Abstract
The r1 and b1 genes of maize, each derived from the chromosomes of two progenitors that hybridized >4.8 million years ago (MYA), have been a rich source for studying transposition, recombination, genomic imprinting, and paramutation. To provide a phylogenetic context to the genetic studies, we sequenced orthologous regions from maize and sorghum (>600 kb) surrounding these genes and compared them with the rice genome. This comparison showed that the homeologous regions underwent complete or partial gene deletions, selective retention of orthologous genes, and insertion of nonorthologous genes. Phylogenetic analyses of the r/b genes revealed that the ancestral gene was amplified independently in different grass lineages, that rice experienced an intragenomic gene movement and parallel duplication, that the maize r1 and b1 genes are descendants of two divergent progenitors, and that the two paralogous r genes of sorghum are almost as old as the sorghum lineage. Such sequence mobility also extends to linked genes. The cisZOG genes are characterized by gene amplification in an ancestral grass, parallel duplications and deletions in different grass lineages, and movement to a nonorthologous position in maize. In addition to gene mobility, both maize and rice regions experienced recent transposition (<3 MYA).
THE grass family (Poaceae) is well known for the economic importance of its members, such as rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), oats (Avena sativa), sorghum (Sorghum spp.), wheat (Triticum spp.), and rye (Secale cereale). Even though nuclear DNA content among members of the family varies extensively, >40-fold (Bennett and Smith 1976; Bennett et al. 2000), comparative mapping studies (Hulbert et al. 1990; Ahn and Tanksley 1993; Moore et al. 1995; Gale and Devos 1998; Feuillet and Keller 2002) have demonstrated that significant portions of the genomes are conserved (collinear) and that, aside from polyploidization, the large size of a genome is mostly due to a high content of repetitive elements (SanMiguel and Bennetzen 1998; Meyers et al. 2001). Large-scale studies using genetic maps based on DNA markers are limited to the detection of large-scale chromosomal rearrangements, such as translocations, intrachromosomal inversions, genome replication, chromosomal fusion, and rearrangements that characterize specific clades. For instance, three translocations marked the ancestral lineage of Panicoideae (Gale and Devos 1998) and genome replication, chromosomal fusion, and rearrangements occurred on the Brassica lineage after its divergence from the Arabidopsis thaliana lineage (Lagercrantz 1998). However, evolutionary forces that lead to speciation of closely related species work locally within the genomes. Recently, to uncover characteristics of local evolution of plant genomes, several researchers compared the genomic content and organization of orthologous regions at the sequence level across various grass species. Regions analyzed to date are the sh2/a1 region (Chen et al. 1997, 1998), the Lrk region (Feuillet and Keller 1999), the adh1/2 region (Tikhonov et al. 1999; Ilic et al. 2003), the 22-kD Zein cluster region (Song et al. 2002), the Vrn1 region (Ramakrishna et al. 2002a), the Rp1 region (Ramakrishna et al. 2002b), the Rph7 region (Brunner et al. 2003), and the lrs/lg region (Langham et al. 2004). Although these studies generally confirmed gene conservation (microcollinearity) among orthologous segments of various grass taxa, they also reported small-scale genic rearrangements, such as gene insertions, deletions, amplifications, inversions, and translocations.
These studies are too few in number to produce a general picture of genome evolution. Only a few of them compare sequences from more than two species and very few studied duplicated (homeologous) regions of polyploid species. Furthermore, evolutionary analytical methods accompanied only a small number of them. Phylogenetic analyses, providing interpretation of the pattern and direction of genome evolution and facilitating reconstruction of the common ancestor, when coupled with small-scale comparative genomic studies, can provide more accurate insight into the history of gene duplication and genic rearrangements. In this respect, rice (with 430 Mb of nuclear DNA per haploid genome), sorghum (750 Mb), and maize (2400 Mb) represent suitable species to study the patterns of local genomic evolution because of their moderate difference (5.8-fold) in genome size and known phylogenetic relationships. Because the divergence of a rice ancestor from the lineage that gave rise to sorghum and maize represents the largest temporal distance within the grass family, ∼50–70 million years ago (MYA; Wolfe et al. 1989), tracing genomic rearrangements between these taxa may allow us to reconstruct the genomic structure of the common ancestor of grasses. The time of sorghum and maize divergence from a common ancestor has been estimated to be 11.9 MYA (Swigoňová et al. 2004). An additional benefit is that the rice genome has largely been sequenced, with its chromosomes now available as 12 pseudomolecules (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml).
Here, we focus on the r/b chromosomal region mostly because the r1 and b1 genes of maize have been subjects of many genetic studies (Neuffer et al. 1997) and a comprehensive analysis of molecular evolution of the gene family in grasses was lacking. The r/b genes, together with members of the c/pl gene family, coregulate transcription of structural genes in the anthocyanin biosynthetic pathway (Ludwig and Wessler 1990; Dooner et al. 1991). Red and purple anthocyanins, the common plant pigments, are produced in a wide variety of tissues where they serve many different functions, such as attracting pollinators and dispersal agents and protecting against insects, pathogens, and UV light. Biochemical and molecular studies showed that one functional allele from each gene family is required for activation of the structural genes of the anthocyanin biosynthetic pathway and that the pigmentation pattern of a plant tissue reflects allelic constitution at both of the regulatory loci (Dooner 1983; Cone et al. 1986; Goff et al. 1991, 1992; Bodeau and Walbot 1992; Tuerck and Fromm 1994; Sainz et al. 1997; Grotewold et al. 1998; Lesnick and Chandler 1998).
The r/b gene family in maize is represented by the R locus (Dellaporta et al. 1988) on the long arm of chromosome 10 (here referred to as maize-10L), the B locus (Chandler et al. 1989) on the short arm of chromosome 2 (maize-2S), and displaced genes, such as the lc (Ludwig et al. 1989) and sn (Consonni et al. 1992) genes located ∼2 cM distal to the R locus (Dooner and Kermicle 1976; Gavazzi et al. 1990) and the hopi gene located 4.5 cM distal to the R locus (Petroni et al. 2000). (To obtain consistency throughout the manuscript, despite the previously published form, we use uppercase letters for loci and lowercase letters for genes.) Depending on the maize variety, the R locus may contain one (Dellaporta et al. 1988) or up to four r genes (Eggleston et al. 1995). Unlike the R locus, the B locus contains only one gene, the b1 gene (Patterson et al. 1991). Both the r1 and the b1 genes in maize can induce tissue pigmentation in other plants, such as wheat, barley, sorghum, petunia, Arabidopsis, Nicotiana, and Lycopersicon (Lloyd et al. 1992; Bilang et al. 1993; Casas et al. 1993; Quattrocchio et al. 1993; Galway et al. 1994).
In this study, we present a comparison of genomic content of orthologous chromosomal regions containing the r/b gene homologs from rice, maize, and sorghum. We examine which types of genome evolution resulted in the observed genomic structure of these regions across the three species and between the two duplicated (homeologous) regions of maize. We present a detailed analysis of molecular evolution of the r/b gene family in grasses and examine the evolutionary pattern of gene duplication of the tightly linked cis-zeatin O-glucosyltransferase (cisZOG) gene. We also examine the content and history of retrotransposition within these regions.
MATERIALS AND METHODS
Bacterial artificial chromosome isolation and mapping of r/b homologs:
An r1 probe obtained from maize by PCR of the lc gene sequence (GenBank accession no. M26227) was used to screen high-density filters for maize (Zea mays L. cv B73) and sorghum (Sorghum bicolor cv BTx623) bacterial artificial chromosome (BAC) libraries (Yim et al. 2002; http://www.tamu.edu/bacindex.html). Positive clones were digested by NotI and HindIII for fingerprint analysis and further analyzed by restriction mapping. The purified BAC DNA, isolated using a BAC DNA isolation kit (QIAGEN, Valencia, CA), was physically sheared and ligated into a pUC vector for shotgun libraries, as previously described by Song et al. (2001). Sequencing was done on an ABI 3700 capillary sequencer using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied BioSystems, Foster City, CA). Base calling and assembly were based on phred/phrap programs (Ewing et al. 1998). About 10× coverage was generated for all of the BACs and sequence gaps were finished by specific primers identified with the help of consed (Gordon et al. 1998) or by transposon minilibraries, constructed according to the manufacturer's instructions (Finnzyme, Helsinki).
Sequence analysis:
Genes predicted by FGENESH (http://www.softberry.com/berry.phtml? topic=gfind&prg=FGENESH) and GENSCAN (http://genes.mit.edu/GENSCAN.html), using the monocots option, were further analyzed by homology searches using a variety of BLAST searches (Altschul et al. 1997) against GenBank nucleotide, protein, and EST databases. The intergenic regions were analyzed separately for the presence of transposable elements using homology-based searches against GenBank and TIGR Gramineae repeat database (http://tigrblast.tigr.org/euk-blast/index.cgi?project=plant.repeats).
Genomic sequences of the r/b homologs identified within the studied genomic intervals were aligned against the published cDNA of the lc gene (Ludwig et al. 1989) using the BESTFIT program of the Wisconsin GCG package (Devereux et al. 1984). Predicted ORFs of the cisZOG gene were aligned against the published cDNA of cisZOG1 and cisZOG2 genes of maize (AF318075 and AY082660, respectively). Multiple sequence alignment of full-length coding sequences performed by ClustalX (Thompson et al. 1997) was manually adjusted with reference to amino acid alignment using MacClade 4 (Maddison and Maddison 2000). In addition to the r/b genes identified within the studied intervals, 8 full-length and 15 partial sequences of r/b homologs were used from GenBank: maize lc (M26227), sn (X60706), hopi (AJ251719), rs (X15806), and b-Peru (X57276); rice plw-OSB1 (AB021079), plw-OSB2 (AB021080), ra (U39860–65), and rb (AP003310, U39867, U39868, U39866); Phyllostachys acuta r gene (U11448); Pennisetum glaucum r genes (U11444–47); and Tripsacum australe r gene (U11449).
Phylogenetic analyses and rate estimation:
Coding sequences of the r/b homologs corresponding to part of the second through part of the ninth exons of the lc gene and the entire coding sequence of the cisZOG homologs were used for phylogenetic analyses and rate estimations. Phylogenetic analyses were performed using PAUP* 4.0b10 (Swofford 1999). Maximum parsimony (MP) analyses were performed with branch-and-bound search, and maximum likelihood (ML) analyses were performed using heuristic searches with 100 random taxon addition replicates with TBR branch swapping. Evolutionary models used in ML analyses were selected using Modeltest 3.06 (Posada and Crandall 1998) with the Akaike Information Criterion. In the case of the r/b homologs, we also performed Bayesian analysis using MrBayes v3.0b4 (Huelsenbeck and Ronquist 2001) on data partitioned according to variable and conserved regions (see below). Support for individual nodes within tree topologies was evaluated by a nonparametric bootstrap analysis (Felsenstein 1985) performed with 100 pseudoreplicates with 10 random taxon addition heuristic searches in each pseudoreplicate. Synonymous and nonsynonymous distances were estimated by the maximum likelihood method implemented in codeml of PAML (Yang 1997) under the F3x4 model (Goldman and Yang 1994). Gene rates at synonymous sites were calculated as described by Swigoňová et al. (2004). For the cisZOG genes, the program r8s (Sanderson 2002a), which implements the semiparametric method using penalized likelihood (Sanderson 2002b), was used to assess variance in evolutionary substitution rates and incorporate such variance into the estimation of ages of lineages. The absolute ages of gene duplications were estimated using two calibration points. One was set to 50 MYA for the divergence of rice, sorghum, and maize (Wolfe et al. 1989) and another to 11.9 MYA for the divergence of sorghum and maize (Swigoňová et al. 2004).
Dating of retrotransposon insertion:
Both long terminal repeats (LTRs) of each identified LTR-retrotransposon were aligned using ClustalX (Thompson et al. 1997) and the resulting alignment was manually reviewed. Distance estimations between pairs of LTRs were based on Kimura's (1980) two-parameter model (K2P). According to the recent study by Ma and Bennetzen (2004) showing that nucleotide substitutions in the LTR-retrotransposons of rice (and, presumably, other grasses) occur at rates at least twofold higher than synonymous substitutions in functioning grass genes, the proposed rate of 1.3 × 10−8 substitutions/site/year was used for dating LTR-retrotransposon insertion times.
RESULTS
Isolation and sequencing of r/b orthologous regions in maize and sorghum:
Analysis of r/b homologs derived from a common ancestral chromosome requires the isolation of genomic clones of BACs from closely related chromosomal regions that are orthologous. Screening of the sorghum and maize BAC libraries with the r1 probe yielded such clones that provided a single BAC contig in sorghum and two BAC contigs in maize by fingerprint analysis. This result was expected, given the duplicated nature of the r/b region within the maize genome. To extend the sequence window for comparative analyses, we selected and fully sequenced one additional BAC clone corresponding to the 3′ extension of the maize-10L region (Table 1). To examine if conserved genes are located in a single BAC size extension of the sequenced contig, another BAC clone, representing the 3′ extension of the maize-2S region, was only partially sequenced (assembly ended in phase 2, 9 ordered contigs, 134,220 bp). Two additional conserved genes were identified in this extension. See Table 1 for descriptions of the BAC contigs used in our study.
TABLE 1.
BAC clones used in this study
| Organism | BAC ID | Size (bp) | Accession no. |
|---|---|---|---|
| Oryza sativa japonica (Nipponbare) | OSJNBa0065O17 (4)a | 167,269 | AL606682 |
| OSJNBb0012E24 (4) | 127,506 | AL606647 | |
| Sorghum bicolor | SB20O07 (?) | 157,237 | AY542311 |
| Zea mays mays (B73) | ZMMBBb0138BO4b and ZMMBBc0333J11c (10L) |
290,350 | AF466202 |
| ZMMBBb0092E12b (2S) | 147,198 | AF466203 | |
| ZMMBBc0566K20c,d (2S) | ∼134,000 | AY542310 |
Chromosomal location of a BAC contig.
Referred to as 5′ BAC in the text.
Referred to as 3′ BAC in the text.
This clone was partially sequenced; present assembly is in phase 2 (nine ordered pieces, 134,220 bp).
Genomic comparison of orthologous regions:
Genes identified within the studied segments are listed in Table 2. Three of the 15 candidate genes in rice and 3 of the 13 candidate genes in sorghum are represented by multiple copies. All 11 genes recognized in the maize-10L segment and all 5 genes within the maize-2S interval are present in single copies. The rice and sorghum regions (Figure 1) share 9 consecutive genes in the same order (collinear). The observed gene density in the rice segment, 11.4 kb/gene, is slightly lower than estimates for the entire genome (7–9 kb/gene; Song et al. 2001; Feng et al. 2002; Goff et al. 2002; Sasaki et al. 2002; Yu et al. 2002; Rice Chromosome 10 Sequencing Consortium). The sorghum r/b region with 7.9 kb/gene exhibits a density similar to those previously reported from other sorghum regions (Chen et al. 1997; Tikhonov et al. 1999; Ramakrishna et al. 2002a). The gene density significantly differed between the two duplicated regions of maize (26.4 kb/gene in maize-10L and 49.1 kb/gene in the maize-2S segment). Within the window of 9 genes shared by rice and sorghum, the microcollinearity of the two regions is disrupted by the presence of genes 3A, 3B, 6, and 8 in the sorghum segment (Table 2).
TABLE 2.
Genes identified within the studiedr/b chromosomal segments of rice, sorghum, and maize
| Organism | Genea | Gene homologyb | Accession no. |
e-value | EST (% identity; source organism)c |
|---|---|---|---|---|---|
| Rice | 1A | Subtilisin-like proteinase (L.e.)d | T06580 | 1.20E-174 | No EST |
| 1B | Subtilisin-like proteinase (L.e.) | T07184 | 1.50E-154 | AU163615 (99; O.s.j.) | |
| 2 | Putative acyl-CoA thioesterase (A.t.) | BAC42016 | 9.40E-109 | BQ906059 (99; O.s.) | |
| 3 | Hypothetical protein (A.t.) | NP_171887 | 5.00E-29 | No EST | |
| 4A | Transcription activator Ra (O.s.) | S65802 | 0 | No EST | |
| 4B | R-type basic helix-loop-helix protein (O.s.) | BAB64302 | 2.00E-131 | No EST | |
| 4C | R-type basic helix-loop-helix protein (O.s.) | BAB64302 | 8.90E-151 | No EST | |
| 5 | Isr (genetic modifier; Z.m.) | AAG17894 | 1.40E-125 | CA759443 (99; O.s.i.) | |
| 6 | Conserved unknown expressed protein (f; A.t.) | NP_973513 | NA | No EST | |
| 7A | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 3.20E-177 | BM418761 (98; O.s.) | |
| 7B | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 0 | No full-size EST | |
| 7C | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 7.30E-98 | BI811986 (93; O.s.) | |
| 8 | Glutathione peroxidase (T.m.) | AAQ64633 | 2.80E-84 | BI800245 (96; O.s.) | |
| 9 | Putative copper-transporting ATPase 3 (A.t.) | Q9SH30 | 0 | CA766376 (96; O.s.i.) | |
| 10 | Probable threonine ammonia-lyase (A.t.) | T04211 | 8.70E-163 | No EST | |
| 11 | Actin depolymerizing factor (A.t.) | P30175 | 1.80E-51 | CA759593 (89; O.s.i.) | |
| 12 | beige/BEACH domain-containing protein (A.t.) | NP 182078 | 0 | CB000865 (100; O.s.j.) | |
| 13 | Pre-mRNA processing RNP-binding region-containing protein (A.t.) |
T02269 | 3.20E-127 | CA765686 (99; O.s.i.) | |
| 14 | Putative glycerol-3-transporter (A.t.) | NP_566891 | 0 | CB647889 (100; O.s.) | |
| 15 | Scl1 protein (O.s.; fragment) | T51243 | 3.90E-106 | BQ906849 (93; O.s.) | |
| Sorghum | 1 | Hypothetical protein (O.s.) | BAA99512 | 4.01E-36 | No EST |
| 2A | Regulatory protein B-Peru (Z.m.) | S16594 | 0 | BE357449 (100; S.b.) | |
| 3A | Kelch repeat-containing F-box family protein (A.t.) | NP_974481 | 1.50E-117 | BI351048 (99; S.b.) | |
| 3B | Kelch repeat-containing F-box family protein (A.t.) | NP_564347 | 3.10E-09 | BI351261 (96; S.b.) | |
| 2B | Regulatory protein B-Peru (Z.m.) | S16594 | 0 | BI096615 (90; Z.m.) | |
| 4 | Isr (genetic modifier; Z.m.) | AAL75477 | 4.80E-45 | AW283838 (97; S.b.) | |
| 5 | Conserved unknown expressed protein (A.t.) | NP_973513 | 1.30E-22 | BG412201 (100; S.b.) | |
| 6 | Putative NAM (O.s.) | NP_912423 | 4.10E-18 | BG557594 (100; S.b.) | |
| 7A | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 0 | BG464169 (98; S.b.) | |
| 7B | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 3.90E-138 | BE594517 (98; S.b.) | |
| 7C | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 6.70E-138 | BE594517 (85; S.b.) | |
| 7D | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 4.40E-150 | BE594517 (85; S.b.) | |
| 8 | Probable small nuclear ribonucleoprotein prp4 (A.t.) | O22212 | 5.67E-24 | No EST | |
| 7E | cis-zeatin O-glucosyltransferase (Z.m.) | AAK53551 | 1.20E-161 | BG241630 (97; S.p.) | |
| 9 | Glutathione peroxidase-like protein (H.v.) | CAB59893 | 9.22E-88 | BG559955 (95; S.b.) | |
| 10A | Potential copper-transporting ATPase 3 (A.t.) | Q9SH30 | 0 | BM331692 (98; S.b.) | |
| 10B | Potential copper-transporting ATPase 3 (A.t.) | Q9SH30 | 0 | BM325963 (83; S.b.) | |
| 11 | Probable threonine dehydratase (A.t.) | T04211 | 1.82E-113 | BI098438 (98; S.b.) | |
| 12 | Actin depolymerizing factor (L.l.) | P30175 | 6.57E-63 | BQ528336 (78; Z.m.) | |
| 13 | beige/BEACH domain-containing protein (A.t.) | NP 182078 | 5.50E-71 | No EST | |
| Maize-10L | 1 | NADP-dependent malic enzyme (f; Z.m.) | AAL75485 | 3.30E-163 | AA979812 (Z.m.) |
| 2 | Pinhead protein (A.t.) | Q9XGW1 | 5.69E-59 | No EST | |
| 3 | Anthocyanin regulatory R-S protein (Z.m.) | P13027 | 0 | BI096615 (100; Z.m.) | |
| 4 | Isr (genetic modifier; Z.m.) | AAG17894 | 0 | CA452435 (98; Z.m.) | |
| 5 | Putative S-domain receptor-like protein kinase (O.s.) | BAA94516 | 6.25E-42 | No EST | |
| 6 | Aldose reductase-related protein (H.v.) | P23901 | 2.39E-92 | AW191412 (98; Z.m.) | |
| 7 | Conserved unknown expressed protein (A.t.) | NP_973513 | 5.40E-19 | CF054723 (96; Z.m.) | |
| 8 | Glutathione peroxidase-like protein (H.v.) | CAB59895 | 2.89E-82 | BG842591 (100; Z.m.) | |
| 9 | Putative glycerol-3-transporter (A.t.) | NP_566891 | 0 | BM259481 (99; Z.m.) | |
| 10 | Response regulator (Z.m.) | T50859 | 3.14E-12 | No EST | |
| 11 | Probable 4-coumarate-CoA ligase-like protein (A.t.) | T46131 | 2.89E-09 | BM317765 (78; S.b.) | |
| Maize_2S | 1 | Anthocyanin regulatory R-S protein (Z.m.) | P13027 | 2.80E-69 | BI096615 (91; Z.m.) |
| 2 | zeatin O-glucosyltransferase (P.v.) | Q9ZSK5 | 1.47E-62 | AW065903 (94; Z.m.) | |
| 3 | Conserved unknown expressed protein (f; A.t.) | A84619 | 1.00E-14 | BQ744651 (99; Z.m.) |
NA, not applicable.
Genes are listed according to their occurrence at the genomic sequences, from 5′ to 3′ end, as depicted in Figure 1.
Genes showing significant sequence similarity with the predicted genes. The accession numbers of these genes are listed in the fourth column and the e-values in the fifth column. f, fragmented genes.
ESTs showing significant sequence similarity to the predicted genes.
Source organisms are abbreviated as follows: A.t., Arabidopsis thaliana; H.v., Hordeum vulgare; H.v.v., H. v. vulgare; L.e., Lycopersicon esculentum; L.l., Lilium longiflorum; O.s., Oryza sativa; O.s.j., O. s. japonica; O.s.i., O. s. indica; P.l., Phaseolus lunatus; S.b., Sorghum bicolor; S.p., S. propinquum; T.m., Triticum monococcum; Z.m. Zea mays.
Figure 1.—
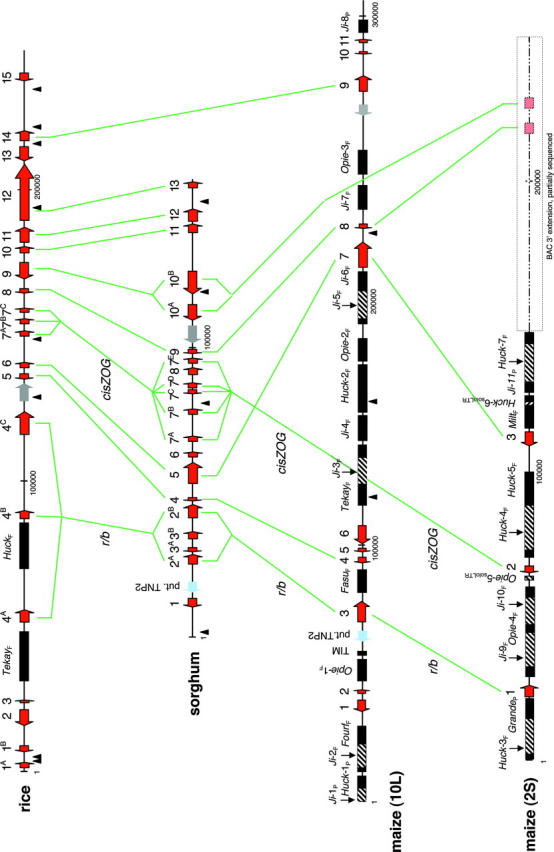
Graphical representation of microcollinearity at the r/b loci of rice, sorghum, and maize. Genes and putative genes are numbered by the order of their appearance from the 5′ to 3′ end of a contig (as listed in Table 2). Copies of an amplified gene are marked by the gene number and a letter in alphabetical order corresponding to their appearance on a contig. Red arrows refer to genes (arrows depicting proposed directions of transcription), light blue arrows refer to DNA transposons, dark rectangles show LTR-retrotransposons (hatching shows nested LTR-retrotransposons), and gray arrows refer to polyproteins. The dashed line indicates the partially sequenced Z566K20 BAC. Lines connecting the contigs show conserved genes. Small triangles show the positions of miniature inverted repeat transposable elements. P, partial; F, full-length LTR-retrotransposon.
Six of the consecutive genes shared by rice and sorghum (corresponding to rice genes 4, 5, 6, 7, 8, and 9) were also detected within the homeologous regions of maize; however, only the genes corresponding to rice genes 4, 6, and 8 are contemporaneously present in both of the maize regions as homeologous duplicates. Two of these shared genes, the r/b gene homolog and the glutathione peroxidase, are of full length. The third conserved gene (corresponding to rice gene 6) is truncated in both the rice and the maize-2S segments. Both maize segments experienced gene deletions, but the pattern of deletion is different on the maize-10L and -2S segments. Disregarding gene amplifications, the maize-10L region experienced deletion of five genes (corresponding to rice 7, 9, 10, 11, and 12) and the maize-2S experienced deletion of at least one gene (corresponding to rice gene 5). Microcollinearity on maize-10L is interrupted by the presence of two genes (5 and 6) not found in this region in rice and sorghum and by the absence of the cis-ZOG gene (rice gene 7). On maize-2S, genes 2 and 3 (corresponding to rice genes 7 and 6, respectively) are inverted relative to their positions in rice and sorghum (Figure 1).
The rice region was found to contain many fragmented LTR-retrotransposons (not shown) and two intact LTR-retrotransposons (Figure 1). We did not find any fragmented or intact retrotransposons in the sorghum segment, in accord with results for the adh1/2 (Tikhonov et al. 1999) and the orp1/2 regions (Lai et al. 2004). One DNA transposon, the putative TNP2, is shared by the maize-10L region and sorghum, suggesting that its insertion predated the time of sorghum and maize divergence. Both maize chromosomal segments are rich in LTR-retrotransposons. The identified retrotransposons constitute 55% of the maize-10L and 76% of the maize-2S genomic sequences. Of the 17 LTR-retrotransposons identified within the maize-10L region, only 4 are nested. In the maize-2S region, 5 LTR-retrotransposons are nested and 1 is present as a solo LTR within another LTR-retrotransposon. In both maize regions the identified LTR-retrotransposons seem to be full length.
Molecular analysis of the r/b homologs:
To investigate the relationship among the r/b gene homologs of sorghum and maize, we used rice as the outgroup. To extend the taxon sampling, we also included full-length sequences of previously described r/b homologs from maize (lc, sn, hopi, rs, and b-Peru) and rice (ra, rb, plw-OSB1, and plw-OSB2). The aligned coding sequences, containing 2016 nucleotides, correspond to part of the second exon through part of the ninth exon of the lc gene of maize (http://pgir.rutgers.edu/).
The rice region contains three r/b homologs (further referred to as r/b-O1, r/b-O2, and r/b-O3), the sorghum region contains two (r/b-S1 and r/b-S2), and each maize region contains one r/b homolog [r1 on maize-10L (representing a simplex R-P allele) and b1 in the maize-2S region]. [In this study, names of genes identified within the studied segments are written as a name of a gene homolog (such as r/b or cisZOG) followed by a hyphen, the first letter of the genus name of the organism (O for Oryza, S for Sorghum, Z for Zea), and a number indicating its occurrence on the studied genomic region from the 5′ to the 3′ end. In the case of the r/b genes of maize, we follow the traditional nomenclature. The r1 gene is from the maize-10L region (representing a simplex R-P allele) and the b1 gene is from the maize-2S region.] The nucleotide sequences of the two r/b homologs of sorghum are 89.8% identical and on average they share 86.8 and 88.2% nucleotide identity with the maize r1 (also as R-P in Figure 2) and b (as B-b in Figure 2) genes, respectively. The two maize r/b homologs are 87.5% identical in their nucleotide sequences. Rice r/b-O1 contains a premature stop codon at position 520–522 in the nucleotide alignment (http://pgir.rutgers.edu/) and is 99.2% identical to plw-OSB1 and 98.8% identical to the ra gene. Gene r/b-O2 is 99.8% identical to plw-OSB2 and also lacks a C terminus, as previously reported for plw-OSB2 (Sakamoto et al. 2001). Both genes have premature stop codons at their sequence, the first one at position 1648–1650 in the alignment. Therefore, substitution rates were estimated for all pairs of genes excluding r/b-O1, r/b-O2, and plw-OSB2, since these three are expected to evolve under different constraints. Rice r/b-O3 exhibits 70.1 and 60.9% nucleotide identity with r/b-O1 and r/b-O2, respectively.
Figure 2.—
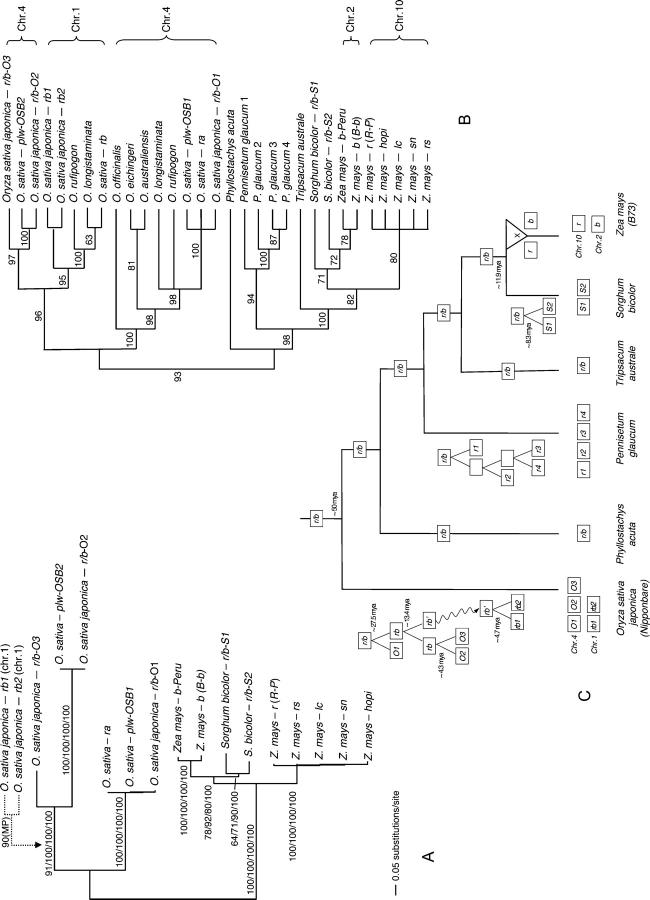
Phylogenetic relationships among the r/b gene homologs. (A) Phylogram resulting from ML analysis with a GTR + I + G model of full-length r and b sequences (2016 nt). Numbers at the internodes refer to the bootstrap proportions from MP (amino acids), MP (nucleotides), ML (nucleotides), and Bayesian-posterior probabilities, respectively. (B) Cladogram resulting from MP analysis of partial sequences (434 nt); numbers show bootstrap values. (C) Proposed history of r/b gene duplication in grasses. The three diagrams located near terminal branches of Oryza, Pennisetum, and Sorghum show the duplication pattern of r/b gene in the corresponding lineage. The wavy line in the Oryza diagram depicts the transfer of one of the r/b homologs to the rice chromosome 1 (the so-called Rb locus). X in the Zea lineage represents the tetraploidization event.
Phylogenetic analyses of amino acid sequences as well as nucleotide sequences recovered the same topology for the gene tree (as shown in Figure 2A). All rice r/b sequences were recovered in a monophyletic group with two clades, one containing r/b-O2, plw-OSB2, and r/b-O3 sequences and the other containing the r/b-O1, plw-OSB1, and ra genes. The two sorghum r/b homologs were recovered as a sister clade to the clade containing maize b-Peru and the maize r/b homolog from chromosome 2S of B73 (b, also known as B-b). The r/b gene homolog from maize chromosome 10L (r1, also known as R-P) belongs to a clade that contains the r/b homologs rs, lc, sn, and hopi, all mapped to the long arm of chromosome 10.
To further examine the stability of the recovered relationships among the 15 full-length sequences of the r/b homologs (as shown in Figure 2A) and to investigate the phylogenetic relationship of the r/b homologs among other members of the grass family, we extended our data for an additional 15 partial sequences of r/b homologs that were already available in GenBank. Those sequences, covering only the conserved basic helix-loop-helix domains and the 5′ end of the C-terminal domains (exons 8 and 9 of the lc gene of maize), are from wild species of rice and also from P. acuta (tribe Bambuseae), P. glaucum (tribe Paniceae), and T. australe (tribe Andropogoneae). The recovered topology is largely in agreement with the topology recovered from the full-length sequences. In addition, the rice sequences were recovered in two well-supported clades (Figure 2B). One clade contains only sequences from the rice Ra locus (chromosome 4), while the other clade includes two sister clades, one containing r/b homologs mapping to chromosome 4 (the Ra locus) and another containing sequences from chromosome 1 (the Rb locus). According to the recent study by Paterson et al. (2004), who reported that the ancestral lineage leading to cereal taxa experienced whole-genome duplication at ∼70 MYA, we examined whether the Ra and Rb regions of rice are indeed the product of the ancient duplication by examining the surrounding genic structure of the Rb locus. Using the Rice FPC map of the Arizona Genomic Institute (http://www.genome.arizona.edu/fpc/rice/) we identified three overlapping BAC clones (AP003432, AP003310, and AP002908) that produced a 361,635-bp-long contig covering the Rb locus. Within this contig we identified two r/b homologs, here called rb1 and rb2, that were included in additional analyses. Sequence annotation did not recover any additional shared genes between the regions of rice chromosome 4 (Ra locus) and chromosome 1 (Rb locus), indicating that those two loci are not the products of a genomic duplication. Phylogenetic analysis using partial sequences of r/b homologs recovered rb1 and rb2 as sister taxa at the base of the r/b homologs from the Rb locus of other rice species (as shown in Figure 2B). Similarly, analysis of the full-length sequences placed the two Rb genes as a sister clade to the r/b-O2, r/b-O3, and plw-OSB2 clade (Figure 2A), indicating that the ra and rb genes are paralogs resulting from duplication of one single ancestral r/b gene at the lineage leading to rice. Using the r/b sequence from Antirrhinum majus delila (M84913) as the outgroup we estimated that the duplication of the ancestral r/b gene of rice occurred at ∼27.5 MYA.
In addition, all four r/b homologs of P. glaucum are from a monophyletic clade, indicating their origin by duplication of a single ancestral gene. Furthermore, the Tripsacum r/b gene is basal to sorghum and maize r/b genes. However, the two sorghum r/b homologs were recovered in polyphyletic position at the base of maize genes from maize chromosome 2S (Figure 2B). Yet, one has to keep in mind that this data set contains shorter sequences and a larger number of terminal taxa. Therefore, we performed additional, Bayesian, analysis of the full-length sequence data. We divided the data set according to five variable and three conserved regions, in which parameters of each partition were estimated independently. Nevertheless, the recovered topology supported the sister relationship of the two sorghum r/b sequences, with the support of Bayesian posterior probability of 100, further justifying the hypothesis that they represent paralogous genes that originated from a duplication of an ancestral gene after sorghum and maize lineages diverged from a common ancestor.
Molecular analysis of the cisZOG gene:
Given the range of mobility exhibited by the R/B locus in the last 50 million years, one wonders whether it might relate to its function and position in its chromosomal region. However, another locus in this region, just 43 kb downstream of the b1 gene, with even greater mobility, has a completely different function. The CisZOG locus encodes a protein that promotes cell division and plant growth (Mok 1994). Using sequence divergence we investigated the history of its gene duplication across the three grass species. All identified cisZOG ORFs consist of one 1377- to 1407-bp exon. Three copies of the cisZOG gene were predicted in the rice chromosomal segment (see genes 7A, 7B, and 7C in Figure 1, hereafter referred to as cisZOG-O1, cisZOG-O2, and cisZOG-O3, respectively). Genes cisZOG-O1 and cisZOG-O2 are 95.7% identical. The cisZOG-O3 gene shares 73.95 and 74.95% nucleotide identity with cisZOG-O1 and cisZOG-O2, respectively. Translation of the nucleotide sequences to the amino acid sequences revealed the presence of premature in-frame stop codons within the cisZOG-O3 gene at positions 1141–1143 and 1405–1407 of the sequence alignment (http://pgir.rutgers.edu/). A homology search using BLAST recovered another putative cisZOG gene on rice chromosome 7 (GenBank accession no. AP004378). This gene is 72.2% identical at the nucleotide sequence level to cisZOG-O1, 72.7% to cisZOG-O2, and 70% to the cisZOG-O3 gene. Phylogenetic analyses as well as a rate heterogeneity test showed that this gene, from rice chromosome 7, evolves at a significantly increased rate compared to all other cisZOG genes from the four studied regions. Moreover, its relative position to the other cisZOG homologs varied in different analyses. Thus, the chromosome 7 cisZOG gene is suspected to evolve under different constraints and was not used in further analyses.
Five cisZOG genes were identified within the sorghum interval (further referred to as cisZOG-S1 through cisZOG-S5), one of which is in an inverted position (7C in Figure 1). While the maize-2S region contains one cisZOG gene (cisZOG-Z1), the orthologous region of maize-10L lacks this gene. Martin et al. (2001) reported isolation of the cisZOG1 gene from maize that matches exactly cisZOG-Z1 from the maize-2S region. Veach et al. (2003) reported isolation of another gene, cisZOG2, from the maize genome but its chromosomal location was not known. The nucleotide sequences of cisZOG-Z1 and cisZOG2 are 98.1% identical. We included sequences of both maize genes in our analyses. Maximum parsimony analyses of nucleotides and amino acid sequences resulted in gene trees with the same topology as the maximum likelihood tree shown in Figure 3A. The estimated phylogram shows several long branches, for instance lineages of cisZOG-O3 and cisZOG-S5. A rate heterogeneity test revealed that rates of substitution vary significantly among lineages. To decrease the effect of long branches on the resulting phylogeny, we examined the number of variable sites in nonoverlapping segments of 100 sites. In the following analyses, we excluded the three most variable regions from the data. Analyses of this reduced data set (1098 characters) resulted in a tree topologically identical to that shown in Figure 3A. However, the rate heterogeneity test still uncovered significant differences for several lineages of the gene tree. Thus we performed several additional analyses with more extensive exclusion of variable characters and also exclusion of sequences that showed significant increases in their rate of change. The resulting trees, although often yielding less resolution (lineages were collapsing to polytomies), all had the same backbone topology as shown in Figure 3A.
Figure 3.—
Phylogenetic relationship of cis-zeatin O-glucosyltransferase genes and a proposed history of their amplification and removal. (A) An unrooted phylogram of cisZOG homologs from orthologous regions of rice, sorghum, and maize resulting from ML(GTR + G) analysis. Numbers at the internodes show bootstrap values from MP (amino acids), MP (nucleotides), and ML (nucleotides) analyses, respectively. The hexagon on the terminal branch of cisZOG-S5 depicts the position of the root of the gene tree (see text). (B) Schema of a proposed pattern of cisZOG gene duplication and deletion in rice, sorghum, and maize. Squares refer to genes. The dashed line between cisZOG-Z1 and cisZOG-Z2 illustrates that the two copies are not within the single orthologous region of maize.
Using the estimated phylogenetic relationship among the cisZOG genes, as shown in Figure 3A, we traced the history of gene duplication. For estimation of duplication times we used the cisZOG homologs from soybean (AF489873 and AF489874), Phaseolus (AF489876 and AF489877), and Lycopersicon (AY082661) as the outgroups. Rooting the gene tree at the outgroup species (as marked in Figure 3A) the cisZOG-S5 was recovered at the base of the ingroup genes. Hence, the cisZOG-S5 represents the oldest gene (here marked as a descendant of the A1 gene; Figure 3B) that was deleted independently on the maize and rice lineages. Furthermore, rice cisZOG-O1 and cisZOG-O2 are close relatives to cisZOG-S1, cisZOG-Z1, and cisZOG-Z2; therefore, they must have shared a common ancestor, which we will call the A2 gene (Figure 3B). The ancestral A2 gene duplicated independently on the lineage leading to rice (∼32 MYA) and on the lineage leading to maize (∼6.7 MYA). While rice cisZOG-O1 and cisZOG-O2 are located within the same chromosomal region, one maize gene, cisZOG-Z2, was transferred to a nonorthologous location in the maize genome. Since rice cisZOG-O3 and sorghum cisZOG-S2 form a clade in all analyses performed, they must have shared a common ancestor, which we will call the A3 gene. Because the common ancestry of cisZOG-S2, -S3, and -S4 genes can be traced to a later date than the split of rice and sorghum lineages, there must have been a duplication on the ancestral lineage of grasses that gave rise to genes A3 and A4. The ancestral gene A3 gave rise to cisZOG-O3 and cisZOG-S2 and gene A4 gave rise to sorghum cisZOG-S3 and cisZOG-S4 genes. The duplication of the A4 gene, however, happened before the time when the two progenitors of the maize genome diverged from sorghum (∼40 MYA). Thus, the immediate ancestor of rice, sorghum, and maize had four copies of cis-zeatin O-glucosyltransferase (A1, A2, A3, and A4; see Figure 3B). While the rice lineage experienced a duplication and two deletions of the ancestral genes, the sorghum still contains descendants of all four ancestral genes. The most extensive gene deletion occurred on the maize lineage that contains only two duplicates of one ancestral gene, and nine genes were removed from its genome.
Retrotransposons and their times of insertion:
While gene mobility has occurred throughout the evolution of this chromosomal region even before speciation, sequence mobility of transposable elements in this region occurred mostly during very recent times. Two full-length LTR-retrotransposons, closely related to maize Tekay and Huck, were identified in the rice segment (Figure 1). They inserted in the rice genomic sequence at ∼0.8 and 0.02 MYA, respectively. Large numbers of fragmented retrotransposons were also found in the rice region (not shown). The maize-10L region contains 17 LTR-retrotransposons, representing 7 different families, and the maize-2S region contains 12 LTR-retrotransposons from 5 families. The most common were Ji (38%), Huck (24%), and Opie (17%). Eight LTR-retrotransposons in the maize-10L regions are intact and not in a nested organization. Both LTRs were identified for 22 retrotransposons. Six of the 22 LTR-retrotransposons were inserted within another LTR-retrotransposon (see Figure 1). In all six cases, the two LTRs of a recipient retrotransposon showed greater nucleotide variation than the LTRs of an inserted retrotransposon. Using an intergenic retrotransposon nucleotide substitution rate of 1.3 × 10−8 substitutions/site/year (Ma and Bennetzen 2004), we calculated the time of insertion of the 22 LTR-retrotransposons (Table 3). All inserted in the maize genomic sequences within the last 3 million years. The most pronounced activity has occurred within the last 1 million years, when >80% of the LTR-retrotransposons inserted.
TABLE 3.
Estimated distances (k) between pairs of LTRs and times of LTR-retrotransposon insertions
| Name | k (±SE)a | Time (±SE)b |
|---|---|---|
| Chromosome 10L | ||
| Fourf | 0.0094 (0.0029) | 0.36 (0.11) |
| Ji-2 | 0.0006 (0.0006) | 0.02 (0.02) |
| Opie-1 | 0.0230 (0.0045) | 0.88 (0.17) |
| TIM | 0.0699 (0.0174) | 2.69 (0.67) |
| Fasu | 0.0000 (0.0000) | 0 |
| Tekay-like | 0.0230 (0.0029) | 0.88 (0.11) |
| Ji-3 | 0.0023 (0.0013) | 0.09 (0.05) |
| Ji-4 | 0.0169 (0.0037) | 0.65 (0.14) |
| Huck-2 | 0.0316 (0.0043) | 1.22 (0.17) |
| Opie-2 | 0.0216 (0.0049) | 0.83 (0.19) |
| Ji-5 | 0.0000 (0.0000) | 0 |
| Ji-6 | 0.0232 (0.0045) | 0.89 (0.17) |
| Ji-7 | 0.0172 (0.0037) | 0.66 (0.14) |
| Opie-3 | 0.0049 (0.0020) | 0.19 (0.08) |
| Chromosome 2S | ||
| Huck-3 | 0.0136 (0.0028) | 0.52 (0.11) |
| Opie-4 | 0.0532 (0.0066) | 2.05 (0.25) |
| Ji-9 | 0.0192 (0.0036) | 0.74 (0.14) |
| Ji-10 | 0.0086 (0.0026) | 0.33 (0.10) |
| Huck-4 | 0.0110 (0.0025) | 0.42 (0.10) |
| Huck-5 | 0.0076 (0.0023) | 0.29 (0.09) |
| Milt | 0.0432 (0.0081) | 1.66 (0.31) |
| Huck-7 | 0.0081 (0.0022) | 0.31 (0.08) |
The estimated number of substitutions per nucleotide site (k), based on the K2P model, and its standard deviation (SE).
The estimated time of LTR-retrotransposon insertion in millions of years ago and its standard deviation.
DISCUSSION
Microcollinearity at the R and B loci in rice, sorghum, and maize:
While there was already genetic and molecular evidence that the r1 gene could exist in multiple copies even in distant positions, we have now found through comparison of genomic sequences that its dynamic properties have already existed throughout the evolution of the grass family. Moreover, the larger window of sequences surrounding this locus also shows that such properties are far more general than anticipated. The orthologous r/b regions available for this analysis span a window of 9 genes for comparison of rice and sorghum and a window of 6 genes for simultaneous comparison of all four genomic segments. Microcollinearity of the rice and sorghum genomic regions is disrupted by 4 genes, here labeled 3A, 3B, 6, and 8. They are not present in any of the other orthologous segments, suggesting that they have been inserted within the sorghum interval after sorghum diverged from maize. The duplicated gene 3 shows sequence similarity to the kelch repeat-containing F-box family protein whose closest homologs are found on rice chromosomes 10 and 1, respectively. Gene 6 shows similarity to a no apical meristem (NAM) protein of rice and Arabidopsis and other NAC domain proteins. In a recent study, Ooka et al. (2003) found that NAM proteins are members of NAC gene families that have at least 75 predicted genes in the rice genome and 105 genes in the Arabidopsis genome. Gene 8 is similar to a probable small nuclear ribonucleoprotein, and homologous sequences were recovered from rice chromosomes 2, 3, 7, 9, and 12. The maize-10L region contains 2 inserted genes, 5 and 6, that are not shared with any other of the studied intervals. Gene 5 shows sequence similarity to an S-domain receptor-like protein kinase gene and similar sequences were recovered from rice chromosomes 1 and 7. This gene family in maize contains at least 4 genes (Zhang and Walker 1993) and multiple members were also reported from Arabidopsis and Brassica (Braun and Walker 1996). Gene 6 is similar to the aldose reductase-related protein and this gene was also predicted from rice chromosomes 1 and 10. In summary, the genic interruption of microcollinearity between rice, sorghum, and maize is caused mostly by insertion of high-copy-number genes in sorghum and maize that probably occurred after sorghum diverged from the maize progenitors over the past 11.9 million years (Swigoňová et al. 2004). The fact that gene movement may be associated with gene amplification was previously pointed out by other studies (Song et al. 2002; Lai et al. 2004).
In maize, deletion of homeologous copies of duplicated genes appears to be randomized across the homeologous regions of maize, since both segments experienced gene deletions. While the r/b region of maize-10L lost 56% (five of nine) of the genes that can be traced to the common ancestor of grasses, the maize-2S region lost only 17% (one of six) of the genes. This is in contrast to the 40% gene loss reported from each of the homeologous regions surrounding the Adh locus of maize (Ilic et al. 2003). This nonuniform deletion of duplicated genes from the homeologous loci of maize was also observed in a recent report by Lai et al. (2004). Deletions of redundant homeologous copies of maize genes over the last 12 million years has been accompanied by retention of at least one copy of each gene within the two r/b regions, similar to that observed in the maize adh region (Ilic et al. 2003). Lai et al. (2004) found only one gene that was conserved between sorghum and rice and was lost from both duplicated regions of maize. It is not yet known whether this gene was deleted from the maize genome or was transferred to a nonorthologous position.
The potential of homeologous duplicates to develop new functions seems to be rarely realized, since the most common fates of duplicated genes are gene silencing of one copy (Walsh 1995), interlocus recombination and concerted evolution of duplicates (Wendel et al. 1995; Wendel 2000), or retention of subfunctional duplicates (Cronn et al. 1999; Adams et al. 2003). Within the studied r/b region, three genes are present as homeologous duplicates. Both copies of the r/b gene and glutathione peroxidase gene (gp) are full length. However, the third gene, which shows similarity to an unknown conserved expressed protein from A. thaliana (corresponding to rice gene 6), is present only as a fragment in the maize-2S region. For the r/b genes, the estimated nonsynonymous substitution rate (Ka) was 0.088 (±0.010) and the synonymous substitution rate (Ks) was 0.309 (±0.042), yielding a Ka / Ks ratio of 0.284. For the gp gene, Ka was 0.035 (±0.010) and Ks was 0.128 (±0.038), giving a Ks /Ka ratio of 0.273. Relative rate tests (Muse and Gaut 1994) performed on the r/b and gp duplicated genes from the maize B73 genome that we studied, using rice as the outgroup, did not recover a significant difference in rates between the homeologous duplicates. Therefore, within the r/b region, one gene encountered possible silencing of one of the homeologous copies by gene fragmentation and two genes appear to be under selection to retain their function, i.e., purifying selection (Yang and Bielawski 2000).
While the r/b region of sorghum has a low content of transposable elements, as similarly reported for adh (Tikhonov et al. 1999), orp (Lai et al. 2004), and zein (Song et al. 2002) regions, other chromosomal segments contain up to 15–20% of retrotransposons (Ramakrishna et al. 2002a,b). Unlike sorghum, the rice r/b genomic sequence contains, in addition to two full-length LTR-retrotransposons, numerous fragmented LTR-retrotransposons in various states of truncation. Recently, Ma et al. (2004) reported analysis of 1000 LTR-retrotransposons from the rice genome, concluding that >75% of them are fragmented. They suggested that fragmentation of LTR-retrotransposons in rice is primarily due to unequal homologous and/or illegitimate recombination that leads to removal of retrotransposons from the rice genome. In sorghum, one solo LTR-retrotransposon was reported from the sh2/a1 (Chen et al. 1998) region and additional fragmented LTR-retrotransposons have been also found within the vrn1-homologous (Ramakrishna et al. 2002a) and rp1-homologous (Ramakrishna et al. 2002b) regions, indicating that similar events that formed the rice genome also influenced the evolution of the sorghum genome, although the dynamics of such rearrangements seem to significantly differ between the two species. For maize, it has been previously estimated that 60–80% of its genome is composed of repetitive DNA (SanMiguel et al. 1996; SanMiguel and Bennetzen 1998; Meyers et al. 2001). Consistent with reports from other regions of maize (SanMiguel et al. 1998), the r/b region contains ∼65% LTR-retrotransposons that have inserted within the last 2.7 million years. Unlike rice, most of the LTR-retrotransposons in the maize r/b region are full length, suggesting that maize may be on the route to genome expansion.
Evolution of the r/b gene family:
Purugganan and Wessler (1994) analyzed partial nucleotide sequences of r/b gene homologs (474 nt in total), covering the most conserved region of the genes, the bHLH domains and the C-terminal domains (exons 8 and 9), from six grass species. They reported that the two maize r/b homologs, the r1 (chromosome 10L) and b1 (chromosome 2S) genes, are as old as sorghum and maize. Hu et al. (1996), analyzing the amino acid sequences of the same region from maize and different rice species, reported that the r/b gene families arose recently and independently in different grasses. Furthermore, they concluded that in rice there are two clades, Ra (grouping sequences mapping to rice chromosome 4) and Rb (grouping sequences from rice chromosome 1), which evolved by duplication of an ancestral gene. Having full genomic sequences of the r/b genes from orthologous regions of rice, sorghum, and maize, we reinvestigated the pattern of molecular evolution of the r/b gene family in grasses and examined the relationship of the maize r/b homologs with the r/b genes of sorghum.
Phylogenetic analyses of full-length coding sequences of the r/b genes, based on nucleotide as well as on amino acid sequences (Figure 2A), showed that genes form clades according to the grass taxon they belong to, indicating that the common ancestor of grasses had a single r/b gene and that this ancestral r/b gene duplicated independently in the lineages leading to rice, sorghum, and maize. Including r/b sequences from additional grass taxa (Figure 2B), we recovered a gene tree topology similar to that observed by Hu et al. (1996), confirming the independent amplification of an ancestral r/b gene in different grasses. Unlike Hu and colleagues' study, our study included multiple r/b gene homologs from both rice loci. Within the studied rice region we identified three r/b homologs at the Ra locus. Two additional genes were found on chromosome 1, the Rb locus. We also included the two genes identified at the Ra locus of O. sativa T65-Plw, the plw-OSB1 and plw-OSB2 genes (Sakamoto et al. 2001). Interestingly, the rice genes are not recovered in monophyletic clades on the basis of their chromosomal position. The r/b homologs mapping to rice chromosome 1, the Rb locus, were recovered as a sister clade to a group of several r/b homologs mapping to rice chromosome 4, the Ra locus (Figure 2, A and B). According to the recent findings that the cereal genomes descended from a lineage that experienced a whole-genome duplication at ∼70 MYA (Paterson et al. 2004), we further examined whether the rice Rb locus is in synteny with the Ra locus. Annotation of a >360-kb contig surrounding the Rb locus did not recover any additional genes shared with the Ra locus, suggesting that the genomic region bearing the Rb locus is not a product of genomic duplication of the Ra locus. Unlike the Rb region, the Ra region of rice chromosome 4 is syntenic with the r/b regions of sorghum and maize; this finding indicates that the ancestor of rice had a single r/b gene at the Ra locus of chromosome 4 that later duplicated (∼27.5 MYA). One of the duplicates gave rise to the Ra-like paralogs and another to the Rb-like paralogs, one of which moved to a new chromosomal location (in the window of 5–13 MYA), the so-called Rb locus of chromosome 1 (shown in Figure 2C) where it further duplicated (∼4.7 MYA).
Our data also show that while the r/b gene complex at the maize R locus (maize chromosome 10L) arose by recent amplification of a single ancestral r gene (within the last 3 million years), the age of the two paralogous copies in sorghum can be traced to the base of the sorghum lineage (∼8.3 MYA). In a study based on systematic analyses of 11 homeologous duplicates from five duplicated chromosomal regions of maize, Swigoňová et al. (2004) showed that the node of the two maize progenitors and sorghum could not be unequivocally resolved; however, they also pointed out that only the r/b gene tree differed significantly from the unresolved tree in several tests. In this study, we performed analyses of nucleotides as well as amino acid sequences on a data set containing additional sequences from other maize varieties and several additional grass species (Figure 2). Our results consistently show that the maize b genes (maize chromosome 2S) are more closely related to both sorghum paralogs than to any of the maize r genes (chromosome 10L). Therefore, analyses of the r/b genes support the hypothesis that maize arose as an allotetraploid, as previously suggested from distance analyses of mdh and wx genes by Gaut and Doebley (1997) and that the R and B loci are allelic descendants of two different diploid maize progenitors.
The maize r and b genes are homeologous duplicates and both are selected to retain their function. The r and b genes differ in their tissue specificity and developmental timing (Styles et al. 1973). The r genes specify anthocyanin pigmentation in the aleurone, anthers, or coleoptiles, and the b genes determine pigmentation in leaves, sheaths, and tassel (Styles et al. 1973). In seed tissues, the r and b genes can functionally substitute for one another to determine pigment accumulation (Styles et al. 1973; Goff et al. 1990). Because the translated amino acid sequences of the r and b genes are highly similar (>91%) while the 5′ controlling regions are significantly heterogeneous, it has been postulated that the protein products are functionally equivalent and that the allelic diversity is inherent in differential expression of the r and b genes. The strongest evidence for this argument comes from the finding that the elements controlling tissue specificity reside within the 5′ variable region (Radicella et al. 1992). Additional functional differentiation occurred at the R locus, where various alleles exhibit a diverse array of tissue-specific expression patterns.
History of the cisZOG gene duplication:
Because the cisZOG locus is intimately associated with the r/b genes and has undergone extensive gene duplication and deletion, we felt that an analysis of its evolutionary history would provide an interesting comparison with the dynamics of the adjacent r/b genes. The cis-zeatin O-glucosyltransferase genes encode for cytokinins, naturally occurring plant hormones promoting cell division and plant growth through regulation of tissue differentiation, bud formation, senescence, and seed germination (Mok 1994). Two cisZOG genes were previously identified in maize; the first was the cisZOG1 gene, reported by Martin et al. (2001), which is 100% identical to the one identified within our maize-10L region, and the other was the unmapped gene cisZOG2, reported by Veach et al. (2003). The two genes are 98.1% identical in nucleotide sequences and 98.8% identical in their amino acid sequences. Both genes were found to be highly expressed in roots, while their expression pattern in kernels differed (high for cisZOG1 and low for cisZOG2). The observed difference in gene expression in kernels was suggested to be the result of promoter sequence heterogeneity (Veach et al. 2003).
Within the studied intervals, we identified three cisZOG genes in tandem triplication in the rice interval, five genes in sorghum, and one gene in the maize-2S region. Using phylogenetic analyses of nucleotide and amino acid sequences, we investigated the evolutionary pattern of cisZOG gene duplication in the three grass taxa. The reconstructed history of cisZOG duplication (Figure 3B) showed that the most immediate common ancestor of grasses contained four copies of the cisZOG gene. In the rice lineage, one ancestral gene duplicated (∼32 MYA) and two were lost. The sorghum lineage retains descendants of all four ancestral genes, one of which duplicated before the time when sorghum diverged from maize progenitors (∼40 MYA). The two maize cisZOG genes are the result of a recent duplication of one ancestral gene (∼6.7 MYA) that was followed by a transfer of one of the paralogous copies to a nonorthologous location. The ancestor of the two maize progenitors each had at least five copies of the cisZOG gene. Nevertheless, all copies but one were deleted, indicating extensive gene loss in maize. Hence, for both the r/b genes and the cisZOG genes in this region, extensive gene duplication has occurred. Interestingly, the timing of the events and the regions duplicated appear to be completely different, indicating that neither large-scale tandem duplication nor large deletions occur at a detected rate in this region. It is not clear whether the numerous small rearrangements in this region are an indication of an unusual lability of this chromosomal segment in several grass species or whether it might be an outcome of selective pressures for amplification and diversification of these gene families.
Timing and nature of LTR-retrotransposons in maize:
As in all other studied maize segments, the exceptional dynamism of transposon accumulation over the last few million years has led to tremendous differentiation of the chromosomal segments, as shown by the unique configurations in the two homeologous regions and in comparison to the apparently retrotranposon-free sorghum segment. In our study, the recognized LTR-retrotransposons represented 55 and 76% of the maize-10L and maize-2S segments, respectively, with Ji, Opie, and Huck the most common types. Although the oldest retrotransposon activity in these regions can be traced to 2.7 MYA (Table 3), the majority (81%) of the LTR-retrotransposons inserted within the last 1 million years. Unlike the multilevel nested LTR-retrotransposons reported from other maize regions (SanMiguel et al. 1998; Ramakrishna et al. 2002a; Ma et al. 2004), nesting of the LTR-retrotransposons was minimal in both maize regions (see Figure 1). About 47% (8/17) of the LTR-retrotransposons within the maize-10L region contain no retrotransposon inserts, indicating that the nested nature of retrotransposons is heterogeneous in the maize genome.
Composition and structure of the r/b region of the common ancestor of grasses:
Since the rice r/b region contains only nine genes, all shared with sorghum and maize, it is simplest to assume that the gene content of the rice region represents the gene content of the common ancestor of grasses, with a few exceptions caused by subsequent gene family duplication and deletion in the rice lineage. First, the common ancestor of grasses contained only one r/b homolog, which later amplified independently in different grass lineages. Second, the cisZOG gene was represented by at least four copies (the three copies in the rice segment represent only two of the ancestral genes, see above) and these experienced parallel amplifications and deletions along different grass lineages. Third, the single ancestral copper-transporting ATPase gene duplicated in the ancestral lineage that gave rise to maize and sorghum. We cannot judge the repetitive DNA content of this region in an ancestral grass, because the rate of removal of these sequences is so high that all traces of such elements are lost within 5–10 million years (Ma et al. 2004). While the rice region contains numerous fragmented retrotransposons, sorghum is lacking recognizable traces of retrotransposon activity and maize contains mostly full-size or nested retrotransposons with rather few solo LTR-retrotransposons. Therefore, different grass lineages show lineage-specific patterns of local evolution in genic and intergenic sequences. Recent studies comparing the genomic structure at the bz region and the region containing the 22-kD zein gene cluster of two different maize inbred lines (Fu and Dooner 2002; Song and Messing 2003) demonstrated that severe structural polymorphism can be observed even within haplotypes of a single species. To understand the full dynamics of plant genome evolution, finer-scale studies, investigating the pattern of local evolution among closely related taxa and different subspecies, will be needed.
Acknowledgments
We thank H. K. Dooner for valuable comments on the manuscript. We also thank W. Ramakrishna and V. Llaca for technical assistance in sequencing. This work was supported by National Science Foundation Plant Genome Program grant no. 9975618.
References
- Adams, K. L., R. Cronn, R. Percifield and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S., and S. D. Tanksley, 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., and J. B. Smith, 1976. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 274: 227–274. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., P. Bhandol and I. J. Leicht, 2000. Nuclear DNA amounts in angiosperms and their modern uses: 807 new estimates. Ann. Bot. 86: 859–909. [Google Scholar]
- Bilang, R., S. Zhang, N. Leduc, V. A. Iglesias, A. Gisel et al., 1993. Transient gene expression in vegetative shoot apical meristems of wheat after ballistic microtargeting. Plant J. 4: 735–744. [Google Scholar]
- Bodeau, J. P., and V. Walbot, 1992. Regulated transcription of the maize Bronze-2 promoter in electroporated protoplasts requires the C1 and R gene products. Mol. Gen. Genet. 233: 379–387. [DOI] [PubMed] [Google Scholar]
- Braun, D. M., and J. C. Walker, 1996. Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Bio. Sci. 21: 70–73. [PubMed] [Google Scholar]
- Brunner, S., B. Keller and C. Feuillet, 2003. A large rearrangement involving genes and low-copy DNA interrupts the microcollinearity between rice and barley at the Rph7 locus. Genetics 164: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas, A. M., A. K. Kononowicz, U. B. Zehr, D. T. Tomes, J. D. Axtell et al., 1993. Transgenic sorghum plants via microprojectile bombardment. Proc. Natl. Acad. Sci. USA 90: 11212–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V. L., J. P. Radicella, T. P. Robbins, J. Chen and D. Turks, 1989. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., P. SanMiguel, A. C. de Oliveira, S.-S. Woo, H. Zhang et al., 1997. Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum. Proc. Natl. Acad. Sci. USA 94: 3431–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., P. SanMiguel and J. L. Bennetzen, 1998. Sequence organization and conservation in sh2/a1-homologous region of sorghum and rice. Genetics 148: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., F. A. Burr and B. Burr, 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83: 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni, G., A. Viotti, S. L. Dellaporta and C. Tonelli, 1992. cDNA nucleotide sequence of Sn, a regulatory gene in maize. Nucleic Acids Res. 20: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn, R. C., R. L. Small and J. F. Wendel, 1999. Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S. L., I. M. Greenblatt, J. L. Kermicle, J. B. Hicks and S. R. Wessler, 1988 Molecular cloning of the R-nj gene by transposon tagging with Ac, pp. 263–281 in Chromosome Structure and Function: Impact of New Concepts, edited by J. P. Gustafson and R. Appels. Plenum Press, New York.
- Devereux, J., P. Haeberli and O. Smithies, 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., 1983. Coordinate genetic regulation of flavonoid biosynthetic enzymes in maize. Mol. Gen. Genet. 189: 136–141. [Google Scholar]
- Dooner, H. K., and J. L. Kermicle, 1976. Displaced and tandem duplications in the long arm of chromosome 10 in maize. Genetics 82: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., T. P. Robbins and R. A. Jorgensen, 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Eggleston, W. B., M. Alleman and J. L. Kermicle, 1995. Molecular organization and bases for germinal instability in R-stippled maize. Genetics 141: 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Feng, Q., Y. Zhang, P. Hao, S. Wang, G. Fu et al., 2002. Sequence and analysis of rice chromosome 4. Nature 420: 316–320. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96: 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 2002. Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann. Bot. 89: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implication in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95: 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway, M. E., J. D. Masuggi, A. M. Lloyd, V. Walbot, R. W. Davis et al., 1994. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavazzi, G., M. Mereghetti, G. Consonni and C. Tonelli, 1990. Sn, a light-dependent and tissue-specific gene of maize: the genetic basis of its instability. Genetics 125: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., T. M. Klein, B. A. Roth, M. E. Fromm, K. C. Cone et al., 1990. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 9: 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., K. C. Cone and M. E. Fromm, 1991. Identification of functional domains in the maize activator C1: comparison of wildtype and dominant inhibitor proteins. Genes Dev. 5: 298–309. [DOI] [PubMed] [Google Scholar]
- Goff, S. A., K. C. Cone and V. L. Chandler, 1992. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6: 864–875. [DOI] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T.-H. Han, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., M. Chamberlin, M. Snook, B. Siame, L. Butler et al., 1998. Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740. [PMC free article] [PubMed] [Google Scholar]
- Hu, J., B. Anderson and S. R. Wessler, 1996. Isolation and characterization of rice R genes: evidence from distinct evolutionary path in rice and maize. Genetics 142: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hulbert, S. H., T. E. Richter, J. D. Axtell and J. L. Bennetzen, 1990. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. USA 87: 4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., P. J. SanMiguel and J. L. Bennetzen, 2003. A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice. Proc. Natl. Acad. Sci. USA 100: 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., J. Ma, Z. Swigoňová, W. Ramakrishna, V. Llaca et al., 2004. Gene loss and movement in the maize genome. Genome Res. 14: 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham, R. J., J. Walsh, M. Dunn, C. Ko, S. A. Goff et al., 2004. Genomic duplication, fractionalization and the origin of regulatory novelty. Genetics 166: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick, M. L., and V. L. Chandler, 1998. Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A. M., V. Walbot and R. W. Davis, 1992. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258: 1773–1775. [DOI] [PubMed] [Google Scholar]
- Ludwig, S. R., and S. R. Wessler, 1990. Maize R gene family: tissue specific helix-loop-helix proteins. Cell 62: 849–851. [DOI] [PubMed] [Google Scholar]
- Ludwig, S. R., L. F. Habera, S. L. Dellaporta and S. R. Wessler, 1989. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86: 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, D. R., and W. P. Maddison, 2000 MacClade 4, Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA.
- Martin, R. C., M. C. Mok, J. E. Habben and D. W. S. Mok, 2001. A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc. Natl. Acad. Sci. USA 98: 5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., S. V. Tingey and M. Morgante, 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, M. C., 1994 Cytokinins and plant development: an overview, pp. 155–166 in Cytokinins: Chemistry, Activity, and Function, edited by D. W. S. Mok and M. C. Mok. CRC Press, Boca Raton, FL.
- Moore, G., K. M. Devos, Z. Wang and M. D. Gale, 1995. Cereal genome evolution: grasses, line up and form a circle. Curr. Biol. 5: 737–739. [DOI] [PubMed] [Google Scholar]
- Muse, S. V., and B. S. Gaut, 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates with application to the chloroplast genome. Mol. Biol. Evol. 11: 715–724. [DOI] [PubMed] [Google Scholar]
- Neuffer, M. G., E. H. Coe and S. R. Wessler, 1997 Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ooka, H., K. Satoh, K. Doi, T. Nagata, Y. Otomo et al., 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bowers and B. A. Chapman, 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 29: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G. I., L. J. Harris, V. Walbot and V. L. Chandler, 1991. Genetic analysis of B-Peru, a regulatory gene in maize. Genetics 126: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni, K., E. Cominelli, G. Consonni, G. Gusmaroli, G. Gavazzi et al., 2000. The developmental expression of the maize regulatory gene Hopi determines germination-dependent anthocyanin accumulation. Genetics 155: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Purugganan, M. D., and S. R. Wessler, 1994. Molecular evolution of the plant R regulatory gene family. Genetics 138: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., J. F. Wing, H. T. C. Leppen, J. N. M. Mol and R. E. Koes, 1993. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella, J. P., D. Brown, L. A. Tolar and V. L. Chandler, 1992. Allelic diversity of the maize B regulatory gene: different leader and promoter sequences of two B alleles determine distinct tissue specificities of anthocyanin production. Genes Dev. 6: 2152–2164. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., J. Dubcovsky, Y.-J. Park, C. Busso, J. Emberton et al., 2002. a Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., J. Emberton, P. SanMiguel, M. Ogden, V. Llaca et al., 2002. b Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz, M. B., E. Grotewold and V. L. Chandler, 1997. Evidence for a direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related MYB domain proteins. Plant Cell 9: 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W., T. Ohmori, K. Kageyama, C. Miyazaki, A. Saito et al., 2001. The purple leaf (Pl) locus of rice: the Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42: 982–991. [DOI] [PubMed] [Google Scholar]
- Sanderson, M. J., 2002a r8s, Version 1.50, User's Manual (August 2002). University of California, Davis (http://ginger.ucdavis.edu/r8s/).
- Sanderson, M. J., 2002. b Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19: 101–109. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., and J. L. Bennetzen, 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44. [Google Scholar]
- SanMiguel, P., A. Tikhonov, Y. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima and J. L. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., T. Matsumoto, K. Yamamoto, K. Sakata, T. Baba et al., 2002. The genome sequence and structure of rice chromosome 1. Nature 420: 312–316. [DOI] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., V. Llaca, E. Linton and J. Messing, 2001. Sequence, regulation, and evolution of the maize 22-kD zein gene family. Genome Res. 11: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., V. Llaca and J. Messing, 2002. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 13: 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., O. Ceska and K. T. Seah, 1973. Developmental differences in action of R and B alleles in maize. Can. Genet. Cytol. 15: 59–72. [Google Scholar]
- Swigoňová, Z., J. Lai, J. Ma, W. Ramakrishna, V. Llaca et al., 2004. Close split of sorghum and maize progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 1999 PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer, Sunderland, MA.
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A. P., P. J. SanMiguel, Y. Nakajima, N. M. Gorenstein, J. L. Bennetzen et al., 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96: 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerck, J. A., and M. E. Fromm, 1994. Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6: 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach, Y. K., R. C. Martin, S. W. S. Mok, J. Malbeck, R. Vankova et al., 2003. O-glucosylation of cis-Zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 131: 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J. B., 1995. How often do duplicated genes evolve new functions? Genetics 139: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wendel, J. F., A. Schnabel and T. Seelanan, 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 92: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., M. Gouy, Y.-W. Yang, P. M. Sharp and W.-H. Li, 1989. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86: 6201–6205.2762323 [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comp. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and J. P. Bielawski, 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim, Y. S., G. L. Davis, N. A. Duru, T. A. Musket, E. W. Linton et al., 2002. Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol. 130: 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., S. N. Hu, J. Wang, K. W. Gane, S. G. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
- Zhang, R., and J. C. Walker, 1993. Structure and expression of the S-locus related genes of maize. Plant Mol. Biol. 21: 1171–1174. [DOI] [PubMed] [Google Scholar]