Abstract
135 years ago Ferdinand Cohn, the founder of bacteriology, microscopically observed a conspicuous filamentous bacterium with a complex life cycle and described it as Crenothrix polyspora. This uncultured bacterium is infamous for mass developments in drinking water systems, but its phylogeny and physiology remained unknown. We show that C. polyspora is a gammaproteobacterium closely related to methanotrophs and capable of oxidizing methane. We discovered that C. polyspora encodes a phylogenetically very unusual particulate methane monooxygenase whose expression is strongly increased in the presence of methane. Our findings demonstrate a previously unrecognized complexity of the evolutionary history and cell biology of methane-oxidizing bacteria.
Keywords: drinking water, methane, oxidizing bacteria
During the nineteenth century, description of microorganisms experienced its first peak. Among others, several morphologically striking bacteria including Thiothrix nivea, Leucothrix mucor, and Zoogloea ramigera (1) and important pathogens, such as Bacillus anthracis, Vibrio cholerae, and Mycobacterium tuberculosis (1), were discovered and named. During this period, in 1870, Ferdinand Cohn observed for the first time an unusual filamentous, sheathed organism in a drinking water well and referred to it as Crenothrix polyspora (2). This organism was described as having a developmental cycle with micro- and macrogonidia, and its morphological description was refined in 1977 by electron microscopy (3). Based on its ultrastructure, it was hypothesized that C. polyspora might be a methane oxidizer (3). However, despite its worldwide importance as a problem organism in drinking water production and supply (4, 5), C. polyspora remained, in contrast to the other bacteria described in this era, phylogenetically and physiologically uncharacterized.
Results and Discussion
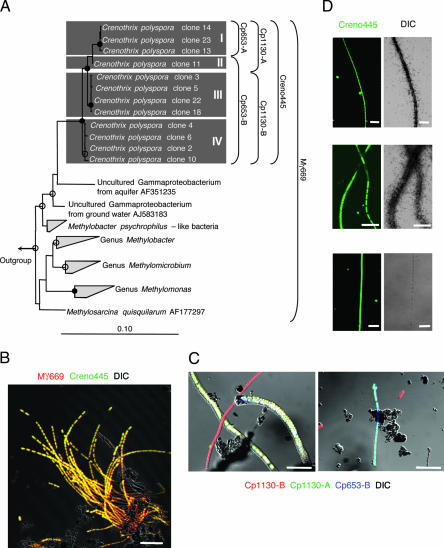
We observed, by differential interference contrast microscopy (DIC), filamentous organisms with the typical C. polyspora morphology (3) in backwash water from rapid sand filters from a German drinking water treatment plant. From the diverse microbial community present in the backwash water, we were able to physically enrich C. polyspora because of its filamentous shape by filtering several hundred liters of water through 200- to 400-μm sieves. Eighteen 16S rRNA gene clones were retrieved from the obtained biomass, and 12 of these clones showed >98% sequence similarity with each other and formed a well supported monophyletic cluster within the gammaproteobacterial type I methanotrophs. Within this cluster, four subclusters (Fig. 1A, I–IV) were identified. No other 16S rRNA gene sequences closely related to known methanotrophs were detected (see Table 1, which is published as supporting information on the PNAS web site). By using newly designed specific FISH probes together with published probes, we demonstrated that this 16S rRNA sequence cluster represents C. polyspora and that the enriched biomass consisted almost exclusively of this species (Fig. 1B; quantification of the FISH data revealed that >99% of the cellular biovolume stained with the nucleic acid-binding dye SYBR green I could be assigned to C. polyspora) and did not contain detectable numbers of Eukarya(probe Euk516) or Archaea(probe Arch915). The phylogenetic assignment of C. polyspora to type I methanotrophs is consistent with the occurrence of intracellular membrane stacks (ICM) and their arrangement as vesicular discs in the cytoplasma (see Fig. 4, which is published as supporting information on the PNAS web site; and see ref. 3). In addition, the FISH hybridization patterns indicated that the four subclusters represent different 16S rRNA genes of one organism that are differentially expressed during the developmental cycle of C. polyspora. Microgonidia were detected in thick filaments, expressed all probe-targeted 16S rRNA genes, and showed a high ribosome content per cell, comparable to that observed in other C. polyspora cells. In contrast, cells found in thinner filaments transcribed only subsets of the 16S rRNA gene repertoire, despite showing bright FISH signals (Fig. 1C).
Fig. 1.
Phylogeny, in situ detection, and physiology of C. polyspora. (A) In the maximum likelihood tree, the 16S rRNA clones of C. polyspora cluster together with clones retrieved from ground water (24). The four subclusters are indicated as I–IV. Methylobacter psychrophilus is the closest cultured relative of C. polyspora (94.2% sequence similarity). Maximum parsimony bootstrap values >75% and 90% are indicated by white and black circles, respectively. Bar shows 10% estimated sequence divergence. Virtually identical tree topologies were retrieved by using maximum parsimony and neighbor-joining analyses. The specificity of applied 16S rRNA-targeted probes is indicated by brackets. (B) In situ detection of C. polyspora with probes Mγ669 and Creno445 (yellow color indicates simultaneous binding of both probes). (C) Differential expression of 16S rRNA genes in C. polyspora. After application of three different probes, three different hybridization patterns were observed. Microgonidia-bearing filaments simultaneously expressed all targeted 16S rRNA genes and, thus, appear white. Some thinner filaments expressed only two or one 16S rRNA gene types and, thus, are labeled turquoise (probes Cp1130-A and Cp653-B) or red (probe Cp1130-B), respectively. The red-labeled cells can be explained only by the presence of a fifth 16S rRNA gene type in C. polyspora (or microdiversity within one of the four types which affects probe-binding sites), which was not retrieved in our gene library and binds probe Cp1130-B but not probes Cp653-B and Cp1130-A. All filaments also hybridized with probes Cp653-A and Creno445 (data not shown). (D) FISH-MAR experiments show that C. polyspora incorporates methane (Top) and methanol (Middle) but not formate (Bottom). (Scale bars, 20 μm.)
The presence of ICM in the cytoplasm of C. polyspora also provides some guidance to its physiology. ICM are either found in phototrophs or are indicative of methane-oxidizing and nitrifying bacteria (6, 7). Because the investigated filaments thrived in a habitat without light, it is unlikely that C. polyspora is a phototroph. Therefore, we used previously developed PCR assays (8–10) to screen the C. polyspora enrichment for genes encoding subunits of the ammonia monooxygenase (amoA) and the particulate methane monooxygenase (pmoA), which represent key catabolic enzymes of the ammonia- and methane-oxidizing bacteria, respectively. In addition, we tested for the presence of the gene encoding a subunit of the soluble methane monooxygenase (mmoX), an enzyme used for methane oxidation by a subset of methanotrophs (11, 12). No mmoX was detectable, but we retrieved several amoA and pmoA fragments. All amoA sequences were identical with each other and phylogenetically closely related with the amoA of the betaproteobacterial ammonia oxidizer Nitrosomonas sp. Nm 47 (AY123830; 87.6% amino acid sequence similarity). In addition, we obtained three types of pmoA. The first pmoA type was closely related to the pmoA of the gammaproteobacterial type I methanotroph Methylomicrobium album (U31654; 89.9% amino acid-sequence similarity). The second pmoA sequence clustered together with some pmoA sequences of putative alphaproteobacterial methanotrophs. The third pmoA type clustered with some unusual pmoA/amoA-like sequences obtained from various other environments and formed a distinct lineage within the pmoA/amoA tree (Fig. 2). This lineage is much more closely related to the amoA of betaproteobacterial ammonia oxidizers than to the pmoA of described methanotrophs (Fig. 2), and this clustering is well supported in all treeing methods applied. We used quantitative, real-time PCR (Q-PCR) to determine whether one of these monooxygenase genes originated from C. polyspora. Comparison of the copy numbers of the respective monooxygenase genes in the enrichment with the number of small subunit rRNA genes of C. polyspora, of all bacteria, and of all organisms, respectively, revealed that the unusual pmoA type belongs to C. polyspora (Fig. 3). The other monooxygenase genes are from other organisms with very low abundance in the enrichment. This finding is consistent with the absence of detectable betaproteobacterial ammonia oxidizers (probe Nso1225), type I (probes Mγ669, Mγ84, and Mγ705), and type II (probe Mα450) methanotrophs other than C. polyspora in the enrichment by using specific FISH probes (data not shown).
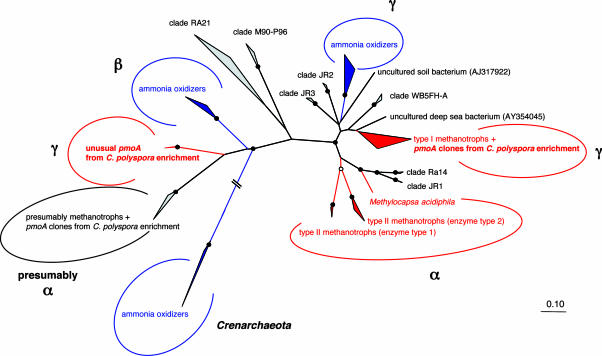
Fig. 2.
Unusual PmoA from the C. polyspora enrichment. Phylogenetic relationships among deduced PmoA and AmoA proteins were inferred by fitch distance analysis. In total, 164 amino acid positions were considered. Application of maximum likelihood, maximum parsimony, and neighbor-joining methods revealed congruent tree topologies for both protein and nucleic acid sequences (data not shown). Maximum parsimony bootstrap values >75% and 90% are indicated by white and black circles, respectively. Bar represents 10% estimated sequence divergence. Lineages containing AmoA or PmoA sequences of known ammonia or methane oxidizers are labeled in blue and red, respectively. It should be noted that, within the alphaproteobacterial PmoA sequence cluster, some organisms are represented by two nonidentical PmoA (25, 26). Environmentally retrieved sequence groups, for which no information on substrate specificity is available, are depicted in gray. The unusual PmoA cluster of C. polyspora also contains sequences retrieved from soil and a drinking water biofilm (27, 28). For the environmental sequence cluster most closely related to the C. polyspora cluster, indirect evidence exists that these enzymes originate from alphaproteobacterial methanotrophs (27, 29). An unfolded version of this tree is given in Fig. 6, which is published as supporting information on the PNAS web site.
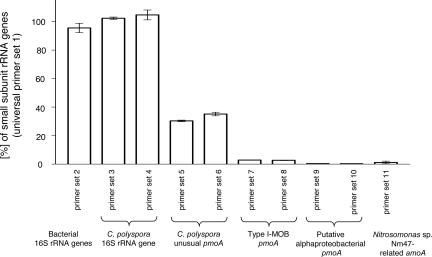
Fig. 3.
Quantification of small-subunit rRNA genes and monooxygenase genes in the C. polyspora enrichment by Q-PCR. The measured number of universal small-subunit rRNA genes determined with primer set 1 was set to 100%. With the universal, the bacterial, and the C. polyspora-specific small-subunit rRNA gene-targeted primer sets, almost equal copy numbers were detected in DNA extracted from the enrichment, confirming that the biomass consisted almost exclusively of C. polyspora. The detected high abundance of the unusual pmoA can, thus, be explained only by its origin from the C. polyspora genome. The absolute 16S rRNA gene and unusual pmoA numbers suggest that these genes occur in three and one copies (or multiples of these numbers), respectively, in the C. polyspora genome. The other pmoA gene types and the amoA occurred in much lower numbers and, thus, do not originate from C. polyspora. However, we cannot exclude that a very-low-abundance subpopulation of C. polyspora, which carries one or more of these genes in its genome, is additionally present in the enrichment. The numbers of primer sets are the same as in Table 3. n = 3. MOB, methane-oxidizing bacteria.
Although the unusual pmoA of C. polyspora is much more closely related to amoA of recognized betaproteobacterial ammonia oxidizers than to the pmoA of described methanotrophs, its sequence contains some signposts of the methane-oxidizer enzyme (see Fig. 5, which is published as supporting information on the PNAS web site). Consistent with this finding, after incubation of C. polyspora-enriched biomass (which did not contain noteworthy numbers of other bacteria, as demonstrated by FISH and Q-PCR) with methane, we observed rapid consumption of methane and oxygen and a simultaneous increase in the amount of CO2. The biomass had a low affinity to methane and oxidized methane at rates which are in the range of rates reported for pure cultures of methanotrophs (data not shown). Using a combination of FISH and microautoradiography (FISH-MAR) (13), we also proved, on a single-cell level, the uptake and incorporation of methane and methanol by all C. polyspora filaments analyzed (Fig. 1D). In addition, C. polyspora cells took up some other organic compounds (see Table 2, which is published as supporting information on the PNAS web site). The uptake of acetate and glucose by C. polyspora in the absence of methane is noteworthy, because described aerobic methanotrophic bacteria assimilate substrates containing carbon–carbon bonds only during exponential growth on one-carbon substrates (14) and could indicate that C. polyspora is a facultative methanotroph, similar to members of the genus Methylocella (15). Furthermore, C. polyspora did not incorporate 14CO2 in the presence of ammonia, demonstrating that this bacterium is not able to transform this compound for noteworthy energy generation (Table 2). Consistent with these findings, reverse-transcriptase Q-PCR data showed that expression of the unusual pmoA in starved C. polyspora cells was enhanced at least 30-fold after 3.5 h of methane incubation, whereas the cellular 16S rRNA content remained almost constant (see Fig. 7, which is published as supporting information on the PNAS web site).
Our findings solved the C. polyspora conundrum by identifying this as yet uncultured microorganism, first described by Ferdinand Cohn in 1870, as a gammaproteobacterial methane oxidizer. This finding suggests that C. polyspora is a biological indicator for methane in drinking water wells and could act as primary producer in oligotrophic groundwater environments, where, under oxic conditions, methane may support an important subset of the microbial community. C. polyspora is not only a filamentous member of this guild and possesses the most complex morphological life cycle of all known methane oxidizers but also carries a very unusual PmoA, which is induced in the presence of methane. Phylogeny inference of this gene and all known PmoA and AmoA (Fig. 2) revealed that the evolutionary history of these key enzymes of methane and ammonia oxidizers is far more complex than had been assumed (8, 16) and does not simply reflect the organismal 16S rRNA phylogeny. There is a clear need for experimental analysis of structural and functional differences of the unusual PmoA of C. polyspora compared with the classical particulate methane monooxygenases of methanotrophs (17). Understanding the selective advantages conferred by the different enzymatic repertoires of cultured and yet uncultured methane oxidizers is a key requisite to deepening our knowledge of the ecology of these unique microorganisms, their significance for drinking water production and supply, and their contribution to global carbon cycling.
Materials and Methods
Sampling.
Samples containing high proportions of C. polyspora filaments were taken from the backwash water of rapid sand filters for removal of iron, manganese, and ammonium of the Wolfenbüttel waterworks (Germany), which treats a mixture of aerobic and anaerobic groundwater. During sampling, C. polyspora filaments were retained from 600 to 850 liters of backwash water by either sedimentation or filtration through a fine-meshed sieve (200 or 400 μm). The combined and thoroughly rinsed samples were diluted to an appropriate volume in raw water.
DNA Extraction, PCR, Cloning, Sequencing, and FISH.
Whole cells were used for 16S rRNA gene amplification according to ref. 18 to minimize chimera formation. Instead of template DNA, 2 μl of ethanol-fixed, enriched C. polyspora biomass were added to the PCR reaction. However, because C. polyspora forms sheaths, the use of whole cells for PCR amplification might have caused underrepresentation of C. polyspora in the 16S rRNA gene library. For all other PCR and Q-PCR assays, total genomic DNA was isolated by using the FastDNA kit according to the instructions of the manufacturer (QBiogene, Irvine, CA). The 16S rRNA genes were PCR-amplified by using the primers 616F (19) and 1492R (20). PCR was carried out by an initial denaturation step at 94°C for 4 min, followed by 27 cycles of denaturation at 94°C for 40 s, annealing at 52°C for 1 min, and elongation at 72°C for 1 min 45 s. Cycling was completed by a final elongation step at 72°C for 10 min. Three different primer pairs and PCR conditions (10, 11, 19) were used for amoA and/or pmoA amplification, but the primer pair described in ref. 11 was slightly modified to pmof1 5′-AAC TTC TGG GGN TGG AC-3′ and pmor 5′-RCN ACG TCN TTA CCG AA-3′ and used under the following conditions: Thermal cycling was carried out by an initial denaturation step at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 40 s, annealing at 56°C for 40 s, and elongation at 72°C for 40s. Cycling was completed by a final elongation step at 72°C for 10 min. mmoX screening was performed by using two different PCR-approaches (11, 12). Cloning and sequencing was carried out as described in ref. 19. FISH was performed as described in ref. 21 by using the probes listed in Table 3, which is published as supporting information on the PNAS web site. More information on these probes can be found at probeBase (22) (www.microbial-ecology.net/probebase). For quantification of the C. polyspora relative biovolume abundance in the enriched biomass, the image analysis program daime (23) was used. For this purpose, the biomass was stained with a C. polyspora-specific probe and was counterstained with the nucleic acid-binding dye SYBR green I. Accurate determination of cell numbers was not possible, because individual cells could not be distinguished in all filaments.
Q-PCR.
Q-PCR was used to determine the copy numbers of the small subunit rRNA genes of all organisms, all Bacteria, and C. polyspora and the copy numbers of the amoA and the different pmoA genes in DNA extracted from the enrichment of C. polyspora. The applied primer sets are listed in Table 3. All reactions were carried out in an I-Cycler (Bio-Rad) by using the IQ SYBR green supermix (Bio-Rad) according to the instructions given by the manufacturer. Thermal cycling was carried out by an initial denaturation step at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 40 s, annealing at 60°C (small-subunit rRNA genes) or 58°C (all amoA/pmoA genes) for 30 s, and elongation at 72°C for 40 s. Plasmids carrying the respective cloned genes were used as standards for calibration of the assay. For each Q-PCR product, a single band of the expected size was observed by agarose gel electrophoresis (data not shown). The specificity of each Q-PCR reaction was further confirmed by (i) comparative melting-curve analyses of the sample-derived PCR products and the respective reference clone-derived PCR products (see Fig. 8, which is published as supporting information on the PNAS web site) and (ii) cloning and comparative sequence analyses of 12 randomly chosen clones of each Q-PCR reaction (data not shown).
RNA Isolation and Transcription Analysis.
Physically enriched C. polyspora biomass suspended in 18 ml of sterile filtered (0.2 μm) raw water was incubated for 5 days at room temperature and without addition of any carbon source in a 50-ml glass vessel. Subsequently, a 3.5-ml aliquot of the suspended biomass was removed and stored in RNAlater (Ambion) until RNA extraction. The vessel was tightly sealed, and the remaining biomass suspension was incubated for 3.5 h with 9% (vol/vol) CH4 in the headspace under agitation. Following this incubation, another 3.5-ml aliquot was sampled and stored for RNA extraction. Total RNA was isolated from the biomass aliquots by using the High pure RNA isolation kit (Roche) according to the instructions of the manufacturer. Subsequently, reverse transcription was carried out with the Revert Aid First Strand cDNA kit (Fermentas, St. Leon-Rot, Germany) according to the instructions of the manufacturer and by using primers Creno445 and pmoA-474R (Table 3) for the 16S rRNA and pmoA mRNA, respectively. The obtained cDNA was quantified by Q-PCR with primers Creno139f/Creno445 (16S rRNA cDNA) and pmoA-241F/pmoA-474R (pmoA cDNA; Table 3).
Electron Microscopy.
Transmission electron microscopy was performed as described in ref. 3, with the modification that a glass knife was used for cutting the cells.
FISH-MAR.
FISH-MAR experiments were performed as described by Lee et al. (13). Briefly, biomass samples were incubated aerobically for 5 h with various radioactively labeled electron donors before fixation with paraformaldehyde, which was performed as described in ref. 21. All incubations were carried out on a rotary table (250 rpm) kept at 20°C, with flasks placed horizontally to ensure sufficient mixing. The following radioactive chemicals were used: [14C]bicarbonate (59 mCi·mmol−1) (1 Ci = 37 GBq), [14C]formate (56 mCi·mmol−1), [14C]methane (57 mCi·mmol−1), and [3H]glucose (36 mCi·mmol−1) (all from Amersham Pharmacia Biosciences) and [3H]methanol (20 mCi·mmol−1) and [3H]acetate (8,000 mCi·mmol−1 (ARC, St. Louis). A total amount of 10 μCi was used in all experiments. Unlabeled glucose, acetate, formate, or methanol were added to a final concentration of 1 mM. Unlabeled methane was added to a final concentration of 16% (vol/vol). After incubation, the fixed biomass was collected by centrifugation, washed three times with filtered (0.22 μm) sample water, applied onto a gelatin-coated coverslip, and dried at 50°C before FISH probing. The hybridized samples were then dipped in preheated (43°C) LM-1 emulsion (Amersham Pharmacia Biosciences), exposed at 4°C for 6–14 days, and developed in Kodak D19 before being examined microscopically. Each MAR experiment was checked for chemography by running a control with a pasteurized sample (heated to 70°C for 10 min before incubation). No uptake was observed in the controls with any of the radiochemicals used in this study. Microscopic examinations of FISH-MAR samples were carried out by using an LSM 510 scanning confocal microscope (Zeiss).
Supplementary Material
Acknowledgments
We thank Ian Head, Paul Bodelier, and Peter Roslev for critically reading earlier versions of this manuscript and for providing many valuable comments; Waltraud Klepal and the team of the Ultrastructure Laboratory for advice and assistance with electron microscopy; Niels Iversen (Aalborg University, Aalborg, Denmark) for providing purified radiolabeled methane; Frank Köneke, Thorsten Dorsch, Jens Stoß, Dorota Bruniecka-Sulewski, Jürgen Dartmann, and Heike Temme for assistance during sampling; Klaus Johannsen for advice with calculations of gas–water equilibria; Purena (Braunschweig, Germany) for cooperation and support during sampling; and Christoph Czekalla for recommending this waterworks. B.B. and B.S. were supported by the German Gas- and Waterworks Association (Deutsche Vereinigung des Gas- und Wasserfaches). This work was supported, in part, by Wiener Wissenschafts-, Forschungs- und Technologiefonds Grant LS 216 (to H.D. and M.W.).
Glossary
Abbreviations:
- MAR
microautoradiography
- Q-PCR
quantitative PCR.
Footnotes
References
- 1.Garrity G. M., Bell J. A., Lilburn T. G. Taxonomic Outline of the Prokaryotes. Bergey’s Manual of Systematic Bacteriology. New York: Springer; 2004. [DOI] [Google Scholar]
- 2.Cohn F. Beitr. Biol. Pflanz. 1870;1:108–131. [Google Scholar]
- 3.Völker H., Schweisfurth R., Hirsch P. J. Bacteriol. 1977;131:306–313. doi: 10.1128/jb.131.1.306-313.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bumb F., Schweisfurth R. Zusammenfassende Darstellung der Kenntnisse über Crenothrix polyspora Cohn und eigene Untersuchungen. Freiburg, Germany: HochschulVerlag; 1981. [Google Scholar]
- 5.Wolfe R. S. J. Am. Water Works Assoc. 1960;52:915–918. [Google Scholar]
- 6.Koops H. P., Purkhold U., Pommerening-Röser A., Timmermann G., Wagner M. In: The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., editors. New York: Springer; 2003. Release 3.13. http://link.springer-ny.com/link/service/books/10125/ [Google Scholar]
- 7.Davies S. L., Whittenbury R. J. Gen. Microbiol. 1970;61:227–232. doi: 10.1099/00221287-61-2-227. [DOI] [PubMed] [Google Scholar]
- 8.Holmes A. J., Costello A., Lidstrom M. E., Murrell J. C. FEMS Microbiol. Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y. S., Halsey J. L., Fode K. A., Remsen C. C., Collins M. L. Appl. Environ. Microbiol. 1999;65:648–651. doi: 10.1128/aem.65.2.648-651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotthauwe J.-H., Witzel K.-P., Liesack W. Appl. Environ. Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auman A. J., Stolyar S., Costello A. M., Lidstrom M. E. Appl. Environ. Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miguez C. B., Bourque D., Sealy J. A., Greer C. W., Groleau D. Microb. Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 13.Lee N., Nielsen P. H., Andreasen K. H., Juretschko S., Nielsen J. L., Schleifer K.-H., Wagner M. Appl. Environ. Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egli T., Mason C. A. In: Biology of Methylotrophs. Goldberg I., Rokem J. S., editors. Boston: Butterworth–Heinemann; 1991. pp. 173–201. [Google Scholar]
- 15.Dedysh S. N., Knief C., Dunfield P. F. J. Bacteriol. 2005;187:4665–4670. doi: 10.1128/JB.187.13.4665-4670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes A. J., Roslev P., McDonald I. R., Iversen N., Henriksen K., Murrell J. C. Appl. Environ. Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman R. L., Rosenzweig A. C. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 18.Snaidr J., Amann R., Huber I., Ludwig W., Schleifer K.-H. Appl. Environ. Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juretschko S., Timmermann G., Schmid M., Schleifer K.-H., Pommerening-Röser A., Koops H.-P., Wagner M. Appl. Environ. Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D. J. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E., Goodfellow M., editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 21.Daims H., Stoecker K., Wagner M. In: Molecular Microbial Ecology. Osborn A. M., Smith C. J., editors. Abingdon, U.K.: Bios–Garland; 2005. pp. 213–239. [Google Scholar]
- 22.Loy A., Horn M., Wagner M. Nucleic Acids Res. 2003;31:514–516. doi: 10.1093/nar/gkg016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daims H., Lücker S., Wagner M. Environ. Microbiol. 8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 24.Bakermans C., Madsen E. L. Microb. Ecol. 2002;44:95–106. doi: 10.1007/s00248-002-0005-8. [DOI] [PubMed] [Google Scholar]
- 25.Ricke P., Erkel C., Kube M., Reinhardt R., Liesack W. Appl. Environ. Microbiol. 2004;70:3055–3063. doi: 10.1128/AEM.70.5.3055-3063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tchawa Yimga M., Dunfield P. F., Ricke P., Heyer J., Liesack W. Appl. Environ. Microbiol. 2003;69:5593–5602. doi: 10.1128/AEM.69.9.5593-5602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radajewski S., Webster G., Reay D. S., Morris S. A., Ineson P., Nedwell D. B., Prosser J. I., Murrell J. C. Microbiology. 2002;148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- 28.Lipponen M. T., Suutari M. H., Martikainen P. J. Water Res. 2002;36:4319–4329. doi: 10.1016/s0043-1354(02)00169-0. [DOI] [PubMed] [Google Scholar]
- 29.Pacheco-Oliver M., McDonald I. R., Groleau D., Murrell J. C., Miguez C. B. FEMS Microbiol. Lett. 2002;209:313–319. doi: 10.1111/j.1574-6968.2002.tb11150.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.