Abstract
Glycosylphosphatidylinositol (GPI) acts as a membrane anchor of many cell surface proteins. Its structure and biosynthetic pathway are generally conserved among eukaryotic organisms, with a number of differences. In particular, mammalian and protozoan mannosyltransferases needed for addition of the first mannose (GPI-MT-I) have different substrate specificities and are targets of species- specific inhibitors of GPI biosynthesis. GPI-MT-I, however, has not been molecularly characterized. Characterization of GPI-MT-I would also help to clarify the topology of GPI biosynthesis. Here, we report a human cell line defective in GPI-MT-I and the gene responsible, PIG-M. PIG-M encodes a new type of mannosyltransferase of 423 amino acids, bearing multiple transmembrane domains. PIG-M has a functionally important DXD motif, a characteristic of many glycosyltransferases, within a domain facing the lumen of the endoplasmic reticulum (ER), indicating that transfer of the first mannose to GPI occurs on the lumenal side of the ER membrane.
Keywords: endoplasmic reticulum/mannosyltransferase/post-translational modification
Introduction
Glycosylphosphatidylinositol (GPI) is a glycolipid required for anchoring many eukaryotic cell surface proteins to the membrane (Tiede et al., 1999; McConville and Menon, 2000). In humans, >100 different proteins are GPI anchored. A lack of GPI biosynthesis in mice results in death due to abnormal embryogenesis (Nozaki et al., 1999) but, at the single cell level, GPI is not essential, i.e. many GPI-deficient mutant cell lines have been established (Hyman, 1988). GPI-anchored proteins are abundant in protozoan parasites, such as Trypanosoma brucei (McConville and Ferguson, 1993). The surface of the blood stream form of T.brucei is covered by 107 molecules of GPI-anchored variant surface glycoproteins (Ferguson, 1999). Biosynthesis of GPI is essential to this form of T.brucei (Nagamune et al., 2000).
The structure of the core of the GPI anchor, EtNP-6Manα1–2Manα1–6Manα1–4GlcNα1–6inositol-phospholipid (where EtNP, Man and GlcN are ethanolaminephosphate, mannose and glucosamine, respectively), is conserved in all eukaryotic cells (Kinoshita et al., 1997). The core is modified variously in different organisms. Biosynthesis of GPI basically involves the sequential addition of sugars and other components to phosphatidylinositol (PI). Pre-assembled GPI is transferred to proteins bearing the GPI attachment signal sequence at the C-terminus. The biosynthetic pathway for GPI is generally conserved in organisms, but a number of significant differences have been found (Ferguson, 1999).
Three mannoses in the GPI core are all derived from dolichol-phosphate-mannose (Dol-P-Man) and are linked through different bonds. Therefore, three Dol-P-Man-dependent mannosyltransferases, GPI-MT-I, -MT-II and -MT-III for the first, second and third mannoses, respectively, are required for generation of the GPI core. Mammalian PIG-B and its yeast ortholog Gpi10p are most probably GPI-MT-III. They are membrane proteins in the endoplasmic reticulum (ER) that are essential for transfer of the third mannose (Takahashi et al., 1996; Canivenc-Gansel et al., 1998; Sutterlin et al., 1998), and have amino acid sequence homology to three other Saccharomyces cerevisiae proteins, Alg9p, Smp3p and Alg12p. Alg9p (Burda et al., 1996) and Smp3p (Taron et al., 2000) are involved in the generation of Dol-P-Man-dependent α1–2 mannosyl linkages, whereas Alg12p generates Dol-P-Man-dependent α1–6 mannosyl linkages (Burda et al., 1999), suggesting that PIG-B and Gpi10p are members of a family of Dol-P-Man-dependent mannosyltransferases.
GPI-MT-I and -MT-II have not been molecularly characterized. Characterization of GPI-MT-I is important for two reasons. First, it is critical for determining the membrane orientation of GPI biosynthesis. The initial two reactions that generate GlcN-PI occur on the cytoplasmic side of the ER membrane, whereas the assembled GPI is transferred to proteins on the lumenal side (Kinoshita et al., 1997). Therefore, flipping of GPI should occur. Mammalian PIG-N and yeast Mcd4p, which are involved in a side chain modification of the first mannose, would transfer EtNP on the lumenal side immediately after the first mannosylation (Gaynor et al., 1999; Hong et al., 1999). So, flipping of GPI would occur before or after transfer of the first mannose. Determination of the membrane topology of the functional sites of GPI-MT-I thus is important. Secondly, the GPI intermediate that accepts the first mannose is different in mammalian cells and T.brucei. In T.brucei, the first mannose is transferred to GlcN-PI, whereas in mammalian cells it is transferred to GlcN-acyl-PI, which has an acyl chain on the inositol ring (Smith et al., 1997). Ferguson’s group demonstrated, using a cell-free system, that some synthetic analogs of GlcN-PI inhibit first mannosylation in T.brucei but not in mammalian cells, implying that GPI-MT-I is a good target of African trypanosomiasis chemotherapy (Smith et al., 1999).
Here, we report the molecular cloning of human PIG-M, which encodes GPI-MT-I, and evidence that the first mannosylation occurs on the lumenal side of the ER. We also show that T.brucei has a PIG-M homolog with a conserved DXD motif, suggesting that the first mannosyl ation also occurs on the lumenal side in T.brucei.
Results
Ramos517 represents a new complementation group, class M, of GPI anchor-deficient mutants
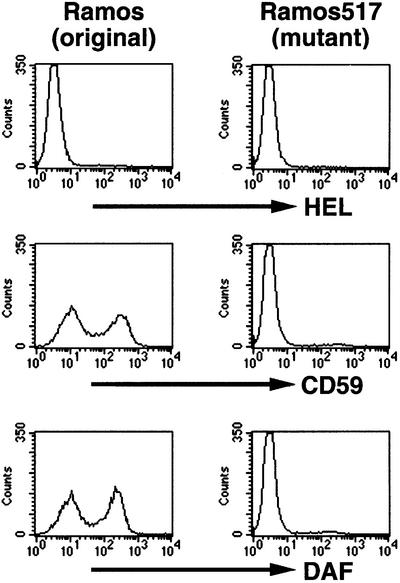
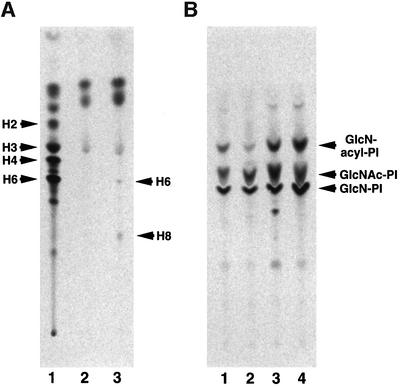
The culture of Ramos cells obtained from the European Type Culture Collection consisted of CD59/decay accelerating factor (DAF)-positive and -negative populations (Figure 1, left panels). The CD59/DAF-negative cells were isolated by a cell sorter and termed Ramos517 (right panels). Since the surface expressions of two GPI-anchored proteins were defective on Ramos517 cells, we tested for biosynthesis of the GPI anchor by metabolic labeling with mannose. Ramos517 cells did not synthesize H8, a mature form of GPI (Hirose et al., 1992), and an intermediate, H6 (Figure 2A, lane 2), whereas original Ramos cells synthesized both mannolipids (lane 3). It appeared, therefore, that Ramos517 cells are GPI deficient.
Fig. 1. FACS analysis showing the defective surface expression of GPI-anchored proteins on mutant Ramos517 cells. Original Ramos (left panels) and mutant Ramos517 (right panels) cells were stained with isotype-matched control anti-hen egg lysozyme (HEL) (upper panels), anti-CD59 (middle panels) and anti-DAF (lower panels) antibodies.
Fig. 2. Defective GPI anchor biosynthesis in Ramos517 cells. (A) In vivo labeling with [3H]mannose. Class F mutant EL4 (lane 1), mutant Ramos517 (lane 2) and original Ramos (lane 3) cells were cultured with d-[2-3H]mannose in the presence of tunicamycin. Mannolipids were analyzed by TLC with a solvent system of chloroform:methanol:H2O (10:10:3) and an image analyzer. H2, H3 and H4, GlcN-acyl-PI bearing one, two and three mannoses; H6, GlcN-acyl-PI bearing three mannoses with ethanolaminephosphate on the first mannose; H8, a mature form of GPI. (B) In vitro analysis of the first three steps. Membranes of wild-type Ramos (lane 1), mutant Ramos517#17 (lane 2), wild-type BW5147 (lane 3) and class E mutant (lane 4) cells were incubated with UDP-[6-3H]GlcNAc. The radiolabeled lipids were analyzed by TLC with a solvent system of chloroform:methanol:1 M NH4OH (10:10:3).
We fused Ramos517 cells with other GPI-deficient cells of known complementation groups and assessed restoration of the surface expression of GPI-anchored proteins. Ramos517 complemented all known mutant cells, indicating that it represents a new complementation group (data not shown). We assigned it class M and termed the gene responsible PIG-M.
We obtained a clone of Ramos517, termed Ramos517#17, by limiting dilution and used it in subsequent experiments. Ramos517#17 had almost no non-specific staining upon flow cytometric analysis, which is critical for expression cloning of PIG-M.
Ramos517 cells are defective in transfer of the first mannose in GPI biosynthesis
To determine which step in GPI biosynthesis is defective in Ramos517 cells, we first tested early reaction steps. We incubated cell lysates with a radiolabeled donor of GlcNAc, UDP-GlcNAc, and assessed the generation of the first three intermediates, GlcNAc-PI, GlcN-PI and GlcN-acyl-PI (Figure 2B). Lysates of mouse lymphoma BW5147 (lane 3) and its Dol-P-Man synthase-deficient class E mutant (lane 4) (used as references) generated the three GPI intermediates (Urakaze et al., 1992). Lysates of original Ramos (lane 1) and mutant Ramos517#17 (lane 2) cells also generated all three intermediates, indicating that the Ramos517 mutant has enzymes for the first three steps. The amount of GlcN-acyl-PI generated by lysates of Ramos517#17 was smaller than that generated by lysates of the original Ramos (lanes 1 versus 2). The reason for this is unclear.
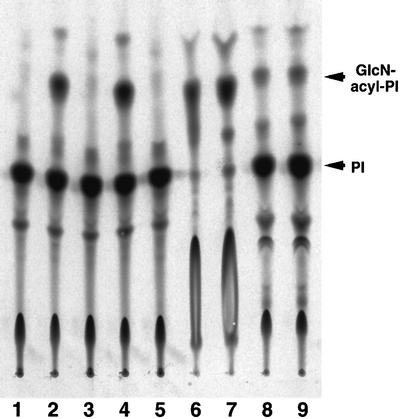
Next, we tested later reaction steps by metabolically labeling the cells with radioactive inositol. To obtain a pure population of wild-type Ramos cells for comparison with the mutant, we separated the CD59/DAF-positive cell population using a cell sorter, but found the phenotype to be unstable, producing CD59/DAF-negative cells spontaneously. Therefore, as a source of wild-type Ramos cells, we generated cells expressing rat PIG-M cDNA, termed RamosR#19 cells, which maintained the surface CD59/DAF expression stably (see below for cloning of PIG-M). RamosR#19, mutant Ramos517#17 and PIG-M-transfected Ramos517#17 cells, as well as wild-type lymphoma BW5147 and its class E mutant, were cultured in a medium containing radioactive inositol, and the inositol-containing lipids generated were analyzed (Figure 3). As reported previously (Urakaze et al., 1992), the class E mutant (lane 2) accumulated GlcN-acyl-PI due to a lack of mannosyl donor Dol-P-Man whereas parental BW5147 cells did not (lane 1). Like class E cells, Ramos517 mutant (lane 4) but not wild-type RamosR#19 (lane 3) cells accumulated GlcN-acyl-PI. The accumulation of GlcN-acyl-PI in the mutant was eliminated by transfection of PIG-M cDNA (lane 5). The identity of GlcN-acyl-PI was confirmed by its resistance to PI-specific phospholipase C (PI-PLC) (lanes 6 and 7) and its sensitivity to GPI-specific phospholipase D (GPI-PLD) (lanes 8 and 9). These results indicate that mannosylation of GlcN-acyl-PI is defective in class M Ramos517 cells.
Fig. 3. Accumulation of GlcN-acyl-PI in Ramos517 cells revealed by in vivo labeling with inositol. Wild-type BW5147 (lane 1), class E mutant (lanes 2, 6 and 8), wild-type RamosR#19 (lane 3), Ramos517V#17 mutant (lanes 4, 7 and 9) and PIG-M-transfected Ramos517#17 (Ramos517R#4) (lane 5) cells were cultured in a medium containing myo-[2-3H(N)]inositol. Labeled lipids were treated either with PI-PLC (lanes 6 and 7) or GPI-PLD (lanes 8 and 9), extracted again and analyzed by TLC with a solvent system of chloroform:methanol:0.25% KCl (55:45:10).
To confirm the defective mannosylation, we used a synthetic substrate, GlcN-PI(C8), i.e. GlcN-PI bearing dioctanoyl-PI. It was reported that GlcN-PI(C8) was acylated by an enzyme that generates GlcN-acyl-PI from GlcN-PI upon incubation with membranes of mammalian cells (Doerrler et al., 1996). Further, when the membrane contained radioactive Dol-P-Man, the GlcN-acyl-PI(C8) generated was in turn mannosylated to become radiolabeled Man-GlcN-acyl-PI(C8). We incubated membranes of wild-type RamosR#19 and mutant Ramos517#17 cells with GDP-[3H]mannose and GlcN-PI(C8) (Figure 4). We also used Lec35 cells as a reference. It was reported that GlcN-acyl-PI accumulates in Lec35 cells due to a defect in proper compartmentalization of Dol-P-Man, and is mannosylated efficiently upon disruption of the cells and incubation with GDP-[3H]mannose (Doerrler et al., 1996). Radiolabeled Dol-P-Man was generated from GDP-[3H]mannose and endogenous dolichol-phosphate by all membranes (lanes 1–7). Membranes of the Lec35 mutant generated Man-GlcN-acyl-PI using endogeneously accumulated GlcN-acyl-PI and radiolabeled Dol-P-Man generated in situ (lane 3). When GlcN-PI(C8) was also included, it was converted to Man-GlcN-acyl-PI(C8) (lane 5). Membranes of wild-type Ramos cells did not generate Man-GlcN-acyl-PI because wild-type cells do not accumulate GlcN-acyl-PI (lane 2) (Hong et al., 1999). When GlcN-PI(C8) was included, Man-GlcN-acyl-PI(C8) was generated (lane 6), showing GPI-MT-I activity. In contrast, membranes of mutant Ramos517 cells did not generate Man-GlcN-acyl-PI(C8) (lanes 4 and 7).
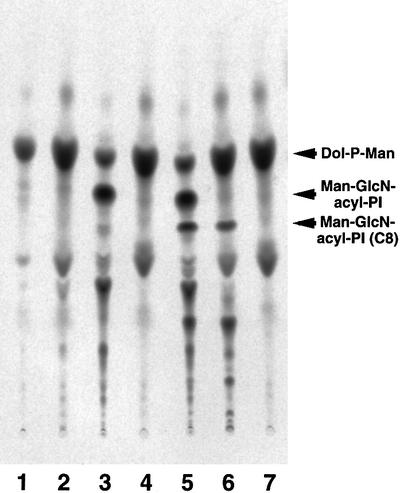
Fig. 4. Ramos517 cells are defective in transfer of the first mannose as revealed by using a synthetic substrate GlcN-PI(C8). The membranes of wild-type CHO (lane 1), Lec35 mutant (lane 3 and 5), wild-type RamosR#19 (lanes 2 and 6) and Ramos517V#17 mutant (lanes 4 and 7) cells were incubated with GDP-[3H]mannose and palmitoyl-CoA in the presence (lanes 5–7) or absence (lanes 1–4) of GlcN-PI(C8). After 1 h incubation, lipids were extracted with n-butanol and analyzed by TLC with a solvent system of chloroform:methanol:0.25% KCl (55:45:10).
Mannosylation of GlcN-acyl-PI can be impaired in a number of ways: (i) a lack of Dol-P-Man, as in class E cells; (ii) an incorrect orientation of Dol-P-Man, as in Lec35 cells (Camp et al., 1993); (iii) a lack of GPI-MT-I; and (iv) an improperly oriented GlcN-acyl-PI. The first possibility was excluded because Dol-P-Man was synthesized in Ramos517 cells (Figure 4). If the second possibility is true, biosynthesis of N-glycan precursors should also be affected, as seen in the Lec35 mutant (Camp et al., 1993). We assessed the surface expression of complex-type oligosaccharides and its sensitivity to swainsonine as a measure of the proper biosynthesis of N-glycan in the ER. The surface expression of complex-type oligosaccharides as measured by binding of phytohemagglutinin-E4 on Ramos517 mutant was fully sensitive to swainsonine, like the wild-type Ramos (data not shown). Therefore, N-glycan presursors were synthesized properly in Ramos517 cells so that trimming by swainsonine-sensitive α-mannosidase II was necessary for the generation of complex-type oligosaccharides. This result indicates that mannoses from Dol-P-Man were used normally, hence the second possibility is not true. Therefore, Ramos517 cells are defective in either GPI-MT-I itself or the orientation of the acceptor substrate GlcN-acyl-PI.
Cloning rat and human PIG-M cDNAs
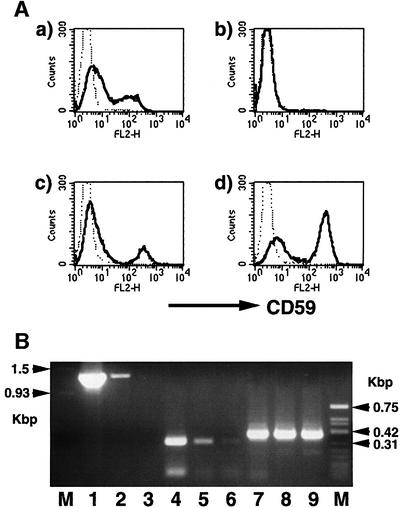
We isolated a rat cDNA that restored the surface expression of GPI-anchored proteins on Ramos517 mutant cells by means of expression cloning. This cDNA consisted of 3700 bp and contained an open reading frame spanning nucleotides 87–1358, which encode 423 amino acids. A sequence around the ATG codon at nucleotides 87–89 was compatible with the Kozak consensus sequence for translational initiation. At nucleotides 15–18 there was an in-frame upstream stop codon TGA. Based on the predicted amino acid sequence, in databases we identified sequences of a human homolog that also encodes 423 amino acids and has 91% identity to the rat homolog. Both rat and human cDNAs restored the surface expression of CD59 on Ramos517 cells (Figure 5A).
Fig. 5. (A) Restoration of the surface expression of GPI-anchored proteins on mutant Ramos cells with PIG-M cDNA. Original Ramos (a), Ramos517#17 transfected with a control vector (b), Ramos517#17 transfected with rat PIG-M cDNA (c) and Ramos517#17 transfected with human PIG-M cDNA (d) were analyzed 2 days after transfection. Solid lines, anti-CD59; dotted lines, isotype-matched control antibody. (B) Decreased PIG-M mRNA expression in mutant Ramos517#17 cells. RNA from JY25 (lanes 1, 4 and 7), wild-type Ramos (lanes 2, 5 and 8) and mutant Ramos517#17 (lanes 3, 6 and 9) cells were examined by RT–PCR using two sets of primers for human PIG-M (lanes 1–3 and 4–6) and primers for human GPI1 (lanes 7–9) as a control. M, molecular size markers.
Using primers designed from the sequence of the human cDNA, mRNA in Ramos517 cells was analyzed by RT–PCR (Figure 5B). No cDNA spanning the full coding region was amplified (lane 3) but a cDNA spanning a shorter region in the coding region was amplified weakly (lane 6) from samples of Ramos517 cells, strongly suggesting that this cDNA corresponds to the gene defective in this mutant, namely PIG-M. The DDBJ/EMBL/GenBank accession Nos of rat and human PIG-M cDNAs are AB028127 and AB028128, respectively.
Characteristics of PIG-M proteins
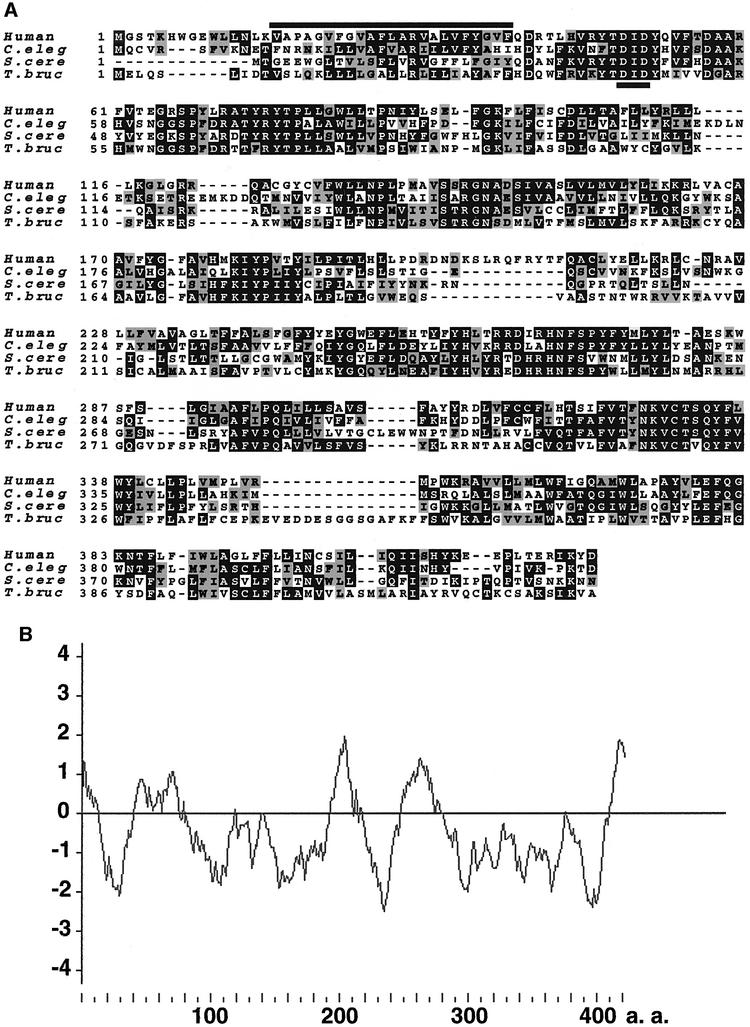
We found yeast and nematode PIG-M homologs in the databases (accession Nos Z49513 and Z49907, respectively). Caenorhabditis elegans PIG-M and S.cerevisiae PIG-M are of 417 and 403 amino acids, respectively, and have 38 and 35% amino acid identity to human PIG-M, respectively (Figure 6A). We also found a fragment of T.brucei PIG-M (TbPIG-M) in a database, and cloned its full-length cDNA (DDBJ/EMBL/GenBank accession No. AB050105). TbPIG-M consists of 430 amino acids and has 30% identity to human PIG-M (Figure 6A). Mammalian PIG-M had no typical ER retention signal whereas S.cerevisiae and C.elegans PIG-Ms had KKXX and KXKXX sequences, respectively, at the C-terminus (Jackson et al., 1990), consistent with their roles in GPI biosynthesis in the ER (Figure 6A). The hydropathy profile of human PIG-M shows multiple hydrophobic regions (Figure 6B). Analysis with the TMpred program (Smith et al., 1996) suggested the presence of 10 transmembrane domains.
Fig. 6. (A) Alignment of human, C.elegans, S.cerevisiae and T.brucei PIG-M amino acid sequences. Black and gray boxes indicate identical and conserved amino acids, respectively. Overline, the first transmembrane domain; underline, a DXD motif. (B) Hydropathy profile of human PIG-M. The plot was drawn using the Kyte–Doolittle method of calculating hydrophilicity over a window length of 17 (Kyte and Doolittle, 1982).
The PIG-M proteins from various organisms all had a DXD motif (Figure 6A, underlined) within a hydrophilic region C-terminal to the first predicted transmembrane domain (overlined). The DXD motif found in many glycosyltransferases is involved in binding a manganese ion that plays a role in binding a nucleotide sugar substrate (Wiggins and Munro, 1998; Gastinel et al., 1999).
PIG-M is GPI-MT-I
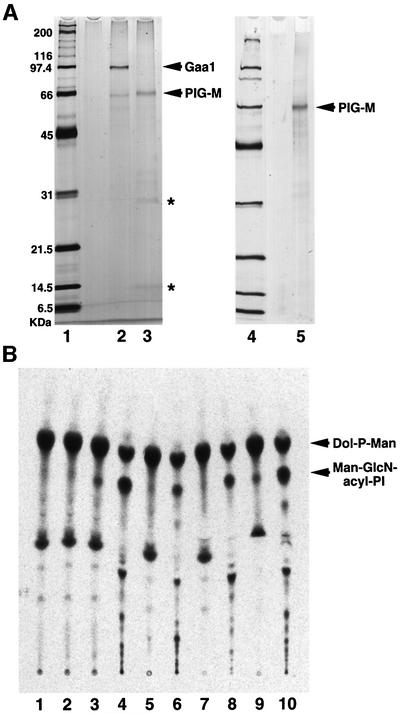
To test whether PIG-M is GPI-MT-I, we isolated tandem-tagged PIG-M from CHO cells transfected with a cDNA of PIG-M tagged with GST and FLAG. As a control, Gaa1 bearing the same tags was affinity purified similarly from CHO cell transfectants (Figure 7A). When proteins were eluted from the second affinity beads, glutathione beads, with an SDS–PAGE sample buffer, a major band of tagged PIG-M was seen with several other minor bands at positions of 32–35, 28 and 14 kDa (lane 3). When proteins were eluted specifically with glutathione, the bands at 28 and 14 kDa (indicated by asterisks in lane 3) were not seen (lane 5), suggesting that they associated with the beads non-specifically.
Fig. 7. The PIG-M gene encodes GPI-MT-I. (A) GST-FLAG-tagged-PIG-M (lanes 3 and 5) and -Gaa1 (lane 2) were isolated by two-step affinity purification from the lysates of CHO cells transiently transfected with their expression plasmids. Molecular weight markers are in lanes 1 and 4. Two different preparations of PIG-M proteins are shown in lanes 3 and 5 (see Results). Asterisks, non-specific bands. (B) The purified GST-FLAG-tagged-PIG-M (lanes 3, 5, 7 and 9) and -Gaa1 (lane 2) proteins were incubated with the substrate lipids. Lipids were extracted again with n-butanol and analyzed by TLC, as described in the legend to Figure 4. The lipids without incubation with proteins are shown in lane 1. The lipids obtained from Lec35 membranes were used as controls (lanes 4, 6, 8 and 10). Lipids were treated with GPI-PLD (lanes 5 and 6), Jack bean α-mannosidase (lanes 7 and 8) and PI-PLC (lanes 9 and 10), extracted again, and analyzed by TLC.
We measured GPI-MT-I activity of PIG-M using proteins still bound to the glutathione beads (Figure 7B) (we did not use the eluted proteins because reduced glutathione inactivated GPI-MT-I). As the substrates, we used lipids extracted from Ramos517 membranes that contained GlcN-acyl-PI and Dol-P-[3H]Man (lane 1). A spot with the same mobility as the authentic Man-GlcN-acyl-PI (generated by membranes of Lec35 cells) (lane 4) appeared after incubation with PIG-M (lane 3) but not Gaa1 (lane 2). Like Man-GlcN-acyl-PI from Lec35 cells, this mannolipid was sensitive to GPI-PLD (lanes 5 and 6) and Jack bean α-mannosidase (lanes 7 and 8) but not to PI-PLC (lanes 9 and 10), indicating that it is Man-GlcN-acyl-PI. PIG-M is, therefore, GPI-MT-I, and Ramos517 cells are defective in GPI-MT-I.
A DXD motif of PIG-M is functionally important
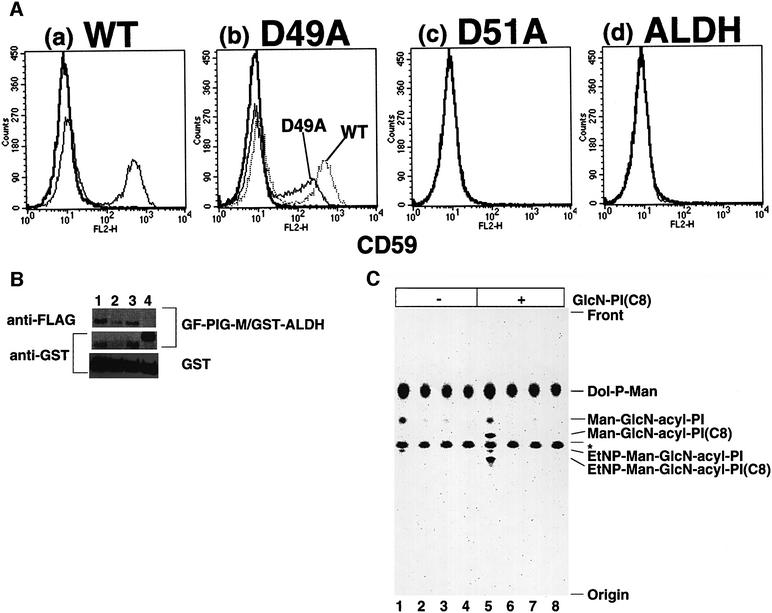
To determine whether a DXD motif found in PIG-M is essential for mannosyltransferase activity, we mutagenized each of the aspartic acid residues to alanine and assessed the abilities of mutants to restore Ramos517 cells (Figure 8). A D51A mutant, having alanine in place of the second aspartic acid at position 51, did not restore the surface expression of CD59 on the transiently transfected Ramos517 cells (Figure 8A,c) like a negative control ALDH (A,d), whereas wild-type PIG-M restored the CD59 expression on ∼30% of cells (A,a). The western blot analysis with anti-tag antibodies (Figure 8B) showed that D51A mutant protein (lane 3) was expressed at a level similar to that of wild-type PIG-M (lane 1), indicating that the D51A mutant lacks GPI-MT-I activity. This was confirmed by an in vitro enzyme assay using microsomes prepared from the same set of cells (Figure 8C). Microsomes from Ramos517 cells transfected with the D51A mutant generated only a trace amount of Man-GlcN-acyl-PI (lane 3), whereas those from wild-type PIG-M transfectants generated Man-GlcN-acyl-PI and its further processed form EtNP-Man-GlcN-acyl-PI (lane 1). Microsomes from wild-type PIG-M transfectants (lane 5) but not D51A mutant transfectants (lane 7) processed exogenously added substrate analog GlcN-PI(C8). Therefore, the second aspartic acid within the DXD motif is critical.
Fig. 8. Importance of a DXD motif in PIG-M assessed by site-directed mutagenesis. (A) FACS analysis of Ramos517 cells transfected with GF-tagged, wild-type PIG-M (a), D49A mutant PIG-M (b), D51A mutant PIG-M (c) and GST-tagged ALDH (d). Cells were stained by anti-CD59 (thin lines) and control (thick lines) antibodies. The dotted line in (b) indicates CD59 staining of the same wild-type PIG-M transfectant shown in (a). (B) Assessment of levels of PIG-M proteins. Ramos517 cells co-transfected with free GST, as a control, plus either GF-tagged, wild-type PIG-M (lane 1), D49A mutant (lane 2), D51A mutant (lane 3) or GST-tagged ALDH (lane 4) were analyzed by western blotting against anti-FLAG (top) and anti-GST (middle and bottom) antibodies. (C) In vitro GPI-MT-I assay. Microsomes of Ramos517 cells transfected with GF-tagged, wild-type PIG-M (lanes 1 and 5), D49A mutant PIG-M (lanes 2 and 6), D51A mutant PIG-M (lanes 3 and 7) and GST-tagged ALDH (lanes 4 and 8) were incubated with GDP-[3H]mannose in the absence (lanes 1–4) or presence (lanes 5–8) of GlcN-PI(C8). Labeled lipids were analyzed by TLC as described in the legend to Figure 4. Asterisk, an unknown mannolipid.
The D49A mutant, having an alanine at position 49, restored subnormal levels of CD59 on Ramos517 cells (Figure 8A,b). Mean relative fluorescence intensities of CD59-positive cells were 20 and 80 for D49A and wild-type PIG-M transfectants, respectively. The expression of D49A PIG-M protein (Figure 8B, lane 2), however, was only 30% of that of wild-type PIG-M (lane 1). Restoration of the surface expression of GPI-anchored protein is a sensitive but not linear measure of the enzyme activity, because the expression of CD59 restored by transfection of wild-type PIG-M cDNA was most probably saturated. The function of D49A was then assessed by in vitro enzyme assay (Figure 8C). The D49A mutant had very little activity to generate Man-GlcN-acyl-PI (lane 2) and no activity to process the substrate analog (lane 6), indicating that the first aspartic acid of the DXD motif is also important for PIG-M.
Cytoplasmic orientation of the N-terminus of PIG-M indicates the lumenal orientation of the DXD motif
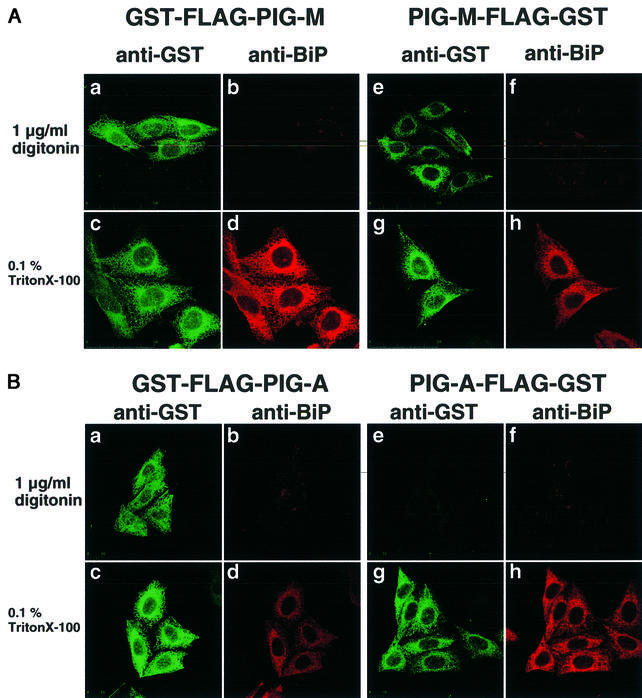
We then determined the membrane orientation of the N- and C-termini of PIG-M. The DXD motif exists within a hydrophilic region between the first and second transmembrane domains (Figure 6A), so it should be oriented on the opposite side to the N-terminus. PIG-M tagged at either the N- or the C-terminus was transfected into CHO cells (Figure 9A). After selective permeabilization of only the plasma membrane by digitonin, or permeabilization of both the plasma and ER membranes by Triton X-100, cells were stained for GST tag (green) and an authentic ER lumenal protein BiP (red). To confirm the reliability of the method, we used similarly tagged PIG-A (Figure 9B), for which orientations of the N- and C-termini were established (Watanabe et al., 1996).
Fig. 9. Membrane orientation of the N- and C-termini of PIG-M. (A) Orientation of PIG-M. Human PIG-M tagged at the N-terminus (GST-FLAG-PIG-M) (a–d) or the C-terminus (PIG-M-FLAG-GST) (e–h) was expressed in CHO cells. After treatment with 1 µg/ml digitonin for selective permeabilization of the plasma membrane (a, b, e and f), or with 1% Triton X-100 for permeabilization of both the plasma membrane and the ER membrane (c, d, g and h), cells were stained for PIG-M by anti-GST antibody (a, c, e and g) and for endogenous BiP, an ER lumenal protein (b, d, f and h). (B) Orientation of PIG-A, an ER membrane protein with known orientation. Human PIG-A was tagged and expressed similarly to PIG-M shown in (A). The cells were also permeabilized and stained similarly.
The GST tags at the N- and C-termini of PIG-M were both accessed by antibody after permeabilization of only the plasma membrane (Figure 9A,a and e). Further permeabilization of the ER membrane did not increase their staining intensity significantly (A,c and g). BiP was stained only after permeabilization of the ER membrane (A,d and h, versus b and f) as expected. The N-terminus of PIG-A, which is on the cytoplasmic side of the ER, was accessed by antibody after permeabilization of the plasma membrane only (Figure 9B,a). The C-terminus of PIG-A, which is on the lumenal side, was accessed by antibody only after permeabilization of the ER membrane (Figure 9B,e and g), indicating that the method was appropriate for determination of the membrane orientation of ER proteins. Therefore, it is shown that both the N- and C-termini of PIG-M reside on the cytoplasmic side of the ER. We concluded that the DXD motif of PIG-M is oriented to the lumenal side of the ER. Taking this together with the above result that the DXD motif is important for PIG-M activity, we also concluded that the transfer of the first mannose occurs on the lumenal side.
Discussion
PIG-M encodes GPI-MT-I
In the present study, we isolated a GPI-anchor-deficient cell line Ramos517 and demonstrated that it is defective in transfer of the first mannose. Using Ramos517 cells as recipients of a cDNA library in expression cloning, we obtained rat PIG-M cDNA that restored GPI anchor biosynthesis in Ramos517 cells. Based on sequence homology, we cloned human PIG-M cDNA and demonstrated that PIG-M mRNA is defective in Ramos517 cells. Human PIG-M is a multimembrane-spanning ER protein consisting of 423 amino acids. The epitope-tagged PIG-M, affinity-purified from transfected human cells, had GPI-MT-I activity in vitro. We, therefore, identified previously uncharacterized GPI-MT-I.
PIG-M has no significant amino acid sequence similarity to PIG-B, which is most probably GPI-MT-III. Although GPI-MT-I and -MT-III use Dol-P-Man, GPI-MT-I generates an α1–4 mannosyl linkage to GlcN whereas GPI-MT-III generates an α1–2 mannosyl linkage to the second mannose. PIG-B belongs to a family of Dol-P-Man-dependent mannosyltransferases. PIG-M has no homology to these family proteins and other glycosyltransferases (except for a DXD motif), therefore, it represents a new type of mannosyltransferase.
PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars (Wiggins and Munro, 1998). It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. It is also possible that the motif is required for glycosyltransferases in other ways (Munro and Freeman, 2000). PIG-M is the first glycosyltransferase bearing a DXD motif that uses a lipid-linked sugar donor rather than a nucleotide sugar. The DXD motif in PIG-M is functionally important because alanine mutations of either aspartic acid residue resulted in a loss of GPI-MT-I activity; however, how it functions is unclear at present.
Membrane orientation of the first mannose transfer and other reactions in GPI biosynthesis
Because the DXD motif is important for GPI-MT-I activity of PIG-M, it is suggested that transfer of the first mannose occurs on the side of the ER membrane where the DXD motif is present. Since the DXD motif is located immediately to the C-terminal side of the first transmembrane domain, we determined the membrane orientation of the N-terminus of PIG-M in order to determine the membrane orientation of the DXD motif. The N-terminus was on the cytoplasmic side, indicating that the DXD motif is on the lumenal side. We concluded that transfer of the first mannose occurs on the lumenal side.
Other known mannose transfers from Dol-P-Man also occur, or are very likely to occur on the lumenal side, i.e. addition of the last four mannosyl residues to the lipid-linked oligosaccharide for N-glycosylation; O-mannosylation of serines and threonines in many yeast proteins (Herscovics and Orlean, 1993); C-mannosylation of specific tryptophans in some proteins (Doucey et al., 1998); and transfer of the third mannose to GPI (Takahashi et al., 1996).
The first two reactions in the pathway of GPI biosynthesis occur on the cytoplasmic side. This was established by determining the orientation of the first two GPI intermediates, GlcNAc-PI and GlcN-PI (Vidugiriene and Menon, 1993), and the first two enzymes in the biosynthesis, GPI-GlcNAc transferase (Watanabe et al., 1996) and GlcNAc-PI deacetylase (Nakamura et al., 1997). Because the first mannose is transferred on the lumenal side, the acceptor substrate of GPI-MT-I, GlcN-acyl-PI in mammalian cells, should be oriented towards the lumen. Therefore, either GlcN-PI or GlcN-acyl-PI translocates to the lumenal side. It was reported that in mouse T-lymphoma cells, >70% of GlcN-PI was accessible to PI-PLC acting from the cytoplasm (Vidugiriene and Menon, 1993). If PI-PLC is in fact membrane impermeable, this means that the large majority of GlcN-PI is oriented towards the cytoplasm. Translocation of GlcN-PI must be a slow process if it occurs.
After transfer of the first mannose, the later reactions in GPI biosynthesis and attachment of GPI to proteins would proceed on the lumenal side. PIG-N and PIG-O, involved in transfers of EtNP to the first and third mannose, respectively, have functionally important conserved motifs in their lumenal domains (Hong et al., 1999, 2000). PIG-B also transfers the third mannose on the lumenal side. GPI8, a catalytic subunit of GPI-transamidase, is also lumenally oriented (Ohishi et al., 2000). In summary, GPI biosynthesis in mammalian cells is initiated on the cytoplasmic side of the ER, changes orientation to the lumenal side immediately before or after generation of GlcN-acyl-PI, and proceeds on the lumenal side all the way through to the transfer to proteins.
PIG-M from T.brucei
The first mannose is transferred to GlcN-PI, rather than to GlcN-acyl-PI, in T.brucei. The basis of the different structures of acceptors for the first mannose in mammalian cells and T.brucei could be either that the two GPI-MT-Is are very different or that they are similar but have different substrate specificity. If the former is true, it would be possible that even the membrane orientation is different. The present finding that T.brucei has a PIG-M homolog that has 30% overall amino acid identity and several well-conserved regions throughout the molecule supports the latter case. TbPIG-M has a conserved DXD motif that is predicted to be lumenally oriented, suggesting that the first mannose is also transferred on the lumenal side in T.brucei. In conclusion, it is likely that mammalian PIG-M and TbPIG-M have different substrate specificities. This point should be determined experimentally when TbPIG-M is expressed as a functional form in a sufficient amount.
Materials and methods
Cells and culture
Wild-type Ramos and Ramos517 cells were cultured in RPMI 1640 medium supplemented with 1 mM pyruvate and 10% fetal calf serum (FCS). CHO-K1 and Lec35.2 mutants (gifts from Dr M.A.Lehrman, Texas Southwestern Medical Center, Dallas, TX) (Camp et al., 1993) were cultured in Ham’s F-12 medium supplemented with 10% FCS. The mouse lymphoma BW5147, Thy-1-negative mutant cells of class E and class F (gifts from Dr R.Hyman, Salk Institute, San Diego, CA) (Hyman, 1988) and human B-lymphoblastoid JY25 cells (Miyata et al., 1993) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. Ramos517V#17, a control transfected line, was established from Ramos517#17, a clone of Ramos517, by transfection of pME-Hyg. Ramos517R#4 was established from Ramos517#17 by transfection of pME-Hyg-rPIG-M, followed by limiting dilution. RamosR#19 was established from original Ramos by transfection of pME-Hyg-rPIG-M followed by limiting dilution.
Plasmids
To construct pME-Hyg, a hygromycin resistance gene excised from pCDNA3.1/Hygro (Invitrogen) was blunt-ligated into the HindIII site of pME18Sf+ (a gift from Dr K.Maruyama, University of Tokyo, Japan). Rat and human PIG-Ms were subcloned into pME-Py (Maeda et al., 1998) and pME-Hyg, and the resulting plasmids were termed pME-Py-rPIG-M, pME-Py-hPIG-M, pME-Hyg-rPIG-M and pME-Hyg-hPIG-M. pMEEB-GF-hPIG-M, pMEEB-GF-hGaa1 and pMEEB-GF-PIG-A were constructed by replacing DPM1 cDNA of pMEEB-GST-FLAG-hDPM1 (Maeda et al., 2000) with human PIG-M, GAA1 (Ohishi et al., 2000) and PIG-A (Watanabe et al., 1996) cDNAs, respectively. pMEEB-PIG-M-FLAG-GST and pMEEB-PIG-A-FLAG-GST were prepared by subcloning XhoI–XbaI fragments bearing PIG-M-FLAG-GST and PIG-A-FLAG-GST, respectively (derived from pBS-PIG-M-FLAG-GST and pBS-PIG-A-FLAG-GST), into pMEEB. pBS-PIG-M-FLAG-GST and pBS-PIG-A-FLAG-GST were generated by inserting XhoI–MluI fragments bearing PIG-M and PIG-A, respectively, into pBS-FLAG-GST cut with XhoI and MluI.
Transfection
Ramos, Ramos517 and their transformants (107 cells in 0.4 ml of HEPES-buffered saline; Miyata et al., 1993) were electroporated at 250 V and 960 µF with 20–25 µg of DNA. CHO cells (107) suspended in 0.4 ml of culture medium with 25 µg of plasmid were electroporated at 260 V and 960 µF. Electroporations were done in a Gene Pulser (Bio-Rad).
Fluorescence staining and FACS analysis
Cells were stained for DAF and CD59 with biotinylated anti-DAF and anti-CD59 monoclonal antibodies followed by phycoerythrin-conjugated streptoavidin (Biomeda) (Hong et al., 1999). To evaluate the biosynthesis of N-glycan, cells were cultured with or without 5 µg/ml swainsonine (Wako Chemicals, Japan) for 4 days and then stained with 25 µg/ml fluorescein isothiocyanate (FITC)-conjugated phytohemagglutinin-E4 (Seikagaku Co., Japan). Stained cells were analyzed in a FACScan cytometer (Becton Dickinson).
Cloning PIG-M cDNA
Ramos517#17 cells (1.6 × 108) were mixed with 200 µg each of the plasmids of the rat glioma cDNA library (Nakamura et al., 1997) and pBSII-SV40T(ori–) in 6.4 ml of HEPES-buffered saline, and were electroporated in 16 cuvettes. Two days later, transfected cells were stained with anti-CD59 antibody, and 2700 cells with restored surface CD59 expression were collected by a cell sorter (FACS-Vantage, Becton Dickinson). Plasmid clones (1.2 × 104) were recovered from the cells. Pooled plasmids (32 µg) were retransfected with 160 µg of pBSII-SV40T(ori–) plasmid into 8 × 107 Ramos517#17 cells in eight cuvettes. After another cycle of cell sorting and recovery, 1152 independent plasmid clones were analyzed and two positive clones were obtained. Both clones had the same restriction pattern and length (4 kbp) of cDNA. We sequenced one of them. Based on this rat sequence, we found two clones of a human homolog in the expressed sequence tag (EST) database. One was a clone with DDBJ/EMBL/GenBank accession No. Z45029, which contained the 5′-untranslated and coding regions, and the other was a clone with accession No. R39406, which contained the 3′-coding and untranslated regions. We amplified a missing portion of the coding region from a human cDNA library by PCR using hMF1 (5′-CACCGTCGACATGGGCTCCACCAAGCACT) and hMR1 primers (5′-CACAAGAGGCAGTAAGCAGAGGTAC), and determined the sequence of human PIG-M.
Metabolic labeling of GPI intermediates
Cells were labeled with d-[2-3H]mannose (Takahashi et al., 1996) or myo-[2-3H(N)]inositol. For the latter, 106 cells were washed with inositol-free DMEM (Gibco-BRL) and then cultured in the same medium supplemented with 10% dialyzed FCS and 20 µCi of myo-[2-3H(N)]inositol (American Radiolabeled Chemicals) for 1 day. Lipids in the radiolabeled cells were extracted twice with 0.3 ml of water-saturated n-butanol. Pooled butanol extracts were backwashed with 0.3 ml of butanol-saturated water and then evaporated. The dried materials were extracted with 30 µl of chloroform:methanol (2:1) and separated by thin layer chromatography (TLC) on Kieselgel 60 (Merck). The radiolabeled lipids were analyzed by an Image Analyzer BAS 1500 (Fuji Film Co., Tokyo) after 2–4 days exposure.
In vitro labeling of GPI intermediates
Cells incubated with 5 µg/ml tunicamycin (Sigma) for 2 h were suspended in a hypotonic buffer [20 mM Tris–HCl pH 7.4, 2 µg/ml leupeptin and 0.1 mM 1-chloro-3-tosylamido-7-l-2-heptanone (TLCK)] on ice for 20 min, before being destroyed with a Teflon homogenizer. After removal of cell debris and nuclei by centrifugation at 10 000 g for 10 min, membranes were collected by centrifugation at 100 000 g for 1 h, suspended in a buffer (50 mM HEPES–NaOH pH 7.4, 25 mM KCl, 2 µg/ml leupeptin and 0.1 mM TLCK) and stored at –80°C.
The membranes obtained from Ramos and its derivative cells (2 × 107), and CHO and Lec35 cells (3.5 × 106) were incubated in 0.15 ml of a buffer containing 2 µCi of GDP-[3H]mannose (American Radiolabeled Chemicals), 50 mM HEPES–NaOH pH 7.4, 25 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 1 mM 5′AMP, 1 µg/ml tunicamycin, 10 µM palmitoyl-CoA, 2 µg/ml leupeptin and 0.1 mM TLCK at 37°C. Some reactions also contained 0.06 µg of GlcN-PI(C8) (a gift from Dr M.A.Lehrman) (Doerrler et al., 1996) added as a solution in 0.03% Triton X-100. After a 1 h incubation, lipids were extracted and analyzed by TLC.
For in vitro analysis of the first three steps, the lysates of cells (107) were incubated with 0.2 µM UDP-[6-3H]GlcNAc (14 µCi/ml, American Radiolabeled Chemicals) for 60 min at 37°C in a buffer containing 50 mM HEPES–NaOH pH 7.4, 25 mM KCl, 5 mM MgCl2, 1 mM ATP, 0.5 mM dithiothreitol (DTT), 1 mM GTP, 2 µM CoA, 0.2 µg/ml tunicamycin, 0.1 mM TLCK and 1 µg/ml leupeptin. Lipids were extracted and analyzed by TLC (Hirose et al., 1992).
Enzyme treatments of labeled lipids
Lipids were dissolved in 0.15 ml of a buffer containing either 100 mM Tris–HCl pH 7.4, 0.1% Triton X-100 and 5 µl of Bacillus thuringiensis PI-PLC (TOAGOSEI, Japan); 100 mM sodium acetate pH 5.0, 1 mM ZnCl2, 0.1% sodium taurodeoxycholate and 15 µl (1.7 U) of Jack bean α-mannosidase (Sigma); or 50 mM Tris–HCl pH 7.4, 10 mM NaCl, 2.5 mM CaCl2, 0.1% Triton X-100 and 15 µl of human serum as a source of GPI-PLD. After incubation for 12–16 h at 37°C, the lipids were extracted with n-butanol and analyzed as described above.
RT–PCR
Total RNA was extracted from 106 cells using TRIzol reagent (Gibco-BRL). Reverse transcription was done with random primers and SuperscriptII (Gibco-BRL). PCRs were hot started and cycled 35 times under conditions of 94°C for 30 s, 62°C for 30 s, 68°C for 2 min, using two sets of primers for PIG-M and one set of primers for human GPI1 as a control. The primers used for amplification of the full coding region of PIG-M were upper primer hMF1 and lower primer hMR4 (5′-GACGCGTGTCATATTTGATTCTCTCTGTCAGGG). The primers used for amplification of a middle portion of the coding region were hMF3 (5′-CACCGGATCCTACTATGAGTACGGCTGGGAATTT) and hMR1. For amplification of GPI1, upper primer F1 (5′-GGGAGAAGCCGCATCGGGCATCT) and lower primer R1 (Watanabe et al., 1998) were used.
GPI-MT-I assay using purified PIG-M protein
To obtain substrates for GPI-MT-I, we used membranes of Ramos517#17 cells in which GlcN-acyl-PI is accumulated. We incubated the membranes from 9 × 107 cells with 15 µCi/ml GDP-[3H]mannose, for 1 h at 37°C in 0.9 ml of the same buffer as described above for in vitro labeling. Lipids were extracted with n-butanol, dried up and resuspended in 900 µl of 50 mM HEPES–NaOH pH 7.4, 25 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 2 µg/ml leupeptin and 0.1 mM TLCK, for use in enzyme reactions.
PIG-M and Gaa1, as a control, were prepared as follows. CHO cells (107) were transfected with 25 µg of pMEEB-GF-hPIG-M or pMEEB-GF-hGaa1, and cultured for 2 days. The cells were washed with phosphate-buffered saline (PBS) and solubilized in 2.5 ml of a lysis buffer consisting of 1% digitonin (Wako Chemicals), 20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 µg/ml leupeptin and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (Wako Chemicals) at 4°C for 1 h. After the removal of insoluble materials by centrifugation at 13 000 g for 10 min, the soluble fraction was mixed with 25 µl of M2 anti-FLAG beads (Sigma) and agitated for 3 h. The beads were collected, washed with 1 ml of the lysis buffer four times and incubated in 25 µl of the lysis buffer containing 1 mg/ml FLAG peptide (Sigma) for 30 min to elute bound proteins. The elution was repeated three more times, and four eluates collected into one tube were then incubated with 10 µl of glutathione beads (Pharmacia) for further purification. After 2 h, the beads were washed with 0.5 ml of the lysis buffer three times and then once with 0.5 ml of a buffer consisting of 20 mM Tris–HCl pH 7.4, 150 mM NaCl and 1 mM EDTA. A fraction of the beads was used for SDS–PAGE and the rest was used to assess the GPI-MT-I activity. The beads were suspended in 150 µl of suspension of the substrate lipids and incubated at 37°C for 90 min. Lipids were extracted with n-butanol and analyzed by TLC. To prepare radiolabeled standard Man-GlcN-acyl-PI, membranes from 2 × 107 Lec35 cells were incubated in 0.6 ml of a reaction buffer containing 16.7 µCi/ml GDP-[3H]mannose in a similar way to that described above.
TbPIG-M cDNA
We found a partial sequence of TbPIG-M in the TIGR database (The Institute for Genomic Research, Rockville, MD). We amplified this sequence from a cDNA library of blood stage cells of T.brucei brucei strain 427 (a gift from Dr R.T.Schwarz, University of Marburg, Germany) by nested PCR using two sets of primers: upper primers (U-1 and U-2) designed within the coding region and lower primers (Cyc1-4 and Cyc1-2) within the vector sequence. U-1 (5′-TGGTCAAAACAGCAGTCGTCGTG) and Cyc1-4 (5′-GAGCGTCCCAAAACCTTCTCAAGCAAG) were used for the first reaction, and U-2 (5′-ACGGTCCTTTGTTACATGAAGTATGG) and Cyc1-2 (5′-CTTCCTTTTCGGTTAGAGCGGATGTGG) for the second reaction. To obtain the N-terminal coding sequence, we used RT–PCR. Reverse transcription was done with primer L-2 (5′-ATACGGTGAAAAGTTGTGACGATGAT), specific to TbPIG-M. PCR was done using an upper primer designed within the trans-splicing sequence (5′-AACGCTATTATTAGAACAGTTTCTGTACTA) and lower primer L-3 within the coding region (5′-CATCCACTTCGCACTTCTTTCCTTG). Both PCR products were subcloned into pBS and sequenced. Based on the sequence obtained, we designed the primers U-3 (5′-GCTCGGCTCGAGACCATGGAATTGCAGTCGCTTATAGACAC) and L-1 (5′-GCTCGGTCTAGAGTAAGACATCGACAACACTGCGCTA), and amplified the full coding region of TbPIG-M from the genomic DNA of T.b.brucei strain 427. All PCRs were done with a Long-template PCR kit (Boehringer).
Membrane topology of PIG-M protein
We transfected pMEEB-GF-hPIG-M, pMEEB-PIG-M-FLAG-GST, pMEEB-GF-PIG-A and pMEEB-PIG-A-FLAG-GST into CHO cells, selected and maintained in a medium containing 200 µg/ml hygromycin. Cells were cultured on 14 mm diameter glass coverslips for 1 day, permeabilized and stained as described previously (Eckhardt et al., 1999). In brief, they were washed with PBS containing Ca2+ and Mg2+ [PBS(+)], fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, again washed with PBS(+), and incubated for 20 min in 50 mM NH4Clto neutralize residual paraformaldehyde. To permeabilize the plasma membrane selectively, the cells were incubated in a buffer consisting of 1 µg/ml digitonin, 0.3 M sucrose, 0.1 M KCl, 2.5 mM MgCl2, 1 mM EDTA and 10 mM HEPES pH 6.9, for 15 min at 4°C. Thereafter, cells were washed three times with PBS(+), and non-specific binding sites were blocked with 1% bovine serum albumin (BSA) in PBS for 30 min at room temperature. For complete permeabilization, 0.1% Triton X-100 was added to the blocking solution. Cells were then incubated with 2.5 µg/ml goat anti-GST antibody (Pharmacia) or 2.5 µg/ml rabbit anti-BiP antibody (ABR, Inc.) in 0.1% BSA in PBS for 2 h. After four washes in PBS, cells were incubated with FITC-conjugated donkey anti-goat IgG or rhodamine-conjugated donkey anti-rabbit IgG antibodies (Chemicon International) in 0.1% BSA in PBS for 1 h. Slides were mounted in moviol and studied under a confocal laser scanning microscope (Bio-Rad).
Acknowledgments
Acknowledgements
We thank N.Inoue for discussion, B.Paul Morgan for information about CD59/DAF-deficient cell lines, and K.Nakamura for help in cell sorting. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Burda P., te Heesen,S., Brachat,A., Wach,A., Dusterhoft,A. and Aebi,M. (1996) Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: identification of the ALG9 gene encoding a putative mannosyl transferase. Proc. Natl Acad. Sci. USA, 93, 7160–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., Jakob,C.A., Beinhauer,J., Hegemann,J.H. and Aebi,M. (1999) Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology, 9, 617–625. [DOI] [PubMed] [Google Scholar]
- Camp L.A., Chauhan,P., Farrar,J.D. and Lehrman,M.A. (1993) Defective mannosylation of glycosylphosphatidylinositol in Lec35 Chinese hamster ovary cells. J. Biol. Chem., 268, 6721–6728. [PubMed] [Google Scholar]
- Canivenc-Gansel E., Imhof,I., Reggiori,F., Burda,P., Conzelmann,A. and Benachour,A. (1998) GPI anchor biosynthesis in yeast: phosphoethanolamine is attached to the α1,4-linked mannose of the complete precursor glycophospholipid. Glycobiology, 8, 761–770. [DOI] [PubMed] [Google Scholar]
- Doerrler W.T., Ye,J., Falck,J.R. and Lehrman,M.A. (1996) Acylation of glucosaminyl phosphatidylinositol revisited. J. Biol. Chem., 271, 27031–27038. [DOI] [PubMed] [Google Scholar]
- Doucey M.A., Hess,D., Cacan,R. and Hofsteenge,J. (1998) Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell, 9, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M., Gotza,B. and Gerardy-Schahn,R. (1999) Membrane topology of the mammalian CMP-sialic acid transporter. J. Biol. Chem., 274, 8779–8787. [DOI] [PubMed] [Google Scholar]
- Ferguson M.A. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors and the contributions of trypanosome research. J. Cell Sci., 112, 2799–2809. [DOI] [PubMed] [Google Scholar]
- Gastinel L.N., Cambillau,C. and Bourne,Y. (1999) Crystal structures of the bovine β4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J., 18, 3546–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor E.C., Mondesert,G., Grimme,S.J., Reed,S.I., Orlean,P. and Emr,S.D. (1999) MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell, 10, 627–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A. and Orlean,P. (1993) Glycoprotein biosynthesis in yeast. FASEB J., 7, 540–550. [DOI] [PubMed] [Google Scholar]
- Hirose S., Prince,G.M., Sevlever,D., Ravi,L., Rosenberry,T.L., Ueda,E. and Medof,M.E. (1992) Characterization of putative glycoinositol phospholipid anchor precursors in mammalian cells. Localization of phosphoethanolamine. J. Biol. Chem., 267, 16968–16974. [PubMed] [Google Scholar]
- Hong Y., Maeda,Y., Watanabe,R., Ohishi,K., Mishkind,M., Riezman,H. and Kinoshita,T. (1999) Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem., 274, 35099–35106. [DOI] [PubMed] [Google Scholar]
- Hong Y., Maeda,Y., Watanabe,R., Inoue,N., Ohishi,K. and Kinoshita,T. (2000) Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem., 275, 20911–20919. [DOI] [PubMed] [Google Scholar]
- Hyman R. (1988) Somatic genetic analysis of the expression of cell surface molecules. Trends Genet., 4, 5–8. [DOI] [PubMed] [Google Scholar]
- Jackson M.R., Nilsson,T. and Peterson,P.A. (1990) Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J., 9, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Ohishi,K. and Takeda,J. (1997) GPI-anchor synthesis in mammalian cells: genes, their products and a deficiency. J. Biochem., 122, 251–257. [DOI] [PubMed] [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Tomita,S., Watanabe,R., Ohishi,K. and Kinoshita,T. (1998) DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1 and binding of dolichol phosphate. EMBO J., 17, 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Tanaka,S., Hino,J., Kangawa,K. and Kinoshita,T. (2000) Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J., 19, 2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville M.J. and Ferguson,M.A.J. (1993) The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J., 294, 305–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville M.J. and Menon,A,K. (2000) Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol., 17, 1–16. [DOI] [PubMed] [Google Scholar]
- Miyata T., Takeda,J., Iida,Y., Yamada,N., Inoue,N., Takahashi,M., Maeda,K., Kitani,T. and Kinoshita,T. (1993) Cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science, 259, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Munro S. and Freeman,M. (2000) The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr. Biol., 10, 813–820. [DOI] [PubMed] [Google Scholar]
- Nagamune K. et al. (2000) Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 97, 10336–10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Inoue,N., Watanabe,R., Takahashi,M., Takeda,J., Stevens,V.L. and Kinoshita,T. (1997) Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem., 272, 15834–15840. [DOI] [PubMed] [Google Scholar]
- Nozaki M., Ohishi,K., Yamada,N., Kinoshita,T., Nagy,A. and Takeda,J. (1999) Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab. Invest., 79, 293–299. [PubMed] [Google Scholar]
- Ohishi K., Inoue,N., Maeda,Y., Takeda,J., Riezman,H. and Kinoshita,T. (2000) Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell, 11, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.F., Wiese,B.A., Wojzynski,M.K., Davison,D.B. and Worley,K.C. (1996) BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res., 6, 454–462. [DOI] [PubMed] [Google Scholar]
- Smith T.K., Sharma,D.K., Crossman,A., Dix,A., Brimacombe,J.S. and Ferguson,M.A. (1997) Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J., 16, 6667–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.K., Sharma,D.K., Crossman,A., Brimacombe,J.S. and Ferguson,M.A. (1999) Selective inhibitors of the glycosylphosphatidylinositol biosynthetic pathway of Trypanosoma brucei. EMBO J., 18, 5922–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Escribano,M.V., Gerold,P., Maeda,Y., Mazon,M.J., Kinoshita,T., Schwarz,R.T. and Riezman,H. (1998) Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem. J., 332, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Inoue,N., Ohishi,K., Maeda,Y., Nakamura,N., Endo,Y., Fujita,T., Takeda,J. and Kinoshita,T. (1996) PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J., 15, 4254–4261. [PMC free article] [PubMed] [Google Scholar]
- Taron C.H., Wiedman,J.M., Grimme,S.J. and Orlean,P. (2000) Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell, 11, 1611–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede A., Bastisch,I., Schubert,J., Orlean,P. and Schmidt,R.E. (1999) Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol. Chem., 380, 503–523. [DOI] [PubMed] [Google Scholar]
- Urakaze M., Kamitani,T., DeGasperi,R., Sugiyama,E., Chag,H.M., Warren,C.D. and Yeh,E.T.H. (1992) Identification of a missing link in glycosylphosphatidylinositol anchor biosynthesis in mammalian cells. J. Biol. Chem., 267, 6459–6462. [PubMed] [Google Scholar]
- Vidugiriene J. and Menon,A.K. (1993) Early lipid intermediates in glycosyl-phosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol., 121, 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Kinoshita,T., Masaki,R., Yamamoto,A., Takeda,J. and Inoue,N. (1996) PIG-A and PIG-H, which participate in glycosylphosphatidylinositol anchor biosynthesis, form a protein complex in the endoplasmic reticulum. J. Biol. Chem., 271, 26868–26875. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue,N., Westfall,B., Taron,C.H., Orlean,P., Takeda,J. and Kinoshita,T. (1998) The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J., 17, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins C.A.R. and Munro,S. (1998) Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl Acad. Sci. USA, 95, 7945–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]