Abstract
The human 8-oxoguanine-DNA glycosylase 1 (OGG1) is the major DNA glycosylase responsible for repair of 7,8-dihydro-8-oxoguanine (8-oxoG) and ring-opened fapyguanine, critical mutagenic DNA lesions that are induced by reactive oxygen species. Here we show that OGG1 is acetylated by p300 in vivo predominantly at Lys338/Lys341. About 20% of OGG1 is present in acetylated form in HeLa cells. Acetylation significantly increases OGG1's activity in vitro in the presence of AP-endonuclease by reducing its affinity for the abasic (AP) site product. The enhanced rate of repair of 8-oxoG in the genome by wild-type OGG1 but not the K338R/K341R mutant, ectopically expressed in oxidatively stressed OGG1-null mouse embryonic fibroblasts, suggests that acetylation increases OGG1 activity in vivo. At the same time, acetylation of OGG1 was increased by about 2.5-fold after oxidative stress with no change at the polypeptide level. OGG1 interacts with class I histone deacetylases, which may be responsible for its deacetylation. Based on these results, we propose a novel regulatory function of OGG1 acetylation in repair of its substrates in oxidatively stressed cells.
Reactive oxygen species (ROS), produced in vivo either as by-products of normal oxidative metabolism or induced by exogenous agents, include superoxide radical O2·−, H2O2, and OH· radical (1, 9, 16). ROS induce a variety of genomic lesions including oxidatively damaged bases, AP sites, and DNA strand breaks, among which 7,8-dihydro-8-oxoguanine (8-oxoG) and ring-opened fapyguanine (FapyG) are the major base lesions (40). 8-oxoG is highly mutagenic because of its preferential mispairing with adenine during DNA replication, thereby generating G·C to T·A transversion mutations (12, 23, 54, 63). In order to prevent mutations induced by 8-oxoG, organisms, ranging from bacteria to humans, efficiently repair this lesion in their genomes (41). The base excision repair (BER) pathway is primarily responsible for repair of 8-oxoG in all organisms. In Escherichia coli, Fpg (also named MutM), a DNA glycosylase/AP lyase, catalyzes excision of ROS-induced purine-derived lesions including 8-oxoG and FapyG (6, 57). Earlier studies suggested that nuclear and mitochondrion-specific isoforms, 8-oxoguanine-DNA glycosylase 1-α (OGG1-α) and OGG1-β, respectively, responsible for repair of 8-oxoG in these organelles, are generated via alternative splicing such that the mitochondrial isoform lacks the C-terminal segment of OGG1-α which contains the nuclear localization signal (47). However, Hashiguchi et al. have recently shown that OGG1-β lacks DNA glycosylase activity and that excision of 8-oxoG from the mitochondrial genome is also catalyzed by OGG1-α (28). We identified a second 8-oxoG-specific DNA glycosylase/AP lyase activity, distinct from OGG1, in mammalian cells and named it OGG2 (29). Recently NEIL1, an ortholog of E. coli Fpg/Nei and not identical to OGG2, has been characterized. This enzyme excises various oxidized bases including 8-oxoG and Fapys (18, 30, 45). In spite of the presence of these additional activities, the main enzyme for repairing 8-oxoG and FapyG in mammalian cells is OGG1, which is structurally unrelated but functionally similar to Fpg (7, 50, 52, 53). Accumulation of 8-oxoG in the genome of OGG1-null mice, and cells derived therefrom, provides strong evidence for the key role of OGG1 in repairing 8-oxoG, at least from the bulk of the genome (46). OGG1 excises 8-oxoG and other damaged base substrates from DNA by attacking the N-glycosidic bond with Lys249 as the active site nucleophile to form a transient Schiff base. After removal of the base lesion, the bound enzyme carries out the lyase reaction via β-elimination to cleave the DNA strand at the damage site and generates 3′-phospho-α-β unsaturated aldehyde and 5′-phosphate termini (50, 53). While all oxidized base-specific DNA glycosylases have intrinsic AP lyase activity, an unusual property of human OGG1 (hOGG1) is its poor turnover in vitro after base release (5, 32, 43, 66). It remains preferentially bound to the resulting AP site without carrying out β-elimination. While other glycosylases also have affinity for the AP site product, it is particularly strong in the case of OGG1 (32, 66). X-ray crystallographic structures of both wild-type (WT) OGG1 and the inactive K249Q point mutant, bound to an 8-oxoG·C-containing oligonucleotide, showed that, upon binding to DNA, the enzyme undergoes extensive local conformational change (10). However, relatively little is known about the mechanism of OGG1's affinity for the AP site. We have shown that the OGG1 activity is significantly stimulated by APE1 without physical interaction, and APE1, the only AP-endonuclease present in human cells, acts subsequently to OGG1 (32).
p300 and closely related CBP (p300/CBP), discovered as transcriptional coactivators, as well as their associated factor (P/CAF), have intrinsic histone acetyltransferase (HAT) activity (48, 64). These proteins were subsequently shown to acetylate Lys residues not only in histones but also in many transcription factors and hence were later named factor acetyltransferases (56). p300/CBP act as components of chromatin-remodeling complexes (2, 19, 21, 48). More recently, p300 has been implicated in DNA replication and repair, based on the observation that it acetylates 5′ flap endonuclease 1 (FEN1), responsible for removing the RNA primer of nascent Okazaki fragments and for processing the 5′ termini at DNA strand breaks during BER (27). p300 also interacts with proliferating cell nuclear antigen (PCNA), which in turn stimulates FEN1. Thus, together with other proteins, they play a central role in DNA replication and BER (26, 27). Several BER proteins including APE1, G·T-specific thymine-DNA glycosylase (TDG), NEIL2, and DNA polymerase β were subsequently shown to be acetylated by p300 (3, 4, 25, 27, 58). Acetylation modulates the activity of DNA replication/repair proteins, either positively or negatively. For example, acetylation reduces the nuclease activity of FEN1, presumably as a result of reduced DNA affinity (27). On the other hand, acetylation of TDG does not change its DNA glycosylase activity, which is, on the other hand, stimulated by sumoylation, another covalent modification (24, 58). We showed earlier that acetylation of human NEIL2, an oxidized pyrimidine-specific DNA glycosylase, could inhibit its activity (3, 31).
In spite of several recent in vitro studies showing altered DNA repair and substrate affinity of repair proteins due to acetylation, the physiological relevance of acetylation of BER and other proteins involved in DNA transactions has not been addressed so far. In this study, we show that OGG1 is acetylated by p300 both in vivo and in vitro and identify Lys338 and Lys341 as the major acetyl acceptor sites. Acetylation of OGG1 increases its in vitro turnover in the presence of APE1. We separated unmodified and acetylated OGG1 (AcOGG1) from HeLa cells by ion-exchange chromatography and showed that the endogenous AcOGG1 is biochemically similar to the acetylated recombinant protein. Moreover, we observed modulation of genomic 8-oxoG repair by changing the level of AcOGG1, which strongly suggests a regulatory role of reversible acetylation in DNA repair.
MATERIALS AND METHODS
Cell culture, transfection, and plasmids.
Human HCT116 colon carcinoma cells (a gift from B. Vogelstein) were grown at 37°C in McCoy 5A (Gibco Life Technologies) medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) in the presence of 5% CO2. Primary MRC5 fibroblasts were grown in minimal essential medium (MEM; Gibco Life Technologies). Mouse embryonic fibroblasts (MEFs) isolated from OGG1+/+ or OGG1−/− mice (a gift from D. Barnes) were grown in Dulbecco's modified Eagle's medium (Gibco BRL). HCT116 cells were transfected with plasmids using LipofectAMINE 2000 (Life Technologies), according to the manufacturer's instructions. Cells were transfected with duplex p300 short interfering RNA (siRNA; 5′-AAC CCC UCC UCU UCA GCA CCA-3′; Dharmacon Research Inc., Colorado) at 100 nM in a 60-mm plate. HeLa cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics at 37°C. In order to induce oxidative stress, the cells at 50% confluence were treated with 25 milliunits/ml glucose oxidase (GO; Roche Applied Science) for 1 h followed by washing with phosphate-buffered saline (PBS) and subsequent incubation in fresh medium (15, 35).
The cDNAs encoding WT hOGG1 and the K249Q mutant were cloned into E. coli expression plasmid pRSETB (32). The pCMV-N-FLAG expression plasmid (Sigma) encoding N-terminal FLAG-tagged WT OGG1 or the K338R/K341R mutant was generated by PCR, and its identity was confirmed by direct sequencing. The purification of recombinant FLAG-p300 (HAT domain) was described earlier (11).
In vitro acetylation of OGG1.
Recombinant WT OGG1 or the K249Q mutant (5 μg) was incubated with 0.2 μg recombinant human p300 (HAT domain) in the presence of 1 mM acetyl coenzyme A (CoA; Sigma) or 1 μCi of [3H]acetyl-CoA (200 mCi/mM; NEN) in 50 μl HAT buffer (50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 10 mM sodium butyrate) at 30°C for various times. The extent of acetylation was monitored either by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis followed by fluorography with enhancing solution (Amplify; Amersham) or by immunoblotting.
In vivo acetylation of OGG1.
HCT116 cells (2 × 106 to 4 × 106 cells/dish) were transfected with the expression plasmid for FLAG-tagged WT OGG1 or K338R/K341R mutant (1 μg) and LipofectAMINE 2000 (3 μl); 40 h later, the cells were incubated in McCoy 5A medium (Gibco Life Technologies) containing 1 mCi/ml [3H]sodium acetate (5 Ci/mmol; NEN) for 1 h. The cells were then lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM NaF, 1 mM sodium orthovanadate, 10 mM sodium butyrate, and a protease inhibitor cocktail, followed by immunoprecipitation at 4°C with anti-FLAG M2 antibody (Sigma) cross-linked to agarose beads for 3 h. After the beads were washed with cold TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl), the FLAG-OGG1 was eluted by gently shaking the beads in TBS containing 300 ng/μl FLAG peptide (Sigma) for 30 min, and the eluate was analyzed by SDS-PAGE (12% polyacrylamide) and fluorography.
Partial purification of AcOGG1 from HeLa cell extract.
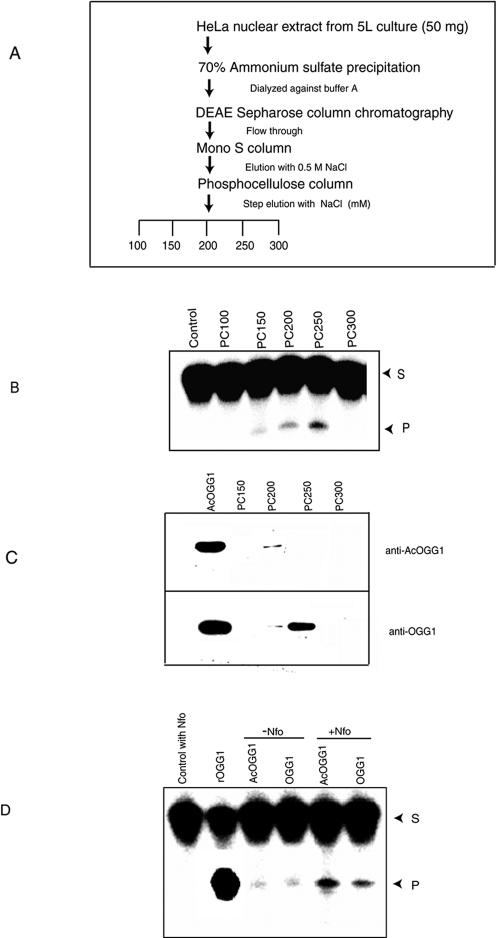
OGG1 and AcOGG1 were partially purified, as schematically shown in Fig. 5A, from HeLa nuclear extract at 4°C in a buffer containing 20 mM Tris, pH 7.5, 0.1 mM EDTA, 1 mM dithiothreitol, protease inhibitor cocktail, 10% glycerol, and 10 mM sodium butyrate (buffer A).
FIG. 5.
Partial purification and separation of acetylated and nonacetylated OGG1 from HeLa nuclear extract. (A) Outline of purification steps of OGG1 from HeLa nuclear extract. (B) Eluted fractions from the phosphocellulose column were assayed for 8-oxoG excision activity as described in Materials and Methods. S, substrate; P, product. (C) Western analysis for AcOGG1 in phosphocellulose fraction (upper panel). The same blot was reprobed with OGG1 antibody (lower panel). (D) Specific activity and analysis of trapped complexes of endogenous AcOGG1 (PC200) and unmodified OGG1 (PC250). OGG1 (10 nM) was incubated at 37°C in 15 μl with 100 nM 8-oxoG·C oligonucleotide in the presence or absence of 100 nM of E. coli Nfo for 10 min, as described in Materials and Methods. S, substrate; P, product.
Identification of acetyl acceptor Lys residues in OGG1.
Recombinant hOGG1 (50 μg) was acetylated with p300 HAT domain (1 μg) and 0.5 mM acetyl-CoA together with 4 μCi of [3H]acetyl-CoA (200 mCi/mM; NEN) at 30°C for 1 h. After digestion, followed by reverse-phase chromatography, the fractions containing 3H-labeled peptides were dried and subjected to N-terminal sequencing. Acetyl Lys (AcLys) residues were unambiguously identified as PTH-AcLys, as described earlier (4).
DNA glycosylase/AP lyase and trapping assay.
We assayed DNA glycosylase/AP lyase activity of OGG1 with a 5′ 32P-labeled duplex oligonucleotide (32, 44, 49). A 31-mer oligonucleotide, 5′-GAA GAG AGA AAG AGA XAA GGA AAG AGA GAA G-3′ (Midland Certified Reagent Co., Midland, TX), containing either 8-oxoG or an AP site at position X and 32P labeled at the 5′ terminus, was used (32). DNA trapping reactions were performed by incubating 3 to 4 fmol 32P-labeled 8-oxoG-containing oligonucleotide with PC200 and PC250 fractions or 5 ng recombinant OGG1in a reaction mixture (10 μl) containing 25 mM HEPES, pH 7.9, 2 mM dithiothreitol, 50 mM KCl, 2.5 mM EDTA, 50 mM NaCNBH3 at 37°C for 30 min (17). All DNA glycosylases with AP lyase activity form a transient Schiff base between the amino group of the active site nucleophile (Lys249 in the case of OGG1) and the aldehyde group of the free AP site after base excision. The Schiff base could be reduced by NaBH3 (or NaCNBH3) to form a stable covalent “trapped complex.” The trapped complexes were separated by SDS-PAGE (12% polyacrylamide) after being heated at 100°C for 5 min, and the gels were dried on DE-81 paper for PhosphorImager analysis of radioactivity. The mobility of trapped complexes of various glycosylases with the same oligonucleotide depends on the size of the enzyme and hence could be used diagnostically.
Quantitation of genomic 8-oxoG in cell nuclei.
OGG1−/− MEFs were transfected with FLAG-tagged WT OGG1, K338R/K341R mutant, or empty vector using LipofectAMINE 2000. Thirty-six hours after transfection, cells growing on microscope coverslips were treated with trichostatin A (TSA; 100 ng/ml) for 12 h or with glucose oxidase (25 milliunits/ml) for 1 h, and the cells were washed and fed fresh medium. At various times after being washed with PBS, air dried, and fixed in (1:1) acetone-methanol, the cells were rehydrated in PBS for 15 min followed by sequential treatment with 100 μg/ml pepsin in 0.1 N HCl for 15 to 30 min at 37°C, 1.5 N HCl for 15 min, and sodium borate for 5 min. After finally being washed with PBS, the cells were incubated with nonimmune immunoglobulin G (0.1 μg per ml) for 30 min and washed in PBS containing 0.5% bovine serum albumin, 0.1% Tween 20 (PBS-T [8]). Following incubation with anti-8-oxoG antibody (Trevigen Inc.; 1:200 dilution) for 30 min, the cells were washed three times with PBS-T for 15 min and then exposed to fluorescein-conjugated secondary antibody (Santa Cruz Biotechnology) for 30 min. After being triple washed with PBS-T, the DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride; 10 ng/ml) for 15 min. The coverslips were finally mounted in antifade medium (Dako Inc.) on a microscope slide for confocal microscopy (Zeiss LSM510 META system). Fluorescence intensities of a minimum of 40 fluorescent cells per plate were determined using MetaMorph software version 5.0 (Universal Imaging).
Two-way analysis of variance (ANOVA) with time and treatment as independent variables was used for data analysis. For significant correlation, step-down, one-way ANOVAs were performed followed by a comparison of means using Dunnett's test.
Analysis of binding of OGG1 and AcOGG1 to AP·C and 8-oxoG·C oligonucleotides.
32P-labeled duplex oligonucleotide (100 fmol) containing AP·C or 8-oxoG·C was incubated with indicated amounts of unmodified inactive K249Q OGG1 mutant or its acetylated form in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.15 μg/μl bovine serum albumin, 1 mM EDTA, and 15% glycerol (10 μl) at 4°C for 30 min, followed by electrophoresis in 6% polyacrylamide to separate the DNA protein complex.
In vitro deacetylation assay.
HCT116 cells were transfected with FLAG-tagged histone deacetylase 1 (HDAC1; 2 μg) or empty vector using LipofectAMINE 2000 (6 μl; Gibco Life Technologies); 48 h later, the cell extracts were immunoprecipitated for 3 h at 4°C with anti-FLAG M2 antibody (Sigma) cross-linked to agarose beads. The beads were then washed with cold TBS, and the FLAG-HDAC1 was eluted as before and then incubated with 2 μg in vitro-acetylated [3H]OGG1 at 30°C in the HAT buffer for 45 min. Deacetylation of OGG1 was analyzed by SDS-PAGE and fluorography.
Coimmunoprecipitation analysis.
HCT116 cells were transfected with 1 μg each of OGG1-FLAG, p300, or FLAG-tagged HDACs (HDAC1 through HDAC6) expression plasmids used individually. After 40 h, the cells were lysed and the extracts (2 mg/ml) were immunoprecipitated with either anti-FLAG M2 antibody (Sigma) or p300 antibody (N-15; Santa Cruz Biotechnology) as before, for SDS-PAGE (6% polyacrylamide for p300 and 12% for FLAG-OGG1) after suspension of the immunoprecipitate in 2× Laemmli buffer. Western analysis was carried out with antibodies against p300 (Santa Cruz Biotechnology), FLAG (M2; Sigma), or OGG1 (Alpha Diagnostic, Texas).
Generation of AcOGG1-specific antibody.
The AcLys-containing peptide, PAKRRAcKGGAcKGPEC, corresponding to amino acid residues 333 to 344 of hOGG1, with an additional C-terminal Cys, was synthesized and purified by high-pressure liquid chromatography at the UTMB Biomolecular Resources Facility and then used for production of antibodies in rabbits after being coupled to hemocyanin (Alpha Diagnostic). The rabbit antisera were enriched for anti-AcOGG1 immunoglobulin G by affinity purification with recombinant AcOGG1.
RESULTS
Interaction of OGG1 with p300 in vivo.
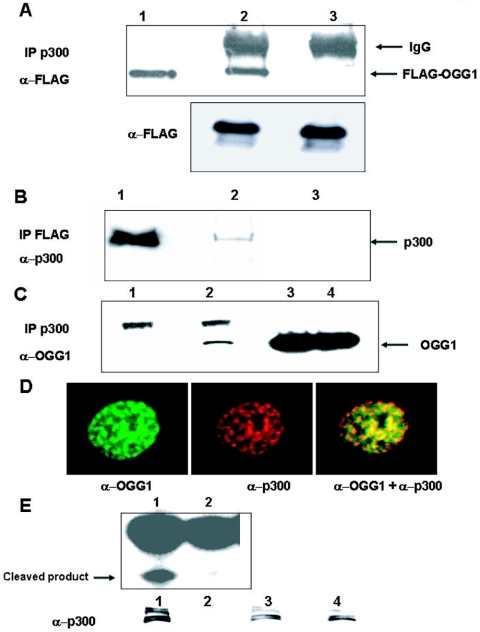
The involvement of p300 in DNA replication and repair, due to its association with PCNA, FEN1, and polymerase β, suggested that this acts as a cofactor for enzymes involved in BER (25-27). To confirm this possibility, we performed coimmunoprecipitation analysis of p300 from extracts of HCT116 transfected with FLAG-tagged OGG1. Western analysis showed that OGG1 was present in the p300 immunoprecipitate (Fig. 1A, lane 2) but not in the control immunocomplex (Fig. 1A, lane 3), while FLAG-OGG1 levels in the starting extracts were comparable (Fig. 1A, lower panel). To further confirm stable interaction between p300 and OGG1, we showed the presence of p300 in FLAG-OGG1 immunocomplex (Fig. 1B, lane 2) but not in the immunoprecipitate with preimmune sera (lane 3). To exclude the possibility that OGG1-p300 interaction could be an artifact of OGG1 overexpression, we immunoprecipitated endogenous p300 from cell extracts and showed the presence of endogenous OGG1 in the immunoprecipitate with p300-specific antibody (Fig. 1C, lane 2) but not with the control antibody (Fig. 1C, lane 1). To provide additional evidence for their in vivo interaction, we examined colocalization of OGG1 and p300 in MRC5 primary human fibroblasts immunostained with anti-OGG1 (green) and anti-p300 (red) antibodies (Fig. 1D), using confocal microscopy. Both proteins were found to be mostly nuclear. More importantly, superimposition of these images showed significant subnuclear regions with overlapping OGG1 and p300 staining as indicated by yellow speckles (Fig. 1D).
FIG. 1.
In vivo interaction of OGG1 and p300. A. Extracts of HCT116 cells cotransfected with expression plasmid for FLAG-tagged OGG1 and p300 were immunoprecipitated with p300 antibody (lane 2) or preimmune sera (lane 3) and then blotted with FLAG antibody. Lane 1, FLAG-tagged OGG1 in cell extract as marker. Lower panel, Western analysis with anti-FLAG antibody of cell extracts used in upper panel. B. Extracts of cells transfected with FLAG-tagged OGG1 were immunoprecipitated with FLAG antibody (lane 2) or preimmune sera (lane 3), and the immunoprecipitates were analyzed for p300 by Western blotting. Lane 1, cell extracts used as marker. C. Extracts of HCT116 cells were immunoprecipitated with p300 antibody (lane 2) or preimmune sera (lane 1) and then immunoblotted with OGG1 antibody; lanes 3 and 4, input controls of cell extracts. D. Colocalization of OGG1 and p300. MRC5 cells were immunostained with OGG1 (green) and p300 (red). E. Incision activity of p300 immunoprecipitates (lane 1) or preimmune sera (lane 2) with 32P-labeled 8-oxoG·C oligonucleotide. Lower panel, Western analysis of immunoprecipitates with p300 antibody and input controls of cell extracts (lanes 3 and 4).
We showed that immunoprecipitated OGG1-p300 complex possessed 8-oxoG excision activity, using a 5′ 32P-labeled 8-oxoG-containing oligonucleotide (Fig. 1E, lane 1), while OGG activity was absent in the immunocomplex as shown by using a control antibody (Fig. 1E, lane 2). Thus, the p300-OGG1 complex binds to substrate DNA and is enzymatically active.
OGG1 is acetylated both in vivo and in vitro.
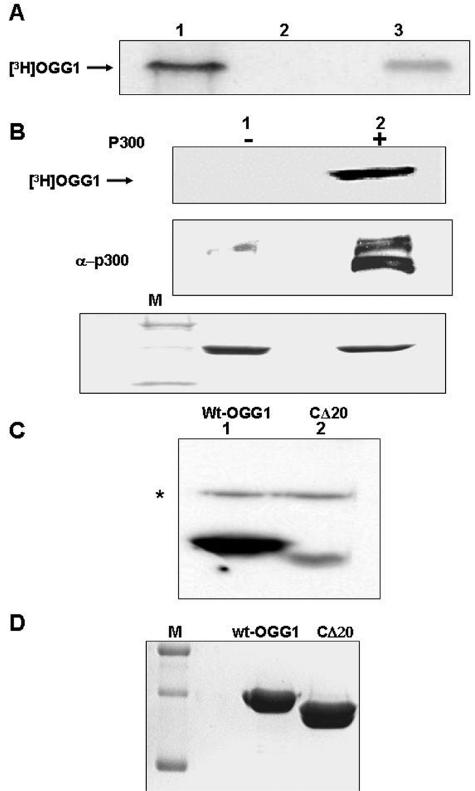
Because of strong interaction of OGG1 with p300 and the presence of 8-oxoG excision activity in the p300-OGG1 immunocomplex, it appeared likely that OGG1 is a target for acetylation by p300 during 8-oxoG repair. We first tested for the presence of AcOGG1 in vivo. HCT116 cells were transfected with either empty vector or FLAG-tagged WT OGG1. After pulse-labeling (1 h) with [3H]sodium acetate and immunoprecipitation of the cell extract with an anti-FLAG antibody followed by SDS-PAGE and fluorography, we observed the presence of radioactivity in the FLAG-OGG1 band (Fig. 2A, lane 3) in the FLAG-tagged OGG1-transfected cells but not in empty FLAG vector-transfected cells (Fig. 2A, lane 2). This provided the first evidence for in vivo acetylation of OGG1.
FIG. 2.
In vivo and in vitro acetylation of OGG1. A. Extracts of HCT116 cells transfected with FLAG-tagged OGG1 (lane 3) or empty vector-transfected cells (lane 2) and then labeled with [3H]sodium acetate were immunoprecipitated with FLAG antibody and analyzed by SDS-PAGE and fluorography. Lane 1, in vitro-acetylated 3H-labeled OGG1 marker. B. In vitro acetylation of full-length OGG1 with immunoprecipitated p300 (lane 2) or preimmune sera (lane 1) followed by SDS-PAGE and fluorography. Middle panel, Western analysis of immunoprecipitate with p300 antibody. Lower panel, Coomassie blue staining of the input OGG1 in a duplicate gel after SDS-PAGE. C. In vitro acetylation of full-length OGG1 (lane 1) or OGG1 with the C-terminal 20 amino acids deleted (CΔ20; lane 2) with purified p300 HAT domain and [3H]acetyl-CoA followed by SDS-PAGE and fluorography. * indicates autoacetylated p300 HAT. D. Coomassie blue staining of the duplicate gel after SDS-PAGE. M, molecular weight markers.
We then examined in vitro acetylation of OGG1 by incubating the immunoprecipitate of WT p300, isolated from the extracts of WT p300 plasmid-transfected HCT116 cells, with recombinant OGG1 in the presence of [3H]acetyl-CoA. Radiolabeling of OGG1 indicated that it was acetylated by the p300 immunoprecipitate but not by a control immunoprecipitate (Fig. 2B, lanes 1 and 2). These initial results were confirmed by acetylation studies of OGG1 with recombinant HAT domain of human p300 (Fig. 2C, lane 1). Parallel incubation with the same amount of an OGG1 mutant lacking 20 C-terminal amino acid residues (CΔ20) showed nearly 90% reduction of OGG1 acetylation compared to the full-length OGG1 (Fig. 2C, lane 2). Coomassie blue staining confirmed that comparable amounts of full-length OGG1 and CΔ20 mutant were used (Fig. 2D). This indicated that the major acetyl acceptor Lys residues are located within the 20 C-terminal residues.
Identification of acetyl acceptor Lys residues in OGG1.
In order to identify the acetyl acceptor Lys residues in OGG1, a peptide corresponding to residues 326 to 345 near the C terminus of OGG1 was chemically synthesized, purified, and then used as a substrate for the recombinant human p300 (HAT domain). Mass spectrometric analysis of in vitro-acetylated peptide confirmed the presence of both monoacetylated and diacetylated species with 42 and 84 more mass units than that of the unmodified peptide (data not shown). N-terminal sequencing of the peptide indicated that Lys338 and Lys341 were preferentially acetylated, although a low level of acetylation was also observed at Lys335 (data not shown). We then confirmed Lys338 and Lys341 residues as acetylation sites in the full-length OGG1 after acetylating OGG1 with p300 and [3H]acetyl-CoA as before, followed by digestion with trypsin. After separation of the peptides by reverse-phase high-pressure liquid chromatography, N-terminal sequencing of 3H-labeled peptides indicated that Lys338 and Lys341 were predominantly acetylated (data not shown). Thus, the major in vitro acetylation sites in OGG1 are Lys338 and Lys341.
Lys338 and Lys341 in OGG1 are the major acetyl acceptor residues in vivo.
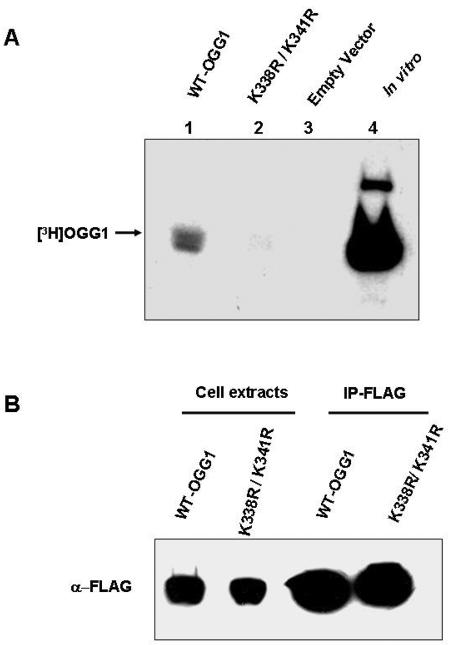
To test whether Lys338 and Lys341 are also in vivo acetyl acceptor sites, we mutated Lys338 and Lys341 to Arg in the FLAG-OGG1 expression plasmid. HCT116 cells, transfected with expression plasmids for empty vector, WT OGG1, or K338R/K341R mutant, were pulse-labeled (1 h) with [3H]sodium acetate. Immunoprecipitation of cell extracts with anti-FLAG antibody followed by SDS-PAGE and fluorography showed a drastically reduced amount of radioactivity in the K338R/K341R mutant band (Fig. 3A, lane 2) relative to the WT OGG1 band (Fig. 3A, lane 1). Western analysis with FLAG antibody confirmed the presence of comparable amounts of OGG1 in the immunoprecipitates and similar levels of ectopic expression of WT and mutant OGG1 in independently transfected cells (Fig. 3B). These results confirm that Lys338 and Lys341 are also the major acetylation sites in vivo.
FIG. 3.
In vivo acetylation of wild-type OGG1 and K338R/K341R mutant. A. Extracts of HCT cells transfected with FLAG-tagged WT OGG1 (lane 1) or FLAG-tagged K338R/K341R OGG1 (lane 2) or empty vector (lane 3) and then labeled with [3H]sodium acetate were immunoprecipitated with FLAG antibody and analyzed by SDS-PAGE and fluorography. Lane 4, in vitro-acetylated 3H-labeled OGG1 marker. B. Western analysis with FLAG antibody for the WT and K338R/K341R OGG1 in the immunoprecipitates and cell extracts used in panel A.
Detection and localization of AcOGG1 in vivo.
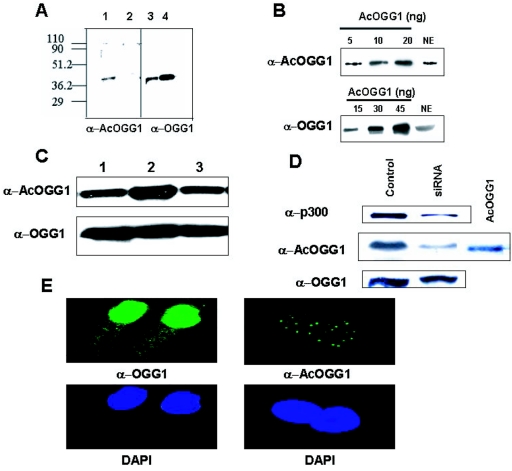
To further confirm the presence of endogenous AcOGG1, we generated and affinity purified AcOGG1-specific antibody by immunizing rabbits with a synthetic peptide corresponding to the amino acid residues 333 to 344 of OGG1 in which Lys338 and Lys341 were acetylated. Western analysis showed that the AcOGG1 antibody was highly selective for AcOGG1 (Fig. 4A, lane 1) and did not detectably cross-react with a threefold-higher level of unmodified OGG1 (Fig. 4A, lane 2). The identity of endogenous AcOGG1 in HeLa nuclear extract was subsequently confirmed by Western analysis with AcOGG1 antibody (Fig. 4B, upper panel). This result provided the first evidence for the presence of AcOGG1 in cells under normal physiological conditions. We then used quantitative immunoblots of nuclear extracts, in parallel with known amounts of recombinant AcOGG1 or OGG1 as standards, to quantitate endogenous levels of the unmodified and AcOGG1 in HeLa cells (Fig. 4B). By matching band intensities of extracts with the standard curve for both AcOGG1 and OGG1, we estimated their amounts in HeLa nuclear extract (75 μg) to be 6 and 24 ng, respectively. Thus, AcOGG1 constitutes about 20% of the total OGG1 in HeLa cells. Because the recombinant p300 HAT domain can strongly acetylate OGG1 in vitro, we tested whether p300 is involved in in vivo acetylation of OGG1. HCT116 cells were cotransfected with expression plasmids for WT full-length p300 or p300 HAT mutant, deficient in histone acetyltransferase activity. Western analysis with AcOGG1 antibody showed a significant increase in the level of AcOGG1 in WT p300-transfected nuclear extracts (Fig. 4C, lane 2) but not in the p300 HAT mutant-transfected extracts (Fig. 4C, lane 3). Comparison of the Western blot with OGG1-specific antibody showed similar amounts of OGG1 in different samples (Fig. 4C, lower panel). We then carried out a complementary experiment by lowering the cellular p300 level by downregulating p300. The p300 level in HCT116 was reduced by fivefold after transfection with p300-specific siRNA with a concomitant decrease in the amount of AcOGG1 without affecting the total OGG1 level (Fig. 4D). We therefore conclude that p300 contributes significantly to in vivo acetylation of OGG1. Because of the specificity of our AcOGG1 antibody, we were able to specifically examine intracellular distribution of AcOGG1 in MRC5 cells by immunofluorescence. AcOGG1 was found to be localized exclusively in the nucleus, mostly distributed in discrete foci (Fig. 4E, right panel), while the unmodified OGG1 diffusely distributed mostly in the nucleus (Fig. 4E, left panel). Similar, discrete subcellular distribution of AcOGG1 was also observed in HCT116 cells (data not shown).
FIG. 4.
Identification and localization of AcOGG1 in vivo. A. Western analysis of in vitro-acetylated OGG1 (15 ng, lane 1) or unmodified OGG1 (45 ng, lane 2) with AcOGG1 antibody. Right panel, Western blot analysis of the same blot with OGG1 antibody (lanes 3 and 4). Numbers at left are molecular masses in kilodaltons. B. Western blot analysis of HeLa nuclear extract (75 μg) with AcOGG1 antibody (upper panel) or OGG1 antibody (lower panel) with known amounts of recombinant AcOGG1. NE, nuclear extract. C. Nuclear extracts (100 μg) of HCT116 cells transfected with pcDNA3 (lane 1) or WT p300 expression plasmid (lane 2) or p300 HAT mutant (lane 3) were immunoblotted with AcOGG1 antibody (upper panel) or OGG1 antibody (lower panel). D. HCT116 cells were transfected with 100 nM control siRNA (lane 1) or p300-specific duplex siRNA (lane 2); 48 h later, cell extracts were analyzed by Western blotting using p300 (upper panel)-, AcOGG1 (middle panel)-, or OGG1 (lower panel)-specific antibodies. E. Immunofluorescence studies of MRC5 cells with AcOGG1 antibody (right panel) or OGG1 antibody (left panel). Lower panels, cells were stained with DAPI.
Purification and characterization of endogenous AcOGG1 and OGG1.
We separated AcOGG1 and OGG1 from HeLa cell extracts via several chromatographic steps and partially purified these for biochemical characterization (Fig. 5A). 8-oxoG excision assay indicated that OGG activity was distributed in three distinct fractions after chromatography on phosphocellulose (Fig. 5B). The fractions eluted at 200 (PC200) and 250 (PC250) mM NaCl with most enzymatic activity containing AcOGG1 or OGG1, respectively, as detected by Western analysis using OGG1 or AcOGG1 antibodies (Fig. 5C). In order to test for cross-contamination of AcOGG1 and OGG1 in these fractions, we used quantitative immunoblot assays as before (44) and calculated the same amount of OGG1 in PC200 using either OGG1 or AcOGG1 antibody (data not shown). This indicates that OGG1 was present in PC200 exclusively as AcOGG1.
Acetylation stimulates OGG1 activity.
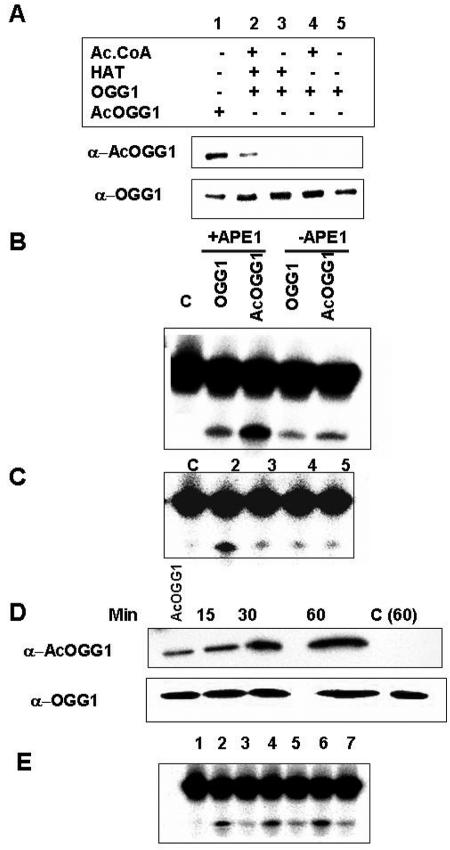
To probe the functional consequence of OGG1 acetylation, we compared DNA glycosylase activity of AcOGG1 and OGG1 with 8-oxoG·C-containing duplex oligonucleotide substrate (29, 32). In this conventional assay which measures both base excision and AP lyase activity, the assay mixture after incubation was treated with alkali, and the cleaved DNA product was separated from the substrate by denaturing gel electrophoresis (17). We acetylated recombinant OGG1 with p300 (HAT domain) and confirmed acetylation by Western analysis with AcOGG1 antibody (Fig. 6A, lane 2). For use as controls, we incubated recombinant OGG1 in the absence of either AcCoA or p300 HAT domain (Fig. 6A, lanes 3 and 4). The nicking assay with 8-oxoG-containing oligonucleotide substrate revealed moderate enhancement (1.5-fold) of 8-oxoG excision and strand cleavage by AcOGG1 in the absence of APE1, whereas in the presence of equimolar amounts of APE1 about fourfold-higher enzymatic activity was observed for AcOGG1 than for unmodified enzyme (Fig. 6B). To further confirm that enhanced 8-oxoG cleavage was due to acetylation of OGG1, we incubated OGG1 in the absence of AcCoA or p300 and did not observe any increase in activity relative to OGG1 alone (Fig. 6C, lanes 3 to 5). Furthermore, we incubated OGG1 with p300 (HAT domain) and AcCoA for various times and analyzed the extent of acetylation with AcOGG1 antibody (Fig. 6D). Figure 6E shows that the 8-oxoG·C oligonucleotide strand incision activity in the presence of APE1 was proportional to the level of acetylation.
FIG. 6.
Incision activity of OGG1 and AcOGG1. A. OGG1 (500 ng) was incubated with p300 HAT domain (0.1 μg) with or without 1 mM AcCoA (lanes 2 and 3) or heat-inactivated HAT domain (lane 4) for 45 min at 30°C, and then 30 ng of protein was used for immunoblotting with either acetylated OGG1 (upper panel) or OGG1 antibody (lower panel). Lanes 1 and 5, AcOGG1 and OGG1 markers. B. Unmodified OGG1 (1 ng) or AcOGG1 as in panel A was incubated with 500 nM 32P-labeled oligonucleotide at 37°C for 20 min in the presence of equimolar amounts of APE1, and the cleaved products were analyzed in an 18% urea-polyacrylamide gel. Lane C, no protein. C. OGG1 incubated with p300 HAT domain with or without AcCoA (lanes 2 and 3, respectively) or inactivated HAT domain (lane 4), in panel A, was used for 8-oxoG incision assay in the presence of APE1. D. OGG1 (500 ng) was incubated at 30°C with p300 HAT domain (0.1 μg) together with1 mM AcCoA for various times and used for Western analysis with AcOGG1 antibody. AcOGG1, marker; C (60), OGG1 without p300 for 1 h. Lower panel, Western blot analysis with OGG1 antibody. E. Incision activity of OGG1 incubated with p300 and AcCoA for 15 min (lane 2), 30 min (lane 4), and 60 min (lane 6) or without p300 (lanes 3, 5, and 7, respectively). Lane 1, no protein.
The stimulation of glycosylase activity due to acetylation of OGG1 was further supported by comparing activities of acetylated and nonmodified OGG1 purified from HeLa cells. Based on quantitative analysis of OGG1 and AcOGG1 levels in PC200 and PC250 fractions as described before, equal amounts and OGG1 or AcOGG1 were used for measuring OGG1 activity after adjusting NaCl concentration to the same level. In view of potential complications due to contaminating APE1 in these phosphocellulose fractions, which would stimulate OGG1 activity to a variable extent, we carried out 8-oxoG excision/strand incision assays in the presence of a 10-fold molar excess of E. coli Nfo and 2 mM EDTA (32, 62). EDTA inactivates endogenous APE1 but not Nfo (36). A significantly higher base excision activity was again observed for AcOGG1 relative to unmodified OGG1 (Fig. 5D). AP lyase activity of glycosylases forms a transient Schiff base adduct with deoxyribose at the AP site, which could then be converted into a stable “trapped complex” by reduction with NaCNBH3 (17). DNA glycosylases can be distinguished from one another in a mixture by the characteristic mobility of their trapped complexes in SDS-PAGE (29). In order to confirm that the excision activity was solely due to OGG1, we performed a trapping assay by adding NaCNBH3 to the reaction mixture and omitting Nfo (17). Formation of a single trapped complex with the PC200 or PC250 which has the same mobility as that of recombinant OGG1 confirmed that OGG1 is the only detectable AP lyase in either fraction (data not shown). At the same time, a smaller amount of trapped complex was observed for AcOGG1 than for OGG1. Because the trapped complex is generated by reduction of the transient Schiff base formed between the enzyme and the AP site aldehyde, it appears likely that acetylation reduced OGG1's AP lyase activity (17; data not shown). These results were further supported by the weaker affinity of AcOGG1 than of the unmodified enzyme for AP site-containing DNA as described below.
Acetylation reduces OGG1's affinity for both product and substrate.
Stimulation of OGG1 activity after its acetylation suggests two possible mechanisms: acetylation could reduce OGG1's affinity for the product AP·C site or increase its affinity for the 8-oxoG·C substrate. To distinguish between these possibilities, we quantitated relative affinities of catalytically inactive OGG1 or AcOGG1 for the AP site versus 8-oxoG, using electrophoretic mobility shift assay. Acetylation reduced the affinity of K249Q OGG1 mutant for the AP·C oligonucleotide and 8-oxoG·C oligonucleotide by five- and twofold, respectively (data not shown). Similar results were obtained with endogenous active enzymes (data not shown).
Impact of OGG1 acetylation on 8-oxoG repair in the cellular genome.
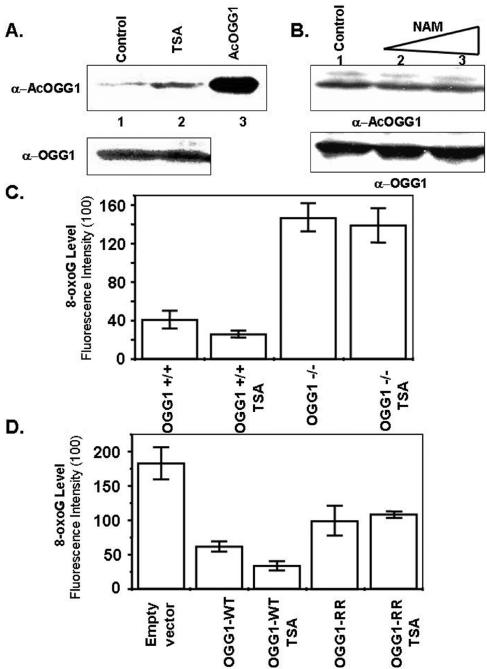
As a follow-up to the above studies, we used two approaches to test whether OGG1 acetylation enhances repair of 8-oxoG (and other substrates) in cellular genomes. First, we treated HCT116 cells with TSA, a specific HDAC inhibitor, which caused a ∼4-fold increase in the amount of AcOGG1 (Fig. 7A) at 12 h after treatment, without changing the total OGG1 level (65) (Fig. 7A, lower panel). The yeast silent information regulator 2 (Sir2) protein belongs to a family of histone deacetylases whose activity is NAD dependent and cannot be inhibited by TSA (34). Human SIRT1 is homologous to yeast Sir2 and was shown to deacetylate p53 and promote survival of cells exposed to stress (37). To test whether mammalian SIRTs are responsible for OGG1 deacetylation in vivo, we treated the cells with nicotinamide (NAM), an inhibitor of SIRT1 (37, 61). Western analysis of extracts of HCT116 cells treated with different doses of NAM did not show any significant change in the AcOGG1 level (Fig. 7B). This indicates that TSA-sensitive HDACs and not SIRTs are involved in deacetylation of AcOGG1 in vivo. We quantitated the relative abundance of 8-oxoG in cell nuclei based on immunofluorescence with an 8-oxoG-specific antibody (8, 13). TSA treatment caused a modest reduction in the basal 8-oxoG level in the genome (data not shown), suggesting that enhanced acetylation of OGG1 was associated with an increased rate of repair of endogenous 8-oxoG. Because TSA is known to stimulate gene expression globally, it is possible that enzymes other than OGG1, also activated by TSA, were responsible for enhanced repair of 8-oxoG (60). We eliminated this possibility by incubating OGG1+/+ and OGG-null MEFs with TSA and quantitated the relative abundance of 8-oxoG in cell nuclei as before. Figure 7C shows that the 8-oxoG level in OGG1-null MEFs was significantly higher than that in WT MEFs. This supports a previous observation showing that OGG1 is the major enzyme for 8-oxoG repair (46). This was further supported by the observation that TSA treatment reduced the basal 8-oxoG level in OGG1+/+ MEFs without affecting its level in OGG1-null MEFs. To show conclusively that the enhanced rate of 8-oxoG removal in TSA-treated cells is mediated by AcOGG1, we transfected OGG1−/− MEFs with FLAG-tagged WT OGG1 or its nonacetylable K338R/K341R mutant and showed enhanced repair of 8-oxoG in WT OGG1-transfected cells relative to the mutant-expressing MEFs (Fig. 7D). Additionally, TSA treatment caused a further reduction in the level of 8-oxoG in WT OGG1-transfected cells without a similar effect in K338R/K341R-transfected cells (Fig. 7D). This provided direct evidence that acetylation of OGG1 increases its 8-oxoG repair activity in vivo.
FIG. 7.
Effect of TSA on the level of AcOGG1 and 8-oxoG repair in OGG+/+ and OGG−/− MEF cells. A. HCT116 cells were either treated with TSA (100 ng/ml) for 12 h (lane 2) or mock treated (lane 1), and then cell extracts were immunoblotted with AcOGG1 (upper panel) or OGG1 (lower panel) antibody. Lane 3, AcOGG1 marker. B. HCT116 cells were treated with NAM (1 mM, lane 2; 5 mM, lane 3) for 12 h or mock treated (lane 1), and then cell extracts were immunoblotted with AcOGG1 (upper panel) or OGG1 (lower panel) antibodies. C. OGG1-null and WT MEFs were treated with TSA (12 h), and immunofluorescence of 8-oxoG in the genome was quantitated with 8-oxoG-specific antibody conjugated with fluorescein isothiocyanate. D. OGG1−/− MEFs were transfected with FLAG WT OGG1 or FLAG K338R/K341R mutant (OGG1 RR) or empty vector. Thirty-six hours after transfection, cells were mock treated or treated with TSA (12 h) and immunofluorescence of 8-oxoG was quantitated as before with fluorescein isothiocyanate. Other details are described in Materials and Methods.
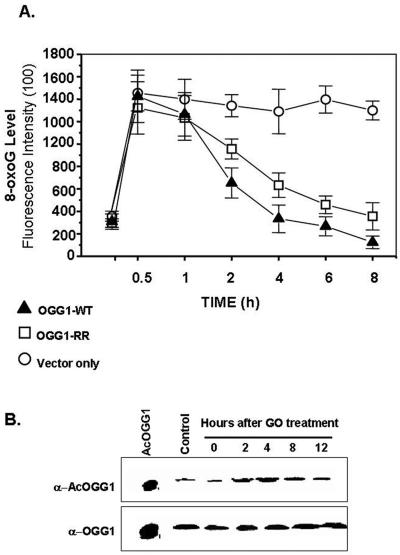
We then transfected OGG1−/− MEFs with expression plasmids for FLAG-tagged WT OGG1, K338R/K341R mutant, or empty vector; incubated the cells with glucose oxidase for 1 h; and then measured 8-oxoG levels at various times. GO induces cellular oxidative stress due to generation of O2·− radical (55). Although GO treatment increased nuclear 8-oxoG levels in both WT OGG1- and K338R/K341R mutant-transfected OGG-null MEFs, the rate of removal of 8-oxoG in WT OGG1-expressing cells was significantly higher than that in the mutant OGG1-expressing cells (Fig. 8A). As expected the 8-oxoG level in the cells was unchanged after transfection with the empty vector. We conclude from these data that acetylation of OGG1 was responsible for the higher rate of repair of genomic 8-oxoG in oxidatively stressed cells.
FIG. 8.
Effect of acetylation of OGG1 on 8-oxoG repair in OGG1−/− MEFs after oxidative stress. A. Immunofluorescence in OGG1−/− cells transfected with FLAG-tagged WT OGG1 (▴) or FLAG-tagged K338R/K341R OGG1 (□) or empty vector (○). B. Effect of GO treatment. AcOGG1 or OGG1 levels were quantitated by Western analysis with 200 μg nuclear extract of GO-treated cells after treatment with GO.
We then tested whether oxidative stress enhances OGG1 acetylation. Western analysis of GO-treated HeLa cells revealed no significant change in OGG1 polypeptide level (Fig. 8B, lower panel). However, the AcOGG1 level was increased 2.5-fold at 4 h after GO treatment and reverted to the basal value after 12 h (Fig. 8B, upper panel), indicating that OGG1 acetylation is enhanced transiently due to oxidative stress. Such enhancement could be due to ROS-induced increase of the HAT activity of p300 (51).
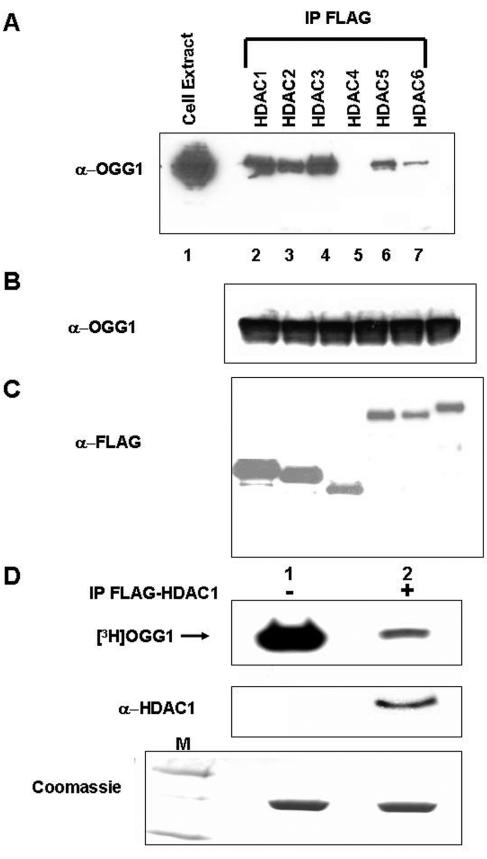
Class I histone deacetylases (HDAC1 to -3) interact with OGG1 in vivo.
Histone acetyltransferases and HDACs acting in opposite directions regulate acetylation levels in target proteins. Several mammalian HDACs have been shown to be responsible for removing acetyl groups from various transcription factors (22). With the expectation that AcOGG1 is deacetylated by a HDAC, we examined association between OGG1 and the classical HDACs. After cotransfecting HCT 116 cells with expression plasmids of OGG1 and FLAG-tagged human HDAC1 through -6, one at a time, we immunoprecipitated HDACs with FLAG antibody. The levels of ectopically expressed HDACs and OGG1 were determined in individually transfected cells (Fig. 9B and C). Western analysis showed a significant presence of OGG1 in the immunoprecipitates of class I HDACs (HDAC1 to -3), but only weak interaction was observed with class II HDACs (HDAC4 to -6) (Fig. 9A). We therefore conclude that OGG1 forms stable complexes primarily with class I HDACs which are likely to be responsible for its deacetylation. We transfected HCT116 cells with FLAG-tagged HDAC1 and immunoprecipitated HDAC1 with FLAG antibody cross-linked to agarose beads. Incubation of immunoprecipitated FLAG-HDAC1 (eluted by FLAG peptide) with in vitro-acetylated [3H]OGG1 followed by SDS-PAGE and fluorography showed significant reduction of radioactivity in the [3H]OGG1 band (Fig. 9D, lane 2) with the immunoprecipitate of HDAC1 but not with the immunoprecipitate from empty FLAG vector-transfected cells (Fig. 9D, lane 1). Coomassie blue staining of the gel confirmed that comparable amounts of [3H]OGG1 were used in two cases (lower panel). This provided direct evidence that HDAC1 can deacetylate OGG1 in vitro.
FIG. 9.
In vivo interaction of OGG1 and HDACs. A. HCT116 cells were transfected with expression plasmids for OGG1 and FLAG-tagged HDAC1 through HDAC6. FLAG immunoprecipitates of FLAG antibody were analyzed for OGG1. Lane 1, cell extract. B. Western analysis for OGG1. C. Western analysis for HDACs. D. In vitro-acetylated [3H]OGG1 (2 μg) was incubated with FLAG immunoprecipitates of FLAG-tagged HDAC1-transfected cells (lane 2) or empty vector-transfected cells (lane 1) at 30°C in HAT assay buffer for 45 min and analyzed by SDS-PAGE and fluorography. Middle panel, immunoblotting with HDAC1 antibody. Lower panel, Coomassie blue staining of input OGG1in a duplicate gel. Lane M, molecular weight markers.
DISCUSSION
Posttranslational modifications are being increasingly documented for a variety of proteins. While some modifications, e.g., polyubiquitination, may target proteins for degradation, others (e.g., phosphorylation) modulate proteins' enzymatic activities, their interaction with other partner proteins or nucleic acids, or nuclear targeting. The histones were first shown to be acetylated by transcription coactivators p300/CBP, and consequent unfolding of the chromatin is a prerequisite to activation of transcription. p300/CBP were subsequently shown to acetylate a number of transcription factors as well as proteins involved in DNA replication/repair (25, 27). We and others have found that several BER enzymes are acetylated by p300/CBP (3, 4, 58). Here we show that OGG1, the major DNA glycosylase responsible for repair of 8-oxoG and FapyG, is also acetylated by p300 (and possibly CBP). The evidence for OGG1 acetylation by p300 includes (i) their stable interaction, as indicated by nuclear colocalization and coimmunoprecipitation; (ii) the observation that overexpression of p300 increases the AcOGG1 level; and (iii) the observation that reduction of cellular AcOGG1 level occurs after downregulation of p300 by siRNA. While other proteins with HAT activity, e.g., P/CAF, could also acetylate OGG1 to a small extent in vitro (data not shown), it appears that OGG1 is acetylated in vivo primarily by p300.
We have shown that the same Lys residues in the OGG1 polypeptide are acetylated both in vivo and in vitro. Our results further indicate that about a fifth of OGG1 is present in the acetylated form in HeLa cells under normal conditions. This characterization became possible because of our success in generating AcOGG1 antibody with high specificity and strong discrimination against unmodified OGG1. We were thus able to identify and quantitate acetylated and unmodified OGG1 after its chromatographic separation from HeLa cell extracts. Nearly comparable specific activity of partially purified, endogenous AcOGG1 and in vitro-acetylated recombinant OGG1 provides further support for physiological relevance of OGG1 acetylation.
It is interesting that the acetyl acceptor residues Lys338 and Lys341, which we identified via site-directed mutagenesis of OGG1, were absent in the C-terminally deleted OGG1 whose structure was solved by X-ray crystallography (10). This suggests that the C-terminal domain including the acetyl acceptor Lys residues, which is dispensable for enzymatic activity, exists in an unstructured form. Such flexibility may be important for acetylation which could affect interaction of OGG1 with substrate DNA and/or other proteins in the repair pathway. The structures of both active OGG1 and catalytically inactive mutant bound to an 8-oxoG·C pair indicate that the enzyme undergoes extensive local conformational change after DNA binding (10, 20). Reduction of DNA affinity associated with deletion of C-terminal residues 328 to 345 suggests interaction of these residues with DNA (10). We tested whether enhanced activity of OGG1 after acetylation is due to its altered affinity for the substrate or to that for product DNA. Although acetylation reduced OGG1's affinity for both the substrate and product, the effect was much higher for the AP·C product than for the 8-oxoG·C substrate. Because the rate-limiting step in 8-oxoG excision is dissociation of OGG1 from the product AP site (32), it is reasonable to postulate that acetylation enhances OGG1's turnover by weakening its interaction with the AP product. Consistently, we observed 1.5-fold-higher specific activity of AcOGG1 than of the unmodified enzyme. However, the activity was further stimulated in the presence of APE1 (Fig. 6B). These results suggest that the unstable AP site after 8-oxoG excision remains bound to OGG1 in order to prevent its nonspecific degradation. Displacement of OGG1 from the bound AP site by APE1 is enhanced when OGG1 is acetylated and its affinity for the AP site is reduced. Similar stimulation of base excision activity due to enhanced turnover after posttranslational sumoylation of TDG was reported earlier (24).
The physiological significance of OGG1 acetylation became evident from the in vivo repair studies. Proficient repair of 8-oxoG in OGG1+/+ MEFs and significant accumulation of 8-oxoG in the genomes of OGG1-null MEFs confirmed earlier studies (46) and are consistent with the general belief that OGG1 is the major repair enzyme for 8-oxoG in mammalian cells (Fig. 7C). Enhanced repair of 8-oxoG, observed in the genome of OGG1−/− MEFs after ectopic expression of WT OGG1 or nonacetylable R338/R341 mutant, indicates that the mutant OGG1 retains 8-oxoG repair activity in vivo. However, when the transfected cells were subsequently treated with TSA, which enhanced the acetylation level of only the wild-type enzyme, a significant increase in the 8-oxoG repair rate was observed in cells expressing WT OGG1 but not the mutant enzyme (Fig. 7D), while the levels of total OGG1 were similar. These results support our conclusion that increased 8-oxoG repair is due to enhanced AcOGG1 level. We quantitated the cellular 8-oxoG level by immunofluorescence using 8-oxoG-specific antibody. Although 8-oxoG fluorescence was mostly nuclear, this analysis might have some inherent error because of the presence of 8-oxoG in the mitochondrial DNA. However, this error should be rather small because of the way we treated cells as described in Materials and Methods, which was also used by others (13, 38, 59).
A large and rapid increase in the 8-oxoG level was observed in MEFs after GO treatment (Fig. 8A). The low rate of its removal in OGG1-null MEFs, transfected with an empty vector, provides further evidence for the key role of OGG1 in repairing both endogenous and induced 8-oxoG in cellular genomes. Complete repair of 8-oxoG took about 8 h in cells expressing WT OGG1 or R338/R341 mutant, similar to that observed in other studies (38, 39, 59). More importantly, the higher rate of 8-oxoG repair in WT OGG1-expressing cells than in nonacetylable mutant-expressing cells after GO treatment further confirms the role of OGG1 acetylation in 8-oxoG repair in oxidatively stressed cells (Fig. 8A). Interestingly, we observed transient enhancement of OGG1 acetylation following oxidative stress (Fig. 8B), which is likely to be due to oxidative stress-mediated enhancement of HAT activity of p300, as reported earlier (51). Thus, oxidative stress raised the level of AcOGG1, concurrently with an increase in the level of 8-oxoG (and other damaged bases). We should point out that the OGG1 polypeptide level was not affected by oxidative stress (Fig. 8B) (42).
Taking these results together, we propose that OGG1-mediated repair of 8-oxoG (and FapyG or other substrate base lesions) is regulated by acetylation of the enzyme, in response to exogenous oxidative stress (which induces additional base damage) and not by changing the polypeptide level. Our surprising observation of discrete AcOGG1 distribution in nuclear foci, compared to homogenous distribution of unmodified OGG1 (Fig. 4E), suggests that the modified enzyme is preferentially localized in large repair complexes.
The lack of acetyl acceptor Lys residues in the mitochondrial OGG1-β variant implies absence of acetylation-dependent regulation of 8-oxoG repair in the mitochondrial genome. However, recent studies showing the presence of nuclear OGG1 in mitochondria raise the possibility of acetylation-dependent regulation of OGG1 activity in the mitochondria as well (28).
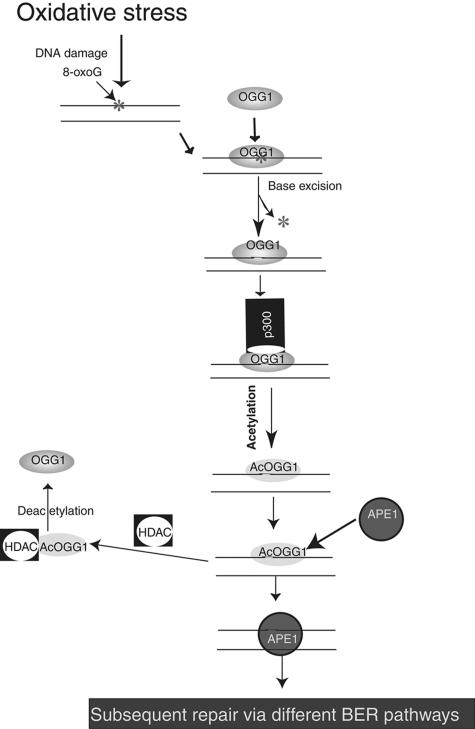
Based on the above studies, we propose a model for regulation of OGG1 acetylation in response to oxidative stress (Fig. 10), by postulating that OGG1 acetylation is not critical for repairing the basal, endogenous level of 8-oxoG. However, in the event of ROS-induced excess production of 8-oxoG and other damaged bases, an immediate increase in 8-oxoG repair activity is warranted which could be effected by acetylating OGG1, without increasing the amount of the protein. In this scenario, OGG1 remaining bound to the product AP site is preferentially acetylated by p300 whose HAT activity is in turn enhanced by ROS. Acetylation of OGG1 increases its turnover due to reduced affinity for the product. Eventually, homeostasis is restored when AcOGG1 is deacetylated by a HDAC. Stable interaction between OGG1 and group I HDACs (Fig. 9) is consistent with this possibility.
FIG. 10.
Model for regulation of OGG1's activity by acetylation in oxidatively stressed cells.
Finally, we would like to stress that our studies unravel complex regulation of oxidative damage repair at multiple levels. We have shown the potential role of acetylation in maintaining cellular homeostasis, particularly in response to oxidative stress. Although other modifications including phosphorylation may also modulate activity and other functions of OGG1 (14, 33), we have provided conclusive evidence that acetylation of OGG1 acts as a regulatory switch in responding to oxidative stress.
Acknowledgments
We thank B. Vogelstein for the HCT116 line, D. Barnes for OGG1-null MEFs, D. Chakravarti for p300 HAT expression plasmid, J. Wang for p300 HAT mutant expression plasmid, and D. Liebenthal in the UTMB NIEHS Center Cell Biology Core for p300 recombinant baculovirus and Sf9 cells. We are grateful to J. S. Smith and A. Kurosky of UTMB's Biomolecular Resources Facility for peptide synthesis and characterization of proteins. We also thank Wanda Smith for expert secretarial assistance.
This work was supported in part by USPHS grants R01 CA 81063, R01 CA53791, and p50 ES 06676.
REFERENCES
- 1.Ames, B. N., M. K. Shigenaga, and T. M. Hagen. 1993. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 90:7915-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 3.Bhakat, K. K., T. K. Hazra, and S. Mitra. 2004. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 32:3033-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhakat, K. K., T. Izumi, S.-H. Yang, T. K. Hazra, and S. Mitra. 2003. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 22:6299-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjoras, M., L. Luna, B. Johnsen, E. Hoff, T. Haug, T. Rognes, and E. Seeberg. 1997. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 16:6314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux, S., T. R. O'Connor, and J. Laval. 1987. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 6:3177-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux, S., and J. P. Radicella. 2000. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 377:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Boldogh, I., D. Milligan, M. S. Lee, H. Bassett, R. S. Lloyd, and A. K. McCullough. 2001. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 29:2802-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breen, A. P., and J. A. Murphy. 1995. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 18:1033-1077. [DOI] [PubMed] [Google Scholar]
- 10.Bruner, S. D., D. P. Norman, and G. L. Verdine. 2000. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403:859-866. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, K. C., D. S. Cahill, H. Kasai, S. Nishimura, and L. A. Loeb. 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J. Biol. Chem. 267:166-172. [PubMed] [Google Scholar]
- 13.Conlon, K. A., D. O. Zharkov, and M. Berrios. 2003. Immunofluorescent localization of the murine 8-oxoguanine DNA glycosylase (mOGG1) in cells growing under normal and nutrient deprivation conditions. DNA Repair (Amsterdam) 2:1337-1352. [DOI] [PubMed] [Google Scholar]
- 14.Dantzer, F., L. Luna, M. Bjoras, and E. Seeberg. 2002. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 30:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, A., T. K. Hazra, I. Boldogh, S. Mitra, and K. K. Bhakat. 2005. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J. Biol. Chem. 280:35272-35280. [DOI] [PubMed] [Google Scholar]
- 16.Dizdaroglu, M. 1991. Chemical determination of free radical-induced damage to DNA. Free Radic. Biol. Med. 10:225-242. [DOI] [PubMed] [Google Scholar]
- 17.Dodson, M. L., R. D. Schrock III, and R. S. Lloyd. 1993. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry 32:8284-8290. [DOI] [PubMed] [Google Scholar]
- 18.Dou, H., S. Mitra, and T. K. Hazra. 2003. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278:49679-49684. [DOI] [PubMed] [Google Scholar]
- 19.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 20.Fromme, J.C., S. D. Bruner, W. Yang, M. Karplus, and G. L. Verdine. 2003. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol. 10:204-211. [DOI] [PubMed] [Google Scholar]
- 21.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 22.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 23.Grollman, A. P., and M. Moriya. 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9:246-249. [DOI] [PubMed] [Google Scholar]
- 24.Hardeland, U., R. Steinacher, J. Jiricny, and P. Schar. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21:1456-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasan, S., N. El-Andaloussi, U. Hardeland, P. O. Hassa, C. Burki, R. Imhof, P. Schar, and M. O. Hottiger. 2002. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol. Cell 10:1213-1222. [DOI] [PubMed] [Google Scholar]
- 26.Hasan, S., P. O. Hassa, R. Imhof, and M. O. Hottiger. 2001. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature 410:387-391. [DOI] [PubMed] [Google Scholar]
- 27.Hasan, S., M. Stucki, P. O. Hassa, R. Imhof, P. Gehrig, P. Hunziker, U. Hubscher, M. O. Hottiger. 2001. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell 7:1221-1231. [DOI] [PubMed] [Google Scholar]
- 28.Hashiguchi, K., J. A. Stuart, N. C. de Souza-Pinto, and V. A. Bohr. 2004. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 32:5596-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazra, T. K., T. Izumi, L. Maidt, R. A. Floyd, and S. Mitra. 1998. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 26:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazra, T. K., T. Izumi, I. Boldogh, B. Imhoff, Y. W. Kow, P. Jaruga, M. Dizdaroglu, and S. Mitra. 2002. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA 99:3523-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazra, T. K., Y. W. Kow, Z. Hatahet, B. Imhoff, I. Boldogh, S. K. Mokkapati, S. Mitra, and T. Izumi. 2002. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277:30417-30420. [DOI] [PubMed] [Google Scholar]
- 32.Hill, J. W., T. K. Hazra, T. Izumi, and S. Mitra. 2001. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 29:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, J., S. Z. Imam, K. Hashiguchi, N. C. de Souza-Pinto, and V. A. Bohr. 2005. Phosphorylation of human oxoguanine DNA glycosylase (α-OGG1) modulates its function. Nucleic Acids Res. 33:3271-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. A., A. Dritschilo, and M. Jung. 2001. Role of ATM in oxidative stress-mediated c-Jun phosphorylation in response to ionizing radiation and CdCl2. J. Biol. Chem. 276:11783-11790. [DOI] [PubMed] [Google Scholar]
- 36.Ljungquist, S. 1977. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem. 252:2808-2814. [PubMed] [Google Scholar]
- 37.Luo, J., A. Y. Nikolaev, S. Imai, D. L. Chen, F. Su, A. Shilh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 38.Masalimov, Z. H. K., L. V. Malakhova, V. I. Bruskov, V. S. Bogatova, and A. I. Gaziev. 2003. Study of formation and elimination of 8-oxoguanine in DNA of the liver and brain of mice after gamma-irradiation. Radiatsionnaya Biol. Radioekol. 43:658-661. (In Russian.) [PubMed] [Google Scholar]
- 39.Mei, N., K. Tamae, N. Kunugita, T. Hirano, and H. Kasai. 2003. Analysis of 8-hydroxydeoxyguanosine 5′-monophosphate (8-OH-dGMP) as a reliable marker of cellular oxidative DNA damage after gamma-irradiation. Environ. Mol. Mutagen. 41:332-338. [DOI] [PubMed] [Google Scholar]
- 40.Melvin, T., S. M. Cunniffe, P. O'Neill, A. W. Parker, and T. Roldan-Arjona. 1998. Guanine is the target for direct ionisation damage in DNA, as detected using excision enzymes. Nucleic Acids Res. 26:4935-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mistry, P., and K. E. Herbert. 2003. Modulation of hOGG1 DNA repair enzyme in human cultured cells in response to pro-oxidant and antioxidant challenge. Free Radic. Biol. Med. 35:397-405. [DOI] [PubMed] [Google Scholar]
- 43.Mitra, S., T. K. Hazra, R. Roy, S. Ikeda, T. Biswas, J. Lock, I. Boldogh, and T. Izumi. 1997. Complexities of DNA base excision repair in mammalian cells. Mol. Cells 7:305-312. [PubMed] [Google Scholar]
- 44.Mokkapati, S. K., L. Wiederhold, T. K. Hazra, and S. Mitra. 2004. Stimulation of DNA glycosylase activity of OGG1 by NEIL1: functional collaboration between two human DNA glycosylases. Biochemistry 43:11596-11604. [DOI] [PubMed] [Google Scholar]
- 45.Morland, I., V. Rolseth, L. Luna, T. Rognes, M. Bjoras, and E. Seeberg. 2002. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 30:4926-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura, S. 2002. Involvement of mammalian OGG1(MMH) in excision of the 8-hydroxyguanine residue in DNA. Free Radic. Biol. Med. 32:813-821. [DOI] [PubMed] [Google Scholar]
- 47.Nishioka, K., T. Ohtsubo, H. Oda, T. Fujiwara, D. Kang, K. Sugimachi, and Y. Nakabeppu. 1999. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 49.Porello, S. L., A. E. Leyes, and S. S. David. 1998. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry 37:14756-14764. [DOI] [PubMed] [Google Scholar]
- 50.Radicella, J. P., C. Dherin, C. Desmaze, M. S. Fox, and S. Boiteux. 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:8010-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman, I. 2002. Oxidative stress, transcription factors and chromatin remodelling in lung inflammation. Biochem. Pharmacol. 64:935-942. [DOI] [PubMed] [Google Scholar]
- 52.Roldan-Arjona, T., Y. F. Wei, K. C. Carter, A. Klungland, C. Anselmino, R. P. Wang, M. Augustus, and T. Lindahl. 1997. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 94:8016-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenquist, T. A., D. O. Zharkov, and A. P. Grollman. 1997. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 94:7429-7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349:431-434. [DOI] [PubMed] [Google Scholar]
- 55.Song, J. Y., J. W. Lim, H. Kim, T. Morio, and K. H. Kim. 2003. Oxidative stress induces nuclear loss of DNA repair proteins Ku70 and Ku80 and apoptosis in pancreatic acinar AR42J cells. J. Biol. Chem. 278:36676-36687. [DOI] [PubMed] [Google Scholar]
- 56.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tchou, J., H. Kasai, S. Shibutani, M. H. Chung, J. Laval, A. P. Grollman, and S. Nishimura. 1991. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. USA 88:4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tini, M., A. Benecke, S. J. Um, J. Torchia, R. M. Evans, and P. Chambon. 2002. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9:265-277. [DOI] [PubMed] [Google Scholar]
- 59.Tsuruya, K., M. Furuichi, Y. Tominaga, M. Shinozaki, M. Tokumoto, T. Yoshimitsu, K. Fukuda, H. Kanai, H. Hirakata, M. Iida, and Y. Nakabeppu. 2003. Accumulation of 8-oxoguanine in the cellular DNA and the alteration of the OGG1 expression during ischemia-reperfusion injury in the rat kidney. DNA Repair (Amsterdam) 2:211-229. [DOI] [PubMed] [Google Scholar]
- 60.Van Lint, C. S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 61.Vaziri, H., S. K. Dessain, E. E. Ng, S. Imai, R. A. Frye, T. K. Panditia, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 62.Vidal, A. E., I. D. Hickson, S. Boiteux, and J. P. Radicella. 2001. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 29:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood, M. L., M. Dizdaroglu, E. Gajewski, and J. M. Essigmann. 1990. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry 29:7024-7032. [DOI] [PubMed] [Google Scholar]
- 64.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 66.Zharkov, D. O., T. A. Rosenquist, S. E. Gerchman, and P. Grollman. 2000. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 275:28607-28617. [DOI] [PubMed] [Google Scholar]