Abstract
As with transcription from DNA templates, RNA synthesis from viral RNA templates must initiate accurately. RNA sequences named specificity and initiation determinants allow recognition of and coordinated interaction with the viral replication enzyme. Using enriched replicase from brome mosaic virus (BMV)-infected plants and variants of the promoter template for minus-strand and subgenomic RNA initiation, we found that a specificity determinant for minus-strand initiation could function at variable distances and positions from the 3′ initiation site in a manner similar to enhancers of transcription from DNA templates. This determinant's addition could convert a cellular tRNA into a template for RNA synthesis by the BMV replicase in vitro. Furthermore, the same specificity element could direct internal initiation, which occurred at a highly preferred site in a manner distinct from initiation at the 3′ terminus of the template. These results document two distinct modes of initiation site recognition by a viral RNA replicase.
DNA promoters direct transcription by positioning the RNA polymerase complex at the site of initiation (33, 47). Similarly, RNA viruses need to specifically replicate and transcribe their genomes. How the replication enzymes recognize their cognate RNA promoters and initiation sites is poorly understood in part because of the myriad structures formed by RNA molecules and the difficulty in obtaining functional enzymes.
Replication and transcription in positive-strand RNA viruses uses a complex of viral and cellular proteins called the replicase. We seek to understand RNA promoter-replicase recognition by using an enzyme complex enriched from barley infected by brome mosaic virus (BMV). The BMV genome is composed of three RNAs and a subgenomic RNA, each capped at the 5′ terminus and each containing a common 3′ sequence of approximately 200 nucleotides (nt) (22). The replicase must produce three classes of RNAs during the infection: minus-strand and plus-strand RNAs of full genome length, and subgenomic RNA. Genome-length RNAs are initiated de novo from the 3′ termini of the template, while the subgenomic RNA is initiated internally in the template RNA. Minimal sequences for all three promoters needed to direct initiation by the BMV replicase in vitro have been characterized and shown to duplicate the in vivo requirements (reviewed in reference 21). Each promoter template functionally contains at least two determinants: one for specificity and one for initiation (21).
Genomic minus-strand initiation requires the 3′ 186 nt of the BMV RNA (10, 11), a stretch that mimics a tRNA-like structure (12, 13). Extensive mutational analysis demonstrated that stem-loop C (SLC) within the tRNA-like structure is necessary and sufficient to bind the BMV replicase (7, 10). The solution conformation of SLC was solved using nuclear magnetic resonance and found to contain two functional motifs: an internal loop of ca. 10 nt and a stable stem with a 5′-AUA-3′ triloop (26) (see Fig. 1A). The flexible internal loop facilitates RNA synthesis but is not essential in vitro, while the stable stem and triloop provide a critical motif for the recognition by BMV replicase. The 5′ A of the triloop is exposed to solution by being rigidly fixed to the stem helix through electrostatic and stacking interactions in the loop and nearby stem nucleotides in a structure called a clamped adenine motif (CAM) (24, 26). The CAM is a key recognition element that interacts with a subunit of the replicase complex. BMV RNAs with single nucleotide replacements disrupting the CAM were severely impaired for RNA synthesis both in inoculated plants and in vitro (7, 10, 24, 39).
FIG. 1.
Spatial requirements between the CAM and the initiation site for minus-strand RNA synthesis. (A) Sequence of SLC +8 and derivatives. Only the affected sequences of SLC derivative are shown, and their names are in parentheses. Nucleotides inserted are next to the names, and the deletions in the stem are boxed. Initiation cytidylates are represented as outlined C's, and the adenine of the CAM is in bold. Replacements of the single-stranded 3′ sequence are shown at the top of the upper stem. (B) Effects of the changes in SLC +8 on RNA synthesis. Qualitative results for all reactions were run in duplicate. Quantification of these and other reactions are shown below the autoradiogram. The results in lanes 1 to 4 and 8 to 11 were normalized to SLC +8 and those in lanes 5 to 7 were normalized to SLC +4. All quantifications were normalized for the number of radiolabeled cytidylates incorporated relative to the control and are the results of at least three independent replicates of the RNA synthesis reactions. The size of the replicase products, in nucleotides, obtained with SLC +4 is shown to the left of the autoradiogram. Products of the BMV replicase that terminated at the 5′ end of the template and those that contain one or a few nontemplated nucleotides appear as doublets in the autoradiogram (43). (C) Effects of changes in the stems relative to SLC +6. The layout of the figure is as described in the legend for panel B.
The addition of an 8-nt sequence containing the 3′ CCA, resulting in the so-called SLC +8, allowed de novo-initiated RNA synthesis with requirements that mimic those for BMV replication (7) (see Fig. 1A and B). Unexpectedly, a version of SLC +8 with a 5′-AUG-3′ triloop, known as S-AUG, which should disrupt the CAM, directed RNA synthesis efficiently (26). The high-resolution nuclear magnetic resonance structure of this S-AUG showed that the 5′-most adenylate of the triloop indeed formed a CAM, but one with a significantly altered angle and loop conformation compared to the wild-type AUA triloop (25).
Previously, our laboratory showed that SLC lacking the initiation sequence could bind to the BMV replicase (7) and that a single-stranded initiation sequence could direct RNA synthesis (41). Therefore, the specificity and initiation elements can function independently, in the absence of one another. However, when both are present within one RNA, mutations in either motif prevented synthesis, indicating coordination of their activities. The present investigation employs variants of SLC +8 to examine the functional relationship between the replicase and the specificity and initiation determinants with variants of SLC +8. Results from RNAs with changes that perturb the spacing between the CAM and an initiation site suggest that replicase-promoter recognition occurs in at least two distinct modes.
MATERIALS AND METHODS
Synthesis of RNA templates for RNA synthesis assays.
DNA oligonucleotides that can direct transcription by T7 RNA polymerase were synthesized by MWG Biotech, Inc. (High Point, N.C.). The oligonucleotides were annealed to a 17-nt DNA containing the top strand of the T7 RNA promoter and were transcribed as specified by Kao et al. (23). Products of the transcription reaction were electrophoresed in a denaturing polyacrylamide gel electrophoresis gel, and the bands of correct molecular mass were identified by their shadows over a UV-fluorescent plate and were excised with a razor. The gel was then crushed to elute the RNA in a solution of 0.3 M ammonium acetate. The eluted RNAs were concentrated by ethanol precipitation and quantified by spectrometry.
Replicase assays.
BMV replicase was prepared from infected barley as described by Sun et al. (45). Replicase activity assays were carried out as described by Adkins et al. (1). Briefly, each assay consisted of a 40-μl reaction mixture containing 20 mM sodium glutamate, pH 8.2, 12 mM dithiothreitol, 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP, 500 μM GTP, 200 μM UTP, 242 nM [α-32P]CTP (400 Ci/mmol; 10 mCi/ml; Amersham), the desired amount of template, and 7 μl of RNA-dependent RNA polymerase (RdRp). Following incubation for 90 min at 30°C, the reaction products were extracted with phenol-chloroform (1:1, vol/vol) and precipitated with six volumes of ethanol, 10 μg of glycogen, and a 0.4 M final concentration of ammonium acetate. Loading buffer (45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue, and 0.04% [wt/vol] xylene cyanol) was added to the products and denatured by heating at 90°C for 3 min prior to electrophoresis on 12% acrylamide-7 M urea denaturing gels unless stated otherwise. All gels were wrapped in plastic and exposed to film at −80°C, and the amount of label incorporated into newly synthesized RNAs was determined with a PhosphorImager (Molecular Dynamics, Inc.).
RESULTS
Spatial requirements in SLC.
We sought to use the well-characterized SLC as the basis to probe for requirements in the recognition of specificity and initiation determinants by the BMV replicase. First, we examined the number of nucleotides within the single-stranded sequence needed for RNA synthesis. Previously, a 3′ single-stranded sequence of 8 nt was found to be able to direct RNA synthesis while a 4-nt sequence could not (7). The structures of the recombinant RdRps suggest a regulatory role for the single-stranded initiation sequence (5, 17, 31). The hepatitis C virus (HCV) RdRp requires a single-stranded sequence of approximately 5 nt to direct RNA synthesis (20). We made SLCs that contained 4-, 5-, 6-, or 8-nt single-stranded sequences (Fig. 1A). A 4-nt sequence was ineffective in directing RNA synthesis (Fig. 1B, lane 1), while 5-, 6-, or 8-nt sequences increased RNA synthesis significantly (lanes 2 to 4). These results confirm and extend previous findings that a single-stranded sequence of longer than 4 nt is needed for efficient RNA synthesis by the viral RdRps.
Next, we sought to define the requirements within the two stems of SLC that would allow efficient initiation of RNA synthesis. The stem located between the bulge and the 3′ single-stranded initiation sequence will be referred to as the upper stem, while the one between the bulge and the triloop will be called the lower stem (Fig. 1A). Insertions and deletions to the upper stem were made in SLC +4 and SLC +8. A mutant named In1 had three additional base pairs in the upper stem, while the mutant named Δ2 had three fewer. In1 had only moderate effects on synthesis in the context of both SLC +4 and SLC +8, while Δ2 increased synthesis by two- to threefold (Fig. 1B, lanes 5 to 7). It is possible that Δ2 destabilized the upper stem of SLC +8 and increased the length of the single-stranded sequence. Next, the lower stem was altered by insertions and deletions in the context of SLC +6. Insertions of 1, 2, or 3 bp in the middle of the lower stem decreased RNA synthesis by the BMV replicase to 72, 35, and 14% relative to the synthesis directed by SLC +6. The length of the lower stem thus appears to be more important than the upper stem. A deletion of one A-U base pair in the middle of the lower stem, resulting in RNA Δ3, directed a similar level of synthesis as SLC +6. Deletions of the 3 bp closer to the bulge resulted in RNA Δ1, which directed synthesis at 56% while deletion of 3 bp closer to the CAM (Δ4) directed synthesis at only 11% (Fig. 1C).
Decreased synthesis observed with RNA Δ4 could be due to an effect on the structure of the CAM or the spacing between the CAM and the initiation site. We believe that the first possibility is more likely since Δ1 and Δ4, both deleting 3 bp in the lower stem, would have similar effects on the spacing and yet only Δ4 shows such a drastic negative effect. Also, should the spacing between the CAM and the initiation site be affected, a longer single-stranded sequence could compensate for the Δ4 deletion. SLC +8 with the Δ4 deletion was also severely affected in RNA synthesis (Fig. 1B, lane 11), indicating that the change likely affected the formation of the CAM and not the spacing between the triloop and the initiation sequence.
A minimal RNA for minus-strand RNA synthesis.
RNA SLD3 could direct RNA synthesis by the HCV RdRp. It consists of an 8-bp stem with an AUA triloop and a 6-nt single-stranded initiation sequence (Fig. 2) (20). Despite being derived from SLC +8, SLD3 cannot direct RNA synthesis by the BMV replicase (C. Kao, unpublished results), possibly due to the lack of one or more of the following: (i) a defined bulge; (ii) a 3′-most adenylate (Fig. 2A); and/or (iii) a proper spatial relationship between the specificity CAM and the initiation site. The first two possibilities are less likely, since SLC lacking a defined bulge was competent for RNA synthesis (26) and the addition of a 3′-terminal adenylate to SLD3, resulting in SLD3A, did not result in RNA synthesis (Fig. 2A and B, lanes 1). The third possibility was tested by inserting an increasing number of AU dinucleotides into the junction between the stem and single-stranded sequence in SLD3A to increase the spacing between the CAM and the initiation site (Fig. 2A). Addition of two sets of AU dinucleotides, (AU)2, did not improve RNA synthesis, while three to five AU dinucleotides, (AU)3, (AU)4, and (AU)5, increased RNA synthesis significantly (Fig. 2B, lanes 3 to 5). RNAs with insertions of AU dinucleotides were predicted by the mfold program to not form alternative structures (data not shown). Also, it is unlikely that the addition of four AU repeats, rather than the increase in spatial relationship between the CAM and the initiation site, led to an increase in RNA synthesis. However, we cannot directly rule out this possibility, since other nucleotide combinations are predicted to form alternative structures that would complicate the interpretation of our experiments.
FIG. 2.
A minimal-length promoter template for minus-strand RNA synthesis. (A) The sequence of SLD3A and its derivatives. Only the affected nucleotides in the derivatives of SLD3A are shown. The names of all of the derivatives are in parentheses. The shaded triangle denotes the position where two or more A-U dinucleotides are inserted. Nucleotide substitutions are indicated with straight arrows. The outlined letter C shows the initiation cytidylate, and the adenine of the CAM is in bold. (B) RNA synthesis directed by derivatives of SLD3A. The sizes of the replicase products, in nucleotides, are shown to the left of the autoradiogram. The percentages of synthesis are normalized to SLD3A and are from at least three replicates.
RNA synthesis from (AU)4 initiated at the CCA initiation site, since a change to UUA decreased synthesis to background levels (Fig. 2B, lanes 6). The CAM was also required for specific recognition of SLD3A, since changes from the wild-type 5′-AUA-3′ to 5′-UAU-3′ or 5′-UUA-3′ reduced RNA synthesis to background levels (Fig. 2B, lanes 7 and 8). The CAM formed by the AUG triloop directed synthesis at 149% relative to (AU)4. Since the AUG triloop makes the sequence of the triloop asymmetric, we constructed RNA AUG-Swi1, which has Watson-Crick transversion of the closing base pair and triloop nucleotides of (AU)4. AUG-Swi1 was unable to direct synthesis, indicating that a specific configuration of the CAM is required for RNA synthesis (Fig. 2B, lane 10). These results suggest that some minimal length between the specificity and initiation determinants and specific RNA stereochemistry are required for RNA synthesis by the BMV replicase.
Recognition of the minus-strand initiation site.
To examine further how replicase binding affects the use of the initiation site, (AU)4 RNA was engineered to have a second potential initiation site added to the original 3′ terminus, resulting in RNA Init2 (Fig. 3A). If it used the original initiation site, it would be predicted to produce a 33-nt RNA, while the 3′-most initiation site would produce a 41-nt RNA (Fig. 3B, lanes 1 and 2). Init2 produced primarily the 41-nt RNA, indicating a strong preference for the 3′-most initiation site (Fig. 3B, lane 2). A change of the 3′-most initiation site from CCA to GGA in an RNA named 3′GG resulted in a much-reduced level of synthesis of the 41-nt RNA (Fig. 3B, lane 3) but still did not allow initiation from the internal CCA site, confirming that initiation took place primarily from the 3′-most initiation site (Fig. 3A and B, lanes 3). A change of the triloop from 5′-AUA-3′ to 5′-UUA-3′ in the context of Init2 reduced RNA synthesis to background (Fig. 3B, lane 4). Also, Sg(AU)4 with a bulge in the stem that should disrupt the CAM was found to be unable to direct RNA synthesis. These results indicate that the BMV replicase will preferentially use a 3′ initiation sequence despite an aberration in the fixed spatial relationship with the specificity determinant.
FIG. 3.
Minus-strand RNA synthesis prefers to initiate from a 3′ initiation site. (A) Schematic of RNA Init2, which has two potential initiation cytidylates (in outlined letters) and Sg(AU)4, which contains a putative stem-loop from the BMV subgenomic promoter, as proposed by Haasnoot et al. (15). Arrows identify nucleotide substitutions. (B) Results of RNA synthesis using the RNAs shown in panel A. All lanes are in duplicate, and quantitative results are from these and other independently assayed reactions.
The SLC specificity determinant can work at variable positions relative to the initiation site.
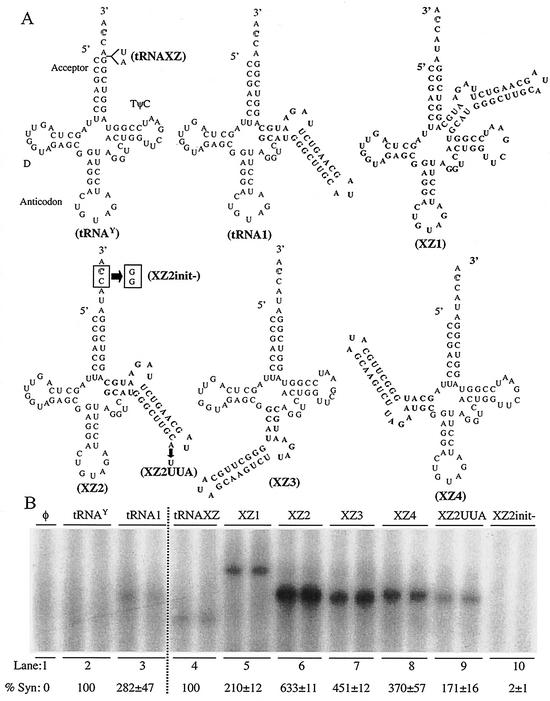
The seemingly flexible relationship between the specificity element and the initiation sequence prompted further examination using constructs based on the tobacco tyrosine tRNA (Fig. 4A). Despite the BMV 3′ sequence having a tRNA-like structure, cellular tRNAs are poor templates for RNA synthesis (Fig. 4B, lane 2, and data not shown). The replacement of the TψC stem-loop with SLC in RNA, tRNA1, increased RNA synthesis relative to tRNATyr (Fig. 4B, lane 3). RNA synthesis, however, remained approximately 20-fold lower than that of SLC +8 (data not shown). As we have already demonstrated, the low level of synthesis suggests that the 4-nt 3′ single-stranded sequences in tRNA1 and in tRNATyr are insufficient for the initiation sites to reach the RdRp active site. Therefore, 2 nt (U-A) were added immediately 5′ of the CCA sequences of tRNATyr and tRNA1, resulting in RNA tRNAXZ and XZ2, respectively (Fig. 4A). tRNAXZ remained a relatively poor template for the BMV replicase, perhaps due to the lack of a replicase specificity determinant (Fig. 4A and B, lanes 4). In support of this hypothesis, XZ2, with both SLC and a 6-nt single-stranded sequence, increased RNA synthesis by the BMV replicase 6.3-fold relative to tRNAXZ, after adjustment for the number of radiolabeled cytidylates (Fig. 4A and B, lanes 6). The addition or replacement of SLC to other portions of the tRNA increased synthesis to all constructs by at least 2.1-fold (Fig. 4B, lanes 5 to 8). The ability of XZ2 to direct RNA synthesis by the BMV replicase is dependent on the CAM and on the 3′ CCA sequence, since mutations of these two motifs in XZ2UUA and XZ2init− decreased synthesis significantly (Fig. 4B, lanes 9 and 10). These results support our model that the specificity determinant for minus-strand RNA synthesis can direct RNA synthesis despite being at variable positions relative to the 3′ initiation site.
FIG. 4.
The specificity determinant SLC can function at different positions relative to the initiation site. (A) The tobacco tyrosine tRNA and its derivatives. While some RNAs are shown in their entirety, only the affected regions of the RNAs derived from tRNATyr and XZ2 are shown. The names of the tRNAs are shown in parentheses. The sequence for SLC is in bold letters, and the initiation cytidylates are in outlined font. Nucleotide substitutions are identified by arrows. (B) Autoradiogram of the RNA synthesis by the BMV replicase using the RNAs diagrammed in panel A. The symbol φ denotes a reaction performed in the absence of exogenously provided RNA. A dashed line is placed between the two sets of RNAs that are normalized to different standards. Synthesis from tRNA1 is normalized to tRNATyr. Syntheses from XZ1 to -4 are normalized to tRNAXZ, since all contain two additional nucleotides added to the 3′ single-stranded sequence. The quantifications, made from at least three independently processed samples, were also normalized for the potential number of radiolabeled CMPs incorporated.
SLC and internal initiation of RNA synthesis.
Haasnoot et al. (15) claimed that the BMV subgenomic promoter is functionally homologous to the SLC. While both the subgenomic promoter and the SLC position the replicase to allow initiation, extensive mutational analyses of the SLC suggest that the sequence of the subgenomic core promoter cannot form the highly specific CAM (7, 25, 26). In fact, RNA SLC +6 Δ4, which happens to have the four key residues of the BMV subgenomic promoter near the loop (Fig. 1A), was able to direct RNA synthesis by only 11% (Fig. 1C, lane 6). To address the functional interchangeability of the subgenomic specificity determinant and SLC as reported by Haasnoot et al. (15), RNA Sg(AU)4 with the putative BMV subgenomic stem-loop structure replacing the triloop stem was synthesized (Fig. 3A). Sg(AU)4 was unable to direct RNA synthesis (Fig. 3B, lane 5), indicating that SLC and the putative subgenomic stem-loop are not interchangeable specificity elements. Haasnoot et al. (15) reported that a replacement of the SLC with the wild-type subgenomic stem-loop also decreased RNA synthesis to 10% of wild type. Thus, despite differences in interpretations, our results are consistent with those of Haasnoot et al. (15).
In spite of the differences in wild-type specificity determinants for minus-strand and internal initiation, we wanted to determine whether SLC could direct the use of an internal initiation site in a template lacking a 3′ single-stranded sequence. Toward that end, a 13-nt template for the BMV subgenomic RNA was attached 5′ of the SLC in RNA iSLC (Fig. 5A). RNA iSLC produced a 13-nt replicase product and one of ca. 67 nt (Fig. 5B, lane 1). After adjusting for the number of radiolabeled CMPs incorporated, the primer-extension product was 1.7-fold more abundant than the de novo-initiated 13-mer. The 67-nt RNA is most likely generated by the replicase extending from the 3′ terminus of the RNA using the 5′ single-stranded sequence as the template, an activity of the viral RdRp. Three observations are consistent with this hypothesis. First, the synthesis of the 67-mer RNA, but not the 13-mer, could take place at 1 μM GTP, a concentration insufficient for de novo initiation (19). Second, the 67-nt RNA was made even though the first 2 bp of the upper stem were changed from 5′-G-C to 5′-C-G in RNA 3′GG (Fig. 5A). This RNA lacks nucleotides at the 3′ terminus that could direct de novo initiation but should, and did, permit primer extension (Fig. 5B, lane 2). Some internal initiation apparently did take place from one or more of the two internal cytidylates, accounting for the 16- and 17-nt replicase products (Fig. 5B, lane 2). Third, Init2− produced the 67-nt product in the absence of GTP, consistent with primer extension, and RNase T1 digestion resulted in the product expected from primer extension (data not shown).
FIG. 5.
SLC can direct internal initiation of RNA synthesis. (A) Schematics of iSLC, which lacks a 3′ initiation sequence but has a potential initiation site in the sequence 5′ of the upper stem. The potential initiation cytidylate is in an outlined font. Only the affected sequences in RNAs derived from iSLC are shown, and their names are in parentheses. Nucleotides inserted are indicated by shaded triangles, while the deleted nucleotides are boxed. Nucleotide substitutions are denoted by arrows. (B) Products of RNA synthesis directed by iSLC and its derivatives. The products were separated on a denaturing 20% polyacrylamide gel. Primer extension (PE) and de novo-initiated 13-nt products of the BMV replicase are identified to the left of the autoradiogram. In lanes 7 and 8, RNA iΔ3 was made to contain either the iΔ2 deletion or the In3 insertion. The differences in the mobilities of the BMV replicase products are expected and are due to changes in the lengths of the templates in each reaction. Quantification of the products of RNA synthesis was performed from at least three independent reactions.
A mutation of the putative internal initiation site from a C to a G in RNA Init2− abolished the production of the 13-nt RNA, consistent with de novo initiation occurring from the internal cytidylate (Fig. 5B, lane 9). Internal initiation was also dependent on the CAM, since a change of the triloop to UUA abolished RNA synthesis (Fig. 5B, lane 10). These results clearly indicate that the CAM can be used for internal initiation of RNA synthesis, consistent with the previous observations of Haasnoot et al. (15, 16). Quite interestingly, iSLC with a UUA triloop had a decrease in primer-extension product (Fig. 5B, lane 10), indicating that the ability to recruit the replicase affects both de novo initiation and primer extension.
To examine whether the recognition of the internal initiation site has spatial requirements relative to the CAM, several changes were made to iSLC. Two deletions, iΔ1 and iΔ2, that decreased the distance from the initiation nucleotide to the CAM increased internal initiation by approximately fourfold (Fig. 5B, lanes 4 and 5). The removal of the two nucleotides in the single-stranded sequence immediately 3′ of the initiation cytidylate in RNA iΔ3 also increased RNA synthesis (Fig. 5B, lane 6). These results indicate that a highly preferred spatial relationship exists between the specificity determinant and the internal initiation site. As further support for this hypothesis, iΔ3 with 2 bp deleted in the upper stem to bring the initiation cytidylate closer to the CAM increased internal initiation fivefold over that in iΔ3 (Fig. 5B, lane 7). Finally, when 3 bp were inserted into the upper stem of iΔ3 RNA, iΔ3-In3 (Fig. 5A), this mutant directed initiation at 49% of that of iSLC and 23% of iΔ3 (Fig. 5B, lane 8), supporting our observation that there is a required distance between the CAM and the internal initiation site. Consistent with this, the addition of a cytidylate within the bulge in bC RNA did not allow initiation to take place from this cytidylate, as judged by the absence of the expected 44-nt RNA (Fig. 5B, lane 3).
Preferred subgenomic initiation is spatially constrained relative to SLC.
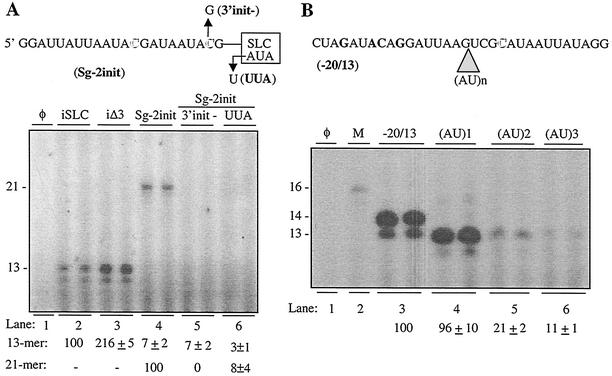
The use of an internal initiation site appears to be spatially more constrained relative to the CAM than initiation from the 3′ penultimate initiation site. This is surprising given that the identical specificity determinant is used. The difference suggests that the protein, which binds the initiation site, likely the 2a RdRp, can adjust its structure and/or function in response to a terminal or an internal initiation site. To examine this further, we added a second potential initiation site to iSLC in an RNA named Sg-2init (Fig. 6A). The 5′-distal initiation site is identical in position to the CAM, as is the one in iΔ3, and it should direct the synthesis of a 21-nt de novo-initiated product. Should the 5′-proximal initiation site be used, it would produce a 13-nt RNA. Sg-2init directed the synthesis of only the 21-nt RNA (Fig. 6A, lane 4). When the initiation cytidylate for the distal site was changed to a guanylate, neither a 13- nor 21-nt product was observed, indicating that the position of the internal initiation site cannot be changed (Fig. 6A, lane 5). The synthesis of the 21-nt product was dependent on the CAM, since a change of the clamped adenine to a uridine virtually abolished RNA synthesis (Fig. 6A, lane 6).
FIG. 6.
Spatial requirements for BMV subgenomic initiation. (A) RNA synthesis from a version of iSLD named Sg-2init, which has two potential internal initiation sites (in outlined letters). The SLC portion of Sg-2init is represented by a box. Nucleotide substitutions in Sg-2init and the names of these derivatives are shown with arrows. The products were separated on a denaturing 20% polyacrylamide gel. The bottom of the panel contains an autoradiogram of the RNAs synthesized from Sg-2init and derivatives as controls (only the de novo-initiated RNA products are shown). RNAs iSLC and iΔ3 produce predominantly a 13-nt product RNA. Quantifications were from at least three independently assayed samples. (B) The subgenomic initiation site cannot be moved relative to the promoter nucleotides. The sequence of the subgenomic proscript −20/13 is shown. The four specificity nucleotides are in bold and the initiation cytidylate is in the outlined font. The site of insertion for one or more A-U dinucleotides is indicated by the gray triangle. RNAs containing one or more A-U insertions are named by the number of dinucleotides inserted. In the bottom of the panel is an autoradiogram of the products from −20/13 and its derivatives. The lane marked with φ contained no exogenously provided template. M denotes a reaction synthesizing the 15- and 16-nt molecular weight markers previously described by Tayon et al. (46).
To examine the spatial requirements of subgenomic initiation, we used the BMV subgenomic proscript −20/13, which contains both a nontemplated promoter sequence and a 13-nt template RNA (41-43). In our previous analysis of the spatial requirements for subgenomic initiation, some insertions and deletions between nt −4 and −5 of the promoter sequence were tolerated (44). We now added one or more AU dinucleotides at this position to determine whether alterations in the subgenomic initiation site and the 4 nt critical for promoter recognition could be tolerated (Fig. 6B, top). Proscript −20/13 produced both a 13-nt and a 14-nt product, the latter of which contains a nontemplated nucleotide at the 3′ terminus of the newly synthesized RNA. (AU)1, with one dinucleotide insertion, retained initiation at the original cytidylate but produced an RNA of 13 nt that lacked a nontemplated nucleotide (Fig. 6B, lane 4). RNAs of 16 and 17 nt were also observed at lower abundances, indicating that some initiation took place from the cytidylate originally at the −1 position despite this site not having a template sequence preferred for efficient RNA synthesis (1) (Fig. 6B, lane 4). Insertions of two or three AU nucleotides reduced the amount of the 13-nt product to less than 21% of that from −20/13 (Fig. 6B, lanes 5 and 6), indicating that subgenomic initiation, in contrast to minus-strand initiation, cannot efficiently take place at alternative positions.
DISCUSSION
Core DNA promoters are generally recognized by specific contacts between the polymerase and the nucleotide moieties in the major and sometimes the minor grooves of the DNA (9, 18, 32, 47). These interactions allow the polymerase to identify the initiation site for transcription, which is positioned at a relatively fixed position relative to the specificity determinants (49; reference 30 and references therein). For BMV RNA synthesis in vitro, one specificity determinant, SLC, could direct internal and end initiations. For the internal initiation of subgenomic RNA synthesis, we found that the specificity and initiation determinants have more limited spatial requirements, in a manner more similar to DNA promoters. However, the spacing between the specificity determinant and the minus-strand initiation site is more flexible. In fact, the specificity determinant for BMV minus-strand RNA synthesis can function at multiple positions of a modified cellular tRNA and lead to the initiation of RNA synthesis from the 3′-terminal CCA motif in vitro. These results suggest that the protein interacting with the initiation site will change the mode of RNA synthesis in response to the context of the initiation site.
The main determinant of promoter specificity in BMV minus-strand synthesis is provided by SLC, which features a clamped adenine motif. Results with various insertions and deletions of SLC and with the SLD3A derivatives suggest that there must be a minimal spatial distance within the SLC. These requirements likely indicate that some steric compatibility must be maintained between the factor(s) that binds SLC or different portions within SLC. There is also a required spatial interaction between SLC and the initiation site. These results indicate that factors that interact with SLC must maintain proper positioning with the RdRp that binds the initiation site.
For both end and internal initiation by the BMV replicase, a single-stranded sequence is required. This may be a general requirement for initiation by RdRps (6, 20, 34). For minus-strand initiation, a minimal length of 5 nt is needed. Similar requirements were also found for the HCV RdRp (20). The initiation sequence may be needed to interact with the template channel of the RdRp. While the structure of DNA-dependent RNA polymerases are likened to an open right hand (8), the interactions between the thumb and finger domains of RdRps from HCV, calicivirus, and φ6 bacteriophage form structures that resemble a closed hand (5, 6, 31, 37), creating a well-defined template channel that could regulate the recognition of the initiation site (4, 6). The channel and additional structures near the active site (17) ensure that initiation of minus-strand RNA synthesis takes place at or near the end of the 3′ termini of the HCV and φ6 RNAs. We postulate that a similar mechanism ensures the initiation of the BMV minus-strand RNA. Furthermore, a narrow channel could also require that subgenomic initiation occurred in a single-stranded sequence, a feature recognized in subgenomic initiation sites (21, 34). Lastly, the template channel can also discriminate against the use of cellular RNAs, which would likely lack a 3′ sequence of sufficient length and of the proper sequence to direct initiation by the viral replicase.
The BMV 3′ tRNA-like sequence actually contains a 4-nt single-stranded sequence (13). This is not incompatible with our result demonstrating that a minimum of a 5-nt single-stranded sequence is needed for efficient initiation. The CCA initiation site is adjacent to a highly labile pseudoknot that exists in equilibrium with an alternative structure that lacks the pseudoknot. Hence, the BMV 3′ tRNA-like structure should be accessible for initiation when the pseudoknot melts. This change in RNA conformation could provide another level of regulation for viral RNA replication. Previously, Kolk et al. (27) proposed that the pseudoknot at the 3′ terminus of plant viral RNAs could provide a highly flexible gate to allow initiation of minus-strand RNA synthesis in a subset of the viral RNA molecules.
A striking result concerning SLC is that it can function at different positions relative to the initiation site for minus-strand initiation; SLC could act in place of, or in addition to, the stem-loops within a cellular tRNA to stimulate initiation of RNA synthesis from the 3′ CCA sequence. Our laboratory previously demonstrated that SLC could interact with the BMV replicase in the absence of the initiation site (7). Altogether, these results suggest that after the BMV replicase complex binds SLC, the complex can recognize an initiation site near the 3′ terminus. The sequence present between the SLC and the initiation site must thus be able to loop out in an interaction analogous to the one between enhancer-binding proteins and RNA polymerase (48; reference 3 and references therein). The relative lack of constraints between the specificity element and the end initiation site will facilitate the evolutionary acquisition of promoter-specific determinants to mediate replicase-viral RNA recognition.
Previously, Nagy et al. (35) reported that turnip crinkle virus (TCV)-associated satellite RNA contains a hairpin motif that can increase the frequency of RNA recombination and RNA replication. We note with interest that while the hairpin in the TCV structure does not contain a CAM, it is also composed of two stems separated by a bulge sequence (35). The TCV motif, however, appears to be less specific, since a transversion of the entire motif also resulted in RNA synthesis while many changes in the BMV SLC, including single-nucleotide changes in the loop and a switch of the triloop sequence to one that is in a mirror image, abolished RNA synthesis (Fig. 1B and 2B). Different viral replicases thus appear to have a range of specificities for their cognate promoters, with the BMV replicase being highly specific. The mechanistic basis for this specificity in promoter recognition remains to be determined.
Initiation from the 3′ terminus of the template obeys different rules than does initiation of subgenomic RNA. For the BMV subgenomic proscript, insertions and deletions between the specificity determinants and the initiation site led to significant initiation from the nucleotide that occupies the position of the original initiation cytidylate (44). This result is confirmed and extended in this work, where the insertion of more than one AU dinucleotide decreased initiation since no suitable initiation nucleotides were readily available (Fig. 6B). The spatial relationship between the specificity and initiation determinants for internal initiation is thus more constrained than is initiation from the 3′ terminus of the template.
There are two major possible factors that could account for the differences in the spatial relationship for end and internal initiation: (i) different proteins bind to the specificity determinants in minus-strand and subgenomic initiation, and/or (ii) RdRp can adjust its properties when an end or internal initiation site is encountered. While we do not yet have results concerning the first possibility, we have shown that internal initiation using SLC has different spatial constraints relative to end initiation, suggesting that the RdRp adjusts to the template sequence containing an initiation site. In fact, the presence of additional sequence 3′ of the initiation site may induce the RdRp to interact with the initiation site in a manner different from that of terminal initiation.
Haasnoot et al. (15) proposed that the BMV subgenomic promoter and the minus-strand promoter are highly similar, if not identical. This claim is based on the observation that the subgenomic promoter can form a stem-loop structure and that a stem-loop containing a CAM can direct internal initiation. Their results provide circumstantial evidence that the factors recognizing the two promoters are the same. In fact, using chemically synthesized RNAs, at least two of the nucleotides forming the putative stem-loop of Haasnoot et al. (15) are not required for efficient RNA synthesis (42, 43), while a stable stem-loop containing the CAM is absolutely required for minus-strand RNA synthesis (7) (Fig. 1C). We show here that the subgenomic stem-loop cannot replace SLC for minus-strand RNA synthesis (Fig. 3B, lane 5). Whether the two promoters bind the same or different factors remains to be determined.
Finally, it is interesting that the viral replicase exhibits many of the features of promoter recognition found in other polymerases. Conversely, DNA-dependent RNA polymerases could also transcribe specifically from RNA templates (2, 14, 28, 29, 38, 40). RdRps have been proposed as one of the oldest forms of polymerases (36). The shared mechanisms of promoter specificity and the identification of the initiation sites could reflect an evolutionary relationship between different classes of polymerases.
Acknowledgments
We thank members of the IU Cereal Killer group for helpful discussions during the course of this work.
Funding was provided by the U.S. Department of Agriculture and by the National Science Foundation. C. C. Kao acknowledges a Linda and Jack Gill fellowship.
REFERENCES
- 1.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. Kao. 1998. Mechanistic analysis of RNA synthesis RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud-Barbe, N., V. Cheynet-Sauvion, G. Oriol, B. Mandrand, and F. Mallet. 1998. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 26:3550-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:61-63. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli, S., L. Tomei, F. A. Rey, and R. DeFrancesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., I. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, and R. DeFrancesco. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bramford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, M., and C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 8.Cheetham, G. M., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117-123. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham, G. M., D. Jeruzalmi, and T. A. Steitz. 1999. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 399:80-83. [DOI] [PubMed] [Google Scholar]
- 10.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus-strand promoter activity. J. Mol. Biol. 201:31-40. [DOI] [PubMed] [Google Scholar]
- 11.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. J. Mol. Biol. 201:41-55. [DOI] [PubMed] [Google Scholar]
- 12.Fechter, P., R. Giege, and J. Rudinger-Thirion. 2001. Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J. Mol. Biol. 309:387-399. [DOI] [PubMed] [Google Scholar]
- 13.Felden, B., C. Florentz, R. Geige', and E. Westhof. 1994. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 235:508-531. [DOI] [PubMed] [Google Scholar]
- 14.Fu, T. B., and J. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 67:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haasnoot, P. C. J., R. C. Olsthoorn, and J. F. Bol. 2002. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8:110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haasnoot, P. C. J., F. T. Brederode, R. C. L. Olsthoorn, and J. F. Bol. 2000. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 18.Joyce, C. M., and T. A. Steitz. 1995. Polymerase structures and function: variations on a theme? J. Bacteriol. 177:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, C., and J. H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao, C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao, C. 2002. Lessons learned from the core RNA promoters of brome mosaic virus and cucumber mosaic virus. Mol. Plant Pathol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 22.Kao, C., and K. Sivakumaran. 2000. Brome mosaic virus, good for a virologist's basic needs. Mol. Plant Pathol. 2:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Kao, C., M. Zheng., and S. Ruedisser. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5:1268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, C.-H., and I. Tinoco. 2001. Structural and thermodynamic studies of mutant RNA motifs that impair the specificity between a viral replicase and its promoter. J. Mol. Biol. 307:827-839. [DOI] [PubMed] [Google Scholar]
- 25.Kim, C.-H., and C. Kao. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA 7:1476-1485. [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, C.-H., C. Kao, and I. Tinoco. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7:415-423. [DOI] [PubMed] [Google Scholar]
- 27.Kolk, M. H., M. van der Graaf, S. S. Wijmenga, C. W. Pleij, H. A. Heus, and C. W. Hilbers. 1998. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science 280:434-438. [DOI] [PubMed] [Google Scholar]
- 28.Konarska, M. M., and P. A. Sharp. 1989. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell 57:423-431. [DOI] [PubMed] [Google Scholar]
- 29.Konarska, M. M., and P. A. Sharp. 1990. Structure of RNAs replicated by the DNA-dependent T7 RNA polymerase. Cell 63:609-618. [DOI] [PubMed] [Google Scholar]
- 30.Landick, R. 2001. RNA polymerase clamps down. Cell 105:567-570. [DOI] [PubMed] [Google Scholar]
- 31.Lesberg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 32.Maslak, M. M., D. Jaworski, and C. T. Martin. 1993. Tests of a model for promoter recognition by T7 RNA polymerase: thymine methyl group contacts. Biochemistry 32:4270-4274. [DOI] [PubMed] [Google Scholar]
- 33.McClure, W. R. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54:171-204. [DOI] [PubMed] [Google Scholar]
- 34.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, T. M., and T. R. Cech. 1998. Reversing time: origin of telomerase. Cell 92:587-590. [DOI] [PubMed] [Google Scholar]
- 37.Ng, K., M. Cherney, A. López-Vázquez, A. Machín, J. Alonso, F. Parra, and M. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 38.Rackwitz, H. R., W. Rhode, and H. L. Sanger. 1981. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 291:297-301. [DOI] [PubMed] [Google Scholar]
- 39.Rao, A. L., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson, H. D., and A. D. Branch. 1987. The viroid replication process, p. 49-69. In J. S. Semancik (ed.), Viroids and viroid-like pathogens. CRC Press, Boca Raton, Fla.
- 41.Siegel, R. W., L. Bellon, L. Beigelman, and C. Kao. 1999. Use of DNA, RNA, and chimeric templates by a viral RNA-dependent RNA polymerase: evolutionary implications for the transition from the RNA to the DNA world. J. Virol. 73:6424-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegel, R., L. Bellon, L. Beigelman, and C. Kao. 1998. Identification of the base moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 95:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegel, R., S. Adkins, and C. Kao. 1997. Sequence-specific recognition of an RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stawicki, S. S., and C. Kao. 1999. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRp) reveal that RdRp can adjust its promoter binding sites. J. Virol. 73:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, J., S. Adkins, G. Faurote., and C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the brome mosaic virus RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 46.Tayon, R., M. J. Kim, and C. Kao. 2001. Completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 29:3576-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Temiakov, D., P. Mentesana, K. Ma, A. Mustaev, S. Borukhov, and W. T. McAllister. 2000. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 97:14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]
- 49.Young, B. A., T. M. Gruber, and C. A. Gross. 2002. Views of transcription initiation. Cell 10:417-420. [DOI] [PubMed] [Google Scholar]