Abstract
A growing body of evidence supports the coordination of pre-mRNA processing and transcriptional regulation. We demonstrate here that mammalian PRP4 kinase (PRP4K) is associated with complexes involved in both of these processes. PRP4K is implicated in pre-mRNA splicing as the homologue of the Schizosaccharomyces pombe pre-mRNA splicing kinase Prp4p, and it is enriched in SC35-containing nuclear splicing speckles. RNA interference of Caenorhabditis elegans PRP4K indicates that it is essential in metazoans. In support of a role for PRP4K in pre-mRNA splicing, we identified PRP6, SWAP, and pinin as interacting proteins and demonstrated that PRP4K is a U5 snRNP-associated kinase. In addition, BRG1 and N-CoR, components of nuclear hormone coactivator and corepressor complexes, also interact with PRP4K. PRP4K coimmunoprecipitates with N-CoR, BRG1, pinin, and PRP6, and we present data suggesting that PRP6 and BRG1 are substrates of this kinase. Lastly, PRP4K, BRG1, and PRP6 can be purified as components of the N-CoR-2 complex, and affinity-purified PRP4K/N-CoR complexes exhibit deacetylase activity. We suggest that PRP4K is an essential kinase that, in association with the both U5 snRNP and N-CoR deacetylase complexes, demonstrates a possible coordination of pre-mRNA splicing with chromatin remodeling events involved in transcriptional regulation.
The regulation of eukaryotic gene expression involves the modulation of chromatin structure and the coordinated transcription and splicing of mRNA. Both histone modification (61) and chromatin remodeling modulate the access of the transcriptional apparatus to chromatin (14, 44). The SWI/SNF proteins are the archetypal family of ATPases involved in remodeling chromatin, and members of this family in both yeast (Snf2) and mammals (BRG1) have been isolated in association with the RNA polymerase II holoenzyme complex (45, 59). Once transcription has been initiated it is thought that pre-mRNA splicing occurs cotranscriptionally (4, 22). A number of recent studies have also shown links between corepressor and coactivator complexes and pre-mRNA splicing (e.g., PSF [40] and PGC1 [42], respectively), which suggests yet another level of coordination between the regulation of gene expression and pre-mRNA splicing.
If chromatin remodeling and pre-mRNA splicing occur in coordination with transcription, there may be proteins that either play a direct role in both chromatin structure and splicing and/or interact with factors involved in each process. Previously, we isolated a murine gene-trap protein, designated CT143 (56), which is similar to Schizosaccharomyces pombe Prp4 kinase (17). Prp4p is essential for pre-mRNA splicing, as demonstrated by the accumulation of unspliced pre-mRNA at the restrictive temperature in yeast carrying a temperature-sensitive (ts) mutation in prp4 (2). Prp4p interacts genetically with other S. pombe splicing proteins, including the non-SR splicing proteins Prp1p (a homologue of Saccharomyces cerevisiae Prp6p) and Prp5p (51). Multiple rounds of phosphorylation and dephosphorylation of spliceosomal components are thought to occur during spliceosomal maturation (reviewed in reference 54) and at least one snRNP-associated kinase activity has been described (U1 snRNP-associated kinase) (60). The mammalian homologues of the Prp4-interacting proteins Prp5p (PRL1) (46) and Prp1p (PRP6) (38) associate with the spliceosome. Although it is not known whether Prp4p associates directly with the snRNPs in S. pombe, it has been suggested that it may regulate the formation of the active spliceosome complex by phosphorylating non-SR components (51).
Here we have characterized the human and mouse full-length PRP4 kinase (PRP4K) proteins that correspond to the gene trapped in the murine CT143 cell line (56). The protein contains a C-terminal dual specificity kinase domain similar to that found in the CLK/STY protein kinase family that is highly conserved between mammals and fission yeast. In addition, mammalian PRP4K has an extended N terminus that contains subdomains rich in lysine-histidine (KH) or arginine-serine (RS) dipeptides that are similar to those found in several splicing factors. This basic N-terminal region is present in metazoan PRP4K homologues, including those of Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana, but is not present in S. pombe Prp4p. Disruption of C. elegans PRP4K by RNA interference (RNAi) results in early embryonic lethality, indicating that this kinase is essential in metazoans. Antibodies directed against the N terminus of mammalian PRP4K detect some endogenous protein on chromosomes at mitosis, whereas in interphase nuclei the protein is concentrated in speckles that colocalize with the splicing factor SC35. We show that that the N-terminal RS-like domain of PRP4K is sufficient for targeting to speckles. Consistent with a role in pre-mRNA splicing, we demonstrate that PRP4K can interact with pre-mRNA splicing factors, including PRP6 and Suppressor-of-white apricot (SWAP), and that it copurifies with the U5 snRNP. Surprisingly, we also identified BRG1 and N-CoR, components of large multiprotein complexes involved in gene regulation mediated by the nuclear hormone receptors (8, 21, 23, 43), as PRP4K-interacting proteins via yeast two-hybrid analysis. We demonstrate that N-CoR and BRG1 interact in vivo with human PRP4K and copurify together as components of N-CoR containing deacetylase complexes. Our findings provide a novel and unexpected connection between chromatin-mediated regulation of transcription and pre-mRNA splicing and suggest that PRP4K, via its interaction with both the U5-snRNP and deacetylase complexes containing N-CoR, may coordinate these processes in metazoans.
MATERIALS AND METHODS
Cloning of human and murine PRP4K.
CT143 (AF033663) (56) sequences were used to probe a λgt11 11-day-old mouse embryo cDNA library (Clontech). Overlapping clones were isolated that represented nearly full-length murine PRP4K (MmPRP4K). 5′-Rapid amplification of cDNA ends was used to extend these sequences as previously described (56).
Human PRP4K (HsPRP4K) was assembled from expressed sequence tags (ESTs) (AF074687, AU123782, AU120988, and AI032816) corresponding to portions of MmPRP4K. Intervening sequences between these ESTs were obtained by reverse transcription-PCR from total HeLa RNA prepared by using Bio/RNA-X-cell (Bio/Gene, Ltd.).
Bioinformatic analysis of PRP4K and its related homologues.
Homologues of mammalian PRP4K in other species were identified by using the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST/), and protein domain and motif analysis was done by using PROSITE (www.isrec.isb-sib.ch). Phylogeny trees were constructed by using the PAUPSEARCH program (GCG 10 package [www.hgmp.mrc.ac.uk]).
dsRNA interference in C. elegans.
Double-stranded RNA (dsRNA) preparation and microinjection were performed as previously described (37). Briefly, templates for dsRNA fragments were generated by PCR from C. elegans genomic DNA with gene-specific primers containing T3 and T7 sequences added onto forward and reverse primers, respectively (exon 2F22D6-1 [T3 promoter], ATTAACCCTCACTAAAGGGAAGAATGGAAAATGATGGTCCTCCAGC; exon 2F22D6-2 [T7 promoter], TAATACGACTCACTATAGGAATTCGTCGCTTTCTTGCCGCTTC). PCR products were gel purified and used as templates for in vitro RNA synthesis by using T3 and T7 RNA polymerase (Roche). RNA was dissolved in sterile water with 0.4 U of RNase inhibitor (Roche)/μl to a final concentration of 0.5 μg/μl. dsRNA was assembled by mixing equal amounts of sense and antisense RNA, followed by incubation at 68°C for 10 min and then 37°C for at least 30 min. Approximately 10 to 15 young adult hermaphrodites (Bristol strain N2) were injected with dsRNA (in the gonads and guts) and were left to recover overnight. Animals were transferred to individual plates from which the phenotype was observed in F1 progeny. Affected progeny were examined by using differential interference contrast microscopy and DAPI (4′,6′-diamidino-2-phenylindole) staining.
Production of affinity-purified antibodies recognizing PRP4K.
Two peptides (MRC1 [RRAKSRSLERKRREPERRRLC] and MRC2 [SKERTRHRSDKRKSKC]; Fig. 1A) were generated and coupled to keyhole limpet hemocyanin (Severn Biotech). Both peptides were used to generate sheep antibodies (Diagnostic Scotland). In addition, a glutathione S-transferase (GST) fusion protein containing sequences encoding 504 to 688 amino acids (aa) of human PRP4K (GST-H143 fusion) cloned into the vector pGEX-5x1 (Pharmacia) was also used to generate sheep antibody (H143). To affinity purify antibodies specific for each peptide, sheep immune serum was diluted 1:10 in phosphate-buffered saline (PBS) and passed over peptide affinity columns, which were produced by coupling either peptide to 2 ml of Sulfolink Gel (Pierce) according to the manufacturer's specifications. In the case of sheep antibodies directed against the GST-H143 fusion protein, serum was affinity purified against the GST-H143 protein-coupled CNBr-activated Sepharose 4B (Pharmacia) according to the manufacturer's protocol. Antibody specificity was determined by Western analysis against a GST fusion protein containing the MRC1 or MRC2 peptide sequence cloned into the expression vector pGEX-5x1 or directly against the GST-H143 protein. GST fusion proteins were purified from BL21(DE3) cells (Stratagene).
FIG. 1.
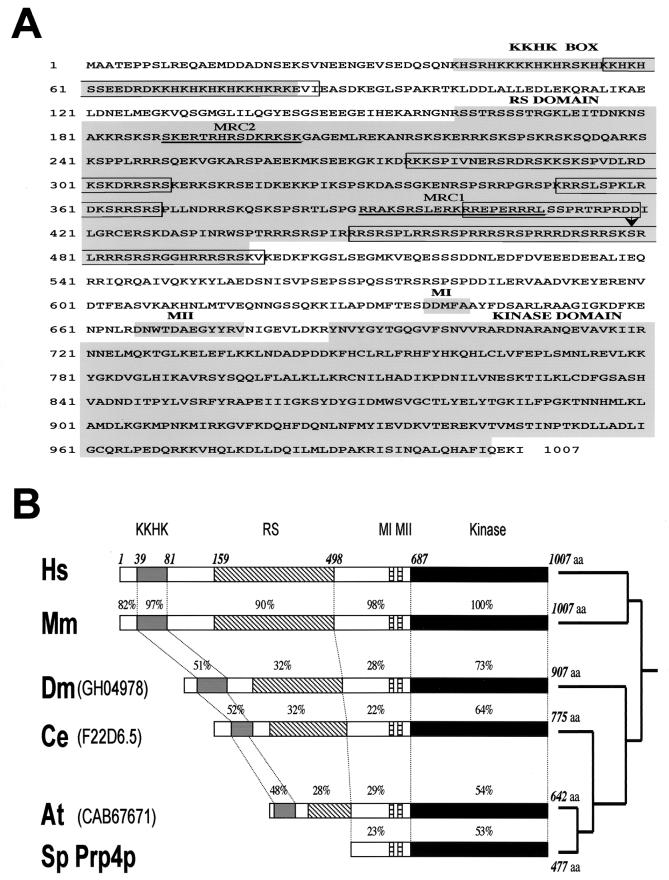
Primary structure of murine PRP4K and phylogenic comparison of PRP4K homologues. (A) Amino acid sequence and domain structure of murine PRP4K. The mouse and human proteins are 1,007 aa in length and share 95% identity and 97% similarity (accession numbers HsPRP4K, AF283465, MmPRP4K, and AF283466). Conserved domains (KKHK box, RS-like domain, MI and MII) and the kinase domain of MmPRP4K are shaded in gray. Unlike the canonical RS domain of splicing proteins such as SC35, the RS-like domain of PRP4K is extended and shares similarities with RS-like domains of the U1 70K and SRp75 splicing factors. Bipartite nuclear localization signals are boxed. Peptides MRC1 and MRC2, used to produce antibodies, are underlined, and the arrow indicates the site of gene-trap integration in cell line CT143 (56). (B) Domain structure and phylogenic comparison of S. pombe Prp4 (Sp Prp4p) and PRP4K homologues in humans (Hs), mice (Mm), D. melanogaster (Dm), C. elegans (Ce), and A. thaliana (At). The amino acid position of each domain is shown above the human sequence only. The position of each conserved domain within these homologues is shown relative to HsPRP4K. The length of each protein is shown at the right, and the percentage identity to human PRP4K is shown above each domain. The evolutionary relationship between these proteins is depicted as a phylogeny tree at the far right.
Subcellular fractionation and Western blot analysis.
Cytoplasmic and nuclear fractions were prepared from tissue culture cells according to the protocol of Andrews and Faller (3). Purified nuclei were either lysed directly in 2× Laemmli sample buffer or extracted with 200 mM, 400 mM, or 1 M KCl as previously described (3). Briefly, nuclei were washed once in buffer NB (20 mM HEPES [pH 7.9], 25% glycerol, 85 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 200 μg of phenylmethylsulfonyl fluoride [PMSF]/ml, 0.5 mM dithiothreitol [DTT]) and then resuspended in buffer NB containing 200 mM, 400 mM, or 1 M KCl, followed by incubation on ice for 30 min at 4°C (with gentle agitation by using a rotary mixer). Nuclei were then centrifuged for 30 min at 12,000 × g, after which nuclear pellet (fraction P) and the extracted protein in the supernatant (fraction S) were resuspended in 2× Laemmli sample buffer and boiled.
For Western blot analysis of immunoprecipitation (IP) reactions or protein fractions, samples were boiled in 2× Laemmli sample buffer prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and semidry transfer onto nitrocellulose membranes (Hybond C-Extra; Amersham). The primary antibodies used for Western blot analysis included anti-PRP4K (MRC2, 1:5,000 dilution), anti-BRG1 (N-15, 1:500; Santa Cruz), anti-hPRP6 (MukaJ, 1:500; M. Yamada and coworkers [47]), anti-N-CoR (1:2,500 [57]), anti-pinin (1:500; S. Sugrue), and anti-U5-102 K (i.e., anti-hPRP6) (1:500 [38]). The secondary antibodies used were horseradish peroxidase-conjugated anti-sheep (Jackson Immunoresearch Lab) and anti-rabbit and anti-mouse (Sigma) antibodies. Western blots were developed by enhanced chemiluminescence detection (Amersham) according to the manufacturer's instructions.
Cell culture, transfections, and indirect immunofluorescence.
Cells and chromosomes were prepared for immunofluorescence as previously described (56). For coimmunofluorescence with transfected epitope-tagged proteins, cells were transfected in six-well plates with 3 to 5 μg of each plasmid DNA (GFP-PRP4K (full-length MmPRP4K), GFP-RS (containing the RS domain and motifs MI and MII of MmPRP4K [188 to 780 aa]), pCGT7-PRP4K (T7-tagged HsPRP4K), GFP-PRP6 (M. Yamada), GFP-SWAP (R. Lafyatis), or myc-tagged pinin (S. Sugrue) by using Lipofectamine 2000 (Roche) according to the manufacturer's instructions. EGFP-N1 and EGFP-C1 plasmids (Clontech) were used to construct all green fluorescent protein (GFP)-tagged expression constructs. Plasmid pCGT7-PRP4K contains the full-length human PRP4K cDNA cloned into the XbaI/BamHI sites of pCGT7 (J. Caceres). Inhibition of transcription was accomplished by treating cells with either 5 μg of actinomycin D/ml for 2 h or by heat shock for 30 min at 45°C.
The primary antibodies used were anti-PRP4K (MRC2 [1:50 to 1:150] or H143 [1:100]), anti-BRG1 (H-88 [1:100]; Santa Cruz), anti-β-galactosidase (1:100; 5 Prime-3 Prime), anti-SC35 (1:1,000; Sigma), and anti-T7.tag (1:500; Novagen). All secondary antibodies (anti-sheep, -rabbit, and -mouse antibodies labeled with fluorescein isothiocyanate [FITC] or Texas red; Jackson Immunoresearch Lab) were used according to the manufacturer's specifications. Image acquisition was carried out by using a Zeiss Axioplan fluoresence microsope fitted with a Chroma 8300 filter set. Images were captured by using a Photometrics cooled charge-coupled device camera and IPLab software (Scanalytics).
Yeast two-hybrid analysis.
All yeast two-hybrid procedures were carried out with the PJ696 strain of yeast (J. Nasir) as outlined in the Yeast Protocols Handbook (document PT3024-1 [Clontech]). Either the N terminus (pGBN143, aa 2 to 644) or the C-terminal kinase domain (pGBKinase, aa 637 to 1007) of human PRP4K was cloned into the bait plasmid pGBT9 (Clontech), and strains carrying each plasmid were mated with a strain pretransformed with a human adult brain cDNA library cloned into pACT2 (Clontech). β-Galactosidase assays and confirmation of each bait and library clone interaction were carried out as previously described (PT3024-1 [Clontech]). The sequence of each positive library clone was determined by PCR-cycle sequencing (Big Dye; PE-Applied Biosystems). Only peptide sequences that were in frame with the activation domain of the GAL4 gene in pACT2 were considered true interacting clones.
In vitro dephosphorylation and IP.
Purified HeLa nuclei were prepared from a 10-cm tissue culture dish and then lysed in 1 ml of dephosphorylation buffer (50 mM Tris-HCl [pH 8.8], 300 mM KCl, 0.5% Triton X-100, 200 μg of PMSF/ml, 1× protease inhibitor cocktail [Roche]), incubated on ice for 30 min, and centrifuged at 12,000 × g for 20 min. Next, 25 μl of the supernatant (nuclear extract) was dephosphorylated with 50 U of calf intestinal alkaline phosphatase (CIAP; New England BioLabs) overnight at 37°C. Mock dephosphorylation was carried out with heat-inactivated CIAP and in the presence of 10 mM NaF, 1 mM sodium vanadate, and 40 mM β-glycerol phosphate. The dephosphorylation reactions were stopped by boiling in 2× Laemmli buffer for 5 min.
For IPs, HeLa cells were scraped from 10-cm tissue culture plates, collected in 1 ml of cold PBS, centrifuged at 270 × g for 3 min in a 1.5-ml Eppendorf tube, resuspended in 800 μl of IP buffer (50 mM Tris-HCl, 300 mM KCl, 5 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 200 μg of PMSF/ml, and 1× protease inhibitor cocktail), and then incubated for 30 min at 4°C (with continuous mixing on a rotary mixer). IP extracts were also generated from isolated nuclei by resuspending ∼107 HeLa nuclei (prepared as described above) in 800 μl of IP buffer. The IP extract was centrifuged at 12,000 × g for 20 min, after which the pellet was discarded; the supernatant (IP extract) was then divided into 200-μl aliquots and used immediately or snap-frozen for later use. IP extracts were precleared with 10 μl of rabbit or sheep serum prebound to 25 μl of protein A or G agarose, respectively, for 2 h. To 200 μl of IP extract, 2 to 5 μg of affinity purified antibody (MRC2, anti-PRP4K; H-88, anti-BRG1 [Santa Cruz]) or 10 μl of immune serum (anti-PRP4K antiserum; anti-102 K [PRP6] [38]) was added. The IP extract was then incubated at 4°C for 1 to 2 h, 25 μl of protein A or G agarose (Roche) was added, and incubation was continued for 3 h to overnight. IP reactions were then washed twice in 1 ml of IP buffer and four times in 1 ml of PBS (with 200 μg of PMSF/ml). Mock IPs were carried out with 10 μg of an unrelated rabbit polyclonal immunoglobulin G (IgG)/ml (rabbit anti-sheep IgG; Jackson Immunoresearch Lab). Peptide block IPs were carried out by preblocking the affinity-purified anti-PRP4K antibody, MRC2, with 2 μg of the MRC2 peptide/ml.
IP kinase assay.
For IP kinase assays, PRP4K, BRG1, and PRP6 IPs were prepared as described above in the presence of phosphatase inhibitors (10 mM NaF, 40 mM β-glycerol phosphate, 1 mM sodium vanadate [Sigma], 200 μg of PMSF/ml). Kinase immunoprecipitates were washed once in kinase buffer (25 mM Tris-HCl [pH 7.6], 10 mM MgCl2) and resuspended in 25 μl of kinase buffer supplemented with 10 μM ATP and 20 μCi of [γ-32P]ATP, followed by incubation at 30°C for 30 to 60 min. Control kinase reactions were carried out by mock IP and peptide block IP as described above. The kinase reaction was stopped by boiling in 2× Laemmli sample buffer. The phosphorylated proteins were then separated by SDS-PAGE and visualized by autoradiography.
Isolation of human snRNPs and glycerol gradient centrifugation.
Nuclear extracts were prepared from HeLa cells according to the method of Dignam et al. (11). Total snRNPs, containing U1, U2, U4/U6, and U5 snRNPs, were purified from nuclear extract by affinity chromatography at 250 mM NaCl with the monoclonal antibody H20 raised against the m3G cap according to the method of Will et al. (58) and then separated into 12S U1/U2, 20S U5, and 25S U4/U6.U5 snRNPs by glycerol gradient centrifugation (31). For fractionation, 0.3 mg of total snRNPs was layered onto a linear 4-ml, 10 to 30% (wt/wt) glycerol gradient prepared with buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithioerythreitol) and then centrifuged at 29,000 rpm in a Sorval TH660 rotor for 14 h 30 min at 4°C. Twenty-three fractions of 175 μl each were harvested manually from top to bottom. Then, 50-μl aliquots were directly subjected to SDS-10% PAGE, followed by staining with Coomassie blue. For immunostaining experiments, the proteins from each fraction were separated by SDS-8% PAGE, transferred electrophoretically to Hybond-P membranes (Amersham), and probed with anti-PRP4K (MRC2, 1:4,000) and anti-PRP6 (anti-U5-102K, 1:3,000) (38).
Purification of N-CoR containing complexes from HeLa cells.
N-CoR-1 and -2 complexes were purified from nuclear extract prepared from 40 liters of HeLa cells grown to mid-log phase (National Cell Culture Center, Minneapolis, Minn.) as previously described (57). In the case of the PRP4K/N-CoR complex, nuclear extract was first dialyzed against buffer A (20 mM Tris-HCl [pH 7.9]; 0.5 mM EDTA; 0.5 mM EGTA; 10% glycerol; 0.5 mM DTT; 0.2 mM PMSF; 5 μg each of leupeptin, aprotinin, and pepstatin/ml) containing 100 mM KCl and was loaded onto an 80-ml P11 phosphocellulose column preequilibrated with the same buffer. The flowthrough was collected (100 mM fraction), and the column was washed with 2 column volumes of buffer A containing 100 mM KCl prior to the collection of the protein peak by step elution with buffer A containing 1 M KCl. This material was then passed on a DEAE-Sepharose column and eluted with an increasing KCl gradient from 0.1 to 1 M. Fractions were analyzed by Western blot, and fractions containing both N-CoR and PRP4K were pooled and concentrated to a volume of 5 ml by using a BioMax centrifugal filter with a 10-kDa molecular mass cutoff (Millipore) before being applied to a Sephacryl S300 column preequilibrated with buffer A containing 100 mM KCl. The column was then washed with buffer A at a flow rate of 0.4 ml/min, and 5-ml fractions were collected. The fractions corresponding to the protein peak by UV absorbance were analyzed by Western blotting for PRP4K, BRG1, PRP6, and N-CoR. Fractions containing PRP4K and N-CoR were pooled (S300 fraction) and precleared by using protein G-Sepharose before being subjected to affinity purification on an anti-PRP4K Sepharose column prepared by covalently coupling the anti-PRP4K antibody H143 to protein G-Sepharose (Roche) by using dimethylpalmilidate as previously described (57). The anti-PRP4K-Sepharose column was then washed consecutively with 10 bed volumes of wash buffer A (buffer A without DTT and supplemented with 150 mM KCl and 0.1% Triton X-100), wash buffer B (buffer A with 300 mM KCl and 0.2% Triton X-100), and finally wash buffer C (buffer A with 500 mM KCl and 0.5% Triton X-100). To remove excess salt, the column was then washed with buffer A containing 100 mM KCl. The affinity-purified PRP4K/N-CoR complex was then eluted from the PRP4K-Sepharose with elution buffer (100 mM glycine [pH 2.8], 150 mM KCl, 10% glycerol) and analyzed by Western blot analysis. Alternately, the PRP4K/N-CoR complex was affinity purified at analytical scale by IP with 20 μl of PRP4K-Sepharose from 100 μl of S300 fraction diluted to 1 ml in buffer A containing 150 mM KCl and 0.1% Triton X-100, followed by incubation for 2 h at 4°C. PRP4K-Sepharose IPs were then washed four times with 1 ml of wash buffer A and twice with 1 ml of wash buffer B or C prior to deacetylase assays or elution by boiling in 2× Laemmli buffer for 5 min and analysis by Western blotting as described above.
HDAC assay.
Histone deacetylase (HDAC) activity was monitored as described previously (57). Briefly, 100-μl aliquots of affinity-purified PRP4K/N-CoR complex were retained on 25 μl of PRP4K-Sepharose and washed as described for the IPs above, followed by two washes with 1 ml of deacetylase buffer (20 mM Tris-HCl [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 0.2 mM DTT, 0.2 mM PMSF). The PRP4K immunoprecipitates were then resuspended in deacetylase buffer containing 3H-labeled histones (20,000 cpm/reaction), prepared from HeLa nuclei (57), to a final volume of 200 μl and incubated at 37°C for 90 min. The reaction was then stopped by the addition of 50 μl of 0.1 M HCl-0.7 M acetic acid. Released [3H]acetate was extracted with 600 μl of ethyl acetate. After centrifugation, 300 μl of the upper organic phase was used for liquid scintillation counting. Total deacetylase activity was expressed in counts per minute (cpm) and normalized for the total volume of ethyl acetate (600 μl). Mock IPs (using protein G-Sepharose alone) were carried out from the pooled S300 fraction (containing PRP4K and N-CoR [see above]) and used for deacetylase assays to control for nonspecific HDAC activity.
Nucleotide sequence accession numbers
The sequences of the full-length human and murine PRP4Ks have been submitted to the GenBank database under accession numbers AF283465 and AF283466, respectively.
RESULTS
Cloning and analysis of murine PRP4K and its homologues.
We cloned and sequenced the full-length gene trapped in the murine embryonic stem cell line CT143 (56) (Fig. 1A). This gene encodes a basic protein (pI 10.26) of 1,007 aa with a predicted molecular mass of 117 kDa. This is larger than the previously described murine Prp4 homologue (Q61136; 496 aa), indicating that the original sequence was truncated at the N terminus (17). Consistent with previous nomenclature, we have chosen to call this protein PRP4K for Prp4 kinase.
We also cloned the human homologue, which is similar to the protein KIAA0536 (AB011108) and shares partial homology to the putative human homologue of Prp4 kinase described by Gross et al. (17). PRP4K homologues in D. melanogaster, C. elegans, and A. thaliana were also identified (Fig. 1B), but none could be found in S. cerevisiae.
Mammalian PRP4K contains a dual-specificity kinase domain that is 53% identical to that of S. pombe Prp4p (Fig. 1). There are other motifs in the basic N terminus that are conserved among metazoan PRP4Ks but absent in S. pombe Prp4p. Among these is the sequence KKHK (KKHK box, Fig. 1). This K/H-rich motif is also present in PACT, a snRNP-associated protein that interacts with both p53 and Rb (52), and in POP101 (Srm160), a component of the splicing-associated interchromatin granules (41, 55). The second conserved region is an RS-like domain rich in arginine and serine (RS) (Fig. 1). Lastly, two conserved sequence motifs, MI (DDMFA) and MII (DNWTDAEGYYRV), are found adjacent to the kinase domain of all Prp4 kinases including that of S. pombe (Fig. 1).
PRP4K is an essential gene in C. elegans.
Prp4 is essential for growth in S. pombe. The ts allele prp4-73 has both a cell cycle defect and accumulates pre-mRNA at the restrictive temperature (2). To determine whether PRP4K is essential in metazoans, we disrupted the expression of the C. elegans homologue (identifier F22D6.5) by RNA interference (RNAi) by using a 512-bp dsRNA fragment corresponding to the second exon of the predicted protein. This led to a highly penetrant early embryonic lethality in the F1 progeny, and no hatched larvae were observed (data not shown). Thus, PRP4K is essential in metazoans.
Subcellular localization of mammalian PRP4K.
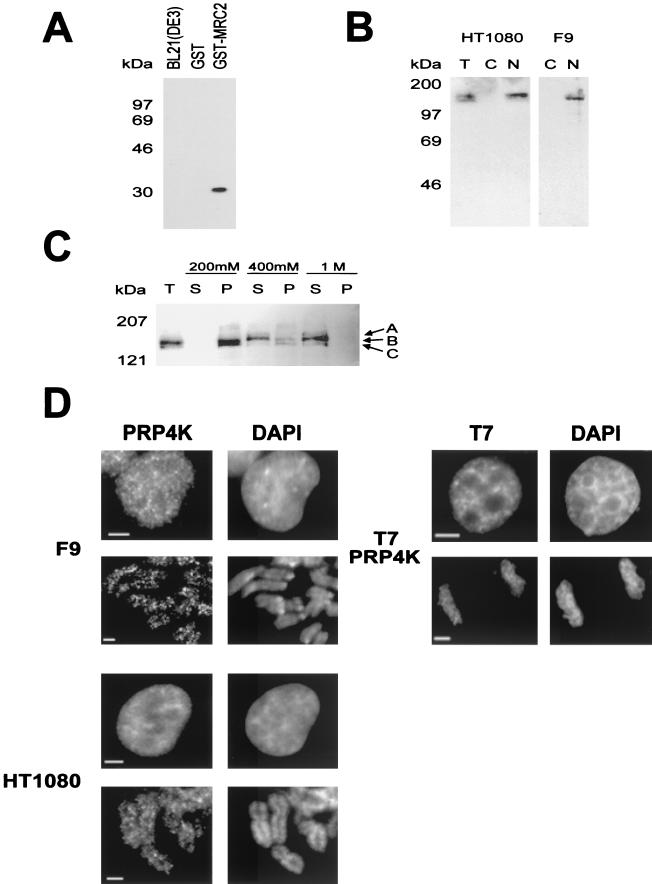
To determine the subcellular localization of endogenous PRP4K, we generated antibodies against two N-terminal peptides conserved between the murine and human proteins (MRC1 and MRC2; Fig. 1A). The specificity of the affinity-purified antibodies was determined by Western blot analysis of GST fusion proteins containing each peptide epitope (Fig. 2A). A third antibody (H143), this one directed against sequences between the RS-like and the kinase domains of PRP4K (aa 504 to 688), was also generated.
FIG. 2.
Characterization of PRP4K by Western blot analysis and indirect immunofluorescence. (A) Specificity of the anti-PRP4K antibody MRC2. Western blot analysis shows that affinity purified anti-PRP4K antibody MRC2 can detect the MRC2 epitope fused to GST (GST-MRC2) but does not cross-react with GST alone or with total bacterial protein extract. (B) Western blot analysis with the anti-PRP4K antibody MRC2 on total (T), cytoplasmic (C), or nuclear (N) extracts prepared from the human HT1080 or murine F9 embryocarcinoma cells indicates that PRP4K is a nuclear protein. (c) Salt extraction of PRP4K from human HT1080 nuclei was performed. Western blot analysis with the MRC2 antibody of soluble (S) and pellet (P) fractions of nuclei extracted with 200 mM, 400 mM, or 1 M KCl was then carried out. Total nuclei are shown for comparison (T). Three bands were detected: band A, 167 kDa; band B, 152 kDa; and band C, 147 kDa. Identical results were obtained with both the MRC1 and the H143 anti-PRP4K antibodies (data not shown). (D) Subcellular localization of PRP4K in murine F9 and human HT1080 cells by indirect immunofluorescence. Immunofluorescence of PRP4K (FITC) is shown in cells costained with DAPI to visualize the DNA. For each cell type, nuclei are shown in the first row of images, followed by mitotic chromosomes shown in the second row. In interphase cells, PRP4K is nucleoplasmic but enriched in foci that do not correspond to regions of concentrated DNA (DAPI). At mitosis, some PRP4K is also localized to mitotic chromosomes. Immunodetection of T7-tagged recombinant PRP4K (T7-PRP4K) in HT1080 cells produces a localization pattern similar to that of endogenous PRP4K in interphase cells and on mitotic chromosomes. Scale bars, 5 μm.
The predicted molecular mass of PRP4K is 117 kDa, but Western blot analysis with anti-PRP4K antibodies detects bands of ca. 152 kDa in nuclear extracts from human and mouse cells (Fig. 2B). Most PRP4K is extracted from nuclei by 400 mM KCl (Fig. 2C). Three major bands of 167, 152, and 147 kDa (Fig. 2C, bands A, B, and C, respectively) are differentially extracted from nuclei with various salt concentrations. HeLa cell extracts containing recombinant T7 epitope-tagged PRP4K also produce these three bands when analyzed by Western blotting with anti-T7 antibodies (data not shown). These data suggest that PRP4K may be posttranslationally modified.
The subcellular distribution of endogenous PRP4K in human and mouse cells was analyzed by indirect immunofluorescence (Fig. 2D). PRP4K is distributed throughout the nucleus, excluding the nucleolus, but is enriched in multiple speckles. In addition, some endogenous PRP4K is associated with unfixed mitotic chromosomes (Fig. 2D; F9 and HT1080). In contrast, the splicing factor SC35 was not associated with mitotic chromosomes (data not shown). Detection of recombinant T7 epitope-tagged PRP4K in HT1080 cells produced a similar pattern of localization to endogenous PRP4K in both nuclei and on mitotic chromosomes (Fig. 2D; T7-PRP4K).
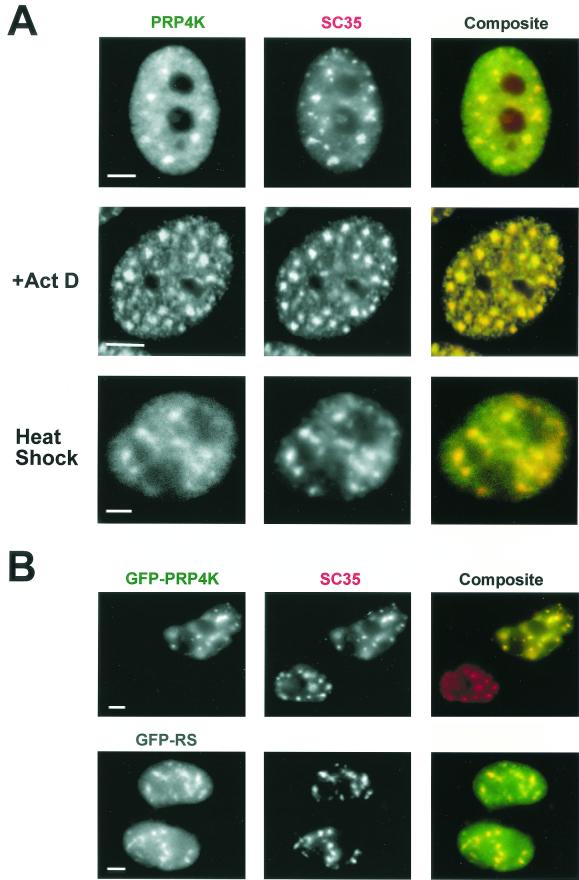
PRP4K is enriched in structures resembling splicing speckles, which colocalize with both SC35 (Fig. 3A) and with Sm antigens (data not shown). Upon the inhibition of transcription by actinomycin D or heat shock, SC35 concentrates into fewer, but larger, foci within the nucleus (53). Under these conditions, PRP4K-enriched speckles continue to localize with SC35 (Fig. 3A).
FIG. 3.
PRP4K is concentrated in SC35-containing splicing speckles. (A) Coimmunofluorescence with antibodies detecting PRP4K (H143) (FITC) and SC35 (Texas red) in human HT1080 cells alone, in the presence of actinomycin D (+Act D), or subjected to heat shock. PRP4K is nucleoplasmic but also concentrates in speckles that colocalize with SC35 in both transcriptionally active and transcriptionally inactive cells (i.e., heat shock and actinomycin D treated). (B) The RS-like domain of PRP4K is sufficient for localization to splicing speckles. HT1080 cells were transfected with either GFP-tagged murine PRP4K (GFP-PRP4K) or the RS domain of PRP4K fused to GFP (GFP-RS). Localization of GFP-tagged proteins was assessed by immunofluorescence in combination with the detection of endogenous SC35 (Texas red). GFP-tagged PRP4K forms distinct and large nuclear foci that colocalize with SC35. Scale bars, 5 μm.
To confirm the localization of PRP4K and to determine the sequences necessary for localization to splicing speckles, cells were transfected with cDNAs encoding full-length protein (GFP-PRP4K) or part of the N terminus containing the RS-like domain (aa 188 to 780) (GFP-RS) tagged with GFP. Both proteins colocalize with SC35 (Fig. 3B). In contrast, constructs containing sequences between the RS-like and kinase domains (aa 512 to 1007), including motifs MI and MII, fail to localize to splicing speckles (data not shown). Thus, sequences within the RS-like domain of PRP4K alone are sufficient for colocalization with SC35-containing splicing speckles. T7 epitope-tagged PRP4K also localized to splicing speckles in interphase nuclei (data not shown).
PRP4K interacts with both chromatin and splicing factors in yeast two-hybrid analysis.
The subcellular localization of PRP4K suggests that this kinase might interact with splicing factors. To identify interacting proteins, yeast two-hybrid screens were carried out with the N terminus (aa 2 to 644) or C-terminal kinase domain (aa 637 to 1007) of human PRP4K as bait (Fig. 4).
FIG. 4.
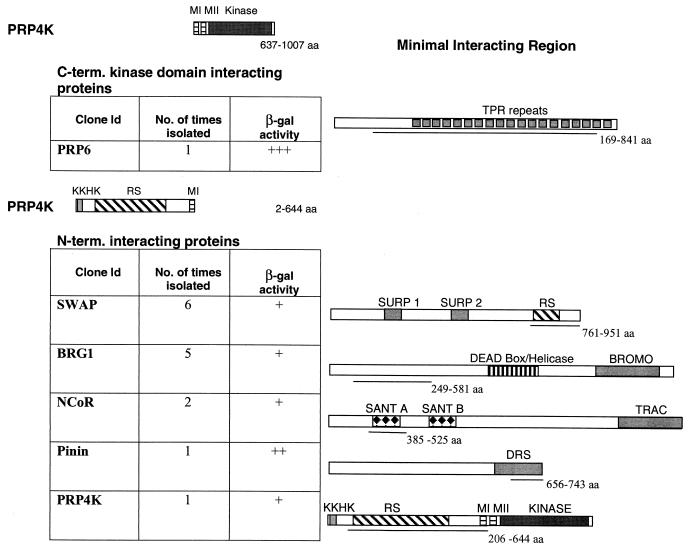
Yeast two-hybrid analysis of human PRP4K. Yeast two-hybrid analysis of the N terminus or the C-terminal kinase domain of human PRP4K was carried out by using an adult human brain library. The region of PRP4K used in each screen is represented above each table. The identity of each protein and the number and relative strength of each interaction (β-galactosidase activity) is shown in the table. Each interacting protein is depicted graphically to the right, with the minimal PRP4K-interacting region underlined.
The PRP4K domain interacted strongly with the U5 snRNP-associated protein, PRP6 (40, 49). The interacting region of PRP6 (aa 169 to 841) contains 17 of the 19 tetratricopeptide repeats (Fig. 4). A ts allele of prp1, the S. pombe PRP6 homologue, accumulates pre-mRNA at the restrictive temperature and interacts genetically with prp4 (51). Furthermore, S. pombe Prp1p has been shown to interact via yeast two-hybrid with Prp4p and is a target of this kinase both in vivo and in vitro (51). Thus, the interaction between PRP6 and the kinase domain of PRP4K appears to be conserved.
In contrast, using the N terminus of PRP4K, which is not present in S. pombe Prp4p, we identified a number of interacting proteins that play roles in pre-mRNA splicing, as well as those involved in chromatin remodeling and/or gene regulation. One N-terminal interacting protein was the human homologue of suppressor-of-white apricot (SWAP), which regulates the alternative splicing of the CD45 and fibronectin genes (50). SWAP can autoregulate splicing of its own transcript and is the prototype of a family of splicing proteins that share a SURP domain (65). The minimal PRP4K-interacting region is C-terminal to the SURP domains and encompasses the RS domain of SWAP (aa 761 to 951) (Fig. 4). We also isolated pinin, a desmosome-associated protein (48). Although no functional connection between pinin and splicing has been demonstrated, pinin localizes to splicing speckles in both Xenopus spp. and in mammals (5). The PRP4K interacting peptide encodes the last 97 aa of pinin (aa 654 to 743), including approximately two-thirds of the DRS domain (domain rich in serines) (5). Interestingly, PRP4K itself was also isolated in this screen, suggesting that it can dimerize through this N-terminal region (aa 206 to 644).
Another PRP4K N-terminal interacting protein, BRG1, is a mammalian homologue of the Drosophila protein Brahma, which regulates homeotic gene expression in the fly and shares homology with the yeast chromatin remodeling factor Swi2/Snf2 (12). BRG1 is the catalytic component of a mammalian Swi/Snf complex and has been implicated in the activation of several hormone-regulated genes (8, 43). The minimal interacting region of BRG1 (aa 249 to 581) is similar to that involved in interaction with HP1α (34). Surprisingly, we also isolated two clones of N-CoR capable of interacting with the N terminus of PRP4K. The interacting region of N-CoR contains the first of two SANT domains (aa 385 to 525) (1).
These data suggest that mammalian PRP4K is capable of interacting both with proteins involved in the regulation of splicing and also proteins that regulate gene expression through the modulation of chromatin structure. Recently, an N-CoR complex has been isolated that also contains BRG1 and the splicing factors SAP130 and SF3a120 (57). Our two-hybrid data support the existence of such complexes, which may link transcriptional regulation, via changes in chromatin structure, to pre-mRNA splicing.
PRP4K interacts in vivo with N-CoR, BRG1, pinin, and PRP6.
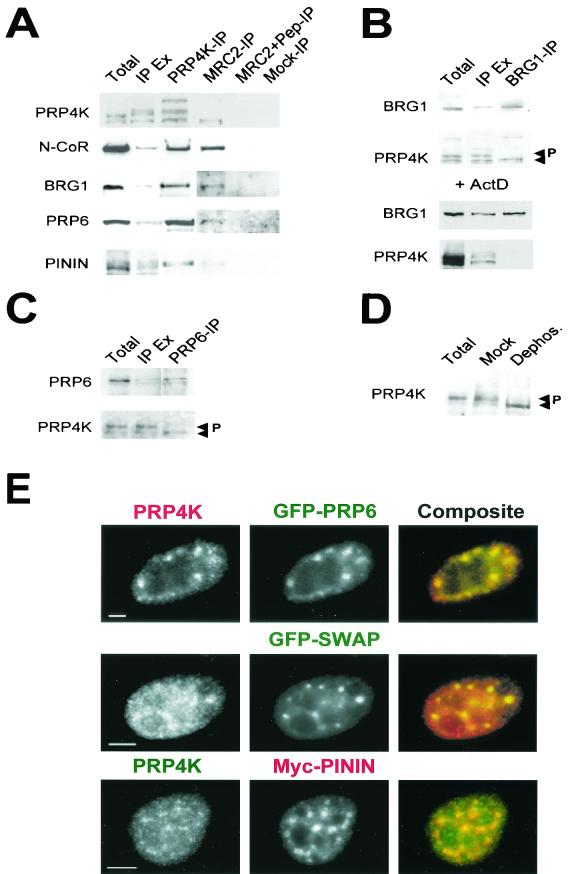
To determine whether PRP4K interacts in vivo with the proteins isolated by yeast-two hybrid analysis, we performed coimmunoprecipitation (Co-IP) experiments from HeLa cell extracts. N-CoR, BRG1, PRP6, and pinin were each coimmunoprecipitated by anti-PRP4K antibody (Fig. 5A). Interestingly, when the reciprocal IPs were carried out by using anti-PRP6 or anti-BRG1 antibodies, only the fastest-migrating form (∼147 kDa) of PRP4K could be coimmunoprecipitated (Fig. 5B and 5C). The pinin antibody did not work efficiently for IP.
FIG. 5.
In vivo interactions of PRP4K. (A to C) Co-IP of PRP4K with pinin, N-CoR, BRG1, and PRP6. Complexes containing PRP6, BRG1, or PRP4K were immunoprecipitated from HeLa nuclear extracts with antibodies against each protein coupled to protein A- or protein G-agarose. Total, total nuclei; IP, IP of the indicated protein; IP Ex, IP extract. (A) Western blot analysis with antibodies recognizing each protein shows that N-CoR, BRG1, PRP6, and pinin coimmunoprecipitated with PRP4K by using anti-PRP4K immune serum against the MRC1 and two epitopes (PRP4K-IP) or the affinity-purified anti-PRP4K antibody MRC2 (MRC2-IP). Control IPs were carried out with MRC2 antibody preblocked with the MRC2 peptide (MRC2+Pep-IP) and with rabbit anti-sheep IgG (Mock-IP). (B) Western blot analysis of proteins immunoprecipitated with BRG1 by using anti-BRG1 antibodies (BRG1-IP). Only the fastest-migrating form of PRP4K coimmunoprecipitated with BRG1. In the presence of actinomycin D (+ActD), PRP4K fails to coimmunoprecipitate with BRG1, suggesting that the interaction between these proteins is transcription dependent. Arrows indicate two prominent forms of PRP4K; the slower-migrating form is phosphorylated (P). (C) Western blot analysis of proteins immunoprecipitated with PRP6 with anti-PRP6 antibodies (PRP6-IP). Only the fastest-migrating form of PRP4K coimmunoprecipitated with PRP6. (D) Dephosphorylation of HeLa nuclear extracts by using CIAP. Two major forms of PRP4K are indicated by the arrows; P indicates the hyperphosphorylated and slower-migrating form. Mock dephosphorylation results are shown for comparison. These results indicate that BRG1 and PRP6 interact primarily with the hypophosphorylated form of PRP4K. (E) Colocalization of PRP4K with GFP-tagged human PRP6 (GFP-PRP6), GFP-tagged murine SWAP (GFP-SWAP), or myc-tagged pinin (Myc-PININ) in HeLa cells. Endogenous PRP4K was detected with either Texas red-labeled (for GFP-PRP6 and GFP-SWAP transfections) or FITC-labeled secondary antibodies (for myc-pinin transfections). Myc-tagged pinin (Myc-PININ) was detected with Texas red-labeled secondary antibodies. Scale bars, 5 μm.
The differentially migrating forms of PRP4K detected by Western blot analysis (Fig. 2C) suggest that it is posttranslationally modified. Phosphatase treatment of nuclear extracts reduces most PRP4K to the 147-kDa fast-migrating form (Fig. 5D). Thus, the slower-migrating forms of PRP4K are phosphorylated, and it is only the hypophosphorylated (147 kDa) form that is coimmunoprecipitated by BRG1 and PRP6 antibodies (Fig. 5B and C). PRP4K fails to coimmunoprecipitate with BRG1 from cells treated with the transcriptional inhibitor actinomycin D (Fig. 5B), suggesting that the interaction between BRG1 and PRP4K is transcription dependent.
PRP6, SWAP, and pinin are known to localize in splicing speckles (5, 36, 38) and so are expected to colocalize with PRP4K in the nucleus. Indeed, GFP-tagged PRP6, GFP-SWAP, and myc-tagged pinin extensively colocalize with endogenous PRP4K, supporting the notion that these proteins can interact in vivo (Fig. 5E).
Immunoprecipitated PRP4K exhibits kinase activity in vitro.
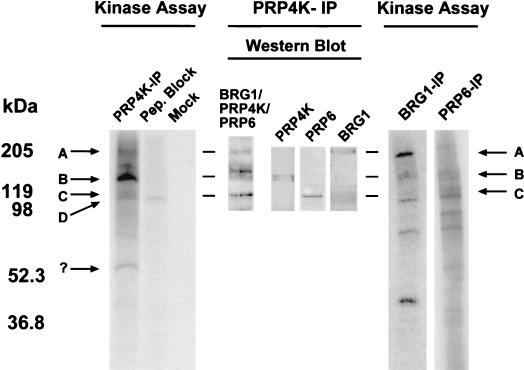
To determine whether immunoprecipitated PRP4K is catalytically active, we carried out in vitro kinase assays. Immunoprecipitated PRP4K was able to phosphorylate both itself and several coimmunoprecipitated proteins (Fig. 6). This kinase activity was absent if the immunoprecipitate was preincubated with MRC2 peptide, suggesting that it is due to PRP4K. A strongly labeled doublet (Fig. 6, band B) corresponds to the relative position of PRP4K, a finding consistent with the reported ability of mammalian PRP4K homologues to autophosphorylate (19, 27). Two other prominently labeled proteins correspond in size to BRG1 and PRP6 (Fig. 6, bands A and C, respectively). To confirm that these proteins were indeed BRG1 and PRP6, we carried out kinase assays with BRG1 and PRP6 IP complexes, which also contain coimmunoprecipitated PRP4K (Fig. 5). These immunoprecipitates do exhibit kinase activity, and both phosphorylated PRP6 and BRG1 are much more intense in their respective IP reactions, reflecting the relative increase in each substrate for PRP4K. Although a contaminating kinase could be coimmunoprecipitating with PRP4K, our data suggest that PRP4K is the kinase primarily responsible for this activity. Furthermore, Prp1p, the fission yeast homologue of PRP6, is a PRP4K substrate both in vitro and in vivo (51). We suggest that PRP6 and BRG1 are substrates of PRP4K in mammalian cells.
FIG. 6.
In vitro kinase activity of PRP4K in immunoprecipitates. IPs with antibodies against PRP4K (MRC2 antibody [PRP4K-IP]), BRG1 (BRG1-IP), or PRP6 (PRP6-IP) were subjected to an in vitro kinase assay and resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography (Kinase Assay). Control reactions were carried out with either peptide-blocked MRC2 antibody (Pep. Block) or rabbit anti-sheep IgG (Mock). Western blot analysis of the PRP4K-IP complex was also carried out. Bands corresponding to known proteins are designated with arrows (arrow A, BRG1; arrow B, PRP4K; arrow C, PRP6). A contaminating band (arrow D) is present in the peptide-blocked kinase assay, and a strong band of ca. 54 kDa (arrow “?”) may correspond to phosphorylated IgG. In the PRP4K-IP, the strongly labeled band (arrow B) probably corresponds to autophosphorylated PRP4K.
PRP4K specifically associates with the U5 snRNP.
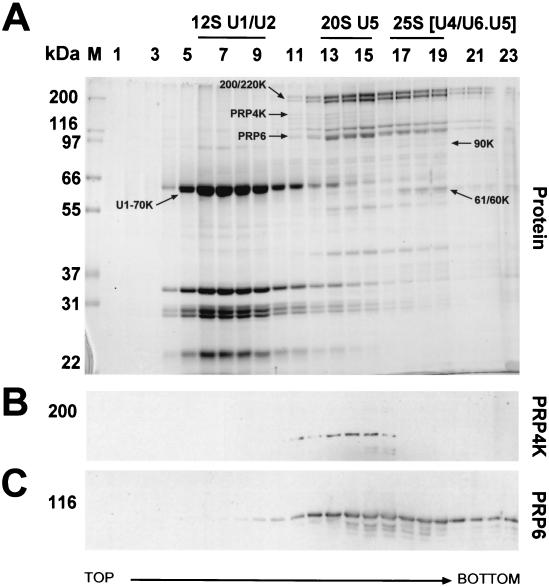
PRP6 is believed to play an important role in bridging the U4/U6 and U5 snRNPs within the tri-snRNP particle during spliceosomal maturation (38). Our Co-IP data suggest that PRP4K and PRP6 interact in vivo. Therefore, we sought to determine whether PRP4K, perhaps through interaction with PRP6, was an snRNP-associated protein. PRP4K cosediments specifically with the 20S U5 snRNP particle containing PRP6 and the U5 snRNP-specific 200- and 220-kDa proteins (Fig. 7). PRP4K is present in the U5 snRNP fractions as a nonstoichiometric component (Fig. 7A) compared to PRP6 or the 200K and 220K proteins. This suggests that only a subset of U5 snRNP particles contain PRP4K or that some PRP4K is lost under our purification conditions. This is perhaps not unexpected since many kinase-substrate interactions are transitory in nature. In contrast to PRP4K, PRP6 is also found in the fractions corresponding to the 25S tri-snRNP particle (38) (Fig. 7C), which is delineated by the appearance of the U4/U6 snRNP-specific 90-, 61-, and 60-kDa proteins (Fig. 7A). These data demonstrate that PRP4K can be copurified with total snRNPs from HeLa cells and that it specifically associates with the U5 snRNP. Consistent with these results, PRP4K was also found to specifically coimmunoprecipitate the U5 snRNA from HeLa nuclear extracts (data not shown). Previously, a snRNP-associated kinase activity, called the U1 snRNP 70K protein kinase, was identified that predominantly associates with the U1 snRNP (60). It is significant to note that PRP4K does not cosediment with the U1 snRNP (Fig. 7), suggesting that it is a novel snRNP-associated kinase that is distinct from the U1 snRNP 70K protein kinase.
FIG. 7.
PRP4K specifically cosediments with the 20S U5-snRNP during glycerol gradient centrifugation. (A) Proteins from each fraction of the glycerol gradient were separated by SDS-10% PAGE and visualized by Coomassie blue staining (Protein). Fractions that cosediment with the 12S U1/U2 snRNP, 20S U5-snRNP, and the U4/U6.U5 snRNP were determined by the position of the corresponding snRNAs (data not shown) as indicated above the panels. The positions of PRP4K, PRP6, the U1-specific U1-70K protein, the U5 snRNP-specific 200K and 220K proteins, and the U4/U6 snRNP-specific 90K, 61K, and 60K proteins are arrowed. (B and C) Western blot analysis of fractions with antibodies to PRP4K (B) or PRP6 (C).
PRP4K is a component of the N-CoR-2 complex.
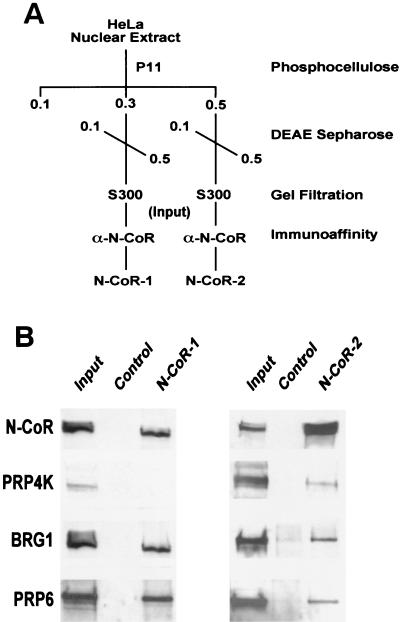
It has recently been demonstrated that N-CoR exists as part of two multiprotein complexes which exhibit deacetylase activity and contain distinct subunits (57). One of the complexes contains both BRG1 and the splicing factors SAP130 and SF3a120 (57). Based on our identification of N-CoR, BRG1, and several splicing factors as proteins interacting with PRP4K by yeast two-hybrid analysis, it was of interest to determine whether PRP4K is a constituent of the biochemically purified N-CoR-1 and N-CoR-2 complexes (Fig. 8). Western blot analysis indicated that PRP4K coeluted from a gel filtration column with N-CoR, and after immunoaffinity chromatography, it was found to be a constituent of the N-CoR-2 complex. In contrast, BRG1 and PRP6 were found in both the N-CoR-1 and the N-CoR-2 complexes. These results suggest that endogenous PRP4K can be isolated as a component of an N-CoR complex containing N-CoR, BRG1, and PRP6.
FIG. 8.
Detection of the presence of PRP4K, BRG1, and PRP6 in the N-CoR-1 and -2 complexes. (A) Schematic representation of the purification of the N-CoR-1 and N-CoR-2 complexes (57). (B) Western blot analysis of the N-CoR-1 and N-CoR-2 complexes after elution from the N-CoR affinity column with antibodies against N-CoR, PRP4K, BRG1, and PRP6. The control lane is an aliquot from the mock affinity purification, and the input lane is the pooled S300 fractions containing N-CoR prior to immunoaffinity chromatography. Whereas BRG1 and PRP6 are components of both the N-CoR-1 and the N-CoR-2 complexes, PRP4K is primarily a component of the N-CoR-2 complex.
The affinity-purified PRP4K/N-CoR complex exhibits HDAC activity.
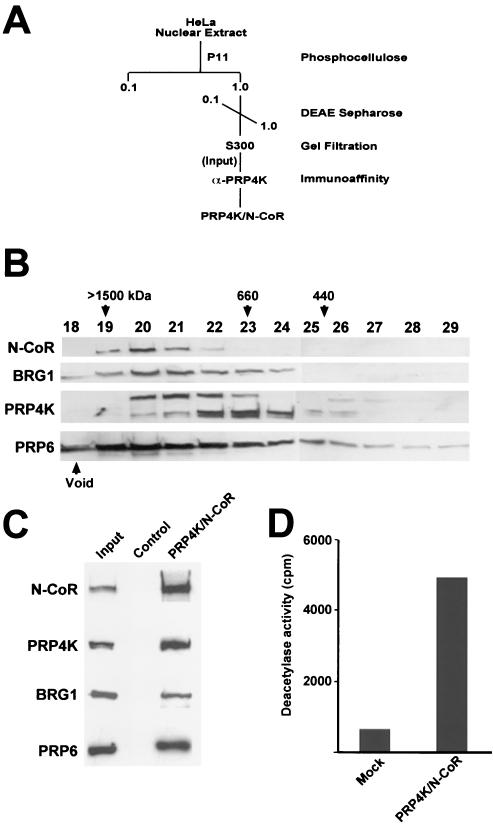
The presence of PRP4K in the N-CoR-2 complex suggested that PRP4K/N-CoR complexes exist in vivo. Such complexes might bridge or coordinate the regulation of transcription, via chromatin modification, and pre-mRNA splicing. To determine whether N-CoR, PRP4K, BRG1, and PRP6 are components of the same complex, we optimized the purification strategy to specifically isolate PRP4K/N-CoR-containing complexes from HeLa nuclei (Fig. 9A). After two rounds of conventional column chromatography, fractions containing both N-CoR and PRP4K were run on an S300 Sepharose gel filtration column to both size the complex and determine whether N-CoR and PRP4K would coelute as single or multiple complexes (Fig. 9B). The majority of N-CoR eluted from the sizing column in complexes ranging from 1.0 to 1.5 MDa, and several of these fractions also contained BRG1, PRP4K, and PRP6 (Fig. 9B, fractions 20 to 22). Thus, it appears that large-molecular-weight complexes exist in vivo that contain N-CoR, BRG1, PRP4K, and PRP6. To further purify the complex, fractions 20 to 22 from the S300 column were pooled and subjected to immunoaffinity purification for PRP4K complexes by using an anti-PRP4K antibody column. PRP4K complexes were then acid eluted from the immunoaffinity column and subjected to Western blot analysis (Fig. 9C). N-CoR, PRP4K, BRG1, and PRP6 were coeluted from the immunoaffinity column, confirming the association of these proteins within the PRP4K/N-CoR complex.
FIG. 9.
Affinity purification of the PRP4K/N-CoR deacetylase complex. (A) Purification scheme for the isolation of complexes enriched in both N-CoR and PRP4K. At each stage, N-CoR- and PRP4K-containing fractions were determined by Western blot analysis and pooled before we continued with the next chromatography step. Affinity-purified PRP4K/N-CoR complex was then subjected to both Western blot analysis and deacetylase assays. (B) S300 gel filtration profile of N-CoR, BRG1, PRP4K, and PRP6. Fractions corresponding to the protein peak from the S300 gel filtration column (fractions 18 to 29) were analyzed by Western blotting for the presence of each protein. Approximate molecular masses of the various complexes eluting from the S300 column are indicated in kilodaltons above the corresponding fraction numbers. N-CoR, BRG1, PRP4K, and PRP6 coelute from the gel filtration column in a large-molecular-mass complex of between 660 and 1,500 kDa. (C) Western blot analysis of the PRP4K/N-CoR complex. The PRP4K/N-CoR complex was affinity purified from the S300 fractions 20 to 22 by using an anti-PRP4K column. Western blot analysis indicates the PRP4K/N-CoR complex contains N-CoR, PRP4K, BRG1, and PRP6. (D) Deacetylase activity of the PRP4K/N-CoR complex. Deacetylase activity of affinity-purified PRP4K/N-CoR complex was monitored by the release of [H3]acetate from in vitro-tritiated histones in the presence of immunoprecipitated complex with anti-PRP4K antibody coupled to protein G-Sepharose. The activity is presented as the total counts per minute, and a mock deacetylase assay (Mock) was carried out by using mock-affinity-purified PRP4K/N-CoR complex immunoprecipitated with protein G-Sepharose alone. The PRP4K/N-CoR complex exhibits strong deacetylase activity compared to the nonspecific activity found associated with the mock-affinity-purified complex.
A critical function of N-CoR as a corepressor is the recruitment of HDAC activity. If PRP4K is truly a component of a functional N-CoR complex, one would expect that such complexes would exhibit HDAC activity. To determine whether the affinity-purified PRP4K/N-CoR complex was associated with HDAC activity, affinity-purified PRP4K/N-CoR complex or mock-affinity-purified complex was subjected to a liquid deacetylase assay with 3H-labeled histones prepared from HeLa nuclei (Fig. 9D). The affinity-purified PRP4K/N-CoR complex exhibited strong HDAC activity compared to the mock-affinity-purified complex, suggesting the PRP4K/N-CoR complex is indeed a deacetylase complex.
DISCUSSION
Characterization of a mammalian PRP4K.
We have characterized human and mouse PRP4K. This dual-specificity kinase is the mammalian homologue of S. pombe Prp4p. Previously, putative mammalian homologues of Prp4p were isolated by Kaufer and coworkers but were truncated in their N termini (17). Recently, a full-length human Prp4 protein, identical to human PRP4K, has been reported and was termed hPRP4 (30). Because there is a WD-repeat containing splicing factor in mammals already called PRP4 (33), we suggest that the alternate name of PRP4K be used for this family of Prp4-like kinases.
We show that PRP4K is a 152-kDa phosphoprotein (Fig. 5) capable of autophosphorylation (Fig. 6). The N terminus contains two motifs, MI (DDMFA) and MII (DNWTDAEGYYRV), found in all PRP4K homologues (Fig. 1). Mutations in these motifs, or in the residues immediately flanking them, abolish Prp4p function in S. pombe (17). PRP4K also contains motifs and domains that are conserved only among the metazoan proteins (Fig. 1B). In particular, the basic N terminus contains both a histidine-lysine-rich (KKHK) box and an RS domain. Both an RS domain and the KKHK motif are found in the splicing-associated proteins POP101 (Srm160) and PACT. RS domains are targets for phosphorylation by SR protein kinases such as CLK and SRPK and are found in many splicing factors (reviewed in reference 16). The RS domain is thought to mediate protein-protein interactions between splicing factors and both the spliceosomal machinery and the transcriptional machinery (16). We have shown that some PRP4K localizes to SC35-containing splicing speckles (Fig. 3A) and that the RS-like domain is sufficient for this localization (Fig. 3B). Similarly, the RS domains of several other splicing factors are sufficient for splicing speckle localization in vivo (6, 20).
PRP4K is a U5 snRNP-associated kinase that interacts with proteins involved in splicing.
PRP4K is implicated in constitutive pre-mRNA splicing by its homology to the S. pombe pre-mRNA kinase Prp4p. Furthermore, the kinase domain of PRP4K can functionally replace that of S. pombe Prp4p (17). Concentrations of PRP4K colocalize with SC35-containing splicing speckles in nuclei, indicating that this kinase also associates with subnuclear compartments involved in pre-mRNA splicing (Fig. 3). We have demonstrated that PRP4K is an snRNP-associated kinase that is specific for the U5 snRNP (Fig. 7) and therefore distinct from the U1 snRNP 70K protein kinase (60). Mammalian PRP6 is a stably associated component of both the U5 and tri-snRNP particles (38). Prp1p, the fission yeast homologue of PRP6 interacts with, and is a substrate of, S. pombe Prp4p (51). Here we have shown that mammalian PRP4K interacts with human PRP6 (Fig. 4), that PRP4K and PRP6 colocalize and coimmunoprecipitate from human cells (Fig. 5), and that they interact in vivo as components of at least two distinct biochemical complexes (Fig. 7 to 9). It is likely that the association of PRP4K with the U5-snRNP is primarily mediated through interaction with PRP6. We suggest that human PRP6 is a substrate of PRP4K (Fig. 6) and argue that S. pombe and mammalian PRP4K may share a conserved role in pre-mRNA splicing. Although the specific function of these kinases in pre-mRNA splicing is unknown, Kaufer and coworkers suggest that these kinases might play a role in the formation of active spliceosomes through interaction and/or phosphorylation of PRP6 and, possibly, other non-SR spliceosome proteins (51). Our data would support this hypothesis.
PRP4K may also play a role in alternative splicing. The splicing factor SF2 can interact with and is a substrate of mammalian PRP4K in vitro (17, 30), and we have shown that the N-terminal domain of PRP4K interacts with SWAP (Fig. 4). SWAP can regulate its own alternative splicing, as well as the splicing of the CD45 and fibronectin genes (50, 65). However, preliminary in vivo splicing data suggest that the overexpression of PRP4K or PRP6 has only a minor affect on alternative splicing of reporter gene constructs (data not shown). In fact, we observed a partial inhibition of splicing, which would be more consistent with a role for these proteins in constitutive rather than alternative splicing.
PRP4K interacts with proteins involved in nuclear hormone-regulated chromatin remodeling.
By using a combination of yeast-two hybrid screening, IP, and biochemical purifcation, we have demonstrated that PRP4K can interact and copurify with BRG1 and N-CoR, components of nuclear hormone-regulated chromatin remodeling complexes (8, 21, 23, 44). In mammals, few splicing proteins appear to have any demonstrated association with chromatin or chromatin remodeling proteins. The N terminus of PRP4K interacts with BRG1, the ATPase subunit of SWI/SNF chromatin remodeling complexes. BRG1 coimmunoprecipitates with the hypophosphorylated form of PRP4K (Fig. 5) in a transcription-dependent manner (Fig. 5B) and appears to be a PRP4K substrate in vitro (Fig. 6). Although the observed interaction may be a consequence of cotranscriptional splicing, these data may also suggest a direct role for PRP4K in chromatin remodeling through the phosphorylation of BRG1.
The N terminus of PRP4K also interacts with N-CoR. N-CoR, and its related family member SMRT, were initially identified by yeast two-hybrid screens with unliganded thyroid hormone receptor or retinoic acid receptor, respectively, as bait (7, 23). Both N-CoR and SMRT are large proteins that mediate their repressive effects by functioning as molecular scaffolds for the recruitment of a number of proteins, including Sin3A and the class I and II HDACs (19, 29; reviewed in reference 27). We have identified the SANT A domain of N-CoR by yeast two-hybrid analysis as the putative interaction region between this corepressor and PRP4K. The SANT domain, a putative regulatory region that may control recruitment or activation of HDACs, is highly conserved among a number of chromatin-associated proteins, including the closely related SMRT corepressor (18), CoREST (26, 64), and MTA1 (1). Several recent biochemical studies have isolated both N-CoR and SMRT as components of large multiprotein complexes. In one such study, N-CoR was found in two biochemically distinct complexes, designated N-CoR-1 and N-CoR-2. N-CoR-1 was found to contain the core constituents of the SWI/SNF complex, including BRG1, as well as the splicing factors SAP130 and SF3a120 (57). Here, we also demonstrate the presence of PRP6 in both the N-CoR-1 and N-CoR-2 complexes (Fig. 8B). The N-CoR-2 complex also contains PRP4K (Fig. 8B), as well as components of the Sin3 repressor complex (Sin3A, HDAC1 to -3, and SAP30) (57). It was not clear from our previous work whether or not N-CoR-2 represented a homogeneous complex. Therefore, we modified our purification procedure to enrich for PRP4K/N-CoR-containing complexes. We report the affinity purification of a megadalton PRP4K/N-CoR complex from HeLa cells that contains BRG1 and PRP6 and exhibits strong deacetylase activity (Fig. 9). Thus, our two-hybrid data and the results of our biochemical analysis strongly suggest the existence of a large-molecular-mass PRP4K-containing complex that may couple transcriptional regulation, via the modulation of chromatin structure, to pre-mRNA splicing.
Why do splicing factors associate with corepressor and coactivator complexes?
Pre-mRNA splicing occurs cotranscriptionally (reviewed in reference 22), and the activation of transcription is coincident with changes in chromatin structure (14, 44). This study and a number of recent studies have also demonstrated links between pre-mRNA splicing and both transcriptional coactivator and corepressor complexes. For example, several coactivators have been implicated in pre-mRNA splicing, including PGC1, which is involved in the oxidative stress response and in adaptive thermogenesis (42); p52, a general transcriptional activator that interacts with SF2 (15); WT1, a coactivator and corepressor of gene expression that has an isoform (+KTS) thought to be involved in mRNA processing (reviewed in reference 35); and the TLS oncoprotein, a potent transactivator that can interact with a number of SR splicing factors (62). In addition, the mammalian STAGA coactivator complex was recently shown to contain the splicing factor SAP130, arguing for the possible recruitment of pre-mRNA splicing machinery to actively transcribing genes (39).
Prior to this study, at least two other corepressor complexes have been described that contain proteins involved in splicing. One of these is the CIR corepressor complex involved in Notch signaling and Epstein-Barr virus-induced immortalization (24). The CIR corepressor interacts with SKIP (66), a component of both splicing speckles and the spliceosome (44). The splicing factor PSF has also been shown to play a role in mediating the repression of genes regulated by type II nuclear hormone receptors (40). Moreover, we have recently isolated a thyroid hormone receptor-associated protein, F9/21B2 (TRAP150), that colocalizes with splicing speckles (55).
Why might splicing factors associate with corepressor or coactivator complexes? The coordination of both transcription and splicing of mRNA may be necessary to allow for rapid changes in protein expression, for instance, as a result of hormone induction or during viral gene expression. The human papillomavirus type 5 uses this strategy through the E2 transactivator protein, which can associate with SR splicing factors and is capable of enhancing the splicing of pre-mRNAs whose transcription is induced by this transactivator (32). Similarly, splicing factors found in association with coactivator complexes could help facilitate the efficient coupling of pre-mRNA splicing to transcription of the target gene or may aid in transcriptional elongation (13). Interestingly, PRP4K truncated at the N terminus can phosphorylate Elk-1 in vitro and can transactivate the expression of an Elk-1 reporter gene, suggesting that the C-terminal kinase domain can act as a coactivator in this system by phosphorylating Elk-1 (25). Although the kinase domain also interacts with the U5 snRNP-associated protein PRP6, we cannot rule out a direct role for PRP4K in transcriptional activation through this domain.
To understand why splicing proteins should associate with corepressors is less intuitive. Corepressor-associated splicing proteins might represent a pool of splicing factors around the gene promoter, perhaps held in an inactive form but ready for use once transcription is activated. In this way, the complement of promoter-associated coactivators and/or repressors, splicing proteins and components of the RNA polymerase II holoenzyme could affect both transcriptional initiation and splicing. Indeed, it has been recently shown that promoter structure can alter the alternative splicing of the fibronectin gene, in part by modulating the interaction of exonic splicing enhancers with splicing factors such as 9G8 and SF2 (9, 10).
In the context of the PRP4K/N-CoR complex, it is tempting to speculate that PRP4K might regulate the deacetylase activity of this complex either by affecting the association of N-CoR with the HDACs or by directly phosphorylating the HDACs themselves. The recent observation that HDAC1 phosphorylation can promote both activity and deacetylase complex formation (49) is consistent with this hypothesis. Furthermore, preliminary analysis of the affinity-purified PRP4K/N-CoR complex indicates the presence of class I HDACs (G. Dellaire and J. Torchia, unpublished observations), which would account for the strong deacetylase activity of this complex. The region of N-CoR that interacts with PRP4K by yeast two-hybrid assay, which contains the SANT A domain, corresponds to the deacetylase activating domain (DAD) region recently found to activate HDAC3 activity (18). These data suggest that, in addition to its direct role in regulating HDAC activity, the SANT/DAD region serves as an interface for the recruitment of PRP4K. We are currently investigating the possibility that PRP4K may modulate HDAC activity through this region of N-CoR. The HDACs are also implicated in the deacetylation of proteins other than histones (e.g., YY1 [63] and p53 [28]). Therefore, it is plausible that PRP4K may also help modulate the acetylation status of proteins other than the histones.
Collectively, the data presented here, combined with several recent studies, argue for the direct recruitment of splicing factors to gene loci through interaction with corepressor and coactivator complexes. Our findings provide a novel and unexpected connection between transcriptional regulation and pre-mRNA splicing through PRP4K, which may modulate the coordination of these processes via phosphorylation of proteins within the U5 snRNP and the PRP4K/N-CoR deacetylase complex.
Acknowledgments
We thank Iain Johnstone (University of Glasgow) for use of his C. elegans injection facility, as well as Javier Caceres and Nick Hastie for comments on the manuscript. We acknowledge the aid of Jamal Nasir and Marie-Jose Lafuente in the yeast two-hybrid analysis. Lastly, we thank Robert Lafyatis (Boston University School of Medicine), Masayasu Yamada (Kyoto University), and Stephen Sugrue (University of Florida College of Medicine) for antibodies and plasmid reagents.
G.D. was supported by a fellowship from the Canadian Institutes of Health Research (CIHR), J.T. is supported by an operating grant from the CIHR, H.G.E.S. is supported by a fellowship from the Association for International Cancer Research, and W.A.B. is a Centennial Fellow of the James S. McDonnell Foundation.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Alahari, S. K., H. Schmidt, and N. F. Kaufer. 1993. The fission yeast prp4+ gene involved in pre-mRNA splicing codes for a predicted serine/threonine kinase and is essential for growth. Nucleic Acids Res. 21:4079-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauren, G., and L. Wieslander. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76:183-192. [DOI] [PubMed] [Google Scholar]
- 5.Brandner, J. M., S. Reidenbach, C. Kuhn, and W. W. Franke. 1998. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur. J. Cell Biol. 75:295-308. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 8.Chiba, H., M. Muramatsu, A. Nomoto, and H. Kato. 1994. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall, A. K., S. J. Beek, C. M. McCallum, J. W. Tamkun, G. V. Kalpana, S. P. Goff, and M. P. Scott. 1995. The Drosophila Snr1 and Brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929-933. [DOI] [PubMed] [Google Scholar]
- 14.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 15.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 16.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross, T., M. Lutzelberger, H. Weigmann, A. Klingenhoff, S. Shenoy, and N. F. Kaufer. 1997. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 25:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 20.Hedley, M. L., H. Amrein, and T. Maniatis. 1995. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA 92:11524-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 22.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 23.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, and C. K. Glass. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Y., T. Deng, and B. W. Winston. 2000. Characterization of hPRP4 kinase activation: potential role in signaling. Biochem. Biophys. Res. Commun. 271:456-463. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 27.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 28.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically downregulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 29.Kao, H. Y., M. Downes, P. Ordentlich, and R. M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55-66. [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima, T., T. Zama, K. Wada, H. Onogi, and M. Hagiwara. 2001. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 276:32247-32256. [DOI] [PubMed] [Google Scholar]
- 31.Laggerbauer, B., J. Lauber, and R. Luhrmann. 1996. Identification of an RNA-dependent ATPase activity in mammalian U5 snRNPs. Nucleic Acids Res. 24:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator: the interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 33.Lauber, J., G. Plessel, S. Prehn, C. L. Will, P. Fabrizio, K. Groning, W. S. Lane, and R. Luhrmann. 1997. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA 3:926-941. [PMC free article] [PubMed] [Google Scholar]
- 34.Le Douarin, B., A. L. Nielsen, J. M. Garnier, H. Ichinose, F. Jeanmougin, R. Losson, and P. Chambon. 1996. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701-6715. [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, S. B., and D. A. Haber. 2001. Wilms tumor and the WT1 gene. Exp. Cell Res. 264:74-99. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., and P. M. Bingham. 1991. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67:335-342. [DOI] [PubMed] [Google Scholar]
- 37.Longman, D., I. L. Johnstone, and J. F. Caceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makarov, E. M., O. V. Makarova, T. Achsel, and R. Luhrmann. 2000. The human homologue of the yeast splicing factor Prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 298:567-575. [DOI] [PubMed] [Google Scholar]
- 39.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mintz, P. J., S. D. Patterson, A. F. Neuwald, C. S. Spahr, and D. L. Spector. 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18:4308-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 43.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodeling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 45.Neish, A. S., S. F. Anderson, B. P. Schlegel, W. Wei, and J. D. Parvin. 1998. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 26:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 47.Nishikimi, A., J. Mukai, N. Kioka, and M. Yamada. 1999. A novel mammalian nuclear protein similar to Schizosaccharomyces pombe Prp1p/Zer1p and Saccharomyces cerevisiae Prp6p pre-mRNA splicing factors. Biochim. Biophys. Acta 1435:147-152. [DOI] [PubMed] [Google Scholar]
- 48.Ouyang, P., and S. P. Sugrue. 1996. Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J. Cell Biol. 135:1027-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pflum, M. K., J. K. Tong, W. S. Lane, and S. L. Schreiber. 2001. Histone deacetylase I phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276:47733-47741. [DOI] [PubMed] [Google Scholar]
- 50.Sarkissian, M., A. Winne, and R. Lafyatis. 1996. The mammalian homolog of suppressor-of-white-apricot regulates alternative mRNA splicing of CD45 exon 4 and fibronectin IIICS. J. Biol. Chem. 271:31106-31114. [DOI] [PubMed] [Google Scholar]
- 51.Schwelnus, W., K. Richert, F. Opitz, T. Gross, Y. Habara, T. Tani, and N. F. Kaufer. 2001. Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO Rep. 2:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simons, A., C. Melamed-Bessudo, R. Wolkowicz, J. Sperling, R. Sperling, L. Eisenbach, and V. Rotter. 1997. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene 14:145-155. [DOI] [PubMed] [Google Scholar]
- 53.Spector, D. L., X. D. Fu, and T. Maniatis. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10:3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stojdl, D. F., and J. C. Bell. 1999. SR protein kinases: the splice of life. Biochem. Cell Biol. 77:293-298. [PubMed] [Google Scholar]
- 55.Sutherland, H. G., G. K. Mumford, K. Newton, L. V. Ford, R. Farrall, G. Dellaire, J. F. Caceres, and W. A. Bickmore. 2001. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 10:1995-2011. [DOI] [PubMed] [Google Scholar]
- 56.Tate, P., M. Lee, S. Tweedie, W. C. Skarnes, and W. A. Bickmore. 1998. Capturing novel mouse genes encoding chromosomal and other nuclear proteins. J. Cell Sci. 111(Pt. 17):2575-2585. [DOI] [PubMed] [Google Scholar]
- 57.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 58.Will, C. L., B. Kastner, and R. Luhrmann. 1994. Analysis of ribonucleoprotein interactions, p. 141-177. In D. Rickwood and B. D. Hames (ed.), RNA processing: practical approach. IRL Press, Oxford, England.
- 59.Wilson, C. J., D. M. Chao, A. N. Imbalzano, G. R. Schnitzler, R. E. Kingston, and R. A. Young. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84:235-244. [DOI] [PubMed] [Google Scholar]
- 60.Woppmann, A., C. L. Will, U. Kornstadt, P. Zuo, J. L. Manley, and R. Luhrmann. 1993. Identification of an snRNP-associated kinase activity that phosphorylates arginine/serine rich domains typical of splicing factors. Nucleic Acids Res. 21:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 62.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273:27761-27764. [DOI] [PubMed] [Google Scholar]
- 63.Yao, Y. L., Yang, W. M., and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zachar, Z., T. B. Chou, J. Kramer, I. P. Mims, and P. M. Bingham. 1994. Analysis of autoregulation at the level of pre-mRNA splicing of the suppressor-of-white-apricot gene in Drosophila. Genetics 137:139-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, and S. D. Hayward. 2000. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol. 74:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]