Abstract
GIY-YIG homing endonucleases are modular proteins, with conserved N-terminal catalytic domains connected by linkers to C-terminal DNA-binding domains. I-TevI, the T4 phage GIY-YIG intron endonuclease, functions both in promoting td intron homing, and in acting as a transcriptional autorepressor. Repression is achieved by binding to an operator, which is cleaved at 100-fold reduced efficiency relative to the intronless homing site. The linker includes a zinc finger, which functions in distance determination, to constrain the catalytic domain to cleave the homing site at a fixed position. Here we show that I-BmoI, a related GIY-YIG endonuclease lacking a zinc finger, also possesses some cleavage distance discrimination. Furthermore, hybrid endonucleases constructed by swapping the domains of I-BmoI and I-TevI are active, precise and demonstrate that features other than the zinc finger facilitate distance determination. Most importantly, I-TevI zinc finger mutants cleave the operator more efficiently than the homing site, the converse of wild-type protein. These results are consistent with the zinc finger acting as a measuring device, directing efficient cleavage of the homing site to promote intron mobility, while reducing cleavage at the operator to ensure transcriptional autorepression and phage viability.
INTRODUCTION
Modular assembly of proteins is thought to be a significant source of evolutionary diversity, facilitating the rapid evolution of proteins with new biochemical activities (1,2). GIY-YIG homing endonucleases, commonly encoded within self-splicing group I introns, are an example of a modular protein family consisting of two functional domains connected by a variable length linker (3–5). Alignments of GIY-YIG endonucleases define a ∼90 amino acid N-terminal domain that contains the conserved GIY-YIG motif and amino acids implicated in catalysis, but with little or no sequence conservation beyond a conserved asparagine residue (Asn90 of the GIY-YIG homing endonuclease I-TevI) (4,6). The GIY-YIG module has a compact, globular structure, as determined crystallographically with the free catalytic domain of I-TevI (6), and should be an ideal candidate for domain shuffling. Indeed, the GIY-YIG catalytic module is found in a variety of proteins, such as the nucleotide excision repair protein UvrC of eubacteria (4,7,8), the endonuclease/reverse transcriptase of eukaryotic retrotransposable elements (9), and a family of eukaryotic enzymes that repair stalled replication forks (10,11). The broad phylogenetic and functional distribution of the GIY-YIG catalytic module attests to its success in promoting a variety of DNA-processing events.
In contrast, the C-terminal DNA-binding domain of I-TevI, and perhaps other GIY-YIG homing endonucleases, is an extended structure consisting of small structural units bound to its DNA substrate (12). The I-TevI C-terminal domain consists of a zinc finger that is part of the linker and is not required for DNA binding (13), a minor-groove binding α-helix, and a helix–turn–helix domain. I-BmoI is a related GIY-YIG endonuclease that binds a homologous stretch of thymidylate synthase (TS)-encoding DNA (14), but lacks a zinc finger and instead possesses additional copies of putative α-helices, termed nuclease-associated modular DNA-binding domains (NUMODs) (15).
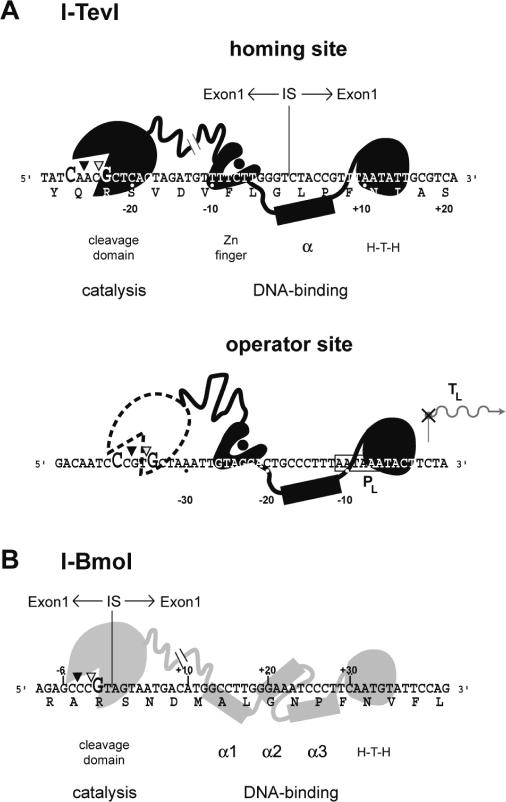
Like other intron endonucleases, GIY-YIG enzymes promote the mobility of their host intron, by binding and cleaving cognate alleles that lack the intron. Double-strand break (DSB) repair is thereby facilitated, using the intron-containing allele as a template [reviewed in Ref. (16)]. The recognition site of a homing endonuclease is usually centered on the intron insertion site (IS) of intronless alleles (Figure 1A). I-TevI's preferred cleavage sites are 23 and 25 nt upstream from the intron IS (17,18), which are defined by a 5′-CX∧XX∨G-3′ sequence (18,19), where ‘∧’ and ‘∨’ represent bottom- and top-strand cleavage sites, respectively (Figure 1A). When this preferred sequence is moved >4 bp away from the intron IS, or >12 bp closer, I-TevI defaults to cleave at the correct distance rather than at the preferred sequence (18). Mutation of the zinc finger relieves this distance preference and allows the catalytic domain to cleave at the correct sequence on substrates where it is moved closer to, or further from, the intron IS (13).
Figure 1.
GIY-YIG endonucleases are two-domain proteins. (A) Schematic representation of the two-domain structure of I-TevI (black) and its interactions with the homing and operator sites. Representations of the structure of I-TevI are based on biochemical and structural data, and computational predictions (3,4,12). The hatch mark between the catalytic domain and the zinc finger on I-TevI represents an unknown structure of the linker. Nucleotides that are cleavage determinants for I-TevI are enlarged. The amino acid sequence of TS is also indicated. Top- and bottom-strand cleavage sites are depicted on the DNA sequence by open and closed triangles, respectively. The intron IS is indicated by a vertical line. Operator substrate is numbered according to the transcriptional start site (TL). The phage T4 late promoter (PL) that regulates expression of I-TevI is indicated by a box. (B) I-BmoI (gray) interactions with its homing site substrate (14). The positions of the three putative α-helices of I-BmoI were assigned as described in Materials and Methods. Labeling is as in (A).
I-TevI possesses a second biologically relevant binding site, the operator, located upstream of the I-TevI coding sequence and overlapping the T4 late promoter that regulates expression of I-TevI (Figure 1A) (20). I-TevI binds the operator and homing sites with the same affinity, but cleaves the operator site ∼100-fold less efficiently than the homing site, at a ‘misplaced’ 5′-CXXXG-3′ sequence that is not at the optimal distance from the operator-binding site. I-TevI interactions with homing and operator substrates highlight how regulation of I-TevI cleavage can promote two distinct biological outcomes: I-TevI cleavage on substrates with the 5′-CXXXG-3′ at an optimal spacing results in a DSB and initiation of intron homing, whereas reduced cleavage on substrates with a misplaced 5′-CXXXG-3′ allows I-TevI to act as a transcriptional autorepressor.
Here, we have examined the modular structure of GIY-YIG endonucleases as it relates to constraining the catalytic domains' ability to cleave at a fixed site. We show that I-BmoI, with a different linker region than I-TevI, also possesses a distance determinant for cleavage, albeit one that is less strict than that of I-TevI. By creating novel I-TevI/I-BmoI hybrid endonucleases, we demonstrate that features other than the zinc finger also facilitate distance determination. Most importantly, we show that I-TevI zinc finger mutants that have lost the distance constraint undergo a switch compared with wild-type enzyme in relative cleavage activity on the homing site versus the operator site. This observation supports the hypothesis that the distance constraint for cleavage by I-TevI is an adaptation to promote cutting at the homing site, while limiting spurious DSBs at the operator and at other potentially deleterious sites.
MATERIALS AND METHODS
Sequences and alignments
I-TevI (Gen Bank accession number P13299) and I-BmoI (Gen Bank accession number AAK09365) were aligned using T-COFFEE (21), and the resulting alignment was edited by hand. The positions of the three putative α-helices were mapped onto the I-BmoI sequence by amino acid similarity to the known I-TevI α-helix, which extends from Ser183 to Met194. A critical histidine residue (His182) in I-TevI immediately precedes the α-helix, and makes two base-specific hydrogen bond contacts with substrate (12). We thus assigned the start of each putative I-BmoI α-helix at a serine or threonine residue that was preceded by a histidine residue. The positions are as follows: α-helix1, residues 149–160; α-helix 2, residues 176–187; α-helix3, residues 203–214. These positions differ from those of the NUMOD repeat units defined by computational analysis (15), which do not start at a serine or threonine residue preceded by a histidine.
Construction, overexpression and purification of hybrid endonucleases
To construct hybrid endonucleases, we separately amplified the N-terminal DNA sequence of I-BmoI or I-TevI, and the C-terminal region of I-BmoI or I-TevI using gene-specific primers, listed in Supplementary Table 1. Plasmid pSP6-716, containing a wild-type version of I-TevI, and Bacillus mojavensis s87-18 genomic DNA were used as templates for PCRs. The individual N- and C-terminal DNA fragments were gel purified, and combined in a SOEing reaction (22) to generate an uninterrupted coding region. The H-TevBmoV1 and H-BmoTevV1 constructs were initially cloned as blunt-end PCR products into pBS (Stratagene) for use in in vitro transcription and translation reactions. Subsequent H-TevBmo constructs were digested with XbaI and BamHI, and were cloned into pET28a (Novagen) digested with the same enzymes. The resulting plasmids were transformed into Escherichia coli ER2566 (New England Biolabs), and the hybrid proteins were overexpressed and purified as described previously for I-TevI (3). I-BmoI was purified according to published protocols (14).
Construction of hybrid homing sites
I-BmoI homing-site derivatives, pBSBmoHS (54 bp insert), pUCBmoHS+5 (59 bp insert) and pUCBmoHS+10 (64 bp insert) were constructed by annealing two complementary oligonucleotides corresponding to the I-BmoI homing site sequence. Oligonucleotides were annealed by heating to 95°C in T4 DNA ligase buffer (New England Biolabs), cooled to room temperature, and then ligated with pBS or pUC19 cut with BamHI and XbaI or EcoRI. A similar strategy was used to construct plasmids containing the I-TevI (TevHS) and hybrid homing sites (TevBmoHS and BmoTevHS) (all 54 bp).
In vitro transcription and translation
Plasmids containing the H-TevBmoV1 and H-BmoTevV1 coding regions were linearized with HindIII, and 1 µg of DNA was used in in vitro transcription reactions with T7 RNA polymerase (New England Biolabs) as described previously (13). RNA was extracted with phenol:chloroform, and precipitated with ethanol. Aliquots of the RNA were incubated with wheat germ extract (Promega), supplemented with 35S-methionine and incubated at room temperature for 2 h. The relative amounts of each protein were determined by comparison of radioactive counts, taking into account the number of methionine residues in each protein, using a Typhoon 9400 phosphorimager (GEHealthcare) and ImageQuant software (Molecular Dynamics).
Gel mobility-shift analysis
All DNA substrates were amplified from plasmid DNA using −20 and M13 reverse primers end-labeled with [γ32P]ATP. Binding reactions were performed at room temperature by incubating ∼0.01 pmol of DNA with 1 µl of in vitro translated proteins. Alternatively, 5 µl, 1 µl and then 1 µl of 5-fold serial dilutions of purified hybrid endonucleases were used. The protein concentrations were H-TevBmoV1, 0.43 mg/ml; H-TevBmoV2, 0.25 mg/ml; H-TevBmoV3, 0.2 mg/ml; H-TevBmoV4, 0.3 mg/ml; and H-TevBmoV5, 0.57 mg/ml. Binding reactions in 20 µl of 50 mM Tris–HCl (pH 8.0), 20 µg/ml poly(dI/dC), 5 mM EDTA and 10 µg/ml BSA were incubated for 10 min. Those reactions that were supplemented with 10 mM MgCl2 did not contain EDTA. Loading dye containing 50% glycerol, 50 mM Tris–HCl (pH 8.0), 20 µg/ml poly(dI/dC), 5 mM EDTA, 10 µg/ml BSA and 0.01% bromophenol blue was added (5 µl), and the bound complexes were separated from unbound DNA at 4°C on 12% 29:1 polyacrylamide:bisacrylamide gels in TBE buffer (90 mM Tris–HCl, 90 mM boric acid and 2 mM EDTA).
Cleavage assays
Cleavage assays were performed as described (13) using 250 ng of ScaI-linearized plasmid substrate, and equivalent amounts of in vitro synthesized proteins. Incubations were performed at 37°C for 15 min for single time point assays and were stopped by addition of loading dye. For time-course assays, 20 µl aliquots were taken from a pooled reaction at the appropriate time and stopped by the addition of loading dye. All reactions were treated with Proteinase K and RNaseA prior to electrophoresis. Agarose gels were stained with SybrGold (Molecular Probes), imaged on Typhoon 9400 phosphorimager, and analyzed using ImageQuant.
Cleavage-site mapping
Plasmids containing the I-BmoI homing site, its +5 and +10 derivatives, the TevBmoHS, and the I-TevI operator sequence, were used in PCR reactions with 5′ end-labeled primers to generate strand-specific substrates. The same primers were used in cycle sequencing reactions (USB) to generate a sequencing ladder. Cleavage-site mapping was performed as described previously (23). Briefly, the cleavage reactions were performed at 37°C by incubating 5000 c.p.m. of 5′ end-labeled DNA substrate with the appropriate enzymes in 30 µl volumes in the presence of 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2 and 100 mM NaCl, for 15 min. The reaction mixtures were extracted and precipitated, and resuspended in stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol). Aliquots of each reaction were separated on 6 or 8% denaturing polyacrylamide (8.3 M urea) gels alongside the corresponding sequencing ladder.
RESULTS
Evidence for attenuated distance determination in endonuclease I-BmoI
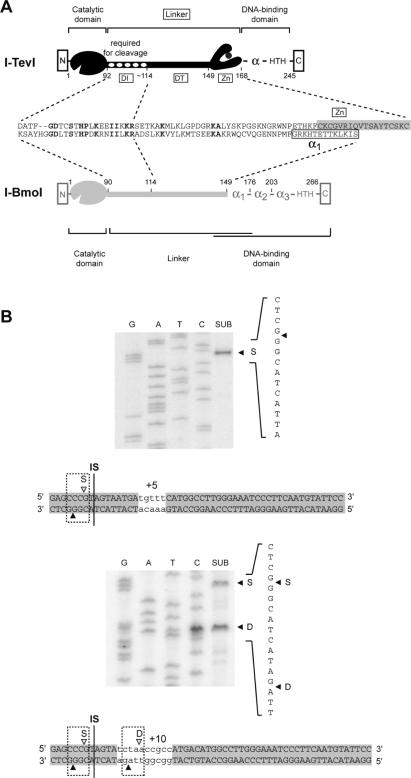
Although there is little amino acid conservation among a broad phylogenetic sampling of GIY-YIG family endonucleases (4,6), the linker region connecting the N- and C-terminal domains of I-BmoI and I-TevI is >40% identical over 23 residues (Figure 2A). This region corresponds to the I-TevI deletion intolerant (DI) region, where amino acid deletions abolished cleavage of intronless substrate (4). Identity between the deletion tolerant (DT) region of I-TevI (4) and the corresponding region of I-BmoI is low, owing in part to a variation in length, and also because the I-TevI DT region includes a zinc finger that is absent from I-BmoI (Figure 2A). Instead, I-BmoI possesses a putative α-helix (α1, Figure 1B) that was identified by computational methods (15), and is similar to the α-helix of I-TevI.
Figure 2.
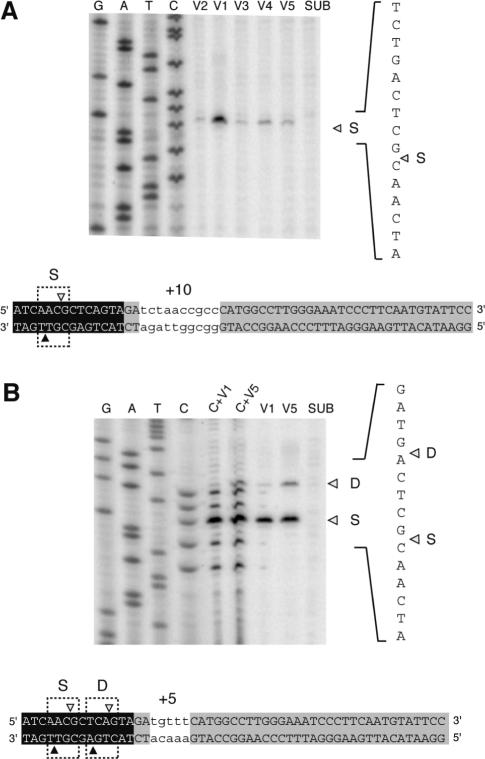
Sequence (S) versus distance (D) preference of I-BmoI. (A) Schematic representation of the modular structures of I-TevI (black) and I-BmoI (gray), with functions assigned to the modules of each protein. Also shown is an amino acid alignment of the I-TevI linker with the corresponding region of I-BmoI, for which it is not known precisely where the linker ends and the DNA-binding domain begins. Identical amino acids are in bold type. DI and DT are the deletion intolerant and deletion tolerant regions of the I-TevI linker, respectively. (B) Mapping of I-BmoI cleavage sites on +5 and +10 insertion substrates. Shown is a sequencing gel of cleavage-site mapping for bottom-strand labeled substrate. The position of the bottom-strand cleavage site is indicated by a closed triangle. The 5 and 10 nt insertion, which are indicated by lower case letters, are patterned on those used in previous studies of distance determination by I-TevI (13,18). Top- and bottom-strand cleavage sites are indicated as in Figure 1. SUB, substrate reacted with I-BmoI; S, cleavage at the correct sequence; D, cleavage at the correct distance.
As the zinc finger of I-TevI has been shown to regulate distance determination of the catalytic domain by constraining it to cleave at a fixed distance from the intron IS (13), we wished to determine if I-BmoI possesses a similar property despite the absence of a zinc finger. I-BmoI cleaves intronless substrate 2 and 4 nt upstream of the intron IS, in identical positions to the I-TevI cleavage sites on its intronless substrate (Figure 1B) (14). To establish if I-BmoI possesses a distance preference (‘D’ in Figure 2B), we constructed DNA substrates that moved the preferred I-BmoI cleavage sites 5 or 10 nt further upstream from the primary binding site of the C-terminal domain, and mapped the position of the I-BmoI cleavage sites on these substrates (Figure 2B). On the +5 substrate, I-BmoI cleaved at the correct sequence (‘S’ in Figure 2B), unlike I-TevI which has a 7:1 bias towards distance (Table 1) (13). However, on +10 substrate, I-BmoI cleaved equally well at sequence as at distance from the intron IS. These data suggest not only that I-BmoI possesses an attenuated distance preference in comparison to I-TevI, which has a 9:1 distance bias on +10 substrate (Table 1), but also implies that I-BmoI can sense distance, despite the absence of a zinc finger.
Table 1.
Cleavage preference of I-TevI, I-BmoI and hybrid endonucleases
| Enzyme | Substratea | |
|---|---|---|
| +5 | +10 | |
| I-TevIb | D > S (7:1) | D > S (9:1) |
| I-BmoIb | S | D = S |
| H-TevBmoV1c | D < S (1:10) | S |
| H-TevBmoV2 | ndd | S |
| H-TevBmoV3 | nd | S |
| H-TevBmoV4 | nd | S |
| H-TevBmoV5 | D < S (1:3) | S |
aFor I-TevI and I-BmoI, the +5 and +10 substrates have an insertion in the TevHS or BmoHS substrate, respectively. For the hybrid enzymes, the insertions are in the TevBmoHS substrate.
cData for H-TevBmoV1- V5 are shown in Figure 5.
dnd, not determined.
Hybrid endonucleases retain binding and cleavage specificity of the parental enzymes
To probe the modular nature of GIY-YIG endonucleases, we swapped the N-terminal catalytic domains of I-BmoI and I-TevI to create the hybrid H-BmoTevV1 (V1 = variant 1) and the reciprocal enzyme, H-TevBmoV1 (Figure 3A). We initially chose as the fusion point the amino acid residue used for cloning and overexpression of the C-terminal DNA-binding domain of each protein, residue 129 for I-BmoI (14) and residue 130 for I-TevI (3), as these modules bind their respective homing sites with the same affinity as the full-length enzymes. To assay the cleavage activity of each hybrid enzyme, we constructed hybrid DNA substrates (Figure 3B), fusing the portion of the I-BmoI sequence required for cleavage by the I-BmoI catalytic domain to the portion of the I-TevI sequence required for DNA-binding (BmoTevHS), and vice-versa (TevBmoHS).
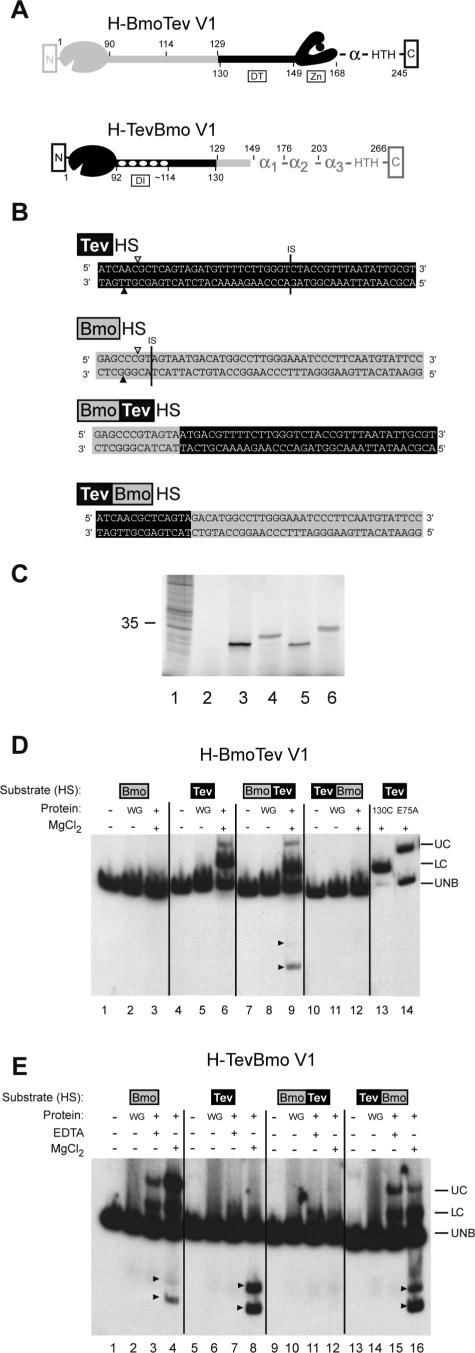
Figure 3.
Hybrid endonucleases retain DNA-binding and DNA-cleavage functions. (A) Schematic representation of H-BmoTevV1 and H-TevBmoV1 hybrid enzymes. I-TevI is black, I-BmoI is gray. Numbers below each hybrid refer to amino acid positions in I-TevI, while numbers above each hybrid refer to amino acid positions in I-BmoI. DI and DT as for Figure 2. (B) I-BmoI, I-TevI and hybrid DNA substrates. Positions of the top- and bottom-strand cleavage sites are depicted as in Figure 1, as is the position of the intron IS. Sequences of the I-TevI and I-BmoI homing sites are on black and gray backgrounds, respectively. (C) Phosphorimager analysis of in vitro translation of hybrid endonucleases fractionated on a polyacrylamide gel. Lane 1, Broome Mosaic Virus RNA positive control with 35 kDa protein indicated; lane 2, unprogrammed wheat germ (WG) extract; lane 3, H-TevBmoV1; lane 4, H-BmoTevV1; lane 5, I-TevI; lane 6, I-BmoI. (D) Gel-shift analysis of H-BmoTevV1. Complexes were formed with the DNA substrates described in (B). WG, unprogrammed wheat germ extract; UC, upper complex; LC, lower complex; UNB, unbound substrate. Cleavage products are indicated by arrowheads. (E) Gel-shift analysis of H-TevBmoV1. The gel is labeled as in (D).
Because cytotoxic I-TevI is extremely difficult to clone and overexpress in wild-type form in vivo, we used in vitro transcription and translation to synthesize each hybrid endonuclease (Figure 3C). Equal amounts of protein, as determined by 35S-methionine incorporation, were incubated with DNA substrates end-labeled on both strands. As seen previously in gel-shift experiments with full-length I-TevI (24) or I-BmoI preparations (14), we consistently observed two protein–DNA complexes with both H-BmoTevV1 and H-TevBmoV1. An upper (UC) complex corresponds to interactions of full-length protein with substrate (Figure 3D, lane 14), and a lower (LC) complex corresponds to interactions of proteolyzed DNA-binding domain fragments with substrate (Figure 3D, lane 13). We also consistently observed an enhancement of H-TevBmoV1 binding to BmoHS DNA upon addition of divalent metal ion to the reaction (Figure 3E, lane 4), which may reflect a conformational change in the DNA substrate or protein, facilitating DNA binding. Their ability to form complexes shows that the DNA-binding activity of the hybrid proteins is determined by the parental origin of the DNA-binding domain and not of the catalytic domain, consistent with previous reports that the C-terminal domain of I-TevI and I-BmoI impart both the binding energy and specificity of the interactions (3,12,14).
Addition of divalent metal ion allowed us to judge the cleavage activity of the hybrid proteins. Although both an upper and lower complex formed when H-BmoTevV1 was incubated with either TevHS or BmoTevHS substrates, cleavage products were only observed with the BmoTevHS substrate (Figure 3D, lane 9). No cleavage products were observed when divalent metal ions were omitted from binding reactions, although protein–DNA complexes were formed (not shown). In contrast, cleavage products were observed with a variety of substrates when H-TevBmoV1 binding reactions were supplemented with MgCl2, consistent with previous studies demonstrating the sequence tolerance of I-TevI to multiple substitutions throughout its recognition site, and its high specific activity (Figure 3E, lanes 4, 8 and 16) (17,25). TevHS substrate was extensively cleaved, although we did not observe an H-TevBmoV1-TevHS complex by gel-shift analysis (Figure 3E, lane 8). As the DNA-binding domain of I-BmoI does have weak affinity for the TevHS substrate (not shown), it is likely that labile H-TevBmoV1-TevHS complexes formed under the binding conditions, but were rapidly converted to product because the catalytic domain of I-TevI is extremely efficient (see below). These cleavage patterns were supported by further assays of the hybrid enzymes showing that H-TevBmoV1 cleaved 80% of TevBmoHS substrate after 60 min, whereas H-BmoTevV1 cleaved <5% of BmoTevHS substrate after 60 min (not shown). Although we have not eliminated the possibility that the reduced activity of H-BmoTevV1 is a consequence of the position of the fusion between the domains, the reduction of activity is fully consistent with the inherently lower cleavage activity of full-length I-BmoI relative to I-TevI (Table 2) (26).
Table 2.
Specific activity of H-TevBmo variants
| Protein | Specific activitya (U/µg) | Relative specific activityb |
|---|---|---|
| I-TevIc | 1.4 × 104 | 1 |
| I-BmoIc | 1.8 × 101 | ∼10−3 |
| H-TevBmoV1 | 1.8 × 100 | ∼10−4 |
| H-TevBmoV2 | 4.0 × 10−1 | ∼10−5 |
| H-TevBmoV3 | 2.8 × 10−3 | ∼10−7 |
| H-TevBmoV4 | 2.1 × 10−3 | ∼10−7 |
| H-TevBmoV5 | 1.5 × 101 | ∼10−3 |
aCleavage assays were performed as described in Materials and Methods. One unit of activity is defined as the amount of enzyme required to cleave 250 ng of ScaI-linearized plasmid to 50% completion in 1 min at 37°C.
bActivity relative to I-TevI, which was assigned a value of 1.
cSpecific activities of I-TevI and I-BmoI were taken from Ref. (26).
Hybrid enzymes with variable linkers are functional
Encouraged by the retention of function of the hybrid endonucleases, we made a series of H-TevBmo enzymes with the I-TevI catalytic domain and differing lengths of the linker region fused to the I-BmoI DNA-binding domain (Figure 4A). Surprisingly, we were able to express all of the hybrid constructs in a T7 RNA polymerase-regulated expression vector in E.coli, presumably because fusion of the N-terminal I-TevI catalytic domain to a different DNA-binding domain reduced the catalytic activity to a level tolerated by E.coli expression strains. To assay the binding and cleavage activities of the hybrid enzymes, we performed gel-shift and cleavage assays on labeled TevBmoHS substrate with purified enzyme preparations. All of the hybrid enzymes bound the TevBmoHS, albeit with different affinities (Figure 4A), and they all cleaved the TevBmoHS substrate in the same position (at the wild-type cleavage site on both the top and bottom strands) again with different efficiencies (Figure 4B).
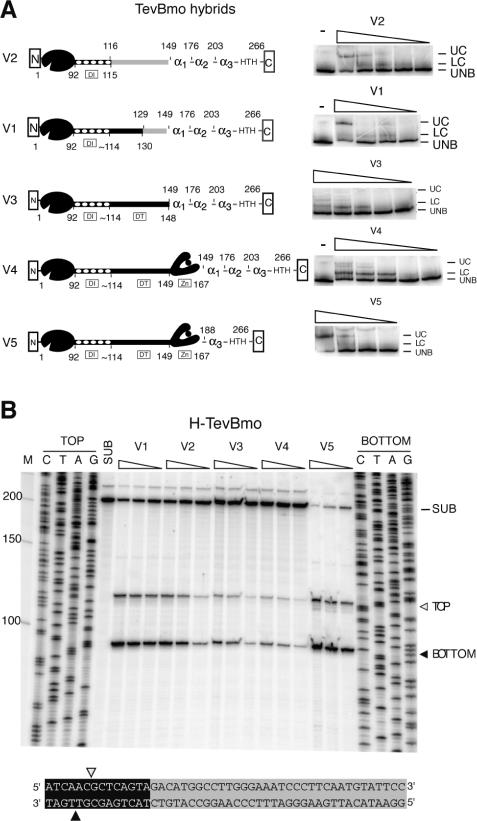
Figure 4.
H-TevBmo variants bind and cleave TevBmoHS substrate. (A) Schematic representation of H-TevBmo hybrid enzymes and DNA-binding assays. The H-TevBmo enzymes are labeled as in Figure 3A. The purified hybrid enzymes were subjected to gel-shift assays with TevBmoHS substrate, with decreasing amounts of the enzymes as indicated in Materials and Methods. The gels are labeled as in Figure 3D. (B) Cleavage assay and mapping of H-TevBmo variants on TevBmoHS substrate. TevBmoHS substrates labeled on top and bottom strands were incubated with decreasing concentrations of H-TevBmo variants, and separated on an 8% denaturing gel. Unreacted substrate is indicated by SUB, the top-strand cleavage product by TOP and an open triangle, and the bottom-strand product by BOTTOM and a closed triangle. Top- and bottom-strand cleavage sites on TevBmoHS substrate are indicated below.
To determine the effect of the different linkers on cleavage efficiency, we measured the specific activity of each of the hybrids on linearized TevBmoHS plasmid substrate (Table 2). We compared the activity of each hybrid to that of I-TevI, as this enzyme was the source of the catalytic domain. As indicated in Table 2, H-TevBmoV5 is the most active of the hybrids, with a specific activity approximately equal to that of full-length I-BmoI, but ∼1000-fold less active than I-TevI. We could not reliably determine the specific activity of the other hybrids, but estimate that their activity is between 104- and 107-fold reduced compared with that of I-TevI. The higher specific activity of H-TevBmoV5 correlates with the highest ratio of upper complex to unbound DNA in gel-shift assays compared with that observed with the other hybrid enzymes (Figure 4A). The upper complex results from interactions of full-length protein with substrate, rather than proteolyzed fragments that retain DNA-binding activity (14,24). The higher activity of H-TevBmoV5 is also manifest in the cleavage site mapping assay (Figure 4B), where both substrate disappearance and product appearance is greatest for this hybrid. These results indicate that putative α-helix1 and α-helix2 are not required for I-BmoI DNA binding and may be components of the I-BmoI linker.
Hybrid enzymes prefer to cleave at the correct sequence rather than distance
The hybrid TevBmo enzymes contain varying amounts of the I-TevI linker, allowing us to probe the role of different segments of the I-TevI linker in distance determination. To this end, we constructed two hybrid DNA substrates where the preferred I-TevI cleavage sites were moved further upstream from the I-BmoI primary binding sequence (Figure 5). On the +10 TevBmoHS substrate, where the cleavage site is moved by one turn to the same face of the helix, all of the hybrid enzymes cleaved at the correct sequence (Figure 5A), rather than at the correct distance as for I-TevI (Table 1). Only the H-TevBmoV1 and V5 hybrids cleaved the +5 substrate, presumably because insertion of 5 nt moves the preferred cleavage site to the opposite face of the DNA helix. As might have been predicted, H-TevBmoV5, with the zinc finger intact, retained the ability to cleave at the correct distance, with a distance to sequence preference of ∼1:3 (Table 1 and Figure 5B). Interestingly, H-TevBmoV1, with no zinc finger, also retained the ability to cleave at the correct distance (Table 1 and Figure 5B), although with a distance to sequence preference of ∼1:10. These results confirm elements other than the zinc finger to be involved in distance determination.
Figure 5.
Cleavage by H-TevBmo variants on hybrid insertion substrates. The cleavage sites of H-TevBmo variants were on +10 (A) and +5 (B) insertion substrates, with the top-strand mapping result shown. Substrates as in Figure 3, with +10 and +5 insertions indicated by lower case letters. SUB, unreacted substrate; S, cleavage at the correct sequence; D, cleavage at the correct distance. H-TevBmoV1 and V5 cleavage products on +5 substrate were co-resolved with the C sequencing reaction, as indicated by C + V1 and C + V5.
I-TevI zinc finger mutants have a switched cleavage preference on homing versus operator sites
I-TevI possesses two biologically relevant binding sites, the homing and operator sites, for which it has equal affinity. I-TevI, however, has reduced cleavage activity on the operator site, facilitating its function as an autorepressor (Figure 1A) (20). We reasoned that if the distance constraint of I-TevI is related to the protein's dual function, then I-TevI zinc finger mutants that no longer possess a distance determinant might cleave operator substrate more efficiently than does wild-type I-TevI with an intact zinc finger.
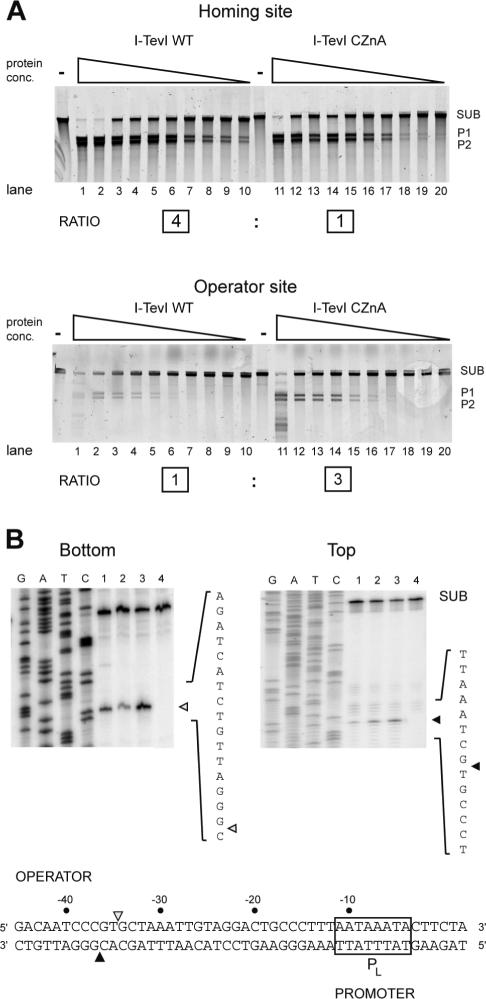
We compared the cleavage efficiencies of wild-type I-TevI and a zinc finger mutant, CZnA, with cysteine-to-alanine substitutions in the zinc finger (13), on homing and operator sites (Figure 6A). We used two assays to determine cleavage activity: a 3-fold dilution series of the proteins with cleavage measured at a single 15 min time point (Figure 6), and a time-course assay using equivalent amounts of protein (Supplementary Figure 1). Although the data preclude rigorous kinetic analyses because the protein remains bound to its downstream cleavage product (27), we are able to compare reactions with similar extents of cleavage to determine relative cleavage activities for the two enzymes on the two substrates. As demonstrated previously, wild-type I-TevI cleaved the homing site substrate more efficiently than does the zinc finger mutant (13) (Figure 6A, compare lanes 8–10 with 18–20). The time-course experiments were consistent with the 4-fold difference reported by Dean et al. (13) (wild-type protein cleaved 45% of the homing site substrate in 0.5 min, while CZnA protein cleaved 25% after 1 min, Supplementary Figure 1). Strikingly, this cleavage preference was reproducibly reversed on operator site substrate as the zinc finger mutant cleaved more efficiently than wild-type protein (Figure 6A; compare lanes 4–6 with lanes 14–16 to see that the limit of detection for CZnA cleavage is with 3-fold less protein). The time-course data were consistent with the 3-fold difference seen in the dilution experiments as wild-type I-TevI cleaved 9% of the operator site substrate after 20 min while CZnA cleaved 12% after 10 min (Supplementary Figure 1). The net result is a 12-fold switch in cleavage activity between the wild-type I-TevI and zinc finger mutants on the homing versus operator sites.
Figure 6.
I-TevI zinc finger mutants cleave the operator more efficiently than does wild-type I-TevI. (A) Cleavage of homing or operator site by I-TevI wild-type (WT) and the CZnA variant. For both substrates agarose gels show cleavage assays using equivalent starting amounts of WT or the CZnA protein in a 3-fold dilution series. Lanes 1–10 have the same amount of protein as lanes 11–20, respectively. SUB, substrate; P1 and P2, products. The relative cleavage activity on homing versus operator substrates is shown by the boxed numbers below cleavage gels. (B) Cleavage mapping of WT, CZnA and ΔZn on bottom and top strands of operator substrate. Lanes 1, WT; lanes 2, CZnA mutant; lane 3, ΔZn mutant; lane 4, no protein. SUB, unreacted substrate. Shown below is the I-TevI operator region with the top- and bottom-strand cleavage sites indicated by open and closed triangles, respectively. The T4 late promoter (PL) is boxed.
We also mapped the cleavage sites of the wild-type protein, the CZnA mutant, and a mutant with a complete deletion of the zinc finger (ΔZn), on operator substrate. The cleavage site mapped precisely to the 5′-CXXXG-3′ sequence at positions −34 and −36 (Figure 6B). Collectively, these data show that the I-TevI zinc finger mutants cleave operator substrate more efficiently than does wild-type protein, whereas this preference is reversed on homing site substrate.
DISCUSSION
Modular structure and distance determination
GIY-YIG endonucleases are unusual among characterized homing and restriction endonucleases in that their cleavage sites are distant from their primary binding site. The separation of cleavage and binding sites is a consequence of the modular structure of the GIY-YIG endonucleases, whereby a complex linker connects an N-terminal catalytic domain to a C-terminal DNA-binding domain (5). Although the I-TevI-specific zinc finger regulates the positioning of the catalytic domain on substrates, we show here that distance determination can occur in I-BmoI and in an I-TevI/I-BmoI hybrid in the absence of a zinc finger (Figures 2–4). Furthermore, although we found that the hybrid enzymes preferred to cleave at the correct sequence rather than at the correct distance, some ability to cleave at the correct distance was maintained, particularly by H-TevBmoV1, which lacks the I-TevI zinc finger (Figure 3A and Table 1). These data suggest that amino acids 116–130 of the I-TevI linker region may also be involved in distance determination. Additionally, some portion of the I-BmoI C-terminal domain may function to regulate distance preference, consistent with the ability of I-BmoI to sense distance (Figure 2). In I-BmoI, the zinc finger is replaced by a putative α-helix (α1, Figure 1B). Although we have shown that putative α-helix1 and α-helix2 of I-BmoI are not required for DNA binding or cleavage (Figure 4), it is unclear at this time if these α-helices of I-BmoI function in an analogous manner to the I-TevI zinc finger in distance determination.
Another well-known example of a modular endonuclease is the type IIS restriction enzyme FokI, a two-domain enzyme consisting of an N-terminal DNA-binding domain and a C-terminal catalytic domain (28–30). Interestingly, FokI cleaves at a distance from its primary recognition site (31), as do I-TevI and I-BmoI, although FokI displays little sequence preference. The FokI cleavage sites can be moved relative to the enzyme's primary DNA-binding site by the insertion of amino acids in the linker region connecting the binding and catalytic domains (32). In contrast, the linkers of I-TevI (13,18) and I-BmoI (Figure 2) have an innate flexibility, with the ability to extend and retract so as to position the catalytic domain at the preferred cleavage sites.
A biological rationale for distance determination by GIY-YIG intron endonucleases
A question raised by our data is the biological rationale for cleavage at a fixed distance from the intron IS by both I-TevI (13,18) and I-BmoI (Figure 2). Previous studies have shown that I-BmoI and I-TevI bind a homologous stretch of TS-encoding DNA, and cleave their respective substrates in the same positions (14). The top-strand cleavage site of both enzymes is 5′ to a G-C base pair that is conserved in all TS genes and corresponds to the second position of a critical arginine residue in the TS active site (Figure 1) (26). We suggest that constraining the catalytic domain of I-TevI and I-BmoI to cleave at a fixed position of intronless TS substrate would facilitate efficient cleavage, since the required base pair will always be present at the same position of phage-encoded or bacterial TS genes targeted by either enzyme.
Another rationale for constraining cleavage, particularly in the case of highly active I-TevI (Table 2), relates to minimizing deleterious cleavage at secondary sites. I-TevI is extremely tolerant of nucleotide substitutions within its recognition site, as evidenced by mutational analyses (18,25), suggesting that the distance constraint might act to minimize spurious and deleterious DSBs. A case in point is I-TevI binding to its operator and homing sites with equivalent affinity but cleaving the operator with ∼100-fold less efficiency than the homing site, because the preferred 5′-CXXXG-3′ sequence is not at the correct distance (20). Although there is a related sequence that is equivalent to positions −15 and −17 of the homing site substrate, this sequence is cleaved extremely inefficiently by wild-type I-TevI. We have shown here, however, that zinc finger mutants of I-TevI, which no longer possess a distance constraint, cleave operator substrate at the ‘misplaced’ 5′-CXXXG-3′ sequence three times more efficiently than does wild-type protein, which, conversely, cleaves the homing site 4-fold more efficiently than the zinc finger mutants (Figure 5). This 12-fold switch in cleavage efficiency, based on which action is being taken by this bi-functional protein, leads us to suggest that another biological advantage to maintaining a distance constraint in I-TevI is to minimize spurious DSBs at the operator site. Thus, the linker region acts as a modular measuring device facilitating both functions of I-TevI, by regulating and positioning the catalytic domain on homing substrate for efficient cleavage and intron mobilization, and on the operator for inefficient cleavage and phage viability. In this respect, it is interesting to note that I-BmoI, which is ∼103-fold less active that I-TevI, does not have a readily identifiable operator site immediately upstream of its coding region, and may thus not require a mechanism to constrain the catalytic domain to cleave at a fixed distance, as does I-TevI.
Supplementary Material
Acknowledgments
We thank John Dansereau for critical reading of the manuscript, and for help with the figures, and Maryellen Carl for assistance with formatting the manuscript. DNA sequencing was provided by the Molecular Genetics Core at the Wadsworth Center. We also thank David Shub for providing laboratory space and reagents during construction of H-BmoTevV1 and H-TevBmoV1 (supported by NIH grant GM37746). D.R.E. was supported by a postdoctoral fellowship from the Canadian Institutes for Health Research, and M.B. and V.D. by NIH grant GM44844 and M.B. by NIH grant GM39422. Funding to pay the Open Access publication charges for this article was provided by NIH GM44844.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doolittle R.F. The multiplicity of domains in proteins. Annu. Rev. Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 2.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 3.Derbyshire V., Kowalski J.C., Dansereau J.T., Hauer C.R., Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski J.C., Belfort M., Stapleton M.A., Holpert M., Dansereau J.T., Pietrokovski S., Baxter S.M., Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Roey P., Derbyshire V. GIY-YIG Homing Endonucleases—Beads on a String. In: Belfort M, editor. Homing endonucleases and Inteins. Springer-Verlag: 2005. pp. 67–83. [Google Scholar]
- 6.Van Roey P., Meehan L., Kowalski J.C., Belfort M., Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nature Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 7.Verhoeven E.E., van Kesteren M., Moolenaar G.F., Visse R., Goosen N. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem. 2000;275:5120–5123. doi: 10.1074/jbc.275.7.5120. [DOI] [PubMed] [Google Scholar]
- 8.Truglio J.J., Rhau B., Croteau D.L., Wang L., Skorvaga M., Karakas E., Dellavecchia M.J., Wang H., Van Houten B., Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyatkov K.I., Arkhipova I.R., Malkova N.V., Finnegan D.J., Evgen'ev M.B. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc. Natl Acad. Sci. USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricke W.M., Brill S.J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Roey P., Waddling C.A., Fox K.M., Belfort M., Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean A.B., Stanger M.J., Dansereau J.T., Van Roey P., Derbyshire V., Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc. Natl Acad. Sci. USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgell D.R., Shub D.A. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc. Natl Acad. Sci. USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sitbon E., Pietrokovski S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 2003;28:473–477. doi: 10.1016/S0968-0004(03)00170-1. [DOI] [PubMed] [Google Scholar]
- 16.Belfort M., Derbyshire V., Cousineau B., Lambowitz A. Mobile Introns: Pathways and Proteins. In: Craig N., Craigie R., Gellert M., Lambowitz A., editors. Mobile DNA II. NY: ASM Press; 2002. pp. 761–783. [Google Scholar]
- 17.Bell-Pedersen D., Quirk S.M., Bryk M., Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc. Natl Acad. Sci. USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryk M., Belisle M., Mueller J.E., Belfort M. Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J. Mol. Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 19.Edgell D.R., Stanger M.J., Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J. Mol. Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Edgell D.R., Derbyshire V., Van Roey P., LaBonne S., Stanger M.J., Li Z., Boyd T.M., Shub D.A., Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nature Struct. Mol. Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- 21.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 22.Horton R.M. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 1997;67:141–149. doi: 10.1385/0-89603-483-6:141. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q., Belle A., Shub D.A., Belfort M., Edgell D.R. SegG endonuclease promotes marker exclusion and mediates co-conversion from a distant cleavage site. J. Mol. Biol. 2003;334:13–23. doi: 10.1016/j.jmb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 24.Mueller J.E., Smith D., Bryk M., Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryk M., Quirk S.M., Mueller J.E., Loizos N., Lawrence C., Belfort M. The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgell D.R., Stanger M.J., Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr. Biol. 2003;13:973–978. doi: 10.1016/s0960-9822(03)00340-3. [DOI] [PubMed] [Google Scholar]
- 27.Mueller J.E., Smith D., Belfort M. Exon coconversion biases accompanying intron homing: battle of the nucleases. Genes Dev. 1996;10:2158–2166. doi: 10.1101/gad.10.17.2158. [DOI] [PubMed] [Google Scholar]
- 28.Li L., Wu L.P., Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc. Natl Acad. Sci. USA. 1992;89:4275–4279. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wah D.A., Hirsch J.A., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 30.Wah D.A., Bitinaite J., Schildkraut I., Aggarwal A.K. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugisaki H., Kanazawa S. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI) Gene. 1981;16:73–78. doi: 10.1016/0378-1119(81)90062-7. [DOI] [PubMed] [Google Scholar]
- 32.Li L., Chandrasegaran S. Alteration of the cleavage distance of Fok I restriction endonuclease by insertion mutagenesis. Proc. Natl Acad. Sci. USA. 1993;90:2764–2768. doi: 10.1073/pnas.90.7.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.