appear to undergo normal embryonic development and are represented at the expected Mendelian frequency at birth (207). However, the homozygous Bin1 knockout mice do not feed and die in the first 24 h after birth. Anatomical characterization of homozygous Bin1 knockout mice revealed obvious cardiac myopathies, in particular a ventricular wall so severely expanded that both ventricular chambers of the heart were occluded. The ventricular cardiomyocytes are packed more tightly than those in wild-type mice, but do not appear to undergo abnormal proliferation. In ventricular cardiomyocytes of wild-type mice Bin1+10+13 is localized to the nucleus (207). This is in contrast to skeletal muscle cells, where Bin1+10+13 localizes to T-tubules that line myofibrils in the cytoplasm (23, 162).
Ultrastructural examination shows that the myofibrils in the ventricular cardiomyocytes are not arranged in closely packed regular arrays in Bin1+10+13-deficient cardiomyocytes as they are in normal cardiomyocytes. T-tubules are normally positioned either side of the thin desmin-containing Z-line in muscle. In Bin1+10+13-deficient ventricular cardiomyocytes the Z-line is more diffuse than in normal cardiomyocytes, suggesting a possible defect in the T-tubule system. Although skeletal muscle is present and gross abnormalities are not apparent in homozygous Bin1 knockout mice, ultrastructural examination reveals that the myofibrils in skeletal muscle cells also appear less well organized than in skeletal muscle from wild-type mice. In addition, the T-tubules appear to be shifted to the Z-line rather than being aligned on either side of it as in normal skeletal muscle (207). Hence, Bin1+10+13 is not essential for muscle development, but is important for T-tubule organization in cardiomyocytes and, to a lesser extent, in skeletal muscle.
Bin1+10+13 and lipid rafts.
The membranes of T-tubules have a distinctive lipid composition, being enriched in sterols and glycosphingolipids (26). Treatment of C2C12 cells with agents such as amphotericin B and methyl-β-cyclodextrin, which deplete cholesterol from membranes, interferes with Bin1+10+13 distribution and prevents T-tubule formation. The same treatments also prevent the induction of membrane tubules in CHO cells upon ectopic expression of Bin1+10+13, suggesting these tubules are also enriched in cholesterol (162). During muscle differentiation the lipid raft marker caveolin-3 localizes to T-tubules (26). Caveolins are a family of proteins that localize to flask-shaped, lipid-raft-enriched cell surface invaginations known as caveolae. Caveolae have been shown to function in endocytosis and in signal transduction (113, 159). As caveolae and T-tubules are both lipid-raft-enriched invaginations of the plasma membrane and newly forming T-tubules contain caveolin-3, it has been proposed that T-tubules may form in the same way as caveolae (26). Interestingly, in CHO cells the endogenous isoform of caveolin (caveolin 1) localizes to Bin1+10+13-induced tubules (162). The localization of Bin1+10+13 to lipid-raft-enriched T-tubules in muscle is interesting in light of the observation that Rvs161p and Rvs167p in yeast fractionate with detergent-insoluble lipid rafts (10, 97).
Bin1+10+13 binds and tubulates liposomes in vitro.
Recombinant Bin1+10+13 has been shown to bind liposomes in vitro via direct interaction with lipids. When incubated in vitro with liposomes recombinant Bin1+10+13 generates narrow membrane tubules by a process of evagination (162). This is similar to what was shown earlier for recombinant amphiphysin 1 in vitro (316). The lipid composition of the liposomes influences the efficiency of binding of Bin1+10+13. The presence of PtdIns(4,5)P2 strongly enhances binding and PtdIns(4)P has a similar but slightly weaker effect. While the Bin1+10+13 N-terminal amphipathic α-helix and BAR domain are sufficient for binding to liposomes in vitro, enhancement of binding by PtdIns(4,5)P2 or PtdIns(4)P is dependent on the domain encoded by exon 10. Neither PtdIns(4,5)P2 nor PtdIns(4)P affect binding of fragments comprising the N-terminal amphipathic α-helix and BAR domains of either amphiphysin 1 or Bin1+6a+12+13 to liposomes (162). Drosophila amphiphysin also has the ability to evaginate tubules from liposomes composed of brain lipids in vitro, showing that this property has been conserved during evolution (258).
The importance of the Bin1+10+13 BAR domain for liposome binding in vitro is consistent with the results of in vivo studies. The Bin1+10+13 BAR domain and the sequence encoded by exon 10 are both essential for ectopically expressed Bin1+10+13 to localize to the plasma membrane in CHO cells. The plasma membrane is the major site of PtdIns(4,5)P2 in cells. Hence, PtdIns(4,5)P2 binding may be important for Bin1+10+13 localization and membrane tubulation in vivo (162).
Bin1−10−12+13/SH3p9
A splice variant of Bin1 that lacks the putative nuclear localization sequence and sequences required for lipid binding encoded by exon 10 compared to muscle Bin1+10+13 but that still retains the central third of the MBD encoded by exon 13 was identified in a screen for novel SH3 domain proteins and named SH3p9 (Fig. 1) (306). SH3p9 is referred to here as Bin1−10−12+13. This Bin1 isoform is expressed in many tissues, although it may be absent in some specialized cell types such as macrophages (100) (see below). The function of Bin1−10−12+13 remains unknown.
Bin1−10−12−13/ALP1/Amphiphysin IIm
While Bin1 was identified as a c-Myc-interacting protein, a distinct splice variant was identified by its ability to bind another oncoprotein, c-Abl, and named Amphiphysin-Like Protein 1 (ALP1) (141). The same splice variant was identified as the sole amphiphysin and Bin1 isoform in macrophages and named amphiphysin IIm (100). Bin1−10−12−13 is the most ubiquitous splice variant of Bin1. This splice variant is distinct from the original Bin1 isoform (Bin1+10+13) because it lacks the 15-residue sequence containing the putative nuclear localization sequence and sequences required for lipid binding encoded by exon 10 and furthermore is distinct from both Bin1−10−12+13 (SH3p9) and Bin1+10+13 in that it lacks exon 13, which encodes the central third of the MBD (note that although the initial report by Kadlec et al. [141] described ALP1 as essentially identical to SH3p9 in lacking exon 13, ALP1 in fact differs from SH3p9 in lacking exon 13) (100, 141, 278, 306, 338, 357).
Despite lacking the putative nuclear localization sequence encoded by exon 10, when transiently expressed in NIH 3T3 fibroblasts Bin1−10−12−13 localizes to the nucleus in a high percentage of cells. However, in some transfected NIH 3T3 cells Bin1−10−12−13 is found predominantly in the cytoplasm (141). Nuclear import of Bin1−10−12−13 is presumably mediated by the alternative nuclear localization sequence present in the BAR-C domain, which is common to all Bin1 splice variants (69). What cellular mechanism regulates the nuclear/cytoplasmic distribution of Bin1−10−12−13 is unknown.
c-Abl is a nonreceptor tyrosine kinase that localizes to both the nucleus and the actin cytoskeleton, and mutations in c-Abl have the ability to transform cells. c-Abl and ALP1 coimmunoprecipitate from cotransfected cells, showing they associate in vivo. Moreover, in vitro protein binding experiments showed that the C-terminal SH3 domain of ALP1 mediates interaction and binds at least two distinct proline-rich motifs within the c-Abl C-terminal domain. The c-Abl SH3 does not bind ALP1. Unlike Bin1+10+13, Bin1−10−12−13 does not bind c-Myc in vitro, presumably due to the absence of MBD sequences encoded by exon 13 (141).
Bin1−10−12−13 in regulation of the actin cytoskeleton.
Transient coexpression of c-Abl and Bin1−10−12−13 induces a dramatic change in cell morphology in which actin stress fibers are lost and the cells adopt a more rounded and “spindly” appearance. High-level expression of c-Abl inhibits cell proliferation and coexpression of Bin1−10−12−13 does not reverse this inhibition. Bin1−10−12−13 expression does not activate c-Abl tyrosine kinase activity, however, induction of the morphological change by coexpression requires an intact c-Abl kinase domain as well as the Bin1−10−12−13 SH3 domain and the proline-rich motifs in c-Abl with which it interacts. The change in cell morphology induced by coexpression of c-Abl and Bin1−10−12−13 resembles those that accompany oncogenic transformation, but is not accompanied by oncogenic transformation. The altered BCR-Abl form of c-Abl is associated with leukemia. Expression of Bin1−10−12−13 somewhat increases the ability of BCR-Abl to oncogenically transform cells in culture (141). This is in contrast to Bin1+10+13, whose high-level expression suppresses oncogenic transformation by the oncoprotein c-Myc (278).
Bin1−10−12−13 functions in phagocytosis in macrophages.
Murine macrophages express Bin1−10−12−13 as their only Bin1 splice variant and this has been named amphiphysin IIm but is identical to ALP1 (100, 141). Like amphiphysin 1 and other splice variants of Bin1, Bin1−10−12−13 binds dynamin (dynamin 2 in macrophages) via its C-terminal SH3 domain. The mutant form of Bin1−10−12−13 that lacks an SH3 domain has dominant negative effects on receptor-mediated endocytosis of low-density lipoprotein (100).
Is Bin1−10−12−13 also required for phagocytosis of particles? Phagocytosis of large particles (>0.5 μm) does not utilize clathrin-coated pits regulated by clathrin, AP-2, and dynamin. Instead, phagocytosis (literally, “cell eating”) involves the spreading of plasma membrane projections around a particle to be internalized in a process dependent on the cortical actin cytoskeleton. Macrophages function in phagocytosis of pathogens and other particles and express cell surface receptors that specifically recognize particles to be phagocytosed (1). Binding of particles to phagocytic receptors is followed by recruitment of actin, Bin1−10−12−13, and dynamin 2 to the site of particle binding. Actin filament assembly then generates an “actin pedestal” beneath the bound particles. Finally, membrane ruffles and projections form that eventually engulf the receptor-bound particles. Both actin and Bin1−10−12−13 are gradually shed from the surface of phagosomes during maturation of phagosomes into digestive compartments (100).
Expression of a dominant negative mutant form of Bin1−10−12−13 lacking the C-terminal SH3 domain (Bin1−10−12−13SH3−) prevents recruitment of dynamin 2 to the site of particle binding and formation of membrane ruffles and projections, and as a consequence results in a severe block in particle engulfment and internalization (Fig. 12). However, expression of Bin1−10−12−13SH3− does not affect cell viability, expression of the phagocytic receptors, particle binding, cell spreading, actin pedestal formation, or secretion to the cell surface (100). Interestingly, in these experiments the recruitment of Bin1−10−12−13 to the site of particle binding was not affected by deletion of the SH3 domain. This suggests that the BAR domain mediates the subcellular localization of Bin1−10−12−13. This is consistent with the finding that in yeast the BAR domain mediates subcellular localization of Rvs167p and that Rvs161p localizes similarly despite comprising only a BAR domain (11, 144). Subcellular localization of Bin1−10−12−13 may be achieved by binding specific membrane lipids. In support of this, treatment of macrophages with the phosphatidylinositol 3-kinase inhibitor wortmannin to deplete PtdIns(3)P abolishes the recruitment of Bin1−10−12−13 to the site of particle binding (100). Interestingly, Bin1−10−12−13 lacks exon 10 that encodes the PtdIns(4,5)P2 binding domain of Bin1+10+13, so other domains in Bin1−10−12−13 may bind phosphoinositides (162).
FIG. 12.
Bin1−10−12−13 functions in phagocytosis in macrophages. RAW-TT10 (murine macrophage-derived) cells were transiently cotransfected with a construct that expresses GFP and with either an empty vector (Control) or a construct that expresses a dominant negative form of Bin1−10−12−13 lacking the C-terminal SH3 domain (AmphIImSH3−). The cells expressing the highest levels of GFP were recovered by flow cytometry and incubated with immunoglobulin G-coated sheep red blood cells for 10 min (panels A and B) or for 1 h (panels C and D) before visualization by scanning electron microscopy. The control cells formed actin pedestals beneath the bound sheep red blood cells, then formed membrane ruffles (A), and then phagocytosed the bound sheep red blood cells (C). In contrast, cells expressing the dominant negative mutant Bin1−10−12−13 lacking the C-terminal SH3 domain formed actin pedestals beneath the bound sheep red blood cells (B), but did not form membrane ruffles, and did not phagocytose the bound sheep red blood cells which remained on the cell surface (D). The scale bar in panel B applies to each panel. (Reprinted from reference 100 with permission from Elsevier.)
In these experiments an acute perturbation of Bin1−10−12−13 function was achieved using a dominant negative construct. Interestingly, however, macrophages derived from homozygous Bin1 knockout mice do not exhibit obvious defects in phagocytosis (207). The reason for the difference between the results obtained by dominant negative constructs and gene knockout is not yet clear. Expression of a mutant construct can have off-target effects, so it is not certain that the defects in phagocytosis seen using the Bin1−10−12−13SH3− construct arise from a role of amphiphysin IIm in phagocytosis. Another possibility is that Bin1−10−12−13 function is critical for phagocytosis but long-term compensatory mechanisms are induced during the development of homozygous knockout mice lacking Bin1−10−12−13 that bypass the normal requirement for Bin1−10−12−13 in phagocytosis. Further experiments will be necessary to distinguish between these possibilities. A possible endocytic or phagocytic role is supported by the finding that Bin1−10−12−13 interacts via its BAR-C domain with SNX4, such as neuronal Bin1+6a+12+13. SNX4 plays a key role in membrane traffic through early endosomes (167).
Originally, it was assumed that the membrane that engulfs bound particles during phagocytosis is derived from the plasma membrane and that the process is one of plasma membrane invagination. However, both in the slime mold Dictyostelium discoideum and in murine macrophages a role for the endoplasmic reticulum in phagocytosis has recently emerged. Dictyostelium mutations that affect the ER-resident proteins calnexin and calreticulin severely compromise phagocytosis (211). In murine macrophages, direct fusion between the ER and the plasma membrane occurs at sites of particle engulfment and the membrane that surrounds the particles being phagocytosed is largely derived from the ER (14, 88). The role of Bin1−10−12−13 in phagocytosis is not yet well understood. Bin1−10−12−13 lacks an NTID that has been shown to be critical for plasma membrane targeting of Bin1+6a+12+13 (257). Perhaps Bin1−10−12−13 localizes to an intracellular compartment such as the ER that moves to the plasma membrane only after engagement of phagocytic receptors.
A potential role for Bin1−10−12−13 in phagocytosis is particularly interesting in light of parallels between endocytosis in yeast and phagocytosis in mammalian cells. Important parallels include the dependence on a functional cortical actin cytoskeleton and the lack of strict dependence on cytoplasmic coat proteins such as clathrin and AP-2 (95, 213, 266). A role for the ER in yeast endocytosis is possible, but has not been fully explored. Some mutations that block the secretory pathway at the step of ER exit also compromise endocytosis (118, 265). However, it has been difficult to assess how direct these effects on endocytosis are (118).
Rvs161p and Rvs167p have recently been shown to physically interact with proteins involved in both membrane traffic from the ER and also post-Golgi apparatus membrane traffic to the cell surface (e.g., Gyp5p and Gyl1p) (33, 84, 317). Gyp5p and Gyl1p regulate the Rab GTPase Ypt1p, which has been implicated in export of lipid rafts from the ER as well as vesicle targeting (202). In mammals, the Ypt1p ortholog Rab1 has also been implicated in export from the ER as well as vesicle targeting (227, 238, 239, 248). It would be interesting if these Rab proteins have additional functions in endocytosis in yeast and phagocytosis in mammalian cells.
AMPHIPHYSIN-RELATED PROTEIN Bin2
Bin2 is perhaps the most enigmatic of the amphiphysin-related proteins. It was identified as a protein in cells not expressing amphiphysin 1 that is also distinct from Bin1 but cross-reacts with polyclonal antisera to Bin1+10+13. The cross-reactive antibodies specifically recognize the BAR-C region of Bin1+10+13. A homology search of the expressed sequence tag database identified a B-lymphocyte cDNA encoding a protein with high sequence identity to Bin1+10+13 in the BAR-C region and this protein was named Bridging INtegrator 2 (Bin2). Bin2 is the product of a distinct gene known as BIN2. Both a major 2.6-kb and a minor 3.5-kb transcript are encoded by the BIN2 gene, suggesting that Bin2, like amphiphysins 1 and Bin1, is differentially spliced (94).
Bin2 has a predicted N-terminal amphipathic helix (residues 23 to 45) (see below) and BAR domain (residues 45 to 249) homologous to those in amphiphysin 1 and Bin1 (residues 1 to 249 of Bin2 exhibit 61% amino acid sequence identity to Bin1) (Fig. 1). Bin2 has a large C-terminal acidic domain enriched in serine and proline residues, but lacks a central insert domain homologous to those in amphiphysins 1 and Bin1+12 that bind clathrin heavy chain and AP-2/α-adaptin or to that in Bin1+10+13 that binds c-Myc. Bin2 also lacks an SH3 domain. As found for other amphiphysin-like proteins it migrates anomalously during electrophoresis, giving an apparent size of 80 kDa, while its sequence predicts a size of 61.7 kDa. When transiently expressed at high levels in COS-7 fibroblasts, Bin2 was found to be cytosolic (94). It is possible that endogenous Bin2 associates with membranes in lymphoid cells.
Bin2 is highly expressed in lymphoid cells and its expression increases during induced differentiation of a granulocyte precursor cell line (HL60) in vitro (granulocytes are a class of lymphoid cell that includes neutrophils, basophils, and eosinophils). Bin2 transcripts are abundant in cultured lymphoid cell lines, but are not found in cell lines derived from brain, liver, lung, breast, prostate, connective tissue (fibroblast), or colon, suggesting that Bin2 may be lymphoid cell specific (94).
Bin2 forms 1:1 heterodimers in vitro with Bin1+10+13. The association is specific because Bin2 cannot form heterodimers in vitro with amphiphysin 1. Bin2 coimmunoprecipitates with Bin1+10+13 (or with the neuron-specific isoform Bin1+6a+12+13) from extracts of COS-7 cells transiently expressing both proteins, suggesting that Bin2 may also form heterodimers in vivo. Association requires the N-terminal part of the Bin1+10+13 BAR domain (residues 1 to 122), but not the BAR-C part (residues 124 to 207). Heterodimer formation does not require the Bin1+10+13 MBD or C-terminal SH3 domain (94). However, heterodimer formation has not yet been demonstrated for endogenous Bin2 in lymphoid cells.
Unlike amphiphysin 1 and Bin1, Bin2 is not implicated in endocytosis. Transient high-level expression of Bin2 in COS-7 cells, unlike high-level expression of neuronal Bin1+6a+12+13, does not inhibit receptor-mediated endocytosis of transferrin. BIN2 knockout mice have not yet been reported, so the effect of loss of Bin2 expression on endocytosis is not yet known. High-level expression of Bin2, unlike that of Bin1+10+13, does not inhibit growth of the tumor cell lines HepG2 (hepatoma), MCF-7 (breast carcinoma), A549 (lung carcinoma), or DU145 and PC3 (prostate carcinoma), nor does Bin2 affect the ability of Bin1+10+13 to inhibit growth of these tumor cells in coexpression experiments (94). The physiological role of Bin2 remains to be determined.
AMPHIPHYSIN-RELATED PROTEIN Bin3
For a time it appeared that proteins comprising a BAR domain only are unique to yeast (42). However, a human protein containing only a BAR domain was eventually identified by homology search of the expressed sequence tag database using the amino acid sequence of S. cerevisiae Rvs161p as the query and named Bin3 (Fig. 1). Bin3 is encoded by a gene distinct from those that encode amphiphysin 1 (AMPH1), Bin1 (BIN1), and Bin2 (BIN2) and has been named BIN3. Human Bin3 exhibits 28 and 29% amino acid sequence identity to S. cerevisiae Rvs161p and S. pombe Hob3p, respectively, but exhibits significantly lower homology to the BAR domains of S. cerevisiae Rvs167p, S. pombe Hob1p, and human Bin1. Unlike Bin1, which generates a large number of distinct transcripts, there appears to be only a single major Bin3 transcript. This Bin3 transcript is expressed ubiquitously in embryos and all adult tissues tested with the sole exception of the brain. Unlike Bin1, whose expression is lost in a significant percentage of cultured cancer cell lines and primary tumors, all tumor cell lines tested to date retain Bin3 expression, suggesting that Bin3 is not a tumor suppressor (276). There has not yet been a thorough characterization of Bin3 and its cellular functions remain to be identified.
ENDOPHILIN FAMILY OF PROTEINS
Identification of Endophilin A1/SH3p4/SH3GL2
Endophilin A1 (also called SH3p4 and SH3GL2) is a 40-kDa BAR domain protein identified as a major synaptojanin 1 binding protein in the brain (28, 51, 192, 269, 270), as a novel SH3 domain protein (SH3p4) that binds a synthetic peptide known to be a ligand of the Src SH3 domain (306), as a protein containing a GRB2-like SH3 domain (SH3GL2) (99), and as an SH3 domain protein that binds a proline-rich motif in the β1-adrenergic receptor cytoplasmic tail (323) (Fig. 1 and Table 2). The name endophilin derives from the affinity this protein displays for several different endocytic proteins (51, 192, 193).
Endophilin A1 possesses an N-terminal amphipathic α-helix, a BAR domain that mediates homodimer formation, and a C-terminal SH3 domain that binds dynamin 1, synaptojanin 1, amphiphysin 1, and Bin1+12 (Fig. 1). Although the endophilin A1 SH3 domain binds the C-terminal PRDs of both synaptojanin 1 and dynamin 1, its affinity for the former is considerably greater (51, 192, 270). Unlike amphiphysin 1, endophilin A1 has only a short central domain and does not bind clathrin heavy chain or AP-2/α-adaptin (28, 51, 73, 192, 193, 255, 269) (note: Cestra et al. [28] refer to endophilin A1/SH3p4 as endophilin 2). Endophilin A1 has been reported to exhibit lysophosphatidic acid acyltransferase activity in vitro (284). However, a very recent study found this activity is due to a contaminant (89).
The endophilin A1 SH3 domain binds the PRD of dynamin 1 at a site distinct from the amphiphysin 1 SH3 domain (302). Similarly, the endophilin A1 SH3 domain binds a motif in the synaptojanin 1 PRD (PKRPPPPR) distinct from the two motifs recognized by the amphiphysin 1 SH3 domain (PIRPSR and PTIPPR) (28, 269). The endophilin A1 SH3 domain also binds germinal center kinase-like kinase (GLK). Endophilin A1 binding to GLK is implicated in Jun N-terminal kinase (Jnk) activation (254). This interaction is interesting, as fission yeast Hob1p also interacts via its SH3 domain with a kinase of the GLK family (Nak1p) (131).
Like amphiphysin 1, endophilin A1 is highly enriched in the brain, where it colocalizes with synaptojanin 1 in presynaptic nerve terminals. Endophilin A1 does not stably associate with the plasmalemma or with synaptic vesicles but is predominantly soluble in the nerve terminal cytoplasm. Endophilin A1 and synaptojanin 1 coimmunoprecipitate from brain extract, indicating that they form a stable complex in vivo. Synaptojanin 1 in the brain also forms complexes with amphiphysin 1/Bin1+6a+12+13 heterodimers that do not contain endophilin A1. Hence, synaptojanin 1 in the brain is present in two separate complexes, one containing endophilin A1 and the other containing amphiphysin 1 and Bin1+6a+12+13. Interestingly, while amphiphysin 1, dynamin 1, and synaptojanin 1 are all phosphorylated in resting neurons and rapidly dephosphorylated upon neuron depolarization, endophilin A1 is not phosphorylated in resting neurons (51, 192, 193).
Two very closely related proteins with the same domain structure as endophilin A1 that are also enriched in the brain are endophilin A2 (also called SH3p8, SH3GL1, and EEN) and endophilin A3 (also called SH3p13 and SH3GL3) (Fig. 1 and Table 2) (28, 34, 99, 133, 269, 270, 306, 323). Endophilin A2 is a 45-kDa protein that localizes to presynaptic nerve terminals and forms heterodimers via BAR domain interactions with endophilin A1 (269). The endophilin A2 SH3 domain, like the endophilin A1 SH3 domain, binds the PRD of dynamin 1 via a site distinct from that recognized by the amphiphysin 1 SH3 domain and also binds the synaptojanin 1 PRD (270, 302). The endophilin A2 SH3 domain also binds the short central domain of endophilin A2 and this homotypic interaction may also occur in the case of endophilin A1 (31). The endophilin 2 SH3 domain also binds a proline-rich motif in the β1-adrenergic receptor cytoplasmic tail (323). Although enriched in the brain, endophilin A3 is also expressed at low levels in the cervical carcinoma HeLa cells, where it localizes to the cytoplasm and also the nucleus (133). The endophilin A3 SH3 domain, like those of endophilins A1 and A2, binds the C-terminal PRDs of dynamin 1 and synaptojanin 1 (270) and a proline-rich motif in the β1-adrenergic receptor cytoplasmic tail (323). Endophilins A1, A2, and A3 are each encoded by a distinct gene (99).
A second subfamily of endophilins comprises endophilins B1 (also called SH3GLB1) and B2 (SH3GLB2), which are also encoded by distinct genes (196, 244) (Fig. 1 and Table 2). Endophilins B1 and B2 have a domain structure similar to that of endophilin A family members, with an N-terminal BAR domain and C-terminal SH3 domain (Fig. 1). Endophilins B1 and B2 form homo- and heterodimers via BAR domain interactions (244). Endophilin B1 interacts with amphiphysin 1, Bin1+12+13, and dynamin 1, but not synaptojanin 1. Endophilin B1 was reported to possess lysophosphatidic acid acyltransferase activity (196), but as with endophilin A1 this is probably due to a contaminant (89). Endophilins B1 and B2 colocalize to structures in the cytoplasm and are not found in the nucleus (244). Endophilin B1 has three known splice variants, of which one is ubiquitously expressed and two are expressed mainly in the brain (196).
Endophilin A and B Family Proteins Bind and Tubulate Membranes
Recombinant endophilin A1 can bind directly to liposomes in vitro. Binding results in evagination of membrane tubules 20 to 100 nm in diameter similar to those formed by dynamin 1 or amphiphysin 1. The tubules formed by endophilin A1 display a pattern of fine tightly packed transverse striations that resembles that seen on the tubules formed by amphiphysin 1. Membrane binding and tubule formation by endophilin A1 are not dependent on the presence of the substrates for its reported lysophosphatidic acid acyltransferase activity, consistent with a recent report that this activity is due to a contaminant enzyme (89). Indeed, liposome evagination appears to be nonenzymatic. When clathrin coat proteins are also present, most of the endophilin A1-coated tubules that form terminate in a clathrin-coated bud (73). This is similar to what is observed for incubation of liposomes with clathrin coat proteins and amphiphysin 1 (316).
Structure-function analysis reveals that the N-terminal domain (residues 1 to 125) of endophilin A1 is both necessary and sufficient for in vitro liposome binding and tubulation activity. This domain is predicted to have a high content of α-helices and to form coiled coils. One endophilin A1 sequence in particular (residues 1 to 35) exhibits particularly high sequence homology to residues 1 to 41 of the BAR domain of amphiphysin 1. Deletion of this sequence in either endophilin A1 or amphiphysin 1 abolishes binding to liposomes and membrane tubulation. This sequence is predicted to contain an amphipathic helix with a hydrophobic face and a highly basic face and may interact with the phospholipid headgroups as well as the hydrophobic interior of the bilayer. Consistent with this idea, replacement of hydrophobic residues with charged residues within the hydrophobic face of this predicted helix abolishes liposome binding and tubulation activity (73).
Chemical cross-linking shows that endophilin A1 assembles into high-molecular-weight oligomers in the presence of liposomes in vitro. Moreover, endophilin A1 assembles with dynamin 1 to form a coat on membrane tubules that has a different physical appearance from those coats formed by endophilin A1 alone. The coat formed by endophilin A1 and dynamin 1 has thicker and more widely spaced striations and resembles the coat formed by brain cytosol or by a mixture of purified dynamin 1 and amphiphysin 1. To form this distinctive coat requires not only the BAR domain but also the C-terminal SH3 domain of endophilin A1 that binds dynamin 1. Unexpectedly, coated membrane tubules formed by dynamin 1 and endophilin A1 do not vesiculate in vitro upon addition of GTP (73). This is in contrast to tubules that are coated by dynamin 1 and amphiphysin 1, which do vesiculate efficiently upon GTP addition (316). Endophilin A1 may therefore prevent dynamin 1-mediated vesiculation of membrane tubules. Both endophilin A1 and amphiphysin 1 have been reported to inhibit dynamin 1 GTPase activity in vitro (73). However, stimulation of dynamin 1 GTPase activity was subsequently shown in the case of amphiphysin 1 and this stimulation is dependent on liposome size (380). Perhaps this will also be the case for endophilin A1, but this is not known.
Interestingly, an amphipathic membrane binding and tubulating domain homologous to that in endophilin A1 is also found in endophilin B1. In neurons a small pool of endophilin B1 localizes to presynaptic termini, similar to endophilin A1, but there is also a large pool that localizes to the Golgi apparatus. Recombinant endophilin B1 binds liposomes in vitro and evaginates membrane tubules such as endophilin A1. Moreover, deletion of the short amphipathic α-helix in the N-terminal BAR domain abolishes membrane tubulation activity, suggesting that endophilin B1 binds and tubulates membranes in a manner similar to endophilin A1 (73).
Endophilin A Family Proteins Function in Receptor-Mediated Endocytosis
Evidence in support of an endocytic role for endophilin A1 has come from studies similar to those that first implicated amphiphysin 1 in endocytosis (293, 369). In permeabilized neurons treated with nonhydrolyzable guanosine 5′-O-(3-thio-triphosphate) (GTPγS) to inhibit dynamin 1 GTPase activity and induce the accumulation of deeply invaginated clathrin-coated pits, endophilin A1 colocalizes with amphiphysin 1 and dynamin 1 in the electron-dense rings at the neck of each clathrin-coated pit (270). Transient overexpression of the endophilin A1 SH3 domain inhibits receptor-mediated endocytosis of transferrin in intact cells (295). In the case of the endophilin A1 SH3 domain the target is more likely to be synaptojanin 1 than dynamin 1 due to the higher affinity for synaptojanin 1. These results from SH3 domain overexpression experiments should be interpreted with caution, however, due to the possibility of off-target interactions and nonspecific effects. As yet, there are no reports of endophilin A1 knockout mice. It will be interesting to see how similar the phenotype associated with loss of endophilin A1 is to the phenotype reported for amphiphysin 1 knockout mice (55).
The endophilin A1 SH3 domain also inhibits transferrin internalization in in vitro receptor-mediated endocytosis assays that utilize either permeabilized 3T3-L1 adipocytes or A431 adenocarcinoma cells (half-maximal inhibition in 3T3-L1 adipocytes occurs at 11 μM) (295). The endophilin A1 SH3 domain is therefore somewhat less potent an inhibitor than the Bin1 SH3 domain. The inhibition may be specific because the c-Abl SH3 domain (which does not bind dynamin 1 or synaptojanin 1) does not inhibit transferrin internalization in this in vitro assay, even at high concentrations. Treatment of permeabilized cells with the endophilin A1 SH3 domain prior to assay gave stronger inhibition than treatment of the cytosol (120, 295). This suggests that the endophilin A1 SH3 domain target relevant to endocytosis is membrane associated. Unexpectedly, addition of neither dynamin 1 nor synaptojanin 1 to the assay relieved inhibition by the endophilin A1 SH3 domain. This suggests that these known endophilin A1 SH3 domain interactors may not be the targets whose inhibition blocks endocytosis (120). These results with SH3 domain inhibition should be considered suggestive rather than conclusive due to possible off-target interactions and nonspecific effects.
At what stage of endocytosis does endophilin A1 function? Endophilin A1 may have multiple roles in clathrin-dependent endocytosis. Microinjection into living synapses of a peptide that binds the endophilin A1 SH3 domain and inhibits its interaction with dynamin 1 and synaptojanin 1 induces the accumulation of both clathrin-coated vesicles and deeply invaginated clathrin-coated pits, suggesting a dual role in membrane fission and in uncoating of clathrin-coated vesicles. Accumulation of deeply invaginated clathrin-coated pits was also induced by microinjection of the endophilin A1 SH3 domain (87). Microinjection of antibodies to endophilin A1 into synapses has been reported to block the conversion of flat clathrin lattices to invaginated clathrin-coated pits, suggesting a role for endophilin A1 in the earliest steps of clathrin-coated vesicle formation (268).
Using in vitro assays that reconstitute the sequential steps of clathrin-dependent endocytosis in permeabilized 3T3-L1 adipocytes and A431 adenocarcinoma cells, one study found that the endophilin A1 SH3 domain specifically inhibits the late step of membrane fission and clathrin-coated vesicle release (295). In contrast, in another study using permeabilized A431 cells, the endophilin A1 SH3 domain was observed to inhibit the formation of deeply invaginated and constricted clathrin-coated pits as well as inhibiting clathrin-coated vesicle fission (120). Immunoelectron microscopy of synaptic membranes after incubation with brain extract, ATP, and GTPγS in vitro revealed that endophilin A1 is not a stoichiometric component of clathrin coats. Instead, endophilin A1 specifically localizes to dynamin 1-coated tubules. Consistent with this (and in contrast to treatment with the endophilin A1 SH3 domain) immunodepletion of endophilin A1 from brain extract does not affect the formation of deeply invaginated clathrin-coated pits on synaptic membranes incubated with ATP and GTPγS in vitro, but does reduce dynamin 1-coated tubule formation (268).
Interestingly, in Drosophila flies endophilin A is highly expressed in neurons and localizes to the presynaptic side of neuromuscular junctions. Like mammalian endophilin A1, Drosophila endophilin A was reported to possess lysophosphatidic acid acyltransferase activity in vitro (106), although, as recently shown for mammalian endophilin A1, this activity may be due to a contaminant (89). Drosophila endophilin A colocalizes with dynamin, suggesting that it may function in endocytosis (267). Mutation of endophilin A perturbs endocytosis of membrane-soluble endocytic dyes and blocks synaptic vesicle recycling in neurons, resulting in lethality at an early larval stage (106, 267, 347). As in mammalian cells, Drosophila endophilin A appears to function in early stages of clathrin-coated pit invagination as well as fission of invaginated pits (106). Drosophila endophilin A also functions in concert with synaptojanin in uncoating of clathrin-coated vesicles (348). Hence, in Drosophila flies amphiphysin is specialized in muscle function, while endophilin A is specialized in neuronal function.
Ubiquitination plays an important role in the sorting of plasma membrane proteins for endocytosis in yeast (117, 327). More recently it has become apparent that ubiquitination is also critical for the sorting of plasma membrane receptors into clathrin-coated pits and endocytosis in mammalian cells (103, 308, 310, 345). Endophilins A1, A2, and A3 form complexes with CIN85 (an SH3 domain adaptor protein similar to yeast Sla1p) and function with the RING domain ubiquitin ligase Cbl in ubiquitin-dependent sorting during endocytosis of growth factor receptors in mammalian cells (240, 305, 313).
A further role of endophilins A1 and A2 in synaptic vesicle endocytosis is in regulating Ca2+ influx into neurons. Both endophilin A1 and A2 exhibit direct binding to voltage-gated Ca2+ channels (VGCCs). This interaction is regulated by Ca2+ binding to a site in the central domain of endophilins A1 and A2. The endophilin A1 and A2 SH3 domains interact with a proline-rich motif in the central domain adjacent to the Ca2+-binding site. Ca2+ binding to the central domain of endophilins A1 and A2 breaks this intramolecular SH3 interaction and allows interaction of the central domain with VGCCs. Endophilin A2 association with VGCCs is important in vivo since transient expression in hippocampal neurons of a mutant endophilin A2 construct that constitutively binds VGCCs inhibits endocytosis (possibly by preventing Ca2+ influx) (31). This regulation of Ca2+ influx is somewhat reminiscent of the role of Rvs161p in low-affinity Ca2+ influx during mating in yeast (210).
In yeast, Rvs161p and Rvs167p are implicated in de novo actin filament assembly. Rvs167p interacts via its SH3 domain with the WASp-related protein Las17p and with Abp1p, both of which are activators of the Arp2/3p actin filament nucleation complex (18, 38, 101, 176, 181, 372). The neuronal isoform of mammalian WASp (N-WASp) is a key activator of the mammalian Arp2/3 actin filament nucleation complex and has been implicated in clathrin-dependent endocytosis (119, 151, 280). Yet evidence of a direct role for amphiphysin 1 or Bin1 in actin polymerization in vertebrate cells is lacking.
Interestingly, there is evidence that endophilin A3 may regulate Arp2/3-dependent actin filament assembly during endocytosis in mammalian cells. The endophilin A3 SH3 domain directly binds proline-rich motifs in N-WASp. Moreover, endophilin A3 enhances the ability of N-WASp to activate the Arp2/3 complex for actin polymerization in vitro. Endophilin A3 and N-WASp coimmunoprecipitate from cell extracts, showing that they form complexes in vivo. Complex formation and accumulation of F-actin at intracellular sites where endophilin A3 and N-WASp colocalize are both enhanced by treatment of cells with epidermal growth factor (230). Endophilin A family members may therefore have a role in actin-dependent movement during endocytosis potentially analogous to that of Rvs161p and Rvs167p (which have been proposed to bear more resemblance to mammalian endophilins than to amphiphysins (307).
These studies show that endophilin A family members have multiple roles in endocytosis. They not only have the ability to tubulate membranes, but also function in Ca2+ influx to trigger dephosphorylation and activation of the endocytic machinery, in ubiquitin-dependent sorting of plasma membrane proteins into forming vesicles, in actin filament assembly which may propel endocytic vesicles from the plasma membrane, and in uncoating of clathrin-coated vesicles for fusion with endosomes in concert with synaptojanin family proteins.
OTHER BAR DOMAIN PROTEINS
Rich-1/Nadrin 1
The RhoGAP Interacting with CIP4 Homologs (RICH, subsequently renamed RICH-1) protein was identified in a screen for proteins that interact with the SH3 domain-containing adaptor protein CDC42-Interacting Protein 4 (CIP4) and found to have GTPase-activating protein activity on Rho family GTPases (263). RICH-1 is also known as nadrin 1. RICH-1 possesses an N-terminal amphipathic α-helix homologous to that in endophilin A1 and B1 that mediates high-affinity liposome binding and tubulation as well as a BAR domain and has been confirmed to have liposome-binding and tubulating activity in vitro (73, 237, 264). RICH-1 localizes to the ER: in contrast to amphiphysin 1 and Bin1 isoforms, RICH-1 does not localize to either the plasma membrane or endosomes. An interesting feature of RICH-1 is the ability of its isolated BAR domain to directly bind phosphoinositides in vitro, including PtdIns, PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, PtdIns(3,5)P2, and PtdIns(4,5)P2 as well as other phospholipids, including phosphatidylserine and phosphatidic acid on a solid support (264). The specificity of binding to these lipids has not yet been tested using more physiological assays such as liposome binding, and it is possible that the BAR domain is recognizing predominantly negative charge rather than these specific lipids.
SNX1
Sorting nexin 1 (SNX1) also has a BAR domain which has the ability to sense membrane curvature and mediate the formation of SNX1 dimers. SNX1 functions in membrane traffic of the cation-independent mannose 6-phosphate receptor from the early endosome to the trans-Golgi network. SNX1 has been shown to drive tubulation of the early endosome in vivo and the purified protein also efficiently tubulates liposomes in vitro (25).
Other BAR Domain Proteins
Other BAR domain proteins recently shown to tubulate liposomes in vitro include arfaptin2, RICH-2/nadrin 2, centaurin β2, and oligophrenin (237). Other proteins have been predicted on the basis of sequence homology to possess BAR domains, e.g., Tuba (279), SNX2, SNX4-9, and SNX18 (25, 325a), Insulin Receptor Substrate protein of 53 kDa (IRSp53), and Adaptor protein containing PH domain, PTB domain, and Leucine zipper motif 1 (APPL1) and 2 (APPL2) (109). These proteins are predicted to have liposome binding and tubulating activity in vitro, but this has not yet been experimentally verified. Liposome binding and tubulation appear to be common features of BAR domain proteins.
CRYSTAL STRUCTURE OF THE AMPHIPHYSIN BAR DOMAIN
Structure of the BAR Domain
The crystal structure of the BAR domain of Drosophila amphiphysin was solved by Peter et al. (237) and the structure has been the topic of several excellent recent reviews (90, 109, 164, 390). Using the structural data together with liposome-binding and tubulation experiments with wild-type and mutant BAR domain proteins, the authors propose a general mechanism for membrane binding and/or bending by BAR domains in different proteins. Structural analysis revealed a banana-shaped dimer formed by monomers, each comprising three α-helices in a coiled-coil arrangement (Fig. 13). The kinks in two of the α-helices together with the angled arrangement of the monomers generate the curved shape of the dimer. Basic residues in the concave face of the dimer (K137 and R140) as well as basic residues at each end of the dimer (K161 and K163) were shown to be critical for membrane binding and tubulation.
FIG. 13.
Three-dimensional crystal structure of the Drosophila amphiphysin BAR domain. A, Two amphiphysin monomers (depicted in purple and green) associate in antiparallel orientation to form a banana-shaped homodimer represented here in ribbon format. The residues that form the basic patches on the concave membrane-binding surface of the dimer are highlighted. The Protein Data Bank identifier is 1URU. Note that the Drosophila amphiphysin BAR domain lacks an NTID, but sequence alignment with mammalian Bin1 suggests the NTID would be positioned within the unstructured loop between helices 2 and 3 as seen in the Drosophila BAR domain. This loop is positively charged and two conserved lysine residues have been shown to be important for membrane association of both Drosophila amphiphysin and rat amphiphysins 1 and 2 (237). The presence of the NTID would contribute extra positive charges to the unstructured loop and may further strengthen membrane association. B, The homodimer represented in electrostatic surface format and viewed with the same orientation as in A. Electrostatic potential is indicated: −10 kTe−1 in red; +10 kTe−1 in blue). C, The homodimer represented in ribbon format but viewed at an angle perpendicular to that in panel A to highlight details of the concave face. D, The homodimer represented in electrostatic surface format and viewed with the same orientation as in panel C. (Reprinted from reference 237 with permission of the publisher. Copyright 2004 American Association for the Advancement of Science.)
The electrostatic interactions between the dimer and membrane coupled with the rigidity of the BAR domain are predicted to be sufficient for membrane bending (390). However, the BAR domain alone does not tubulate liposomes as efficiently as the full-length protein, which also contains the N-terminal amphipathic helix (residues 1 to 41). Also, binding of the BAR domain alone to liposomes is dependent on liposome size, displaying enhanced binding to smaller liposomes with more curvature. In contrast, the full-length protein binds efficiently to liposomes independently of their size. Based on these two observations, Peter et al. propose that the BAR domain itself is primarily a sensor of membrane curvature (237).
More recently, the crystal structure of the endophilin A1 BAR domain was solved (359). This BAR domain also forms a banana-shaped dimer. However, a striking difference between this structure and that of the Drosophila amphiphysin BAR domain is the presence of a disordered region (amino acids 60 to 88), within helix 1. The disordered regions in each monomer are adjacent and in the middle of the dimer, where they extend out of the concave face of the dimer. It is not clear whether they extend perpendicularly out of the concave face of the dimer or to the side of the dimer or whether they become ordered upon membrane binding. The role of these regions and their effect on the membrane binding properties of the BAR domain have not been determined. In addition, the endophilin A1 BAR domain was bound to 11 cadmium ions and is proposed to bind to Ca2+ in vivo and provide a basis for the role of intracellular Ca2+ in endocytosis.
Coupling of the BAR Domain with Additional Lipid-Binding Sequences
BAR domains frequently exist together with a second membrane binding sequence such as an amphipathic α-helix, a pleckstrin homology (PH) domain, or a phox homology (PX) domain (109, 237). The N-terminal amphipathic α-helix found in amphiphysin 1, arfaptin, and RICH-1/nadrin 1 increases the ability of the BAR domain protein to bend membranes. These proteins are efficient at tubulation, and binding of the N-terminal amphipathic α-helix and BAR domain of these proteins to liposomes is independent of membrane curvature (237). This may be due to deformation of the membrane by the amphipathic α-helix in a manner similar to the amphipathic α-helix of epsin (81). In amphiphysin 1 the BAR domain and N-terminal amphipathic α-helix are thought to mediate membrane invagination upon recruitment of the protein to the membrane. The BAR domain then drives accumulation of the protein at the neck of the invagination, where the curvature is optimal for binding. This is followed by recruitment of dynamin to the neck of the invagination, where it oligomerizes and allows fission of the vesicle from the membrane (380).
BAR domain proteins with adjacent PH domains, which lack the N-terminal amphipathic α-helix, tubulate liposomes less efficiently and binding of these BAR domains and their adjacent PH domains to liposomes is sensitive to membrane curvature (237). The PH and PX domains, which bind to specific phosphoinositides, are thought to increase the specificity of membrane targeting of BAR domain proteins since the BAR domain together with the PH/PX domain would allow targeting to a membrane subdomain with a specific membrane curvature and lipid composition.
Several members of the sorting nexin family of proteins, which are characterized by the presence of PX domains, are predicted to contain BAR domains adjacent to PX domains. The BAR and PX domains of SNX1 have been shown to target SNX1 to a highly curved microdomain in early endosomes that contains PtdIns(3)P (25). SNX1 mediates tubulation of the early endosome and via association with the retromer complex (107) drives transport from the early endosome to the trans-Golgi network. A PtdIns(4,5)P2 binding site encoded by exon 10 is thought to recruit Bin1+10+13 to plasma membrane sites enriched in this phosphoinositide in muscle cells. The BAR domain and N-terminal amphipathic α-helix then bend the membrane and drive the formation of T-tubules (162).
Membrane Binding and GTPase Binding of BAR Domains: Independent or Associated?
The structure of the BAR domain is highly homologous to the structure of a fragment of Arfaptin2 GTPase, which binds to Rac (341), and Arf GTPase (147). Arfaptin2 is proposed to modulate membrane ruffling through interactions with these GTPases (324). Arfaptin2 is now recognized to possess a BAR domain and the concave face of the Arfaptin2 BAR domain dimer contains the binding site for Rac. APPL1 and APPL2 bind the Rab5 endosomal GTPase via sequences predicted to form BAR domains. APPL1 and APPL2 colocalize and are associated with tubular and vesicular elements of a membrane compartment that is likely to be an endosome. The binding of these proteins to GTPases via their BAR domains has raised the question of whether membrane binding and GTPase binding are mutually exclusive or common functions of the BAR domain (109). The BAR domain of Arfaptin2 is also able to tubulate liposomes in vitro, suggesting that it retains the membrane-binding function. However, an in vivo role for arfaptin2 in membrane binding or bending has not yet been demonstrated. Further studies are required to establish whether other BAR domain proteins bind small GTPases and whether membrane binding and GTPase binding are mutually exclusive or common features of BAR domains.
BAR Domain and IMD Structure
IRSp53 is a Rac GTPase binding protein that was also predicted to contain a BAR domain (109). The N-terminal 250 amino acids form a domain that is homologous to the B isoform of Missing In Metastasis (MIM). Thus, this region, which resembles a BAR domain, has been independently defined as an IMD (IRSp53 MIM homology Domain) (374). The IMD alone has been shown to play a role in actin bundling and contains the Rac1 binding site. The recent crystal structure of the IRSp53 IMD revealed that it is a cylindrically shaped dimer with tapered ends (195). Nevertheless, there is a striking homology between the IMD and BAR domain. The IMD monomer is composed of a coiled-coil arrangement of α-helices similar to that of the BAR domain monomers, however, an additional shorter fourth α-helix is present and the resulting dimer is flat, unlike the curved BAR domain. Basic residues present at the ends of the IMD dimer have been shown to be critical for the function of the IMD in actin filament bundling. The structures of the BAR domain and IMD illustrate how the coiled-coil motif can be used to build protein domains with diverse functions, including membrane binding, binding to small GTPases, and actin filament bundling.
MEMBRANE TUBULATION AND BAR DOMAIN PROTEIN FUNCTION IN VIVO
We now know that BAR domains bind and tubulate membranes both in vitro and also in vivo (73, 264, 316, 380). Furthermore, crystallography has revealed that BAR domains are banana-shaped dimers able to sense and (depending on the presence of other membrane-binding domains) create membrane curvature (237). The challenge now is to use this new knowledge to gain insight into the diverse roles of BAR domain proteins in vivo. Can a role in membrane binding and tubulation help explain the roles of BAR domain proteins in clathrin-dependent endocytosis, muscle development, sporulation, tumor suppression, phagocytosis, mitochondrial function, transmembrane ion flux, cell-cell fusion, and viability upon starvation?
The ability of BAR domain proteins to tubulate the plasma membrane readily fits with current understanding of clathrin-dependent endocytosis. In clathrin-dependent endocytosis invagination of the clathrin-coated pit leads to the formation of a deeply invaginated pit in which the formed vesicle is still attached to the plasma membrane by a narrow neck comprising a membrane tubule (121, 283, 290, 293, 315). Formation of the tubular neck by amphiphysin 1/Bin1+12 heterodimers combined with SH3 domain interactions may facilitate the recruitment of dynamin 1 to the neck of deeply invaginated pits. Curvature sensing by amphiphysin 1/Bin1+12 dimers may also play an important role in positioning the associated dynamin 1 correctly at the neck, which has the highest membrane curvature (380). Dynamin 1 mediates GTP-dependent fission of membrane tubules in vitro (312). Moreover, dynamin 1 is implicated in fission of the necks of deeply invaginated clathrin-coated pits and release of clathrin-coated vesicles in vivo (45, 116, 121, 155, 344).
The ability of Bin1+10+13 to bind and tubulate membranes via its BAR domain and PtdIns(4,5)P2-binding domain encoded by exon 10 also readily explains the role of this protein in T-tubule biogenesis. T-tubules are believed to form by progressive invagination of the plasma membrane (26, 162). In this case dynamin-dependent fission does not occur and the tubules remain as a type of extensive network of deeply invaginated plasma membrane pits.
Membrane tubulation by BAR domain proteins is not a phenomenon specific to the plasma membrane. For example, RICH-1 has been shown to localize to the ER and to play a role in tubule formation at this organelle (73, 264). Endophilin B1 tubulates Golgi apparatus membranes (73) while SNX1 plays a role in tubule formation at the early endosome (25). Bin1+6a+12+13 and Bin1−10−12−13 both interact with SNX4 and may therefore function with SNX4 in membrane tubulation events in endosomes (167). Endophilin A3 has been shown to localize to a fine network of cytoplasmic tubules as well as to sites of endocytosis at the plasma membrane (133). Hence, the phenomenon of membrane tubulation by BAR domain proteins may not be specific to clathrin-coated pits or T-tubules.
Is membrane tubulation a process that also occurs in the nucleus? The observation that Bin1+10 and Bin1−10−12 (each isoform with or lacking exon 13) localize to the nucleus in at least some cell types suggests a possible role for BAR domain proteins in nuclear events (69, 141, 278, 355). A role for BAR domain proteins in the nucleus is further supported by studies of APPL1 and APPL2 (191). APPL1 and APPL2 are predicted BAR domain proteins that bind the endosomal Rab5 GTPase and localize with Rab5 to a subset of early endosomes (109, 191). APPL1 and APPL2 play a role in signaling to the nucleus in response to changes in the extracellular environment. Upon exposure of cells to epidermal growth factor or oxidative stress, APPL1 has been shown to dissociate from the early endosome and to be imported into the nucleus. In the nucleus APPL1 interacts with the multisubunit histone deacetylase complex NuRD/MeCP1. NuRD/MeCP1 is a key factor in chromatin remodeling and regulation of gene expression. Both APPL1 and APPL2 are known to be important for cell proliferation. Hence, APPL1 and APPL2 have been proposed to link events occurring at the cell surface to nuclear changes in gene expression required for cell proliferation (191).
The role of Bin1+10+13 in the nucleus (if this splice variant is indeed found in the nucleus) is still unclear. Bin1+10+13 regulates c-Myc function, but whether this regulation is imposed in the cytoplasm or nucleus is a question that remains unresolved. A recent study suggests the role of Bin1+10+13 may be related to that of APPL1. As mentioned above, overexpression of Drosophila c-Myc in select cells within a tissue induces the high-expressor cells to become supercompetitors and to proliferate at the expense of their neighboring cells in the tissues that express lower levels of Drosophila c-Myc (199). Intriguingly, the same study showed that overexpression of Rab5 also induces a supercompetitor state. This finding supports the existence of links between Rab5 function in early endosomes and c-Myc signaling pathways that regulate cell proliferation and survival. Moreover, Bin1 has been shown to interact via its C-terminal SH3 domain with Rin3 (142). Rin3 displays Rab5-specific GTP/GDP exchange factor activity (i.e., is a Rab5-GEF) and colocalizes with Rab5 to a subset of early endosomes (142). This study implicates Bin1 in Rab5 GTPase activation via interaction with Rin3.
An intriguing possibility is that the tumor suppression and apoptotic functions of Bin1+10+13 reflect an involvement in the process known as autophagy, in which membranes form around internal organelles as a prelude to their degradation (see below). There are strong emerging links between autophagy in mammalian cells and tumor suppression and apoptosis (66).
The picture that is emerging for the BAR domain proteins in some ways resembles that which has emerged for dynamin. The classical dynamin (dynamin 1) was first implicated in endocytosis via clathrin-coated pits (45, 116, 121, 155, 290, 344). However, the discovery and characterization of diverse dynamin-like proteins has revealed that the membrane fission activity of this family of proteins extends to other membrane compartments, including the ER, mitochondria, and peroxisomes (154, 174, 246, 378, 379). Different BAR domain proteins may function in different organelles in processes related to membrane binding, tubulation, fission, and fusion. It is also possible that the same BAR domain protein may function in more than one cellular location and process. This possibility is especially likely in the case of Rvs161p and Rvs167p, since these are the only BAR domain proteins in yeast and therefore expected to provide a more diverse range of functions than the vertebrate BAR domain proteins, of which there are many.
How can a role in membrane tubule formation explain the phenotypes of yeast rvs mutants? Loss of Rvs161p or Rvs167p reduces the efficiency of endocytic internalization in yeast (21, 38, 180, 215). This endocytic defect may be explained by an inability of the rvs mutants to tubulate the plasma membrane to form deeply invaginated endocytic pits or to form endocytic vesicles by membrane fission. Rvs161p and Rvs167p heterodimers may be sufficient for plasma membrane tubulation during endocytosis, since clathrin, AP-2, and dynamin are not strictly required for endocytosis in yeast (37, 92, 130, 226, 235, 318, 377, 383). Alternatively, by analogy to the putative role of amphiphysin IIm in phagocytosis in macrophages, Rvs161p and Rvs167p may be required for fusion of the cortical ER with sites of endocytosis at the plasma membrane in order to provide additional membrane material (14, 88, 100, 211). Indeed, some mutations that block traffic from the ER also lower the efficiency of endocytosis, although the molecular basis for this link remains unclear (118, 265).
Three lines of evidence suggest that a model in which Rvs proteins regulate direct fusion of cortical ER with plasma membrane is worth consideration. First, electron microscopy studies reveal regions of contact between the cortical ER and plasma membrane in yeast known as plasma membrane-associated membranes (named by analogy to mitochondrion-associated membranes [MAMs]) (see below) (243). Second, Rvs161p and Rvs167p physically and genetically interact with proteins involved in membrane traffic from the ER (Gyp5p, Gyl1p, Rud3p, Sec22p, Sec21p, and Sec27p) (84, 123, 317, 332) and with proteins that function in vesicle fusion with the plasma membrane (Exo70p, Sec8p, Sur4p, Fen1p/Sur5p, and possibly also Gyp5p and Gyl1p) (10, 18, 33, 48, 53, 262). Third, a very recent study found that at least one yeast integral membrane protein (Ist2p) is transported directly from the cortical ER to the plasma membrane without passing through intermediate membrane compartments, suggesting the ER and plasma membrane are able to undergo localized fusion (140).
Can a role in membrane tubulation provide an explanation for why loss of Rvs161p and Rvs167p perturbs the polarized distribution of cortical actin patches (12, 215, 298)? Cortical actin patches are associated with plasma membrane invaginations (205). Electron microscopy studies suggested that these invaginations are distinct from the invaginations associated with receptor-mediated endocytosis (204). However, several subsequent real-time imaging studies have shown that membrane structures containing endocytosed material colocalize with cortical actin patches and that actin filament nucleation correlates with movement of endocytic cargo (71, 132, 139, 143). Therefore, a defect in plasma membrane tubulation might directly impact on the formation, movement, and distribution of cortical actin patches. Alternatively, defects in lipid delivery to the plasma membrane from the ER may perturb the membrane dynamics of the plasma membrane or perturb signal transduction pathways involving lipid second messengers. These changes in rvs mutants may then affect plasma membrane tubulation and cortical actin patch formation, movement, and distribution.
Although the role of Rvs161p and Rvs167p in mitochondrial function is poorly understood it is intriguing that some Rvs-interacting proteins are implicated directly or indirectly in mitochondrial import of hydrophobic proteins, e.g., Pse1p/Kap121p (18, 39). Moreover, it is interesting that the dynamin-like protein 1 (DLP1) in mammalian cells has been shown to localize to sites of contact between the ER and mitochondria known as MAMs and to play a role in regulation of mitochondrial morphology (378). MAMs are thought to play a role in lipid transfer from the site of lipid synthesis in the ER to mitochondria, in particular the transfer of phosphatidylserine (291, 342, 343, 378). The current view is that transfer is achieved by lipid flipping at a site of contact. Perhaps membrane tubule formation also plays a role in transfer of lipids to mitochondria and defects in this process may perturb mitochondrial function. Consistent with a role for BAR domain proteins in lipid transfer to mitochondria, overexpression or down-regulation of endophilin B1 in mammalian cells has severe consequences for mitochondrial morphology (148).
It is interesting that the lipid transferred from the ER to mitochondria by the MAM, phosphatidylserine, is also a lipid enriched in yeast plasma membrane-associated membranes (see above) (243). Moreover, phosphatidylserine strongly stimulates dynamin GTPase activity and is essential for dynamin-dependent liposome fission in vitro (312, 380). Finally, given the roles of some Bin1 splice isoforms in caspase-independent apoptosis, it is interesting that elevated phosphatidylserine in the outer leaflet of the plasma membrane is a hallmark of apoptosis (342). It is possible that BAR domain proteins will be found to play a role in delivery of phosphatidylserine to the outer leaflet of the plasma membrane during Myc-induced apoptosis and in phosphatidylserine delivery from the ER to mitochondria via MAMs in concert with dynamin family proteins, although this is conjecture and experimental evidence for such postulated roles is lacking.
A role for membrane tubulation in lipid transfer between the ER and other organelles might also contribute to sporulation. After sporulation of a diploid cell to form four haploid spores, the spores are held together in a cluster (ascus) by the surrounding plasma membrane contributed by the diploid parent. Each haploid nucleus formed during meiosis requires de novo membrane biogenesis to construct a membrane (prospore membrane) that will become the plasma membrane of the spore (198). The process of membrane assembly around haploid nuclei is not well understood but involves recruitment of membrane material from elsewhere in the cell. A role for another cortical actin patch component (End3p) in membrane traffic to the prospore membrane has very recently been demonstrated (201). Perhaps the Rvs proteins play a role in membrane recruitment via in vivo tubule formation. Mutations in the ER-to-Golgi apparatus Rab GTPase Ypt1p that confer reduced viability upon starvation and block sporulation as rvs mutations do have been isolated (288). Rvs161p and Rvs167p both interact with proteins that regulate Ypt1p, suggesting Rvs161p and Rvs167p may regulate Ypt1p function in sporulation (18, 33, 49, 84, 85, 317).
Finally, it is interesting to consider the phenotype of reduced viability upon starvation exhibited by rvs mutants. During nutrient starvation, cells induce a process known as autophagy in which nonessential internal structures and organelles are degraded (145, 351). Autophagy is a way of providing nutrients for use in essential cellular processes when external nutrients are no longer available. A classic phenotype of mutants defective in autophagy is reduced viability upon starvation. Like sporulation, autophagy is a process that relies on de novo membrane biogenesis. During autophagy, internal structures and organelles are surrounded by a double-layer membrane derived, at least in part, from the ER. The newly formed compartment is known as the autophagosome. The autophagosome eventually fuses with the vacuole, and its contents are degraded by resident hydrolases.
The formation of an autophagosome requires the recruitment of membrane material from donor compartments such as the ER. This process is still very poorly understood. One possibility is that membrane is recruited via membrane tubules that connect the donor compartment (e.g., the ER) to the autophagosome. If this is correct then BAR domain proteins such as Rvs161p and Rvs167p may play a crucial role in generating the membrane tubules that connect the donor compartment to the nascent autophagosome. According to this model, loss of Rvs161p or Rvs167p would block the formation of the autophagosome and thereby prevent efficient turnover of nonessential structures and organelles. This would result in an acute shortage of nutrients for essential processes and reduced viability upon starvation. Intriguingly, elements of the ER itself are turned over by autophagy in yeast. Although it is not known if Rvs161p and Rvs167p specifically are required for this process, the actin cytoskeleton has recently been demonstrated to play a critical role (110). Mutations that affect GTP hydrolysis by Ypt1p and lock it in the active GTP-bound state appear to induce cellular changes characteristic of autophagy (49). The Rvs161p and Rvs167p interactors Gyp5p/Ypl249cp and Gyl1p/Ymr192wp are important regulators of Ypt1p (18, 33, 49, 84, 85, 317).
CONCLUSIONS AND FUTURE PERSPECTIVES
The BAR domain proteins provide a function so fundamental and important it has been used repeatedly in different contexts, including synaptic vesicle recycling in neurons, fusion of myoblasts into myotubes in developing muscle, tumor suppression, and possibly regulation of ion flux across membranes. Genetic studies in yeast have shown that even in a simple unicellular eukaryote BAR domain proteins are important for a bewildering range of processes, including regulation of cell cycle progression, cell polarity, endocytosis, mitochondrial function, response to nutrient starvation, sensitivity to Na+, cell-cell fusion during mating, actin cytoskeletal organization, exocytosis, and differentiation into spores. The underlying molecular function of the BAR domain as a generator and a sensor of membrane curvature has been demonstrated. The structure of the BAR domain has revealed the structural basis for this molecular function. The major challenge now is to define how the fundamental processes of membrane curvature generation and sensing are adapted in vivo to support such a tremendous range of important physiological processes from yeast to humans.
Acknowledgments
We apologize to those whose important contributions to this field unfortunately could not be included in this review because of space and time constraints. We thank H. Riezman, B. Andrews, H. Friesen, A. Gammie, A. Breton, F. Bauer, J. Nickels, P. Robinson, G. Prendergast, P. De Camilli, R. Parton, and P. McPherson for invaluable discussions and a special thanks to G. Prendergast, B. Andrews, P. Robinson, and H. Riezman for sharing unpublished data and/or providing manuscripts prior to publication. We thank A. Gammie and M. Rose for permission to reproduce original images in this review. R. Teasdale is kindly acknowledged for performing advanced bioinformatic searches of sequence databases for novel BAR domains.
Financial support from the National Health and Medical Research Council of Australia (Project Grant 252750) and the Queensland State Government (to A.L.M.) as well as from UMR7156 and CNRS, France (to B.W.), is gratefully acknowledged. G.R. acknowledges the support of a UQ Graduate Student Scholarship.
REFERENCES
- 1.Aderem, A., and D. M. Underhill. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17:593-623. [DOI] [PubMed] [Google Scholar]
- 2.Ahle, S., and E. Ungewickell. 1986. Purification and properties of a new clathrin assembly protein. EMBO J. 5:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amatruda, J. F., J. F. Cannon, K. Tatchell, C. Hug, and J. A. Cooper. 1990. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature 344:352-354. [DOI] [PubMed] [Google Scholar]
- 4.Amberg, D. C., E. Basart, and D. Botstein. 1995. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2:28-35. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, B. L., I. Boldogh, M. Evangelista, C. Boone, L. A. Greene, and L. A. Pon. 1998. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J. Cell Biol. 141:1357-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, B., and V. Measday. 1998. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 14:66-72. [DOI] [PubMed] [Google Scholar]
- 7.Ashrafi, K., T. A. Farazi, and J. I. Gordon. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273:25864-25874. [DOI] [PubMed] [Google Scholar]
- 8.Badour, K., J. Zhang, and K. A. Siminovitch. 2003. The Wiskott-Aldrich syndrome protein: forging the link between actin and cell activation. Immunol. Rev. 192:98-112. [DOI] [PubMed] [Google Scholar]
- 9.Bagnat, M., S. Keranen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balguerie, A., M. Bagnat, M. Bonneu, M. Aigle, and A. M. Breton. 2002. Rvs161p and sphingolipids are required for actin repolarization following salt stress. Eukaryot. Cell 1:1021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balguerie, A., P. Sivadon, M. Bonneu, and M. Aigle. 1999. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112:2529-2537. [DOI] [PubMed] [Google Scholar]
- 12.Bauer, F., M. Urdaci, M. Aigle, and M. Crouzet. 1993. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13:5070-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauerfeind, R., K. Takei, and P. De Camilli. 1997. Amphiphysin I is associated with coated endocytic intermediates and undergoes stimulation-dependent dephosphorylation in nerve terminals. J. Biol. Chem. 272:30984-30992. [DOI] [PubMed] [Google Scholar]
- 14.Becker, T., A. Volchuk, and J. E. Rothman. 2005. Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl. Acad. Sci. USA 102:4022-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belmont, L. D., G. M. Patterson, and D. G. Drubin. 1999. New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J. Cell Sci. 112:1325-1336. [DOI] [PubMed] [Google Scholar]
- 16.Bi, E., P. Maddox, D. J. Lew, E. D. Salmon, J. N. McMillan, E. Yeh, and J. R. Pringle. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bleazard, W., J. M. McCaffery, E. J. King, S. Bale, A. Mozdy, Q. Tieu, J. Nunnari, and J. M. Shaw. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bon, E., P. Recordon Navarro, P. Durrens, M. Iwase, A. Toh, and M. Aigle. 2000. A network of proteins around Rvs167p and Rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast 16:1229-1241. [DOI] [PubMed] [Google Scholar]
- 19.Breton, A. M., and M. Aigle. 1998. Genetic and functional relationship between Rvsp, myosin and actin in Saccharomyces cerevisiae. Curr. Gen. 34:280-286. [DOI] [PubMed] [Google Scholar]
- 20.Breton, A. M., J. Schaeffer, and M. Aigle. 2001. The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 18:1053-1068. [DOI] [PubMed] [Google Scholar]
- 21.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 23.Butler, M. H., C. David, G. C. Ochoa, Z. Freyberg, L. Daniell, D. Grabs, O. Cremona, and P. De Camilli. 1997. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol. 137:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Care, A., K. A. Vousden, K. M. Binley, P. Radcliffe, J. Trevethick, I. Mannazzu, and P. E. Sudbery. 2004. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlton, J., M. Bujny, B. J. Peter, V. M. Oorschot, A. Rutherford, H. Mellor, J. Klumperman, H. T. McMahon, and P. J. Cullen. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14:1791-1800. [DOI] [PubMed] [Google Scholar]
- 26.Carozzi, A. J., E. Ikonen, M. R. Lindsay, and R. G. Parton. 2000. Role of cholesterol in developing T-tubules: analogous mechanisms for T-tubule and caveolae biogenesis. Traffic 1:326-341. [DOI] [PubMed] [Google Scholar]
- 27.Carozzi, A. J., S. Roy, I. C. Morrow, A. Pol, B. Wyse, J. Clyde Smith, I. A. Prior, S. J. Nixon, J. F. Hancock, and R. G. Parton. 2002. Inhibition of lipid raft-dependent signaling by a dystrophy-associated mutant of caveolin-3. J. Biol. Chem. 277:17944-17949. [DOI] [PubMed] [Google Scholar]
- 28.Cestra, G., L. Castagnoli, L. Dente, O. Minenkova, A. Petrelli, N. Migone, U. Hoffmuller, J. Schneider Mergener, and G. Cesareni. 1999. The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J. Biol. Chem. 274:32001-32007. [DOI] [PubMed] [Google Scholar]
- 29.Chant, J., and J. R. Pringle. 1991. Budding and cell polarity in Saccharomyces cerevisiae. Curr. Opin. Gen. Dev. 1:342-350. [DOI] [PubMed] [Google Scholar]
- 30.Chant, J., and J. R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, Y., L. Deng, Y. Maeno Hikichi, M. Lai, S. Chang, G. Chen, and J. F. Zhang. 2003. Formation of an endophilin-Ca2+ channel complex is critical for clathrin-mediated synaptic vesicle endocytosis. Cell 115:37-48. [DOI] [PubMed] [Google Scholar]
- 32.Chen, Y., and P. W. Piper. 1995. Consequences of the overexpression of ubiquitin in yeast: elevated tolerances of osmostress, ethanol and canavanine, yet reduced tolerances of cadmium, arsenite and paromomycin. Biochim. Biophys. Acta 1268:59-64. [DOI] [PubMed] [Google Scholar]
- 33.Chesneau, L., S. Dupre, A. Burdina, J. Roger, S. Le Panse, M. Jacquet, and M. H. Cuif. 2004. Gyp5p and Gyl1p are involved in the control of polarized exocytosis in budding yeast. J. Cell Sci. 117:4757-4767. [DOI] [PubMed] [Google Scholar]
- 34.Cheung, N., C. W. So, J. W. Yam, C. K. So, R. Y. Poon, D. Y. Jin, and L. C. Chan. 2004. Subcellular localization of EEN/endophilin A2, a fusion partner gene in leukaemia. Biochem. J. 383:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury, S., K. W. Smith, and M. C. Gustin. 1992. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 118:561-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chrebet, G. L., D. Wisniewski, A. L. Perkins, Q. Deng, M. B. Kurtz, A. Marcy, and S. A. Parent. 2005. Cell-based assays to detect inhibitors of fungal mRNA capping enzymes and characterization of sinefungin as a cap methyltransferase inhibitor. J. Biomol. Screening 10:355-364. [DOI] [PubMed] [Google Scholar]
- 37.Chu, D. S., B. Pishvaee, and G. S. Payne. 1996. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J. Biol. Chem. 271:33123-33130. [DOI] [PubMed] [Google Scholar]
- 38.Colwill, K., D. Field, L. Moore, J. Friesen, and B. Andrews. 1999. In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics 152:881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corral Debrinski, M., N. Belgareh, C. Blugeon, M. G. Claros, V. Doye, and C. Jacq. 1999. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol. Microbiol. 31:1499-1511. [DOI] [PubMed] [Google Scholar]
- 40.Cousin, M. A., T. C. Tan, and P. J. Robinson. 2001. Protein phosphorylation is required for endocytosis in nerve terminals: potential role for the dephosphins dynamin I and synaptojanin, but not AP180 or amphiphysin. J. Neurochem. 76:105-116. [DOI] [PubMed] [Google Scholar]
- 41.Cross, F., L. H. Hartwell, C. Jackson, and J. B. Konopka. 1988. Conjugation in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 4:429-457. [DOI] [PubMed] [Google Scholar]
- 42.Crouzet, M., M. Urdaci, L. Dulau, and M. Aigle. 1991. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast 7:727-743. [DOI] [PubMed] [Google Scholar]
- 43.Cuddeback, S. M., H. Yamaguchi, K. Komatsu, T. Miyashita, M. Yamada, C. Wu, S. Singh, and H. G. Wang. 2001. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J. Biol. Chem. 276:20559-20565. [DOI] [PubMed] [Google Scholar]
- 44.D'Adamo, P., A. Menegon, C. Lo Nigro, M. Grasso, M. Gulisano, F. Tamanini, T. Bienvenu, A. K. Gedeon, B. Oostra, S. K. Wu, A. Tandon, F. Valtorta, W. E. Balch, J. Chelly, and D. Toniolo. 1998. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat. Genet. 19:134-139. [DOI] [PubMed] [Google Scholar]
- 45.Damke, H., M. Gossen, S. Freundlieb, H. Bujard, and S. L. Schmid. 1995. Tightly regulated and inducible expression of dominant interfering dynamin mutant in stably transformed HeLa cells. Methods Enzymol. 257:209-220. [DOI] [PubMed] [Google Scholar]
- 46.David, C., P. S. McPherson, O. Mundigl, and P. de Camilli. 1996. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc. Natl. Acad. Sci. USA 93:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David, C., M. Solimena, and P. De Camilli. 1994. Autoimmunity in stiff-Man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161. FEBS Lett. 351:73-79. [DOI] [PubMed] [Google Scholar]
- 48.David, D., S. Sundarababu, and J. E. Gerst. 1998. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 143:1167-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Antoni, A., J. Schmitzova, H. H. Trepte, D. Gallwitz, and S. Albert. 2002. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277:41023-41031. [DOI] [PubMed] [Google Scholar]
- 50.De Camilli, P., A. Thomas, R. Cofiell, F. Folli, B. Lichte, G. Piccolo, H. M. Meinck, M. Austoni, G. Fassetta, G. Bottazzo, et al. 1993. The synaptic vesicle-associated protein amphiphysin is the 128-kD autoantigen of Stiff-Man syndrome with breast cancer. J. Exp. Med. 178:2219-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Heuvel, E., A. W. Bell, A. R. Ramjaun, K. Wong, W. S. Sossin, and P. S. McPherson. 1997. Identification of the major synaptojanin-binding proteins in brain. J. Biol. Chem. 272:8710-8716. [DOI] [PubMed] [Google Scholar]
- 52.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desfarges, L., P. Durrens, H. Juguelin, C. Cassagne, M. Bonneu, and M. Aigle. 1993. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast 9:267-277. [DOI] [PubMed] [Google Scholar]
- 54.Dickson, R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta 1426:347-357. [DOI] [PubMed] [Google Scholar]
- 55.Di Paolo, G., S. Sankaranarayanan, M. R. Wenk, L. Daniell, E. Perucco, B. J. Caldarone, R. Flavell, M. R. Picciotto, T. A. Ryan, O. Cremona, and P. De Camilli. 2002. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron 33:789-804. [DOI] [PubMed] [Google Scholar]
- 56.Dorer, R., C. Boone, T. Kimbrough, J. Kim, and L. H. Hartwell. 1997. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics 146:39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drake, M. T., M. A. Downs, and L. M. Traub. 2000. Epsin binds to clathrin by associating directly with the clathrin-terminal domain. Evidence for cooperative binding through two discrete sites. J. Biol. Chem. 275:6479-6489. [DOI] [PubMed] [Google Scholar]
- 58.Drake, M. T., and L. M. Traub. 2001. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J. Biol. Chem. 276:28700-28709. [DOI] [PubMed] [Google Scholar]
- 59.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, W. Guo, K. G. Kozminski, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie, 3rd, P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. N. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drubin, D. G., K. G. Miller, and D. Botstein. 1988. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107:2551-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DuHadaway, J. B., F. J. Lynch, S. Brisbay, C. Bueso Ramos, P. Troncoso, T. McDonnell, and G. C. Prendergast. 2003. Immunohistochemical analysis of Bin1/Amphiphysin II in human tissues: diverse sites of nuclear expression and losses in prostate cancer. J. Cell. Biochem. 88:635-642. [DOI] [PubMed] [Google Scholar]
- 62.Duncan, M. C., M. J. Cope, B. L. Goode, B. Wendland, and D. G. Drubin. 2001. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3:687-690. [DOI] [PubMed] [Google Scholar]
- 63.Dunn, R., and L. Hicke. 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 65.Durrens, P., E. Revardel, M. Bonneu, and M. Aigle. 1995. Evidence for a branched pathway in the polarized cell division of Saccharomyces cerevisiae. Curr. Genet. 27:213-216. [DOI] [PubMed] [Google Scholar]
- 66.Edinger, A. L., and C. B. Thompson. 2004. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 16:663-669. [DOI] [PubMed] [Google Scholar]
- 67.Elion, E. A., J. Trueheart, and G. R. Fink. 1995. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J. Cell Biol. 130:1283-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elliott, K., K. Ge, W. Du, and G. C. Prendergast. 2000. The c-Myc-interacting adaptor protein Bin1 activates a caspase-independent cell death program. Oncogene 19:4669-4684. [DOI] [PubMed] [Google Scholar]
- 69.Elliott, K., D. Sakamuro, A. Basu, W. Du, W. Wunner, P. Staller, S. Gaubatz, H. Zhang, E. Prochownik, M. Eilers, and G. C. Prendergast. 1999. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene 18:3564-3573. [DOI] [PubMed] [Google Scholar]
- 70.Endow, S. A., and M. A. Titus. 1992. Genetic approaches to molecular motors. Annu. Rev. Cell Biol. 8:29-66. [DOI] [PubMed] [Google Scholar]
- 71.Engqvist Goldstein, A. E., and D. G. Drubin. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19:287-332. [DOI] [PubMed] [Google Scholar]
- 72.Evangelista, M., B. M. Klebl, A. H. Tong, B. A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D. Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farsad, K., V. Slepnev, G. Ochoa, L. Daniell, V. Haucke, and P. De Camilli. 2003. A putative role for intramolecular regulatory mechanisms in the adaptor function of amphiphysin in endocytosis. Neuropharmacology 45:787-796. [DOI] [PubMed] [Google Scholar]
- 75.Fishbein, J. D., R. T. Dobrowsky, A. Bielawska, S. Garrett, and Y. A. Hannun. 1993. Ceramide-mediated growth inhibition and CAPP are conserved in Saccharomyces cerevisiae. J. Biol. Chem. 268:9255-9261. [PubMed] [Google Scholar]
- 76.Fitch, P. G., A. E. Gammie, D. J. Lee, V. B. de Candal, and M. D. Rose. 2004. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics 168:733-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Floyd, S., M. H. Butler, O. Cremona, C. David, Z. Freyberg, X. Zhang, M. Solimena, A. Tokunaga, H. Ishizu, K. Tsutsui, and P. De Camilli. 1998. Expression of amphiphysin I, an autoantigen of paraneoplastic neurological syndromes, in breast cancer. Mol. Med.. 4:29-39. [PMC free article] [PubMed] [Google Scholar]
- 78.Floyd, S. R., E. B. Porro, V. I. Slepnev, G. C. Ochoa, L. H. Tsai, and P. De Camilli. 2001. Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2. J. Biol. Chem. 276:8104-8110. [DOI] [PubMed] [Google Scholar]
- 79.Flucher, B. E., M. E. Morton, S. C. Froehner, and M. P. Daniels. 1990. Localization of the alpha 1 and alpha 2 subunits of the dihydropyridine receptor and ankyrin in skeletal muscle triads. Neuron 5:339-351. [DOI] [PubMed] [Google Scholar]
- 80.Folli, F., M. Solimena, R. Cofiell, M. Austoni, G. Tallini, G. Fassetta, D. Bates, N. Cartlidge, G. F. Bottazzo, G. Piccolo, et al. 1993. Autoantibodies to a 128-kd synaptic protein in three women with the stiff-man syndrome and breast cancer. N. Engl. J. Med. 328:546-551. [DOI] [PubMed] [Google Scholar]
- 81.Ford, M. G., I. G. Mills, B. J. Peter, Y. Vallis, G. J. Praefcke, P. R. Evans, and H. T. McMahon. 2002. Curvature of clathrin-coated pits driven by epsin. Nature 419:361-366. [DOI] [PubMed] [Google Scholar]
- 82.Friant, S., R. Lombardi, T. Schmelzle, M. N. Hall, and H. Riezman. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friant, S., B. Zanolari, and H. Riezman. 2000. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 19:2834-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friesen, H., K. Colwill, K. Robertson, O. Schub, and B. Andrews. 2005. Interaction of the Saccharomyces cerevisiae cortical actin patch protein Rvs167p with proteins involved in ER to Golgi vesicle trafficking. Genetics 170:555-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84a.Friesen, H., C. Humphries, Y. Ho, O. Schub, K. Colwill, and B. Andrews. 2006. Characterization of the yeast amphiphysins Rvs161 and Rvs167 reveals roles for the Rvs heterodimer in vivo. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 85.Friesen, H., K. Murphy, A. Breitkreutz, M. Tyers, and B. Andrews. 2003. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol. Biol. Cell 14:3027-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fukushima, N. H., E. Brisch, B. R. Keegan, W. Bleazard, and J. M. Shaw. 2001. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell 12:2756-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gad, H., N. Ringstad, P. Low, O. Kjaerulff, J. Gustafsson, M. Wenk, G. Di Paolo, Y. Nemoto, J. Crun, M. H. Ellisman, P. De Camilli, O. Shupliakov, and L. Brodin. 2000. Fission and uncoating of synaptic clathrin-coated vesicles are perturbed by disruption of interactions with the SH3 domain of endophilin. Neuron 27:301-312. [DOI] [PubMed] [Google Scholar]
- 88.Gagnon, E., S. Duclos, C. Rondeau, E. Chevet, P. H. Cameron, O. Steele Mortimer, J. Paiement, J. J. Bergeron, and M. Desjardins. 2002. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119-131. [DOI] [PubMed] [Google Scholar]
- 89.Gallop, J. L., P. J. Butler, and H. T. McMahon. 2005. Endophilin and CtBP/BARS are not acyl transferases in endocytosis or Golgi fission. Nature 438:675-678. [DOI] [PubMed] [Google Scholar]
- 90.Gallop, J. L., and H. T. McMahon. 2005. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem. Soc. Symp. 72:223-231. [DOI] [PubMed] [Google Scholar]
- 91.Gammie, A. E., V. Brizzio, and M. D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gammie, A. E., L. J. Kurihara, R. B. Vallee, and M. D. Rose. 1995. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ge, K., J. DuHadaway, W. Du, M. Herlyn, U. Rodeck, and G. C. Prendergast. 1999. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl. Acad. Sci. USA 96:9689-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ge, K., and G. C. Prendergast. 2000. Bin2, a functionally nonredundant member of the BAR adaptor gene family. Genomics 67:210-220. [DOI] [PubMed] [Google Scholar]
- 95.Geli, M. I., and H. Riezman. 1998. Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 111:1031-1037. [DOI] [PubMed] [Google Scholar]
- 96.Geli, M. I., and H. Riezman. 1996. Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272:533-535. [DOI] [PubMed] [Google Scholar]
- 97.Germann, M., E. Swain, L. Bergman, and J. T. Nickels, Jr. 2005. Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast amphiphysin-like orthologs, Rvs161p and Rvs167p. J. Biol. Chem. 280:4270-4278. [DOI] [PubMed] [Google Scholar]
- 98.Gerst, J. E., L. Rodgers, M. Riggs, and M. Wigler. 1992. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc. Natl. Acad. Sci. USA 89:4338-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giachino, C., E. Lantelme, L. Lanzetti, S. Saccone, G. Bella Valle, and N. Migone. 1997. A novel SH3-containing human gene family preferentially expressed in the central nervous system. Genomics 41:427-434. [DOI] [PubMed] [Google Scholar]
- 100.Gold, E. S., N. S. Morrissette, D. M. Underhill, J. Guo, M. Bassetti, and A. Aderem. 2000. Amphiphysin IIm, a novel amphiphysin II isoform, is required for macrophage phagocytosis. Immunity 12:285-292. [DOI] [PubMed] [Google Scholar]
- 101.Goode, B. L., A. A. Rodal, G. Barnes, and D. G. Drubin. 2001. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goodson, H. V., B. L. Anderson, H. M. Warrick, L. A. Pon, and J. A. Spudich. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133:1277-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Govers, R., P. van Kerkhof, A. L. Schwartz, and G. J. Strous. 1997. Linkage of the ubiquitin-conjugating system and the endocytic pathway in ligand-induced internalization of the growth hormone receptor. EMBO J. 16:4851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Govindan, B., R. Bowser, and P. Novick. 1995. The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol. 128:1055-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grabs, D., V. I. Slepnev, Z. Songyang, C. David, M. Lynch, L. C. Cantley, and P. De Camilli. 1997. The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. J. Biol. Chem. 272:13419-13425. [DOI] [PubMed] [Google Scholar]
- 106.Guichet, A., T. Wucherpfennig, V. Dudu, S. Etter, M. Wilsch Brauniger, A. Hellwig, M. Gonzalez Gaitan, W. B. Huttner, and A. A. Schmidt. 2002. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 21:1661-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gullapalli, A., T. A. Garrett, M. M. Paing, C. T. Griffin, Y. Yang, and J. Trejo. 2004. A role for sorting nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting nexin 1 and 2 in protein trafficking. Mol. Biol. Cell 15:2143-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo, W., A. O. Jorgensen, and K. P. Campbell. 1994. Characterization and ultrastructural localization of a novel 90-kDa protein unique to skeletal muscle junctional sarcoplasmic reticulum. J. Biol. Chem. 269:28359-28365. [PubMed] [Google Scholar]
- 109.Habermann, B. 2004. The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 5:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hamasaki, M., T. Noda, M. Baba, and Y. Ohsumi. 2005. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6:56-65. [DOI] [PubMed] [Google Scholar]
- 111.Hannun, Y. A. 1996. Functions of ceramide in coordinating cellular responses to stress. Science 274:1855-1859. [DOI] [PubMed] [Google Scholar]
- 112.Hannun, Y. A., and R. M. Bell. 1989. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science 243:500-507. [DOI] [PubMed] [Google Scholar]
- 113.Harder, T., and K. Simons. 1997. Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol. 9:534-542. [DOI] [PubMed] [Google Scholar]
- 114.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates 3rd, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 115.Heinemeyer, W., J. A. Kleinschmidt, J. Saidowsky, C. Escher, and D. H. Wolf. 1991. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 10:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herskovits, J. S., C. C. Burgess, R. A. Obar, and R. B. Vallee. 1993. Effects of mutant rat dynamin on endocytosis. J. Cell Biol. 122:565-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 118.Hicke, L., B. Zanolari, M. Pypaert, J. Rohrer, and H. Riezman. 1997. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol. Biol. Cell 8:13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Higgs, H. N., and T. D. Pollard. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70:649-676. [DOI] [PubMed] [Google Scholar]
- 120.Hill, E., J. van Der Kaay, C. P. Downes, and E. Smythe. 2001. The role of dynamin and its binding partners in coated pit invagination and scission. J. Cell Biol. 152:309-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hinshaw, J. E., and S. L. Schmid. 1995. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374:190-192. [DOI] [PubMed] [Google Scholar]
- 122.Hitchcock, A. L., K. Auld, S. P. Gygi, and P. A. Silver. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100:12735-12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 124.Hoepfner, D., M. van den Berg, P. Philippsen, H. F. Tabak, and E. H. Hettema. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Holtzman, D. A., K. F. Wertman, and D. G. Drubin. 1994. Mapping actin surfaces required for functional interactions in-vivo. J. Cell Biol. 126:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Holtzman, D. A., S. Yang, and D. G. Drubin. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Horvath, A., C. Sutterlin, U. Manning Krieg, N. R. Movva, and H. Riezman. 1994. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 13:3687-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hosoya, O., and K. Tsutsui. 2004. Localized expression of amphiphysin Ir, a retina-specific variant of amphiphysin I, in the ribbon synapse and its functional implication. Eur. J. Neuroscie. 19:2179-2187. [DOI] [PubMed] [Google Scholar]
- 129.Huang, D., G. Patrick, J. Moffat, L. H. Tsai, and B. Andrews. 1999. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA 96:14445-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang, K. M., K. D'Hondt, H. Riezman, and S. K. Lemmon. 1999. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 18:3897-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang, T. Y., M. Renaud Young, and D. Young. 2005. Nak1 interacts with Hob1 and Wsp1 to regulate cell growth and polarity in Schizosaccharomyces pombe. J. Cell Sci. 118:199-210. [DOI] [PubMed] [Google Scholar]
- 132.Huckaba, T. M., A. C. Gay, L. F. Pantalena, H. C. Yang, and L. A. Pon. 2004. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167:519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hughes, A. C., R. Errington, R. Fricker Gates, and L. Jones. 2004. Endophilin A3 forms filamentous structures that colocalise with microtubules but not with actin filaments. Brain research. Mol. brain research. 128:182-192. [DOI] [PubMed] [Google Scholar]
- 134.Huttner, W. B., and A. Schmidt. 2000. Lipids, lipid modification and lipid-protein interaction in membrane budding and fission—insights from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr. Opin. Neurobiol. 10:543-551. [DOI] [PubMed] [Google Scholar]
- 135.Iida, H., and I. Yahara. 1984. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J. Cell Biol. 98:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnston, G. C., C. W. Ehrhardt, A. Lorincz, and B. L. Carter. 1979. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol. 137:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Johnston, G. C., J. A. Prendergast, and R. A. Singer. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113:539-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jonsdottir, G. A., and R. Li. 2004. Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr. Biol. 14:1604-1609. [DOI] [PubMed] [Google Scholar]
- 140.Juschke, C., A. Wachter, B. Schwappach, and M. Seedorf. 2005. SEC18/NSF-independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J. Cell Biol. 169:613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kadlec, L., and A. M. Pendergast. 1997. The amphiphysin-like protein 1 (ALP1) interacts functionally with the cABL tyrosine kinase and may play a role in cytoskeletal regulation. Proc. Natl. Acad. Sci. USA 94:12390-12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kajiho, H., K. Saito, K. Tsujita, K. Kontani, Y. Araki, H. Kurosu, and T. Katada. 2003. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 116:4159-4168. [DOI] [PubMed] [Google Scholar]
- 143.Kaksonen, M., Y. Sun, and D. G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115:475-487. [DOI] [PubMed] [Google Scholar]
- 144.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123:305-320. [DOI] [PubMed] [Google Scholar]
- 145.Kamada, Y., T. Sekito, and Y. Ohsumi. 2004. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 279:73-84. [DOI] [PubMed] [Google Scholar]
- 146.Kaminska, J., B. Gajewska, A. K. Hopper, and T. Zoladek. 2002. Rsp5p, a new link between the actin cytoskeleton and endocytosis in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6946-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kanoh, H., B. T. Williger, and J. H. Exton. 1997. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J. Biol. Chem. 272:5421-5429. [DOI] [PubMed] [Google Scholar]
- 148.Karbowski, M., S. Y. Jeong, and R. J. Youle. 2004. Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 166:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Karpova, T. S., J. G. McNally, S. L. Moltz, and J. A. Cooper. 1998. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142:1501-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Karpova, T. S., S. L. Reck Peterson, N. B. Elkind, M. S. Mooseker, P. J. Novick, and J. A. Cooper. 2000. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell 11:1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kessels, M. M., and B. Qualmann. 2004. The syndapin protein family: linking membrane trafficking with the cytoskeleton. J. Cell Sci. 117:3077-3086. [DOI] [PubMed] [Google Scholar]
- 152.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim, D. W. 2003. Characterization of Grp1p, a novel cis-Golgi matrix protein. Biochem. Biophys. Res. Commun. 303:370-378. [DOI] [PubMed] [Google Scholar]
- 154.Koch, A., M. Thiemann, M. Grabenbauer, Y. Yoon, M. A. McNiven, and M. Schrader. 2003. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278:8597-8605. [DOI] [PubMed] [Google Scholar]
- 155.Koenig, J. H., and K. Ikeda. 1989. Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J. Neurosci. the official J. Soc. Neurosci. 9:3844-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kolesnick, R. N., and M. Kronke. 1998. Regulation of ceramide production and apoptosis. Annu. Rev. Physiol. 60:643-665. [DOI] [PubMed] [Google Scholar]
- 157.Krueger, E. W., J. D. Orth, H. Cao, and M. A. McNiven. 2003. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol. Biol. Cell 14:1085-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kurihara, L. J., C. T. Beh, M. Latterich, R. Schekman, and M. D. Rose. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kurzchalia, T. V., and R. G. Parton. 1999. Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11:424-431. [DOI] [PubMed] [Google Scholar]
- 160.Landgraf, C., S. Panni, L. Montecchi Palazzi, L. Castagnoli, J. Schneider Mergener, R. Volkmer Engert, and G. Cesareni. 2004. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee, E., M. Marcucci, L. Daniell, M. Pypaert, O. A. Weisz, G. C. Ochoa, K. Farsad, M. R. Wenk, and P. De Camilli. 2002. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 297:1193-1196. [DOI] [PubMed] [Google Scholar]
- 163.Lee, J., K. Colwill, V. Aneliunas, C. Tennyson, L. Moore, Y. Ho, and B. Andrews. 1998. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8:1310-1321. [DOI] [PubMed] [Google Scholar]
- 164.Lee, M. C., and R. Schekman. 2004. BAR domains go on a bender. Science 303:479-480. [DOI] [PubMed] [Google Scholar]
- 165.Lee, S. Y., M. R. Wenk, Y. Kim, A. C. Nairn, and P. De Camilli. 2004. Regulation of synaptojanin 1 by cyclin-dependent kinase 5 at synapses. Proc. Natl. Acad. Sci. USA 101:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Legesse Miller, A., R. H. Massol, and T. Kirchhausen. 2003. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14:1953-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Leprince, C., E. Le Scolan, B. Meunier, V. Fraisier, N. Brandon, J. De Gunzburg, and J. Camonis. 2003. Sorting nexin 4 and amphiphysin 2, a new partnership between endocytosis and intracellular trafficking. J. Cell Sci. 116:1937-1948. [DOI] [PubMed] [Google Scholar]
- 168.Leprince, C., F. Romero, D. Cussac, B. Vayssiere, R. Berger, A. Tavitian, and J. H. Camonis. 1997. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J. Biol. Chem. 272:15101-15105. [DOI] [PubMed] [Google Scholar]
- 169.Leventis, P. A., B. M. Chow, B. A. Stewart, B. Iyengar, A. R. Campos, and G. L. Boulianne. 2001. Drosophila amphiphysin is a post-synaptic protein required for normal locomotion but not endocytosis. Traffic 2:839-850. [DOI] [PubMed] [Google Scholar]
- 170.Lew, D. J., and S. I. Reed. 1992. A proliferation of cyclins. Trends Cell Biol. 2:77-81. [DOI] [PubMed] [Google Scholar]
- 171.Lew, J., Q. Q. Huang, Z. Qi, R. J. Winkfein, R. Aebersold, T. Hunt, and J. H. Wang. 1994. A brain-specific activator of cyclin-dependent kinase 5. Nature 371:423-426. [DOI] [PubMed] [Google Scholar]
- 172.Li, F. Q., A. Coonrod, and M. Horwitz. 2000. Selection of a dominant negative retinoblastoma protein (RB) inhibiting satellite myoblast differentiation implies an indirect interaction between MyoD and RB. Mol. Cell. Biol. 20:5129-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li, R. 1997. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 136:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li, X., and S. J. Gould. 2003. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278:17012-17020. [DOI] [PubMed] [Google Scholar]
- 175.Lichte, B., R. W. Veh, H. E. Meyer, and M. W. Kilimann. 1992. Amphiphysin, a novel protein associated with synaptic vesicles. EMBO J. 11:2521-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lila, T., and D. G. Drubin. 1997. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8:367-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lillie, S. H., and J. R. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol 143:1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lippincott, J., and R. Li. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lodder, A. L., T. K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lombardi, R., and H. Riezman. 2001. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J. Biol. Chem. 276:6016-6022. [DOI] [PubMed] [Google Scholar]
- 181.Madania, A., P. Dumoulin, S. Grava, H. Kitamoto, C. Scharer Brodbeck, A. Soulard, V. Moreau, and B. Winsor. 1999. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10:3521-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mao, N. C., E. Steingrimsson, J. DuHadaway, W. Wasserman, J. C. Ruiz, N. G. Copeland, N. A. Jenkins, and G. C. Prendergast. 1999. The murine Bin1 gene functions early in myogenesis and defines a new region of synteny between mouse chromosome 18 and human chromosome 2. Genomics 56:51-58. [DOI] [PubMed] [Google Scholar]
- 183.Marks, B., and H. T. McMahon. 1998. Calcium triggers calcineurin-dependent synaptic vesicle recycling in mammalian nerve terminals. Curr. Biol. 8:740-749. [DOI] [PubMed] [Google Scholar]
- 184.Masur, S. K., Y. T. Kim, and C. F. Wu. 1990. Reversible inhibition of endocytosis in cultured neurons from the Drosophila temperature-sensitive mutant shibirets1. J. Neurogenet. 6:191-206. [DOI] [PubMed] [Google Scholar]
- 185.Matsumoto, K., I. Uno, and T. Ishikawa. 1983. Control of cell division in Saccharomyces cerevisiae mutants defective in adenylate cyclase and cAMP-dependent protein kinase. Exp. Cell Res. 146:151-161. [DOI] [PubMed] [Google Scholar]
- 186.McFarlane, E. S. 1980. Ribonuclease activity during G1 arrest of the yeast Saccharomyces cerevisiae. Arch. Microbiol. 124:243-247. [DOI] [PubMed] [Google Scholar]
- 187.McMahon, H. T., P. Wigge, and C. Smith. 1997. Clathrin interacts specifically with amphiphysin and is displaced by dynamin. FEBS Lett. 413:319-322. [DOI] [PubMed] [Google Scholar]
- 188.McPherson, P. S., E. P. Garcia, V. I. Slepnev, C. David, X. Zhang, D. Grabs, W. S. Sossin, R. Bauerfeind, Y. Nemoto, and P. De Camilli. 1996. A presynaptic inositol-5-phosphatase. Nature 379:353-357. [DOI] [PubMed] [Google Scholar]
- 189.McPherson, P. S., K. Takei, S. L. Schmid, and P. De Camilli. 1994. p145, a major Grb2-binding protein in brain, is colocalized with dynamin in nerve terminals where it undergoes activity-dependent dephosphorylation. J. Biol. Chem. 269:30132-30139. [PubMed] [Google Scholar]
- 190.Measday, V., L. Moore, R. Retnakaran, J. Lee, M. Donoviel, A. M. Neiman, and B. Andrews. 1997. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 17:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miaczynska, M., S. Christoforidis, A. Giner, A. Shevchenko, S. Uttenweiler Joseph, B. Habermann, M. Wilm, R. G. Parton, and M. Zerial. 2004. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116:445-456. [DOI] [PubMed] [Google Scholar]
- 192.Micheva, K. D., B. K. Kay, and P. S. McPherson. 1997. Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J. Biol. Chem. 272:27239-27245. [DOI] [PubMed] [Google Scholar]
- 193.Micheva, K. D., A. R. Ramjaun, B. K. Kay, and P. S. McPherson. 1997. SH3 domain-dependent interactions of endophilin with amphiphysin. FEBS Lett. 414:308-312. [DOI] [PubMed] [Google Scholar]
- 194.Miele, A. E., P. J. Watson, P. R. Evans, L. M. Traub, and D. J. Owen. 2004. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. 11:242-248. [DOI] [PubMed] [Google Scholar]
- 195.Millard, T. H., G. Bompard, M. Y. Heung, T. R. Dafforn, D. J. Scott, L. M. Machesky, and K. Futterer. 2005. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 24:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Modregger, J., A. A. Schmidt, B. Ritter, W. B. Huttner, and M. Plomann. 2003. Characterization of endophilin B1b, a brain-specific membrane-associated lysophosphatidic acid acyl transferase with properties distinct from endophilin A1. J. Biol. Chem. 278:4160-4167. [DOI] [PubMed] [Google Scholar]
- 197.Moreau, V., A. Madania, R. P. Martin, and B. Winson. 1996. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J. Cell Biol. 134:117-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Moreno Borchart, A. C., and M. Knop. 2003. Prospore membrane formation: how budding yeast gets shaped in meiosis. Microbiol. Res. 158:83-90. [DOI] [PubMed] [Google Scholar]
- 199.Moreno, E., and K. Basler. 2004. dMyc transforms cells into super-competitors. Cell 117:117-129. [DOI] [PubMed] [Google Scholar]
- 200.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 201.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 170:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Morsomme, P., and H. Riezman. 2002. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell 2:307-317. [DOI] [PubMed] [Google Scholar]
- 203.Muhlberg, A. B., D. E. Warnock, and S. L. Schmid. 1997. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 16:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mulholland, J., J. Konopka, B. Singer Kruger, M. Zerial, and D. Botstein. 1999. Visualization of receptor-mediated endocytosis in yeast. Mol. Biol. Cell 10:799-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mulholland, J., D. Preuss, A. Moon, A. Wong, D. Drubin, and D. Botstein. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mulholland, J., A. Wesp, H. Riezman, and D. Botstein. 1997. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol. Biol. Cell 8:1481-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Muller, A. J., J. F. Baker, J. B. DuHadaway, K. Ge, G. Farmer, P. S. Donover, R. Meade, C. Reid, R. Grzanna, A. H. Roach, N. Shah, A. P. Soler, and G. C. Prendergast. 2003. Targeted disruption of the murine Bin1/amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol. Cell. Biol. 23:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Muller, A. J., J. B. Duhadaway, P. S. Donover, E. Sutanto Ward, and G. C. Prendergast. 2005. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med.. 11:312-319. [DOI] [PubMed] [Google Scholar]
- 209.Muller, A. J., J. B. DuHadaway, P. S. Donover, E. Sutanto Ward, and G. C. Prendergast. 2004. Targeted deletion of the suppressor gene bin1/amphiphysin2 accentuates the neoplastic character of transformed mouse fibroblasts. Cancer Biol. Ther. 3:1236-1242. [DOI] [PubMed] [Google Scholar]
- 210.Muller, E. M., N. A. Mackin, S. E. Erdman, and K. W. Cunningham. 2003. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 278:38461-38469. [DOI] [PubMed] [Google Scholar]
- 211.Muller Taubenberger, A., A. N. Lupas, H. Li, M. Ecke, E. Simmeth, and G. Gerisch. 2001. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 20:6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mundigl, O., G. C. Ochoa, C. David, V. I. Slepnev, A. Kabanov, and P. De Camilli. 1998. Amphiphysin I antisense oligonucleotides inhibit neurite outgrowth in cultured hippocampal neurons. J. Neurosci. 18:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Munn, A. L. 2001. Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta 1535:236-257. [DOI] [PubMed] [Google Scholar]
- 214.Munn, A. L. 2000. The yeast endocytic membrane transport system. Microscopy research and technique 51:547-562. [DOI] [PubMed] [Google Scholar]
- 215.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1721-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Murphy, J. E., I. T. Pleasure, S. Puszkin, K. Prasad, and J. H. Keen. 1991. Clathrin assembly protein AP-3. The identity of the 155K protein, AP 180, and NP185 and demonstration of a clathrin binding domain. J. Biol. Chem. 266:4401-4408. [PubMed] [Google Scholar]
- 217.Na, S., M. Hincapie, J. H. McCusker, and J. E. Haber. 1995. MOP2 (SLA2) affects the abundance of the plasma membrane H+-ATPase of Saccharomyces cerevisiae. J. Biol. Chem. 270:6815-6823. [DOI] [PubMed] [Google Scholar]
- 218.Naqvi, S. N., Q. Feng, V. J. Boulton, R. Zahn, and A. L. Munn. 2001. Vrp1p functions in both actomyosin ring-dependent and Hof1p-dependent pathways of cytokinesis. Traffic 2:189-201. [DOI] [PubMed] [Google Scholar]
- 219.Naqvi, S. N., R. Zahn, D. A. Mitchell, B. J. Stevenson, and A. L. Munn. 1998. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 8:959-962. [DOI] [PubMed] [Google Scholar]
- 220.Navarro, P., P. Durrens, and M. Aigle. 1997. Protein-protein interaction between the RVS161 and RVS167 gene products of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1343:187-192. [DOI] [PubMed] [Google Scholar]
- 221.Nelson, B., A. B. Parsons, M. Evangelista, K. Schaefer, K. Kennedy, S. Ritchie, T. L. Petryshen, and C. Boone. 2004. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics 166:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nemoto, Y., M. Arribas, C. Haffner, and P. DeCamilli. 1997. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J. Biol. Chem. 272:30817-30821. [DOI] [PubMed] [Google Scholar]
- 223.Newpher, T. M., R. P. Smith, V. Lemmon, and S. K. Lemmon. 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell 9:87-98. [DOI] [PubMed] [Google Scholar]
- 224.Nickels, J. T., and J. R. Broach. 1996. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10:382-394. [DOI] [PubMed] [Google Scholar]
- 225.Nishizawa, M., Y. Kanaya, and A. Toh. 1999. Mouse cyclin-dependent kinase (Cdk) 5 is a functional homologue of a yeast Cdk, pho85 kinase. J. Biol. Chem. 274:33859-33862. [DOI] [PubMed] [Google Scholar]
- 226.Nothwehr, S. F., E. Conibear, and T. H. Stevens. 1995. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J. Cell Biol. 129:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nuoffer, C., H. W. Davidson, J. Matteson, J. Meinkoth, and W. E. Balch. 1994. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125:225-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Obeid, L. M., C. M. Linardic, L. A. Karolak, and Y. A. Hannun. 1993. Programmed cell death induced by ceramide. Science 259:1769-1771. [DOI] [PubMed] [Google Scholar]
- 229.Otsuga, D., B. R. Keegan, E. Brisch, J. W. Thatcher, G. J. Hermann, W. Bleazard, and J. M. Shaw. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143:333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Otsuki, M., T. Itoh, and T. Takenawa. 2003. Neural Wiskott-Aldrich syndrome protein is recruited to rafts and associates with endophilin A in response to epidermal growth factor. J. Biol. Chem. 278:6461-6469. [DOI] [PubMed] [Google Scholar]
- 231.Owen, D. J., Y. Vallis, M. E. Noble, J. B. Hunter, T. R. Dafforn, P. R. Evans, and H. T. McMahon. 1999. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell 97:805-815. [DOI] [PubMed] [Google Scholar]
- 232.Owen, D. J., P. Wigge, Y. Vallis, J. D. Moore, P. R. Evans, and H. T. McMahon. 1998. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 17:5273-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Parton, R. G., and J. F. Hancock. 2004. Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 14:141-147. [DOI] [PubMed] [Google Scholar]
- 234.Pawson, T., and G. D. Gish. 1992. SH2 and SH3 domains: from structure to function. Cell 71:359-362. [DOI] [PubMed] [Google Scholar]
- 235.Payne, G. S., D. Baker, E. van Tuinen, and R. Schekman. 1988. Protein transport to the vacuole and receptor-mediated endocytosis by clathrin heavy chain-deficient yeast. J. Cell Biol. 106:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Peng, J., D. Schwartz, J. E. Elias, C. C. Thoreen, D. Cheng, G. Marsischky, J. Roelofs, D. Finley, and S. P. Gygi. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921-926. [DOI] [PubMed] [Google Scholar]
- 237.Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. Butler, P. R. Evans, and H. T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495-499. [DOI] [PubMed] [Google Scholar]
- 238.Peter, F., C. Nuoffer, S. N. Pind, and W. E. Balch. 1994. Guanine nucleotide dissociation inhibitor is essential for Rab1 function in budding from the endoplasmic reticulum and transport through the Golgi stack. J. Cell Biol. 126:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Peter, F., H. Plutner, H. Zhu, T. E. Kreis, and W. E. Balch. 1993. Beta-COP is essential for transport of protein from the endoplasmic reticulum to the Golgi in vitro. J. Cell Biol. 122:1155-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Petrelli, A., G. F. Gilestro, S. Lanzardo, P. M. Comoglio, N. Migone, and S. Giordano. 2002. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature 416:187-190. [DOI] [PubMed] [Google Scholar]
- 241.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Philpott, A., E. B. Porro, M. W. Kirschner, and L. H. Tsai. 1997. The role of cyclin-dependent kinase 5 and a novel regulatory subunit in regulating muscle differentiation and patterning. Genes Dev. 11:1409-1421.9192869 [Google Scholar]
- 243.Pichler, H., B. Gaigg, C. Hrastnik, G. Achleitner, S. D. Kohlwein, G. Zellnig, A. Perktold, and G. Daum. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. European J. Biochem. FEBS. 268:2351-2361. [DOI] [PubMed] [Google Scholar]
- 244.Pierrat, B., M. Simonen, M. Cueto, J. Mestan, P. Ferrigno, and J. Heim. 2001. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics 71:222-234. [DOI] [PubMed] [Google Scholar]
- 245.Pineda Lucena, A., C. S. Ho, D. Y. Mao, Y. Sheng, R. C. Laister, R. Muhandiram, Y. Lu, B. T. Seet, S. Katz, T. Szyperski, L. Z. Penn, and C. H. Arrowsmith. 2005. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J. Mol. Biol. 351:182-194. [DOI] [PubMed] [Google Scholar]
- 246.Pitts, K. R., Y. Yoon, E. W. Krueger, and M. A. McNiven. 1999. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 10:4403-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Plesset, J., J. R. Ludwig, B. S. Cox, and C. S. McLaughlin. 1987. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J. Bacteriol. 169:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Plutner, H., A. D. Cox, S. Pind, R. Khosravi Far, J. R. Bourne, R. Schwaninger, C. J. Der, and W. E. Balch. 1991. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115:31-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Prior, I. A., C. Muncke, R. G. Parton, and J. F. Hancock. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Proszynski, T. J., R. W. Klemm, M. Gravert, P. P. Hsu, Y. Gloor, J. Wagner, K. Kozak, H. Grabner, K. Walzer, M. Bagnat, K. Simons, and C. Walch-Solimena. 2005. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. USA 102:17981-17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Protopopov, V., B. Govindan, P. Novick, and J. E. Gerst. 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74:855-861. [DOI] [PubMed] [Google Scholar]
- 252.Provance, D. W., and J. A. Mercer. 1999. Myosin-V: head to tail. Cell. Mol. Life Sci. 56:233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pruyne, D. W., D. H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931-1945. [DOI] [PubMed] [Google Scholar]
- 254.Ramjaun, A. R., A. Angers, V. Legendre Guillemin, X. K. Tong, and P. S. McPherson. 2001. Endophilin regulates JNK activation through its interaction with the germinal center kinase-like kinase. J. Biol. Chem. 276:28913-28919. [DOI] [PubMed] [Google Scholar]
- 255.Ramjaun, A. R., and P. S. McPherson. 1998. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J. Neurochem. 70:2369-2376. [DOI] [PubMed] [Google Scholar]
- 256.Ramjaun, A. R., K. D. Micheva, I. Bouchelet, and P. S. McPherson. 1997. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J. Biol. Chem. 272:16700-16706. [DOI] [PubMed] [Google Scholar]
- 257.Ramjaun, A. R., J. Philie, E. de Heuvel, and P. S. McPherson. 1999. The N terminus of amphiphysin II mediates dimerization and plasma membrane targeting. J. Biol. Chem. 274:19785-19791. [DOI] [PubMed] [Google Scholar]
- 258.Razzaq, A., I. M. Robinson, H. T. McMahon, J. N. Skepper, Y. Su, A. C. Zelhof, A. P. Jackson, N. J. Gay, and C. J. O'Kane. 2001. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 15:2967-2979.11711432 [Google Scholar]
- 259.Razzaq, A., Y. Su, J. E. Mehren, K. Mizuguchi, A. P. Jackson, N. J. Gay, and C. J. O'Kane. 2000. Characterisation of the gene for Drosophila amphiphysin. Gene 241:167-174. [DOI] [PubMed] [Google Scholar]
- 260.Ren, G., J. Wang, R. Brinkworth, B. Winsor, B. Kobe, and A. L. Munn. 2005. Verprolin cytokinesis function mediated by the Hof one trap domain. Traffic 6:575-593. [DOI] [PubMed] [Google Scholar]
- 261.Ren, R., B. J. Mayer, P. Cicchetti, and D. Baltimore. 1993. Identification of a ten-amino acid proline-rich SH3 binding site. Science 259:1157-1161. [DOI] [PubMed] [Google Scholar]
- 262.Revardel, E., M. Bonneau, P. Durrens, and M. Aigle. 1995. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1263:261-265. [DOI] [PubMed] [Google Scholar]
- 263.Richnau, N., and P. Aspenstrom. 2001. Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J. Biol. Chem. 276:35060-35070. [DOI] [PubMed] [Google Scholar]
- 264.Richnau, N., A. Fransson, K. Farsad, and P. Aspenstrom. 2004. RICH-1 has a BIN/Amphiphysin/Rvsp domain responsible for binding to membrane lipids and tubulation of liposomes. Biochem. Biophys. Res. Commun. 320:1034-1042. [DOI] [PubMed] [Google Scholar]
- 265.Riezman, H. 1985. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell 40:1001-1009. [DOI] [PubMed] [Google Scholar]
- 266.Riezman, H., A. Munn, M. I. Geli, and L. Hicke. 1996. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia 52:1033-1041. [DOI] [PubMed] [Google Scholar]
- 267.Rikhy, R., V. Kumar, R. Mittal, and K. S. Krishnan. 2002. Endophilin is critically required for synapse formation and function in Drosophila melanogaster. J. Neurosci. 22:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ringstad, N., H. Gad, P. Low, G. Di Paolo, L. Brodin, O. Shupliakov, and P. De Camilli. 1999. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron 24:143-154. [DOI] [PubMed] [Google Scholar]
- 269.Ringstad, N., Y. Nemoto, and P. De Camilli. 2001. Differential expression of endophilin 1 and 2 dimers at central nervous system synapses. J. Biol. Chem. 276:40424-40430. [DOI] [PubMed] [Google Scholar]
- 270.Ringstad, N., Y. Nemoto, and P. De Camilli. 1997. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc. Natl. Acad. Sci. USA 94:8569-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Robinson, P. J., J. M. Sontag, J. P. Liu, E. M. Fykse, C. Slaughter, H. McMahon, and T. C. Sudhof. 1993. Dynamin GTPase regulated by protein kinase C phosphorylation in nerve terminals. Nature 365:163-166. [DOI] [PubMed] [Google Scholar]
- 272.Rosales, J. L., M. J. Nodwell, R. N. Johnston, and K. Y. Lee. 2000. Cdk5/p25nck5a interaction with synaptic proteins in bovine brain. J. Cell. Biochem. 78:151-159. [DOI] [PubMed] [Google Scholar]
- 273.Rossanese, O. W., C. A. Reinke, B. J. Bevis, A. T. Hammond, I. B. Sears, J. O'Connor, and B. S. Glick. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rothman, J. H., C. K. Raymond, T. Gilbert, P. J. O'Hara, and T. H. Stevens. 1990. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell 61:1063-1074. [DOI] [PubMed] [Google Scholar]
- 275.Roumanie, O., M. F. Peypouquet, M. Bonneu, D. Thoraval, F. Doignon, and M. Crouzet. 2000. Evidence for the genetic interaction between the actin-binding protein Vrp1 and the RhoGAP Rgd1 mediated through Rho3p and Rho4p in Saccharomyces cerevisiae. Mol. Microbiol. 36:1403-1414. [DOI] [PubMed] [Google Scholar]
- 276.Routhier, E. L., T. C. Burn, I. Abbaszade, M. Summers, C. F. Albright, and G. C. Prendergast. 2001. Human BIN3 complements the F-actin localization defects caused by loss of Hob3p, the fission yeast homolog of Rvs161p. J. Biol. Chem. 276:21670-21677. [DOI] [PubMed] [Google Scholar]
- 277.Routhier, E. L., P. S. Donover, and G. C. Prendergast. 2003. hob1+, the fission yeast homolog of Bin1, is dispensable for endocytosis or actin organization, but required for the response to starvation or genotoxic stress. Oncogene 22:637-648. [DOI] [PubMed] [Google Scholar]
- 278.Sakamuro, D., K. J. Elliott, R. Wechsler Reya, and G. C. Prendergast. 1996. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Gen.. 14:69-77. [DOI] [PubMed] [Google Scholar]
- 279.Salazar, M. A., A. V. Kwiatkowski, L. Pellegrini, G. Cestra, M. H. Butler, K. L. Rossman, D. M. Serna, J. Sondek, F. B. Gertler, and P. De Camilli. 2003. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 278:49031-49043. [DOI] [PubMed] [Google Scholar]
- 280.Schafer, D. A. 2002. Coupling actin dynamics and membrane dynamics during endocytosis. Curr. Opin. Cell Biol. 14:76-81. [DOI] [PubMed] [Google Scholar]
- 281.Schafer, D. A., S. A. Weed, D. Binns, A. V. Karginov, J. T. Parsons, and J. A. Cooper. 2002. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 12:1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Schmid, S. L., and E. Smythe. 1991. Stage-specific assays for coated pit formation and coated vesicle budding in vitro. J. Cell Biol. 114:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Schmidt, A., M. Wolde, C. Thiele, W. Fest, H. Kratzin, A. V. Podtelejnikov, W. Witke, W. B. Huttner, and H. D. Soling. 1999. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature 401:133-141. [DOI] [PubMed] [Google Scholar]
- 285.Schneiter, R., V. Tatzer, G. Gogg, E. Leitner, and S. D. Kohlwein. 2000. Elo1p-dependent carboxy-terminal elongation of C14:1Δ(9) to C16:1Δ(11) fatty acids in Saccharomyces cerevisiae. J. Bacteriol 182:3655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schott, D., J. Ho, D. Pruyne, and A. Bretscher. 1999. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147:791-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Segev, N. 2001. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13:500-511. [DOI] [PubMed] [Google Scholar]
- 288.Segev, N., and D. Botstein. 1987. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol. Cell. Biol. 7:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sesaki, H., and R. E. Jensen. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sever, S., H. Damke, and S. L. Schmid. 2000. Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic 1:385-392. [DOI] [PubMed] [Google Scholar]
- 291.Shiao, Y. J., G. Lupo, and J. E. Vance. 1995. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270:11190-11198. [DOI] [PubMed] [Google Scholar]
- 292.Shpetner, H. S., J. S. Herskovits, and R. B. Vallee. 1996. A binding site for SH3 domains targets dynamin to coated pits. J. Biol. Chem. 271:13-16. [DOI] [PubMed] [Google Scholar]
- 293.Shupliakov, O., P. Low, D. Grabs, H. Gad, H. Chen, C. David, K. Takei, P. De Camilli, and L. Brodin. 1997. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science 276:259-263. [DOI] [PubMed] [Google Scholar]
- 294.Silva, A. J., T. W. Rosahl, P. F. Chapman, Z. Marowitz, E. Friedman, P. W. Frankland, V. Cestari, D. Cioffi, T. C. Sudhof, and R. Bourtchuladze. 1996. Impaired learning in mice with abnormal short-lived plasticity. Curr. Biol. 6:1509-1518. [DOI] [PubMed] [Google Scholar]
- 295.Simpson, F., N. K. Hussain, B. Qualmann, R. B. Kelly, B. K. Kay, P. S. McPherson, and S. L. Schmid. 1999. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat. Cell Biol. 1:119-124. [DOI] [PubMed] [Google Scholar]
- 296.Singer Kruger, B., and S. Ferro Novick. 1997. Use of a synthetic lethal screen to identify yeast mutants impaired in endocytosis, vacuolar protein sorting and the organization of the cytoskeleton. Eur. J. Cell Biol. 74:365-375. [PubMed] [Google Scholar]
- 297.Singer Kruger, B., H. Stenmark, A. Dusterhoft, P. Philippsen, J. S. Yoo, D. Gallwitz, and M. Zerial. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125:283-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Sivadon, P., F. Bauer, M. Aigle, and M. Crouzet. 1995. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol. Gen. Genet. 246:485-495. [DOI] [PubMed] [Google Scholar]
- 299.Sivadon, P., M. Crouzet, and M. Aigle. 1997. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett. 417:21-27. [DOI] [PubMed] [Google Scholar]
- 300.Sivadon, P., M. F. Peypouquet, F. Doignon, M. Aigle, and M. Crouzet. 1997. Cloning of the multicopy suppressor gene SUR7: evidence for a functional relationship between the yeast actin-binding protein Rvs167 and a putative membranous protein. Yeast 13:747-761. [DOI] [PubMed] [Google Scholar]
- 301.Slepnev, V. I., G. C. Ochoa, M. H. Butler, and P. De Camilli. 2000. Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites. J. Biol. Chem. 275:17583-17589. [DOI] [PubMed] [Google Scholar]
- 302.Slepnev, V. I., G. C. Ochoa, M. H. Butler, D. Grabs, and P. De Camilli. 1998. Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science 281:821-824. [DOI] [PubMed] [Google Scholar]
- 303.Smith, D. S., and L. H. Tsai. 2002. Cdk5 behind the wheel: a role in trafficking and transport? Trends Cell Biol. 12:28-36. [DOI] [PubMed] [Google Scholar]
- 304.So, C. W., M. H. Sham, S. L. Chew, N. Cheung, C. K. So, S. K. Chung, C. Caldas, L. M. Wiedemann, and L. C. Chan. 2000. Expression and protein-binding studies of the EEN gene family, new interacting partners for dynamin, synaptojanin and huntingtin proteins. Biochem. J. 348 Pt. 2:447-458. [PMC free article] [PubMed] [Google Scholar]
- 305.Soubeyran, P., K. Kowanetz, I. Szymkiewicz, W. Y. Langdon, and I. Dikic. 2002. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416:183-187. [DOI] [PubMed] [Google Scholar]
- 306.Sparks, A. B., N. G. Hoffman, S. J. McConnell, D. M. Fowlkes, and B. K. Kay. 1996. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat. Biotechnol. 14:741-744. [DOI] [PubMed] [Google Scholar]
- 307.Stamenova, S. D., R. Dunn, A. S. Adler, and L. Hicke. 2004. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J. Biol. Chem. 279:16017-16025. [DOI] [PubMed] [Google Scholar]
- 308.Stang, E., F. D. Blystad, M. Kazazic, V. Bertelsen, T. Brodahl, C. Raiborg, H. Stenmark, and I. H. Madshus. 2004. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell 15:3591-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Strom, M., P. Vollmer, T. J. Tan, and D. Gallwitz. 1993. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature 361:736-739. [DOI] [PubMed] [Google Scholar]
- 310.Strous, G. J., P. van Kerkhof, R. Govers, A. Ciechanover, and A. L. Schwartz. 1996. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15:3806-3812. [PMC free article] [PubMed] [Google Scholar]
- 311.Sutterlin, C., T. L. Doering, F. Schimmoller, S. Schroder, and H. Riezman. 1997. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 110:2703-2714. [DOI] [PubMed] [Google Scholar]
- 312.Sweitzer, S. M., and J. E. Hinshaw. 1998. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93:1021-1029. [DOI] [PubMed] [Google Scholar]
- 313.Szymkiewicz, I., K. Kowanetz, P. Soubeyran, A. Dinarina, S. Lipkowitz, and I. Dikic. 2002. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J. Biol. Chem. 277:39666-39672. [DOI] [PubMed] [Google Scholar]
- 314.Takei, K., V. Haucke, V. Slepnev, K. Farsad, M. Salazar, H. Chen, and P. De Camilli. 1998. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell 94:131-141. [DOI] [PubMed] [Google Scholar]
- 315.Takei, K., P. S. McPherson, S. L. Schmid, and P. De Camilli. 1995. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature 374:186-190. [DOI] [PubMed] [Google Scholar]
- 316.Takei, K., V. I. Slepnev, V. Haucke, and P. De Camilli. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1:33-39. [DOI] [PubMed] [Google Scholar]
- 317.Talarek, N., A. Balguerie, M. Aigle, and P. Durrens. 2005. A novel link between a Rab GTPase and Rvs proteins: the yeast amphiphysin homologues. Cell Biochemistry and Function. 23:253-266. [DOI] [PubMed] [Google Scholar]
- 318.Tan, P. K., N. G. Davis, G. F. Sprague, and G. S. Payne. 1993. Clathrin facilitates the internalization of seven transmembrane segment receptors for mating pheromones in yeast. J. Cell Biol. 123:1707-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tan, T. C., V. A. Valova, C. S. Malladi, M. E. Graham, L. A. Berven, O. J. Jupp, G. Hansra, S. J. McClure, B. Sarcevic, R. A. Boadle, M. R. Larsen, M. A. Cousin, and P. J. Robinson. 2003. Cdk5 is essential for synaptic vesicle endocytosis. Nat. Cell Biol. 5:701-710. [DOI] [PubMed] [Google Scholar]
- 320.Tang, D., J. Yeung, K. Y. Lee, M. Matsushita, H. Matsui, K. Tomizawa, O. Hatase, and J. H. Wang. 1995. An isoform of the neuronal cyclin-dependent kinase 5 (Cdk5) activator. J. Biol. Chem. 270:26897-26903. [DOI] [PubMed] [Google Scholar]
- 321.Tang, H. Y., and M. Cai. 1996. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4897-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tang, H. Y., J. Xu, and M. Cai. 2000. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 20:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Tang, Y., L. A. Hu, W. E. Miller, N. Ringstad, R. A. Hall, J. A. Pitcher, P. DeCamilli, and R. J. Lefkowitz. 1999. Identification of the endophilins (SH3p4/p8/p13) as novel binding partners for the beta1-adrenergic receptor. Proc. Natl. Acad. Sci. USA 96:12559-12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Tarricone, C., B. Xiao, N. Justin, P. A. Walker, K. Rittinger, S. J. Gamblin, and S. J. Smerdon. 2001. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411:215-219. [DOI] [PubMed] [Google Scholar]
- 325.Tatchell, K., L. C. Robinson, and M. Breitenbach. 1985. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc. Natl. Acad. Sci. USA 82:3785-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325a.Teasdale, R. D., D. Loci, F. Houghton, L. Karlsson, and P. A. Gleeson. 2001. A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 358:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Terada, Y., K. Tsutsui, K. Sano, O. Hosoya, H. Ohtsuki, and A. Tokunaga. 2002. Novel splice variants of amphiphysin I are expressed in retina. FEBS Lett. 519:185-190. [DOI] [PubMed] [Google Scholar]
- 327.Terrell, J., S. Shih, R. Dunn, and L. Hicke. 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell 1:193-202. [DOI] [PubMed] [Google Scholar]
- 328.Thanabalu, T., and A. L. Munn. 2001. Functions of Vrp1p in cytokinesis and actin patches are distinct and neither requires a WH2/V domain. EMBO J. 20:6979-6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 330.Tomizawa, K., S. Sunada, Y. F. Lu, Y. Oda, M. Kinuta, T. Ohshima, T. Saito, F. Y. Wei, M. Matsushita, S. T. Li, K. Tsutsui, S. Hisanaga, K. Mikoshiba, K. Takei, and H. Matsui. 2003. Cophosphorylation of amphiphysin I and dynamin I by Cdk5 regulates clathrin-mediated endocytosis of synaptic vesicles. J. Cell Biol. 163:813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Tong, A. H., B. Drees, G. Nardelli, G. D. Bader, B. Brannetti, L. Castagnoli, M. Evangelista, S. Ferracuti, B. Nelson, S. Paoluzi, M. Quondam, A. Zucconi, C. W. Hogue, S. Fields, C. Boone, and G. Cesareni. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321-324. [DOI] [PubMed] [Google Scholar]
- 332.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 333.Torre, E., M. A. McNiven, and R. Urrutia. 1994. Dynamin 1 antisense oligonucleotide treatment prevents neurite formation in cultured hippocampal neurons. J. Biol. Chem. 269:32411-32417. [PubMed] [Google Scholar]
- 334.Traub, L. M., M. A. Downs, J. L. Westrich, and D. H. Fremont. 1999. Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl. Acad. Sci. USA 96:8907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Trueheart, J., and G. R. Fink. 1989. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA 86:9916-9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Tsai, L. H., I. Delalle, V. S. Caviness, Jr., T. Chae, and E. Harlow. 1994. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature 371:419-423. [DOI] [PubMed] [Google Scholar]
- 338.Tsutsui, K., Y. Maeda, S. Seki, and A. Tokunaga. 1997. cDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem. Biophys. Res. Commun. 236:178-183. [DOI] [PubMed] [Google Scholar]
- 339.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 340.Vaduva, G., N. C. Martin, and A. K. Hopper. 1997. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J. Cell Biol. 139:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Van Aelst, L., T. Joneson, and D. Bar Sagi. 1996. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 15:3778-3786. [PMC free article] [PubMed] [Google Scholar]
- 342.Vance, J. E. 2003. Molecular and cellular biology of phosphatidylserine and phosphatidylethanolamine metabolism. Progress Nucleic Acid Res. Mol. Biol. 75:69-111. [DOI] [PubMed] [Google Scholar]
- 343.Vance, J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248-7256. [PubMed] [Google Scholar]
- 344.van der Bliek, A. M., T. E. Redelmeier, H. Damke, E. J. Tisdale, E. M. Meyerowitz, and S. L. Schmid. 1993. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. Cell Biol. 122:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.van Kerkhof, P., M. Sachse, J. Klumperman, and G. J. Strous. 2001. Growth hormone receptor ubiquitination coincides with recruitment to clathrin-coated membrane domains. J. Biol. Chem. 276:3778-3784. [DOI] [PubMed] [Google Scholar]
- 346.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Verstreken, P., O. Kjaerulff, T. E. Lloyd, R. Atkinson, Y. Zhou, I. A. Meinertzhagen, and H. J. Bellen. 2002. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell 109:101-112. [DOI] [PubMed] [Google Scholar]
- 348.Verstreken, P., T. W. Koh, K. L. Schulze, R. G. Zhai, P. R. Hiesinger, Y. Zhou, S. Q. Mehta, Y. Cao, J. Roos, and H. J. Bellen. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 40:733-748. [DOI] [PubMed] [Google Scholar]
- 349.Volchuk, A., S. Narine, L. J. Foster, D. Grabs, P. De Camilli, and A. Klip. 1998. Perturbation of dynamin II with an amphiphysin SH3 domain increases GLUT4 glucose transporters at the plasma membrane in 3T3-L1 adipocytes. Dynamin II participates in GLUT4 endocytosis. J. Biol. Chem. 273:8169-8176. [DOI] [PubMed] [Google Scholar]
- 350.Vollert, C. S., and P. Uetz. 2004. The phox homology (PX) domain protein interaction network in yeast. Mol. Cell. Proteomics 3:1053-1064. [DOI] [PubMed] [Google Scholar]
- 351.Wang, C. W., and D. J. Klionsky. 2003. The molecular mechanism of autophagy. Mol. Med. 9:65-76. [PMC free article] [PubMed] [Google Scholar]
- 352.Wang, L. H., T. C. Sudhof, and R. G. W. Anderson. 1995. The appendage domain of alpha-adaptin is a high-affinity binding-site for dynamin. J. Biol. Chem. 270:10079-10083. [DOI] [PubMed] [Google Scholar]
- 353.Warnock, D. E., J. E. Hinshaw, and S. L. Schmid. 1996. Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 271:22310-22314. [DOI] [PubMed] [Google Scholar]
- 354.Watts, F. Z., G. Shiels, and E. Orr. 1987. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 6:3499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wechsler-Reya, R., K. Elliott, M. Herlyn, and G. C. Prendergast. 1997. The putative tumor suppressor BIN1 is a short-lived nuclear phosphoprotein, the localization of which is altered in malignant cells. Cancer Res. 57:3258-3263. [PubMed] [Google Scholar]
- 356.Wechsler-Reya, R., D. Sakamuro, J. Zhang, J. Duhadaway, and G. C. Prendergast. 1997. Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem. 272:31453-31458. [DOI] [PubMed] [Google Scholar]
- 357.Wechsler-Reya, R. J., K. J. Elliott, and G. C. Prendergast. 1998. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell. Biol. 18:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Weiss, A., and L. A. Leinwand. 1996. The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 12:417-439. [DOI] [PubMed] [Google Scholar]
- 359.Weissenhorn, W. 2005. Crystal structure of the endophilin-A1 BAR domain. J. Mol. Biol. 351:653-661. [DOI] [PubMed] [Google Scholar]
- 360.Wen, K. K., and P. A. Rubenstein. 2005. Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J. Biol. Chem. 280:24168-24174. [DOI] [PubMed] [Google Scholar]
- 361.Wendland, B., and S. D. Emr. 1998. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wendland, B., K. E. Steece, and S. D. Emr. 1999. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18:4383-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wertman, K. F., D. G. Drubin, and D. Botstein. 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Wesp, A., L. Hicke, J. Palecek, R. Lombardi, T. Aust, A. L. Munn, and H. Riezman. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8:2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.White, J. M., and M. D. Rose. 2001. Yeast mating: getting close to membrane merger. Curr. Biol. 11:R16-20. [DOI] [PubMed] [Google Scholar]
- 366.Wicky, S., S. Frischmuth, and B. Singer Kruger. 2003. Bsp1p/Ypr171p is an adapter that directly links some synaptojanin family members to the cortical actin cytoskeleton in yeast. FEBS Lett. 537:35-41. [DOI] [PubMed] [Google Scholar]
- 367.Wigge, P., K. Kohler, Y. Vallis, C. A. Doyle, D. Owen, S. P. Hunt, and H. T. McMahon. 1997. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol. Biol. Cell 8:2003-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wigge, P., and H. T. McMahon. 1998. The amphiphysin family of proteins and their role in endocytosis at the synapse. Trends Neurosci. 21:339-344. [DOI] [PubMed] [Google Scholar]
- 369.Wigge, P., Y. Vallis, and H. T. McMahon. 1997. Inhibition of receptor-mediated endocytosis by the amphiphysin SH3 domain. Curr. Biol. 7:554-560. [DOI] [PubMed] [Google Scholar]
- 370.Wills, C. 1990. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 25:245-280. [DOI] [PubMed] [Google Scholar]
- 371.Winsor, B., and E. Schiebel. 1997. Review: an overview of the Saccharomyces cerevisiae microtubule and microfilament cytoskeleton. Yeast 13:399-434. [DOI] [PubMed] [Google Scholar]
- 372.Winter, D., T. Lechler, and R. Li. 1999. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9:501-504. [DOI] [PubMed] [Google Scholar]
- 373.Wong, E. D., J. A. Wagner, S. V. Scott, V. Okreglak, T. J. Holewinske, A. Cassidy Stone, and J. Nunnari. 2003. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Yamagishi, A., M. Masuda, T. Ohki, H. Onishi, and N. Mochizuki. 2004. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279:14929-14936. [DOI] [PubMed] [Google Scholar]
- 375.Yang, S., K. R. Ayscough, and D. G. Drubin. 1997. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J. Cell Biol. 136:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Yeo, S. C., L. Xu, J. Ren, V. J. Boulton, M. D. Wagle, C. Liu, G. Ren, P. Wong, R. Zahn, P. Sasajala, H. Yang, R. C. Piper, and A. L. Munn. 2003. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 116:3957-3970. [DOI] [PubMed] [Google Scholar]
- 377.Yeung, B. G., H. L. Phan, and G. S. Payne. 1999. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell 10:3643-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Yoon, Y., K. R. Pitts, S. Dahan, and M. A. McNiven. 1998. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140:779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Yoon, Y., K. R. Pitts, and M. A. McNiven. 2001. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12:2894-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Yoshida, Y., M. Kinuta, T. Abe, S. Liang, K. Araki, O. Cremona, G. Di Paolo, Y. Moriyama, T. Yasuda, P. De Camilli, and K. Takei. 2004. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 23:3483-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Young, M. E., T. S. Karpova, B. Brugger, D. M. Moschenross, G. K. Wang, R. Schneiter, F. T. Wieland, and J. A. Cooper. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Yu, H., J. K. Chen, S. Feng, D. C. Dalgarno, A. W. Brauer, and S. L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933-945. [DOI] [PubMed] [Google Scholar]
- 383.Yu, X., and M. Cai. 2004. The yeast dynamin-related GTPase Vps1p functions in the organization of the actin cytoskeleton via interaction with Sla1p. J. Cell Sci. 117:3839-3853. [DOI] [PubMed] [Google Scholar]
- 384.Zanolari, B., S. Friant, K. Funato, C. Sutterlin, B. J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19:2824-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zarrinpar, A., S. H. Park, and W. A. Lim. 2003. Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426:676-680. [DOI] [PubMed] [Google Scholar]
- 386.Zelhof, A. C., H. Bao, R. W. Hardy, A. Razzaq, B. Zhang, and C. Q. Doe. 2001. Drosophila amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis. Development 128:5005-5015. [DOI] [PubMed] [Google Scholar]
- 387.Zeng, G., and M. Cai. 1999. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 144:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Zhang, Q., H. K. Chieu, C. P. Low, S. Zhang, C. K. Heng, and H. Yang. 2003. Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J. Biol. Chem. 278:47145-47155. [DOI] [PubMed] [Google Scholar]
- 389.Zhao, R., M. Davey, Y. C. Hsu, P. Kaplanek, A. Tong, A. B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, C. Boone, A. Emili, and W. A. Houry. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120:715-727. [DOI] [PubMed] [Google Scholar]
- 390.Zimmerberg, J., and S. McLaughlin. 2004. Membrane curvature: how BAR domains bend bilayers. Curr. Biol. 14:R250-252. [DOI] [PubMed] [Google Scholar]