Abstract
The TATA binding protein (TBP) and transcription factor IIB (TFIIB) play crucial roles in transcription of class II genes. The requirement for TBP–TFIIB interactions was evaluated in maize cells by introducing mutations into the Arabidopsis TBP (AtTBP2) within the C-terminal stirrup. Protein binding experiments indicated that amino acid residues E-144 and E-146 of AtTBP2 are both essential for TFIIB binding in vitro. Activation domains derived from herpes simplex viral protein VP16, the Drosophila fushi tarazu glutamine-rich domain (ftzQ), and yeast Gal4 were tested in transient assays. TBP–TFIIB interactions were dispensable for basal transcription but were required for activated transcription. In general, activated transcription was more severely inhibited by TBP mutation E-146R than by mutation E-144R. However, these TBP mutations had little effect on activity of the full-length cauliflower mosaic virus 35S and maize ubiquitin promoters, thus demonstrating that strong TBP–TFIIB contacts are not always required for transcription driven by complex promoters.
INTRODUCTION
The transcription machinery of eukaryotic class II genes consists of two megacomplexes of general factor proteins: transcription factor IID (TFIID) and the RNA polymerase II holoenzyme (reviewed in Burley and Roeder, 1996; Myer and Young, 1998). TFIID provides promoter recognition and binding activity, and the holoenzyme contains the catalytic function for mRNA synthesis. Assembly of the two complexes on the core promoter results in formation of the preinitiation complex, which is the final configuration assumed by RNA polymerase II before transcription is initiated. Mechanisms that facilitate the recruitment of either TFIID or the holoenzyme to the promoter enhance formation of the preinitiation complex.
In yeast and vertebrates, the recruitment of TFIID or holoenzyme to the promoter by directly tethering the TATA binding protein (TBP), TBP-associated factors (TAFIIs), or TFIIB can lead to high amounts of transcription, sometimes comparable with that achieved by a strong activator, such as the human herpes simplex viral protein VP16 (Chatterjee and Struhl, 1995; Klages and Strubin, 1995; Xiao et al., 1995, 1997; Gonzalez-Couto et al., 1997; Majello et al., 1998; Huh et al., 1999). In these artificially tethered systems, recruitment of either TFIID or the holoenzyme complex to the promoter results in the subsequent recruitment of the remaining complex in a process mediated by interactions between components of TFIID and the holoenzyme. In vitro studies indicate that the association of TFIID and the holoenzyme on the promoter involves multiple protein–protein contacts involving TBP, including those with TFIIB (Buratowski and Zhou, 1993; Nikolov et al., 1995), TFIIA (Geiger et al., 1996; Tan et al., 1996), the C-terminal domain of RNA polymerase II (Usheva et al., 1992), and a fraction containing TFIIH (Tang et al., 1996). The TAFs of TFIID also interact with TFIIB (Goodrich et al., 1993), the α subunit of TFIIE (Hisatake et al., 1995), and the large subunit of TFIIF RAP74 (Hisatake et al., 1995; Ruppert and Tjian, 1995). These in vitro studies have revealed the potential for protein–protein interactions; still unknown, however, are the relative importance and strength of individual interactions in the context of the assembled preinitiation complex and activated transcription in vivo.
Protein sequence comparisons suggest that among eukaryotic organisms the TBP–TFIIB interaction is highly conserved, involving eight amino acid residues from TBP and 12 from TFIIB (Nikolov et al., 1995). Four of the eight residues from TBP are located in the C-terminal stirrup, the structure of which is identical among Arabidopsis (Kim and Burley, 1994), humans (Kao et al., 1990), Drosophila (Muhich et al., 1990), and yeast (Horikoshi et al., 1989) proteins. Of these, residue E-146 (amino acid position according to the Arabidopsis TBP2 [AtTBP2] protein) makes the most contacts with TFIIB by forming a strong salt bridge and two hydrogen bonds; van der Waals interactions account for another bond (Nikolov et al., 1995). All 12 TBP binding residues of the conserved core of human TFIIB are conserved in Drosophila, 10 are conserved in Arabidopsis, and eight are conserved in yeast (Baldwin and Gurley, 1996). The importance of the C-terminal stirrup of TBP in the TFIIB interaction has been confirmed by mutational analysis of human and yeast TBPs. Alanine substitution of stirrup residues E-284, E-286, or L-287 of human TBP reduced the affinity for TFIIB in vitro to ∼5% of that of the wild type (Tang et al., 1996). Analogous substitutions in yeast TBP resulted in 100-, 50-, and 10-fold reductions in TFIIB binding, respectively (Lee and Struhl, 1997). These mutations specifically disrupt the interaction of TBP with TFIIB but do not affect its interactions with TFIIA, TFIIF, RNA polymerase II, TFIIE, or TFIIH (Tang et al., 1996).
The functional importance of the TBP–TFIIB interaction has been tested in both human and yeast cells by using the altered-specificity system, in which a mutated TATA (TGTA) reporter is used in combination with the TBPm3 mutant that is able to recognize the TGTA motif of the promoter (Strubin and Struhl, 1992). Transcription of the TGTA reporter gene uses TBPm3 rather than the endogenous TBP. In that system, additional mutations of TBPm3 in the C-terminal stirrup generally suppress transcription in human cells (Bryant et al., 1996; Tansey and Herr, 1997) but not in yeast (Lee and Struhl, 1997), which suggests that the TBP–TFIIB interaction is critical for transcription in human cells but dispensable in yeast.
Although cDNAs for general transcription factors have been isolated in plants (Gasch et al., 1990; Haass and Feix, 1992; Baldwin and Gurley, 1996; Li et al., 1999), no studies have examined their function in detail or addressed questions on the roles of specific protein–protein interactions within the preinitiation complex. Using a maize transient expression system to assess the importance of the TBP–TFIIB interaction in both basal and activated transcription, we found the TBP–TFIIB interaction to be critical for transcription that is activated by transactivators consisting of a single type of activation domain fused to the Gal4 DNA binding domain. However, mutations in the C-terminal stirrup of TBP had little effect on basal transcription or on the activity of natural promoters, such as the cauliflower mosaic virus (CaMV) 35S promoter and the maize ubiquitin promoter.
RESULTS
C-Terminal Stirrup of the Arabidopsis TBP Is Required for Binding to TFIIB
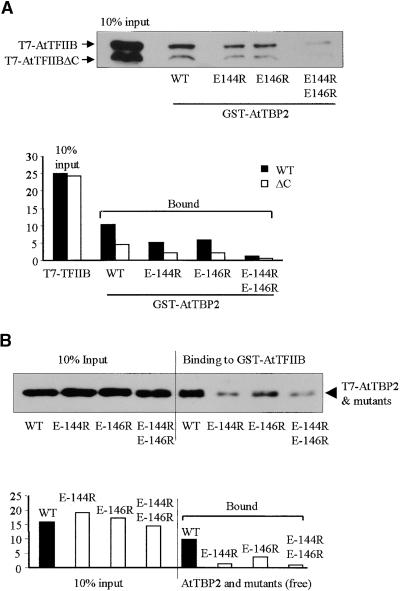
Single or double amino acid substitutions were introduced into the C-terminal stirrup of AtTBP (Kim and Burley, 1994) in two positions (E-144 and E-146) to evaluate the importance of the TBP–TFIIB interaction in transcription. The position and structural relationships of mutations used in this study are shown in Figure 1. Glutathione S-transferase (GST) pull-down assays were used to test in vitro interactions between AtTBP2 and AtTFIIB, in which one of the proteins was immobilized on glutathione–Sepharose beads as a GST fusion and the other was allowed to interact as a free ligand in the absence of DNA (Figure 2). As shown in Figure 2A, the bead-immobilized TBP binds both wild-type TFIIB and a C-terminal truncation of TFIIB inadvertently produced by the in vitro translation system (faster migrating band in Figure 2A). The two single mutations E-144R and E-146R reduced binding by ∼50%, whereas the double mutation E-144R/E-146R reduced binding by more than eightfold. The truncated TFIIB protein bound to TBP and its mutants with ∼50% efficiency (Figure 2A). Although the precise point of truncation is not known for the Arabidopsis TFIIB, a human TFIIB deletion mutant showed a similar reduction in binding TBP (by 60%) after removal of 30 or 80 C-terminal residues (Ha et al., 1993). No binding was evident between either TBP (including mutant forms) or TFIIB with immobilized GST alone (data not shown).
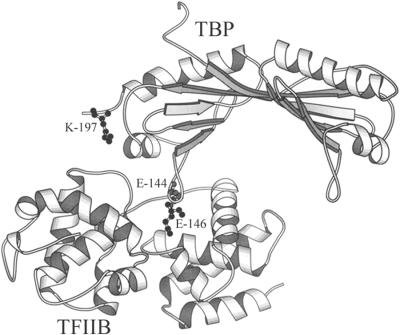
Figure 1.
TBP–TFIIB Complex from Nikolov et al. (1995) Showing the Location of Amino Acid Residues Involved in Close Contacts between TBP and TFIIB.
DNA has been omitted from the ternary complex (TATA box–AtTBP2–hTFIIB) for clarity. Mutations E-144R, E-146R, E-144R/ E-146R, and E-144R/E-146R/K-197E were introduced into AtTBP. The diagram was produced by using MOLSCRIPT software (Kraulis, 1991).
Figure 2.
In Vitro GST Pull-Down Assays Showing Interactions between AtTFIIB and Wild-Type or Mutated AtTBP2.
(A) Protein gel blot of TFIIB bound to GST-TBP beads. T7 epitope–tagged AtTFIIB was synthesized in a coupled transcription/translation (TNT) reaction by using wheat germ lysate. AtTFIIBΔC represents prematurely terminated protein, resulting in an undefined C-terminal truncation. Bands were visualized by probing the protein gel blot with the anti-T7 tag monoclonal antibody. Binding was quantified by analyzing scanned images with Scion Image software. Units on the vertical axis are arbitrary.
(B) Protein gel blot of the TBP bound to GST-TFIIB beads. AtTBP and its mutants were synthesized in a coupled TNT reaction in rabbit reticulocyte lysate. The blotting procedures and quantification of binding are as given in (A).
WT, wild type.
In the reciprocal experiment, which is shown in Figure 2B, GST-TFIIB was immobilized on beads and the TBP was free in solution. Again, similar results were obtained, indicating that the C-terminal stirrup mutations of TBP interfered with TBP–TFIIB interactions. The binding of TBP to GST-TFIIB was markedly inhibited by TBP C-terminal stirrup mutations. TBP mutation E-146R reduced binding to TFIIB by ∼50%, whereas the E-144R mutation showed an 85% reduction in binding. The most severe reduction in binding was exhibited by the double mutation E-144R/E-146R that showed a >88% reduction in affinity for TFIIB (Figure 2B). No bands were detectable in the GST negative control (data not shown). Although substitution of the two glutamic acid residues in the C-terminal stirrup of TBP did not completely abolish interactions with TFIIB, these residues are clearly critical for such connections in vitro, as is also the case with the corresponding human and yeast proteins (Tang et al., 1996; Lee and Struhl, 1997).
TBP–TFIIB Interaction Is Dispensable in Basal Transcription in Vivo
Basal transcription in maize suspension cells was determined by a transient expression assay using the β-glucuronidase (GUS) reporter gene under control of the CaMV 35S minimal promoter (Odell et al., 1985; Figure 3A, construct 3), which contains no activation elements upstream of the TATA motif. Control experiments performed with the promoterless–GUS reporter (Figure 3A, construct 2) indicated that background amounts of GUS activity were very low in the maize cell line (Figure 4). The activity of the null effector (Figure 3B, construct 9 with T7 epitope tag alone) with the minimal promoter was 12-fold greater than the activity with the promoterless reporter, representing basal transcription activity driven by endogenous factors in maize cells (Figure 4).
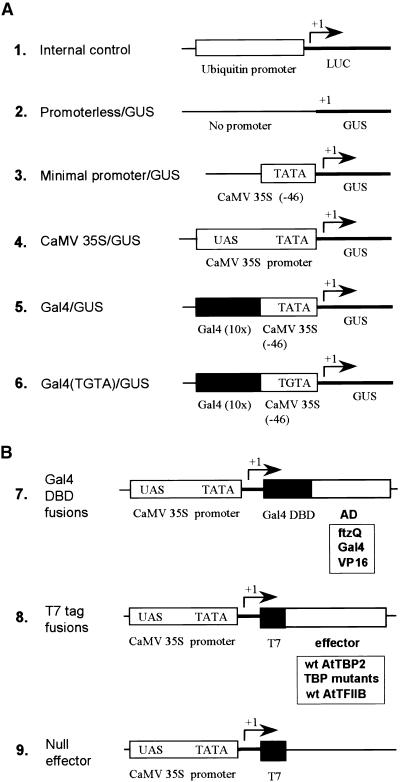
Figure 3.
Diagram of Reporter and Effector Constructs.
(A) Reporter constructs are as follows: (1) maize ubiquitin promoter–LUC reporter used in all transient assays as an internal control to normalize GUS activities; (2) promoterless–GUS reporter used as a control for no transcription; (3) minimal CaMV 35S promoter (−46 bp) used to monitor basal transcription; (4) full-length CaMV 35S promoter used to monitor transcription from a complex natural promoter; (5) Gal4–GUS reporter used to monitor activity of Gal4 DNA binding domain–activator effector proteins; and (6) same as construct 5, except that the promoter contains a mutated TATA motif (TGTA).
(B) Effector constructs are as follows: (7) vector used to express Gal4 DNA binding domain (DBD) fusions with ftzQ, Gal4, and VP16 activation domains; (8) T7 tag vector used to express TBP and TFIIB; and (9) null effector. The T7 vector without an insert was used as a control to keep DNA amounts and promoter concentrations constant between experiments.
wt, wild type.
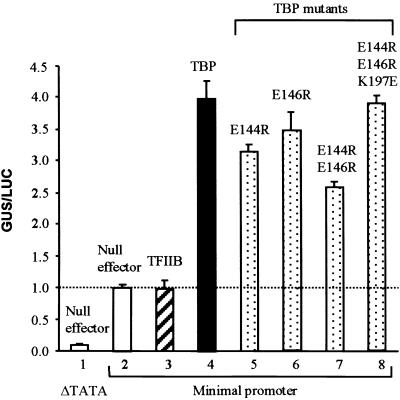
Figure 4.
Basal Transcription in Maize Suspension Cells Is Stimulated by Wild-Type and Mutant TBP, but Not by TFIIB.
Basal transcription was tested with the minimal promoter–GUS reporter and the indicated effectors (10 μg of DNA each). The total amount of DNA used in bombardment for each treatment was kept constant for all combinations of effector and reporter DNAs. The promoterless–GUS reporter (Figure 3A, construct 2) served as a control for no transcription and was coexpressed with the null effector (bar 1). Experiments represented in bars 2 to 8 used the CaMV 35S minimal promoter–GUS vector (Figure 3A, construct 3). Activity is given as a ratio of GUS/LUC units and represents an average of triplicate experiments. Standard errors are indicated.
Coexpression of the wild-type TBP effector with the minimal promoter–GUS reporter stimulated GUS activity fourfold above that obtained when the reporter was coexpressed with the null effector (Figure 4, cf. bars 4 and 2). However, coexpression of TFIIB showed no effect on the basal transcription (Figure 4). These results indicate that TBP concentrations were rate limiting for basal transcription, whereas TFIIB concentrations were not.
This dependence on exogenous TBP for increased amounts of basal activity allowed testing of TBP C-terminal stirrup mutations. Expression of mutant TBPs with single, double, or triple point mutations enhanced basal transcription to levels comparable with those obtained with wild-type TBP (Figure 4, cf. bars 5 to 8 with 4). From the lack of suppression by the C-terminal stirrup mutations in TBP, we conclude that TBP–TFIIB interactions are not critical for supporting basal transcription in plant cells.
TBP–TFIIB Interaction Is Required for Transcription Activated by Single Activation Domains
To address whether the TBP–TFIIB interaction is important for activated transcription, we conducted experiments using the Gal4 reporter system (Figure 3A, construct 5). Transactivator proteins (effectors) were constructed as heterologous fusions between the Gal4 DNA binding domain (Marmorstein et al., 1992) and three different activation domains: the acidic activation domains of VP16 (herpes simplex virus; Cousens et al., 1989), Gal4 (yeast; Ma and Ptashne, 1987), and the glutamine-rich activation domain of fushi tarazu (ftzQ; Drosophila; Fitzpatrick and Ingles, 1989). In separate experiments, all of these transactivators strongly stimulated transcription in maize cells expressing the endogenous TBP, the activity being enhanced >100-fold over that obtained in the presence of the Gal4 DNA binding domain alone (data not shown).
Transcriptional activity of these activation domains was monitored by coexpression of TBP or its C-terminal stirrup mutants with each chimeric activator in three replicate experiments (Figure 5). Coexpression of wild-type AtTBP inhibited activity of ftzQ and VP16 (Figures 5A and 5C, cf. bars 1 and 2) by ∼25%, but enhanced Gal4 activation domain activity by ∼67% (Figure 5B). Compared with wild-type TBP, the E-144R mutation provoked no impairment in ability to enhance transcriptional activity of all activation domains (Figure 5, cf. bars 2 and 3); indeed, the ftzQ activity showed substantial enhancement. A minimal interpretation of these results is that the E-144R mutation of TBP does not impair activated transcription in vivo. The stimulation in Gal4-ftzQ activity by E144R TBP is unexplained.
Figure 5.
Activated Transcription Driven by a Single Type Activation Domain Is Sensitive to TBP C-Terminal Stirrup Mutations in Vivo.
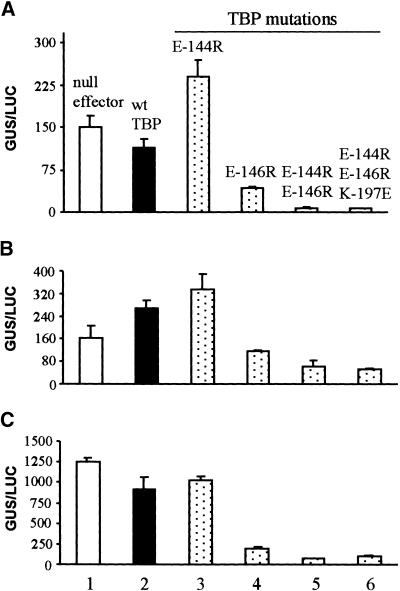
Transcription of the Gal4–GUS reporter (Figure 3A, construct 5) was activated by coexpression with Gal4 DNA binding domain activator fusions in transient assays using maize cells. The effect of TBP C-terminal stirrup mutations on activated transcription was monitored by coexpression of TBP mutants with each Gal4 DNA binding domain activator as indicated. Three types of activation domain were tested as Gal4 DNA binding domain fusion proteins: (A) ftzQ; (B) Gal4; and (C) VP16. The null effector (Figure 3B, construct 9) used in bar 1 was included to show activity of endogenous TBP. Bars 2 to 6 represent expression of wild-type and mutant AtTBP, as indicated. Activity, given as a ratio of GUS/LUC units, is averaged from triplicate experiments. Standard errors are indicated. wt, wild type.
In contrast to the lack of impairment in activity exhibited by TBP mutant E-144R, the E-146R mutation significantly inhibited transcriptional activity for all of the activation domains tested, the degree of reduction ranging from more than two- to approximately fivefold (Figure 5, cf. bars 4 and 2). The double mutation E-144R/E-146R was even less able to support activated transcription, with the extent of inhibition ranging from five- to 16-fold. Although the E-144R mutation either showed no inhibition or appeared to have a stimulatory effect, when combined with E-146R (E-146R/ E-144R), transcriptional activities were less than those exhibited by the E-146R mutation alone.
A third charge–charge interaction between TBP and TFIIB involves K-197, which is near the C terminus of TBP (Nikolov et al., 1995). However, the triple mutation E-144R/E-146R/K-197E showed no further inhibition of transcription compared with E144R/E146R, suggesting that double mutation of the stirrup of TBP is sufficient to abolish the TBP–TFIIB interaction in vivo. Taken together, these results are in strong contrast to those observed for basal transcription, indicating that activated transcription driven by a transactivator protein consisting of the Gal4 DNA binding domain fused to a simple activation domain is highly dependent on the TBP–TFIIB interaction.
The slight inhibition caused by coexpression of the wild-type TBP with VP16 and ftzQ may be a result of transcriptional squelching (Gill and Ptashne, 1988; titration data not shown) because the expression of TBP was not optimized for each activator. In general, the degree of squelching seen in these experiments was not severe and did not alter conclusions regarding the importance of TBP–TFIIB interactions in activated transcription.
Requirement for TBP–TFIIB Interaction in Activated Transcription Confirmed by Using the Altered-Specificity System
The TBP altered-specificity system, which is free of possible interference by endogenous TBP, was used to demonstrate more definitively the effect of TBP stirrup mutations on activated transcription. The TATA element of the Gal4–GUS reporter was mutated to TGTA (Figure 3A, construct 6) so that activity would be dependent on overexpressed TBPm3, which is able to recognize both TGTA and TATA (Strubin and Struhl, 1992). To demonstrate the lack of recognition of the TGTA motif by the endogenous maize TBP, we coexpressed the null effector with the TATA and TGTA GUS reporters. Endogenous activity was much less with the TGTA reporter than with the normal TATA reporter (Figures 6A to 6C, bars 1 and 2). Furthermore, overexpression of wild-type AtTBP produced either no or only low increases in activity compared with the endogenous TBP. In contrast, overexpression of AtTBPm3 resulted in substantially more activity than was exhibited by the wild-type AtTBP, ranging from a very modest increase in activity for VP16 (Figure 6C, bar 4) to a complete restoration of endogenous activity for ftzQ (Figure 6A, bar 4) and yeast Gal4 activation domain (Figure 6B, bar 4).
Figure 6.
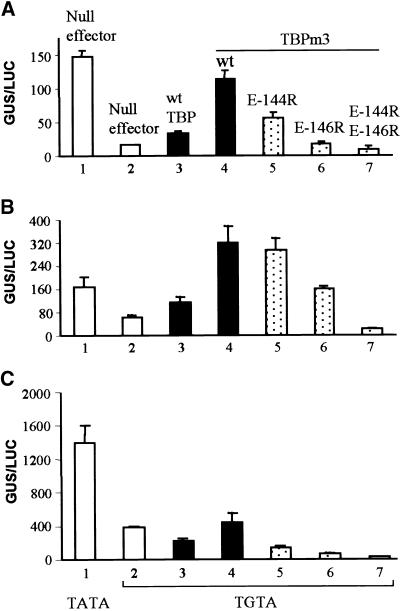
Role of the C-Terminal Stirrup of TBP in Activated Transcription Evaluated by Using the Altered-Specificity System (TGTA/TBPm3).
(A) Transcription was activated by Gal4 DNA binding domain ftzQ. AtTBP2 was mutated by three amino acid substitutions within its concave DNA binding surface to generate TBPm3 (Strubin and Struhl, 1992). Transcriptional activity was monitored with the Gal4 (TGTA)–GUS reporter (Figure 3A, construct 6) in transient assays with maize cells. The activator was coexpressed with wild-type (wt), single, or double point mutations of TBPm3 as indicated. Quantities of DNA used in transformations remained constant. Bar 1 represents the control for normal activity as determined by using the wild-type TATA reporter and the endogenous TBP. The TGTA reporter (Figure 3A, construct 6) was used for experiments in bars 2 to 7. Activity, given as relative GUS/LUC units, is an average of triplicate assays. Standard errors are indicated.
(B) Transcription was activated by the Gal4 DNA binding domain–Gal4 activation domain. Experiments were conducted as described in (A).
(C) Transcription was activated by Gal4 DNA binding domain–VP16. Experiments were conducted as described in (A).
When activated transcription relied on exogenous TBPm3, all three stirrup mutations suppressed activated transcription (Figure 6, bars 5 to 7). The E-144R mutation inhibited activity from two- to threefold for ftzQ and VP16 but only 10% for the Gal4 activation domain (Figures 6A to 6C, cf. bars 5 and 4). In addition, E-144R TBPm3 had no stimulatory effect (Figure 6, bar 5), in contrast to E-144R TBP and the TATA reporter (Figures 5A to 5C, bars 3). The E-146R and E-144R/E-146R mutations of TBPm3 were more severe in their inhibition of transcription than was the E-144R mutation (Figures 6A to 6C, cf. bars 5 to 7). Overall, these results are consistent with those observed for the natural TATA-TBP system, although TBPm3 seems to be much more sensitive to C-terminal stirrup mutations than is wild-type TBP. In both systems, however, residue E-146 was clearly more critical than E-144 for supporting activated transcription in vivo, which is consistent with structural predictions (Nikolov et al., 1995).
Complex Promoters Show Much Less Dependence on the TBP–TFIIB Interaction
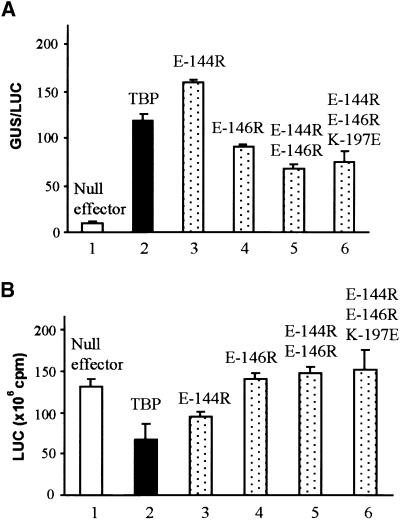
Transcription directed by the full-length CaMV 35S promoter (Odell et al., 1985; Lam et al., 1989; Benfey et al., 1990) and the maize ubiquitin promoter (Christensen and Quail, 1996) also were evaluated regarding the importance of the TBP–TFIIB interaction. Both of these natural promoters are more complex in upstream element composition than is the synthetic Gal4 promoter, and they presumably rely on multiple transactivator proteins for activity. As shown in Figure 7, the activities of the natural promoters were dependent on endogenous transactivators present in maize cells. Note that in bar 1 of Figure 7A, the activity of the CaMV 35S promoter was relatively weak in maize suspension cells. Transcription driven by the CaMV 35S promoter was strongly stimulated by overexpression of wild-type TBP, which suggests that recruitment of TFIID must be rate limiting in maize cells. In addition, all C-terminal stirrup mutants of TBP strongly stimulated 35S activity (Figure 7A, cf. bars 2 to 6 with 1). However, except for the E-144R mutant, the efficiency of activation appeared to be slightly reduced by the mutations in comparison with wild-type TBP.
Figure 7.
Transcription Activated by Multiple Recruitment Pathways Is Much Less Dependent on the TBP–TFIIB Interaction.
(A) CaMV 35S promoter activity. The experiments were conducted as described in Figure 5, except that the GUS reporter was driven by the wild-type CaMV 35S promoter (Figure 3A, construct 4), the activity of which relies on factors endogenous to maize cells. Activity for the CaMV 35S promoter is given as relative GUS/LUC units.
(B) Maize ubiquitin promoter activity. Activity for the maize ubiquitin promoter is given as LUC units (photon emission, counts per minute) without normalization because this reporter (Figure 3A, construct 1) was used for the internal standard for all experiments in AFigures 4 to 7A.
All LUC and GUS activities represent an average of triplicate assays. Standard errors are indicated.
Simultaneous experiments with the maize ubiquitin promoter showed a similar insensitivity to TBP mutations (Figure 7B). In contrast to the CaMV 35S promoter, however, overexpression of AtTBP2 showed a modest inhibition of activity rather than the strong stimulation seen with the 35S promoter. As with the 35S promoter, mutations in TBP showed no indication of inhibiting transcription. Luciferase (LUC) activities were simply averaged rather than normalized, because no internal standard was available.
DISCUSSION
In this study, plants were used to examine in vivo the function of protein–protein interactions within the preinitiation complex. Transient assays using maize cells demonstrated that the TBP–TFIIB interaction can be critically important for activated transcription; however, the functional requirement for this association is clearly sensitive to context. In this study, the activated transcription from a simple promoter driven by a single activation domain was heavily dependent on the TBP–TFIIB interaction, whereas in more complex natural promoters, this interaction made only minor contributions to overall transcription activity. Similarly, interactions between TBP and TFIIB were not important in basal transcription from the minimal CaMV 35S promoter.
In maize suspension cells, basal transcription dependent on the CaMV 35S minimal promoter was stimulated by coexpression of TBP but not by TFIIB (Figure 4). Similar results were observed with Drosophila, in which basal activity was stimulated 20-fold in vivo by overexpression of TBP, whereas TFIIB was totally ineffective (Colgàn et al., 1993). This differential stimulation by TBP suggests that the concentration of TBP or TFIID on the promoter, rather than that of TFIIB or the RNA polymerase II holoenzyme, is limiting for basal transcription in vivo. These results argue that recognition of the TATA motif by TBP or TFIID may be the most important step in determining the amounts of basal transcription in living cells.
Basal transcription in maize cells appears not to require the TBP–TFIIB interaction, as evidenced by the lack of sensitivity to the TBP stirrup mutations (Figure 4). This result contrasts with those obtained in reconstituted in vitro transcription systems using human components, which indicate that mutation of either glutamic acid residue in the C-terminal stirrup of TBP is sufficient to abolish basal transcription (Bryant et al., 1996). These contradictory results suggest that basal transcription may differ between plants and humans or, alternatively, between in vivo and in vitro transcription systems. Regardless of possible differences, alternative interaction pathways that enable assembly of the preinitiation complex to support basal transcription when the TBP–TFIIB interaction is disrupted apparently are available in plant cells.
In contrast to the lack of a requirement for contacts between TBP and TFIIB in basal transcription, this connection appears to be critical for function when a synthetic Gal4 promoter is activated by a single type of activation domain. This conclusion is based on the severe inhibition of transcription that resulted from coexpression of the mutated TBP E-144R/ E-146R with the five activation domains tested (Figures 5 and 6 and two plant activation domains; data not shown). Additional evidence suggests that the reductions in activity obtained with the TBP stirrup mutations result from dysfunctional TBP, not from a squelching event (S. Pan and W.B. Gurley, manuscript in preparation). The suppression of activity seen with the stirrup mutations can be reversed by inserting an additional mutation in another location within TBP. This reversal of suppression is specific to GAL4 DNA binding domain fusions with acidic activation domains from VP16 and GAL4 but is not specific to similar constructs with ftzQ.
In a simple experimental system in which the promoter is driven by a single activation domain, either holoenzyme or TFIID is initially recruited to the promoter. Once this occurs, the rate-limiting step is probably the subsequent recruitment of the second complex (Gonzalez-Couto et al., 1997). Under these conditions, the strength of interaction between TFIID and the holoenzyme may be correlated with the amount of transcriptional activity. Although many documented in vitro interactions among general transcription factors could serve this bridging function, the TBP–TFIIB association clearly has the potential to play a major role in joining the two complexes during transcriptional activation in plant cells.
The roles of the two individual glutamic acids in the C-terminal stirrup of TBP apparently differ between the in vivo and the in vitro transcription systems. Although residue E-144 seems to be as important as E-146 in binding TFIIB in vitro (Figure 2), E-146 is apparently more critical in activated transcription in vivo, as shown by using several different activation domains (Figures 5 and 6; data not shown). Unlike direct interaction between proteins in vitro, the in vivo TBP–TFIIB interaction takes place in the context of the preinitiation complex in association with the TATA motif of the promoter, in which interactions may be complicated by the presence of other components of the preinitiation complex and even upstream activators.
The degree of dependency on the TBP–TFIIB interaction in supporting activated transcription in vivo seems to vary between plants, humans, and yeast. In yeast, the TBP–TFIIB interaction appears to be totally dispensable for transcription that is activated by several different acidic activation domains, including VP16 (Chou and Struhl, 1997; Lee and Struhl, 1997). A variable pattern of dependency on the TBP–TFIIB interaction occurs in HeLa cells, in which VP16-activated transcription is totally dependent on close TBP–TFIIB contacts, whereas SP1-directed transcription shows little requirement for this (Tansey and Herr, 1997). In HeLa, single mutations at either of the two glutamic acid residues totally eliminate VP16 activity in the TGTA/TBPm3 altered-specificity system (Tansey and Herr, 1997). In plants, the pattern is more complicated. Not only does the dependency on the TBP–TFIIB interaction vary between different activation domains and between promoter configurations (synthetic Gal4 promoter versus more complex natural promoters), but also the two glutamic residues of the stirrup differ in importance, with E-146 being more critical than E-144 (Figures 5A to 5C, 6A to 6C, and 7A).
The effect of TBP overexpression on the activity of the full-length CaMV 35S promoter showed striking similarity to that observed with basal expression from the minimal promoter. Both promoters were strongly stimulated by overexpression of wild-type and mutant AtTBP2. The CaMV 35S promoter is poorly expressed in maize and other monocots relative to dicotyledonous plant species (Wilmink et al., 1995). Our results suggest that the defect in expression may be partially a result of the failure of the 35S promoter to efficiently recruit TBP (or TFIID) in maize cells. As occurs with basal expression, the lack of sensitivity to TBP mutants may result from the very low activity of the 35S promoter in which the rate-limiting step is TBP recruitment, not in interactions between TFIID and the holoenzyme. This, again, implies that alternative pathways of interaction exist for the recruitment of the holoenzyme by TFIID, making the TBP–TFIIB contact redundant.
A further prediction is that the strong recruitment of the holoenzyme is also lacking for the 35S promoter in maize cells. The minimal promoter serves as a model for no recruitment. Likewise, transcriptional activation by ftzQ serves as a model for promoter function in which the holoenzyme is strongly recruited through interactions with TFIIB (Colgàn et al., 1993, 1995) and the recruitment of TFIID is presumably absent or very inefficient (S. Pan and W.B. Gurley, manuscript in preparation). The subsequent recruitment of TFIID by the holoenzyme tethered to ftzQ is very sensitive to mutations that disrupt the TBP–TFIIB interaction (Figures 5A and 6A). Therefore, the lack of sensitivity to TBP mutations exhibited by the 35S promoter argues that neither the holoenzyme nor TFIID is strongly recruited to the promoter in maize. This lack of a requirement for close TBP–TFIIB interactions is similar to the conditions for basal transcription, in which recruitment of TFIID and the holoenzyme was eliminated by removing all upstream elements from the 35S promoter to construct the minimal promoter. The prediction that recruitment of both TFIID and the holoenzyme is highly inefficient for the 35S promoter in maize is consistent with the suggestion of others that transactivator proteins that bind efficiently to the upstream elements of the 35S promoter in dicotyledonous plants may be either absent or much less abundant in many monocots (Last et al., 1991). Because major classes of transactivator proteins are expected to be conserved among all plant species, the low activity of the CaMV 35S promoter in maize may reflect a divergence in the specificity of DNA sequence recognition by certain monocot transactivators.
In the case of the maize ubiquitin promoter, TBP is not rate limiting; however, transcription is still relatively insensitive to the TBP mutations (Figure 7B). The strong activity of this promoter suggests that TFIID and the holoenzyme are efficiently recruited to the promoter. The lack of sensitivity to the TBP mutations, as with basal transcription, argues that alternative pathways for interactions between TFIID and the holoenzyme must exist. Transcription from the ubiquitin promoter differs from basal transcription in that the TFIID–holoenzyme interaction may be stabilized by the tethering of both TFIID and the holoenzyme to the promoter through multiple interactions with transactivator proteins bound to upstream elements (Genschik et al., 1994). The strong recruitment of general transcription factors to the ubiquitin promoter by transactivators endogenous to maize cells is predicted to greatly facilitate TFIID–holoenzyme interactions and thus reduce the importance of direct contacts between TBP and TFIIB.
In summary, we interpret our results to indicate that the transcriptional requirement for interactions between TBP and TFIIB can vary widely depending on the context of gene expression. Under circumstances in which recruitment of general transcription factors is low because of a lack of upstream elements (minimal promoters) or an inefficient recruitment of transactivator proteins, the TBP–TFIIB interaction is not critical for transcriptional activity because TFIID–holoenzyme associations still can form by alternative pathways. Under these conditions, presumably it is the recruitment of TFIID that is rate limiting, not the subsequent recruitment of the holoenzyme by TFIID. In strong natural promoters, the TBP–TFIIB interaction also is not critical, but for different reasons. In this case, TFIID–holoenzyme interactions probably are facilitated by the simultaneous tethering of TFIID and holoenzyme through interactions with multiple promoter-bound transactivator proteins.
In special circumstances, however, TBP–TFIIB contacts do play a critical role in supporting transcription, such as with promoters that rely on a limited number of recruitment pathways. For example, when the tethering of TFIID and the holoenzyme to the promoter is limited by the use of single activation domains (e.g., Gal4 DNA binding domain–activator constructs), stabilization of TFIID and holoenzyme contacts is minimal, presumably because only one of the two megacomplexes is strongly recruited to the promoter.
Our results suggest that the importance of the TBP–TFIIB interaction is highly dependent on parameters that determine the efficacy of recruitment of TFIID and the holoenzyme to the promoter. A direct test of these predictions requires further experimentation with natural and synthetic promoters under conditions in which recruitment pathways are better understood.
METHODS
Point Mutagenesis for Arabidopsis thaliana TATA Binding Protein Isoform 2
The Altered Site II mutagenesis system (Promega, Madison, WI) was used to generate amino acid substitution mutations for Arabidopsis thaliana TATA binding protein (TBP) isoform 2 (AtTBP2; Gasch et al., 1990) in the Ex-1 vector according to the manufacturer's protocol. The targeted residues were E-144 and E-146 within the C-terminal stirrup and K197 near the C terminus of the protein. For each of the three residues, the amino acid substituted had an ionic charge opposite that of its wild-type counterpart. Single, double, or triple point mutations were generated as follows: E-144R, E-146R, E-144R/E-146R, and E-144R/E-146R/K-197E. After confirmation by DNA sequencing, both wild-type and mutant TBP cDNAs were excised from the Ex-1 mutagenesis vector by SalI-BamH1 digestion and subcloned into the SalI-BglII sites of the pBI221-derived plant expression vector (Clontech, Palo Alto, CA; E. Czarnecka-Verner, C.-X. Yuan, K.-D. Scharf, G. Englich, and W.B. Gurley, submitted manuscript). The first two amino acid residues of TBP within the N-terminal region were inverted from MT to TM during subcloning. Both AtTBP2 and transcription factor AtTFIIB (Baldwin and Gurley, 1996) recombinant proteins contained the T7 epitope tag (MASMTGGQQMG), followed by two amino acids (RS) at the N-terminal ends, and two additional residues (EI) inserted at the C termini.
Particle Bombardment in Maize Suspension Cells
DNA/gold particles were prepared as described previously (Pan et al., 1999). Each DNA/gold preparation contained 2.5 μg of the maize ubiquitin promoter–luciferase (LUC) reporter plasmid pUbi-LUC (Christensen and Quail, 1996) as the internal control, 2.5 μg of the β-glucuronidase (GUS) reporter driven by the minimal cauliflower mosaic virus (CaMV) 35S promoter (nucleotide −46; 35S-GUS; Odell et al., 1985) with or without Gal4 binding sites, 2.5 μg of the Gal4 DNA binding domain fusion activators, and 10 μg of vector containing the null effector (T7 tag alone), T7-TFIIB, or T7-TBP effectors (wild type or mutants). Reporter and effector plasmids are diagrammed in Figure 3.
The DNA/gold particles were delivered into log-stage maize suspension cells by using a Bio Rad PDS-1000 particle bombardment apparatus as described previously (Pan et al., 1999). Maize cells (Black Mexican Sweet) (Chourey and Zurawski, 1981) were poured into a 50-mL sterile centrifuge tube and allowed to settle by gravity to a volume of 5 to 7 mL. The extra medium was discarded to obtain a cell/medium ratio of 1:1 (v/v). The cells then were well suspended, and 300 μL was pipetted onto a 2.5-cm-diameter circle Whatman filter paper previously placed on a Murashige and Skoog (Murashige and Skoog, 1962) plate (Phytagel; Sigma). After particle bombardment, the filter-immobilized cells were allowed to recover for 22 hr in the dark at 26°C. The cells were harvested, and GUS and LUC activities were determined as described previously (Pan et al., 1999).
Representative experiments shown in Figures 4 to 7 were conducted in triplicate. GUS activity was normalized against LUC activity (Ubi-LUC; Figure 3A, construct 1) and expressed as arbitrary units of relative GUS/LUC (nanomoles of 4-methylumbelliferone per hour per counts per minute). The Gal4 DNA binding domain–activation domain effectors were assayed over a range of DNA concentrations to ensure that transcriptional activities were not subject to squelch effects. We determined that 2.5 μg of effector DNA precipitated onto gold particles was within the range in which transcriptional squelch was minimal.
Protein Expression and Purification from Escherichia coli
Chimeric constructs of glutathione S-transferase (GST)–AtTFIIB and GST-AtTBP2 were expressed in E. coli and purified as described previously (Pan et al., 1999). Expression of the GST fusion proteins was induced by exposure to 10 μM isopropyl thioglactoside for 7 hr at room temperature. The induced cells were collected by centrifugation, washed with cold PBS, and suspended in 1 mL of cold protein binding buffer (PBB): 20 mM Hepes, pH 7.5, 0.1 M KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, and 0.05% Nonidet P-40 (Sigma).
In Vitro Protein Translation
The pGEM-3z vector (Promega) was engineered to have the T7 epitope coding sequence directly after the start codon. Coding sequences of AtTFIIB and AtTBP2 were cloned into the modified pGEM-3z vector to express the T7-tagged proteins in vitro. T7-AtTFIIB was expressed in a transcription/translation (TNT)–coupled wheat germ system, and T7-AtTBP was expressed in the rabbit reticulocyte TNT system (Promega) by using the manufacturer's protocol. The only modification was that TBP constructs were expressed at room temperature.
In Vitro GST Pull-Down Assay
To examine TBP–TFIIB interactions, we incubated T7-AtTBP2 with glutathione bead–immobilized GST-AtTFIIB (15 μg); for the reciprocal experiment, we incubated free T7-AtTFIIB with glutathione bead–immobilized GST-TBP (15 μg). The total amount of beads was kept the same for each binding reaction by adding buffer-washed glutathione beads alone whenever necessary. Binding reactions were promoted for 1 hr at 4°C in 300 μL of PBB containing 0.1% BSA in a continuously rotated tube, after which the beads were extensively washed with PBB (four times, 1 mL each). Bound protein was released by boiling in regular SDS loading buffer, resolved by SDS-PAGE, and detected with anti-T7 monoclonal antibody (Novagen, Madison, WI) used in conjunction with the enhanced chemiluminescence system (Pharmacia, Piscataway, NJ). Binding efficiency was quantified by the Scion Image analysis program (Scion Corp., Frederick, MD).
Acknowledgments
We thank Dr. Nam-Hai Chua for the AtTBP2 clone, Dr. Donald Baldwin for the AtTFIIB clone, Dr. James L. Manley for the ftzQ clone, Dr. Prem Chourey for the maize cell line, Dr. Karen Koch for the null promoter–GUS construct, and the DNA Sequencing Core of the Microbiology and Cell Science Department (University of Florida) for sequencing the TBP mutants. Research support was provided in part by NRICGP/USDA Grant No. 9801167. This article is Florida Agricultural Experiment Station journal series number R-07178.
References
- Baldwin, D.A., and Gurley, W.B. (1996). Isolation and characterization of cDNAs encoding transcription factor IIB from Arabidopsis and soybean. Plant J. 10 561–568. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.-H. (1990). Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 9 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, G.O., Martel, L.S., Burley, S.K., and Berk, A.J. (1996). Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 10 2491–2504. [DOI] [PubMed] [Google Scholar]
- Buratowski, S., and Zhou, H. (1993). Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 90 5633–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S.K., and Roeder, R.G. (1996). Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65 769–799. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S., and Struhl, K. (1995). Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature 374 820–822. [DOI] [PubMed] [Google Scholar]
- Chou, S., and Struhl, K. (1997). Transcriptional activation by TFIIB mutants that are severely impaired in interaction with promoter DNA and acidic activation domains. Mol. Cell. Biol. 17 6794–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey, P.S., and Zurawski, D.B. (1981). Callus formation from protoplasts of a maize cell culture. Theor. Appl. Genet. 59 341–344. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgen. Res. 5 213–218. [DOI] [PubMed] [Google Scholar]
- Colgàn, J., Wampler, S., and Manley, J.L. (1993). Interaction between a transcriptional activator and transcription factor IIB in vivo. Nature 362 549–553. [DOI] [PubMed] [Google Scholar]
- Colgàn, J., Ashali, H., and Manley, J.L. (1995). A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol. Cell. Biol. 15 2311–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens, D.J., Greaves, R., Goding, C.R., and O'Hare, P. (1989). The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 8 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, V.D., and Ingles, C.J. (1989). The Drosophila fushi tarazu polypeptide is a DNA-binding transcriptional activator in yeast cells. Nature 337 666–668. [DOI] [PubMed] [Google Scholar]
- Gasch, A., Hoffmann, A., Horikoshi, M., Roeder, R.G., and Chua, N.-H. (1990). Arabidopsis thaliana contains two genes for TFIID. Nature 346 390–394. [DOI] [PubMed] [Google Scholar]
- Geiger, J.H., Hahn, S., Lee, S., and Sigler, P.B. (1996). Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272 830–836. [DOI] [PubMed] [Google Scholar]
- Genschik, P., Marbach, J., Uze, M., Feuerman, M., Plesse, B., and Fleck, J. (1994). Structure and promoter activity of a stress and developmentally regulated polyubiquitin-encoding gene of Nicotiana tabacum. Gene 148 195–202. [DOI] [PubMed] [Google Scholar]
- Gill, G., and Ptashne, M. (1988). Negative effect of the transcriptional activator GAL4. Nature 334 721–724. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Couto, E., Klages, N., and Strubin, M. (1997). Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 94 8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J.A., Hoey, T., Thut, C.J., Admon, A., and Tjian, R. (1993). Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75 519–530. [DOI] [PubMed] [Google Scholar]
- Ha, I., Roberts, S., Maldonado, E., Sun, X., Kim, L.-U., Green, M., and Reinberg, D. (1993). Multiple functional domains of human transcription factor IIB: Distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7 1021–1032. [DOI] [PubMed] [Google Scholar]
- Haass, M.M., and Feix, G. (1992). Two different cDNAs encoding TFIID proteins of maize. FEBS Lett. 301 294–298. [DOI] [PubMed] [Google Scholar]
- Hisatake, K., Ohta, T., Takada, R., Guermah, M., Horikoshi, M., Nakatani, Y., and Roeder, R.G. (1995). Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc. Natl. Acad. Sci. USA 92 8195–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi, M., Wang, C.K., Fujii, H., Cromlish, J.A., Weil, P.A., and Roeder, R.G. (1989). Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature 341 299–303. [DOI] [PubMed] [Google Scholar]
- Huh, J.R., Park, J.M., Kim, M., Carlson, B.A., Hatfield, D.L., and Lee, B.J. (1999). Recruitment of TBP or TFIIB to a promoter proximal position leads to stimulation of RNA polymerase II transcription without activator proteins both in vivo and in vitro. Biochem. Biophys. Res. Commun. 256 45–51. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Lieberman, P.M., Schmidt, M.C., Zhou, Q., Pei, R., and Berk, A.J. (1990). Cloning of a transcriptionally active human TATA binding factor. Science 248 1646–1650. [DOI] [PubMed] [Google Scholar]
- Kim, J.L., and Burley, S.K. (1994). 1.9 Å resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat. Struct. Biol. 1 638–653. [DOI] [PubMed] [Google Scholar]
- Klages, N., and Strubin, M. (1995). Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature 374 822–823. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Lam, E., Benfey, P.N., Gilmartin, P.M., Fang, R.X., and Chua, N.-H. (1989). Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl. Acad. Sci. USA 86 7890–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last, D.I., Brettell, R.I.S., Chamberlain, D.A., Chaudhury, A.M., Larkin, P.J., Marsh, E.L., Peacock, W.J., and Dennis, E.S. (1991). pEmu: An improved promoter for gene expression in cereal cells. Theor. Appl. Genet. 81 581–588. [DOI] [PubMed] [Google Scholar]
- Lee, M., and Struhl, K. (1997). A severely defective TATA-binding protein–TFIIB interaction does not preclude transcriptional activation in vivo. Mol. Cell. Biol. 17 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.F., Le Gourierrec, J., Torki, M., Kim, Y.J., Guerineau, F., and Zhou, D.X. (1999). Characterization and functional analysis of Arabidopsis TFIIA reveal that the evolutionarily unconserved region of the large subunit has a transcription activation domain. Plant Mol. Biol. 39 515–525. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Ptashne, M. (1987). Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48 847–853. [DOI] [PubMed] [Google Scholar]
- Majello, B., Napolitano, G., De Luca, P., and Lania, L. (1998). Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J. Biol. Chem. 273 16509–16516. [DOI] [PubMed] [Google Scholar]
- Marmorstein, R., Carey, M., Ptashne, M., and Harrison, S.C. (1992). DNA recognition by GAL4: Structure of a protein–DNA complex. Nature 356 408–414. [DOI] [PubMed] [Google Scholar]
- Muhich, M.L., Iida, C.T., Horikoshi, M., Roeder, R.G., and Parker, C.S. (1990). cDNA clone encoding Drosophila transcription factor TFIID. Proc. Natl. Acad. Sci. USA 87 9148–9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Myer, V.E., and Young, R.A. (1998). RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273 27757–27760. [DOI] [PubMed] [Google Scholar]
- Nikolov, D.B., Chen, H., Halay, E.D., Usheva, A.A., Hisatake, K., Lee, D.K., Roeder, R.G., and Burley, S.K. (1995). Crystal structure of a TFIIB-TBP-TATA element ternary complex. Nature 377 119–128. [DOI] [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812. [DOI] [PubMed] [Google Scholar]
- Pan, S., Sehnke, P.C., Ferl, R.J., and Gurley, W.B. (1999). Specific interactions with TBP and TFIIB in vitro suggest 14-3-3 proteins may participate in transcriptional regulation when part of a DNA binding complex. Plant Cell 11 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert, S., and Tjian, R. (1995). Human TAFII250 interacts with RAP74: Implications for RNA polymerase II initiation. Genes Dev. 9 2747–2755. [DOI] [PubMed] [Google Scholar]
- Strubin, M., and Struhl, K. (1992). Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68 721–730. [DOI] [PubMed] [Google Scholar]
- Tan, S., Hunziker, Y., Sargent, D.F., and Richmond, T.J. (1996). Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381 127–151. [DOI] [PubMed] [Google Scholar]
- Tang, H., Sun, X., Reinberg, D., and Ebright, R.H. (1996). Protein–protein interactions in eukaryotic transcription initiation: Structure of the preinitiation complex. Proc. Natl. Acad. Sci. USA 93 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey, W.P., and Herr, W. (1997). Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science 275 829–831. [DOI] [PubMed] [Google Scholar]
- Usheva, A., Maldonado, E., Goldring, A., Lu, H., Houbavi, C., Reinberg, D., and Aloni, Y. (1992). Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell 69 871–881. [DOI] [PubMed] [Google Scholar]
- Wilmink, A., van de Ven, B.C., and Dons, J.J. (1995). Activity of constitutive promoters in various species from the Liliaceae. Plant Mol. Biol. 28 949–955. [DOI] [PubMed] [Google Scholar]
- Xiao, H., Friesen, J.D., and Lis, J.T. (1995). Recruiting TATA-binding protein to a promoter: Transcriptional activation without an upstream activator. Mol. Cell. Biol. 15 5757–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., Lis, J.T., and Jeang, K.T. (1997). Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol. 17 6898–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]