Abstract
Photomorphogenesis is regulated by red/far-red light–absorbing phytochromes and blue/UV-A light–absorbing cryptochromes. We isolated an Arabidopsis thaliana blue light mutant, short hypocotyl under blue1 (shb1), a knockout allele. However, shb1-D, a dominant allele, exhibited a long-hypocotyl phenotype under red, far-red, and blue light. The phenotype conferred by shb1-D was caused by overaccumulation of SHB1 transcript and recapitulated by overexpression of SHB1 in Arabidopsis. Therefore, SHB1 acts in cryptochrome signaling but overexpression may expand its signaling activity to red and far-red light. Consistent with this, overexpression of SHB1 enhanced the expression of PHYTOCHROME-INTERACTING FACTOR4 (PIF4) under red light. PIF4 appears to specifically mediate SHB1 regulation of hypocotyl elongation and CHLOROPHYLL a/b BINDING PROTEIN3 or CHALCONE SYNTHASE expression under red light. Overexpression of SHB1 also promoted proteasome-mediated degradation of phytochrome A and hypocotyl elongation under far-red light. Under blue light, shb1 suppressed LONG HYPOCOTYL IN FAR-RED LIGHT1 (HFR1) expression and showed several deetiolation phenotypes similar to hfr1-201. However, the hypocotyl and cotyledon-opening phenotypes of shb1 were opposite to those of hfr1-201, and HFR1 acts downstream of SHB1. SHB1 encodes a nuclear and cytosolic protein that has motifs homologous with SYG1 protein family members. Therefore, our studies reveal a signaling step in regulating cryptochrome- and possibly phytochrome-mediated light responses.
INTRODUCTION
Plants have evolved red and far-red light–absorbing phytochromes and UV-A/blue light–absorbing cryptochromes and phototropins (Neff et al., 2000; Huq and Quail, 2005). Among the photoreceptors, phytochromes and cryptochromes regulate seedling deetiolation responses, including the inhibition of hypocotyl elongation, the opening of hypocotyl hooks, the opening and expansion of cotyledons, and the development of chloroplasts. In Arabidopsis thaliana, the phytochrome gene family has five members, PHYA through PHYE (Huq and Quail, 2005). phyA is the photoreceptor for far-red light–mediated deetiolation responses, and phyB is the major photoreceptor for red light–mediated deetiolation responses. Purified oat (Avena sativa) phyA preparations have been shown to autophosphorylate and to phosphorylate PHYTOCHROME SUBSTRATE1 (PKS1), cryptochrome1 (cry1), and auxin/indole-3-acetic acid proteins in vitro (Ahmad et al., 1998; Yeh and Lagarias, 1998; Fankhauser et al., 1999; Colón-Carmona et al., 2000). phyA and phyB also translocate to the nucleus in a light-dependent manner and interact with PHYTOCHROME-INTERACTING FACTOR1 (PIF1), PIF3, and PIF4, PKS1, NUCLEOSIDE DIPHOSPHATE KINASE2, ARABIDOPSIS RESPONSE REGULATOR4, EARLY FLOWERING3 (ELF3), PHYTOCHROME-ASSOCIATED PROTEIN PHOSPHATASE, and PHYTOCHROME-SPECIFIC TYPE 5 PHOSPHATASE in vitro (Ni et al., 1998, 1999; Choi et al., 1999; Fankhauser et al., 1999; Kircher et al., 1999; Liu et al., 2001; Sweere et al., 2001; Kim et al., 2002; Huq and Quail, 2005; Ryu et al., 2005).
Mutations in CRY1 impair seedling deetiolation responses under blue/UV-A light (Yang et al., 2000; Cashmore, 2003). CRY1 encodes a flavoprotein with sequence similarity to photolyases but lacks photolyase activity and has a C-terminal extension not found in the photolyases. Dark-grown Arabidopsis seedlings carrying the C-terminal domains of either cry1 or cry2 show phenotypes that are normally associated with light-grown seedlings and are often observed for constitutive photomorphogenic1 (cop1) (Yang et al., 2000). The signaling activity of cry1, therefore, probably involves a direct interaction of its C terminus with COP1, a negative regulator of photomorphogenesis (Wang et al., 2001; Yang et al., 2001). cry2 is involved in the control of photoperiodic flowering in addition to its role in regulating seedling deetiolation responses (Guo et al., 1998; Lin et al., 1998). cry1, when fused with GREEN FLUORESCENT PROTEIN (GFP), was localized in the nucleus in a transient expression system or in dark-grown transgenic Arabidopsis seedlings, but it was primarily in the cytoplasm under continuous white light conditions (Guo et al., 1999; Yang et al., 2000). Although light induces a translocation of cry1 to the cytoplasm, cry1 signaling may involve both nuclear and cytosolic events, and early cry1 signaling may still operate in the nucleus. By contrast, cry2 was localized predominantly to the nucleus under dark or light conditions (Guo et al., 1999; Kleiner et al., 1999; Yang et al., 2000). Both cry1 and cry2 undergo a blue light–dependent phosphorylation, and the phosphorylation status may be closely associated with their regulatory functions (Shalitin et al., 2002, 2003). cry1, when expressed and purified from insect cells, is also phosphorylated in a blue light–dependent manner, consistent with an autophosphorylation function of cry1 (Bouly et al., 2003). cry2 protein is unstable, and this instability may be mediated by its interaction with COP1 (Wang et al., 2001).
Genetic screens have identified many far-red light signaling mutants, and nine of the genes affected in these mutants have been identified (for review, see Huq and Quail, 2005). Genetic screens have also identified mutants with defects in red light responses, such as gigantean (gi), pif4, elf3, and sensitivity to red light-reduced1 (srr1), or in both red and far-red light responses, such as phytochrome signaling early flowering1, pseudoresponse regulator7, and phytochrome signaling2 (Huq and Quail, 2005). Many light signaling components are nuclear proteins, whereas others, such as FAR-RED INSENSITIVE219 (FIN219) and PHYTOCHROME A SIGNAL TRANSDUCTION1 (PAT1), are cytosolic proteins. FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and SRR1 exist in both the nucleus and the cytoplasm (Staiger et al., 2003; Huq and Quail, 2005). A blue light–specific component, PHOSPHATASE7 (PP7), was isolated recently, and PP7 encodes a Ser/Thr protein phosphatase (Møller et al., 2003). SHORT UNDER BLUE1, a cytoplasmic calcium binding protein, has a major function in cryptochrome signaling but also modulates phyA-mediated far-red light responses (Guo et al., 2001). HFR1, a basic helix-loop-helix protein identified previously for its involvement in far-red light signaling, also plays a role in regulating several blue light–mediated responses (Duek and Fankhauser, 2003). In addition, HYPERSENSITIVE TO RED AND BLUE1 (HRB1), PIF4, and OBF BINDING PROTEIN3 have been shown recently to act in both red and blue light signaling (Kang et al., 2005; Ward et al., 2005).
Mutants in a different class, cop/deetiolated/fusca, exhibit a light-grown phenotype even when grown in darkness and identify a total of 11 loci. Many of the proteins encoded are involved in control of the stability of a few key light signaling components, such as LONG HYPOCOTYL IN THE LIGHT5 (HY5), LONG AFTER FAR-RED LIGHT1 (LAF1), and HFR1 (Osterlund et al., 2000; Seo et al., 2003; Jang et al., 2005; Yang et al., 2005). Early biochemical studies using microinjection and pharmacological techniques have also suggested the involvement of cyclic GMP, G-proteins, protein phosphatase, and calcium/calmodulin in phytochrome signaling (Neuhaus et al., 1997; Huq and Quail, 2005). Recent genetic studies have further suggested the involvement of G-protein in light signaling (Okamoto et al., 2001; Jones et al., 2003).
We have isolated a new long-hypocotyl mutant, shb1-D, under red, far-red, and blue light conditions. The long-hypocotyl phenotype of shb1-D is caused by the overexpression of its wild-type gene. By contrast, shb1, a knockout allele, exhibits hypocotyl and cotyledon phenotypes mostly under blue light. SHB1 is localized in the nucleus and the cytoplasm and regulates various light responses either positively or negatively. The transcript of SHB1 is extremely low, and the level of SHB1 protein may be limiting for red and far-red light signaling, or the lack of hypocotyl and cotyledon phenotypes in shb1 under red or far-red light may be attributable to a redundant function of SHB1 homologues. Consistent with this possibility, overexpression of SHB1 activated the expression of PIF4 and PIF4-mediated red light signaling, and also promoted phyA degradation and hypocotyl elongation under far-red light. Moreover, SHB1 signaling under blue light may involve HFR1, and the shb1 mutation indeed suppressed the expression of HFR1 under blue light. Double mutant analysis suggested that hfr1-201 is epistatic to shb1 in hypocotyl growth regulation and cotyledon opening responses under blue light. Interestingly, blue and red light signaling of SHB1 appears to involve two basic helix-loop-helix proteins, PIF4 and HFR1, important for both cryptochrome and phytochrome signaling.
RESULTS
shb1-D Has a Long-Hypocotyl Phenotype under Red, Far-Red, and Blue Light
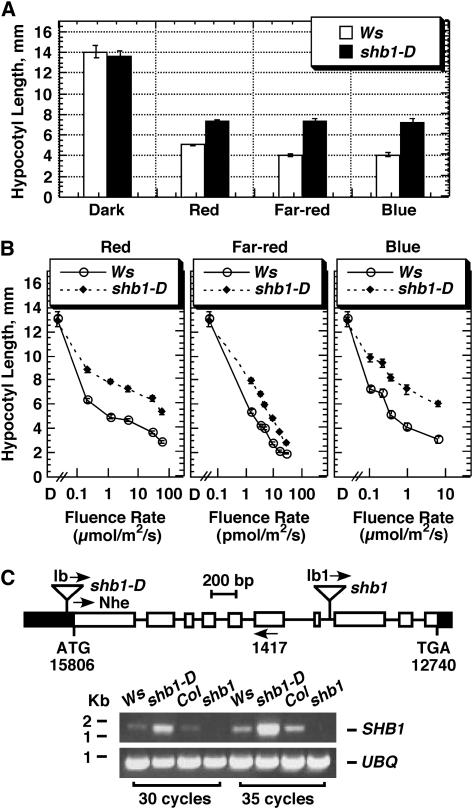
We screened a collection of 40,000 lines containing T-DNA insertions (ABRC) for mutants with either a short- or long-hypocotyl phenotype under red light. One mutant identified, shb1-D (for short hypocotyl under blue1 Dominant), showed a long-hypocotyl phenotype under red and far-red light (Figure 1A). Subsequently, shb1-D was also identified to have a long-hypocotyl phenotype under blue light. The long-hypocotyl phenotype of shb1-D appeared persistent over a broad range of red and blue light intensities (Figure 1B, left and right). By contrast, shb1-D exhibited a stronger hypocotyl phenotype under relatively weak or intermediate intensities of far-red light, ranging from 1 to 7 pmol·m−2·s−1 (Figure 1B, middle).
Figure 1.
shb1-D Shows a Long-Hypocotyl Phenotype under Red, Far-Red, and Blue Light.
(A) Hypocotyl growth responses of Ws and shb1-D to continuous red light (15 μmol·m−2·s−1), far-red light (10 pmol·m−2·s−1), or blue light (3 μmol·m−2·s−1) for 4 d. Data are presented as means ± se.
(B) Hypocotyl fluence responses of Ws and shb1-D to continuous red, far-red, and blue light for 4 d. Data are presented as means ± se.
(C) SHB1 is upregulated in shb1-D but is knocked out in shb1. Top, the SHB1 gene structure shows sequence coordination from BAC clone T30C3. Black boxes represent the 5′ and 3′ untranslated regions. White boxes indicate exons, and lines indicate introns. Bottom, RT-PCR analysis of SHB1 expression using 0.5 μg of total RNA prepared from Ws, shb1-D, Col, and shb1. RT-PCR analysis was run for 30 and 35 cycles using the primer pair Nhe and 1417. Control RT-PCR was run for UBQ10 under identical conditions.
Molecular Cloning of SHB1
By backcrossing shb1-D to Wassilewskija (Ws), we found that shb1-D has a T-DNA insertion at a single genetic locus and a dominant phenotype, and the T-DNA insertion is tightly linked to the mutation. We cloned the mutated gene by walking to the flanking plant genomic sequence using a GenomeWalker kit (see Methods). In shb1-D, the T-DNA left border is inserted at transformation-competent BAC clone T30C3 sequence 15935, 129 bp upstream of the start codon of a predicted gene, At4g25350, on chromosome 4 (Figure 1C). The putative gene is composed of 10 exons and 9 introns. An additional database search identified a second T-DNA insertional allele, shb1, from the SALK T-DNA insertion collection. The T-DNA left border is inserted at transformation-competent BAC clone T30C3 sequence 13588, 8 bp from the beginning of the eighth exon of the putative gene (Figure 1C). Among six plants propagated from the original SALK seeds, four plants were found to be wild type and two plants were found to be heterozygous based on both PCR genotyping and kanamycin resistance. The two heterozygous lines showed a 3:1 segregation ratio of their kanamycin resistance, suggesting a single T-DNA insertion in shb1. Homozygous lines were subsequently identified from the heterozygous parents. Both T-DNA insertions in shb1-D and shb1 were verified using PCR techniques (see Supplemental Figure 1 online).
SHB1 Is Overexpressed in shb1-D but Is Knocked Out in shb1
SHB1 message was extremely low in total RNA preparations from the Ws wild type, and we failed to detect it through RNA gel blot hybridization analysis. Instead, RT-PCR analysis with the primer pair Nhe and 1417 revealed a detectable level of SHB1 mRNA in Ws but a very high level of SHB1 mRNA in shb1-D (Figure 1C, bottom). To exclude a possible difference in the quality of RNA preparations, we performed a control RT-PCR analysis for UBIQUITIN10 (UBQ10) and detected fairly constant levels of UBQ10 messages in Ws and shb1-D (Figure 1C, bottom). However, the reason for the increased level of SHB1 mRNA in shb1-D remains unknown. It is possible that an enhancer element carried on the T-DNA insertion may activate the transcription of SHB1 downstream. Alternatively, the T-DNA insertion may inactivate a negative regulatory element in the SHB1 gene promoter region. To learn the consequence of the T-DNA insertion on SHB1 expression in shb1, we performed an additional RT-PCR analysis using a 5′ primer, Nhe, and a 3′ primer, 1417, positioned before the T-DNA insertion. RT-PCR analysis with this pair of primers failed to detect SHB1 transcript in shb1, and the T-DNA insertion apparently resulted in a loss of SHB1 expression in shb1 (Figure 1C, bottom).
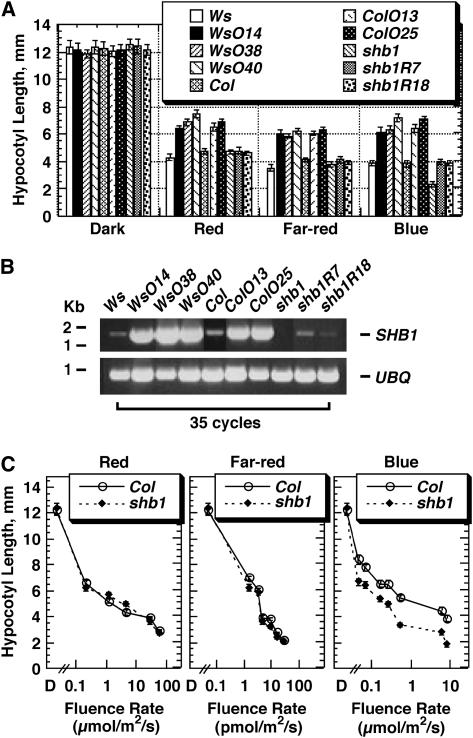
Overexpression of SHB1 Recapitulates the Phenotype Conferred by shb1-D, and shb1 Has a Short-Hypocotyl Phenotype under Blue Light
Given the gain-of-function phenotype of shb1-D, we performed additional experiments to verify the cause of the long-hypocotyl phenotype in shb1-D (Figure 2A). We generated transgenic plants that carry a single T-DNA insertion with a SHB1 coding sequence under the control of the cauliflower mosaic virus (CaMV) 35S promoter in either the Ws or Columbia (Col) background. Several representative lines, homozygous for the transgenes in either the Ws or Col background, all exhibited a long-hypocotyl phenotype under red, far-red, and blue light (Figure 2A). RT-PCR analysis demonstrated that SHB1 was indeed overexpressed in these transgenic lines (Figure 2B).
Figure 2.
Overexpression of SHB1 Recapitulates the Long-Hypocotyl Phenotype of shb1-D, and shb1 Shows a Short-Hypocotyl Phenotype under Blue Light.
(A) Hypocotyl growth responses of Ws, shb1-D, SHB1 overexpression lines in the Ws background (WsOE14, WsOE38, and WsOE40), Col, SHB1 overexpression lines in the Col background (ColOE13 and ColOE25), shb1, and shb1 rescue lines (shb1-2R7 and shb1R18) to continuous red, far-red, or blue light for 4 d under intensities as specified for Figure 1A. Data are presented as means ± se.
(B) RT-PCR analysis of SHB1 expression using 0.5 μg of total RNA isolated from white light-grown Ws, shb1-D, Col, shb1, and various SHB1 overexpression or shb1 rescue lines (top). Controls were performed for UBQ10 under identical conditions (bottom).
(C) Hypocotyl fluence responses of Col and shb1 to continuous red, far-red, or blue light for 4 d. Data are presented as means ± se.
The original wild-type and heterozygous shb1 individuals derived from the SALK seeds, as well as the wild-type, heterozygous, and homozygous decedents of the original heterozygous individuals, were all further tested for their hypocotyl growth responses under various light conditions. Homozygous shb1 individuals showed a short-hypocotyl phenotype only under blue light, and the short-hypocotyl phenotype is particularly pronounced under moderate to strong blue light (Figure 2C). By contrast, heterozygous individuals showed a hypocotyl growth response much similar to wild-type individuals, indicating the recessive nature of the shb1 mutation and the tight linkage of the T-DNA insertion to the phenotype observed. We then introduced a 4.7-kb genomic fragment, containing a 1.2-kb SHB1 promoter sequence, the SHB1 coding sequence, and 0.5 kb of 3′ sequence, into shb1. The introduced genomic fragment resulted in a wild-type level of expression of SHB1 and fully complemented the hypocotyl phenotype of shb1 to the wild type (Figures 2A and 2B).
Mutations in SHB1 Alter Other Light Responses
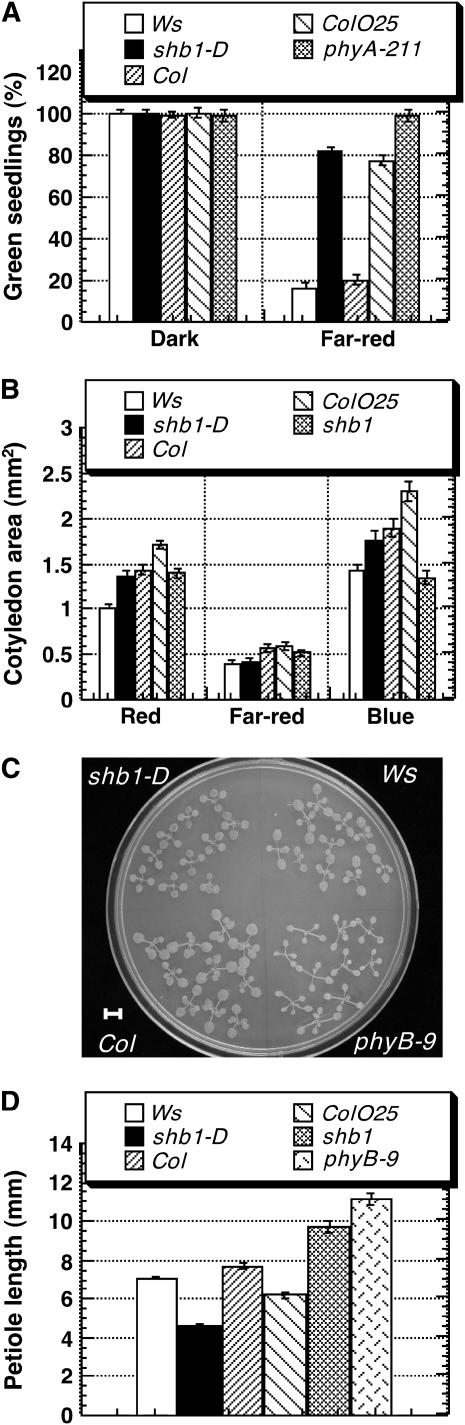
When wild-type seedlings are grown under far-red light for several days, they have low survival success after being transferred to white light, a phenomenon known as far-red light preconditioned block of greening (Neff et al., 2000; Huq and Quail, 2005). Under such conditions, the survival rate for Col wild-type seedlings was 23%, whereas phyA-211 had a survival rate of 99% (Figure 3A). Similar to phyA-211, shb1-D had a significantly higher survival rate (81%) than its Ws wild type (15%). A SHB1 overexpression line in the Col background, ColOE25, also had a significantly higher survival rate (78%) than its Col wild type (23%).
Figure 3.
Mutations in SHB1 Alter Other Light Responses.
(A) Percentage of green seedlings of Ws, shb1-D, Col, SHB1 overexpression line 25 in the Col background (ColO25), and phyA-211, grown either in darkness or under far-red light (1 μmol·m−2·s−1) for 4 d, and then transferred to white light for an additional 6 d. Data are presented as means ± se.
(B) Cotyledon size, measured 6 d after germination, for Ws, shb1-D, Col, line ColO25, and shb1 seedlings grown under continuous red, far-red, or blue light at intensities as indicated for Figure 1A. Data are presented as means ± se.
(C) Ws, shb1-D, Col, and phyB-9 plants, grown under continuous white light (30 μmol·m−2·s−1) for 21 d after germination, and transferred to an agar plate for photography. Bar = 4 mm.
(D) Petiole length, measured 21 d after germination, for Ws, shb1-D, Col, line ColO25, shb1, and phyB-9 plants grown under continuous white light (30 μmol·m−2·s−1). Data are presented as means ± se.
Six days after germination, the rate of light-induced cotyledon expansion in shb1-D was rather enhanced. Both shb1 and line ColOE25 had larger cotyledons than the Ws or Col wild type under red or blue light at this early developmental stage (Figure 3B). The knockout allele, shb1, had much smaller cotyledons under blue light than its Col wild type at the same developmental stage (Figure 3B). Twenty-one days after germination, shb1 and line ColOE25 developed smaller leaves than the Ws or Col wild type under white light, resembling that of phyB-9 (Figure 3C; data not shown). As a result, shb1-D looked quite different from either the wild type or phyB-9 (Figure 3C). At the same developmental stage, shb1-D and line ColOE25 developed short petioles (Figure 3D). By contrast, shb1 plants had longer petioles than did Col wild-type plants (Figure 3D).
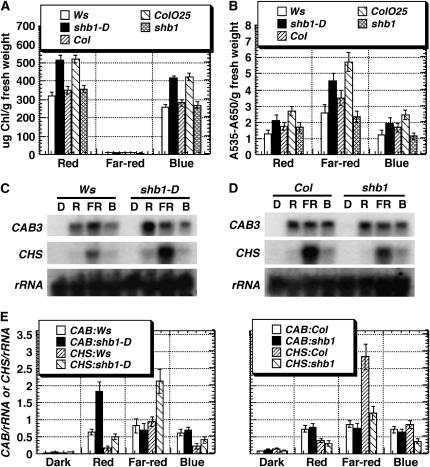
Mutations in SHB1 Affect Pigment Accumulation and CHLOROPHYLL a/b BINDING PROTEIN3 or CHALCONE SYNTHASE Expression
Under red and blue light, 5-d-old seedlings of shb1-D and line ColOE25 accumulated much more chlorophylls than did Ws or Col wild-type plants (Figure 4A). shb1-D and line ColOE25 also accumulated more anthocyanin pigments under red, far-red, and blue light than did Ws or Col wild-type plants (Figure 4B). By contrast, shb1 accumulated a normal amount of chlorophylls under all light conditions tested but less anthocyanin pigments under far-red and blue light (Figures 4A and 4B). Chlorophyll biosynthesis is coordinately regulated with the expression of chlorophyll a/b binding protein genes (McCormack and Terry, 2002), and we further examined the expression of CHLOROPHYLL a/b BINDING PROTEIN3 (CAB3), one member of this gene family, under red, far-red, and blue light.
Figure 4.
Mutations in SHB1 Alter Pigment Accumulation and CAB3 or CHS Expression.
(A) and (B) Chlorophyll (A) and anthocyanin (B) content of 5-d-old Ws, shb1-D, Col, line ColO25, and shb1 seedlings grown under red, far-red, or blue light at intensities as indicated for Figure 1A. Data are presented as means of three independent biological replicates ± se.
(C) and (D) RNA gel blot hybridization analysis of CAB3 and CHS expression in Ws and shb1-D (C) or in Col and shb1 (D). Total RNA was isolated from 5-d-old dark-grown seedlings that received no light treatment (D) or were treated for 4 h with red (R), far-red (FR), or blue (B) light under intensities as specified for Figure 1A.
(E) Normalization of CAB3 and CHS mRNA levels to 18 rRNA signals. The hybridization signals were detected with a PhosphorImager (Molecular Dynamics) screen and quantified using the ImageQuant program. Data are presented as means ± se from three independent experiments.
The level of CAB3 expression in Ws or Col plants is very low in darkness, but it is strongly induced by red, far-red, and blue light. The light-induced expression of CAB3 was enhanced threefold over that of Ws by shb1 under red light, but it remained the same as in the Ws wild type under far-red and blue light (Figure 4C). By contrast, shb1 did not affect the light-induced expression of CAB3 under all light conditions tested (Figure 4D). We also examined the light-induced expression of CHALCONE SYNTHASE (CHS), a gene that encodes an enzyme involved in flavonoid and anthocyanin biosynthesis. The level of CHS expression is very low in darkness, but it was induced by red, far-red, and blue light in either Ws or Col wild-type plants. shb1-D caused a further accumulation of CHS transcripts, 3.0-, 2.4-, and 2.0-fold, compared with that in Ws under red, far-red, and blue light, respectively (Figure 4C). By contrast, shb1 attenuated the light-induced accumulation of CHS transcripts by 2.5-fold under far-red and blue light (Figure 4D). Because CHS encodes the first committed enzyme in anthocyanin biosynthesis, the amount of anthocyanin accumulation may be influenced by the overexpression or underexpression of CHS in shb1-D or shb1.
cry Mutants Are Epistatic to shb1
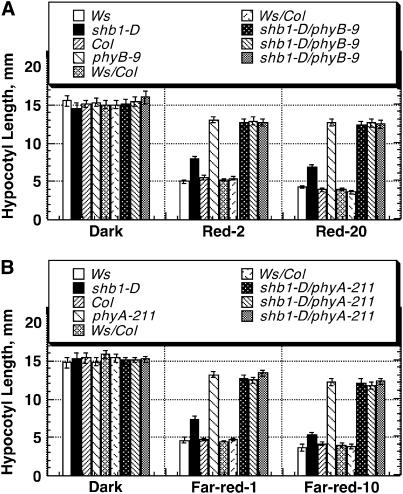
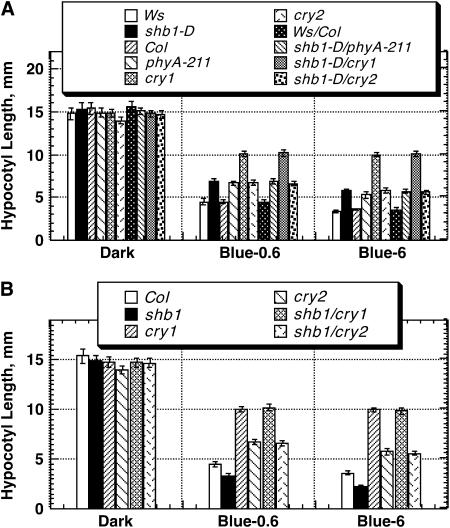
To learn the roles of SHB1 in light signaling in relation to photoreceptors, we generated double mutants of shb1-D with phyA, phyB, cry1, or cry2, and of shb1 with cry1 or cry2. Either shb1-D phyB-9 or shb1-D phyA-211 double mutant showed the severe hypocotyl phenotypes of the phyB-9 or phyA-211 single mutant, respectively, under red and far-red light (Figure 5). Under blue light, shb1-D showed a moderate hypocotyl phenotype very similar to that of phyA-211 or cry2, and shb1-D phyA-211 or shb1-D cry2 exhibited a hypocotyl phenotype indistinguishable from either of their mutant parents (Figure 6A; see Supplemental Figure 2 online). cry1 had the most severe hypocotyl phenotype under blue light, and shb1-D cry1 showed the severe hypocotyl phenotype of cry1 (Figure 6A). Thus, shb1-D does not suppress or enhance the phy and cry hypocotyl phenotypes.
Figure 5.
Double Mutant Analysis of shb1-D with phyB-9 or phyA-211 under Red or Far-Red Light.
(A) Hypocotyl growth responses of Ws, shb1-D, Col, phyB-9, Ws/Col lines, and multiple shb1-D phyB-9 lines to 2 or 20 μmol·m−2·s−1 red light (Red-2 or Red-20) for 4 d. Data are presented as means ± se.
(B) Hypocotyl growth responses of Ws, shb1-D, Col, phyA-211, Ws/Col lines, and multiple shb1-D phyA-211 lines to 1 or 10 pmol·m−2·s−1 far-red light (Far-red-1 or Far-red-10) for 4 d. Data are presented as means ± se.
Figure 6.
Double Mutant Analysis of shb1-D with phyA-211 and cry Mutants and of shb1 with cry Mutants under Blue Light.
(A) Hypocotyl growth responses of Ws, shb1-D, Col, phyA-211, cry1, cry2, Ws/Col, shb1-D phyA-211, shb1-D cry1, and shb1-D cry2 to 0.6 or 6 μmol·m−2·s−1 blue light (Blue-0.6 or Blue-6) for 4 d. Data are presented as means ± se.
(B) Hypocotyl growth responses of Col, shb1, cry1, cry2, shb1 cry1, and shb1 cry2 to 0.6 or 6 μmol·m−2·s−1 blue light (Blue-0.6 or Blue-6) for 4 d. Data are presented as means ± se.
We also generated double mutants of shb1 with cry1 or cry2. Under blue light, shb1 has a shorter hypocotyl phenotype, whereas cry1 or cry2 has a longer hypocotyl phenotype. Either shb1 cry1 or shb1 cry2 double mutant showed a hypocotyl phenotype similar to that of cry1 or cry2 (Figure 6B). This epistasis may indicate that the expression of the phenotype conferred by shb1 requires either functional cry1 or cry2. We noticed that the hypocotyl length of either the wild type or cry2 under 0.6 μmol·m−2·s−1 blue light was shorter than that reported under equivalent light intensity (Lin et al., 1998). The difference between our results and those previous results may be attributed to the different light sources used.
SHB1 Contains an SPX Domain and an EXS Domain Found in the SYG1 Protein Family
SHB1 contains an N-terminal SPX domain and a C-terminal EXS domain found in yeast SYG1 protein (gi 731805) and mouse XENOTROPIC AND POLYTROPIC MURINE LEUKEMIA VIRUSES RECEPTOR1 (XPR1) protein (gi 6093320) (Figure 7A; see Supplemental Figure 3 online). The SPX domain (pfam03105 family) is named after SYG1, PHOSPHATE TRANSPORTER81 or PHO81, and XPR1, whereas the EXS domain (pfam03124 family) is named after ER RETENTION-DEFECTIVE1 or ERD1, XPR1, and SYG1. The SPX domain can also be found in several predicted proteins of uncertain function from Arabidopsis (gi 3548805 and gi 3548806), Drosophila melanogaster (gi 22832470, gi 22946878, and gi 7290442), Saccharomyces cerevisiae (gi 731951), and Schizosaccharomyces pombe (gi 5832411). SHB1 is relatively closer to one SYG1 or XPR1 homologue from yeast (gi 731951) and two SYG1 or XPR1 homologues from Arabidopsis (gi 3548805 and gi 3548806) than to other SYG1 or XPR1 homologues from yeast, fission yeast, Neurospora, worm, fly, mouse, and human (see Supplemental Figures 3 and 4 online). The consensus sequences derived from all SYG1 homologues, and the alignment of SHB1 with SYG1 protein from yeast, are presented for both SPX and EXS domains (see Supplemental Figure 5 online).
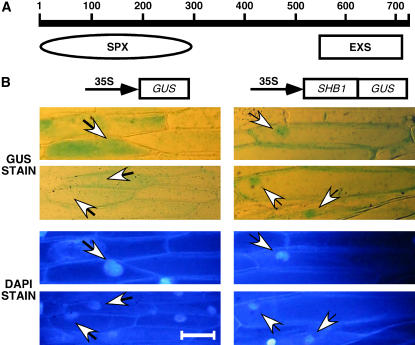
Figure 7.
SHB1 Is Localized to the Nucleus and the Cytosol.
(A) Diagram shows the N-terminal SPX domain and the C-terminal EXS domain in the SHB1 protein.
(B) Subcellular localization of GUS (left) or the SHB1:GUS fusion protein (right) in onion epidermal cells (arrows, top panels), and 4′,6-diamidino-2-phenylindole (DAPI) stain for the location of the nuclei (arrows, bottom panels). The images were taken for onion epidermal layers incubated in darkness. Bar = 100 μm.
The EXS region of similarity contains several predicted transmembrane helices. For example, yeast SYG1 is predicted to contain eight membrane-spanning motifs, and three motifs are predicted in the EXS domain. Another five motifs are located between the SPX domain and the EXS domain. As demonstrated by cell fractionation techniques, yeast SYG1 is localized to a fraction enriched in plasma membrane (Spain et al., 1995). Examination of the SHB1 sequence with the PSORT program also identified six weakly predicted integral membrane motifs (certainty, 0.6) located in amino acid sequences 347 to 363, 429 to 445, 465 to 481, 492 to 508, 559 to 575, and 665 to 681. The fifth and sixth motifs fall within the predicted EXS domain.
SHB1 Is Localized in the Nucleus and the Cytoplasm
Recently, genomic scale profiling has identified SHB1 (gi 7487574 or At4g25350) as PHO1 homologue 4, or H4, a member of the PHO1 family of putative phosphate transporters (Wang et al., 2004). The PHO1 family comprises 11 members, and PHO1 is involved in the loading of inorganic phosphate into the xylem of roots (Hamburger et al., 2002). Phylogenetic analysis also showed that SHB1 (gi 7487574) is more closely related to two Arabidopsis PHO1 homologues (gi 3548805 or At2g03250 and gi 3548806 or At2g03249) and a yeast protein, PHO84 (gi 731951) (see Supplemental Figure 3A online). PHO84 is described as a member of the PHO family involved in low-affinity inorganic phosphate transport and potentially phosphate sensing (Wykoff and O'Shea, 2001). Thus, the homology of SHB1 with the Arabidopsis and yeast proteins may suggest a potential role of SHB1 in phosphate homeostasis.
To investigate the subcellular localization of SHB1, we constructed a SHB1:β-d-glucuronidase (GUS) fusion in pBI221 and conducted a transient transfection assay using onion (Allium cepa) epidermal peels. In darkness, SHB1:GUS was clearly localized in the nucleus and the cytosol in these onion epidermal cells (Figure 7B). Treatment with white, red, far-red, or blue light for 4 and 8 h did not alter the pattern of SHB1:GUS subcellular localization (data not shown). The staining of SHB1:GUS was particular enriched in the nucleus, and the staining in the cytosol therefore may not be as strong as that of GUS alone (Figure 7B). Therefore, the nuclear and cytosolic localization of SHB1 may suggest a role of SHB1 in light signaling rather than in organic phosphate transport.
SHB1 Is Required for the Proper Expression of PIF4 and HFR1
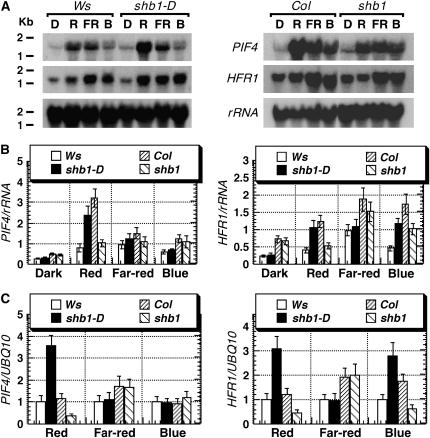
To further explore the role of SHB1 in cryptochrome or phytochrome signaling, we surveyed the expression of many light signaling genes identified to date in shb1-D or shb1 under red, far-red, and blue light using RT-PCR techniques. The genes include PP7, HRB1, PIF3, PIF4, ELF3, ELF4, GI, SRR1, FHY1, FHY3, HFR1, FAR-RED IMPAIRED RESPONSE1, LAF1, PAT1, SUPRESSOR OF PHYTOCHROME-105 1, EMPFINDLICHER IM DUNKELROTEN LICHT1, FIN219, and HY5 (Huq and Quail, 2005). We detected changes in the expression of PIF4 and HFR1 in both shb1-D and shb1 and subsequently verified the changes using RNA gel blot hybridization analysis (Figure 8A). The expression of PIF4 in either the Ws or Col wild type was strongly induced by red, far-red, and blue light (Huq and Quail, 2002) (Figure 8A). Compared with the Ws or Col wild type, overexpression of SHB1 in shb1-D resulted in a further 3.2-fold increase in PIF4 expression under red light, whereas loss of SHB1 expression in shb1 led to a 3.2-fold decrease in PIF4 expression under red light (Figure 8A). By contrast, the effects of shb1-D or shb1 on PIF4 expression were not obvious under either far-red or blue light. Comparable changes in the expression of PIF4 caused by either shb1-D or shb1 under red light were also observed using real-time PCR analysis (Figure 8B, left). Consistent with RNA gel blot hybridization analysis, real-time PCR did not reveal any changes in the expression of PIF4 in either shb1-D or shb1 under far-red or blue light.
Figure 8.
Mutations in SHB1 Affect the Expression of PIF4 and HFR1.
(A) RNA gel blot hybridization analysis of PIF4 and HFR1 in shb1-D and shb1. Total RNA was isolated from 5-d-old dark-grown Ws, shb1-D, Col, and shb1 seedlings that received no light treatment (D) or were treated for 4 h with red (R), far-red (FR), or blue (B) light under the intensities specified for Figure 1A. The gel images for PIF4 and HFR1 expression in Col and shb1 were overexposed to clearly show the decreases in the expression of the two genes.
(B) Normalization of PIF4 and HFR1 mRNA levels to 18 rRNA signals. The hybridization signals were detected with a PhosphorImager and quantified using the ImageQuant program. Data are presented as means ± se from two independent experiments.
(C) Real-time PCR analysis of PIF4 (left) or HFR1 (right) expression in Ws, shb1-D, Col, and shb1 under red, far-red, or blue light at the intensities specified for Figure 1A. PCR was performed on RT reaction using SYBR green PCR master mix and the 7500 Real Time PCR system. The levels of PIF4 or HFR1 expression were normalized to UBQ10. Data are presented as means ± se from two independent experiments.
The expression of another basic helix-loop-helix gene, HFR1, in either the Ws or Col wild type was induced strongly by far-red light and moderately by blue light (Figure 8A) (Duek and Fankhauser, 2003). Under red light, the expression of HFR1 was weakly induced by a 4-h red light treatment (Figure 8A). However, Duek and Fankhauser (2003) only observed a slight increase in HFR1 message up to 2 or 4 h and a sharp reduction in HFR1 message after 4 h of red light treatment. Overexpression of SHB1 in shb1-D caused further 2.6- and 2.5-fold increases in the expression of HFR1 under red and blue light, respectively, compared with the Ws wild type (Figure 8B). By contrast, lack of SHB1 expression in shb1 resulted in 2.4- and 1.7-fold decreases in the expression of HFR1 under red and blue light, respectively (Figure 8B). No changes in HFR1 expression were observed under far-red light in either shb1-D or shb1. Because some of the changes observed were less than twofold, we performed real-time PCR analysis to further verify the RNA gel blot analysis data. The increases caused by shb1-D on HFR1 expression under red and blue light were 3.1- and 2.7-fold, respectively, greater than that in the Ws wild type, whereas shb1 caused 2.6- and 2.7-fold decreases in HFR1 expression under red and blue light, respectively, compared with the Ws wild type (Figure 8C). Real-time PCR analysis revealed slightly larger decreases in the expression of HFR1 by shb1 than those observed through RNA gel blot hybridization analysis (Figures 8B and 8C).
PIF4 Specifically Mediates SHB1 Regulation of Hypocotyl Elongation and CAB3 or CHS Expression under Red Light
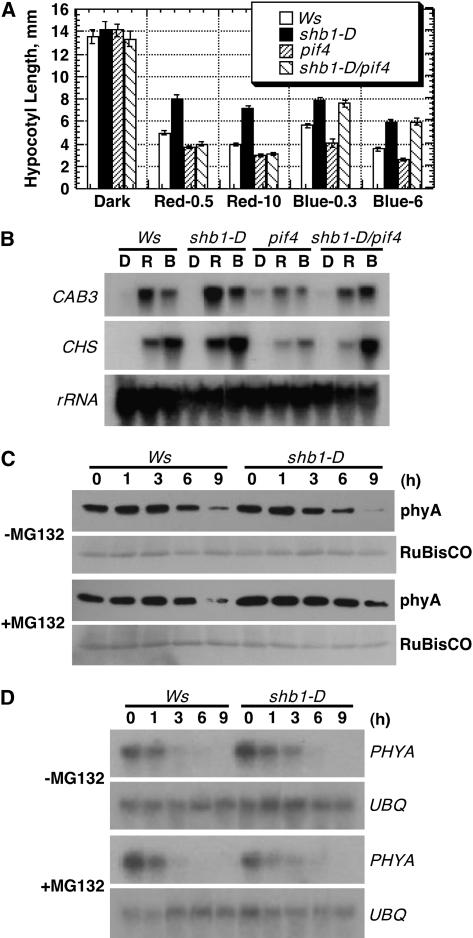
Because the expression of PIF4 was affected in shb1-D or shb1 under red light, we further examined the genetic interactions that control hypocotyl elongation and light-regulated gene expression responses. shb1-D has a longer hypocotyl and pif4 has a shorter hypocotyl under both red and blue light compared with the Ws wild type, whereas the shb1-D pif4 double mutant showed a pif4-like hypocotyl phenotype under red light but an shb1-D–like hypocotyl phenotype under blue light (Figure 9A). Apparently, the expression of the shb1-D hypocotyl phenotype under blue light was independent of the pif4 mutation. Similar genetic interactions were also observed for their light-induced gene expression responses. Under red light, shb1-D enhanced but pif4 reduced the expression of CAB3 and CHS, and shb1-D pif4 behaved like pif4 (Figure 9B). Under blue light, shb1-D enhanced the expression of only CHS and pif4 reduced the expression of both CAB3 and CHS. The shb1-D pif4 double mutant behaved like shb1-D under blue light, and the effects of shb1-D on the expression of these two genes were independent of pif4 (Figure 9B). Therefore, PIF4 may specifically mediate SHB1 regulation on hypocotyl elongation and light-induced gene expression under red light, consistent with the specific effect of shb1-D on the expression of PIF4 under red light but not blue light (Figures 8A and 9A).
Figure 9.
SHB1 Acts Upstream of PIF4, and the shb1-D Mutation Promotes phyA Degradation.
(A) Hypocotyl growth responses of Ws, shb1-D, pif4, and shb1-D pif4 seedlings to 0.5 or 10 μmol·m−2·s−1 red light (Red-0.5 or Red-10) or 0.3 or 6 μmol·m−2·s−1 blue light (Blue-0.3 or Blue-6) for 4 d. Data are presented as means ± se.
(B) RNA gel blot analysis of CAB3 and CHS expression on total RNA isolated from 5-d-old dark-grown Ws, shb1-D, pif4, and shb1-D pif4 seedlings that received no light treatment (D) or were treated for 3 h with red (R) or blue (B) light under the intensities specified for Figure 1A.
(C) PhyA degradation in 5-d-old Ws and shb1-D seedlings that were treated with or without MG132 under far-red light (0.2 μmol·m−2·s−1) for the times indicated. RuBisCO, ribulose-1,5-bis-phosphate carboxylase/oxygenase.
(D) RNA gel blot hybridization analysis of PHYA and UBQ10 mRNA levels in 5-d-old Ws and shb1-D seedlings that were treated with or without MG132 under far-red light (0.2 μmol·m−2·s−1) for the times indicated.
shb1-D Promotes phyA Degradation under Far-Red Light
To explore how the overexpression of SHB1 expands its signaling activity to far-red light, we examined the phyA protein level in dark-grown Ws and shb1-D seedlings after exposure to far-red light for several hours. The experiments also included MG132, a proteasome inhibitor, because phyA can be ubiquitinated by COP1 and undergoes proteasome-mediated degradation (Seo et al., 2004). A normal degradation pattern of phyA was observed in Ws after far-red light treatment, and a noticeable reduction in phyA protein level was observed 6 h after far-red light illumination (Figure 9C). phyA degradation was promoted in shb1-D, and an apparent reduction in phyA protein level was observed 3 h earlier after far-red light treatment. Addition of MG132 partially attenuated the rate of phyA disappearance in the Ws wild type and also restored the rate of phyA degradation in shb1-D to a level comparable to that in the wild type under similar conditions (Figure 9C) (Seo et al., 2004). By contrast, shb1-D did not alter the decrease in PHYA transcription under far-red light, and an even slightly higher level of PHYA mRNA was noticed after far-red light treatment in shb1-D compared with that in the Ws wild type (Figure 9D). MG132 did not affect the level of PHYA mRNA under far-red light in either the wild type or shb1-D.
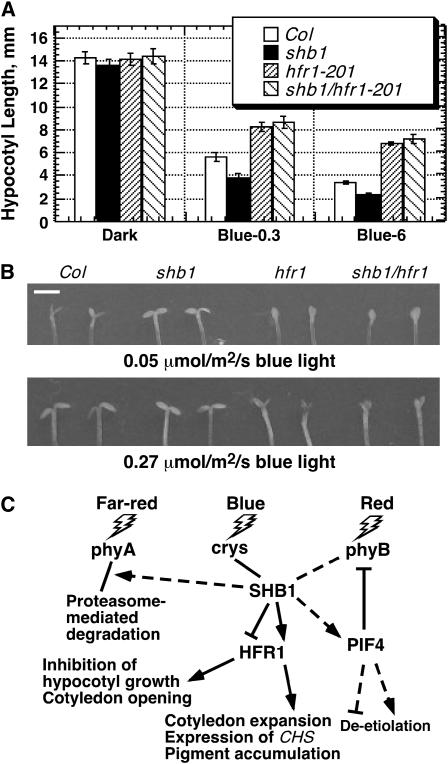
SHB1 Blue Light Signaling Involves HFR1
shb1 affected the expression of HFR1 under both red and blue light (Figure 8A). Indeed, shb1 and hfr1-201 shared several similar phenotypes under blue light, such as cotyledon expansion and pigment accumulation (Figures 3 and 4) (Duek and Fankhauser, 2003). shb1 and hfr1-201 also had phenotypes opposite to each other, such as hypocotyl elongation and cotyledon opening, under blue light (Figures 2 and 10B) (Duek and Fankhauser, 2003). The shb1 hfr1-201 double mutant showed a long-hypocotyl phenotype under blue light, similar to that of the hfr1-201 single mutant. The smaller effects of either shb1 or hfr1-201 on the expression of CAB3 or CHS did not reveal a clear epistasis (see Supplemental Figure 6 online). Therefore, we examined the shb1 hfr1-201 double mutant for the cotyledon-opening response (Figure 10B). Under relatively weak blue light, shb1 seedlings fully opened their cotyledons, whereas Col wide-type seedlings just partially opened their cotyledons (Figure 10B, top). Under similar conditions, the cotyledons of hfr1-201 were fully closed and shb1 hfr1-201 showed a phenotype similar to that of hfr1-201. Under higher blue light intensity, both Col and shb1 seedlings fully opened their cotyledons, whereas hfr1-201 seedlings only partially opened theirs (Figure 10B, bottom). The shb1 hfr1-201 double mutant behaved like hfr1-201, suggesting an epistasis of hfr1-201 to shb1 in their control of the cotyledon-opening responses.
Figure 10.
SHB1 Acts Upstream of HFR1, and the Roles of SHB1 in Light Signaling.
(A) Hypocotyl growth responses of Col, shb1, hfr1-201, and shb1 hfr1-201 seedlings to 0.3 or 6 μmol·m−2·s−1 blue light (Blue-0.3 or Blue-6) for 4 d. Data are presented as means ± se.
(B) Cotyledon-opening responses of 4-d-old Col, shb1, hfr1-201, and shb1 hfr1-201 seedlings to weak blue light (0.05 μmol·m−2·s−1) or moderate blue light (0.27 μmol·m−2·s−1). Bar = 1.5 mm.
(C) SHB1 acts in cry- and possibly phy-mediated light signaling pathways and regulates various light responses either positively (arrows) or negatively (T-bars). SHB1 signaling may involve HFR1 in the control of hypocotyl elongation, cotyledon opening and expansion, expression of CHS, and pigment accumulation (solid lines). Overexpression of SHB1 activates PIF4-mediated red light signaling and also promotes the proteasome-mediated degradation of phyA and hypocotyl elongation under far-red light (dotted lines).
DISCUSSION
We have identified two SHB1 mutant alleles, an overexpression allele, shb1-D, and a knockout allele, shb1. The phenotypes of shb1-D and shb1 are opposing each other in most cases and together suggest the different roles of SHB1 in controlling various light responses. For example, lack of SHB1 expression in shb1 resulted in a short hypocotyl, smaller cotyledons, accelerated cotyledon opening, reduced expression of CHS, and reduced accumulation of anthocyanins under blue light (Figures 2 to 4). By contrast, overexpression of SHB1 in shb1-D led to a longer hypocotyl under red, far-red, and blue light, an enhanced far-red light preconditioned block of greening response, larger cotyledons under red and blue light, enhanced expression of CAB3 and CHS, and enhanced accumulation of pigments (Figures 1, 3, and 4). Loss of SHB1 expression in shb1 also created a long-petiole phenotype under white light, whereas overexpression of SHB1 in shb1-D led to shorter petioles and smaller leaves under white light (Figure 3). Therefore, we propose a model in which SHB1 negatively regulates the inhibition of hypocotyl elongation, cotyledon opening, and leaf expansion but positively regulates cotyledon expansion, the inhibition of petiole elongation, the expression of CHS, and pigment accumulation (Figure 10C). Considering that all of the photoreceptors seem to interact in numerous responses, we still could not rule out a role of SHB1 in phototropin-mediated blue light signaling.
shb1 showed photomorphogenic phenotypes primarily under blue light, although reduced accumulation of anthocyanins and expression of CHS were also observed in shb1 under far-red light (Figure 4). By contrast, overexpression of SHB1 in shb1-D created various photomorphogenic phenotypes under red, far-red, and blue light (Figures 1, 3, and 4). A similar case has been observed for phyA. Mutation in PHYA led to a long hypocotyl only under far-red light, but overexpression of PHYA in Arabidopsis and other dicots created a short-hypocotyl phenotype under red, far-red, and white light (Boylan and Quail, 1991; Nagatani et al., 1991). Interestingly, overproduction of HFR1:GFP also caused a hypersensitive hypocotyl growth response to red light (Yang et al., 2005), although hfr1 exhibited a hyposensitive hypocotyl growth response only to far-red and blue light (Soh et al., 2000; Duek and Fankhauser, 2003; Huq and Quail, 2005). However, these cases may not sufficiently explain the phenotypes of shb1-D under all light wavelengths tested.
Considering the extremely low abundance of SHB1 transcript, the level of SHB1 protein may be limiting for red and far-red light signaling. Alternatively, the lack of hypocotyl or cotyledon phenotype in shb1 under red or far-red light may be attributable to a redundant function of SHB1 homologues. Overexpression of SHB1 enhances the expression of PIF4 under red light, and pif4 is epistatic to shb1-D in the red light–mediated hypocotyl elongation response (Figures 8, 9, and 10C). Overexpression of SHB1 also promotes phyA protein degradation and hypocotyl elongation under far-red light (Figures 1, 9, and 10C). Under blue light, shb1 suppressed the expression of HFR1, and shb1 indeed showed several deetiolation phenotypes similar to those of hfr1-201, such as smaller cotyledons, reduced expression of CHS, and reduced accumulation of anthocyanins (Figures 3 and 4). Therefore, SHB1 and HFR1 have overlapping functions in regulating the blue light–mediated responses. However, the hypocotyl elongation and cotyledon-opening phenotypes of shb1 were opposite to those of hfr1-201 under blue light. Although the altered expression of HFR1 in shb1 does not explain its hypocotyl phenotype, genetic analysis indicates that HFR1 acts downstream of SHB1 (Figure 10).
On the other hand, shb1-D enhanced the expression of HFR1 under blue light and resulted in several deetiolation phenotypes opposite to that of hfr1-201, such as cotyledon size, CHS expression, and pigment accumulation (Figures 3 and 4). As shown recently, overexpression of full-length HFR1 or HFR1:GFP led to hypersensitive phenotypes opposite to that of hfr1-201 under red, far-red, or blue light (Jang et al., 2005; Yang et al., 2005). The responses include hypocotyl and cotyledon growth and CAB3 expression. Thus, several deetiolation phenotypes, but not the hypocotyl phenotype, of shb1-D can be explained well by the increased expression of HFR1 in shb1-D. By contrast, SHB1 may either regulate hypocotyl elongation and cotyledon-opening responses through an HFR1-independent branch or act negatively upstream of HFR1.
Genetic analysis revealed a phy- or cry-like phenotype in shb1-D phy or shb1-D cry double mutants (Figures 5 and 6). However, epistatic interactions may not be revealed clearly because shb1-D is a dominant and gain-of-function allele. phyA and phyB have the most severe hypocotyl phenotypes under far-red or red light and phyA is almost blind to far-red light, whereas shb1-D has a moderate hypocotyl phenotype under these light conditions. Introduction of shb1-D into the phyA or phyB background did not enhance or suppress the severe hypocotyl phenotypes of phyA or phyB. Again, our data may not reveal much genetic interaction because in most cases the phenotype of the stronger mutation is always observed in these double mutants. In addition, the two single mutations cause a similar phenotype, and one may still observe the phenotype of the stronger mutation even if there is no genetic interaction. Under blue light, cry1 has the most severe hypocotyl phenotype, and the situation observed for shb1-D cry1 is similar to that for shb1-D phyA or shb1-D phyB. The hypocotyl phenotype of shb1-D is moderate and similar to that of phyA or cry2 under blue light, and the response of shb1-D phyA-211 or shb1-D cry2 to blue light was indistinguishable from that of either of their parents. By contrast, shb1 has an opposite hypocotyl phenotype to either cry1 or cry2, and the shb1 cry1 or shb1 cry2 double mutant showed the cry1 or cry2 hypocotyl phenotype. Thus, expression of the shb1 hypocotyl phenotype requires blue light, and such an epistasis reflects a dependence of the phenotype conferred by shb1 on functional cryptochromes.
SHB1 contains an N-terminal SPX domain and a C-terminal EXS domain found in SYG1-like and XPR1-like proteins (Figure 7A; see Supplemental Figure 3 online). The human and mouse XPR1 encodes a cell surface receptor for xenotropic and polytropic murine leukemia viruses, conferring susceptibility to infection with murine leukemia viruses (Tailor et al., 1999; Yang et al., 1999). PHO81 has been shown to serve as a putative sensor of phosphate level in yeast and presumably Arabidopsis (Hamburger et al., 2002; Neef and Kladde, 2003). A truncated version of yeast SYG1, containing the N-terminal SPX domain, has been shown to rescue the lethal phenotype of a mutation in the G-protein α-subunit (Spain et al., 1995). The truncated protein does so by interacting directly with the G-protein β-subunit, and the interaction may inhibit the transduction of the mating pheromone signals. Apparently, a major function of the SPX domain in SYG1 is to mediate protein–protein interactions.
The EXS domain is also found in the yeast ERD1 protein (Hardwick et al., 1990). The region of EXS similarity contains several predicted transmembrane helices, suggesting a possible membrane localization of the proteins (Spain et al., 1995; Tailor et al., 1999; Yang et al., 1999). For example, three membrane-spanning motifs out of eight in yeast SYG1 reside in this region. Indeed, SYG1 was enriched in a plasma membrane fraction (Spain et al., 1995). Examination of the SHB1 protein sequence with PSORT also identified six weakly predicted integral membrane motifs. However, SHB1 is localized in the nucleus and the cytoplasm, like two other light signaling proteins, FHY1 and SRR1 (Figure 7B) (Huq and Quail, 2005; Ni, 2005). Thus, the EXS domain in SHB1 may have a distinct function from those in SYG1-like proteins. In summary, SHB1-like proteins exist in a variety of organisms, including yeast, fission yeast, Neurospora, worm, fly, mouse, and human. Although the function of most SHB1-like proteins remains to be determined in other organisms, we demonstrated the involvement of one member, SHB1, in regulating blue light responses specifically and/or possibly red and far-red light responses in Arabidopsis.
METHODS
Plant Growth Conditions, Genetic Screen, and Phenotypic Analysis
The Arabidopsis thaliana shb1-D mutant allele was isolated from a T-DNA insertion population in the Ws background (ABRC), and the shb1 mutant allele was isolated from a SALK T-DNA insertion population in the Col background. Monochromatic red, far-red, or blue light was generated with LED SNAP-LITE (Quantum Devices). Light intensity and peak wavelength were measured with a SPEC-UV/PAR spectroradiometer (Apogee Instruments). For most light experiments, Arabidopsis seeds were surface-sterilized, plated on regular agar growth medium minus sucrose, stratified at 4°C for 3 d, treated with fluorescent white light for 0.5 to 1 h, and allowed to germinate and grow under monochromatic light for 4 d. Hypocotyl, cotyledon, and leaf images were taken using an Olympus digital Camedia C-700, and length and area were measured using ImageJ. Each measurement includes 40 to 50 seedlings. Petiole length and leaf area were measured for the first fully expanded true leaf. The chlorophyll and anthocyanin contents were measured as described (Kim et al., 2003).
Molecular Cloning and Characterization of SHB1
Plant genomic DNA was isolated using the DNeasy plant mini kit (Qiagen). Plant DNA sequences flanking the T-DNA right or left border in shb1-D were obtained using a pair of right border primers, XR2 and nested XR3, and a pair of left border primers, JL202 and nested JL270 (University of Wisconsin Knockout Facility), along with the adapter primers, AP1 and nested AP2, from a GenomeWalker kit (Clontech). The T-DNA insertion also caused a 32-bp deletion of the plant genomic sequence surrounding the insertion site. The phylogenetic analysis on SHB1 and its homologues was conducted using the ClustalX 1.83 software program (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/). Multiple alignment mode and protein weight matrix Gonnet series with gap opening set 20 were chosen for the analysis. The phylogenetic tree was constructed using Bootstrap N-J tree mode with bootstrap trial number 1000 and was viewed using the TreeView software program (taxonomy.zoology.gla.ac.uk/rod/treeview.html). The presence or absence of integral membrane motifs was predicted using PSORT (http://psort.nibb.ac.jp).
Total RNA was isolated from plant tissues using the SV total RNA isolation system (Promega). Ten micrograms of total RNA was loaded on each lane for RNA gel blot analysis as described (Kang et al., 2005). Total RNA was used to perform the RT reaction using SuperScript RNase H− reverse transcriptase and gene-specific primers (Invitrogen). PCR was subsequently performed on 0.5 to 1 μL of RT reaction using Taq polymerase. Real-time PCR was performed on RT reaction using SYBR green PCR master mix and the 7500 Real Time PCR system (Applied Biosystems). Each experiment was repeated twice with completely independent biological samples, and three RT-PCR reactions were performed for each of the samples. Gene expression data were presented relative to average values for the Ws wild type using a relative quantification assay, after normalization to the control UBQ10 (Applied Biosystems).
To examine phyA protein or PHYA mRNA level in Ws or shb1-D, 5-d-old seedlings grown in darkness were treated with far-red light (0.2 μmol·m−2·s−1) for various times on regular agar growth medium. For experiments involving MG132, 5-d-old dark-grown seedlings were collected from regular agar growth medium and incubated in liquid growth medium containing either 50 μM MG132 (dissolved in DMSO) or DMSO in an equivalent volume added to the liquid growth medium under far-red light for the indicated times (Seo et al., 2004; Jang et al., 2005). Total plant proteins were prepared from seedlings, and phyA was detected by protein gel blot analysis using phyA-specific monoclonal antibodies as described previously (Ni et al., 1998).
Overexpression of SHB1 and Complementation of shb1
For overexpression analysis, SHB1 cDNA was amplified from the CD4-22 cDNA library (ABRC) with end-incorporated NheI and XmaI restriction sites and cloned into a modified PBI121 vector, in which the GUS gene was replaced with a GFP gene. A HindIII-EcoRI fragment from the modified PBI121 vector, containing CaMV 35S promoter, SHB1 and GFP coding sequences, and 3′ terminator, was then subcloned into pCAMBIA3300 vector. For phenotypic complementation, a 4.7-kb genomic fragment, spanning 1.2 kb upstream to 0.5 kb downstream of the SHB1 coding sequence, was generated using PCR from BAC clone T30C3 (ABRC) and contained end-incorporated XmaI and PstI restriction sites. The PCR fragment was then cloned into binary vector pCAMBIA3300.
Subcellular Localization Studies
For transient expression assay, SHB1 cDNA was amplified from the CD4-22 cDNA library (ABRC) with end-incorporated NheI and XmaI restriction sites and cloned into PBI221 vector. The resulting vector carries a SHB1:GUS fusion under the control of the CaMV 35S promoter. Transformation of SHB1:GUS into onion (Allium cepa) epidermal cells and 4′,6-diamidino-2-phenylindole staining were performed as described previously (Ni et al., 1998). The transfected onion epidermal peels were incubated in darkness for 24 h and then either received no light treatment or were treated with white, red, far-red, or blue light for 4 or 8 h. GUS staining was performed and nuclear images were acquired using a Nikon Eclipse E800 microscope with a Cool Cam color CCD camera (Cool Camera) and Imago Pro Plus version 3.0 software (Media Cybermetics).
Double Mutant Analysis
phyA-211 (Col), phyB-9 (Col), and cry2-1 (Col) mutants were obtained from the Arabidopsis Stock Center, and cry1-304 (Col) was kindly provided by Chentao Lin. phyB-9 is an ethyl methanesulfonate mutant, whereas phyA-211, cry1-304, and cry2-1 contain large deletions. Homozygous phyA-211 or phyB-9 was selected at the F2 generation based on a striking long-hypocotyl phenotype under far-red or red light, respectively. Homozygous cry1 or cry2 was selected at the F2 generation based on a long-hypocotyl phenotype under blue light but verified by PCR genotyping at F3. Both shb1-D (Ws) and shb1 (Col) contain a T-DNA insertion near or in their coding regions. Homozygous shb1-D or shb1 was selected by a linked resistance to kanamycin from the F3 individuals in a homozygous phyA-211, phyB-9, cry1, or cry2 background. An extra step was taken to generate shb1-D cry1 and shb1 cry1 double mutants, because the mutations are both on chromosome 4 but are 37 centimorgans apart. Recombinants were selected based on their long-hypocotyl phenotype under blue light and their resistance to kanamycin. Ws × Col wild type was also generated to control the difference in hypocotyl growth between crosses of different ecotypes.
The shb1-D pif4 double mutant was generated by genetic cross, and both shb1-D and pif4 are in the Ws background and contain a T-DNA insertion and linked resistance to kanamycin near or in their coding regions (Huq and Quail, 2002). Homozygous shb1-D or pif4 was genotyped using PCR at F2 or F3. hfr1-201 was kindly provided by Pill-Soon Song and was used to generate the shb1 hfr1-201 double mutant. Both shb1 and hfr1-201 are in the Col background and contain a T-DNA insertion and linked resistance to kanamycin near or in their coding regions (Soh et al., 2000). Homozygous shb1 or hfr1-201 was also genotyped using PCR at F2 or F3.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g25350 (SHB1), At2g43010 (PIF4), and At1g02340 (HFR1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Verification of the T-DNA Insertions in shb1-D and shb1 with PCR.
Supplemental Figure 2. Double Mutant Analysis of shb1-D with phyA-211 and cry Mutants under Blue Light.
Supplemental Figure 3. Phylogenetic Analysis of SHB1 with Its Homologues.
Supplemental Figure 4. Alignment of SHB1 with Its Homologues Used for the Phylogenetic Analysis.
Supplemental Figure 5. Sequence Alignment of SHB1 with Yeast SYG1.
Supplemental Figure 6. RNA Gel Blot Analysis of CAB3 and CHS Expression in the shb1 hfr1-201 Double Mutant.
Supplementary Material
Acknowledgments
We thank Neil Olszewski and Paul Lefebvre for comments on the manuscript, David Marks for help with and the use of the Nikon Eclipse E800 microscope, Chentao Lin for cry1-304, Enamul Huq and Peter Quail for pif4, Pill-Soon Song for hfr1-201, and Peter Quail for antibody against phyA. We also thank the Ohio State Stock Center for Arabidopsis mutants, T-DNA insertion collections, and BAC clones. This work was supported in part by University of Minnesota Start-Up and Grant-in-Aid funds (to M.N.) and by a grant from the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (2004-35304-14939 to M.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Min Ni (nixxx008@tc.umn.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037879.
References
- Ahmad, M., Jarrilo, J.A., Smirnova, O., and Cashmore, A.R. (1998). The cry1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1 939–948. [DOI] [PubMed] [Google Scholar]
- Bouly, J.P., Giovani, B., Djamei, A., Mueller, M., Zeugner, A., Dudkin, E.A., Batschauer, A., and Ahmad, M. (2003). Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270 2921–2928. [DOI] [PubMed] [Google Scholar]
- Boylan, M.T., and Quail, P.H. (1991). Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88 10806–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R. (2003). Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114 537–543. [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.R., and Song, P.S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401 610–613. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., Chen, D.L., Yeh, K.-C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., and Fankhauser, C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J. 34 827–836. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, A.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-translational mechanism. Plant J. 19 279–287. [DOI] [PubMed] [Google Scholar]
- Guo, H., Mockler, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291 487–490. [DOI] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hamburger, D., Rezzonico, E., MacDonald-Comber Petetot, J., Somerville, C., and Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K.G., Lewis, M.J., Semenza, J., Dean, N., and Pelham, H.R. (1990). ERD1, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. EMBO J. 9 623–630. [DOI] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2005). Phytochrome signaling. In Handbook of Photosensory Receptors, W.R. Briggs and J.L. Spudich, eds (Weinheim, Germany: Wiley VCH), pp. 151–170.
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M., Ecker, J.R., and Chen, J.G. (2003). A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 131 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, X., Chong, J., and Ni, M. (2005). HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell 17 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Kang, J.G., Yanga, S.S., Chung, K.S., Song, P.S., and Park, C.M. (2002). A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14 3043–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner, O., Kircher, S., Harter, K., and Batschauer, A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19 289–296. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack, A.C., and Terry, M.J. (2002). Light-signaling pathways leading to the coordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32 549–559. [DOI] [PubMed] [Google Scholar]
- Møller, S.G., Kim, Y.S., Kunkel, T., and Chua, N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A., Kay, S.A., Deak, M., Chua, N.H., and Furuya, M. (1991). Rice type I phytochrome regulates hypocotyl elongation in transgenic tobacco seedlings. Proc. Natl. Acad. Sci. USA 88 5207–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef, D.W., and Kladde, M.P. (2003). Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14 257–271. [PubMed] [Google Scholar]
- Neuhaus, G., Bowler, C., Hiratsuka, K., Yamagata, H., and Chua, N.H. (1997). Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 16 2554–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M. (2005). Integration of light signaling with photoperiodic flowering and circadian regulation. Cell Res. 15 559–566. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature 400 781–784. [DOI] [PubMed] [Google Scholar]
- Okamoto, H., Matsui, M., and Deng, X.W. (2001). Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Ryu, J.S., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120 395–406. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 763–767. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yu, X., Maymon, M., Mockler, T., and Lin, C. (2003). Blue light–dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh, M.S., Kim, Y.M., Han, S.J., and Song, P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12 2061–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain, B.H., Koo, D., Ramakrishnan, M., Dzudzor, B., and Colicelli, J. (1995). Truncated forms of a novel yeast protein suppress the lethality of a G protein α subunit deficiency by interacting with the β subunit. J. Biol. Chem. 270 25435–25444. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Schafer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294 1108–1111. [DOI] [PubMed] [Google Scholar]
- Tailor, C.S., Nouri, A., Lee, C.G., Kozak, C., and Kabat, D. (1999). Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 96 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J.M., Cufr, C.A., Denzel, M.A., and Neff, M.M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D.D., and O'Shea, E.K. (2001). Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Tang, R., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Wu, Y., Tang, R., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C-termini of Arabidopsis cryptochrome mediate a constitutive light response. Cell 103 815–827. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.L., Guo, L., Xu, S., Holland, C.A., Kitamura, T., Hunter, K., and Cunningham, J.M. (1999). Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21 216–219. [DOI] [PubMed] [Google Scholar]
- Yeh, K., and Lagarias, C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.