Abstract
Temperature dramatically affects plant–virus interactions. Outbreaks of virus diseases are frequently associated with low temperature, while at high temperature viral symptoms are often attenuated (heat masking) and plants rapidly recover from virus diseases. However, the underlying mechanisms of these well-known observations are not yet understood. RNA silencing is a conserved defence system of eukaryotic cells, which operates against molecular parasites including viruses and transgenes. Here we show that at low temperature both virus and transgene triggered RNA silencing are inhibited. Therefore, in cold, plants become more susceptible to viruses, and RNA silencing-based phenotypes of transgenic plants are lost. Consistently, the levels of virus- and transgene-derived small (21–26 nucleotide) interfering (si) RNAs—the central molecules of RNA silencing-mediated defence pathways—are dramatically reduced at low temperature. In contrast, RNA silencing was activated and the amount of siRNAs gradually increased with rising temperature. However, temperature does not influence the accumulation of micro (mi) RNAs, which play a role in developmental regulation, suggesting that the two classes of small (si and mi) RNAs are generated by different nuclease complexes.
Keywords: heat masking/miRNA/RNA silencing/siRNA/temperature dependence
Introduction
RNA silencing is a sequence-specific post-transcriptional, gene-inactivation mechanism that operates in diverse eukaryotes. Accumulation of short (21–26 nucleotide), complementary guide RNAs corresponding to the sequence of target RNA occurs during RNA silencing in plants and animals (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Parrish et al., 2000). These complementary guide RNAs provide the sequence-specificity of target RNA silencing. The double-stranded (ds) RNA-specific enzyme, DICER, produces two types of guide RNAs, small interfering (si) RNAs and micro (mi) RNAs. siRNAs are ds molecules generated from long dsRNAs and mediate defensive responses against molecular parasites [referred to as post-transcriptional gene silencing (PTGS) in plants and RNA interference (RNAi) in animals] (reviewed in Voinnet, 2001; Zamore, 2001; Hannon, 2002). miRNAs are single-stranded molecules excised by DICER from short hairpin precursors and miRNAs are hypothesized to play a critical role in both animal and plant developmental regulation (Grishok et al., 2001; Hutvagner et al., 2001; Llave et al., 2002; Reinhart et al., 2002). Both siRNAs and miRNAs incorporate into a multicomponent nuclease complex (RISC) and guide RISC to target homologous mRNAs for cleavage at near perfect base-pairing (Elbashir et al., 2001; Hutvagner and Zamore, 2002), or for translational repression at partial complementarity (Reinhart et al., 2000). In plants, siRNAs might also act as the specificity determinants for other complexes, including dsRNA amplification by RNA-dependent RNA polymerase (RdRP), systemic silencing (see below) and RNA-directed methylation of homologous DNA (reviewed in Waterhouse et al., 2001; Mlotshwa et al., 2002).
A good deal of evidence suggests that RNA silencing plays a natural role in plant defense against molecular parasites, including viruses (reviewed in Voinnet, 2001). Virus-induced RNA silencing operates at both single-cell and whole-plant levels. Accumulation of dsRNA intermediates of virus replication in a plant cell induces silencing activity, which degrades the dsRNA itself and the cognate mRNAs (referred to as cell-autonomous silencing). Cell-autonomous silencing also generates a mobile signal that triggers the degradation of homologous mRNAs in distant cells (systemic silencing) (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). Consistent with the antiviral nature of RNA silencing in plants, many plant viruses have evolved silencing-suppressor proteins that block cell-autonomous or systemic silencing (Li and Ding, 2001). Transgene expression can also trigger RNA silencing. Virus- and transgene-induced silencing pathways are similar except that transgene-derived dsRNAs are likely generated by a plant encoded RdRP from aberrant transcripts. Transgene-induced RNA silencing is widely used in plant biotechnology. Phenotypes of virus-resistant plants expressing virus-derived sequences and transgenic plants, in which an endogenous gene is inactivated by ectopic expression of sense or antisense RNAs, are based on transgene-induced RNA silencing (Di Serio et al., 2001; Rovere et al., 2002).
Plant–virus interactions are strongly modified by environmental factors; especially by temperature. High temperature is frequently associated with attenuated symptoms (‘heat masking’) and with low virus content of virus-infected plants (Johnson, 1922; Hull, 2002). In contrast, rapid spread of virus diseases and the development of severe symptoms are frequently associated with cold air temperature (Hine et al., 1970; Gerik et al., 1990). Harrison (1956) has speculated that the virus content of a plant represents an equilibrium between replication and degradation and that the activity of the virus degrading system increases with temperature. However, the underlying molecular mechanisms of these effects of temperature are still missing. We hypothesized that RNA silencing might be the postulated virus degradation system and temperature modifies plant–virus interactions through the regulation of RNA silencing.
Here we report that RNA silencing-mediated plant defence is temperature dependent. At low temperature, both virus- and transgene-induced RNA silencing is inhibited leading to enhanced virus susceptibility and to loss of silencing-mediated transgenic phenotypes. Furthermore, at low temperature, the level of virus- or transgene-derived siRNAs is dramatically reduced. In contrast, RNA silencing is activated and the amount of siRNAs gradually increases with temperature rising. However, accumulation of miRNAs, which play a critical role in developmental regulation, is temperature independent, suggesting that the two classes of small (si and mi) RNAs are generated by different nuclease complexes in plants.
Results and discussion
Temperature regulates RNA silencing-based antiviral defence by the control of siRNA generation
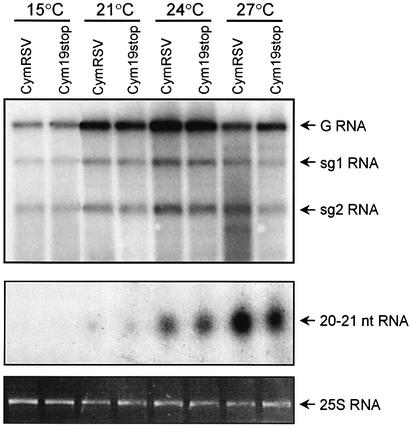
To test whether RNA silencing might correspond to the suggested temperature-dependent virus degradation system (Harrison, 1956), we analysed the temperature dependence of virus-induced RNA silencing at both cellular and whole-plant levels. To define the role of temperature in virus-induced cell-autonomous silencing, accumulation of virus-specific siRNAs was analysed in Nicotiana benthamiana protoplasts transfected with Cymbidium ringspot virus (CymRSV) at different temperatures (15, 21, 24 and 27°C). Cym19stop mutant virus, which is unable to express CymRSV encoded p19 silencing suppressor (Szittya et al., 2002), was also used for protoplasts transfection in order to avoid the effect of silencing suppression. Viral RNA and siRNA levels were comparable in Cym19stop- and CymRSV-infected protoplasts at each temperature (Figure 1) confirming that p19 does not affect virus-induced local silencing (Silhavy et al., 2002). Striking differences were found, however, in virus-derived siRNA accumulation at different temperatures; siRNAs were abundant at 27°C, but undetectable at 15°C. The amount of siRNAs gradually increased with temperature from 21 to 27°C (Figure 1). These findings indicate that virus-induced cell-autonomous silencing is temperature dependent, and suggest that at low temperature the silencing pathway is inhibited at or before the production of siRNAs. Since virus-specific siRNAs mainly derive from the dsRNA intermediates of viral replication (Voinnet, 2001), and since CymRSV still replicates efficiently at 15°C (Figure 1), it is likely that DICER activity is compromised at low temperature.
Fig. 1. Effect of temperature on virus-induced cell-autonomous RNA silencing. RNA gel blot analysis of RNA samples extracted from N.benthamiana protoplasts infected with in vitro transcript of CymRSV and Cym19stop. Transfected protoplasts were incubated for 24 h under a 14 h light and 10 h dark regime at different temperatures. G shows genomic RNA, while sg1 and sg2 refer to subgenomic 1 and subgenomic 2 RNAs, respectively. The size of siRNAs were estimated using labelled 21 nucleotide synthetic RNAs, and 25S RNA was applied as loading control.
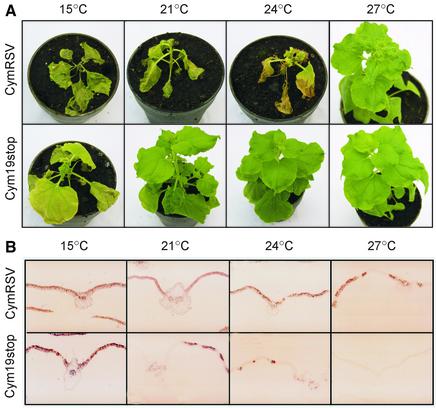
Previously, we showed that the CymRSV-encoded p19 acts as a viral suppressor of systemic silencing, thus Cym19stop-infected plants recover from viral infection at standard temperature (21°C; Figure 2A) (Silhavy et al., 2002). To test the effect of temperature on virus-induced systemic RNA silencing, N.benthamiana plants were inoculated with CymRSV and Cym19stop viruses and grown at different temperatures. As expected, CymRSV-infected plants died within 2 weeks at 15, 21 and 24°C (Figure 2A). Consistent with previous results (Gerik et al., 1990), we found that CymRSV symptoms were ‘heat masked’ at 27°C (Figure 2A) and that the attenuated symptoms were associated with reduced virus level (Figure 2B). Plants infected with the Cym19stop showed the expected recovery phenotype at 21 and 24°C (Szittya et al., 2002). Strikingly, at 27°C, the mutant virus was unable to infect the plants (Figure 2), while at 15°C, Cym19stop-infected plants displayed strong viral symptoms (Figure 2A). This latter result demonstrated that at low temperature, RNA silencing failed to protect the plants even when the virus lacked a silencing suppressor. It is believed that in many incompatible plant–virus interactions, plants are protected by RNA silencing-mediated defence (Voinnet, 2001). However, at low temperature, these interactions might become compatible and allow viruses to spread and accumulate in plants.
Fig. 2. Effect of temperature on symptom development and virus spreading of virus-infected plants. Virus-infected N.benthamiana plants were grown at the indicated temperatures. (A) Symptoms of CymRSV- and Cym19stop-infected plants at 14 dpi. (B) Spread of CymRSV and Cym19stop in the first systemically-infected leaves at 7 dpi. Digoxigenin-11-UTP-labelled RNA probe complementary to the last 900 nucleotides of CymRSV was hybridized to tissue sections and detected with alkaline phophatase-conjugated anti-digoxygenin antibody.
In situ hybridization analysis revealed (Z.Havelda, unpublished data) that at 24°C, CymRSV and Cym19stop viruses accumulated to similar levels in the infected cells of the first non-inoculated leaves that showed viral symptoms (systemically-infected leaves). However, the spread of CymRSV and Cym19stop were dramatically different. CymRSV invaded the whole systemically-infected leaf, while Cym19stop was confined close to the veins, presumably due to systemic silencing. To monitor systemic silencing at different temperatures, we compared the virus extent in the first systemically-infected leaves of both CymRSV- and Cym19stop-inoculated plants. As Figure 2B shows, at 21°C as well as at 24°C, the spread of Cym19stop was blocked whereas CymRSV invaded the whole leaf. By contrast, at low temperature (15°C), both mutant and wild-type viruses invaded the entire first systemically-infected leaves. At 27°C, no virus accumulation was detected in systemic leaves of Cym19stop-inoculated plants. Moreover, RNA gel blot analyses failed to detect Cym19stop RNA in the inoculated leaves (data not shown), presumably because the mutant virus was restricted in the initially-infected cells. The spread of wild-type CymRSV was also limited at 27°C (Figure 2B), suggesting that a hyper-activated systemic silencing can protect plants even from highly invasive viruses.
Thus, virus-induced RNA silencing is a temperature-dependent RNA degradation mechanism that operates inefficiently at low temperature and whose activity increases with temperature. Since the lack of RNA silencing enhances plant susceptibility to viral infection (Mourrain et al., 2000; Dalmay et al., 2001), we suggest that the frequent outbreaks of plant virus diseases in unusually cold seasons (Hine et al., 1970; Gerik et al., 1990) reflect reduced RNA silencing capacity, while heat masking is due to an increased RNA silencing activity. Moreover, the efficiency of heat therapy (growing plants at 35–40°C to free from viruses) could also be explained as reflecting a hyper-activated RNA silencing activity.
Low temperature inhibits transgene-induced RNA silencing
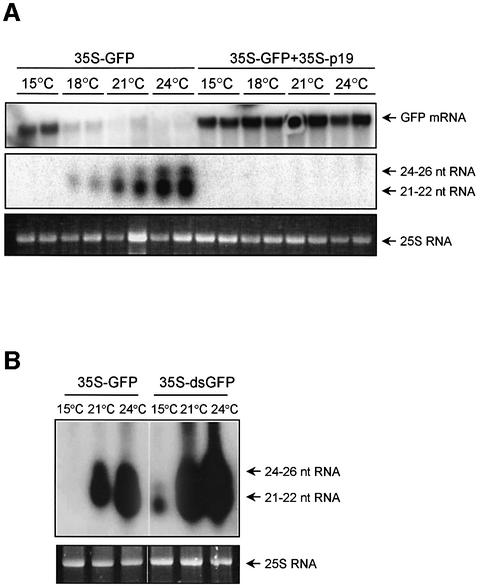
In plants, transgene- and virus-induced RNA silencing pathways are overlapping but not identical (Voinnet, 2001). To test whether transgene-induced silencing is also temperature dependent, Agrobacterium infiltration (agroinfiltration) assays were carried out at different temperatures. Infiltration of non-transgenic N.bentha miana leaves with a strain of Agrobacterium tumefaciens carrying a GFP gene construct (35S-GFP) results in transient GFP expression but also leads to induction of GFP RNA silencing (Johansen and Carrington, 2001; Silhavy et al., 2002). Decline in GFP mRNA levels and accumulation of GFP-specific siRNAs in the infiltrated patches reflect the intensity of transgene-induced cell-autonomous RNA silencing. As Figure 3A shows, at 3 days post-infiltration (dpi), GFP mRNA levels were high at 15°C and decreased with the increase of temperature. To clarify whether the temperature-dependent GFP mRNA levels reflect the different silencing or different transcriptional activity, 35S-GFP was co-infiltrated with p19 (35S-p19), a potent inhibitor of transgene-induced RNA silencing (Silhavy et al., 2002), and the infiltrated plants were incubated at different temperatures. Since in the lack of RNA silencing GFP mRNA levels were equally high at any temperature (Figure 3A), we concluded that temperature-dependent GFP mRNA accumulation reflected the different silencing activities. Consistently, GFP-specific siRNAs did not accumulate at 15°C, but were easily detectable at 18°C, and siRNA levels increased gradually with temperature (Figure 3A). We also repeated the infiltration experiment using GFP-expressing N.benthamiana plants (Voinnet et al., 1998). Similarly to the results obtained by agroinfiltration of wild-type, non-trangenic plants, GFP mRNAs accumulated to a high level, while siRNAs were not detectable in the infiltrated patches at 15°C. However, with the temperature rising, siRNA levels gradually increased displaying correlation with the GFP mRNAs degradation. (see Supplementary figure, available at The EMBO Journal Online). RNA silencing induced by agroinfiltration leads to accumulation of two distinct siRNA size classes, a 21–22 nucleotide short and a 24–26 nucleotide long fraction. We found that levels of short and long siRNAs increased simultaneously with temperature. Short siRNAs might mediate mRNA degradation, while long siRNAs are supposed to play a role in systemic silencing and methylation of homologous DNA (Hamilton et al., 2002). To investigate whether processing of dsRNA to siRNAs in transgene-induced RNA silencing is temperature dependent, plants were infiltrated with an Agrobacterium carrying an inverted repeat of the GFP sequence thereby expressing dsGFP RNA (35S-dsGFP). Since cleavage of dsRNAs into siRNAs does not require RdRP activity (Beclin et al., 2002), low siRNA levels in 35S-dsGFP-infiltrated plants at 15°C (Figure 3B), suggest that cleavage of dsRNA to siRNAs is temperature dependent. However, we still do not know whether RdRP-mediated dsRNA generation is temperature dependent or not. An unlikely, alternative explanation for low siRNA accumulation is that siRNA stability is greatly reduced at low temperature (discussed later).
Fig. 3. Effect of temperature on transgene-induced RNA silencing. Non-transgenic N.benthamiana leaves were infiltrated with an Agrobacterium expressing 35S-GFP (A and B, left panels) or 35S-dsGFP (B, right panel), thereby inducing GFP silencing, or co-infiltrated with 35S-GFP and 35S-p19 silencing suppressor (A). Plants were further grown at different temperatures as indicated. RNA gel blot analysis was carried out to monitor the accumulation of GFP mRNA and GFP-specific siRNAs at 3 dpi. Total RNAs were isolated from infiltrated plants grown at the indicated temperatures under a 14 h light and 10 h dark regime. 25S ribosomal RNA was applied as loading control.
RNA silencing-based transgenic phenotypes are lost at low temperature
RNA silencing has enormous potential in plant biotechnology to confer protection against viruses and to inactivate endogenous genes. Since both virus- and transgene-induced RNA silencings are temperature dependent, we tested the stability of RNA silencing-mediated transgenic phenotypes at different temperatures.
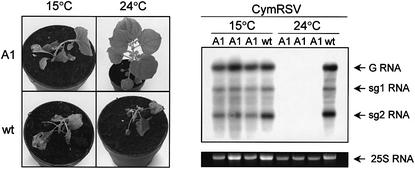
To examine the effect of temperature on engineered virus resistance, transgenic N.benthamiana plants expressing a CymRSV-derived RNA (92KA1; Rubino and Russo, 1995) were inoculated with CymRSV and incubated at different temperatures. We confirmed that transgenic plants were resistant at 24°C and that the resistance was based on RNA silencing (Rubino and Russo, 1995; data not shown). However, at 15°C, strong symptoms developed and CymRSV RNA accumulated to a high level in the inoculated plants (Figure 4), demonstrating that the transgene-mediated virus resistance was broken at low temperature. Our results predict previously unanticipated failure in virus resistance at lower temperature for transgenic crops with RNA silencing-mediated virus resistance.
Fig. 4. Effect of temperature on RNA silencing-mediated transgenic virus resistance. CymRSV 92KA1 N.benthamiana transgenic plants (A1) expressing a CymRSV RNA segment, which confers CymRSV resistance, and control non-transgenic plants (wt) were infected with CymRSV transcript. Virus-infected plants were incubated at the indicated temperatures. Symptoms (left panel) and viral RNA levels (right panel) of infected plants at 10 dpi. G shows genomic RNA, while sg1 and sg2 refer to subgenomic 1 and subgenomic 2 RNAs, respectively. 25S ribosomal RNA was applied as loading control.
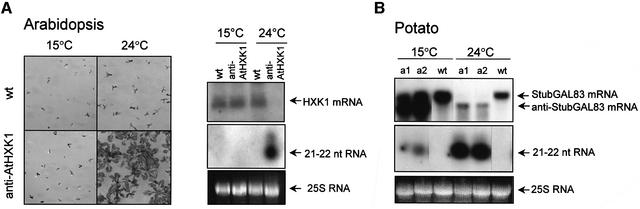
The effect of temperature on antisense-mediated endogen gene inactivation was examined in the model species Arabidopsis and in the economically important potato, in which antisense inhibition of genes involved in carbohydrate metabolism is widely applied. Arabidopsis plants (anti-AtHXK1) expressing antisense hexokinase (HXK1) RNAs are sugar hyposensitive, allowing anti-AtHXK1 plants to grow at limiting glucose (6%) concentration (Jang et al., 1997). Seedlings of anti-AtHXK1 and wild-type plants were grown on agar plates containing 6% glucose at 15°C and 24°C. As expected, at 24°C development of wild-type plants was inhibited, while anti-AtHXK1 plants developed normally (Figure 5A). Accumulation of transgene-derived siRNAs in anti-AtHXK1 plants at 24°C indicated that HXK1 mRNAs were inactivated by RNA silencing (Figure 5A). At 15°C, the development of both wild-type and anti-AtHXK1 plants was inhibited, showing that the antisense inhibition was not effective at low temperature. Consistently, transgene-derived siRNAs could not be detected in anti-AtHXK1 plants at 15°C (Figure 5A).
Fig. 5. Effect of temperature on antisense RNA-mediated inhibition of endogenous genes. Seedlings of transgenic Arabidopsis (anti-AtHXK1) expressing antisense RNA for hexokinase (HXK1) and wild-type plants were grown for 3 weeks on agar plates containing 6% glucose at 15°C and 24°C (A, left panel). Accumulation of mRNAs and specific siRNAs in transgenic anti-AtHXK1 expressing antisense RNA for hexokinase (HXK1) mRNA (A, right panel), and in two transgenic potato lines (aG1 and aG2) expressing antisense RNA (anti-StubGal83 RNA) for Gal83 mRNA (StubGal83 RNA) (B). Plants were incubated at 15°C and 24°C. Northern blots were hybridized with a strand-specific probe to HXK1 mRNA and ds StubGAL83 cDNA, respectively. 25S ribosomal RNA was applied as loading control.
Transgenic potato plants displaying antisense inhibition of StubGAL83 mRNAs (A.Lovas, unpublished data) were also analysed at different temperatures. At 24°C, high levels of StubGAL83-specific siRNA accumulation were associated with extremely low StubGAL83 mRNA levels. In contrast, at 15°C, StubGAL83 mRNA accumulated to a high level and transgene-derived siRNAs were barely detectable (Figure 5B). These observations demonstrate that StubGAL83 inactivation is mediated by RNA silencing in antisense expressing plants, and that this RNA silencing is compromised at low temperature in potato.
Since RNA silencing-mediated transgenic phenotypes could be lost at low temperature, biotechnological application of these transgenic plants has a considerable environmental limit. This constrain could be especially important for perennials and less limiting for annual crops. Inefficient RNA silencing activity at low temperature might be useful to express foreign proteins to a high level in transgenic plants. Moreover, inefficient RNA silencing activity at low temperature could also be utilized in RNA silencing-based genetic screens, since silencing-mediated early lethal phenotypes could be rescued at low temperature. Indeed, in plants, RNA silencing-based male sterility was rescued by lower temperature (Mou et al., 2002).
Accumulation of miRNAs are not affected by temperature
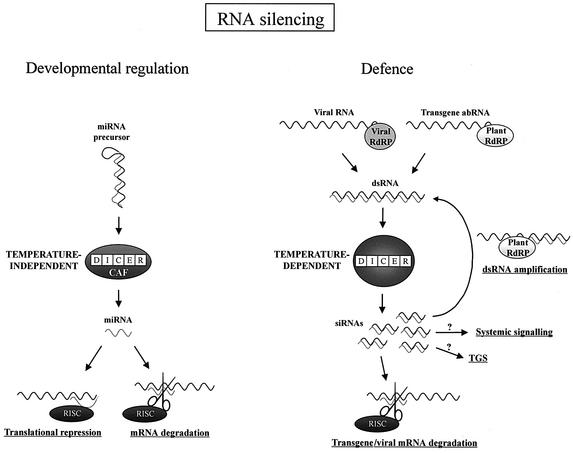
In animals, DICER is required for both siRNA and miRNA production (Grishok et al., 2001; Hutvagner et al., 2001). The DICER homologue, CARPEL FACTORY is required for miRNA generation in plants (Reinhart et al., 2002). Since siRNA accumulation is temperature dependent in plants, we tested whether miRNA accumulation is also controlled by temperature. In contrast to siRNA production, miR157, miR169 and miR171 RNAs accumulated to equal levels at 15, 21 and 24°C in Arabidopsis (Figure 6), probably explaining why the plants developed normally at all three temperatures. These findings suggest that plant siRNAs and miRNAs are generated by different pathways, and probably by distinct DICER-like complexes. At least four DICER-related genes can be identified in the Arabidopsis genome (Reinhart et al., 2002); it is conceivable that a temperature-dependent DICER homologue plays role in siRNA generation and a temperature-independent DICER-related gene is involved in miRNA production. Alternatively, any component of the DICER complex that is specifically required for siRNA generation might be temperature dependent (Tabara et al., 2002). Both siRNAs and miRNAs incorporate into the same RISC complex (Elbashir et al., 2001), therefore we do not favour the alternative explanation that siRNAs are degraded at low temperature while miRNAs are not. Since plant siRNAs and miRNAs might act in an identical silencing pathway by guiding RISC to cleave target genes (Rhoades et al., 2002), the temperature-dependent antiviral role and the temperature-independent developmental regulatory function of RNA silencing machinery may be separated at their respective small RNA-generating steps (Figure 7).
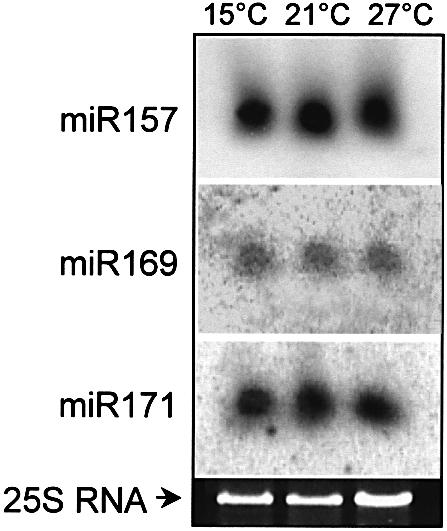
Fig. 6. miRNA accumulation in Arabidopsis. Accumulations of miRNAs were analysed in Arabidopsis seedling grown in vitro at different temperatures. RNA gel blots were hybridized at 37°C using the 5′-labelled anti-sense DNA oligos corresponding to the indicated miRNAs. 25S ribosomal RNA was applied as loading control.
Fig. 7. Model for RNA silencing pathways in plants. RNA silencing is branched into temperature-independent regulatory and temperature-dependent defensive pathways. siRNA generation is temperature dependent, thus RNA silencing-based defence responses are affected by temperature. Since miRNA production is temperature independent, developmental regulation is not affected by low temperature.
Conclusions
We find that siRNA-mediated RNA silencing is temperature dependent in three dicot species, thus inefficient siRNA generation at low temperature is probably a universal feature of higher plants. We speculate that separation of siRNA and miRNA production in plants is a part of a general adaptive response. We suggest a model (Figure 7) in which, at low temperature and perhaps under other unfavourable conditions, the non-essential RNA silencing machinery is inactivated, but miRNA-mediated essential regulatory functions continue to operate, ensuring normal plant growth. As a cost, plants are more susceptible to viruses and to other molecular parasites at low temperature. In animals and Chlamydomonas, RNA silencing-mediated defence pathways are involved in inactivation of mobile elements. In plants, siRNAs are supposed to provide sequence specificity for a complex that mediates transcriptional gene silencing (TGS) by directly methylating homologous DNA segments (Mette et al., 2001; Hamilton et al., 2002) or initiating DNA methylation by imposing histone H3 lysine-9 methylation (Gendrel et al., 2002; Volpe et al., 2002). At low temperature, siRNA-mediated methylation might decrease, leading to the re-expression of transcriptionally silenced genes. Consistently, the general level of DNA methylation is reduced in plants at low temperature and TGS is less efficient (Finnegan et al., 1998). Furthermore, the Antirrhinum Tam3 transposon is excised at 15°C, but not at 25°C, and transposition correlates with reduced methylation status of mobile element (Kitamura et al., 2001). We can also speculate that inefficient siRNA generation at low temperature might be involved in certain developmental regulation steps, for instance, in vernalization cold results in demethylation of regulatory genes (Reeves and Coupland, 2000).
Materials and methods
Plant materials
Wild-type N.benthamiana plants grown in soil under normal growth conditions were used for protoplast preparation, virus infection and agroinfiltration. For temperature treatment, after infection or infiltration, plants were grown in fitotron (Versatile Enviromental Test Chambers; Sanyo, Tokyo Japan) under a 14 h light (50 µE m–2s–1) and 10 h dark regime at different temperatures. The 92KA1 transgenic N.benthamiana plants were grown as described previously (Rubino et al., 1993). For temperature treatment, after infection with CymRSV, 92KA1 transgenic plants were grown in fitotrons under a 14 h light (50 µE m–2s–1) and 10 h dark regime at 15 and 24°C, respectively. For temperature treatment, the AtHXK1 transgenic Arabidopsis (Jang et al., 1997) and the control (wild-type) lines were germinated and grown in fitotrons under constant white light (100 µE m–2s–1) for 3 weeks on half-strength Murashige and Skoog plates containing 6% glucose at 15 and 24°C, respectively. For temperature treatment, stem segments from the anti-StubGAL83 transgenic potato and the control (wild type) lines were grown in fitotrons under a 14 h light (50 µE m–2s–1) and 10 h dark regime for 2 months on RM medium at 15 and 24°C, respectively. To determine the miRNA level, Arabidopsis plants were germinated and grown in fitotrons under a 14 h light (100 µE m–2s–1) and 10 h dark regime for 3 weeks on half-strength Murashige and Skoog plates at 15 and 24°C, respectively.
Agrobacterium tumefaciens infiltration
The A.tumefaciens infiltration method was carried out according to Voinnet et al. (1998) except that both non-transgenic and GFP-expressing transgenic plants were used. For co-infiltration, an equal volume of indicated A.tumefaciens cultures (OD600 = 1) were mixed before infiltration. The RNA silencing suppression assay was carried out as described previously by Silhavy et al. (2002). Visual detection of GFP fluorescence was performed using a 100 W hand-held long-wave ultraviolet lamp (UV products, Upland, CA 91786, Black Ray model B 100AP).
Plasmid constructs
The infectious cDNA clones of CymRSV (Dalmay et al., 1993), Cym19stop mutant (Szittya et al., 2002) and 35S-GFP plasmid were described previously (Silhavy et al., 2002). Primers corresponding to the first 20 nucleotides of 5′ and 3′ ends of GFP were used to amplify GFP from 35S-GFP, and cloned to EcoRV-digested Bluescript SK(+) creating SK-GFP. SK-GFP was digested with HpaI and SmaI, dephosphorylated and then a full-length GFP PCR product was cloned into this vector in inverted orientation to construct SK-dsGFP. KpnI and BamHI fragments were cloned from SK-dsGFP to Bin61S to create 35S-dsGFP.
Protoplast preparation and inoculation
Protoplasts were isolated from N.benthamiana plants and transfected with in vitro-synthesized transcripts of genomic RNAs using the polyethylene glycol method as described previously (Dalmay et al., 1993). After infection protoplasts were incubated in Versatile Enviromental Test Chambers (Sanyo, Tokyo Japan) under a 14 h light (50 µE m–2s–1) and 10 h dark regime at different temperatures.
In vitro RNA transcription and plant inoculation
In vitro transcription of CymRSV and Cym19stop, and inoculation of RNA transcripts onto N.benthamiana plants were performed as described previously (Dalmay et al., 1993).
RNA gel blot analysis
Total RNA extraction and RNA gel blot analysis of high and low molecular weight RNAs was carried out as described previously (Szittya et al., 2002). PCR fragments labelled with random priming method were used for northern analyses of high molecular weight RNAs. Radioactively labelled in vitro transcripts corresponding to the negative strand of virus RNA and antisense strand of GFP were used as probes for northern analyses of low molecular weight RNAs. For miRNA detection, 5′-labelled antisense DNA oligonucleotides corresponding to the analysed miRNAs were used as probes. Labellings were carried out according to Sambrook et al. (1989).
In situ RNA hybridization
Upper leaves of systemically infected plants were fixed and paraffin embedded according to Jackson (1992). Hybridization probe to detect target RNA by in situ hybridization was prepared as described previously (Aranda et al., 1996). Negative-sense RNA probe corresponding to the last 900 nucleotides of viral genomic RNA was prepared after linearizing the pBluescript II vector containing the target sequence with SmaI and transcribing with T7 RNA polymerase. Digoxigenin-11-UTP-labelled probe was hybridized to leaf cross-sections of 10 µm thickness and detected with alakaline phosphatase-conjugated anti-digoxigenin antibody, as described previously (Wang and Maule, 1995).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Phillip D.Zamore for critical reading of manuscript. We are grateful to David Baulcombe for comments on the manuscript and for kindly providing A.tumefaciens carrying 35S-GFP construct. We thank Marcello Russo for providing seeds of 92KA1 plants. We also thank Jyan-Chyun Jang for providing seeds of anti-AtHXK1 plants. This research was supported by grants from the Hungarian Ministry of Education (NKFP 4/023/2001) and ‘VIS’ EU project (QLG2-CT-2002-01673).
Note added in proof
During the review process of this paper, we were informed that temperature-dependent accumulation of siRNAs derived from inverted repeat transgenes was observed and reported by Kalantidis et al. (Mol. Plant Microbe Interact., 2002, 8, 826–833).
References
- Aranda M.A., Escaler,M., Wang,D. and Maule,A.J. (1996) Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl Acad. Sci. USA, 93, 15289–15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beclin C., Boutet,S., Waterhouse,P. and Vaucheret,H. (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol., 12, 684–688. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Rubino,L., Burgyan,J., Kollar,A. and Russo,M. (1993) Functional analysis of cymbidium ringspot virus genome. Virology, 194, 697–704. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Horsefield,R., Braunstein,T.H. and Baulcombe,D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Serio F., Schob,H., Iglesias,A., Tarina,C., Bouldoires,E. and Meins,F.,Jr (2001) Sense- and antisense-mediated gene silencing in tobacco is inhibited by the same viral suppressors and is associated with accumulation of small RNAs. Proc. Natl Acad. Sci. USA, 98, 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E.J., Genger,R.K., Peacock,W.J. and Dennis,E.S. (1998) DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 223–247. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman,Z., Yordan,C., Colot,V. and Martienssen,R.A. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene ddm1. Science, 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gerik J.S., Duffus,J.E., Perry,R., Stenger,D.C. and Van Maren,A.F. (1990) Etiology of tomato plant decline in the California desert. Phytopathology, 80, 1352–1356. [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton A., Voinnet,O., Chappell,L. and Baulcombe,D. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J., 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Harrison B.D. (1956) Studies on the effect of temperature on virus multiplication in inoculated leaves. Annu. Appl. Biol., 44, 215–226. [Google Scholar]
- Hine R.B., Osborne,W.E. and Dennis,R.E. (1970) Elevation and temperature effects on severity of maize dwarf mosaic virus in sorghum in Arizona. Plant Dis. Rep., 54, 1064–1068. [Google Scholar]
- Hull R. (2002) Matthews’ Plant Virology. Academic Press, San Diego, CA.
- Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Jackson D.P. (1992) In situ hybridisation in plants. Molecular Plant Pathology: A Practical Approach. Oxford University Press, Oxford, UK, 163–174.
- Jang J.C., Leon,P., Zhou,L. and Sheen,J. (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell, 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.K. and Carrington,J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the agrobacterium-mediated transient expression system. Plant Physiol., 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. (1922) The relation of air temperature to the mosaic disease of potatoes and other plants. Phytopathology, 12, 438–440. [Google Scholar]
- Kitamura K., Hashida,S.N., Mikami,T. and Kishima,Y. (2001) Position effect of the excision frequency of the Antirrhinum transposon Tam3: implications for the degree of position-dependent methylation in the ends of the element. Plant Mol. Biol., 47, 475–490. [DOI] [PubMed] [Google Scholar]
- Li W.X. and Ding,S.W. (2001) Viral suppressors of RNA silencing. Curr. Opin. Biotechnol., 12, 150–154. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau,K.D., Rector,M.A. and Carrington,J.C. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M.F., Matzke,A.J. and Matzke,M.A. (2001) Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr. Biol., 11, 1119–1123. [DOI] [PubMed] [Google Scholar]
- Mlotshwa S., Voinnet,O., Mette,M.F., Matzke,M., Vaucheret,H., Ding,S.W., Pruss,G. and Vance,V.B. (2002) RNA silencing and the mobile silencing signal. Plant Cell, 14, S289–S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Wang,X., Fu,Z., Dai,Y., Han,C., Ouyang,J., Bao,F., Hu,Y. and Li,J. (2002) Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell, 14, 2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan,T., Pollien,J.M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Fleenor,J., Xu,S., Mello,C. and Fire,A. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell, 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Reeves P.H. and Coupland,G. (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr. Opin. Plant Biol., 3, 37–42. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein,E.G., Rhoades,M.W., Bartel,B. and Bartel,D.P. (2002) MicroRNAs in plants. Genes Dev., 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M., Reinhart,B., Lim,L., Burge,C., Bartel,B. and Bartel,D. (2002) Prediction of plant microRNA targets. Cell, 110, 513. [DOI] [PubMed] [Google Scholar]
- Rovere C.V., del Vas,M. and Hopp,H.E. (2002) RNA-mediated virus resistance. Curr. Opin. Biotechnol., 13, 167–172. [DOI] [PubMed] [Google Scholar]
- Rubino L. and Russo,M. (1995) Characterization of resistance to cymbidium ringspot virus in transgenic plants expressing a full-length viral replicase gene. Virology, 212, 240–243. [DOI] [PubMed] [Google Scholar]
- Rubino L., Capriotti,G., Lupo,R. and Russo,M. (1993) Resistance to cymbidium ringspot tombusvirus infection in transgenic Nicotiana benthamiana plants expressing the virus coat protein gene. Plant Mol. Biol., 21, 665–672. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Silhavy D., Molnar,A., Lucioli,A., Szittya,G., Hornyik,C., Tavazza,M. and Burgyan,J. (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J., 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Molnar,A., Silhavy,D., Hornyik,C. and Burgyan,J. (2002) Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell, 14, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H., Yigit,E., Siomi,H. and Mello,C.C. (2002) The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell, 109, 861–871. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O. and Baulcombe,D.C. (1997) Systemic signalling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wang D. and Maule,A.J. (1995) Inhibition of host gene expression associated with plant virus replication. Science, 267, 229–231. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Finnegan,E.J. (2001) Role of short RNAs in gene silencing. Trends Plant Sci., 6, 297–301. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nat. Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]