Abstract
Nearly all human immunodeficiency virus (HIV) infections are acquired mucosally, and the gut-associated lymphoid tissues are important sites for early virus replication. Thus, vaccine strategies designed to prime virus-specific cytotoxic T lymphocyte (CTL) responses that home to mucosal compartments may be particularly effective at preventing or containing HIV infection. The Salmonella type III secretion system has been shown to be an effective approach for stimulating mucosal CTL responses in mice. We therefore tested ΔphoP-phoQ attenuated strains of Salmonella enterica serovar Typhimurium and S. enterica serovar Typhi expressing fragments of the simian immunodeficiency virus (SIV) Gag protein fused to the type III-secreted SopE protein for the ability to prime virus-specific CTL responses in rhesus macaques. Mamu-A*01+ macaques were inoculated with three oral doses of recombinant Salmonella, followed by a peripheral boost with modified vaccinia virus Ankara expressing SIV Gag (MVA Gag). Transient low-level CTL responses to the Mamu-A*01 Gag181-189 epitope were detected following each dose of Salmonella. After boosting with MVA Gag, strong Gag-specific CTL responses were consistently detected, and tetramer staining revealed the expansion of Gag181-189-specific CD8+ T-cell responses in peripheral blood. A significant percentage of the Gag181-189-specific T-cell population in each animal also expressed the intestinal homing receptor α4β7. Additionally, Gag181-189-specific CD8+ T cells were detected in lymphocytes isolated from the colon. Yet, despite these responses, Salmonella-primed/MVA-boosted animals did not exhibit improved control of virus replication following a rectal challenge with SIVmac239. Nevertheless, this study demonstrates the potential of mucosal priming by the Salmonella type III secretion system to direct SIV-specific cellular immune responses to the gastrointestinal mucosa in a primate model.
Sexual transmission across mucosal barriers is by far the most common mode of human immunodeficiency virus (HIV) infection. Furthermore, the gut-associated lymphoid tissues are a major source of virus replication during the first few weeks of infection (39). These observations suggest that vaccine strategies designed to maximize virus-specific immune responses at mucosal barriers and in mucosal lymphoid tissues may be particularly effective at preventing or containing infections early, before diversification and dissemination of the virus population.
Peripheral vaccine strategies based on prime and boost regimens with recombinant DNA and/or virus vectors can elicit potent cellular immune responses to simian immunodeficiency virus (SIV) and HIV-1 antigens (3, 6, 11, 28, 31, 35). These approaches also afford limited protection, as measured by reductions in peak and postacute viral loads following challenges with pathogenic SIV strains or simian-human immunodeficiency virus (SHIV) chimeras (3, 6, 11, 28, 31, 35). Higher frequencies of virus-specific CD8+ T lymphocytes and the stimulation of CD4+ T helper cell responses were generally associated with better control of viral loads after challenge (3, 6, 31, 35). Thus, while both antibody and cellular immune responses may ultimately be required for optimal protection, these studies indicate that strong cellular immune responses induced by vaccination can serve to limit virus replication.
Since immunization by peripheral routes typically does not induce immune responses in mucosal compartments (7, 10, 12), the protection afforded by prime and boost regimens could potentially be improved by modifications that direct cellular immune responses to mucosal sites. In support of this idea, a direct comparison of intrarectal versus subcutaneous immunization with similar peptide formulations demonstrated better protection of mucosally immunized animals against an intrarectal SHIV challenge (8). In this study, protection, as measured by reduced postchallenge viral loads, correlated with the stimulation of both virus-specific cytotoxic T lymphocyte (CTL) and T helper cell responses (8).
Attenuated strains of Salmonella spp. have been developed as potential vectors for stimulating immune responses in the gastrointestinal mucosa (34). However, the confinement of Salmonella strains to the intracellular vesicular compartments of bacterially infected cells has limited their potential for stimulating major histocompatibility complex (MHC) class I-restricted CTL responses. Indeed, this may explain the inefficient stimulation of virus-specific CTL responses in earlier studies in which macaques were immunized with recombinant Salmonella strains engineered to express SIV antigens (13, 37). To circumvent this problem, we have used an approach that takes advantage of the bacterial type III secretion system to deliver vaccine antigens directly into the cytoplasm and into the MHC class I antigen-processing pathway of Salmonella-infected cells (33).
The Salmonella type III secretion system is a multicomponent system consisting of structural, regulatory, and secreted elements specialized for the delivery of bacterial proteins into eukaryotic host cells (14). Upon contact with mammalian cells, structural elements of the Salmonella type III secretion system assemble into needle-like molecular filaments that span the inner and outer bacterial membranes and traverse the plasma membrane of the mammalian cell. By fusing heterologous protein sequences to type III-secreted bacterial proteins, antigens may be efficiently targeted into the host cell cytoplasm for antigen processing and presentation in association with MHC class I molecules (33). Infection with Salmonella recombinants expressing immunodominant CTL epitopes of influenza virus and lymphocytic choriomeningitis virus as fusions with the bacterial SptP protein has been shown to sensitize cells for CTL recognition in vitro (33). CTL recognition was MHC class I restricted, TAP dependent, and dependent on bacterial translocation of the SptP fusions by the type III secretion pathway (33). Furthermore, immunization of mice with an attenuated Salmonella strain engineered to deliver a single immunodominant CTL epitope of lymphocytic choriomeningitis virus stimulated potent virus-specific CTL responses that protected against a lethal challenge with lymphocytic choriomeningitis virus (33).
Here we explore the potential of the Salmonella type III secretion system to stimulate SIV-specific CTL responses in the gastrointestinal mucosa of rhesus macaques. Our data show for the first time that the Salmonella type III secretion system can be used to deliver large polypeptide antigens rather than minimal CTL epitopes for MHC class I antigen processing and presentation. This study is also the first to demonstrate the ability of the Salmonella type III antigen delivery system to prime T-cell responses in a nonhuman primate model and provides an important proof of principle for the feasibility of mucosal prime/peripheral boost vaccine approaches to direct cellular immune responses to mucosal compartments.
MATERIALS AND METHODS
Salmonella type III secretion constructs.
A panel of constructs expressing fusions between the SIV Gag protein and the bacterial SopE protein were created for the delivery of Gag polypeptides into mammalian cells by the Salmonella type III secretion system. Sequences coding for amino acids 4 to 507, 4 to 439, 4 to 284, 146 to 213, and 267 to 507 of Gag were cloned downstream from the sequence encoding the promoter and the first 104 amino acids of SopE in the low-copy plasmid pWSK30 (30, 41). The H-2b-restricted influenza virus nucleoprotein epitope NP366-374 was included between the SopE and Gag sequences to enable the detection of antigen presentation in murine cells. ΔphoP-phoQ attenuated strains of Salmonella enterica serovar Typhimurium (χ8429) and Salmonella enterica serovar Typhi (χ8521) were transformed with each of these constructs to create SopE-Gag-expressing Salmonella recombinants (15).
Generation of modified vaccinia virus Ankara SIVmac239 gag recombinant.
The parental vaccinia virus was a plaque-purified isolate of a replication-defective strain of vaccinia virus designated modified vaccinia virus Ankara (MVA) (25). Recombinant MVA virus vT338 contains the gag gene from SIVmac239 (30) inserted into the deletion III region of the MVA genome (26) under the control of the vaccinia virus 40K (H5R) promoter (17). The virus also contains the Escherichia coli lacZ gene under the control of the fowlpox C1 promoter (21) for use as a colorimetric screen for recombinant viruses. The genomic structure of the recombinant virus was determined by amplification by PCR. Expression of Gag p55 was demonstrated by Western blot assay with anti-p27 antibody (Advanced Biotechnologies, Inc.).
Animals and housing.
Rhesus macaques (Macaca mulatta) were housed at the New England Regional Primate Research Center in a centralized animal biosafety level 3 containment facility in accordance with standards of the Association for Assessment and Accreditation of Laboratory Animal Care and the Harvard Medical School Animal Care and Use Committee. Research was conducted according to the principles described in the Guide for the Care and Use of Laboratory Animals (4) and was approved by the Harvard Medical School Animal Care and Use Committee. Animals positive for the MHC class I allele Mamu-A*01 were identified by sequence-specific PCR (22) and selected for this study. All animals were also tested and found to be free of simian retrovirus type D, SIV, simian T lymphotropic virus type 1, and herpes B virus prior to assignment. Additionally, animals were screened and found to be negative for Salmonella-specific antibodies in serum. Over the course of the study, animals were monitored closely and euthanized when they were moribund or it was deemed necessary by the veterinary staff.
Salmonella strains and MVA inoculations.
Two Mamu-A*01+ rhesus macaques were inoculated with three intragastric doses of an attenuated Salmonella strain expressing SopE-Gag and boosted peripherally with MVA Gag (vT338; Therion Biologics Corporation, Cambridge, Mass.). Salmonella cultures were grown at 37°C with gentle shaking from an overnight starter culture in modified Luria-Bertani (LB) medium containing 0.3 M NaCl and 100 μg of ampicillin per ml to late log phase (0.8 to 0.9 optical density units at 600 nm [OD600]). The bacteria were then pelleted and resuspended in 2 ml of phosphate-buffered saline. Bacterial suspensions were infused directly into the duodenum of anesthetized animals that had been treated with a solution of 0.5% sodium bicarbonate to prevent the loss of the inoculum due to exposure to stomach acids (36).
Each animal was given two doses of S. enterica serovar Typhimurium, consisting of a 1:1 mixture of bacteria expressing SopE-Gag146-213 and SopE-Gag4-507 (5 × 1010 CFU) on weeks 0 and 10. On week 27, each animal received an additional dose of S. enterica serovar Typhi (5 × 109 CFU) expressing SopE-Gag146-213 and SopE-Gag4-284. On week 40, the two Salmonella strain-primed animals and two additional Mamu-A*01+ control animals were boosted intradermally with 5 × 108 PFU of MVA Gag suspended in 0.5 ml of phosphate-buffered saline as described previously (2, 19).
Analysis of Salmonella shedding.
The duration of Salmonella shedding in the feces was monitored following each bacterial inoculation. Fecal samples were collected and resuspended in selenite broth (1 g per 10 ml). Serial log dilutions (10−1 to 10−9) were spread on duplicate MacConkey agar plates (50 μl/plate) with and without ampicillin (100 μg/ml) and incubated overnight at 37°C. Plates with ampicillin selected for the plasmid-containing Salmonella strain, while plates without ampicillin were permissive for the growth of bacteria that had lost the SopE-Gag expression construct during growth in vivo. Selenite broth cultures were also incubated overnight at 37°C to enrich for Salmonella organisms present at low frequencies. Overnight selenite broth cultures were plated to MacConkey agar when Salmonella colonies were not observed at the lowest dilutions for the initial set of plates. Non-lactose-fermenting colonies were selected for biochemical confirmation as Salmonella strains with the RapiD 20E assay (BioMérieux, Lyon, France). The last dilution positive for Salmonella colonies was scored to obtain a semiquantitative estimate of the number of organisms present in the feces at each time point.
CTL assays. (i) CTL recognition of Salmonella-infected cells.
Mamu-A*01+ B-lymphoblastoid cell lines (B-LCLs) were infected at an multiplicity of infection of 40 CFU/cell with recombinant Salmonella strain grown to 0.8 to 0.9 OD600. Cells (106) were infected for 2 h in 200 μl of antibiotic-free R10 medium (RPMI 1640 with 10% fetal bovine serum) and labeled with 50 μCi of sodium [51Cr]chromate during the second hour of incubation. B-LCLs pulsed with the Mamu-A*01 Gag181-189 peptide (CTPYDINQM) and an irrelevant Nef peptide, 3K (ARGETYGRL), were also labeled as positive and negative controls, respectively. 51Cr-labeled B-LCL targets were then washed three times in R10 medium with gentamicin (100 μg/ml) to kill extracellular bacteria and to remove excess [51Cr]chromium.
CTL lines were derived from SIV-infected Mamu-A*01+ macaques by peptide stimulation as described previously (28) and plated with 51Cr-labeled B-LCLs at effector-to-target cell ratios of 40:1, 20:1, and 10:1. Unlabeled B-LCLs infected with wild-type vaccinia virus (NYCBH) were added at a 15:1 unlabeled-to-labeled target cell ratio to reduce nonspecific 51Cr release. After 4 h at 37°C, 30 μl of supernatant was harvested from each well into the wells of a LumaPlate-96 (Packard), and emitted radioactivity was measured in a 1450 Microβeta Plus Liquid scintillation counter (Wallac, Turku, Finland). The percent specific 51Cr release was calculated by the standard formula percent lysis = [(cpm with effectors − spontaneous cpm)/(maximal cpm − spontaneous cpm)] × 100.
(ii) Gag-specific CTL activity following immunization.
Freshly isolated peripheral blood mononuclear cells (PBMC) (5 × 106/ml) were stimulated directly with the Mamu-A*01 Gag181-189 peptide (1 μg/ml) or by coculture with peptide-pulsed, γ-irradiated (3,000 rad) autologous B-LCLs (10:1 PBMC to B-LCL ratio) in multiple wells of a 24-well plate. After 4 days, recombinant human interleukin-2 (generously provided by M. Gately, Hoffmann-La Roche, Nutley, N.J.) was added to 20 U/ml, and cultures were incubated for 1 week to expand the peptide-specific CTL population. CTL recognition of peptide-pulsed autologous B-LCLs was assessed after 10 to 14 days of culture at three different effector-to-target cell ratios in a 5-h chromium release assay as described above.
Proliferation assays.
PBMC were stimulated in quadruplicate wells of a 24-well plate (105 cells/well) with concanavalin A (5 μg/ml; Sigma, St. Louis, Mo.), p55 Gag (1 μg/ml; provided by Shabbir Ahmad, University of California, Davis), or aldrithiol-2-inactivated SIVmneE11S (E11S; 1 μg of p27 CA equivalent per ml) (5). PBMC controls were also incubated in R10 medium alone or with mock-treated H9 supernatant because E11S was harvested from SIV-infected H9 cells. Five days after concanavalin A stimulation and 7 days after antigen stimulation, cultures were pulsed with [3H]thymidine (1 μCi) for 6 h and then harvested with an automated cell harvester (Tomtec; Wallac). [3H]thymidine incorporation was determined with a Microβeta Plus liquid scintillation counter (Wallac). Antigen-specific proliferation, expressed as the stimulation index, was calculated by dividing the mean [3H]thymidine incorporation in antigen-stimulated wells by the mean incorporation in control wells.
Lymphocyte isolation from colon biopsies.
Endoscope-guided biopsy samples were collected from multiple sites in the colon to monitor for the homing of SIV-specific CD8+ T cells to the gastrointestinal mucosa with previously described techniques (40). Pooled biopsy specimens from 10 to 12 separate sites representing both the epithelium and the underlying lamina propria were incubated in 1 mM EDTA for 30 min, followed by one hour in 0.5 mg of type II collagenase (Sigma) per ml at 37°C with vigorous shaking. Tissues were then mechanically dispersed with a blunt-ended 18-gauge needle and filtered through a 70-μm nylon cell strainer. Cell suspensions were then layered over 35% to 60% discontinuous Percoll gradients and centrifuged at 1,000 × g for 20 min at 4°C to separate the lymphocytes and epithelial cells. Enriched lymphocyte populations were collected from the interface between the Percoll layers, washed, and resuspended in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline with 2% fetal bovine serum).
Flow cytometry.
Peripheral blood and colonic lymphocytes were analyzed by four-color flow cytometry for SIV-specific CD8+ T cells. Whole blood (100 μl) or colonic lymphocytes (1 × 106 to 2 × 106 cells) were stained for 25 min at 37°C with fluorescein isothiocyanate, peridinin chlorophyll protein, or phycoerythrin-conjugated monoclonal antibodies and allophycocyanin-conjugated tetramers. Cells were stained simultaneously with CD3-fluorescein isothiocyanate (Becton Dickinson, San Jose, Calif.), CD8-peridinin chlorophyll protein (Becton Dickinson), and α4β7-phycoerythrin (generously provided by Millennium Pharmaceuticals, Cambridge, Mass.) monoclonal antibodies and either Mamu-A*01 Gag181-189 or Mamu-A*01 Tat28-35 tetramers (generously provided by John Altman, Emory University, Atlanta, Ga.). Whole blood samples were treated for 10 min after antibody staining with FACS lysing solution (Becton Dickinson) to remove erythrocytes. Cells were then washed once in FACS buffer and fixed in 2% paraformaldehyde-phosphate-buffered saline. Samples were analyzed on a FACSCalibur flow cytometer (Becton Dickinson), and data analysis was performed with FlowJo (Tree Star, San Carlos, Calif.).
Intrarectal challenge.
Macaques were challenged intrarectally with a dose of 8.5 ng of p27 equivalents (103.5 50% tissue culture infectious doses) of SIVmac239 (23). The anal area of anesthetized animals was gently cleansed with soap and water, and the pelvis was elevated to an approximately 45° angle. The inoculum was drawn into a 1-ml syringe fitted with a 2-in. (ca. 5-cm) gastric gavage tube lubricated with KY jelly. The tube was gently inserted into the rectum to prevent abrasion or tearing of the epithelium, and the inoculum was gradually infused into the rectal cavity. After infusion, the pelvis was kept in an elevated position for approximately 10 min to prevent drainage of the inoculum.
Viral loads and CD4 counts.
SIV loads in plasma were quantitated with a real-time reverse-transcriptase-PCR assay as previously described (24). CD4 counts were determined by flow cytometry as previously described (43).
RESULTS
CTL recognition of rhesus monkey cell lines infected with SopE-Gag recombinant Salmonella strains.
Attenuated ΔphoP-phoQ strains of S. enterica serovar Typhimurium and S. enterica serovar Typhi were engineered to deliver SIV Gag polypeptides into the MHC class I antigen-processing pathway of mammalian cells via the bacterial type III secretion system. A panel of five type III-secretion constructs expressing fusions between the first 104 amino acids of the Salmonella SopE protein and different segments of the SIV Gag protein were created. Four of the constructs (SopE-Gag4-507, SopE- Gag4-439, SopE-Gag4-284, and SopE-Gag146-213) included the immunodominant Gag181-189 CTL epitope bound by the rhesus macaque MHC class I molecule Mamu-A*01 (1, 27), while a fifth construct (SopE-Gag267-507) did not include this epitope.
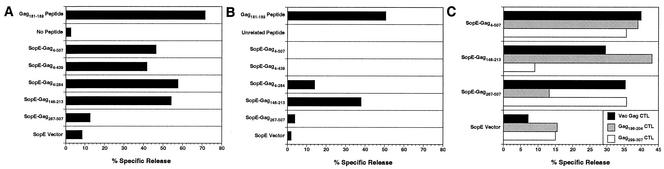
To identify the most promising bacterial strains and constructs for immunizing animals, rhesus B-cell lines were infected with ΔphoP-phoQ attenuated strains of S. enterica serovar Typhimurium and S. enterica serovar Typhi expressing each of the SopE-Gag fusions and tested for recognition by CTL lines derived from SIV-infected, Mamu-A*01+ animals (Fig. 1A and B). Delivery of each of the SopE-Gag fusions spanning the Mamu-A*01/Gag181-189 epitope by S. enterica serovar Typhimurium efficiently sensitized target cells for CTL recognition (Fig. 1A). However, CTL recognition of S. enterica serovar Typhi-infected target cells was considerably less efficient. There was no detectable CTL recognition of SopE-Gag4-507 and SopE-Gag4-439 in S. enterica serovar Typhi-infected cells (Fig. 1B). Only SopE-Gag4-284 and SopE-Gag146-213 were recognized above background levels (Fig. 1B). These results indicate that, at least in vitro, the S. enterica serovar Typhimurium recombinants are better able to sensitize rhesus cells for CTL recognition than the S. enterica serovar Typhi recombinants. These observations are also consistent with assays performed in rodent cells suggesting that the larger SopE-Gag fusions may not be translocated into cells as efficiently as the shorter fusions (data not shown).
FIG. 1.
CTL recognition of rhesus B-cell lines infected with attenuated strains of Salmonella expressing SopE-Gag fusion proteins. Mamu-A*01+ target cells were infected with ΔphoP-phoQ attenuated S. enterica serovar Typhimurium (A) or S. enterica serovar Typhi (B) and tested for recognition by CTL lines derived from an SIV-infected, Mamu-A*01+ animal. Positive (Gag181-189 peptide) and negative (no peptide or unrelated peptide [Nef23-31]) control targets were also included (A and B). CTL lines derived from an SIV-infected, Mamu-A*01− animal by stimulation of PBMC with vaccinia virus Gag (Vac Gag) or previously defined Gag epitopes (Gag196-204 and Gag299-307) were also tested for the ability to recognize target cells infected with SopE-Gag-recombinant, attenuated S. enterica serovar Typhimurium (C).
We also tested CTL lines derived from an SIV-infected Mamu-A*01− animal for the ability to recognize additional, non-Mamu-A*01-restricted Gag epitopes. CTL lines were derived from PBMC by stimulation with a recombinant vaccinia virus expressing the full-length SIV Gag protein (Vac Gag) and with peptides representing previously defined CTL epitopes in this animal (Gag196-204 and Gag299-307). Each of the CTL lines was tested for recognition of autologous B-cell targets infected with S. enterica serovar Typhimurium expressing SopE-Gag4-507, SopE-Gag146-213, SopE-Gag267-507, and the SopE vector. While the Vac Gag-stimulated CTL line killed target cells infected with each of the SopE-Gag recombinants, recognition by the peptide-stimulated CTL lines was more specific. The Gag196-204-stimulated CTL line killed target cells infected with S. enterica serovar Typhimurium expressing SopE-Gag4-507 and SopE-Gag146-213 but not SopE-Gag267-507 (Fig. 1C). Likewise, the Gag299-307-stimulated CTL line killed target cells infected with S. enterica serovar Typhimurium expressing SopE-Gag4-507 and SopE-Gag267-507 but not SopE-Gag146-213 (Fig. 1C). These results demonstrate that the Salmonella type III secretion system is capable of delivering complex polypeptide antigens for MHC class I antigen processing and presentation of multiple epitopes. Thus, CTL responses elicited by immunization with SopE-Gag recombinant Salmonella need not be limited to the Mamu-A*01/Gag181-189 epitope.
Inoculations with recombinant Salmonella strains and MVA.
Based on CTL recognition in vitro, we selected S. enterica serovar Typhimurium as the bacterial platform for the initial priming doses. To maximize Mamu-A*01/Gag181-189-specific CTL responses and to broaden T-cell responses to additional Gag epitopes, a combination of bacteria expressing SopE-Gag146-213 and SopE-Gag4-507 was used for immunization. Two Mamu-A*01+ macaques were inoculated with an equal mixture of ΔphoP-phoQ attenuated S. enterica serovar Typhimurium expressing SopE-Gag146-213 and SopE-Gag4-507 for a total dose of 5 × 1010 CFU. An identical dose of SopE-Gag146-213 and SopE-Gag4-507 recombinant S. enterica serovar Typhimurium was administered 10 weeks later. Reasoning that we might minimize Salmonella-specific immunity and enhance Gag-specific responses by switching bacterial platforms, ΔphoP-phoQ attenuated S. enterica serovar Typhi was used for a second boost on week 27. Since there was no detectable CTL recognition of SopE-Gag4-507 in S. enterica serovar Typhi-infected cells (Fig. 1B), SopE-Gag4-284 was used in place of SopE-Gag4-507. Each animal was inoculated with an equal mixture of bacteria expressing SopE-Gag146-213 and SopE-Gag4-284 for a total of 5 × 109 CFU. At week 40, each of the Salmonella-primed animals and two additional Mamu-A*01+ control animals were boosted intradermally with an MVA recombinant expressing the full-length SIV Gag protein.
Following each dose of recombinant Salmonella cells, bacterial shedding was monitored in the feces to confirm gastrointestinal exposure and to approximate the duration of Salmonella infection. Fecal samples were collected periodically after inoculation, and serial dilutions were plated to MacConkey agar with and without ampicillin to assess the stability of the SopE-Gag expression plasmids in vivo. There was a marked difference between the two animals in clinical signs and duration of shedding following the first S. enterica serovar Typhimurium inoculation (Table 1). Animal 478.99 showed no signs of clinical infection and resolved Salmonella infection within 1 week. However, animal 537.99 developed a severe case of diarrhea that resulted in significant weight loss and continued to shed S. enterica serovar Typhimurium for 4 weeks.
TABLE 1.
Salmonella shedding in feces of macaques postinoculation
| Day | Dilution of fecesa
|
|||||
|---|---|---|---|---|---|---|
|
S. enterica serovar Typhimurium
|
S. enterica serovar Typhi
|
|||||
| Wk 0
|
Wk 10
|
Wk 27
|
||||
| Animal 478.99 | Animal 537.99 | Animal 478.99 | Animal 537.99 | Animal 478.99 | Animal 537.99 | |
| 1 | 5 | 6 | 0 | 6 | 3 | 2 |
| 2 | 5 | 6 | 6 | 5 | 1 | + |
| 3 | 6 | 4 | ND | ND | 0 | 0 |
| 4 | 3 | 7 | 4 | + | ND | ND |
| 6 | ND | ND | ND | ND | 0 | 0 |
| 7 | 0 | 6 | ND | ND | ND | ND |
| 8 | ND | ND | + | ND | 0 | 0 |
| 9 | ND | 2 | ND | + | 1 | 0 |
| 10 | ND | ND | 1 | 0 | 0 | 0 |
| 11 | 0 | 2 | ND | ND | ND | ND |
| 14 | 0 | + | 0 | 0 | ND | ND |
| 16 | ND | 2 | 0 | 0 | ND | ND |
| 18 | ND | + | ND | ND | ND | ND |
| 21 | ND | 1 | ND | ND | ND | ND |
| 28 | ND | + | ND | ND | ND | ND |
| 35 | ND | 0 | ND | ND | ND | ND |
| 42 | ND | 0 | ND | ND | ND | ND |
Fecal samples were collected on the indicated days postinoculation, and serial log dilutions were plated on MacConkey agar with ampicillin (100 μg/ml). Values represent the inverse of the exponent for the last dilution positive for Salmonella (e.g., 5 = 10−5). 0, no Salmonella was recovered. +, positive only after overnight enrichment in selenite broth. ND, no sample was collected.
Following the S. enterica serovar Typhimurium and S. enterica serovar Typhi boosts at weeks 10 and 27, respectively, no adverse clinical signs were observed in either animal. Shedding after the week 10 boost ceased by 2 weeks postinoculation. Likewise, with the exception of a single sample from animal 478.99 on day 9, S. enterica serovar Typhi shedding was resolved within 1 week postinoculation in both animals (Table 1). No differences in the frequency of Salmonella recovery on plates with and without ampicillin were observed (data not shown). Western blot analysis of Salmonella colonies recovered from fecal cultures revealed the expression SopE-Gag146-213 and SopE-Gag4-507 at roughly equivalent frequencies after more than 3 weeks in animal 537.99 (data not shown). Thus, the plasmids encoding the SopE-Gag fusions were maintained well during bacterial replication in vivo in the absence of antibiotic selection.
Salmonella-specific antibody responses also confirmed infection with S. enterica serovar Typhimurium. Preimmune serum from each animal was negative for Salmonella-specific antibodies. However, S. enterica serovar Typhimurium lipopolysaccharide-specific antibody responses were detectable 1 week after the first inoculation in both animals and increased following the second inoculation at week 10 (data not shown). However, there was no increase in antibody responses after the S. enterica serovar Typhi boost at week 27, possibly due to the rather limited course of infection with this strain (data not shown and Table 1).
SIV Gag-specific CD8+ CTL responses.
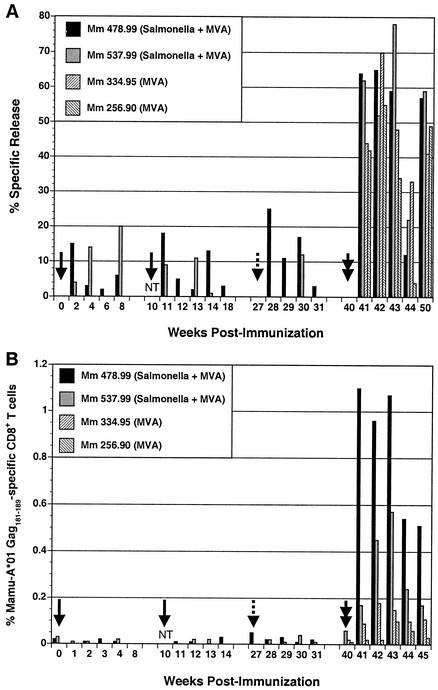
Peptide-stimulated CTL cultures were derived from peripheral blood, and Gag-specific CTL responses were measured by 51Cr release assays. CTL responses directed against the Gag181-189 epitope ranging from 5% to 25% specific release above background were observed following each dose of recombinant Salmonella (Fig. 2A). Staining of these peptide-stimulated T cells with Mamu-A*01/Gag181-189 tetramers confirmed the expansion of a Gag-specific CD8+ T-cell population at multiple time points in animal 478.99 (data not shown). After the MVA Gag boost, CTL activity was consistently observed in both of the Salmonella-primed animals and in control animals immunized with MVA Gag alone. Although CTL responses were somewhat higher at three of five time points in the Salmonella-primed animals compared to the animals that received only MVA Gag, these results did not clearly differentiate between primary and secondary responses in these two groups of animals (Fig. 2A). However, this was not surprising because 51Cr release assays are not a particularly quantitative method for measuring CTL responses in vivo.
FIG. 2.
SIV Gag-specific CD8+ T-cell responses following immunization with recombinant Salmonella and modified vaccinia virus Ankara (MVA) expressing SIV Gag. Gag-specific CD8+ T-cell responses were followed by 51Cr release assays (A) and tetramer staining (B). The solid arrows indicate inoculations with S. enterica serovar Typhimurium given at weeks 0 and 10, the dashed arrow indicates the S. enterica serovar Typhi inoculation at week 27, and the double arrow indicates the MVA Gag boost at week 40. NT indicates that samples were not tested at week 10. (A) CTL cultures were derived from PBMC by peptide stimulation and tested for CTL recognition of autologous B-cell lines pulsed with Gag181-189 in 51Cr release assays. Bars indicate the difference in percent specific release for target cells pulsed with Gag181-189 and targets pulsed with an unrelated Nef peptide at the highest effector-to-target cell ratio tested for each time point (20:1 to 40:1). Differences of greater than 10% specific release are considered positive. (B) PBMC were stained with monoclonal antibodies to CD3 and CD8 and with Mamu-A*01/Gag181-189 tetramers and analyzed by flow cytometry. After gating on the CD3+ CD8+ population, the percentage of tetramer-positive cells was determined at each time point. Values greater than 0.1% are considered significant.
Peripheral blood lymphocytes were also stained directly with Mamu-A*01/Gag181-189 tetramers as a more quantitative method for following the development of Gag-specific CD8+ T-cell responses. During the Salmonella-priming phase, the frequency of Gag-specific CD8+ T cells was below the threshold of detection in both animals 478.99 and 537.99. However, after boosting with MVA Gag, tetramer-positive CD8+ T-cell populations were detectable in all four animals (Fig. 2B). Furthermore, there was a clear difference in the kinetics and magnitude of these responses between the Salmonella-primed animals and the animals that received only MVA Gag. Both of the macaques that were inoculated with recombinant Salmonella, 478.99 and 537.99, were tetramer positive (>0.1% of the CD8+ T-cell population) at week 41, 1 week after the MVA Gag boost. In contrast, tetramer-positive T-cell responses were undetectable until weeks 42 and 43 in animals 334.95 and 256.90, respectively, that had received only MVA Gag (Fig. 2B). The frequency of Gag-specific CD8+ T cells was also considerably higher in the Salmonella-primed animals than in the MVA Gag controls. While frequencies varied from one time point to the next, at week 43, when all four animals had detectable Gag-specific CD8+ T-cell responses, the percentage of tetramer-positive cells was 4- to 10-fold higher in the Salmonella-primed animals than in the animals immunized with MVA Gag alone. Thus, these observations are consistent with the expansion of secondary CD8+ T-cell responses to Gag in the Salmonella-primed animals.
SIV-specific proliferative responses.
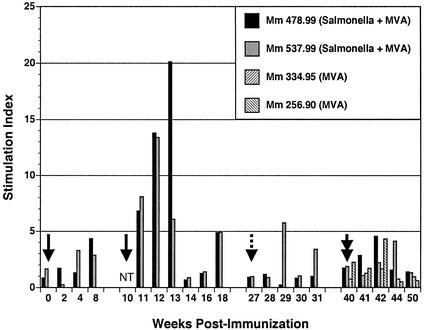
Proliferative responses to exogenous SIV antigens were monitored as an indicator of the development of Gag-specific CD4+ T-cell responses. Fresh PBMC were stimulated with aldrithiol-inactivated preparations of SIV (5), and antigen-specific proliferation was assessed by [3H]thymidine incorporation. Stimulation indexes were calculated by dividing the [3H]thymidine incorporation in response to antigen by the basal level of incorporation for PBMC treated with control medium, and stimulation index values greater than 3 were considered positive (Fig. 3).
FIG. 3.
Gag-specific proliferative responses following immunization with a recombinant Salmonella and MVA. PBMC were stimulated with aldrithiol-2-inactivated SIV or a 1,000-fold concentrate of aldrithiol-2-treated microvesicles prepared from uninfected cells. After 7 days, antigen-specific proliferation was determined in a 6-h [3H]thymidine incorporation assay. Stimulation indexes represent [3H]thymidine incorporation in response to SIV divided by incorporation in response to the microvesicle controls. Stimulation index values greater than 3 are considered positive. Solid arrows indicate the doses of S. enterica serovar Typhimurium given at weeks 0 and 10, the dashed arrow indicates the dose of S. enterica serovar Typhi given at week 27, and the double arrow indicates the MVA Gag inoculation at week 40. NT indicates that SIV-specific proliferation was not tested.
SIV-specific proliferative responses were low in both animals after the initial priming dose of S. enterica serovar Typhimurium. However, after the second S. enterica serovar Typhimurium dose at week 10, robust proliferative responses were observed in both animals for 3 weeks after inoculation (Fig. 3). Stimulation indexes ranged between 5 and 20, which are comparable to T-cell proliferative responses observed in animals chronically infected with live, attenuated strains of SIV (16). Thus, these results clearly demonstrate the stimulation of virus-specific CD4+ T-cell responses by SopE-Gag-recombinant S. enterica serovar Typhimurium. However, with the exception of two time points for animal 537.99, proliferative responses after the S. enterica serovar Typhi boost at week 27 were undetectable (Fig. 3). While the reasons for this are unclear, the decision to use SopE-Gag4-284 rather than SopE-Gag4-507 and to lower the immunizing dose of the Salmonella strain 10-fold may have resulted in less efficient stimulation of Gag-specific CD4+ T-cell responses in animals 478.99 and 537.99. Likewise, proliferative responses after the week 40 MVA Gag boost were also low compared to responses following the second dose of S. enterica serovar Typhimurium in these two animals (Fig. 3). Since MVA Gag potently stimulated CD8+ T-cell responses, the less efficient boosting of CD4+ T-cell responses may reflect a more effective stimulation of CD8+ T-cell responses than CD4+ T-cell responses.
Gag-specific CD8+ T cells induced following Salmonella/MVA vaccination express the mucosal homing receptor α4β7.
As a marker for mucosal homing, peripheral blood lymphocytes were stained with antibodies to α4β7. α4β7 is an integrin expressed on lymphocytes specific for gastrointestinal pathogens and binds to the mucosal addressin cell adhesion molecule (MadCAM1) expressed on high endothelial venules serving the gastrointestinal mucosa (9, 32, 38). Interactions between α4β7 and MadCAM1 participate in the migration of lymphocytes out of the circulatory system and into mucosal tissues (18, 42). Thus, α4β7 represents a convenient surface marker for lymphocytes homing to gastrointestinal compartments.
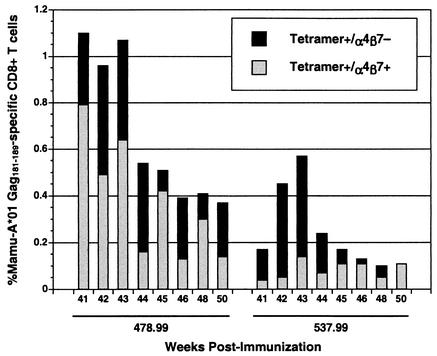
Following the MVA Gag boost, a significant percentage of the Mamu-A*01/Gag181-189 tetramer-positive population for each of the Salmonella-primed animals expressed α4β7. While the α4β7+ tetramer-positive population was more prominent in animal 478.99, the animal that made the strongest Gag-specific CD8+ T-cell response, this population was also observed in animal 537.99 at multiple time points (Fig. 4). In most cases, more than 50% of the tetramer-positive CD8+ T-cell population expressed α4β7. The tetramer-positive CD8+ T-cell responses in the two animals that received a single peripheral immunization with MVA Gag were too low to afford a meaningful comparison. However, Gag-specific CD8+ T-cell responses stimulated by peripheral DNA prime/MVA boost regimens were previously shown to be α4β7 negative (10). Thus, these results further suggest that Gag-specific CD8+ T-cell responses were primed in the gastrointestinal mucosa by the SopE-Gag-recombinant Salmonella strain.
FIG. 4.
Gag-specific CD8+ T cells in the peripheral blood of the Salmonella-primed animals express the mucosal homing receptor α4β7. PBMC were stained with monoclonal antibodies to CD3, CD8, and α4β7 and with Mamu-A*01 Gag181-189 tetramers and analyzed by flow cytometry. The percentage of total Gag181-189-specific CD8+ T cells at each time point is indicated by the black bars, and the fraction of this population that was also α4β7+ is indicated by the overlapping gray bars.
Gag-specific CD8+ T cells induced by Salmonella strain/MVA vaccination home to the colonic mucosa.
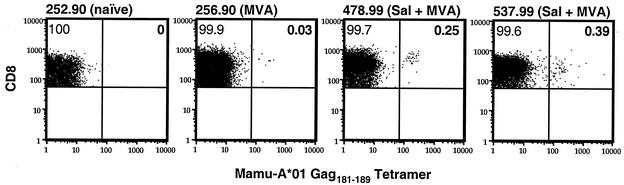
SIV Gag-specific CD8+ T cells were also observed among lymphocytes isolated from the colon of Salmonella strain-primed animals. Lymphocytes were isolated from endoscope-guided pinch biopsies of the colon 4 weeks after the MVA Gag boost (week 44) and stained with Mamu-A*01/Gag181-189 tetramers. A clear elevation in the Gag-specific CD8+ T-cell population was observed in both Salmonella-primed animals (animals 478.99 and 537.99) compared to naïve (animal 252.90) and MVA Gag-immunized (animal 256.90) Mamu-A*01+ control animals (Fig. 5). Thus, gastrointestinal priming with the SopE-Gag-secreting recombinant Salmonella followed by boosting intradermally with MVA Gag induced Gag-specific CD8+ T-cell responses that trafficked to the colonic mucosa.
FIG. 5.
Homing of Gag-specific CD8+ T cells to the colon in Salmonella strain (Sal)-primed macaques. Freshly isolated lymphocytes from endoscope-guided pinch biopsies of the colon were stained with antibodies to CD3 and CD8 in addition to Mamu-A*01 Gag181-189 tetramers and analyzed by flow cytometry. After gating on the CD3+ CD8+ T-cell population, the percentage of CD8+ T cells specific for Mamu-A*01/Gag181-189 was determined, as indicated in the upper right quadrants of each plot. More than 100,000 events and at least 6,000 CD3+ CD8+ T cells were collected for each sample.
Intrarectal challenge with SIVmac239.
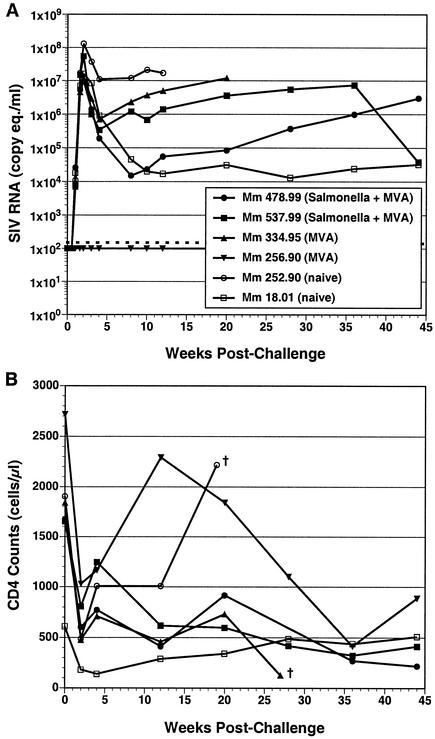
Each of the four immunized animals and two additional Mamu-A*01+, naïve control animals were challenged intrarectally with SIVmac239 at week 50. Both of the Salmonella-primed animals became infected, and neither exhibited significant differences in peak or postacute viral loads compared to naïve control animals (Fig. 6A). Furthermore, both of the Salmonella strain-primed animals exhibited declining CD4+ T-cell counts, similar to naïve and MVA Gag control animals (Fig. 6B). Thus, we did not observe any evidence of protection in this pilot study. Surprisingly, one of the animals immunized with MVA Gag alone (animal 256.90) did not become infected and remained resistant to infection with a threefold-higher dose of SIVmac239 administered intrarectally at week 58 (Fig. 6A).
FIG. 6.
SIV RNA in plasma and CD4+ T-cell counts after intrarectal challenge with SIVmac239. (A) Plasma viral RNA loads were determined with a quantitative reverse transcription-PCR assay (24). This assay has a nominal sensitivity of 100 copy equivalents/ml of plasma (indicated by the dashed line) and an interassay coefficient of variation of <25%. (B) CD4+ T-cell counts were determined by flow cytometry as described previously (43). Time points marked with † indicate the week that an animal was euthanized.
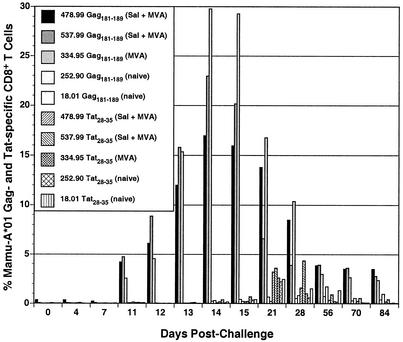
We continued to follow the development of SIV-specific CD8+ T-cell responses in the blood after challenging by staining with Mamu-A*01/Gag181-189 and Mamu-A*01/Tat28-35 tetramers. With the exception of animal 256.90, each of the immunized animals made dramatic secondary CD8+ T-cell responses to Gag. These responses peaked 2 weeks after infection at between 15% and 30% of the CD8+ T-cell population (Fig. 7) and declined with resolution of primary viremia (Fig. 6A). In contrast, primary CD8+ T-cell responses to SIV emerged a week later and were much lower in frequency. The Gag-specific responses in the naïve control animals and the Tat-specific responses in each of the immunized animals peaked between 3 and 4 weeks after infection at less than 5% of the peripheral blood CD8+ T-cell population (Fig. 7).
FIG. 7.
Postchallenge SIV Gag- and Tat-specific CD8+ T-cell responses. PBMC were stained with monoclonal antibodies to CD3 and CD8 and with Mamu-A*01/Gag181-189 or Mamu-A*01/Tat28-35 tetramers and analyzed by flow cytometry. After gating on the CD3+ CD8+ population, the percentage of cells specific for Gag181-189 and Tat28-35 was determined at each time point.
DISCUSSION
Despite recent advances in the development of vaccine approaches able to induce CTL responses, there is a continuing need to develop better methods to induce CTL responses able to home to mucosal sites. We therefore evaluated the potential of the Salmonella type III secretion system to prime SIV-specific T-cell responses in the gastrointestinal mucosa of rhesus macaques. The ability of the Salmonella type III secretion system to deliver SIV Gag polypeptides into rhesus cells for antigen processing and presentation to CTL was first demonstrated in vitro. Infection of rhesus B-cell lines with S. enterica serovar Typhimurium and S. enterica serovar Typhi expressing SopE-Gag fusions sensitized cells for recognition by Gag-specific CTL derived from SIV-infected animals. These results demonstrated that the Salmonella type III secretion system was capable of delivering polypeptide antigens, not just discrete CTL epitopes, into the MHC class I antigen-processing pathway of macaque cells.
Recombinants expressing SopE fusions with shorter Gag polypeptides were generally recognized better than recombinants expressing fusions with longer Gag sequences, suggesting that the larger fusion proteins may not be efficiently translocated into host cells. This was particularly evident in CTL assays with S. enterica serovar Typhi-infected target cells. Indeed, there was no detectable CTL recognition of the two largest fusions, SopE-Gag4-507 and SopE-Gag4-439, when they were expressed by S. enterica serovar Typhi. Similar differences in CTL recognition of the SopE-Gag fusion proteins were observed in preliminary assays with S. enterica serovar Typhimurium recombinants in rodent cells. These experiments revealed a trend toward better T-cell recognition of the shorter SopE-Gag fusion proteins and are consistent with Western blots showing that the larger SopE-Gag fusion proteins were not secreted by the Salmonella type III secretion system as efficiently as the smaller fusion proteins (data not shown). At present, it is unclear whether these differences in the efficiency of secretion and antigen presentation reflect differences in the size of the polypeptide fusions or sequence-specific properties unique to certain regions of the SIV Gag protein.
Intragastric inoculation of rhesus macaques with attenuated Salmonella strains expressing selected SopE-Gag fusions also primed Gag-specific CTL responses in vivo. While CTL responses to Gag were initially low during the Salmonella-priming phase, these responses increased dramatically following the MVA Gag boost. Gag-specific CD8+ T-cell frequencies in the two Salmonella-primed/MVA-boosted animals were similar to Gag-specific CD8+ T-cell frequencies stimulated by early DNA prime/MVA boost regimens (2, 19). Furthermore, these responses were higher in the Salmonella-primed animals than in the animals that received MVA Gag alone. The higher frequency of Gag-specific CD8+ T cells in the two Salmonella-primed animals is characteristic of a secondary immune response and is consistent with the priming of these responses by SopE-Gag-expressing Salmonella strains.
Following the MVA Gag boost, a fraction of the Gag-specific CD8+ T-cell population in the peripheral blood of each of the Salmonella-primed animals also expressed the mucosal homing receptor α4β7. The α4β7+ population was variable but generally represented 30% to 80% of the total tetramer-positive population. α4β7 is expressed on lymphocytes specific for gastrointestinal pathogens, and interactions between α4β7 and MadCAM1 on the endothelium of blood vessels serving the gastrointestinal tract facilitate the trafficking of lymphocytes into effector sites of the gut-associated lymphoid tissues (9, 18, 32, 42). α4β7+ CD8+ T-cell responses to SIV have been observed in macaques infected with wild-type and live attenuated strains of the virus (10). This α4β7+ phenotype probably reflects the preferential replication of SIV in activated CD4+ T cells of the intestine (39, 40). In contrast, SIV-specific CD8+ T responses stimulated by a peripheral DNA prime/MVA boost vaccine regimen were shown to be α4β7 negative (10). The expression of α4β7 on Gag-specific CD8+ T cells of Salmonella-primed/MVA-boosted animals further supports the gastrointestinal priming of these responses by recombinant Salmonella strains. To our knowledge, this represents the first demonstration of SIV-specific, α4β7+ CD8+ T-cell responses stimulated by a recombinant mucosal vaccine approach.
Consistent with the α4β7+ phenotype of Gag-specific CD8+ T cells in the blood, Gag-specific CD8+ T cells were also found in lymphocytes isolated from endoscope-guided colon biopsies. Tetramer staining revealed a small but distinct Gag-specific CD8+ T-cell population that was present in colonic lymphocytes isolated from the Salmonella-primed animals but not from naïve or MVA Gag-immunized Mamu-A*01+ control animals. Thus, mucosal priming with recombinant Salmonella strains followed by an intradermal boost with MVA Gag stimulated CD8+ T-cell responses that homed to mucosal surfaces of the gastrointestinal tract. This analysis did not discriminate between intraepithelial and lamina propria lymphocyte populations, both of which were represented in the biopsy samples. Nevertheless, the presence of Gag-specific CD8+ T cells at such superficial sites is encouraging, since it indicates that the virus-specific T cells trafficked to mucosal tissues relevant to the earliest stages of viral transmission.
Despite having stimulated Gag-specific CD8+ T-cell responses in both peripheral and mucosal compartments, animals immunized with SopE-Gag-recombinant Salmonella were not protected against an intrarectal challenge with SIVmac239. Each of the Salmonella-primed animals became infected and did not differ significantly in peak or postacute viral loads from control animals. The goal of this pilot study was to evaluate the potential of the Salmonella type III secretion system to prime SIV-specific mucosal CTL responses. Thus, the Salmonella and MVA recombinants were designed to express only Gag polypeptides and emphasized responses to a single CD8+ T-cell epitope (Mamu-A*01/Gag181-189) that could be detected easily with available MHC class I tetramers. However, these responses may not have been diverse enough to contain virus replication.
Although animals were immunized with a Salmonella strain expressing both SopE-Gag146-213 and SopE-Gag4-507, the SopE-Gag4-507 fusion protein was secreted much less efficiently than SopE-Gag146-213 in Western blot assays on bacterial culture supernatants (data not shown). Therefore, despite CTL recognition of SopE-Gag4-507 in S. enterica serovar Typhimurium-infected cells in vitro (Fig. 1A), this construct may have contributed little to the priming of Gag-specific CD8+ T-cell responses in vivo. Hence, mucosal priming of CD8+ T-cell responses may have been limited to Gag residues 146 to 213 and to the Mamu-A*01/Gag181-189 epitope alone.
An additional explanation for the lack of protection in this study is the relative weakness of Gag-specific CD8+ T-cell responses. While the responses in the Salmonella strain-primed/MVA-boosted animals were similar in frequency to responses achieved by early DNA prime/MVA boost regimens (2, 19), they were lower than responses stimulated by more recent, better-optimized regimens (3, 35). Finally, SIVmac239 represents a particularly rigorous challenge virus. Protective immunity to SIVmac239 has been extremely difficult to elicit even with multivalent vaccine approaches that stimulate stronger and much broader cellular immunity (20). Thus, it is not surprising that the narrowly focused and relatively weak cellular immune responses stimulated in this pilot study failed to contain virus replication after challenge with SIVmac239.
One of the animals vaccinated with MVA Gag alone (animal 256.90) did not become infected after intrarectal challenge with SIVmac239. Since this animal remained resistant to a threefold-higher dose of virus administered 8 weeks later, it is unlikely that the failure to establish an infection in this animal simply reflects the odds of virus transmission across the rectal mucosa with a low-titer virus inoculum. While it is possible that immunization with MVA Gag may have contributed to the resistance of 256.90 to SIV infection, this is also unlikely to be the sole explanation for this result, since such solid protection against challenge with pathogenic SIVmac strains is rarely, if ever, observed in animals immunized with recombinant poxviruses. Resistance to mucosal SIV infection among control animals has been observed in other vaccine studies and often complicates the interpretation of protection data obtained from mucosal challenges (29). Thus, some animals may have an inherent resistance to mucosal infection that is poorly understood at present (29).
The results of this study suggest that mucosal priming with vectors such as attenuated Salmonella may direct virus-specific cellular immune responses to mucosal sites of virus transmission and early virus replication. However, the T-cell responses stimulated in this study were insufficient to protect against an intrarectal challenge with SIVmac239. Having demonstrated the potential of the Salmonella type III secretion system to prime SIV-specific CTL responses in macaques, future studies with additional Salmonella strains and MVA recombinants and more optimal vaccination protocols can now be designed to more rigorously evaluate the ability of this approach to provide protection against viral challenge.
Acknowledgments
We thank Angela Carville for collecting the intestinal biopsies, Mandy Cromwell for her assistance in analyzing the flow cytometry data for the colon biopsies, and Michael Piatak, Jr., for assistance with the viral load measurements. We also thank John Altman for providing the MHC class I tetramers, Meryl Forman for providing the α4β7 antibody, Maurice Gately for providing the recombinant interleukin-2, and Roy Curtiss for providing the ΔphoP-phoQ Salmonella strains.
This work was supported by Public Health Service grants AI10464, RR00168, AI35365, AI46953, and AI45314 and funded in part with federal funds from the National Institutes of Health under contract no. NO1-CO-12400. R. Paul Johnson is an Elizabeth Glaser Scientist and is supported by the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 2.Allen, T. M., T. U. Vogel, D. H. Fuller, B. R. Mothe, S. Steffen, J. E. Boyson, T. Shipley, J. Fuller, T. Hanke, A. Sette, J. D. Altman, B. Moss, A. J. McMichael, and D. I. Watkins. 2000. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/modified vaccinia virus Ankara boost regimen. J. Immunol. 164:4968-4978. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., R. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1996. The Institute of Laboratory Animal Resources, National Research Council: guide for the care and use of laboratory animals, 86-123. National Institutes of Health, Washington, D.C.
- 5.Arthur, L. O., J. W. Bess, E. N. Chertova, J. L. Rossio, M. T. Esser, R. E. Benveniste, L. E. Henderson, and J. D. Lifson. 1998. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS. Res. Hum. Retroviruses 14:S311-S319. [PubMed]
- 6.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T.-M. Fu, W. Wagner, M. Bliska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M.-E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 7.Belyakov, I. M., M. A. Derby, J. D. Ahlers, B. L. Kelsall, P. Earl, B. Moss, W. Strober, and J. A. Berzofsky. 1998. Mucosal immunization with HIV-1 peptide vaccine induces mucosal and systemic cytotoxic T lymphocytes and protective immunity in mice against intrarectal recombinant HIV-vaccinia challenge. Proc. Natl. Acad. Sci. USA 95:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belyakov, I. M., Z. Hel, B. Kelsall, V. A. Kuznetsov, J. D. Ahlers, J. Nacsa, D. I. Watkins, T. M. Allen, A. Sette, J. Altman, R. Woodward, P. D. Markham, J. D. Clements, G. Franchini, W. Strober, and J. A. Berzofsky. 2001. Mucosal AIDS vaccine reduces disease and viral load in gut reservoir and blood after mucosal infection of macaques. Nat. Med. 7:1320-1326. [DOI] [PubMed] [Google Scholar]
- 9.Berlin, C., E. L. Berg, M. J. Briskin, D. P. Andrew, P. J. Kilshaw, B. Holzmann, I. L. Weissman, A. Hamann, and E. C. Butcher. 1993. α4β7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell 74:185-195. [DOI] [PubMed] [Google Scholar]
- 10.Cromwell, M. A., R. S. Veazey, J. D. Altman, K. G. Mansfield, R. Glickman, T. M. Allen, D. I. Watkins, A. A. Lackner, and R. P. Johnson. 2000. Induction of mucosal homing virus-specific CD8+ T lymphocytes by attenuated simian immunodeficiency virus. J. Virol. 74:8762-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, N. L., I. J. Caley, K. W. Brown, M. R. Betts, D. M. Irlbeck, K. M. McGrath, M. J. Connell, D. C. Montefiori, J. A. Frelinger, R. Swanstrom, P. R. Johnson, and R. E. Johnston. 2000. Vaccination of macaques against pathogenic simian immunodeficiency virus with Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:371-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eo, S. K., M. Gierynska, A. A. Kamar, and B. T. Rouse. 2001. Prime-boost immunization with DNA vaccine: mucosal route of administration changes the rules. J. Immunol. 166:5473-5479. [DOI] [PubMed] [Google Scholar]
- 13.Franchini, G., M. Robert-Guroff, J. Tartaglia, A. Aggarwal, A. Abimiku, J. Benson, P. Markham, K. Limbach, G. Hurteau, J. Fullen, K. Aldrich, N. Miller, J. Sadoff, E. Paoletti, and R. C. Gallo. 1995. Highly attenuated HIV type 2 recombinant poxviruses, but not HIV-2 recombinant Salmonella vaccines, induce long-lasting protection in rhesus macaques. AIDS. Res. Hum. Retroviruses 11:909-920. [DOI] [PubMed] [Google Scholar]
- 14.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 15.Galán, J. E., and R. Curtiss. 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 6:433-443. [DOI] [PubMed] [Google Scholar]
- 16.Gauduin, M.-C., R. L. Glickman, S. Ahmad, R. Yilma, and R. P. Johnson. 1999. Immunization with live attenuated simian immunodeficiency virus induces strong type1 T helper responses and β-chemokine production. Proc. Natl. Acad. Sci. USA 96:14031-14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritz, L., A. Destree, N. Cormier, E. Day, V. Stallard, T. Caiazzo, G. Mazzara, and D. Panicali. 1990. Generation of hybrid genes and proteins by vaccinia virus-mediated recombination: application to human immunodeficiency virus type 1 env. J. Virol. 64:5948-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamann, A., D. P. Andrew, D. Jablonski-Westrich, B. Holzmann, and E. C. Butcher. 1994. Role of α4-integrins in lymphocyte homing to mucosal tissues in vivo. J. Immunol. 152:3282-3293. [PubMed] [Google Scholar]
- 19.Hanke, T., R. V. Samuel, T. J. Blanchard, V. C. Neumann, T. M. Allen, J. E. Boyson, S. A. Sharpe, N. Cook, G. L. Smith, D. I. Watkins, M. P. Cranage, and A. J. McMichael. 1999. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by with a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J. Virol. 73:7524-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wollinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins, S., L. Gritz, C. H. Fedor, E. M. O'Neill, L. K. Cohen, and D. L. Panicali. 1991. Formation of lentivirus particles by mammalian cells infected with recombinant fowlpox virus. AIDS Res. Hum Retroviruses 7:991-998. [DOI] [PubMed] [Google Scholar]
- 22.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, M. G., S. Bellah, K. McKinnon, J. Yalley-Ogunro, P. M. Zack, W. R. Elkins, R. C. Desrosiers, and G. E. Eddy. 1994. Titration and characterization of two rhesus derived SIVmac challenge stocks. AIDS. Res. Hum. Retroviruses 10:213-220. [DOI] [PubMed] [Google Scholar]
- 24.Lifson, J. D., J. L. Rossio, M. Piatak, T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr, A., V. Hochstein-Mintzel, and H. Stickl. 1975. Abstammung, Eigenschaften and Verwendung des attenuierten vaccinia-Stammes MVA. Infection 3:6-14. [Google Scholar]
- 26.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72:1031-1038. [DOI] [PubMed] [Google Scholar]
- 27.Miller, M. D., H. Yamamoto, A. L. Hughes, D. I. Watkins, and N. L. Letvin. 1991. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J. Immunol. 147:320-329. [PubMed] [Google Scholar]
- 28.Murphy, C. G., W. T. Lucas, R. E. Means, S. Czajak, C. L. Hale, J. D. Lifson, A. Kaur, R. P. Johnson, D. M. Knipe, and R. C. Desrosiers. 2000. Vaccine protection against simian immunodeficiency virus by recombinant strains of herpes simplex virus. J. Virol. 74:7745-7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng, B., R. Voltan, L. Lim, Y. Edghill-Smith, S. Phogat, D. S. Dimitrov, K. Arora, M. Leno, S. Than, R. Woodward, P. D. Markham, M. Cranage, and M. Robert-Guroff. 2002. Rhesus macaque resistance to mucosal simian immunodeficiency virus infection is associated with a postentry block in viral replication. J. Virol. 76:6016-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS. Res. Hum. Retroviruses 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 31.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 32.Rott, L. S., J. R. Rose, D. Bass, M. B. Williams, H. B. Greenberg, and E. C. Butcher. 1997. Expression of mucosal homing receptor α4β7 by circulating CD4+ cells with memory for intestinal rotavirus. J. Clin. Investig. 100:1204-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russmann, H., H. Shams, F. Poblete, Y. Fu, J. E. Galán, and R. O. Donis. 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science 281:565-568. [DOI] [PubMed] [Google Scholar]
- 34.Schodel, F., and R. Curtiss. 1995. Salmonellae as oral vaccine carriers. Dev. Biol. Stand. 84:245-253. [PubMed] [Google Scholar]
- 35.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 36.Steger, K. K., and C. D. Pauza. 1997. Immunization of Macaca mulatta with aroA attenuated Salmonella typhimurium expressing the SIVp27 antigen. J. Med. Primatol. 26:44-50. [DOI] [PubMed] [Google Scholar]
- 37.Steger, K. K., P. J. Valentine, F. Heffron, M. So, and C. D. Pauza. 1999. Recombinant attenuated Salmonella typhimurium stimulate lymphoproliferative responses to SIV capsid in rhesus macaques. Vaccine 17:923-932. [DOI] [PubMed] [Google Scholar]
- 38.Streeter, P. R., E. L. Berg, B. T. N. Rouse, R. F. Bargatze, and E. C. Butcher. 1988. A tissue-specific endothelial cell molecule involved in lymphocyte homing. Nature 331:41-46. [DOI] [PubMed] [Google Scholar]
- 39.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rozenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T-cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 40.Veazey, R. S., I. C. Tham, K. G. Mansfield, M. DeMaria, A. E. Forand, D. E. Shvetz, L. V. Chalifoux, P. K. Sehgal, and A. A. Lackner. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 42.Williams, M. B., and E. C. Butcher. 1997. Homing of naive and memory T lymphocyte subsets to Peyer's patches, lymph nodes, and spleen. J. Immunol. 159:1746-1752. [PubMed] [Google Scholar]
- 43.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]