Abstract
The ispF gene product in Escherichia coli has been shown to catalyze the formation of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MEC) in the deoxyxylulose (DOXP) pathway for isoprenoid biosynthesis. In this work, the E. coli gene ispF and its Bacillus subtilis orthologue, yacN, were deleted and conditionally complemented by expression of these genes from distant loci in the respective organisms. In E. coli, complementation was achieved through integration of ispF at the araBAD locus with control from the arabinose-inducible araBAD promoter, while in B. subtilis, yacN was placed at amyE under control of the xylose-inducible xylA promoter. In both cases, growth was severely retarded in the absence of inducer, consistent with these genes being essential for survival. E. coli cells depleted of MEC synthase revealed a filamentous phenotype. This was in contrast to the depletion of MEC synthase in B. subtilis, which resulted in a loss of rod shape, irregular septation, multicompartmentalized cells, and thickened cell walls. To probe the nature of the predominant deficiency of MEC synthase-depleted cells, we investigated the sensitivity of these conditionally complemented mutants, grown with various concentrations of inducer, to a wide variety antibiotics. Synthetic lethal behavior in MEC synthase-depleted cells was prevalent for cell wall-active antibiotics.
Isoprenoids comprise a diverse group of compounds with over 30, 000 members that are unique in both their chemistry and structure (34). All isoprenoids are made from the same basic five-carbon isoprene unit, isopentenyl pyrophosphate (IPP) and its isomer, dimethylallyl pyrophosphate (DMAPP). Initial work on the isoprenoid biosynthetic pathway was done predominantly with yeast and animals, and led to the discovery of the mevalonate pathway, which until recently was believed to be the only route for isoprenoid biosynthesis (14, 23). In 1993, Rohmer et al. showed that 13C-labeled precursors were incorporated into isoprenoids in some eubacteria in a labeling pattern that was inconsistent with the mevalonate pathway (32). Shortly after, a new pathway stemming from 1-deoxy-d-xylulose 5-phosphate (DOXP) was discovered, and it is now known as the DOXP pathway or the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway (33).
In the DOXP pathway, DOXP is made from pyruvate and glyceraldehyde-3-phosphate, a reaction catalyzed by the enzyme DOXP synthase (Dxs) (22, 39). In the first committed step of this pathway, MEP synthase catalyzes the isomerization and reduction of DOXP to form MEP (40). MEP is subsequently converted into 4-diphosphocytidyl-2-C-methylerythritol (CDP-ME) by a cytidyltransferase known as IspD (formerly YgbP) (31). The next step is an ATP-dependent phosphorylation of CDP-ME at the C-2 hydroxy group to form 4-diphosphocytidyl-2-C-methylerythritol-2-phosphate (26). This product is converted by a unique cyclization reaction, catalyzed by IspF (formerly YgbB), into 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MEC) (19). These first steps of the DOXP pathway have been discussed in more detail in some recent reviews (23, 30). The last two steps in this pathway leading to the formation of IPP and DMAPP have only just been identified. Following the formation of the cyclic diphosphate, IspG (formerly designated GcpE) catalyzes a two-electron reduction forming 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (18). In the final step of the DOXP pathway, branching occurs and IPP and DMAPP are formed from 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate by separate reactions catalyzed by the IspH protein (formerly designated LytB) (29).
The distribution of the DOXP pathway has revealed that the majority of eubacteria use this pathway, while archaea, fungi, and animals appear to use only the mevalonate pathway (8, 23, 30). Higher plants, such as Arabidopsis thaliana, have all of the genes for both pathways, with the mevalonate pathway being used exclusively in the cytoplasm and the DOXP pathway being used exclusively in the plastids (2). The DOXP pathway is present in many pathogenic bacteria, plants, and the malaria parasite, making this pathway an intriguing target for the development of antibacterial compounds, herbicides, and antimalarial drugs (28, 30).
Two independent groups have noted the indispensability of the gene ispF in Escherichia coli (7, 15). Campos et al. replaced the chromosomal copies of ispD, ispE, and ispF with chloramphenicol (CM) resistance markers and were able to complement these mutations with a synthetic operon for the mevalonate-dependent pathway coding for yeast 5-diphosphomevalonate decarboxylase, human 5-phosphomevalonate kinase, yeast mevalonate kinase, and E. coli isopentenyl diphosphate isomerase (7). Gene ispF has likewise been listed as an essential gene in a large-scale gene knockout study reported by Freiberg and coworkers (15). To date, however, the physiological consequences of MEC synthase depletion in E. coli have not been examined. Moreover, the dispensability of the DOXP pathway for isoprenoid synthesis in the model gram-positive rod Bacillus subtilis has not been addressed.
Known functions of isoprenoids include the modification of tRNA (5), dolichol production, and the formation of the respiratory quinones (36). Indispensable roles for the latter two molecules underline the likely importance of this pathway in bacteria; however, isoprenoids may well also have unexpected roles in other areas of bacterial physiology.
The physiological consequences of the depletion of IspF and YacN in E. coli and B. subtilis, respectively, were explored for the first time in this work through the construction of precise deletions of ispF and yacN. Conditional complementation of these deletions was achieved by placement of ispF at the araBAD locus under the control of the arabinose promoter and by integration of yacN at the amyE locus under the control of the xylose promoter by using the pSWEET system (4). Phenotypic characterization of the E. coli ispF and the B. subtilis yacN mutants by light microscopy and scanning and transmission electron microscopy revealed distinct phenotypes in these organisms upon depletion of IspF and YacN. We also exploited the principle of synthetic lethal interactions (38, 41) to probe the dominant mechanism for cell death associated with loss of MEC synthase. To do this, we looked for sensitization of MEC synthase-depleted cells to a variety of antibiotics with diverse mechanisms of action. Inhibitors of peptidoglycan biogenesis, in particular, showed antibacterial synergy with depletion of MEC synthase in both E. coli and B. subtilis. We conclude that lesions in the DOXP pathway exert their antibacterial effects primarily through an impact on cell wall synthesis.
MATERIALS AND METHODS
General methods.
Tables 1 and 2 list the strains, plasmids, and primers used. E. coli and B. subtilis strains were grown in rich Luria-Burtani (LB) medium. The following concentrations of antibiotics were used for E. coli: kanamycin (KAN), 25 μg/ml; CM, 20 μg/ml; and ampicillin (AMP), 50 μg/ml. The following concentrations of antibiotics were used for B. subtilis: CM, 5 μg/ml; and spectinomycin (SPEC), 150 μg/ml. When required, sucrose was used at a final concentration of 5% (wt/vol), and unless otherwise indicated, arabinose was used at 0.2% (wt/vol) and xylose was used at 2% (wt/vol). HotStarTaq PCR reagents, gel extraction kits, and plasmid Mini-Prep kits were from Qiagen (Mississauga, Ontario, Canada). Cloning was performed in the E. coli cloning strain Novablue (Novagen, Madison, Wis.) according to established methods (35). Preparation and transformation of E. coli electrocompetent cells were done according to the electroporator manufacturer's instructions (Bio-Rad, Inc., Hercules, Calif.), while B. subtilis competent cells and transformations were done according to established methods (10). Transformations into competent strains were done with 100 ng to 1 μg of DNA. Restriction enzymes and Vent polymerase were obtained from New England Biolabs (Beverly, Mass.), with the exception of SrfI, which was purchased from Stratagene (La Jolla, Calif.).
TABLE 1.
E. coli strains, plasmids, and primers used in this study
| Strain, plasmid, or oligonucleotide | Descriptiona | Reference, source, or site |
|---|---|---|
| Strains | ||
| Novablue | endA1 hsdR17 (rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proA+B+ lacIqZΔM15::Tn10] | Novagen |
| EB68 | MG1655 (F− λ−) | George Church, Harvard Medical School (24) |
| EB356 | MG1655 (F− λ−) araBAD::ispF(optimal RBS) kan | This work |
| EB370 | MG1655 (F− λ−) araBAD::ispF(optimal RBS) kan ΔispF | This work |
| Plasmids | ||
| pKO3 | E. coli gene replacement plasmid described by Link and coworkers | 24 |
| pKO3-ispFflank | pKO3 with ispF flanks from EB68 | This work |
| pKD46 | Red recombinase expression plasmid for transformation of linear DNA in E. coli | 11 |
| pBluescript SKII+ | Cloning vector | Stratagene (La Jolla, Calif.) |
| pBS-araBADflank | pBluescript with araBAD flanks from EB68 | This work |
| pBS-araBADflankkan | pBluescript with Kanr cassette inserted between araBAD flanking sequence | This work |
| pBS-araBADflankispFkan | pBluescript with ispF (optimal RBS) and Kanr cassette inserted between araBAD flanking sequence | This work |
| Oligonucleotides | ||
| BAD-a | 5′-AAGGAAAAAAGCGGCCGCACCGCGAATGGTGAGATTGAGAATATA-3′ | NotI |
| BAD-bb | 5′-CACGCAATAACCTTCACACTCCGCCCGGGCAACCAACGGTATGGAGAAACAGT-3′ | SrfI |
| BAD-cb | 5′-GTTGCCCGGGCGGAGTGTGAAGGTTATTGCGTGCTGTATAAAACCACAGCCAA-3′ | SrfI |
| BAD-d | 5′-CGCACGCATGTCGACTTCAGACGGGCATTAACGATAGTG-3′ | SalI |
| BAD-e | 5′-ATGCAGGATTTTTGCCCAGA-3′ | |
| kan-F | 5′-TGTGTTTAAACGTATGTGGAAGGTGGAAAGCCACGTTGTGTCTC-3′ | PmeI |
| kan-R | 5′-TAACCAATTCTGATTAGAAA-3′ | |
| ispF-Fc | 5′-GGGGTACCAATAAGGAGGAAAAAAAAATGCGAATTGGACACGGT-3′ | KpnI |
| ispF-R | 5′-GCTCTAGATCATTTTGTTGCCTTAAT-3′ | XbaI |
| ispF-a | 5′-AAGGAAAAAAGCGGCCGCGAATCATCCGCAAATCACCGT-3′ | NotI |
| ispF-bd | 5′-ACGCAATAACCTTCACACTCCAAATTTATAACCATTATGTATTCTCCTGATGGATGGTTC-3′ | |
| ispF-cd | 5′-GTTATAAATTTGGAGTGTGAAGGTTATTGCGTGAAATGATTGAGTTTGATAATCTCACTTACT-3′ | |
| ispF-d | 5′-CGCACGCATGTCGACCCCAAAACGTTGGGCACC-3′ | SalI |
Underlined sequences indicate restriction enzyme sites.
Boldface sequence indicates complementarity between BAD-b and BAD-c.
Boldface sequence indicates optimal ribosome binding site (RBS).
Boldface sequence indicates complementarity between ispF-b and ispF-c.
TABLE 2.
B. subtilis strains, plasmids, and primers used in this study
| Strain, plasmid or oligonucleotide | Descriptiona | Reference, source, or site |
|---|---|---|
| Strains | ||
| EB6 | hisA1 argC4 metC3 | L5087 (6) |
| EB315 | hisA1 argC4 metC3 amyE::xylR PxylA yacN cat86 | This work |
| EB323 | hisA1 argC4 metC3 amyE::xylR PxylA yacN cat86 yacN::spec | This work |
| Plasmids | ||
| pBS-yacNflank | pBluescript with yacN flanks from EB6 | This work |
| pBS-yacNflankspec | pBluescript with Specr cassette inserted between yacN flanking sequence | This work |
| pSWEET | B. subtilis gene replacement plasmid described by Bhavsar and coworkers | 4 |
| pSWEET-yacN | pSWEET with wild-type yacN from EB6 | This work |
| pUS19 | pUC19 derivative containing Specr cassette | P. Levin |
| Oligonucleotides | ||
| yacN-Fb | 5′-GGGATTAATTAAGGAGAAGGCGCTGTAAAGGGAGAAGAAACAAATGTTTAGAATTGGACAAG GATTTGATGTG-3′ | PacI |
| yacN-R | 5′-GGGAGCTAGCTTAGCCTTTTTGTATCAGTACTGTCGCCTG-3′ | NheI |
| yacN-a | 5′-GGAAGGATCCGGAGCGGGAGCACTTTCAGC-3′ | BamHI |
| yacN-bc,d | 5′-CACGCAATAACCTGCCCGGGCCAAATTTATAACCTAAACGTGTTTATTCCCACTTTCT GATTCC ATGATAGC-3′ | SrfI |
| yacN-cc | 5′-GTTATAAATTTGGCCCGGGCAGGTTATTGCGTGCTTGTTTTGATGCCGGGTCTATTTGGTGG-3′ | SrfI |
| yacN-d | 5′-GGAAGGATCCGTTACGCTCGTACTGACGGTATGG-3′ | BamHI |
| spec-F | 5′-GGTTTACACTTACTTTAGTTTTATGGAAATGAAAGATC-3′ | |
| spec-R | 5′-TTATAATTTTTTTAATCTGTTATTTAAATAGTTTATAG-3′ |
Underlined sequences indicate restriction enzyme sites.
Boldface sequence indicates sequence upstream of yacM including the ribosome binding site.
Boldface sequence indicates complementarity between yacN-b and yacN-c.
Boldface and italic sequence indicates a conservative histidine mutation in yacM to delete the yacN start codon.
Construction of araBAD deletion plasmid.
A crossover PCR strategy adapted from reference 24 was used to amplify 500 bp upstream and 500 bp downstream of araBAD from E. coli MG1655 chromosomal DNA with primers BAD-a, BAD-b, BAD-c, and BAD-d. The final product (1,000 bp) contained the araBAD flanking sequences with an SrfI site introduced between them and NotI and SalI restriction sites at either end allowing for insertion into pBluescript. This plasmid was named “pBS-araBADflank.” A KAN resistance (Kanr) cassette was Vent amplified from pBAD18-Kan (17), including amplification of the promoter, but not the terminator, with the introduction of an upstream PmeI restriction site. This PCR product was cloned into pBS-araBADflank at the SrfI site, and a plasmid with the Kanr cassette in the forward orientation with respect to the araB promoter was chosen and named “pBS-araBADflankkan.” Amplification of ispF from MG1655 chromosomal DNA was done with primers ispF-F (which places a consensus ribosome binding site upstream of ispF) (42) and ispF-R with Vent polymerase. This PCR product was cloned into the PmeI site of pBS-araBADflankkan, and a plasmid with ispF in the forward orientation with respect to the araB promoter was selected and named “pBS-araBADflankispFkan.”
Construction of ispF deletion plasmid.
As described above, a crossover PCR strategy was used to amplify sequence 500 bp upstream and 500 bp downstream of ispF from MG1655 chromosomal DNA with primers ispF-a, ispF-b, ispF-c, and ispF-d. The final PCR product was cloned into pKO3 (24) at the NotI and SalI restriction sites. This new plasmid was named “pKO3-ispFflank.”
Precise deletion of E. coli ispF.
The deletion of ispF was carried out as described by Datsenko et al. and Link et al. (11, 24) with slight modifications. Linear DNA was PCR amplified from pBS-araBADflankispFkan with primers BAD-a and BAD-d, resulting in an approximately 2,500-bp product. MG1655-pKD46 was transformed with 100 ng of this product and plated on KAN-supplemented LB medium (LB/KAN) at 37°C overnight to select for integrants at araBAD and loss of the temperature-sensitive plasmid. To screen for strains in which the araBAD genes had been replaced by ispF-kan, colony PCR was used with primers BAD-a and BAD-d (positive for chromosomal integrants), as well as BAD-a and BAD-e (positive for wild type). A strain positive for chromosomal integration was selected and named “EB356.” The gene replacement protocol described by Link et al. (24) was conducted with strain EB356 with pKO3-ispFflank. Colony PCR with primers ispF-a and ispF-d confirmed the deletion of ispF, and the resulting strain was named “EB370.”
Construction of pSWEET-yacN plasmid.
Plasmid pSWEET-bgaB (4) was digested with PacI and NheI to remove bgaB. B. subtilis 168 chromosomal DNA was used as a template for the amplification of yacN with primers yacN-F and yacN-R. The upstream primer was designed to place yacN under the control of the ribosome binding site preceding the open reading frame of yacM. (The gene yacM is upstream of and overlaps yacN by 7 bp.) The PCR product was cloned into pSWEET-bgaB at the PacI and NheI restriction sites, and the resulting plasmid was named “pSWEET-yacN.”
Construction of yacN deletion plasmid.
A crossover PCR strategy, as described above, was used to amplify sequence from B. subtilis 168 chromosomal DNA 500 bp upstream of yacN and 350 bp downstream of it. This was done with primers yacN-a, yacN-b, yacN-c, and yacN-d. Primer yacN-b contains a silent mutation in His231 of yacM (CAT to CAG) to disrupt the start codon of yacN without altering the product of yacM. The final PCR product was purified and cloned into pBluescript at the EcoRV site, and the resulting plasmid was named “pBS-yacNflank.” A SPEC cassette was amplified with Vent polymerase from pUS19, amplifying the transcriptional promoter, but not the terminator, and was cloned into pBS-yacNflank at the SrfI site. Restriction mapping facilitated the choice of a clone in which the orientation of the SPEC cassette conserved the transcriptional context of yacN, and the resulting plasmid was named “pBS-yacNflankspec.”
Precise deletion of B. subtilis yacN.
Plasmid pSWEET-yacN was digested with PstI, gel purified, and used to transform B. subtilis 168 cells. Transformants were screened for CM resistance and then restreaked on starch plates to test for disruption of amyE (9). A strain that did not produce halos was selected and named “EB315.” Plasmid pBS-yacNflankspec was digested with PstI and gel purified. The purified fragment was transformed into strain EB315, and the resulting strain was named “EB323.” Genomic DNA was prepared from strain EB323, and PCR was performed with primers yacN-a and yacN-d to verify deletion of yacN.
B. subtilis growth curve.
EB315 and EB323 were grown overnight on LB/CM and LB/CM/SPEC/xylose plates, respectively, at 30°C. The following day, the cells were resuspended in 2 ml of sterile saline, and both cultures were diluted to an optical density at 600 nm (OD600) of 0.5. One milliliter of EB315 was used to inoculate 100 ml of LB/CM/xylose. One milliliter of EB323 was used to inoculate 100 ml of LB/CM/SPEC/xylose (2, 0.2, 0.063, 0.02, and 0%). The samples were incubated at 30°C with shaking at 250 rpm for 1,020 min. Every hour, the OD600 for a 0.5-ml sample was read.
Light microscopy.
Samples to be examined were prepared by growing the cells on plates with the appropriate selection overnight and resuspending the cells in 2 ml of sterile saline the following day. The cells were pelleted at 2,300 × g, and the pellet was resuspended in 1 ml of sterile saline plus 30% (vol/vol) glycerol. Phase-contrast microscopy was used to view live cells with an Olympus BX51 microscope with a ×100 oil immersion lens. Images were captured with a CoolSNAP-Pro 12-bit monochrome camera with Image-Pro Express version 4.0 software.
Transmission electron microscopy.
For transmission electron microscopy, cells were grown as described above for light microscopy and resuspended in 2 ml of sterile saline the following day. The cells were pelleted at 2,300 × g, the saline was removed, and 2% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) was added. Samples were refrigerated at 4°C for 24 h to allowing fixing to occur. Following this time period, the samples were postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4), and dehydration was performed with a graded ethanol series. The samples were then treated with propylene oxide and embedded in Spurr's resin. Sections (70 nm) were cut with a Reichert Ultracut E ultramicrotome (Leica, Inc., Vienna, Austria). The sections were stained for 5 min with uranyl acetate and for 2 min with lead citrate. Samples were visualized with and photographed with a JEOL 1200EX transmission electron microscope (JEOL, Ltd., Tokyo, Japan) operating at 80 kV.
Scanning electron microscopy.
For scanning electron microscopy, cells were grown as described above for light microscopy and resuspended in 2 ml of sterile saline the following day. The cells were pelleted at 2,300 × g, the saline was removed, and the cells were resuspended in 2% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Samples were refrigerated at 4°C for 24 h to allow fixing to occur. Following this time period, the samples were postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4), and dehydration was performed with a graded ethanol series. Samples were visualized and photographed with a PHILIPS 501B scanning electron microscope.
MICs of antibacterial agents for B. subtilis.
Strains EB6, EB315, and EB323 were grown overnight on LB, LB/CM, and LB/CM/SPEC/xylose plates, respectively. Cells were resuspended from the plates in saline and added to LB with the appropriate antibiotics at a final concentration of 25,000 CFU per 100 μl. Medium (100 μl) containing antibiotics, xylose, and cells was added to each well of a 96-well microtiter plate. Strain EB6 was tested in the absence of xylose, strain EB315 was tested in both the presence (2%) and absence of xylose, and strain EB323 was tested over a range of xylose concentrations of 2, 0.2, 0.063, 0.02, and 0%. The MICs of various antibacterial agents specific for gram-positive organisms were determined for each of the three strains at all levels of xylose by adding the drugs to the wells of the microtiter plate in twofold dilutions and incubating the plates at 30°C with shaking at 200 rpm. After 24 h, the OD for each well of the plate was determined with a Spectra-max Plus instrument (Molecular Devices, Sunnyvale, Calif.). The MIC was assigned as the lowest concentration of drug that inhibited growth of a particular strain.
MICs of antibacterial agents for E. coli.
Strains EB68, EB356, and EB370 were grown overnight on LB, LB/KAN, and LB/KAN/arabinose plates, respectively. One colony from each plate was used to inoculate an overnight liquid culture for each strain, with EB370 grown in the absence of arabinose. This step is required to achieve adequate depletion of the deletion strain. The following day, 10 μl of each of the overnight cultures was added to 5 ml of fresh medium (same as that used for the overnight cultures), and the cultures were grown until the cells reached an OD600 of 0.3 to 0.5. The cells were added to fresh media at a final concentration of 37,500 CFU per 150 μl, which was then distributed in a 96-well microtiter plate at 150 μl per well. Strain EB68 was tested in the absence of arabinose, strain EB356 was tested in both the presence (0.2%) and absence of arabinose, and strain EB370 was tested over a range of arabinose concentrations of 0.2, 0.02, 0.0063, 0.002, and 0%. The MICs of a variety of antibacterial agents specific for gram-negative organisms were determined after 16 h of growth for each of the three strains at all levels of arabinose as described above for B. subtilis, except that incubation was at 37°C.
RESULTS
Conditional complementation of E. coli ispF and B. subtilis yacN deletion.
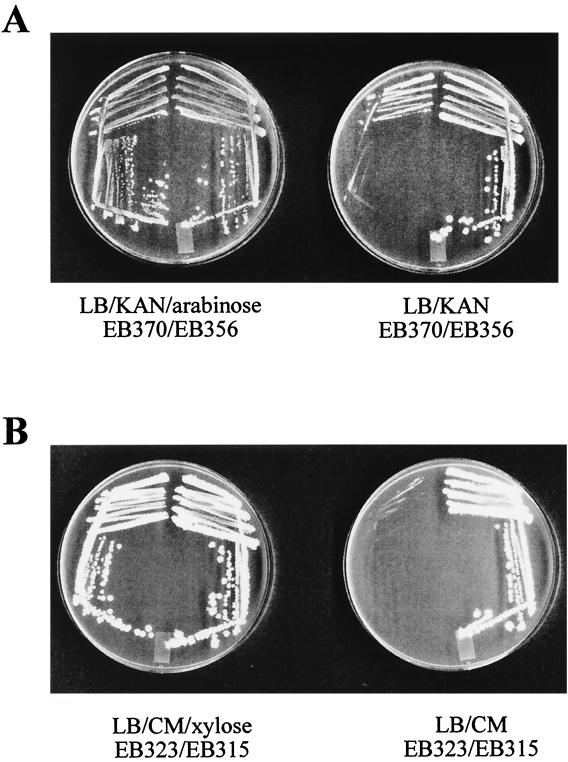
E. coli strains EB356 and EB370 were grown at 37°C on agar plates containing LB/KAN with and without arabinose. A similar experiment was done at 30°C for B. subtilis strains EB315 and EB323 with plates containing LB/CM and either 2% or no xylose. The results from these plate experiments are shown in Fig. 1. The E. coli ispF deletion strain (EB370), as seen in Fig. 1A, was able to grow well in the presence of arabinose, showing a wild-type colony morphology, but upon its removal, there was a distinct decrease in growth and a loss of the ability to form single colonies. The same results were seen for the B. subtilis yacN deletion strain, EB323 (Fig. 1B), which had an even more pronounced decrease in cell growth in the absence of complementation. In both cases, the strains with an additional copy of either ispF or yacN, at the araBAD or amyE loci, respectively, were able to grow equally well in both the presence and absence of inducer. These results show that the poor growth of E. coli strain EB370 and B. subtilis strain EB323 in the absence of inducer is a consequence of the depletion of MEC synthase. The small amount of growth observed with E. coli strain EB370 in the absence of arabinose can be attributed to incomplete depletion of MEC synthase, which requires further passage in the absence of arabinose. These experiments represent the first time that yacN has been conditionally complemented and illustrate the tight control of MEC synthase expression that has been achieved in this work.
FIG. 1.
Conditional complementation of deletions in ispF and yacN. (A) Arabinose dependence of E. coli ispF deletion strain EB370. E. coli strains EB356 and EB370 were plated on LB/KAN in the presence and absence of arabinose and grown overnight at 37°C. (B) Xylose dependence of B. subtilis yacN deletion strain EB323. B. subtilis strains EB315 and EB323 were plated on LB/CM in the presence or absence of xylose and grown overnight at 30°C.
IspF- and YacN-depleted cells show altered cell morphology from the wild type and from one another.
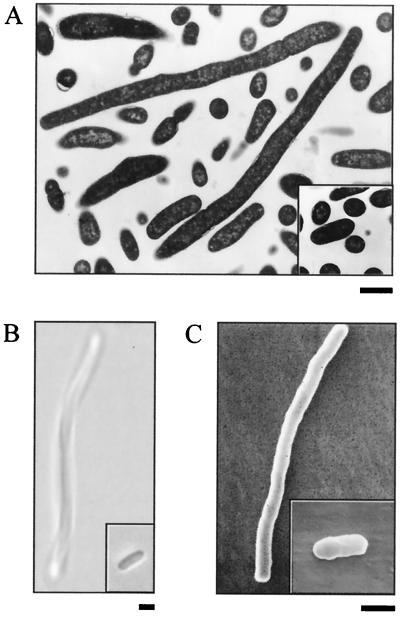
The growth of both deletion strains was minimal in the absence of inducer, but with a heavy inoculum, enough cells could be obtained for microscopic examination. Depletion of the 2-C-methyl-d-erythritol-2,4-cyclodiphosphate synthase (MEC synthase) in E. coli led to a filamentous phenotype (Fig. 2). To examine the ultrastructure of these long filamentous cells, transmission electron microscopy was used (Fig. 2A). Most striking was the finding that there do not appear to be any visible septa within the filaments, although there may be some points of constriction visible near the ends of the cells. It is difficult to determine if the filaments are plurinucleoid, because distinct DNA-containing regions are indiscernible. Further examination by light microscopy revealed cells up to 36 μm in length, 18 times longer than wild-type cells (Fig. 2B). To examine the surface morphology of the cells in detail, scanning electron microscopy was used (Fig. 2C). These micrographs indicate that despite the filamentous shape, IspF-depleted cells have a normal surface appearance comparable to the exterior of wild-type E. coli cells. It is noteworthy that no visible constrictions were observed, and the filaments appeared to be one long continuous cell. In all cases, the appearance of E. coli strain EB370 grown in the presence of arabinose was identical to that of E. coli strain EB356 (data not shown). Overall, the most striking phenotypic characteristic associated with IspF depletion in E. coli was the presence of long filamentous cells that appear to be lacking septa.
FIG. 2.
Characterization of E. coli ispF deletion strain by microscopy. E. coli strains EB370 (ispF deletion) and EB356 (wild-type ispF [inset]) were grown in the absence of arabinose and were visualized by light microscopy (A), transmission electron microscopy (B), and scanning electron microscopy (C). Each size bar represents 1 μm. Cells were grown overnight at 37°C on LB/KAN plates with no arabinose.
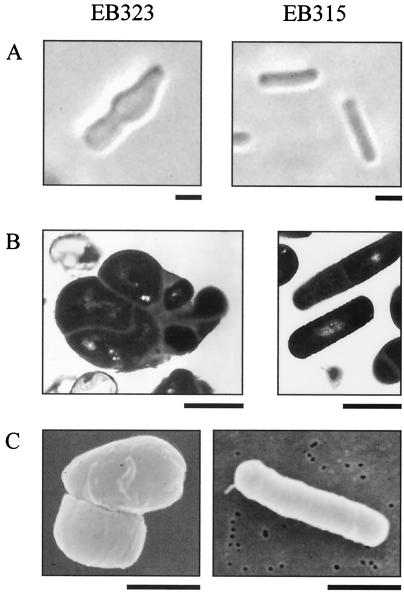
Similar examination of B. subtilis YacN-depleted cells compared with strain EB315 is illustrated in Fig. 3. Light microscopy (Fig. 3A) shows that depletion of MEC synthase resulted in an altered cell shape from the normal rod-shaped cells of B. subtilis. There was bulging around the central region of the cell, and in some cases, there were clumps of almost spherical cells (data not shown). Ultrastructural examination of these cells is shown in Fig. 3B. The MEC synthase-depleted strain (EB323) was a multicompartmentalized cell of irregular shape. The cell walls were of various thicknesses in different regions of the cell, and the septa were not confined to the medial region of the cell, but were located irregularly throughout the entire bacterium. Some of the compartments also appeared to lack a well-defined nucleoid region and likely lack chromatin bodies. Surface examination of MEC synthase-depleted cells is shown in Fig. 3C. Again, what is obvious is the altered cell morphology from the ordinary rod shape, and in addition, there was the appearance of grooves along the surface with a large furrow at the central portion of the cell. In all cases, cells of strain EB315 (right panels) were similar in appearance to wild-type B. subtilis cells, and fully complemented strain EB323 resembled EB315 (data not shown). These micrographs show that depletion of MEC synthase in E. coli and B. subtilis causes an altered cellular morphology from that of cells with normal levels of this protein. Additionally, the phenotype observed in E. coli was markedly different from that seen in B. subtilis, despite the depletion of orthologous proteins.
FIG. 3.
Characterization of B. subtilis yacN deletion strain by microscopy. B. subtilis strains EB323 (yacN deletion [left]) and EB315 (wild-type yacN [right]) were grown in the absence of xylose and were visualized by light microscopy (A), transmission electron microscopy (B), and scanning electron microscopy (C). Each size bar represents 1 μm. Cells (EB323) were grown overnight at 30°C on LB/CM/SPEC plates with no xylose.
YacN-depleted cells show an altered growth rate and morphology depending on the level of inducer.
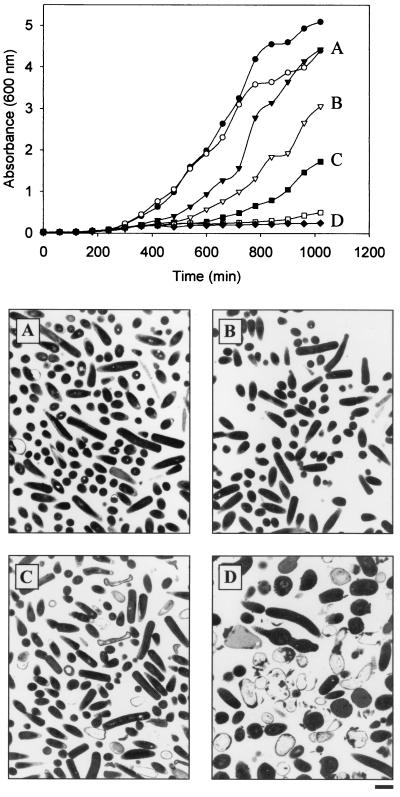
Figure 4 highlights the impact of depletion of MEC synthase in the B. subtilis yacN mutant (EB323), where the growth in liquid media was monitored at various concentrations of inducer relative to that of the control strain (EB315). At 2% xylose, B. subtilis strain EB323 had a slightly increased lag period compared with the control (EB315), but was able to grow at the same rate during the exponential phase and reached a similar final cell density, as determined by the A600. As the level of inducer was decreased, an increase in the lag phase was observed along with a decrease in the growth rate during the exponential phase. In the absence of any inducer, there was almost no discernible growth over the time course of the experiment. These results indicate that the ability of B. subtilis strain EB323 to grow is related to the expression of MEC synthase in the cell.
FIG. 4.
Growth and morphology of YacN-depleted B. subtilis cells. B. subtilis strain EB323 was grown overnight on LB/CM/SPEC/xylose plates and used to inoculate LB/CM/SPEC with 2% (▾, A), 0.2% (▿, B), 0.063% (▪, C), 0.02% (□), or no (⧫, D) xylose. B. subtilis strain EB315 was grown overnight on LB/CM plates and inoculated into LB/CM with 2% xylose (•) or no xylose (○). Growth was monitored at 30°C for 17 h. For the transmission electron micrographs, cells were grown overnight at 30°C on LB/CM/SPEC plates with 2%, 0.2%, 0.063%, or no xylose. The size bar represents 1 μm and applies to all micrographs.
Transmission electron micrographs were taken of B. subtilis strain EB323 at the inducer levels used in Fig. 4. At the 2 and 0.2% levels of inducer, the cells appeared as normal rod-shaped B. subtilis. At 0.063% xylose, the cells were still normal in appearance, but cell lysis was beginning to occur. In the absence of inducer, cells were altered in their morphology and showed a loss of rod shape and multiple septa with irregular wall thickness. Interestingly, as the level of inducer was decreased, there was a corresponding decrease in growth rate, but the cells remained normal in appearance until full depletion was achieved.
In this work, we have confined our microscopic analysis of the incremental depletion of MEC synthase to B. subtilis. In E. coli, using the araBAD promoter, the autocatalytic nature of arabinose transport is understood to lead to a mixed population of fully induced and fully repressed cells at subsaturating concentrations of inducer (37).
IspF- and YacN-depleted strains are sensitive to cell wall-active antibacterial agents.
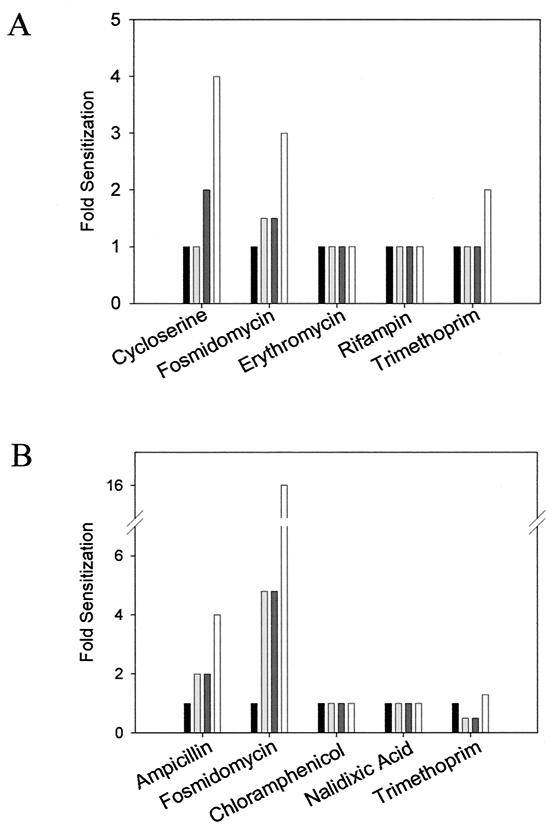
We have exploited the principle of synthetic lethal interactions in an attempt to better understand the mechanism of MEC synthase depletion. The susceptibility of E. coli and B. subtilis strains depleted of MEC synthase to a diverse collection of antibacterial agents was tested. These agents targeted cell wall, protein synthesis, nucleic acid synthesis, metabolic pathways, and the cell membrane (Table 3). For each of these targets, the antibiotics selected were of different chemical classes, where possible, in an effort to establish a consensus by using multiple and diverse probes of each. Table 3 shows that both E. coli and B. subtilis complemented mutants showed an increased sensitivity (represented by a lower MIC) to cell wall inhibitors as the level of inducer was decreased. We show a subset of the graphical data for these experiments in Fig. 5. A similar sensitization was noted with the antibiotic fosmidomycin. Fosmidomycin is a known inhibitor of the DOXP pathway, inhibiting the enzyme MEP synthase, which represents the first committed step in the pathway (20, 21). In contrast, the potency of inhibitors of protein synthesis, nucleic acid synthesis, one-carbon metabolism (trimethoprim), and membrane integrity was unaffected by the level of inducer present. In a few of these cases, however, a slight decrease in MIC was seen at the lowest level of inducer tested. This is presumably due to a more trivial relationship between antibiotic potency and general cell fitness due to extreme depletion of MEC synthase. In aggregate, these experiments demonstrated a synthetic lethal interaction between cell wall-active agents and the DOXP pathway inhibitor fosmidomycin with the ispF and yacN mutants.
TABLE 3.
Fold sensitivity of B. subtilis and E. coli MEC synthase depletion strains to a variety of antibacterial agentsa
| Antibacterial agent | Fold sensitivity of:
|
|
|---|---|---|
| B. subtilis EB323 | E. coli EB370 | |
| Cell wall inhibitors | ||
| Ampicillin | 2 | 4 |
| Bacitracin | 4 | NPb |
| Cycloserine | 4 | 1.5 |
| Fosfomycin | 3 | 5 |
| Vancomycin | 2 | NP |
| Protein synthesis inhibitors | ||
| Chloramphenicol | NP | 1 |
| Tetracycline | 2 | 2 |
| Amikacin | 2 | 1 |
| Neomycin | 2 | NP |
| Spectinomycin | NP | 1.5 |
| Erythromycin | 1 | NP |
| Thiostrepton | 0.8 | NP |
| Puromycin | 1 | NP |
| Nucleic acid synthesis inhibitors | ||
| Nalidixic acid | 0.5 | 1 |
| Ciprofloxacin | 2 | 1 |
| Rifampin | 1 | NP |
| Metabolic inhibitors | ||
| Trimethoprim | 2 | 1.3 |
| Fosmidomycin | 3 | 16 |
| Membrane inhibitor | ||
| Polymyxin B sulfate | NP | 2 |
The range of inducer tested for B. subtilis strain EB323 was 2, 0.2, 0.063, and 0.02% xylose. The range of inducer tested for E. coli strain EB370 was 0.2, 0.02, 0.0063, and 0.002% arabinose. In both cases, the samples with no inducer did not grow, and those data have not been included in the results. Fold sensitization in MIC is calculated from the lowest level of inducer tested that allowed for growth with respect to the fully complemented deletion strain.
NP, not possible.
FIG. 5.
Sensitivity of E. coli and B. subtilis MEC synthase-depleted cells to various antibacterial agents. (A) Change in MIC for the B. subtilis MEC synthase deletion strain (EB323) with various concentrations of inducer to representative antibacterial agents. Fold sensitization in MIC is calculated with respect to the fully complemented deletion strain (2% xylose). From left to right, the amounts of xylose are as follows: 2, 0.2, 0.063, and 0.02%. Cells were grown in a volume of 100 μl in a 96-well microtiter plate for 24 h, and then the A600 was read. (B) Change in sensitivity of E. coli MEC synthase deletion strain (EB370) to antibacterial agents with various concentrations of inducer. Fold sensitization in MIC is calculated with respect to the fully complemented deletion strain (0.2% arabinose). From left to right, the amounts of arabinose are as follows: 0.2, 0.02, 0.0063, and 0.002%. Cells were grown in a volume of 150 μl in a 96-well microtiter plate for 16 h, and then the A600 was read. In all cases, the MIC was determined as the lowest concentration of drug that resulted in no growth (less than 0.1 absorbance unit at 600 nm).
DISCUSSION
In bacteria, isoprenoids are thought to be required for three distinct purposes: the modification of tRNA, believed to serve as a regulatory device (5), the formation of dolichols, and the production of the respiratory quinones (36). It follows that, if nonredundant, the DOXP pathway for isoprenoid biosynthesis ought to be indispensable for the growth of E. coli and B. subtilis. Furthermore, it is possible that isoprenoids also have unrecognized roles in bacterial physiology. We have endeavored in this work to rigorously address the dispensability question for MEC synthase of the DOXP pathway for isoprenoid biosynthesis. In addition, we have probed the mechanism and dominant physiological effects of a lesion in the DOXP pathway with mutants with conditional mutations in the ispF and yacN genes encoding MEC synthase in E. coli and B. subtilis.
We have shown that MEC synthase is essential for the growth of E. coli and B. subtilis. Depletion of these products resulted in a decreased growth rate and altered cell morphology; however, most striking was the finding that MEC synthase-depleted E. coli and B. subtilis have very different cell shapes and ultrastructures. In E. coli, depletion of IspF resulted in long filamentous cells, which appear to lack septa. Although peptidoglycan biosynthesis is required for both elongation and septum formation in E. coli, these processes represent two distinct morphogenic pathways, and so it is possible for one to be functional in the absence of the other (25). Long filamentous E. coli cells typically result from an inability to form septa (13). Multinucleate, filamentous cells with blunt constrictions are seen with FtsI (PBP3) mutants (12, 27), while FtsZ mutants give smooth filaments with no visible constrictions (27). FtsI has dideoxy-transpeptidase activity and is required for septal murein synthesis (13). FtsZ acts before FtsI and is required to form a cytokinetic ring at the division site followed by the assembly of murein biosynthetic and hydrolytic enzymes (27). The E. coli MEC synthase deletion strain created in this work was filamentous, with possible points of constriction. Synthesis of peptidoglycan requires dolichol phosphates in the cell membrane as carriers for assembly of disaccharide pentapeptide (36). Interestingly, it is believed that the amount of dolichol monophosphates available to the bacterium is the rate-limiting step in peptidoglycan biosynthesis (16). It follows, therefore, that depletion of MEC synthase in this work may have impact primarily at the point of septal murein synthesis, while elongation remained relatively unaffected.
In B. subtilis, depletion of YacN resulted in globular, multicompartmentalized cells with inconsistent wall thickness and regions lacking nucleoid bodies. The rod-to-sphere transition observed in this work is typical of the general response in B. subtilis to inhibition of peptidoglycan biosynthesis. Especially striking, however, is the phenotypic similarity to a conditional mutant in teichoic acid biosynthesis, tagD, which has been reported on recently (3). TagD encodes glycerol 3-phosphate cytidylyltransferase, which is required for biosynthesis of cell wall teichoic acid. Both TagD- and YacN-depleted strains showed multicompartmentalization and aberrant septum formation along with wall thickening, implying that these features of the phenotype seen with YacN depletion may be a result of a teichoic acid deficiency. Wall teichoic acids are polymers of substituted glycerol or ribitol linked by phosphodiester bridges that are covalently attached to peptidoglycan (1). The biosynthesis of wall teichoic acids requires dolichol phosphate carriers, and so it is reasonable to speculate that teichoic acid biosynthesis may be affected by depletion of MEC synthase. Additionally, because gram-negative bacteria do not possess teichoic acids, such an effect would not be seen in E. coli, consistent with distinct morphologies observed for the two organisms here.
To further probe the predominant mechanism by which MEC synthase depletion is growth inhibitory, we exploited the principle of synthetic lethal interaction. Synthetic lethal screens allow for the identification of interactions between unlinked genes and facilitate the determination of functional relationships (38, 41). Both ispF and yacN were placed under tight control of inducible promoters, and the expression of each gene was varied in the presence of a variety of antibacterial agents with diverse mechanisms of action. As inducer levels were decreased, both strains showed an increased sensitivity to cell wall inhibitors and to fosmidomycin. Fosmidomycin was a valuable control in this study, because it inhibits MEP synthase, the first enzyme of the DOXP pathway (20, 21), and sensitization to this antibiotic is consistent with the principle of synthetic lethality. That cell wall inhibitors were also synergistic with MEC synthase depletion suggests that the dominant impact of a lesion in this step was in cell wall biosynthesis in both E. coli and B. subtilis.
From this work, it is clear that depletion of MEC synthase in both E. coli and B. subtilis has an early and significant impact on cell wall biosynthesis and leads ultimately to cell death. Cell wall biosynthesis has been and continues to be one of the foremost targets for antibacterial therapeutics, in part, due to the catastrophic outcomes of complete loss of cell shape and lysis that accompany its inhibition. That the DOXP pathway appears to have a particular impact on cell wall biosynthesis in both E. coli and B. subtilis augurs well for the therapeutic potential of this recently discovered route to isoprenoid synthesis.
Acknowledgments
We thank Barry Wanner for pKD46 and for advice and help in creating the E. coli deletion strain and Tamara O'Connor for assistance with the light microscopy. We appreciate the assistance of Amit Bhavsar and Andy Ho in creating the clones for the yacN deletion strain.
This research was supported by a postgraduate fellowship to T.L.C. from the Natural Sciences and Engineering Research Council of Canada. E.D.B. holds a Canada Research Chair in Microbial Biochemistry.
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 2.Arigoni, D., S. Sagner, C. Latzel, W. Eisenreich, A. Bacher, and M. H. Zenk. 1997. Terpenoid biosynthesis from 1-deoxy-d-xylulose in higher plants by intramolecular skeletal rearrangement. Proc. Natl. Acad. Sci. USA 94:10600-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhavsar, A. P., T. J. Beveridge, and E. D. Brown. 2001. Precise deletion of tagD and controlled depletion of its product, glycerol 3-phosphate cytidylyltransferase, leads to irregular morphology and lysis of Bacillus subtilis grown at physiological temperature. J. Bacteriol. 183:6688-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjork, G. R., J. U. Ericson, C. E. Gustafsson, T. G. Hagervall, Y. H. Jonsson, and P. M. Wikstrom. 1987. Transfer RNA modification. Annu. Rev. Biochem. 56:263-287. [DOI] [PubMed] [Google Scholar]
- 6.Briehl, M., H. M. Pooley, and D. Karamata. 1989. Mutants of Bacillus subtilis 168 thermosensitive for growth and wall teichoic acid biosynthesis. J. Gen. Microbiol. 135:1325-1334. [Google Scholar]
- 7.Campos, N., M. Rodriguez-Concepcion, S. Sauret-Gueto, F. Gallego, L. M. Lois, and A. Boronat. 2001. Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate: a novel system for the genetic analysis of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem. J. 353:59-67. [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, F. X., Jr., T. P. Lafond, and E. Gantt. 2000. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 182:5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. V. Horn. 1990. Genetic analysis, p. 27-61. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Toronto, Ontario, Canada.
- 10.Cutting, S. M., and P. Youngman. 1994. Gene transfer in gram-positive bacteria, p. 348-364. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC.
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePedro, M. A., W. D. Donachie, J.-V. Höltje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenreich, W., M. Schwarz, A. Cartayrade, D. Arigoni, M. H. Zenk, and A. Bacher. 1998. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 5:R221-R233. [DOI] [PubMed] [Google Scholar]
- 15.Freiberg, C., B. Wieland, F. Spaltmann, K. Ehlert, H. Brotz, and H. Labischinski. 2001. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 3:483-489. [PubMed] [Google Scholar]
- 16.Fujisaki, S., T. Nishino, and H. Katsuki. 1986. Biosynthesis of isoprenoids in intact cells of Escherichia coli. J. Biochem. 99:1137-1146. [DOI] [PubMed] [Google Scholar]
- 17.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht, S., W. Eisenreich, P. Adam, S. Amslinger, K. Kis, A. Bacher, D. Arigoni, and F. Rohdich. 2001. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA 98:14837-14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herz, S., J. Wungsintaweekul, C. A. Schuhr, S. Hecht, H. Luttgen, S. Sagner, M. Fellermeier, W. Eisenreich, M. H. Zenk, A. Bacher, and F. Rohdich. 2000. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate to 2C-methyl-d-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. USA 97:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Turbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 21.Koppisch, A. T., D. T. Fox, B. S. J. Blagg, and C. D. Poulter. 2002. E. coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry 41:236-243. [DOI] [PubMed] [Google Scholar]
- 22.Lange, B. M., M. R. Wildung, D. McCaskill, and R. Croteau. 1998. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 95:2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenthaler, H. K. 2000. Sterols and isoprenoids. Biochem. Soc. Trans. 28:785-789. [PubMed] [Google Scholar]
- 24.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutkenhaus, J., and E. Bi. 2001. Cell morphogenesis and division. Curr. Opin. Microbiol. 4:621-624. [Google Scholar]
- 26.Luttgen, H., F. Rohdich, S. Herz, J. Wungsintaweekul, S. Hecht, C. A. Schuhr, M. Fellermeier, S. Sagner, M. H. Zenk, A. Bacher, and W. Eisenreich. 2000. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-d-erythritol. Proc. Natl. Acad. Sci. USA 97:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohdich, F., W. Eisenreich, J. Wungsintaweekul, S. Hecht, C. A. Schuhr, and A. Bacher. 2001. Biosynthesis of terpenoids: 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (IspF) from Plasmodium falciparum. Eur. J. Biochem. 268:3190-3197. [DOI] [PubMed] [Google Scholar]
- 29.Rohdich, F., S. Hecht, K. Gartner, P. Adam, C. Krieger, S. Amslinger, D. Arigoni, A. Bacher, and W. Eisenreich. 2002. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. USA 99:1158-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohdich, F., K. Kis, A. Bacher, and W. Eisenreich. 2001. The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr. Opin. Chem. Biol. 5:535-540. [DOI] [PubMed] [Google Scholar]
- 31.Rohdich, F., J. Wungsintaweekul, M. Fellermeier, S. Sagner, S. Herz, K. Kis, W. Eisenreich, A. Bacher, and M. H. Zenk. 1999. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA 96:11758-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohmer, M., M. Knani, P. Simonin, B. Sutter, and H. Sahm. 1993. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohmer, M., M. Seemann, S. Horbach, S. Bringer-Meyer, and H. Sahm. 1996. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 118:2564-2566. [Google Scholar]
- 34.Sacchettini, J. C., and C. D. Poulter. 1997. Creating isoprenoid diversity. Science 277:1788-1789. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sherman, M. M., L. A. Petersen, and C. D. Poulter. 1989. Isolation and characterization of isoprene mutants of Escherichia coli. J. Bacteriol. 171:3619-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simons, A. H., N. Dafni, I. Dotan, Y. Oron, and D. Canaani. 2001. Genetic synthetic lethality screen at the single gene level in cultured human cells. Nucleic Acids Res. 29:E100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprenger, G. A., U. Schorken, T. Wiegert, S. Grolle, A. A. de Graaf, S. V. Taylor, T. P. Begley, S. Bringer-Meyer, and H. Sahm. 1997. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 94:12857-12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, S., T. Kuzuyama, H. Watanabe, and H. Seto. 1998. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95:9879-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong, A. H. Y., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. V. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systemic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 42.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]