Abstract
In plants, double-stranded RNA that is processed to short RNAs ≈21–24 nt in length can trigger two types of epigenetic gene silencing. Posttranscriptional gene silencing, which is related to RNA interference in animals and quelling in fungi, involves targeted elimination of homologous mRNA in the cytoplasm. RNA-directed DNA methylation involves de novo methylation of almost all cytosine residues within a region of RNA–DNA sequence identity. RNA-directed DNA methylation is presumed to be responsible for the methylation observed in protein coding regions of posttranscriptionally silenced genes. Moreover, a type of transcriptional gene silencing and de novo methylation of homologous promoters in trans can occur if a double-stranded RNA contains promoter sequences. Although RNA-directed DNA methylation has been described so far only in plants, there is increasing evidence that RNA can also target genome modifications in other organisms. To understand how RNA directs methylation to identical DNA sequences and how changes in chromatin configuration contribute to initiating or maintaining DNA methylation induced by RNA, a promoter double-stranded RNA-mediated transcriptional gene silencing system has been established in Arabidopsis. A genetic analysis of this system is helping to unravel the relationships among RNA signals, DNA methylation, and chromatin structure.
The term “RNA silencing” refers to epigenetic gene silencing effects that are initiated by double-stranded RNA (dsRNA) (1). Discovered independently in plants, fungi, and animals, RNA silencing phenomena are revealing new ways to repress gene expression and to subdue transposable elements and viruses that produce dsRNA during their replication cycle (2–8). A fundamental step in RNA silencing pathways is cleavage of dsRNA into short RNAs (9), which are believed to act as guides for enzyme complexes that either degrade or modify homologous nucleic acids.
The most familiar type of RNA silencing occurs primarily in the cytoplasm and is termed posttranscriptional gene silencing (PTGS) in plants, quelling in Neurospora, and RNA interference (RNAi) in animals. PTGS/RNAi involves a dsRNA that is processed by an RNase III-like enzyme called Dicer into short interfering (si) RNAs 21–22 nt in length. The antisense siRNAs associate with a ribonuclease complex and guide sequence-specific degradation of complementary mRNAs (5–8).
A second form of RNA silencing involves sequence-specific changes at the genome level. RNA-directed DNA methylation (RdDM) (10), which has been described so far only in plants, leads to de novo methylation of almost all cytosine residues within the region of sequence identity between the triggering RNA and the target DNA. Similarly to PTGS/RNAi, RdDM requires a dsRNA that is cleaved to short RNAs ≈21–24 nt in length (11). It is not yet certain whether the short RNAs or dsRNA guide methylation of homologous DNA sequences, although the length of short RNAs is consistent with the minimum DNA target size of RdDM (≈30 bp) (12).
RdDM is assumed to be the source of methylation observed in protein coding regions in many cases of PTGS, where it can contribute in an unknown way to the maintenance of silencing (13, 14). In addition, RdDM has been implicated in a type of transcriptional gene silencing (TGS) that is initiated by dsRNAs containing promoter sequences. Promoter dsRNAs that trigger TGS and RdDM of homologous promoters in trans can be produced in the nucleus by transcription through inverted repeats (IRs) of promoter sequences (11, 15) or in the cytoplasm by a replicating RNA virus that is engineered to contain sequences identical to the promoter of a nuclear gene (16, 17).
Although the phenomenon of RdDM is well established in plants, a number of questions remain. One concerns the identity of the DNA methyltransferases (MTases) that are required for establishing and maintaining the unusual pattern of methylation characteristic of RdDM. A second issue concerns the relationship between DNA methylation and changes in chromatin structure. Given the close links between DNA methylation, chromatin remodeling (18–20), and histone modifications, such as acetylation (21) and methylation (22, 23), it might be anticipated that alterations in chromatin structure would be required to initiate and/or retain methylation induced by RdDM.
To address these questions, we carried out a genetic analysis of a promoter dsRNA-mediated TGS system that we have established in Arabidopsis. In this paper, we describe this system and the impact of several mutations that impair DNA methylation and/or possible chromatin remodeling processes. We discuss whether RdDM might occur in animals and whether RNA might direct chromatin modifications in organisms that do not methylate their DNA.
Experimental Procedures
T-DNA Constructs and Plant Transformation.
Arabidopsis thaliana plants (ecotype Columbia) were grown at 22°C in a 16 h/8 h day/night cycle. Transformation was done by the floral dip method (24). The nopaline synthase promoter (NOSpro) target construct [NOSpro–NPTII (neomycin phosphotransferase) NOSter–NOSpro–NOS (nopaline synthase) NOSter] was introduced into Arabidopsis by using Agrobacterium strain A208 harboring a disarmed Ti-plasmid (25). A line homozygous for the target locus was retransformed by using Agrobacterium harboring a binary vector with a NOSpro IR (in which the halves were separated by ≈250 bp of the α′ promoter of soybean β-conglycinin; ref. 26) driven by the 35S promoter of cauliflower mosaic virus (35Spro), a pUC18 plasmid vector, and a 19Spro-HPT (hygromycin phosphotransferase) gene as selectable marker (11). The 35Spro was flanked by lox sites in direct orientation to allow removal by Cre recombinase. Selection of transgenic target and silencer plants was done as described (11).
Cre/lox-Mediated Deletion of the 35Spro.
To delete the 35Spro by Cre recombinase, plants doubly homozygous for target and silencer were supertransformed with a third T-DNA construct expressing Cre recombinase from the 35Spro (27). The Cre construct encodes glufosinate resistance (BAR), which allows the herbicide BASTA to be used for selection directly on soil. Soil-grown triple transformants were selected by spraying with BASTA (Celaflor, Hoechst, Vienna, Austria; 400 mg/liter ammonium glufosinate) twice a week for 2 weeks. T2 seeds from BASTA-resistant T1 plants were plated on MS agar containing 40 mg/liter kanamycin (Kan) (Sigma), and/or 40 mg/liter Kan plus 20 mg/liter hygromycin B (Hyg) (Calbiochem). Resistant T2 seedlings were genotyped by PCR using BAR primers to confirm the absence of the Cre construct because the presence of Cre-recombinase interferes with Southern analysis of DNA-fragments containing lox sites (M.F.M., unpublished observations). Genotype-PCR-grade DNA was isolated from Arabidopsis leaves as described (28). BAR primers were 5′-CGAGACAAGCACGGTCAACTTC-3′ and 5′-ACCCACGTCATGCCAGTTCC-3′. BAR-negative plants were allowed to set seeds, and DNA was extracted from T3 progeny plants by using a DNAeasy plant maxi kit (Qiagen, Hilden, Germany). The DNA was subjected to restriction digests and Southern hybridization as described in the Fig. 1 and Fig. 6 legends. Data in Fig. 3C and Fig. 6 were obtained for plants 2 generations after Cre-mediated removal of the 35Spro.
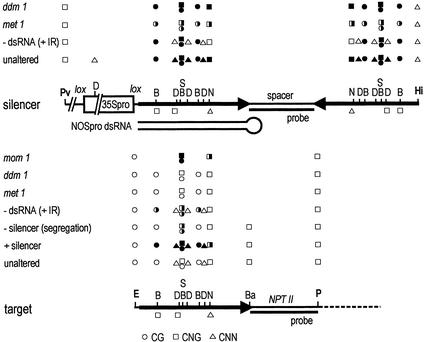
Figure 1.
Structures and methylation analysis of the silencer NOSpro IR (Upper) and target NOSpro–NPTII gene (Lower). NOSpro sequences are depicted as heavy black arrows. Enzymes and probes used for DNA blot analyses are indicated. Abbreviations: Pv, PvuII; D, DdeI; S, SacII; N, NheI; Hi, HindIII; E, EcoRI; B, BstUI; Ba, BamHI; P, PstI. To assess methylation in the target NOSpro, an E and P double digest was performed (the minus lanes in Figs. 4A and 6 A, C, E, and G) and one of several methylation-sensitive enzymes (B, D, S, N, Ba) was added. Methylation in the NOSpro IR was tested by digesting with Pv and Hi (the minus lanes in Figs. 4B and 6 B, D, F, and H), together with either B, D, S, or N. Filled, half-filled, and open circles, squares, and triangles (CG, CNG, and CNN, respectively) indicate >90%, ≈50%, and <10% cytosine (C) methylation, respectively. Open squares or triangles below each map for the enzymes D, N, and B indicate that the top and bottom DNA strands contain C residues in different sequence contexts (e.g., CG and CNG, or CNG and CNN). The NheI site (underlined) is in the sequence context: 5′-CAGCTACGmCAA-3′ (top); and 3′-GTmCGATCGTT-5′ (bottom).
Figure 6.
Methylation analysis in the presence and absence of NOSpro dsRNA. (A, C, E, and G) Target NOSpro. (B, D, F, and H) Silencer NOSpro IR. The enzymes and probes used are described in Fig. 1. The arrows to the left of each blot show the position of the expected unmethylated fragment. Results from two lines containing the Cre-altered silencer are shown. (G) The NOSpro–NPTII bands of interest are flanked by blue dots. The large hybridizing fragment in the minus lanes and the fragment in the D lanes running just below the NOSpro–NPTII band should be disregarded as they are caused by a second NPTII gene (not visible in A, C, and E) used for bacterial selection during cloning (25). The size of the fragment in the minus lanes in B, D, F, and H is shifted after the Cre cross (indicated by red dots) because of removal of the 35Spro and is independent of methylation. Because of an unmethylated DdeI site in the 35Spro (Fig. 1 Upper), the size of the fragment of the unaltered silencer shifts after addition of DdeI (H).
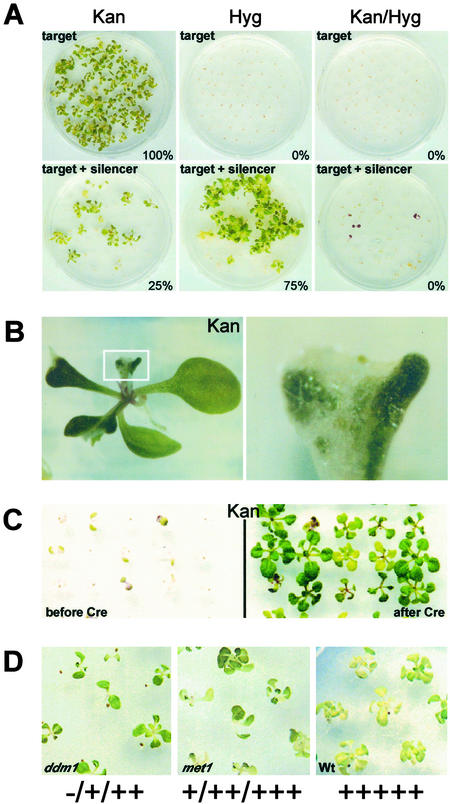
Figure 3.
Phenotypic analysis of silencing. NOSpro–NPTII target gene expression is assayed by KanR; the silencing locus by HygR. (A) Selfing a plant homozygous for an active target gene produces 100% KanR progeny. Selfing a plant homozygous for the target locus and hemizygous for the silencing locus, revealed by 75% HygR, produces only 25% KanR progeny. KanR seedlings lack the silencer, indicated by 0% (Kan/Hyg)R. (B) Mottled KanR seedling in the first generation after crossing out the silencing locus (Right is an enlargement of the boxed region in Left; white and green patches represent KanS and KanR regions, respectively). (C Left) KanS seedlings before removing the 35Spro with Cre recombinase. (Right) KanR seedlings two generations after removing the 35Spro. (D) Ranges of phenotypes on Kan-containing medium (plus signs, different degrees of KanR; minus sign, KanS) in seedlings after three generations of homozygosity for the ddm1 and met1 mutations, based on 5 plus signs for wild-type levels of KanR in seedlings containing the target locus in the unsilenced state.
Mutant Crosses.
The following Arabidopsis mutants were used in this study: ddm1-5/som8 (decrease in DNA methylation/somniferous) (29) in ecotype Zürich; mom1 (Morpheus' molecule) (30) in ecotype Zürich; and met1/ddm2–1 (DNA methyltransferase 1) (E. Richards, personal communication) in ecotype La-er. Because the strength of NOSpro silencing varied somewhat in different ecotypes (La-er > Col-0 > Zürich), control crosses of double homozygous target/silencer plants with the respective wild-type backgrounds were always performed. Reactivation of the NOSpro–NPTII target gene in a given mutant was assessed by survival of seedlings in the F2 generation and/or advanced generations on Murashige and Skoog (MS) medium containing 40 mg/liter Kan alone or 40 mg/liter Kan plus 20 mg/liter Hyg. The mom1 and met1 mutations are not genetically linked to either the target locus or the silencing locus, and could be tested for their effects on NOSpro–NPTII silencing immediately in the F2 generation; ddm1/som8 is linked to the target locus and had to be introgressed into the NOSpro target/silencer line as described below.
ddm1-5/som8 is a fast neutron-generated allele that is distinguishable from the wild-type gene by an 82 bp insertion into the second exon (18). The ddm1-5/som8 mutation was separated from transgene locus A (which harbors 35S-HPT genes) (29) by two outcrosses to ecotype Col-0. Plants homozygous for ddm1-5/som8 and lacking locus A were screened out from selfed progeny of the second outcross. Genotyping reactions were done by using som primers: 5′-AAGCGACGGAGACGACTGTTTG-3′ and 5′-TTTCACAAAGCAACCACACTACG-3′. 35S-HPT primers were 5′-CCCACTATCCTTCGCAAGA-3′and 5′-CGTCTGCTGCTCCATACAAGC-3′. Because the DDM1 gene is linked to the target transgene locus (the physical distance is ≈270 kb), the ddm1-5 mutation was introgressed into the genetic background of the NOSpro target/silencer transgenic plant. A ddm1-5/ddm1-5 plant lacking transgene locus A was crossed with a plant doubly homozygous for the NOSpro target/silencer. An F2 plant homozygous for the NOSpro silencing locus, hemizygous for the NOSpro target locus and heterozygous for the ddm1-5 allele was selected. This plant was allowed to self-pollinate, and 193 F3 seedlings were analyzed for their DDM1 genotype with the som primers described above. Forty-eight F3 seedlings were homozygous for the ddm1-5 allele and were further subjected to PCR-genotyping for the presence of target NOSpro–NPTII sequences by using NOSpro–NPTII primers: 5′-GAGAATTAAGGGAGTCACG-3′and 5′-TCGTCCTGCAGTTCATTC-3′. Two of the 48 ddm1-5/ddm1-5 plants were hemizygous for the target locus indicating a recombination event between the ddm1-5 allele and the NOSpro target locus during meiosis of the parental F2 plant. Progeny seeds from this genotype (i.e., the second generation of homozygosity for ddm1-5; the target is still segregating) were analyzed for reactivation of the NOSpro–NPTII gene on medium containing Kan (n = 308 seeds) or Kan and Hyg (n = 317). Only weak reactivation was observed in a few progeny. Eventually, in progeny that were third generation homozygous for the ddm1-5 allele (and homozygous for the target and silencing loci), resistance generally improved and stronger Kan resistant (KanR) seedlings appeared stochastically in populations of seedlings that also contained moderately and weakly KanR members (Fig. 3D).
Plants homozygous for the mom1 allele without transgene locus A were crossed with a doubly homozygous NOSpro target/silencer plant. F2 seeds were plated on medium containing Kan, Hyg, or both to test for an immediate effect of the mom1 mutant allele. No immediate effect was observed. Among the F3 progeny of this cross, a triple homozygous line for the NOSpro target, NOSpro silencer, and the mom1 allele was selected. The genotype for the NOSpro silencer was determined by selecting seeds on Hyg-containing medium. The genotype with respect to the mom1 allele, which is tagged with a T-DNA conferring BASTA resistance, was determined by spraying seedlings with the herbicide as described above. To assess the genotype for the NOSpro target, PCR genotyping with NOSpro–NPTII primers (see above) was performed. The progeny F4 seeds of the triple homozygous plant (i.e., the third generation of homozygosity for mom1) were plated on medium containing Kan (n = 440) or Kan and Hyg (n = 441). No resistant seedlings were observed.
met1/ddm2–1 was obtained as progeny from a heterozygous plant and homozygous plants were screened out by using the demethylation assay of centromeric and ribosomal DNA (rDNA) repeats (ref. 31; E. Richards, personal communication). A homozygous met1 plant was crossed to plants homozygous for both the NOSpro target locus and the NOSpro silencing locus. Resulting F1 plants were allowed to self-pollinate, and the F2 seeds were plated on medium containing Kan, Hyg, or both antibiotics. The F2 seedlings showed weak to moderate resistance on Kan and Kan–Hyg, consistent with a partial release of silencing. Double resistant seedlings were recovered on soil, and met1 homozygosity was confirmed by using cleaved amplified polymorphic sequence markers (CAPS) markers as described (32). F4 progeny lines that are triple homozygous for met1 (i.e., third generation of met1 homozygosity), the target locus and the silencing locus were selected for further analysis. These seedlings display weak to strong KanR (Fig. 3D).
Methylation Analysis.
Plant genomic DNA was extracted as described (11). For the DNA methylation analyses in met1, ddm1, and mom1 mutants, DNA was isolated from plants that had been homozygous for a given mutation for two generations (i.e., F3 generation for mom1 and met1; F4 generation for ddm1). Methylation in plants containing the Cre-altered, nontranscribed silencing locus were performed after the Cre-expressing locus had been segregated out, eliminating possible background transcription of the Cre locus. Restriction digests were done according to the instructions of the manufacturers. DNA blot analysis using 32P-labeled RNA probes has been described (33). As probes, subcloned 0.19 kb NPTII and 0.25 kb α′ pro fragments were transcribed in vitro. Methylation analysis of centromeric repeats and rDNA repeats was conducted as described by others (32). Bisulfite sequencing was performed as described (12, 34). The following degenerate primers, which allowed for unconverted or converted cytosines, were used to amplify the top strand of the NOS-NPTII target: 5′ primer 5′-YATGAGYGGAGAATTAAGGGAGT-3′ (Y = C or T); 3′ primer 5′-CCRAATARCCTCTCCACCCAA-3′ (R = G or A).
Cloning of Transgene Inserts.
Genomic λ clones from target and silencer transgenes were obtained and sequenced as described (35). The silencer transgene complex comprises a single copy of the T-DNA construct with a complete 35Spro–NOSpro IR that was integrated in chromosome 4, BAC clone F10M10 (GenBank accession no. AL035521) with the right border downstream of nucleotide 21681 and the left border upstream of nucleotide 21693. Between plant sequence and the right border, the filler sequence “TTTTT” was inserted. The target locus was originally screened out genetically as a single transgene locus that was readily inactivated when the silencing locus was introduced, and largely reactivated the first generation after segregating away from the silencing locus. More detailed structural analysis by λ cloning revealed that the target locus contains several complete and incomplete copies of the T-DNA construct flanked by Arabidopsis DNA from chromosome 5 on the left and chromosome 3 on the right. This finding suggests that a rearrangement of plant DNA, which occasionally occurs during T-DNA integration (36, 37), had occurred. All of the bands visualized in Southern blot analyses using an NPTII probe always cosegregated in multiple, independent crosses, which is consistent with a single, multicopy transgene locus. In the absence of the silencing locus, the target locus was stably expressed over multiple homozygous generations. The moderate structural complexity probably enhanced its susceptibility to silencing, as has been observed in a NOSpro silencing system in tobacco (38). Genetic analysis revealing linkage to ddm1 indicated that the actual T-DNA insertion site was on chromosome 5 (data not shown). Nucleotide 143 of the T-DNA right border region (GenBank accession no. J01826) was fused upstream of nucleotide 66153 of BAC clone F2103 (GenBank accession no. AC009853) on chromosome 3. Nucleotide 98 of the left T-DNA border region (GenBank accession no. J01825) was fused to sequences at the distal end of chromosome 5, BAC clone K9I9 (GenBank accession no. AB013390) upstream of nucleotide 4316. The rearrangement had no visible phenotype effects or impact on target NOSpro expression. Mitotic chromosome counts (39) revealed a normal diploid number of 2n = 10.
RNA Analyses.
Total RNA was extracted, electrophoresed, and blotted as described (40). Control hybridization of tobacco with an actin probe was carried out following published procedures (40). Arabidopsis RNA was hybridized with a eukaryotic protein synthesis initiation factor 4A (eIF-4A) fragment from Arabidopsis (41). Analysis of NOSpro dsRNA and small RNA was carried out as described (11). Transcriptional run-on analysis was performed as described in a former report (42).
Results
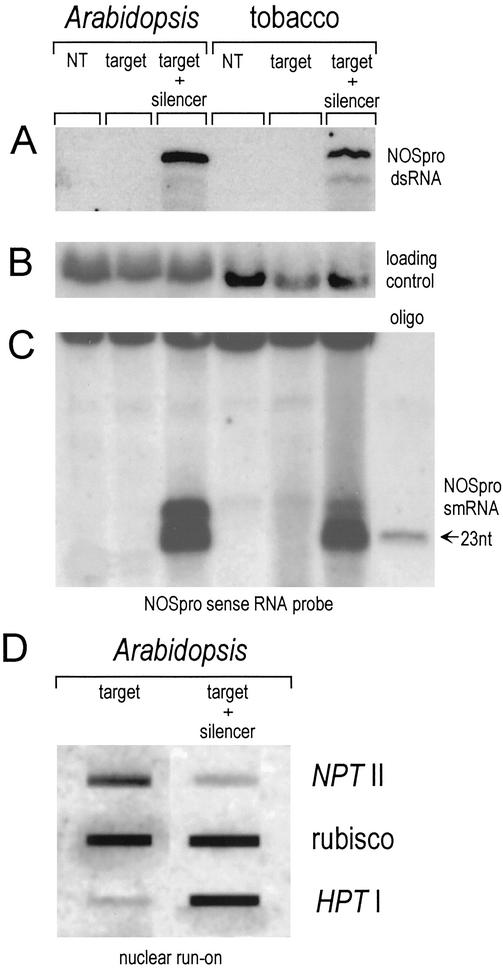
A promoter dsRNA-mediated trans-silencing system based on the NOSpro was originally established in tobacco (11) and has served as a model for setting up a similar system in Arabidopsis. A homozygous line that stably expresses a NOSpro–NPTII target gene encoding resistance to Kan (Fig. 1 Lower) was produced. The target line was then retransformed with a silencing construct, which contains a NOSpro IR under the control of the 35Spro (Fig. 1 Upper) together with a gene encoding resistance to Hyg driven by the 19S promoter of cauliflower mosaic virus (11). The silencing locus produces NOSpro dsRNA (Fig. 2A, Arabidopsis target + silencer) that is processed into short RNAs ≈21–24 nucleotides in length (Fig. 2C, Arabidopsis target + silencer), similar to those observed in tobacco transformed with the same construct (Fig. 2 A and C, tobacco target + silencer) (11).
Figure 2.
RNA analysis. (A) RNase protection reveals the ≈0.3-kb NOSpro dsRNA transcribed from the silencer NOSpro IR. NT, normal untransformed plants. (B) Total RNA used in A probed with an actin probe from tobacco and an eIF-4A probe from Arabidopsis as loading controls. (C) Detection of NOSpro short RNAs (sense probe) produced by means of dsRNA cleavage. Identical results were obtained with an antisense probe. (D) Nuclear run-on analysis demonstrating transcriptional down-regulation of the NOSpro–NPTII target gene in the presence of the silencing locus, which encodes HPT and NOSpro dsRNA. A constitutively expressed ribulose 1,5-bisphosphate carboxylase (rubisco) gene was used as a control. Positive controls in A and C were prepared from tobacco plants transformed with the 35Spro–NOSproIR construct.
In the presence of the silencing locus, the target NOSpro–NPTII gene is inactivated, as revealed by cultivation of seedlings on media containing different antibiotics. When self fertilized, a plant that is homozygous for the active NOSpro–NPTII target gene produces, as expected, 100% KanR progeny (Fig. 3A, target–Kan). When the silencing locus, which encodes NOSpro dsRNA and resistance to Hyg, is introduced into the target line and is present in the hemizygous condition, selfing produces 75% Hyg-resistant seedlings (Fig. 3A, target + silencer-Hyg). Even though the parent is homozygous for the target locus, however, only 25% of the seedlings are KanR (Fig. 3A, target + silencer-Kan). Any seedling that is KanR has not inherited the silencing locus, as indicated by the lack of double resistance (Fig. 3A, target + silencer, Kan/Hyg). Conversely, a seedling that has inherited the silencing locus is Kan-sensitive because of silencing of the NOSpro–NPTII target gene. Silencing of the target gene occurs at the transcriptional level as demonstrated by a nuclear run-on analysis (Fig. 2D).
Transcriptional silencing of the NOSpro–NPTII target gene is accompanied by de novo methylation of the target NOSpro. When active, the target gene is normally unmethylated in the NOSpro region, as indicated by nearly complete digestion with the methylation-sensitive restriction enzymes SacII (mCmCGmCGG), BstUI (mCGmCG), and NheI(GCTAGmC) (Fig. 4A, unmeth. control; a superscript “m” indicates a methylated cytosine that can inhibit cleavage). In the presence of the silencing locus, the NOSpro region specifically becomes methylated in both symmetrical (CG and CNG) and nonsymmetrical (CNN) cytosines as demonstrated, respectively, by negligible digestion with SacII and BstUI, and approximately 50% digestion with NheI (Fig. 4A, target + silencer). This pattern of methylation, in which C residues in any sequence context are modified specifically in the region of RNA-DNA sequence identity, is characteristic of RdDM and was confirmed when bisulfite sequencing was used to examine methylation in more detail (Fig. 5).
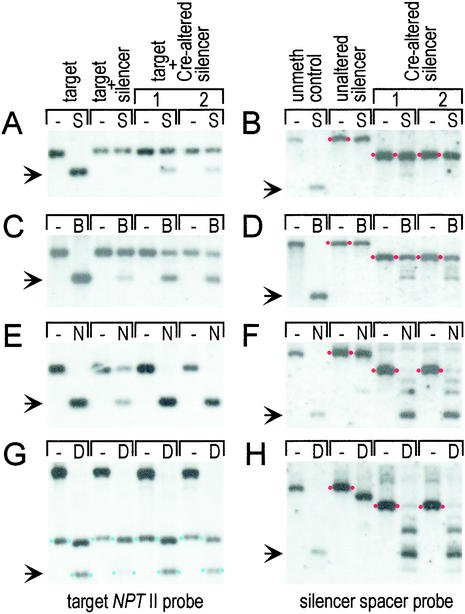
Figure 4.
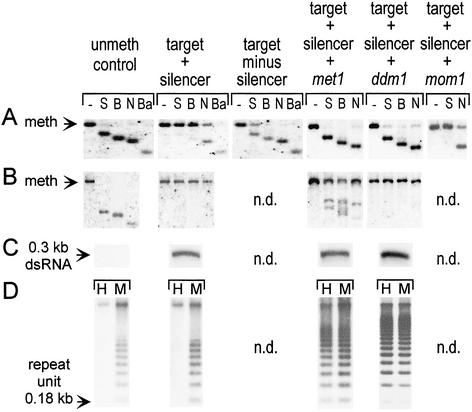
Methylation analysis. (A) Target NOSpro. (B) Silencer NOSpro IR. (C) NOSpro dsRNA. (D) Centromeric repeats. Methylation of the target and silencing loci were analyzed by using the enzymes and probes described in Fig. 1. For met1, ddm1, and mom1 mutants, methylation was analyzed by using DNA isolated from plants that had been homozygous for the respective mutation for two generations. Methylation of centromeric repeats was analyzed by using HpaII (H: mCmCGG) and MspI (M: mCCGG). The unmethylated control for the silencer NOSpro IR consisted of a λ genomic clone containing the silencing locus. Shifts to the smaller fragments indicate no methylation at a particular site. Arrows in A and B indicate position of methylated fragment; in C and D, arrows represent the sizes of the indicated features. n.d., not determined.
Figure 5.
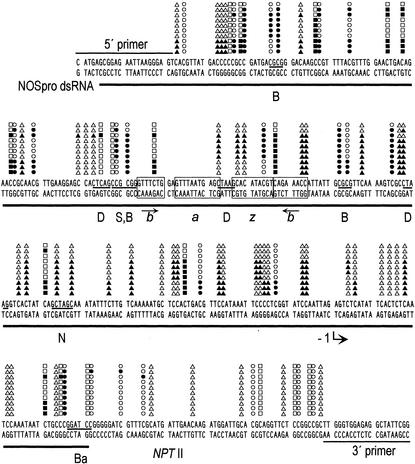
Bisulfite sequencing. The ≈300-bp NOSpro sequence is shown with the region of identity to NOSpro dsRNA underlined. Methylation (filled symbols) in 10 cloned PCR fragments from the upper DNA strand is indicated. Symbols are described in the Fig. 1 legend. The positions of restriction enzyme sites used in the DNA blot analyses are indicated (abbreviations are given in the Fig. 1 legend). The four boxed regions represent transcriptional regulatory elements (61), which contain short IRs (arrows). The transcription start site is indicated by the bent arrow at −1. The sequence of the primers used is indicated. Methylation does not infiltrate significantly into NPTII coding sequences.
Methylation of the target NOSpro–NPTII gene is largely eliminated when the target locus and silencing locus segregate in progeny, as indicated by nearly complete digestion with BstUI and NheI, and about 50% cleavage with SacII (Fig. 4A, target minus silencer). The remaining methylation at the SacII site, which is correlated with the mottled phenotype of many KanR seedlings (Fig. 3B), is presumably caused by maintenance of some CG and/or CNG methylation through meiosis (42).
A requirement for NOSpro dsRNA in silencing and methylation of the target NOSpro–NPTII gene was demonstrated by removing the transcribing 35Spro, which is flanked by lox sites (Fig. 1 Upper), with Cre recombinase. The removal of the 35Spro and retention of the NOSpro IR at the Cre-modified “silencing” locus was monitored by a shift to a smaller band of the expected size with all enzymes tested in a DNA blot analysis (Fig. 6 B, D, F, and H; compare minus lanes in unaltered silencer panels with minus lanes in Cre-altered silencer panels) and confirmed by cloning and sequencing the Cre-modified silencing locus (data not shown).
After deletion of the 35Spro from the silencing locus, NOSpro short RNAs are no longer detectable, even after long exposures of the respective Northern blots (data not shown). Consequently, the target NOSpro–NPTII gene is active in the presence of the nontranscribed NOSpro IR, as indicated by the KanR phenotype of seedlings that are doubly homozygous for the target locus and Cre-modified silencing locus (Fig. 3C, after Cre). Furthermore, methylation of the NOSpro–NPTII target gene is reduced approximately 30% at symmetrical Cs in the SacII (Fig. 6A, target + Cre-altered silencer) and BstUI (Fig. 6C, target + Cre-altered silencer) sites and almost completely at nonsymmetrical C residues in the NheI site (Fig. 6E, target + Cre-altered silencer) and DdeI sites (mCTNAG) (Fig. 6G, target + Cre-altered silencer).
The NOSpro dsRNA not only triggers methylation and silencing of the target NOSpro in trans, it also contributes to methylation in cis of the NOSpro copies in the IR at the silencing locus. This was demonstrated by examining methylation of the NOSpro IR before and after removing the transcribing 35Spro with Cre recombinase. The transcribed NOSpro IR at the unaltered silencing locus is heavily methylated at both symmetrical and nonsymmetrical Cs within the repeated region as indicated, respectively, by lack of digestion with SacII and BstUI (Fig. 6 B and D, unaltered silencer panels), and NheI and DdeI (Fig. 6 F and H, unaltered silencer panels). In contrast, the nontranscribed NOSpro IR at the Cre-altered silencing locus loses methylation at nonsymmetrical C residues, which was revealed by substantial digestion with NheI and DdeI (Fig. 6 F and H; compare unaltered silencer panel with Cre-altered silencer panels). At the same time, methylation at symmetrical C residues is almost completely retained, as indicated by poor digestion with SacII and BstUI (Fig. 6 B and D; compare unaltered silencer panel with Cre-altered silencer panels).
The effects of several mutations that release TGS in other systems (14, 17, 29, 30, 32, 43, 44) were tested on the NOSpro dsRNA-mediated TGS system. For these experiments, the double homozygous target/silencer line was crossed with lines homozygous for the following recessive mutations: the som8 allele (29) of ddm1, which encodes a putative component of a SWI/SNF2 chromatin remodelling complex (18); met1 (ddm2), which encodes a DNA MTase (E. Richards, personal communication) that maintains methylation in CG dinucleotides (45); and mom1, which encodes a possible chromatin remodeling protein (30). F1 progeny obtained from these crosses were selfed and the extent of silencing evaluated in the F2, F3, and F4 generations. If a given mutation has no effect (and assuming no linkage between a mutation and the target locus or silencing locus), the percentages of antibiotic resistance in F2 progeny should be 19% KanR, 75% HygR, and 0% (Kan–Hyg)R. If a mutation releases silencing and is fully penetrant, these percentages would change to 33% KanR, 75% HygR, and 14% (Kan–Hyg)R. In other words, impaired silencing would be indicated by an increase in the percentage of KanR F2 progeny and by the appearance of some F2 progeny that display double resistance.
The mom1 mutation is the only one of the three tested that did not visibly affect NOSpro silencing, as indicated by no recovery of (Kan–Hyg)R progeny, even in F3 and F4 generations (data not shown). Methylation of the target NOSpro is also not reduced in mom1 mutants, as demonstrated by levels of methylation at the SacII and NheI sites that approximate those in the silenced state (Fig. 4A, compare target + silencer + mom1 with target + silencer).
The met1 mutation partially released silencing of the NOSpro–NPTII gene in F2 progeny, as indicated by an increase in the percentage of KanR seedlings (29%, n = 428) and weak resistance of some of these seedlings on medium containing both Kan and Hyg (11% Kan-HygR, n = 912). Because the ddm1 mutation is linked to the target locus on chromosome 5, it had to be introgressed into the double homozygous target/silencer line. In the first generation, when the strength of antibiotic resistance could be tested in ddm1 mutants, sporadic weak reactivation of NOSpro–NPTII gene expression was observed (data not shown). In both met1 and ddm1 mutants, KanR resistance could improve in advanced generations, although the strength of NOSpro–NPTII gene expression continued to be nonuniform in genotypically identical seedlings (Fig. 3D). The strongest KanR plants sustained significant losses of methylation from the target locus, as indicated by substantial digestion with SacII, BstUI, and NheI (Fig. 4A, target + silencer + met1 and + ddm1). These plants are indeed homozygous for the respective mutations, which cause global demethylation, as revealed by the loss of CG methylation at centromeric repeats (Fig. 4D, target + silencer + met1 and + ddm1 panels) and rDNA repeats (data not shown).
In contrast to the substantial reduction of methylation of the NOSpro target locus in plants showing relatively strong KanR in the met1 and ddm1 backgrounds, the NOSpro IR at the silencing locus retains considerable methylation in these mutants. This was particularly evident in ddm1 mutant plants, where—similarly to wild type plants—virtually no digestion by SacII, BstUI, and NheI was observed (Fig. 4B, compare target + silencer + ddm1 with target + silencer). In met1 plants, methylation is reduced ≈20–30% at both symmetrical (SacII, BstUI) and nonsymmetrical (NheI) sites (Fig. 4B, target + silencer + met1 panels). NOSpro dsRNA continues to be synthesized at wild-type levels in the met1 and ddm1 mutant plants (Fig. 4C).
Discussion
To dissect the mechanism of RdDM and dsRNA-mediated TGS, we have established a two component silencing system based on the NOSpro in Arabidopsis. A NOSpro dsRNA transcribed from a NOSpro IR at the silencing locus is processed to short RNAs 21–24 nt in length. Either the dsRNA or the short RNAs can locate and interact with the homologous NOSpro at the unlinked target locus and trigger TGS and promoter methylation. We report here an analysis of methylation of the target NOSpro and the NOSpro IR at the silencing locus in the presence and absence of NOSpro dsRNA, and in several mutants deficient in DNA methylation and/or putative chromatin remodeling proteins. The results are summarized in Fig. 1.
NOSpro dsRNA induces de novo methylation of the target NOSpro at Cs in any sequence context within the region of RNA-DNA sequence identity (Fig. 1 target + silencer). Removing the source of the dsRNA, either by segregating away the silencing locus [Fig. 1 target, − silencer (segregation)] or by removing the transcribing 35Spro via Cre/lox-mediated recombination [Fig. 1 target, − dsRNA (+IR)], results readily in loss of methylation in nonsymmetrical C residues, indicating that they require continuous de novo methylation induced by RdDM. In contrast, methylation in symmetrical CG and CNG nucleotide groups, which can be maintained, respectively, by the DNA MTases MET1 (45) and chromomethylase3 (CMT3) (46, 47), is retained to varying degrees in the target NOSpro after it segregates away from the silencing locus or in the presence of the Cre-altered silencing locus. The persistent target promoter methylation resembles paramutation, which involves meiotically heritable changes in gene expression induced by allelic interactions (48). Indeed, some paramutation-like phenomena are probably caused by maintenance of RNA-induced CG and CNG methylation through meiosis (17), suggesting that MET1 and CMT3 can function during this period. Interestingly, methylation in symmetrical C residues is not lost as readily in the presence of the nontranscribed Cre-altered silencing locus as it is following segregation of the unaltered silencing locus. This difference cannot be ascribed to NOSpro dsRNA, which is not synthesized to detectable levels following Cre-mediated excision of the 35Spro, nor to the somewhat repetitive nature of the target locus, which remains unchanged regardless of the presence of the silencing locus or the Cre-altered derivative. One possibility is that the nontranscribed IR enhances maintenance methylation of the target NOSpro, perhaps through DNA–DNA pairing interactions (49).
The met1 and ddm1 mutations, which reduce global methylation and release silencing to varying extents in other TGS systems (14, 17, 29, 32, 43, 44), partially alleviate silencing and reduce methylation of the NOSpro–NPTII target gene but these effects can only be considered indirect. With met1, partial, nonuniform recovery of Kan resistance was observed in the F2 generation, followed by progressive improvement in subsequent generations. The ddm1 mutation appeared somewhat less efficient at releasing silencing than met1, though it is difficult to make a strict comparison because the ddm1 mutation had to be introgressed into our NOSpro target/silencer line. In both mutants, losses of methylation in the target NOSpro can be substantial in F3 and F4 progeny that show the strongest KanR (Fig. 1 target, met1, ddm1). Any slight methylation that persists is presumably caused by continued de novo methylation induced by NOSpro dsRNA. The lack of effect of mom1 on NOSpro silencing and methylation (Fig. 1 target, mom1) is not unexpected, because this mutation affects a subset of transcriptionally inactivated genes that are silenced by a methylation-independent pathway and, unlike met1 and ddm1, does not cause global demethylation (30, 50).
The two copies of the NOSpro in the IR at the silencing locus are methylated substantially at symmetrical and nonsymmetrical C residues (Fig. 1, unaltered silencer). When dsRNA synthesis terminates following Cre-mediated removal of the transcribing 35Spro, methylation in CG and CNG nucleotide groups is maintained. In contrast, nonsymmetrical CNN methylation is substantially reduced, demonstrating its dependence on transcription of NOSpro dsRNA [Fig. 1 silencer − dsRNA (+IR)]. After withdrawal of the triggering dsRNA, the nontranscribed IR maintains CG and CNG methylation better than singlet copies of NOSpro at target locus, suggesting that some intrinsic feature of the IR helps to maintain methylation independently of dsRNA. One possibility is that pairing of the IR in cis generates an unusual structure (51) that is recognized by the maintenance MTase activities. Consistent with a critical role for maintenance MTases in retaining methylation in the NOSpro IR, reductions in CG and CNG methylation in this region were greater in met1 than in ddm1 mutant plants (Fig. 1 silencer, met1 and ddm1). A similar stronger effect of met1 compared with ddm1 on methylation of an IR has been noted previously on studies with the PAI gene family in Arabidopsis (32).
Overall, the results from the mutant analysis indicate that efficient maintenance of methylation triggered by RdDM requires MET1 and the activity of DDM1, perhaps as part of a chromatin remodeling complex. Despite the continued presence of NOSpro dsRNA, significant losses of target NOSpro methylation were observed after several generations in met1 and ddm1 mutants. This suggests that in the absence of a maintenance MTase and chromatin restructuring activities, which can help to reinforce silencing, methylation induced by RdDM is lost more rapidly than it can be regenerated de novo.
It is not yet known which DNA MTase catalyzes the de novo methylation step of RdDM, though MET1 is considered unlikely because of the somewhat delayed influence of the met1 mutation on our NOSpro silencing system. A CMT was initially a promising candidate for RdDM (1, 52) because of the presence in these enzymes of a chromodomain, which can serve as an RNA-protein interaction module (53). Initial results with the cmt3 mutation, however, suggest negligible effects on NOSpro silencing in F2 generation (W.A., X. Cao, S. and Jacobsen, M.M., unpublished results). NOSpro methylation must still be examined in the cmt3 mutants. Another candidate for RdDM is a member of the domain rearranged (DRM) class, which is the major de novo DNA MTase family in plants (54). Mutants defective in DRM2 (X. Cao and S. Jacobsen, personal communication) are currently being tested with the NOSpro system. A final possibility is a member of the Dnmt2 family, which is also present in vertebrates, Drosophila and—in a mutated form—in Schizosaccharomyces pombe (55). Mutations in this class of putative DNA MTases remain to be assessed in our NOSpro system.
There are so far no reports that RNA directs DNA methylation in animals. This apparent deficiency may reflect differences between plants and animals with respect to specific requirements for RdDM. Factors to consider include whether the unique pattern of methylation triggered by RdDM can be detected at a particular developmental stage in animals, and whether the required DNA MTase is available at that time. In both mammals and Drosophila, non-CG methylation, which is conceivably directed by RNA (56), is present in early embryos (57, 58). This methylation might be catalyzed by Dnmt3a, which is the major de novo DNA MTase active early in mammalian development (57), or by Dnmt2, which is also primarily active during the initial stages of development in mammals and in Drosophila (58). Both of these enzymes have been implicated in the catalysis of non-CG methylation (57, 58), which would be consistent with RdDM. Thus, if RdDM occurs in animals, it might be limited to early stages of development when the appropriate DNA MTase(s) is active. In contrast, the occurrence of RdDM throughout plant development (56) suggests the continuous activity of the necessary DNA MTase, a feature that probably facilitated the detection of RdDM in adult plants.
Even for organisms that do not methylate their DNA, there is growing evidence that chromatin modifications are targeted by components of the RNAi machinery. In Drosophila, transgene TGS and PTGS are both dependent on the piwi protein, which is a member of the Argonaute family required for RNAi (59). TGS is associated with complexes of polycomb-group proteins, which are perhaps directed to the transgene promoter by short RNAs containing transcriptional regulatory sequences. In S. pombe, homologs of three proteins required for RNAi—dicer, a putative RNA-dependent RNA polymerase, and argonaute—are needed for histone methylation and localization of the heterochromatin protein Swi6 at centromeric repeats (S. Grewal, personal communication). RNAi-based genetic screens to find genes required for RNAi in Caenorhabditis elegans identified several ORFs that are predicted to encode chromatin-associated proteins (60).
Genetic screens are required to recover novel mutations affecting NOSpro dsRNA-mediated TGS. We have recently identified one prospective mutant, rts-1 (RNA-mediated transcriptional silencing), in which silencing is substantially alleviated, whereas target NOSpro methylation is only reduced about 50% (W.A., M.F.M., and A.J.M.M., unpublished results). The rts-1 mutation does not map to a region of the Arabidopsis genome known to encode a DNA MTase, suggesting that it might encode a chromatin factor. Identification of the RTS-1 gene and further genetic analyses using the NOSpro dsRNA-mediated TGS system should continue to provide insights into the relationship between RdDM and chromatin modifications.
Acknowledgments
We thank Eric Richards for the met1 (ddm2) mutant and for probes to analyze methylation in centromeric and rDNA repeats; Ortrun Mittelsten Scheid for the som8 allele of ddm1; Jurek Paszkowski for mom1; Ann Depicker for supplying a Cre-expressing T-DNA construct; and Steve Jacobsen and Xiaofeng Cao for a collaboration on cmt3 and drm2 mutants. We are grateful to Michael Wassenegger for helpful advice on bisulfite sequencing. Our work is supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung (Grant Z21-MED).
Abbreviations
- dsRNA
double-stranded RNA
- Kan
kanamycin
- Hyg
hygromycin
- Kanr
Kan resistant
- IR
inverted DNA repeat
- MTase
DNA cytosine methyltransferase
- NOSpro
nopaline synthase promoter
- PTGS
posttranscriptional gene silencing
- RdDM
RNA-directed DNA methylation
- RNAi
RNA interference
- TGS
transcriptional gene silencing
- 35Spro
35S promoter of cauliflower mosaic virus
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Matzke M A, Matzke A J M, Kooter J. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 2.Vance V B, Vaucheret H. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 3.Waterhouse P, Wang M B, Lough T. Nature (London) 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Denli A M, Hannon G J. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 6.Cogoni C. Annu Rev Microbiol. 2001;55:381–406. doi: 10.1146/annurev.micro.55.1.381. [DOI] [PubMed] [Google Scholar]
- 7.Chicas A, Macino G. EMBO Rep. 2001;2:992–996. doi: 10.1093/embo-reports/kve231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutvágner G, Zamore P. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton A, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 10.Wassenegger M. Plant Mol Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- 11.Mette M F, Aufsatz W, van der Winden J, Matzke M A, Matzke A J M. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pélissier T, Wassenegger M. RNA. 2000;6:55–65. doi: 10.1017/s135583820099201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovarik A, Van Houdt H, Holy A, Depicker A. FEBS Lett. 2000;467:47–51. doi: 10.1016/s0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- 14.Morel J B, Mourrain P, Béclin C, Vaucheret H. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 15.Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol J N M, Kooter J. Curr Biol. 2001;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 16.Jones L, Hamilton A J, Voinnet O, Thomas C L, Maule A J, Baulcombe D C. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones L, Ratcliff F, Baulcombe D C. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 18.Jedelloh J, Stokes T, Richards E. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 19.Gibbons R, McDowell T, Raman S, O'Rourke D, Garrick D, Ayyub H, Higgs D. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 20.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobosy J, Selker E. Cell Mol Life Sci. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamaru H, Selker E. Nature (London) 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 23.Jackson J, Lindroth A, Cao X, Jacobsen S. Nature (London) 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 24.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 25.Matzke A J M, Matzke M. Plant Mol Biol. 1986;7:357–365. doi: 10.1007/BF00032565. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z L, Schuler M A, Beachy R N. Proc Natl Acad Sci USA. 1986;83:8560–8564. doi: 10.1073/pnas.83.22.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Buck S, De Wilde C, Van Montagu M, Depicker A. Mol Plant– Microbe Interact. 2000;13:658–665. doi: 10.1094/MPMI.2000.13.6.658. [DOI] [PubMed] [Google Scholar]
- 28.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittelsten Scheid O, Afsar K, Paszkowski J. Proc Natl Acad Sci USA. 1998;95:632–637. doi: 10.1073/pnas.95.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Nature (London) 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 31.Vongs A, Kakutani T, Martienssen R, Richards E. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 32.Bartee L, Bender J. Nucleic Acids Res. 2001;29:2127–2134. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matzke M, Primig M, Trnovsky J, Matzke A J M. EMBO J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pélissier T, Thalmeir S, Kempe D, Sänger H L, Wassenegger M. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakowitsch J, Mette M F, van der Winden J, Matzke M, Matzke A J M. Proc Natl Acad Sci USA. 1999;96:13241–13246. doi: 10.1073/pnas.96.23.13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tax F E, Vernon D M. Plant Physiol. 2001;126:1527–1538. doi: 10.1104/pp.126.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakowitsch J, Papp I, Moscone E, van der Winden J, Matzke M, Matzke A J M. Plant J. 1999;17:131–140. doi: 10.1046/j.1365-313x.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- 39.Jones G H, Heslop Harrison J S. In: Arabidopsis: A Practical Approach. Wilson Z A, editor. Oxford: Oxford Univ. Press; 2000. pp. 105–124. [Google Scholar]
- 40.Mette M F, van der Winden J, Matzke M, Matzke A J M. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metz A M, Timmer R T, Browning K S. Gene. 1992;120:313–314. doi: 10.1016/0378-1119(92)90112-3. [DOI] [PubMed] [Google Scholar]
- 42.Park Y D, Papp I, Moscone E, Iglesias V, Vaucheret H, Matzke A J M, Matzke M. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- 43.Jedelloh J, Bender J, Richards E. Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vielle-Calzada J, Thomas J, Spillane C, Coluccio A, Hoeppner M, Grossniklaus U. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishimoto N, Sakai H, Jackson J, Jacobsen S, Meyerowitz E, Dennis E, Finnegan E J. Plant Mol Biol. 2001;46:171–183. doi: 10.1023/a:1010636222327. [DOI] [PubMed] [Google Scholar]
- 46.Bartee L, Malagnac F, Bender J. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindroth A, Cao X, Jackson J, Zilberman D, McCallum C, Henikoff S, Jacobsen S. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 48.Chandler V L, Eggleston W, Dorweiler J. Plant Mol Biol. 2000;43:121–145. doi: 10.1023/a:1006499808317. [DOI] [PubMed] [Google Scholar]
- 49.Matzke M, Mette M F, Jakowitsch J, Kanno T, Moscone E, van der Winden J, Matzke A J M. Genetics. 2001;158:451–461. doi: 10.1093/genetics/158.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steimer A, Amedeo P, Afsar K, Fransz P, Mittelsten Scheid O, Paszkowski J. Plant Cell. 2000;12:1165–1178. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith S S, Kann J L, Baker D J, Kaplan B E, Dembek P. J Mol Biol. 1991;217:39–54. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- 52.Habu Y, Kakutani T, Paszkowski J. Curr Opin Genet Dev. 2001;11:215–220. doi: 10.1016/s0959-437x(00)00182-9. [DOI] [PubMed] [Google Scholar]
- 53.Akhtar A, Zink D, Becker P B. Nature (London) 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 54.Cao X, Springer N, Muszynski M, Phillips R, Kaeppler S, Jacobsen S. Proc Natl Acad Sci USA. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bestor T H. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 56.Matzke M, Mette M F, Kanno T, Aufsatz W, Matzke A J M. In: Gene Silencing. Hannon G, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 2002. , in press. [Google Scholar]
- 57.Ramsahoye B, Biniszkiewicz D, Lyko F, Clark V, Bird A, Jaenisch R. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyko F. Trends Genet. 2001;17:169–172. doi: 10.1016/s0168-9525(01)02234-x. [DOI] [PubMed] [Google Scholar]
- 59.Pal-Bhadra M, Bhadra U, Birchler J. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 60.Dudley N, Labbé J, Goldstein B. Proc Natl Acad Sci USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitra A, An G. Mol Gen Genet. 1989;215:294–299. doi: 10.1007/BF00339731. [DOI] [PubMed] [Google Scholar]