Abstract
Collagen is the most abundant protein in animals and the major component of connective tissues. Although collagen isolated from natural sources has long served as the basis for some biomaterials, natural collagen is difficult to modify and can engender pathogenic and immunological side effects. Collagen comprises a helix of three strands. Triple helices derived from synthetic peptides are much shorter (<10 nm) than natural collagen (≈300 nm), limiting their utility. Here, we describe the synthesis of short collagen fragments in which the three strands are held in a staggered array by disulfide bonds. Data from CD spectroscopy, dynamic light scattering, analytical ultracentrifugation, atomic force microscopy, and transmission electron microscopy indicate that these “sticky-ended” fragments self-assemble via intermolecular triple-helix formation. The resulting fibrils resemble natural collagen, and some are longer (>400 nm) than any known collagen. We anticipate that our self-assembly strategy can provide synthetic collagen-mimetic materials for a variety of applications.
Keywords: biomaterial, coiled-coil, nanotechnology, cystine knot, peptide
Collagen constitutes one-third of the human proteome, including three-quarters of the dry weight of human skin. Its high natural abundance and intrinsic plasticity have spurred the development of collagen as a biomaterial (1, 2). The most common source of clinical collagen is now Bos taurus, the domestic cow. Unfortunately, bovine collagen can illicit deleterious pathological and immunological effects when transplanted into humans (3–5). Moreover, the preparation of enriched solutions of natural collagen is problematic (6), and its site-specific covalent modification is not feasible. We suspected that synthetic chemistry could offer a solution to these problems.
The quaternary structure of collagen comprises three strands that wrap around one another to form a triple helix (7). Each strand has the repeating sequence XaaYaaGly, with the most abundant triplet being ProHypGly [Hyp = (2S,4R)-4-hydroxyproline] (8). The folding of (XaaYaaGly)n ≤ 10 peptides into blunt-ended triple helices has been investigated thoroughly (7). Although such triple helices are biomaterial candidates (9–12), they are limited to the length of a synthetic peptide (<10 nm), which is much shorter than natural collagen (≈300 nm). The polymerization of (XaaYaaGly)10 peptides has afforded long strands that adopt triple-helical structure but have high polydispersity (13).
Molecular self-assembly underlies the “bottom–up” approach to macromolecular design, wherein a desirable structure forms spontaneously through noncovalent interactions (14, 15). Self-assembling peptides and proteins have been designed to serve as materials for biological and nanotechnological applications (16, 17). Using the self-assembly approach, Woolfson and coworkers (18, 19) have produced fibers with a design based on a dimeric coiled-coil structure. Likewise, fibrous peptides based on the natural protein elastin (20) and de novo building blocks (15, 21–26) have been assembled and implicated as biomaterial candidates. Here, we report on the self-assembly of collagen fragments into an extended trimeric coiled-coil.
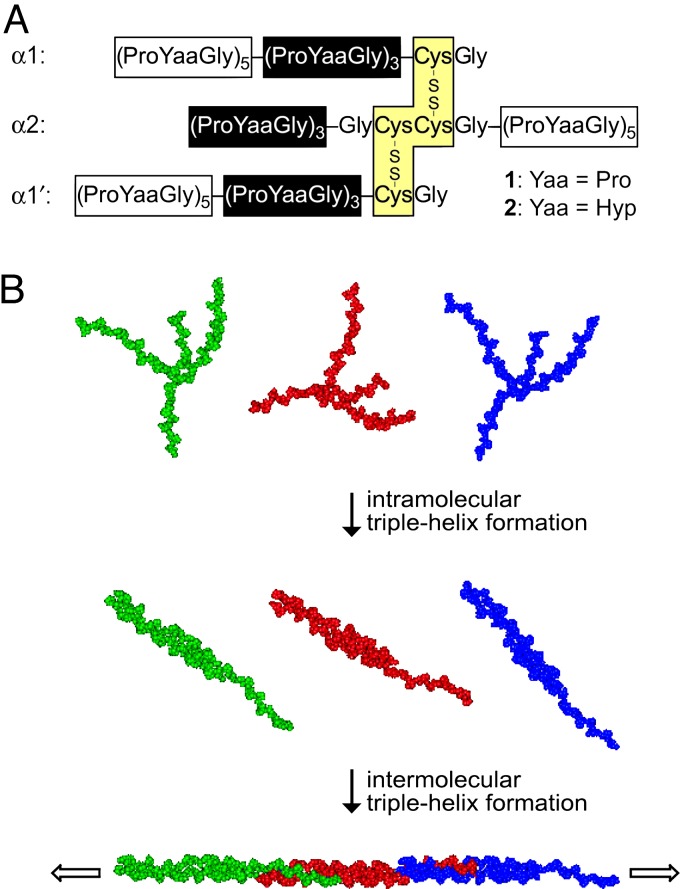
Inspired by the self-assembly of double-helical fragments of DNA, we envisioned that sticky-ended fragments of collagen could self-assemble into long triple helices. Unlike the situation with DNA, there is no “code” for the noncovalent association of collagen strands, other than the need for one Xaa, Yaa, and Gly residue to be in each cross section of a triple helix. Hence, we relied on covalent bonds to tether the strands of our collagen fragments. Specifically, we prepared fragments 1 and 2 in which three helicogenic strands (7) are linked by a pair of disulfide bonds (27) that both offset the strands and set their register (Fig. 1A). In 1 and 2, the (ProYaaGly)3 segments in the two identical strands (α1 and α1′) and third strand (α2) form an intramolecular triple helix (Fig. 1B). The intermolecular assembly of the overhanging (ProYaaGly)5 segments yield collagen triple helices of unprecedented length.
Fig. 1.
Structure and self-assembly of fragments 1 and 2. (A) Amino acid sequence of fragments 1 and 2. The glycine residue preceding the adjacent cysteines in the α2 strand establishes the requisite register for triple-helix formation with the identical α1 and α1′ strands. Hyp, (2S,4R)-4-hydroxyproline. (B) Representation of the self-assembly process; green, red, and blue are used simply to distinguish individual fragments.
Results and Discussion
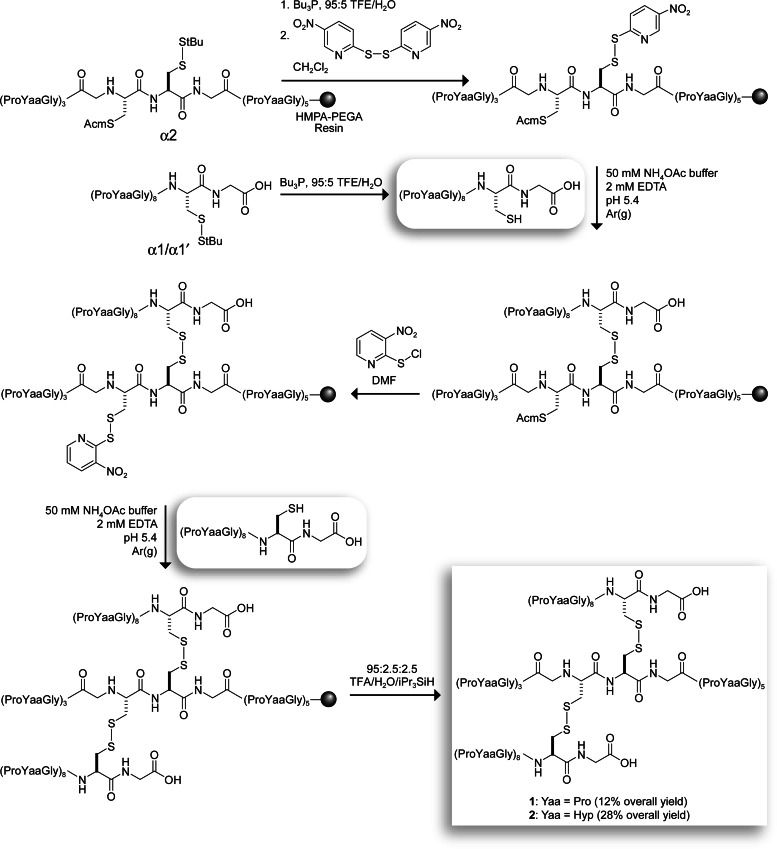
Fragments 1 and 2 were synthesized directly on a solid support by using a strategy based on the orthogonal deprotection of cysteine residues (Fig. 2) (27). Briefly, the thiol of a deprotected α1 strand was reacted with the p-Npys-activated thiol of a resin-bound α2 strand in aqueous buffer to afford an α1α2 heterodimer. (The use of polyethylene glycol acrylamide copolymer resin was critical, presumably because of increased strand accessibility; disulfide formation did not occur on polyethylene glycol polystyrene resin.) The heterodimer was deprotected, and its thiol was activated with o-Npys-Cl and reacted with another deprotected α1 strand (α1′ strand in Fig. 1A). Cleavage from the resin afforded fragments 1 and 2 in 12% and 28% overall yields, respectively, which significantly exceed those reported for analogous solution-phase syntheses (27). The N and C termini in 1 and 2 were left unprotected to mimic natural collagen. Analytical experiments were performed at pH 3 to minimize the scrambling or reduction of disulfide bonds (28).
Fig. 2.
Scheme for the synthesis of self-assembling fragments 1 and 2. tBu, tert-butyl; Acm, acetamidomethyl; Bu, butyl; TFE, 2,2,2-trifluoroethanol; iPr, isopropyl; HMPA, 4-hydroxymethylphenoxyacetyl; PEGA, polyethylene glycol acrylamide copolymer.
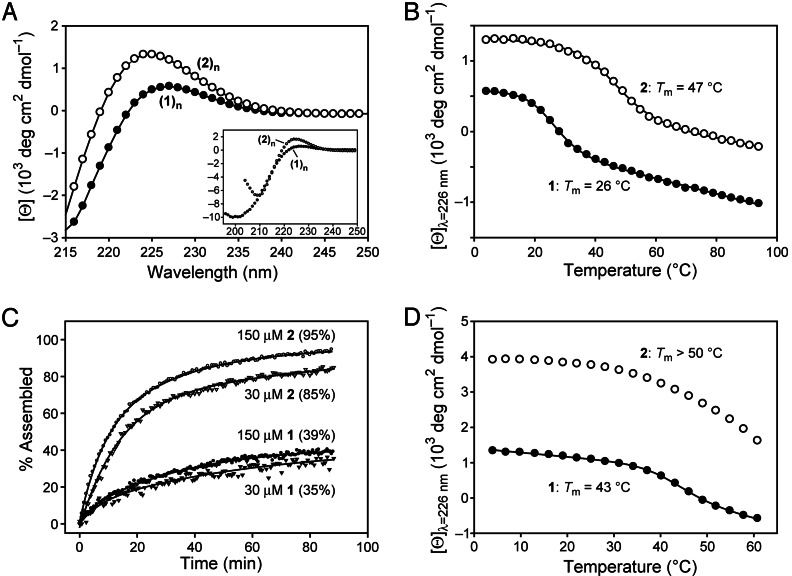
Fragments 1 and 2 form a triple helix. After incubation at 4°C for 48 h, CD spectra of both assemblies were diagnostic of a triple-helical structure, having a positive peak near 225 nm and a strong negative peak at 200–210 nm (Fig. 3A). Heating produced cooperative denaturation characteristic of triple-helix unfolding (Fig. 3B). The thermal stability of assembled 2 [melting temperature (Tm)= 47°C] was greater than that of assembled 1 (Tm = 26°C), as expected from the presence of Hyp residues in the Yaa position of 2 (29). These Tm values exceed those expected for a triple helix of the tethered (ProYaaGly)3 core of fragments 1 and 2, suggesting that the (ProYaaGly)5 overhangs of 1 and 2 assemble to form (1)n and (2)n triple helices, respectively.
Fig. 3.
CD data for assemblies of fragments 1 and 2. (A) Spectra of assembled 1 and 2 (200 μM) at 4°C. (Inset) 1, 70 μM; 2, 100 μM. (B) Thermal denaturation of assembled 1 and 2 (200 μM), monitored at 226 nm. (C) Rate of assembly of fragments 1 and 2 (30 and 150 μM), over 90 min, monitored at 226 nm after a rapid transition from 90°C to 10°C. The [Θ]max for each sample was calculated as the difference between the ellipticity before denaturation (100% assembled) and the first data point at 10°C, thus omitting the initial jump in ellipticity (which was not resolvable) (47). The value of percent assembled was then calculated as the ratio of the observed recovery to [Θ]max. Data in A–C were from solutions in 50 mM HOAc(aq) at pH 2.9. (D) Thermal denaturation of assembled 1 and 2 (200 μM) in 67:33 MeOH/68 mM HOAc(aq) monitored at 226 nm.
The rate of assembly of fragments 1 and 2 depends on their concentration and the identity of their Yaa residue. After 90 min, solutions of fragment 2 displayed 85% (30 μM) and 95% (150 μM) assembly (Fig. 3C). Fragment 1 assembled more slowly, reaching only 35% (30 μM) and 39% (150 μM) assembly after 90 min. The dependence of these yields on fragment concentration is indicative of an intermolecular process, consistent with the sticky-end–directed assembly of 1 and 2. At 200 μM, which was the highest concentration examined herein, the assembly of 2 was much more rapid than that of 1 (Fig. 5, which is published as supporting information on the PNAS web site). This difference is likely caused by the higher concentration of the requisite trans isomer of the Pro–Yaa peptide bonds in the overhangs of 2 (Yaa = Hyp) (30) and the faster cis–trans isomerization of those bonds (31).
The size of (1)n and (2)n in aqueous solution was estimated by using dynamic light scattering (DLS, Table 1). Weight analyses of the DLS data showed an average hydrodynamic radius (Rh) of 3.1 nm for (1)n and 4.0 nm for (2)n at 10°C (Table 1, entries 1 and 2). The lengths of the assemblies were estimated by using the Broersma (32–34) and Tirado and Garcia de la Torre (35, 36) relations, which were applied previously to type I collagen (37). The estimated lengths of 14–18 nm for (1)n and 22–26 nm for (2)n indicate that the most prevalent assemblies consist of two to three monomer units.
Table 1.
DLS data for assemblies of fragments 1 and 2
| Entry | Peptide | Solvent | Temperature, °C | Rh, nm (SDP weight analysis) | Length, nm (32–34) | Length, nm (35, 36) | Estimated monomer units* | Rh, nm (SDP intensity analysis) |
|---|---|---|---|---|---|---|---|---|
| Low temperature | ||||||||
| 1 | 1 | 50 mM HOAc(aq) | 10 | 3.1 ± 0.2 | 14 ± 2 | 18 ± 2 | 2 | 3.3, 72 |
| 2 | 2 | 50 mM HOAc(aq) | 10 | 4.0 ± 0.1 | 22 ± 1 | 26 ± 1 | 3 | 5.2, 65 |
| 3 | 1 | 67:33 MeOH/HOAc(aq) | 10 | 5.4 ± 0.5 | 35 ± 5 | 40 ± 5 | 4–5 | 7.5, 43, 200, 650 |
| 4 | 2 | 67:33 MeOH/HOAc(aq) | 10 | 7.6 ± 1.1 | 58 ± 12 | 63 ± 12 | 7–8 | 14, 600 |
| High temperature | ||||||||
| 5 | 1 | 50 mM HOAc(aq) | 60 | 1.9 ± 0.2 | 5 ± 1 | 8 ± 2 | 1 | 2.3, 78 |
| 6 | 2 | 50 mM HOAc(aq) | 60 | 2.1 ± 0.4 | 7 ± 2 | 10 ± 3 | 1 | 2.4, 92 |
| 7 | 1 | 67:33 MeOH/HOAc(aq) | 55 | 1.6 ± 0.6 | 4 ± 3 | 6 ± 4 | 1 | 6.5, 63, 230 |
| 8 | 2 | 67:33 MeOH/HOAc(aq) | 55 | 3.1 ± 0.5 | 14 ± 4 | 18 ± 4 | 2 | 3.7, 900 |
*Estimates are based on the 8.6-nm length of a (ProProGly)10 segment in a crystalline triple helix (41).
The size of (1)n and (2)n estimated from size distribution processor (SDP) weight analyses of DLS data are consistent with the results of sedimentation equilibrium experiments (Fig. 6, which is published as supporting information on the PNAS web site). Data for (1)n recorded at 4°C at 8.6, 12, and 18 k rpm in an An-60 Ti rotor (Beckman) were fitted as a nonequilibrating mixture (38) of dimers and trimers by using molecular masses of 13,440 and 20,160 Da. The 4°C data for (2)n were fitted to the same model as a dimer–tetramer mixture by using molecular masses of 14,210 and 28,420 Da.
Assemblies of 2 tend to be longer than assemblies of 1 according to DLS data (Table 1), suggesting that the extent of assembly correlates with triple-helix stability. To test this hypothesis, the thermal stabilities and lengths of (1)n and (2)n were measured in aqueous methanol, where collagen triple helices are highly stable (39). Indeed, increases in both thermal stability (Fig. 3D) and length (Table 1, entries 3 and 4) for (1)n and (2)n were observed in the methanolic solution. In addition to confirming that more stable triple helices produce longer assemblies, these data demonstrate that the extent of assembly can be modulated by the choice of solvent.
Thermal denaturation of assemblies was observable during DLS and sedimentation equilibrium experiments, and the trends are in gratifying agreement with CD denaturation data (Figs. 2B and 4). Heating an aqueous solutions of (1)n or (2)n to 60°C (>Tm) resulted in monomeric 1 or 2 (Table 1, entries 5 and 6). Sedimentation equilibrium experiments revealed that (1)n had dissociated to a monomer at 37°C, which is above its Tm (26°C). As expected, (2)n had not completely dissociated at 37°C, which is below its Tm (47°C). DLS data in methanolic solution show that 1 is monomeric at 55°C (>Tm), whereas 2 is dimeric (Table 1, entries 7 and 8). Likewise, CD spectroscopy indicates that assemblies are the prevalent species of 2 at 55°C (Fig. 3D).
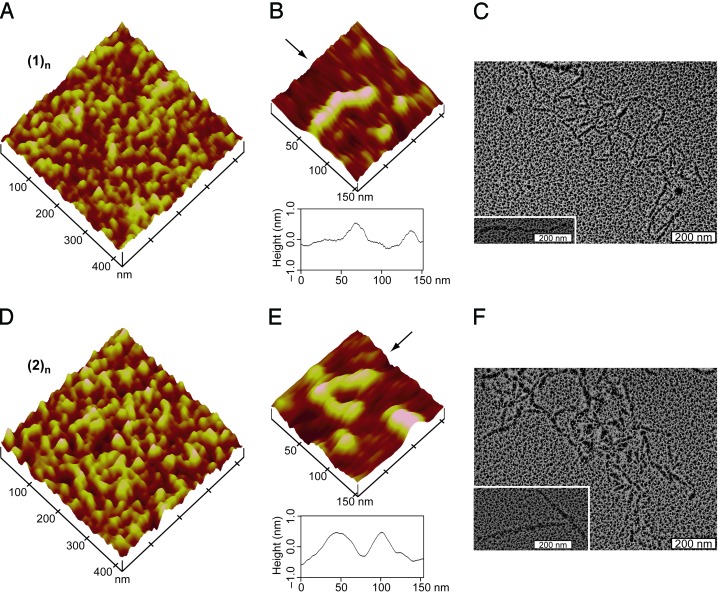
Fig. 4.
AFM and TEM images for assemblies of fragments 1 and 2. (A) AFM height image of (1)n. (B) AFM height image of isolated (1)n (Upper) and section analysis of the data along the direction indicated by the arrow (Lower). (C) TEM image of (1)n after rotary shadowing with platinum. (Inset) An isolated fibril. (D) The same as A, except for (2)n. (E) The same as B, except for (2)n. (F) The same as C, except for (2)n.
The existence of larger assemblies was apparent from SDP intensity analyses of the DLS data, which is weighted to those particles that scatter more light. Assemblies with Rh ≈70 nm were observed for both (1)n and (2)n (Table 1, entries 1 and 2), corresponding to a length of >1 μm (32–34, 37). These longer assemblies were still present in solution at 60°C (Table 1, entries 5 and 6); shorter assemblies were denatured at this temperature. The apparent hyperstability of longer assemblies could be a manifestation of cooperativity.
The morphology of (1)n and (2)n was examined by atomic force microscopy (AFM). AFM height images (tapping mode) of (1)n (Fig. 4 A and B) and (2)n (Fig. 4 D and E) showed one-dimensional fibrils of length 20–120 nm. Section analyses of the AFM data indicated fibril diameters of 0.5–1.0 nm (Fig. 4 B and E), which is in gratifying agreement with the diameter of natural type I collagen observed by us (Fig. 7, which is published as supporting information on the PNAS web site) and others with AFM (40) and x-ray diffraction analysis (41). For both 1 and 2, smaller assemblies (20–40 nm) were abundant in AFM images, consistent with DLS data (Table 1, entries 1 and 2). After thermal denaturation of the assemblies (90°C for 15 min), AFM images revealed no evidence for extended structures (Fig. 8, which is published as supporting information on the PNAS web site).
Transmission electron microscopy (TEM) provided additional morphological characterization of (1)n and (2)n. After rotary shadowing with platinum, TEM images of (1)n and (2)n revealed fibrillar structures of length 30 nm to >400 nm (Fig. 4 C and F). The morphology and diameter of these assemblies strongly resemble rotary-shadowed images of type I collagen (Fig. 7) and other types of natural collagen (42, 43).
Both DLS and TEM data indicate that fragments 1 and 2 self-assemble into fibrils that can exceed the length of natural collagen (≈300 nm). In previous work, we demonstrated how to use stereoelectronic effects to produce collagen triple helices that have greater conformational stability than any found in nature (30, 44, 45). Here, we have complemented that work by demonstrating how to use molecular self-assembly to produce collagen triple helices that are longer than any found in nature.
Conclusion
Our long-term goal is to develop collagen-based biomaterials with tunable attributes that can be used as both collagen surrogates and templates for nanotechnological applications. We have now taken the key step toward that goal. By exploiting triple-helical propensity to direct the assembly of synthetic collagen fragments with sticky ends, we have generated fibrils that mimic the structure and thermal behavior of natural collagen. Control over the stability and, to some extent, the length of assemblies was accomplished by modulating amino acid composition, temperature, and solvent. It is noteworthy that our self-assembly strategy yields collagen-like fibrils of 1-nm diameter and nearly 1-μm length composed from only three or four proteinogenic amino acids. Minimalist fragments like 1 and 2 can be elaborated by chemical synthesis to display motifs that promote cell adhesion for engineering tissues, lateral packing for accessing two- and three-dimensional architectures, and metal coordination for producing nanowires (46), and are thereby templates for a diversity of materials for biomedicine and nanotechnology.
Materials and Methods
Solid-Phase Peptide Synthesis.
Peptides were synthesized on solid phase by using fluorenylmethoxycarbonyl (Fmoc) chemistry with an Applied Biosystems Pioneer instrument. Iterative 1-h couplings used three equivalents of Fmoc-amino acid (or Fmoc-ProYaaGly-OH tripeptide), O-(7-azabenzotriazole-l-yl)-N,N,N′, N′-tetramethyluronium hexafluorophosphate/diisopropylethylamine as the coupling reagent, and 4:1 dimethylformamide/piperidine for Fmoc deprotections. Disulfide bonds between the three strands were formed directly on the solid support as shown in Fig. 2 and described in detail in Supporting Text, which is published as supporting information on the PNAS web site.
CD Spectroscopy.
CD spectra were recorded with an Aviv Associates (Lakewood, NJ) 202SF CD spectrometer. Data were collected on a solution of fragment 1 or 2 that had been incubated at ≤4°C for ≥48 h. Spectra were recorded in 1-nm increments with a 3-s averaging time, 1-nm bandpass, and 0.1-cm pathlength.
Samples for thermal stability experiments were generated by incubating a solution of fragment 1 or 2 (200 μM) at 4°C for ≥48 h. This solution was then heated from 4°C to 60°C or 94°C (depending on the solvent) at 3°C increments with a 5-min equilibration at each step. The ellipticity at 226 nm was monitored with a 5-s averaging time, 1-nm bandpass, and 0.1-cm pathlength. Values of Tm were determined by fitting the data to a two-state model.
Samples for folding rate experiments were generated by heating a solution of fragment 1 or 2 to 90°C for 15 min and then putting that solution into a cuvette that had been cooled to 10°C. After ≈1 min, the ellipticity at 226 nm was monitored continuously at 10°C. The maximal change in ellipticity ([Θ]max) for each sample was calculated as the difference between the ellipticity before denaturation (taken as 100% assembled) and the first data point at 10°C. In this way, the initial jump in ellipticity upon cooling (which reports on an unresolved kinetic phase) was omitted (47). Values of percent assembled were then calculated as the ratio of the observed recovery to [Θ]max.
DLS.
DLS experiments were performed with a Coulter N4 Plus DLS instrument equipped with a 10-mW helium-neon laser (λ = 632.8 nm) and thermoelectric temperature controller. Measurements were taken at a 90° scattering angle in a 3 × 3-mm quartz cuvette on solutions that had been equilibrated at 10°C, 55°C, or 60°C for ≥30 min.
Samples for DLS were prepared by heating a solution of fragment 1 or 2 to 65–70°C for 10 min, filtering through a 0.22-μm filter, and incubating at 4°C for ≥48 h. Solutions in 50 mM HOAc(aq) and 67:33 MeOH/68 mM HOAc(aq) had fragment concentrations of 20 and 4 mg/ml, respectively, resulting in 90°-scattering intensities in the range from 5 × 104 to 1 × 106 counts per s. Assembly size was calculated by averaging the values of 6–15 runs of 300 s at each temperature.
DLS data were analyzed with the contin algorithm (48), which is incorporated into the SDP of the n4 plus software. Intensity distributions were converted into weight distributions by using a Mie scattering theory approximation in the software. Particle sizes were estimated by SDP weight and intensity analyses. For samples in 50 mM HOAc(aq), the values of viscosity (η) and refractive index (r.i.) were η10°C = 1.309 centipoise (cp), r.i.10°C = 1.333, and η60°C = 0.469 cp, r.i.60°C = 1.332. For samples in 67:33 MeOH/68 mM HOAC(aq), the values were η10°C = 1.274 cp, r.i.10°C = 1.331, and η55°C = 0.531 cp, r.i.55°C = 1.312.
The lengths of assemblies (1)n and (2)n were estimated from the Rh values measured with DLS by using the Broersma relations (32–34) and Tirado and Garcia de la Torre relations (35, 36), as described for collagen (37).
Sedimentation Equilibrium.
Sedimentation equilibrium experiments were performed with a Beckman XL-A analytical ultracentrifuge.
Samples for analytical ultracentrifugation were prepared by incubating a solution (30 μM) of fragment 1 or 2 in 50 mM potassium phosphate buffer, pH 2.9, at 4°C for ≥72 h. About 100 μl of a sample was put into a cell with a 12-mm double-sector charcoal-filled centerpiece (Epon); 110 μl of buffer was put into the reference cell. Experiments at 4°C were run at 8.6, 12, and 18 k rpm in an An-60 Ti rotor, and those at 37°C were run at 18 k rpm. Gradients recorded at A230nm were monitored until they became superimposable when recorded ≥4 h apart. The buffer density of 1.0049 g/ml was measured at 4°C with an Anton Paar (Graz, Austria) DMA5000 density meter. The partial specific volumes for 1 and 2 were calculated based on amino acid content to be 0.72 ml/g. The extinction coefficient per mg of fragment was assumed to be identical for all species.
Equilibrium gradients at 4°C were analyzed by fitting the data by using nonlinear least-squares methods to various models, including single species and multiple noninteracting and interacting species, with programs written for igor pro (WaveMetrics, Lake Oswego, OR) by D. R. McCaslin (University of Wisconsin). Nonsedimenting baselines of 0.03 OD for fragment 1 (32 μM), and 0.09 and 0.14 OD for fragment 2 (20 and 32 μM), which were based on high-speed data, were included in the data analysis. The final choice of model was made by analyzing each data set as a combination of species from monomer to hexamer (monomer molecular masses used were 6,721 and 7,105 Da for 1 and 2, respectively) (38). Although the data did not identify a single model uniquely, data for (1)n were described best by a model with a combination of dimers and trimers, and data for (2)n were described best by a model with a combination of dimers and tetramers (Fig. 6).
AFM.
AFM images were recorded with a Digital Instruments (Sterling Heights, MI) Nanoscope IV microscope equipped with an E scanner in tapping mode by using silicon-etched RTESP7 cantilevers (Veeco Nanoprobes), which had a nominal tip radius of <10 nm and spring constant of 20–80 N/m. The drive frequency was set at 250–300 kHz with integral and proportional gains of 0.5–1.5. A scan rate of 1.5 Hz and scan size of 0.5–1.0 μm were used to collect height and amplitude images simultaneously, although only height images are shown herein (Figs. 4, 7, and 8). Images were flattened, low-pass filtered, and zoomed off-line with Digital Instruments nanoscope iii software (version 5.12r5).
Samples for AFM were prepared by incubating solutions (≈300 μg/ml) of 1, 2, or type I calfskin collagen (ICN) in 50 mM HOAc(aq) at 4°C for ≥72 h. For imaging isolated assemblies (Fig. 4 B and E), solutions of (1)n or (2)n were diluted to ≈10 μg/ml. Material from these solutions was adsorbed onto freshly cleaved mica (grade V-4, SPI Supplies, West Chester, PA) at 4°C for 30 s. The mica was then rinsed with 50 mM HOAc(aq) (2 × 30 s) and allowed to dry at 4°C before imaging.
TEM.
TEM images were recorded at 80 kV and ×88,000 with a Philips CM120 transmission electron microscope and documented with a MegaView III digital camera (Soft Imaging System, Lakewood, CO).
Samples for TEM were prepared on freshly cleaved mica with solutions (1.4 mg/ml) of 1, 2, or type I calfskin collagen as described for AFM. The rinsed mica was placed in an Edwards (Wilmington, MA) Auto 306 evaporator, and the chamber was evacuated to <1 × 10−6 Torr. Platinum wire was coiled around a carbon rod and resistance-evaporated at an angle of 4–8° (incident to the mica surface) while the stage holding the mica was rotated at 2 Hz. To cast the support film, carbon was evaporated over the platinum at an angle of 90°. Replicas were floated off the mica onto water and picked up onto 400-mesh nickel grids for TEM imaging. A control sample was also prepared with 50 mM HOAc(aq) alone as a negative control; no structures were observed in this sample by TEM.
Supplementary Material
Acknowledgments
We are grateful for the help and advice of D. R. McCaslin (biophysical measurements, data analysis, and editorial comments), C. L. Jenkins (synthesis), R. M. Murphy (DLS), and R. J. Massey (TEM). This work was supported by National Institutes of Health Grant AR44276, National Science Foundation Instrumentation Grant BIR-9512577, and National Institutes of Health Instrumentation Grant lRR13790. F.W.K. was supported by National Institutes of Health Postdoctoral Fellowship AR50881.
Abbreviations
- Tm
melting temperature
- DLS
dynamic light scattering
- Rh
hydrodynamic radius
- AFM
atomic force microscopy
- TEM
transmission electron microscopy
- r.i.
refractive index
- SDP
size distribution processor
- [Θ]max
maximal change in ellipticity.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ramshaw J. A. M., Werkmeister J. A., Glattauer V. Biotechnol. Genet. Eng. Rev. 1996;13:335–382. doi: 10.1080/02648725.1996.10647934. [DOI] [PubMed] [Google Scholar]
- 2.Lee C. H., Singla A., Lee Y. Int. J. Pharm. 2001;221:1–22. doi: 10.1016/s0378-5173(01)00691-3. [DOI] [PubMed] [Google Scholar]
- 3.Cooperman L., Michaeli D. J. Am. Acad. Dermatol. 1984;10:638–646. doi: 10.1016/s0190-9622(84)80271-6. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi M., Hori H., Hattori S., Irie S., Imai A., Yanagida M., Miyazawa H., Toda M., Inouye S. J. Allergy Clin. Immunol. 1999;104:695–699. doi: 10.1016/s0091-6749(99)70344-1. [DOI] [PubMed] [Google Scholar]
- 5.Lynn A. K., Yannas I. V., Bonfield W. J. Biomed. Mater. Res. B. 2004;71:343–354. doi: 10.1002/jbm.b.30096. [DOI] [PubMed] [Google Scholar]
- 6.Ruszczak Z. In: Biological Matrices and Tissue Reconstruction. Stark G. B., Horch R., Tanczos E., editors. Berlin: Springer; 1998. pp. 21–28. [Google Scholar]
- 7.Jenkins C. L., Raines R. T. Nat. Prod. Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 8.Ramshaw J. A. M., Shah N. K., Brodsky B. J. Struct. Biol. 1998;122:86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 9.Qian J. J., Bhatnagar R. S. J. Biomed. Mater. Res. 1996;31:545–554. doi: 10.1002/(SICI)1097-4636(199608)31:4<545::AID-JBM15>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 10.Fields G. B., Lauer J. L., Dori Y., Forns P., Yu Y. C., Tirrell M. Biopolymers. 1998;47:143–151. doi: 10.1002/(SICI)1097-0282(1998)47:2<143::AID-BIP3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 11.Johnson G., Jenkins M., McLean K. M., Griesser H. J., Kwak J., Goodman M., Steele J. G. J. Biomed. Mater. Res. 2000;51:612–624. doi: 10.1002/1097-4636(20000915)51:4<612::aid-jbm9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Wang A. Y., Mo X., Chen C. S., Yu S. M. J. Am. Chem. Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 13.Paramonov S. E., Gauba V., Hartgerink J. D. Macromolecules. 2005;38:7555–7561. [Google Scholar]
- 14.Whitesides G. M., Mathias J. P., Seto C. T. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S. G. Nat. Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopal K., Schneider J. P. Curr. Opin. Struct. Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Fairman R., Akerfeldt K. S. Curr. Opin. Struct. Biol. 2005;15:453–463. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Pandya M. J., Spooner G. M., Sunde M., Thorpe J. R., Rodger A., Woolfson D. N. Biochemistry. 2000;39:8728–8734. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
- 19.MacPhee C. E., Woolfson D. N. Curr. Opin. Solid State Mater. Sci. 2004;8:141–149. [Google Scholar]
- 20.Wright E. R., Conticello V. P. Adv. Drug Delivery Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 21.Aggeli A., Nyrkova I. A., Bell M., Harding R., Carrick L., McLeish T. C. B., Semenov A. N., Boden N. Proc. Natl. Acad. Sci. USA. 2001;98:11857–11862. doi: 10.1073/pnas.191250198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartgerink J. D., Beniash E., Stupp S. I. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 23.Holmes T. C. Trends Biotechnol. 2002;20:16–21. doi: 10.1016/s0167-7799(01)01840-6. [DOI] [PubMed] [Google Scholar]
- 24.Langer R., Tirrell D. A. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 25.Lutolf M. P., Hubbell J. A. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 26.Wagner D. E., Phillips C. L., Ali W. M., Nybakken G. E., Crawford E. D., Schwab A. D., Smith W. F., Fairman R. Proc. Natl. Acad. Sci. USA. 2005;102:12656–12661. doi: 10.1073/pnas.0505871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottl J., Moroder L. J. Am. Chem. Soc. 1999;121:653–661. [Google Scholar]
- 28.Barth D., Kyrieleis O., Frank S., Renner C., Moroder L. Chem. Eur. J. 2003;9:3703–3714. doi: 10.1002/chem.200304918. [DOI] [PubMed] [Google Scholar]
- 29.Berg R. A., Prockop D. J. Biochem. Biophys. Res. Commun. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 30.Bretscher L. E., Jenkins C. L., Taylor K. M., DeRider M. L., Raines R. T. J. Am. Chem. Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 31.Bächinger H. P., Bruckner P., Timpl R., Engel J. Eur. J. Biochem. 1978;90:605–613. doi: 10.1111/j.1432-1033.1978.tb12641.x. [DOI] [PubMed] [Google Scholar]
- 32.Broersma S. J. Chem. Phys. 1960;32:1626–1631. [Google Scholar]
- 33.Broersma S. J. Chem. Phys. 1960;32:1632–1635. [Google Scholar]
- 34.Broersma S. J. Chem. Phys. 1981;74:6989–6990. [Google Scholar]
- 35.Tirado M. M., Garcia de la Torre J. J. Chem. Phys. 1979;71:2581–2587. [Google Scholar]
- 36.Tirado M. M., Garcia de la Torre J. J. Chem. Phys. 1980;73:1986–1993. [Google Scholar]
- 37.Claire K., Pecora R. J. Phys. Chem. B. 1997;101:746–753. [Google Scholar]
- 38.Schechter N. M., Sharp M., Reynolds J. A., Tanford C. Biochemistry. 1976;15:1897–1904. doi: 10.1021/bi00654a016. [DOI] [PubMed] [Google Scholar]
- 39.Henkel W., Vogl T., Echner H., Voelter W., Urbanke C., Schleuder D., Rauterberg J. Biochemistry. 1999;38:13610–13622. doi: 10.1021/bi9905157. [DOI] [PubMed] [Google Scholar]
- 40.Maeda H. Langmuir. 1999;15:8505–8513. [Google Scholar]
- 41.Kramer R. Z., Vitagliano L., Bella J., Berisio R., Mazzarella L., Brodsky B., Zagari A., Berman H. M. J. Mol. Biol. 1998;280:623–638. doi: 10.1006/jmbi.1998.1881. [DOI] [PubMed] [Google Scholar]
- 42.Bächinger H. P., Dodge K. J., Petschek J. P., Fessler L. I., Fessler J. H. J. Biol. Chem. 1982;257:14590–14592. [PubMed] [Google Scholar]
- 43.Myers J. C., Li D. Q., Amenta P. S., Clark C. C., Nagaswami C., Weisel J. W. J. Biol. Chem. 2003;278:32047–32057. doi: 10.1074/jbc.M304629200. [DOI] [PubMed] [Google Scholar]
- 44.Holmgren S. K., Taylor K. M., Bretscher L. E., Raines R. T. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 45.Hodges J. A., Raines R. T. J. Am. Chem. Soc. 2003;125:9262–9263. doi: 10.1021/ja035881z. [DOI] [PubMed] [Google Scholar]
- 46.Scheibel T., Parthasarathy R., Sawicki G., Lin X. M., Jaeger H., Lindquist S. L. Proc. Natl. Acad. Sci. USA. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boudko S., Frank S., Kammerer R. A., Stetefeld J., Schulthess T., Landwehr R., Lustig A., Bächinger H. P., Engel J. J. Mol. Biol. 2002;317:459–470. doi: 10.1006/jmbi.2002.5439. [DOI] [PubMed] [Google Scholar]
- 48.Provencher S. W. Comp. Phys. Commun. 1982;27:229–242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.