Abstract
Being the largest land mammals, elephants have very few natural enemies and are active during both day and night. Compared with those of diurnal and nocturnal animals, the eyes of elephants and other arrhythmic species, such as many ungulates and large carnivores, must function in both the bright light of day and dim light of night. Despite their fundamental importance, the roles of photosensitive molecules, visual pigments, in arrhythmic vision are not well understood. Here we report that elephants (Loxodonta africana and Elephas maximus) use RH1, SWS1, and LWS pigments, which are maximally sensitive to 496, 419, and 552 nm, respectively. These light sensitivities are virtually identical to those of certain “color-blind” people who lack MWS pigments, which are maximally sensitive to 530 nm. During the day, therefore, elephants seem to have the dichromatic color vision of deuteranopes. During the night, however, they are likely to use RH1 and SWS1 pigments and detect light at 420–490 nm.
VERTEBRATES can be classified roughly into diurnal, nocturnal, and arrhythmic species according to their visual habits. The eyes of diurnal animals contain predominantly cone photoreceptor cells and are specifically designed for high visual acuity during the day, whereas those of nocturnal animals contain predominantly rod cells and are designed to operate mostly under low illuminations at night (Walls 1942; Ali and Klyne 1985). Compared to these, the eyes of the arrhythmic, known also as cathemeral, species seem to function equally in bright light hours of day and dim ones of night (Nowak 1991). The large terrestrial mammals such as ungulates, elephants, and large carnivores have arrhythmic vision (Walls 1942; Ali and Klyne 1985). These animals typically contain abundant populations of rods in their retinas, which are often found in conjunction with a retinal tapetum (Jacobs et al. 1994). To react to changes in light intensities relatively quickly, many arrhythmic species not only elongate the rods and contract the cones in light and execute the opposite movements in darkness but also regulate the amount of light reaching the retina by changing the size of the pupil (Walls 1942; Ali and Klyne 1985).
Despite their fundamental importance in vision, the role of visual pigments in arrhythmic vision is not well understood. Visual pigments consist of a transmembrane (TM) protein, an opsin, and the 11-cis-retinal chromophore. They are classified into rhodopsins (RH1), RH1-like (RH2), short wavelength-sensitive type 1 (SWS1), short wavelength-sensitive type 2 (SWS2), and middle and long wavelength-sensitive (M/LWS) pigment groups (Yokoyama and Yokoyama 1996; Yokoyama 2000a; Ebrey and Koutalos 2001). The RH2 and SWS2 opsin genes became nonfunctional in the early stage of mammalian evolution (Yokoyama and Yokoyama 1996) and the RH1, SWS1, and M/LWS pigments in the mammalian ancestor had the wavelengths of maximal absorption (λmax) of ∼500, ∼360, and ∼560 nm, respectively (Yokoyama 2000a; Yokoyama and Radlwimmer 2001; Ebrey and Takahashi 2002; Shi and Yokoyama 2003). The respective groups of visual pigments in arrhythmic mammals have λmax-values of 497–508, 428–456, and 531–555 nm (Table 1). In the arrhythmic mammals, therefore, the λmax-values of RH1 pigments have been maintained more or less at the ancestral level, but those of SWS1 pigments have increased significantly and those of M/LWS pigments have decreased.
TABLE 1.
The λmax-values for the visual pigments in ungulates and carnivores
| Absorption spectrum (nm)
|
||||
|---|---|---|---|---|
| Species | RH1 | SWS1 | M/LWS | Reference |
| Bovine (Bos taurus) | — | 455 | 554 | Jacobs et al. (1998)a |
| 500 | — | — | Oprian et al. (1987)a | |
| Horse (Equus caballus) | — | 428 | 539 | Carroll et al. (2001)a |
| — | — | 545 | Yokoyama and Radlwimmer (1999)b | |
| Pig (Sus domestica) | — | 439 | 556 | Neitz and Jacobs (1989)a |
| Goat (Capra hircus) | — | 444 | 552 | Jacobs et al. (1998)a |
| — | — | 553 | Radlwimmer and Yokoyama (1997)b | |
| Sheep (Ovis aries) | — | 446 | 552 | Jacobs et al. (1998)a |
| Deer (Odocoileus virginianus) | 497 | 456 | 537 | Jacobs et al. (1994)a |
| — | — | 531 | Yokoyama and Radlwimmer (1999)b | |
| Deer (Dama dama) | — | 454 | 542 | Jacobs et al. (1994)a |
| Cat (Felis catus) | — | 447 | 554 | Loop et al. (1987)a |
| — | — | 553 | Yokoyama and Radlwimmer (1999)b | |
| Dog (Canis familiaris) | 508 | 431 | 555 | Jacobs et al. (1993)a |
| Fox (Urocyon littoralis) | — | 432 | 555 | Jacobs et al. (1993)a |
| Fox (Vulpes vulpes) | — | 438 | 555 | Jacobs et al. (1993)a |
The λmax-values were measured by using an electroretinogram.
The λmax-values were measured by using an in vitro assay (e.g., Yokoyama 2000b).
Elephants can be trained to paint and their paintings have been sold in auction (Gilbert 1990; Tennesen 1998). This “artistic ability” suggests a reasonably well-developed color vision of elephants. Thus, it is of interest to study what types of visual pigments elephants possess and how they compare to those of other arrhythmic animals. To explore the possible roles of visual pigments in the arrythmic vision of mammals, we have cloned the opsin genes from African elephant (Loxodonta africana) and Asian elephant (Elephas maximus), which are evolutionarily distantly related to the arrhythmic mammals studied to date (Eizirik et al. 2001). The results show that the elephants use RH1, SWS1, and LWS pigments, which have λmax-values of 496, 419, and 552 nm, respectively. These spectral sensitivities are virtually identical to those of certain human deuteranopes who lack MWS pigments. Therefore, elephants seem to have the dichromatic color vision of these deuteranopes. The mutagenesis results show that the λmax-values of the elephant RH1, SWS1, and LWS pigments have been attained by D83N, F86S/T93I/L116V, and S180A, respectively.
MATERIALS AND METHODS
The eye samples and RT-PCR amplification:
The eyes of African elephant (L. africana) and Asian elephant (E. maximus) were sampled from necropsied females of 24 and 57 years old, respectively. Their total retinal RNAs were isolated using the procedures of Yokoyama et al. (1995). The entire coding regions of all different types of opsin cDNAs of the African elephant have been cloned in two steps: (1) cloning of the internal segments by using RT-PCR and (2) completion of the cloning by using 5′- and 3′-rapid amplification of cDNA ends (RACE; see the next section). To characterize internal segments, we first cloned the segment between codon positions 248 and 300 of all opsin cDNAs by using the forward and reverse degenerate primers designed by Carleton and Kocher (2001) (Figure 1A, all internal), where the amino acid site numbers are standardized by those of bovine RH1 pigment (GenBank accession nos. K00502, K00503, K00504, K00505, K00506). Note that these degenerate primers have been designed to clone all RH1, RH2, SWS1, SWS2, and M/LWS opsin genes. Using these primers, cDNA was reverse transcribed at 42° for 1 hr and 95° for 5 min and then PCR amplification was carried out for 30 cycles at 94° for 45 sec, 55° for 1.5 min, and 72° for 2 min. The PCR products were gel isolated and subcloned into the T-tailed EcoRV-digested Bluescript plasmid vector with T-overhang attached to 3′ ends (Hadjeb and Berkowitz 1996). Nucleotide sequences of these cDNA clones were determined by cycle-sequencing reactions using the Sequitherm Excel II long-read kits (Epicentre Technologies, Madison, WI) with dye-labeled M13 forward and reverse primers. Reactions were run on a LI-COR (Lincoln, NE) 4200LD automated DNA sequencer. To clone the longer internal fragments, we then designed additional sets of the RH1, SWS1, and M/LWS gene-specific degenerate primers. The forward primers (internal forward) for the RH1, SWS1, and LWS opsin cDNAs are located around codon sites 51, 70, and 51, respectively, while the corresponding reverse primers (internal reverse) are located between codon sites 260 and 280, making the total lengths of RT-PCR products between 573 and 691 nucleotide sites (Figure 1A).
Figure 1.—

Oligonucleotide primers for RT-PCR amplification and 5′- and 3′-RACE of RH1, SWS1, and M/LWS opsin mRNAs. (A) All internal primers are expected to amplify the codon positions between 248 and 300 of all opsin cDNAs and the RH1-, SWS1-, and LWS-specific internal primers are expected to amplify the codon positions between 50–70 and 260–280. (B) The EcoRI and SalI sites are boxed in the forward and reverse primers, respectively, and were used for cloning the amplified products into the expression vector pMT5. A Kozak sequence (CCACC) was inserted between the EcoRI site and the initiation codon to promote translation.
RACE:
To determine the rest of the cDNA sequences, we have constructed additional gene-specific primers (GSPs) (Figure 1A) and conducted 5′- and 3′-RACE analyses according to the manufacturer's protocol (GIBCO BRL, Gaithersburg, MD). In short, for the 3′-RACE, the first-strand cDNA was made using the oligo(dT)-containing adapter primer (AP) provided by the manufacturer and the original mRNA was degraded by RNase H. Then, two sequential PCR amplifications were performed, applying two sets of GSPs and universal adapter primers (UAPs) to these cDNAs, first using GSP1 with UAP and then the nested GSP2 with abridged UAP (Figure 1A), where the UAPs were supplied by the manufacturer. For the 5′-RACE, the cDNAs were first synthesized from total RNA using GSP1 and the mRNA was degraded by RNase H. The cDNA was purified using a spin column and then a poly(C) tail was added to the 3′ end of the cDNAs using dCTP and terminal transferase. Then, the entire coding region was obtained by two sequential PCR amplifications, first using nested GSP2 with abridged AP and then nested GSP3 and abridged UAP (Figure 1A), where the UAPs were again supplied by the manufacturer. The nucleotide sequences of the resulting cDNA products were again determined by using a LI-COR 4200LD automated DNA sequencer.
Expression and spectral analyses of pigments (in vitro assay):
The contiguous full-length opsin cDNAs were then obtained by RT-PCR using primers based on the nucleotide sequences of the 5′ and 3′ end cDNA clones (Figure 1B). The opsin cDNAs of full length were subcloned into the EcoRI and SalI restriction sites of the expression vector pMT5 (Khorana et al. 1988). All of these DNA fragments were sequenced to rule out spurious mutations. These plasmids were expressed in COS1 cells by transient transfection. The pigments were regenerated by incubating the opsins with 11-cis-retinal (Storm Eye Institute, Medical University of South Carolina) and purified using immobilized 1D4 (The Culture Center, Minneapolis) in buffer W1 [50 mm N-(2-hydroxyethyl) piperazine-N′-2-ethanesulfonic acid (HEPES) (pH 6.6), 140 mm NaCl, 3 mm MgCl2, 20% (w/v) glycerol, and 0.1% dodecyl maltoside] (for more details, see Yokoyama 2000b). UV-visible spectra were recorded at 20° using a Hitachi U-3000 dual beam spectrophotometer. Visual pigments were bleached for 3 min using a 60-W standard light bulb equipped with a Kodak Wratten no. 3 filter at a distance of 20 cm. Data were analyzed using Sigmaplot software (Jandel Scientific, San Rafael, CA).
Site-directed mutagenesis:
Mutant opsins were generated by using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All DNA fragments that were subjected to mutagenesis were sequenced to rule out spurious mutations.
RESULTS
The opsin genes of elephants:
The opsin genes cloned by using RT-PCR amplification and 5′- and 3′-RACE show that the African elephant has RH1, SWS1, and M/LWS opsin genes, which consist of 349, 351, and 365 codons, respectively. These numbers are virtually identical to those of the corresponding orthologous genes in human with respective codon lengths of 349 (GenBank accession no. U49742), 349 (M13295), and 365 (M13300). Since it encodes the LWS pigment-specific tyrosine at site 277 (Y277) and threonine at site 285 (T285) (Yokoyama and Yokoyama 1990; Neitz et al. 1991; Asenjo et al. 1994; Yokoyama and Radlwimmer 2001), the elephant M/LWS gene can be classified as LWS-type (see also the next section). When the coding regions of the orthologous RH1, SWS1, and LWS genes in elephant and human are compared, the proportions of identical nucleotides are 0.91, 0.89, and 0.87 per site, respectively. The proportions of identical nucleotides for the three pairs of paralogous elephant opsin genes are much lower and are 0.45–0.46 per site.
When the RH1 opsin gene of the African elephant is compared to that of a phylogenetically relatively closely related manatee (Trichechus manatus; GenBank accession no. AF055319) (e.g., Springer et al. 2003), the proportion of nucleotide sequence identity increases only by 1%. From these data, we evaluated the numbers of synonymous (ds) and nonsynonymous (dn) nucleotide substitutions by using the Nei and Gojobori (NG) method (Nei and Gojobori 1986). Using the divergence time of ∼60 million years (MY) (Springer et al. 2003), the rates of nucleotide substitution at synonymous and nonsynonymous sites of this gene are given by 2.58 (±0.98) × 10−9 and 0.08 (±0.012) × 10−9 site/year, respectively. Thus, the Afrotherian RH1 gene is one of very slowly evolving protein-coding genes in mammals (e.g., Li 1997, Table 7.1; see also discussion).
Using the three pairs of the African elephant opsin gene-specific forward and reverse primers (Figure 1B), we also RT-PCR amplified the coding regions of the RH1 (between codon positions 6 and 342), SWS1 (between codon positions 8 and 346), and LWS (between codon positions 5 and 359) opsin genes of the Asian elephant. The results show that the orthologous RH1, SWS1, and LWS opsin genes between the two elephant species have only 1, 2, and 1 synonymous nucleotide differences, respectively. Thus, during the last 5 MY of their divergence (Maglio 1973), no amino acid replacement has occurred in the visual pigments in the two elephant lineages.
Absorption spectra of the elephant pigments:
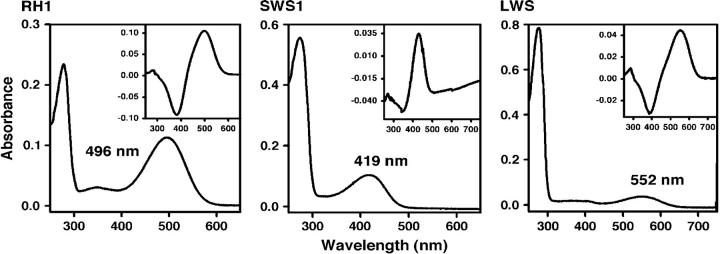
Using an in vitro assay, we have evaluated the absorption spectra of the RH1, SWS1, and LWS pigments in the African elephant. The absorption spectra of these pigments show two peaks, one at ∼280 nm and another at 419–552 nm (Figure 2). When these pigments are exposed to light, the second peak shifts to ∼380 nm (results not shown), indicating the cis-trans isomerization of the chromophore (Hubbard and Kropf 1958). These control experiments demonstrate that the second peaks are due to opsins covalently linked to 11-cis-retinal via a Schiff base bond (Yokoyama 2000b). When measured in the dark, the RH1, SWS1, and M/LWS pigments have λmax-values of 496 ± 1 nm [referred to as elephant (P496)], 419 ± 1 nm [elephant (P419)] and 552 ± 1 nm [elephant (P552)], respectively (Figure 2). As suspected, the elephant M/LWS pigment is, indeed, the LWS type. The respective dark-light difference spectra are given by 499, 430, and 550 nm (Figure 2, insets). The λmax-values of the dark-light spectra of the RH1 and LWS pigments are very close to those of the corresponding dark spectra. However, the dark and dark-light values for the SWS1 pigment differ by 11 nm, which may occur because the pigment peak in the dark and the peak of photobleached SWS1 pigment in the light are too close to determine the correct value (see also Kawamura and Yokoyama 1998).
Figure 2.—
The dark absorption spectra of the elephant RH1, SWS1, and LWS pigments. The dark-light difference spectra are shown in the insets.
The absorption spectra of visual pigments seem to be determined exclusively through the interactions between the 11-cis-retinal chromophore and amino acids in TM1–TM7 helices (Yokoyama 2000a; Ebrey and Takahashi 2002; Shi and Yokoyama 2003). Because of the identical amino acid sequences in the seven TM helices of the orthologous pigments, the RH1, SWS1, and LWS pigments in the Asian elephant should also have λmax-values of 496, 419, and 552 nm, respectively. The λmax-value of the RH1 pigment is slightly lower than that of 500 nm of the ancestral vertebrate pigment. Information on the absorption spectra of the RH1 pigments in arrhythmic animals is limited (Table 1), but the λmax-value of the elephant RH1 pigment does not seem to differ much from those of other arrhythmic mammals. The λmax-value of the LWS pigment is virtually identical to those of bovine, pig, goat, sheep, cat, dog, and foxes (Table 1). Among these, the currently known LWS pigments of bovine, goat, cat, and elephant all have amino acids, A180. Since S180A decreases the λmax-value by ∼7 nm (Yokoyama and Radlwimmer 2001), the slightly decreased λmax-value seems to be widely spread among the arrhythmic mammals. The biological significance of this λmax-value of the LWS pigment in arrhythmic mammals is not immediately clear. In addition, the λmax-value of the SWS1 pigment is >10 nm lower in the elephant than in any other ungulates and carnivores studied to date (Table 1).
Molecular basis of spectral tuning in the elephant visual pigments:
At present, specific amino acid changes are known to be responsible for generating variable λmax-values of RH1 pigments (positions 83, 122, 211, 261, 292, and 295) (Yokoyama 2000a), of SWS1 pigments (positions 46, 49, 52, 86, 90, 93, 97, 114, 116, and 118) (Yokoyama 2000a; Shi and Yokoyama 2003; Fasick et al. 2002), and of M/LWS pigments (positions 180, 197, 277, 285, and 308) (Yokoyama and Radlwimmer 2001). Here the amino acid sites of the RH1 and SWS1 pigments are standardized by those of the bovine RH1 pigment, while those of the M/LWS pigments are those of the human M/LWS pigments (K033490 and M13300). Since these sites are highly conserved, we can easily identify a total of five potentially important amino acid replacements that may have shifted the λmax-values of the elephant pigments: D83N in elephant (496); F86S, T93I, and L116V in elephant (P419); and S180A in elephant (P552).
When reverse mutations N83D and A180S are introduced into elephant (P496) and elephant (P552), the mutant pigments have λmax-values of 498 and 558 nm, respectively (Table 2). If we let θD83N be the effect of D83N on the λmax-shift and ZRh be the λmax value of the ancestral RH1 pigment, respectively, then ZRh = 498 and θD83N = −2 (Table 2). Similarly, if we let θS180A be the effect of S180A on the λmax-shift and ZR be the λmax-value of the ancestral LWS pigment, then ZR = 558 and θS180A = −6 (Table 2). This result is consistent with the observed λmax-shift that is caused by S180A in the human LWS pigment (Merbs and Nathans 1992). The results for the ancestral RH1 and LWS pigments are consistent with previous estimates (Yokoyama 2000a; Yokoyama and Radlwimmer 2001; Shi and Yokoyama 2003).
TABLE 2.
Mutagenesis results and the effects of amino acid changes on λmax-shift
| Pigment | Amino acid change |
λmax (nm) | Estimate (nm) |
|---|---|---|---|
| RH1 | WT | 496 | ZRh = 498 |
| N83D | 498 | θD83N = −2 | |
| SWS1 | WT | 419 | ZB = 360 |
| S86F | 367 | θF86S = 51 | |
| I93T | 413 | θT93I = 0 | |
| V116L | 416 | θL116V = −1 | |
| S86F/I93T | 359 | θF86S×T93I = 5 | |
| S86F/V116L | 360 | θF86S×L116V = 3 | |
| I93T/V116L | 411 | θT93I×L116V = 8 | |
| S86F/I93T/V116L | 360 | θF86S×T93I×L116V = −7 | |
| LWS | WT | 552 | ZR = 558 |
| A180S | 558 | θS180A = −6 |
When S86F, I93T, and V116L are introduced into elephant (P419), the mutant pigments have λmax-values of 367, 413, and 416 nm, respectively (Table 2). These results suggest that the absorption spectrum of the SWS1 pigment was achieved mostly by F86S. This is totally unexpected because F86S in the ancestral avian SWS1 pigment increased its λmax-value only by 17 nm (Shi and Yokoyama 2003). To evaluate the magnitudes of individual and synergistic effects of the three amino acids on the λmax-shift, we also introduced all combinations of these amino acid changes into elephant (P419) (Table 2). To interpret the mutagenesis results, let θF86S, θT93I, θL116V, θF86S×T93I, θF86S×L116V, θT93I×L116V, and θF86S×T93I×L116V be the magnitudes of λmax-shifts caused by F86S, T93I, L116V, F86S/T93I, F86S/L161V, T93I/L116V, and F86S/T93I/L116V, respectively, and ZB be the ancestral λmax-value. Then, from Table 2,
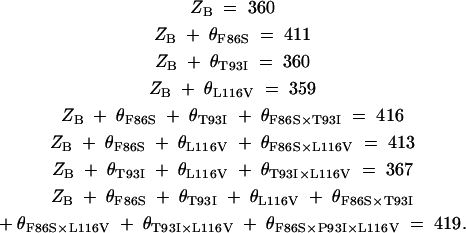 |
Solving these equations, we can see that F86S has, indeed, increased the λmax-value by 51 nm and neither T93I nor L116V has caused any λmax-shift individually, but interactions of F86S, T93I, and L116V, including the three-way interaction, cannot be ignored (Table 2). The ancestral λmax-value of 360 nm agrees with the estimate obtained by engineering the ancestral SWS1 pigment (Shi and Yokoyama 2003).
Since amino acid sites 86 and 93 are located in TM2 helix and site 116 is in the TM3 helix (Palczewski et al. 2000), the cumulative effects of amino acid changes in the TM2 helix, the TM3 helix, and in both TM2 and TM3 helices have increased the λmax-value by 56, −1, and 59 nm, respectively. Thus, F86S explains 86% of the λmax-shift from the ancestral pigment to the contemporary elephant SWS1 pigments, while F86S and T93I together explain 95% of the λmax-shift. Note that the molecular bases of the blue (or violet) sensitivities of orthologous human (Shi et al. 2001), bovine (Fasick et al. 2002), and avian pigments (Shi and Yokoyama 2003) have also been studied. For these pigments, however, mutagenesis analyses are still incomplete and the exact roles of specific amino acid changes in the spectral tuning cannot be determined. Thus, our results of elephant (P419) reveal the first complete molecular characterization of spectral tuning in the SWS1 pigment.
DISCUSSION
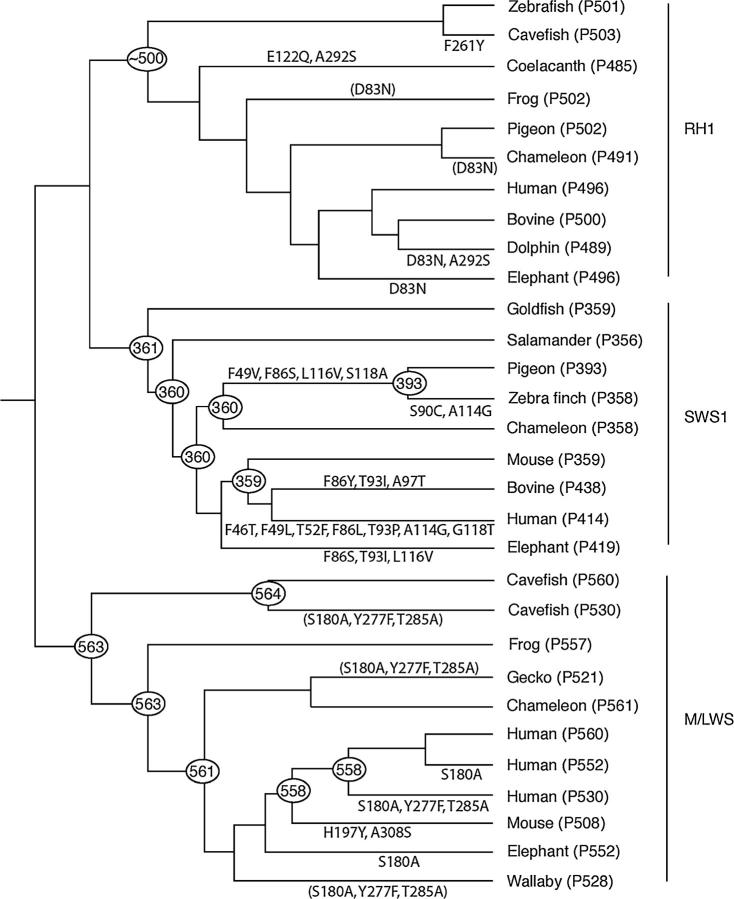
We have seen that the RH1, SWS1, and LWS pigments in the vertebrate ancestor had λmax-values of ∼500, ∼360, and ∼560 nm, respectively. Figure 3 shows a composite tree for elephant RH1, SWS1, and M/LWS pigments and some representative vertebrate pigments, whose tree topology is supported by molecular and paleontological data (Yokoyama 2000a; Yokoyama and Radlwimmer 2001; Madsen et al. 2001; Murphy et al. 2001; Shi and Yokoyama 2003; Springer et al. 2003). As we can see in Figure 3, the λmax-values of the RH1 and LWS pigments in elephant decreased by ∼5 and ∼10 nm, respectively, whereas that of the SWS1 pigment increased by ∼60 nm.
Figure 3.—
The phylogenetic tree of some representative RH1, SWS1, and LWS pigments. The numbers after P and at the nodes refer to λmax-values. Zebrafish (Danio rerio), cavefish (Astyanax fasciatus), goldfish (Carassius auratus), coelacanth (Latimeria chalamnae), frog (Xenopus laevis), salamander (Ambystoma tigrinum), pigeon (Columba livia), zebra finch (Taeniopygia guttata), chameleon (Anolis carolinensis), human (Homo sapiens), bovine (Bos taurus), dolphin (Tursiops truncates), mouse (Mus musculus), and elephant (Loxodonta africana) are shown. The λmax-values of the ancestral SWS1 and M/LWS pigments are taken from Shi and Yokoyama (2003) and Yokoyama and Radlwimmer (2001), respectively. Amino acid changes whose λmax-shifts are not evaluated are shown in parentheses. The branch lengths are not to scale.
Evolutionary rates of the elephant opsin genes:
We have seen that the λmax-values of the elephant pigments are virtually identical to those of certain deuteranope people. The λmax-values of the LWS pigments in elephant and human have been achieved by the same amino acid change, S180A, but those of the SWS1 pigments have been attained independently by entirely different amino acid replacements (Figure 2); F86S/T93I/L116V have occurred in elephant (P419) and F46T/F49L/T52F/F86L/T93P/A114G/G118T in human (P414) (Shi et al. 2001). To study the patterns of evolutionary changes of the opsin genes in the two species, we compared codons between sites 35 and 306 of the opsin genes of elephant, human, and zebrafish (Danio rerio: RH1, GenBank accession no. AF109368; SWS1, AB087810; LWS, AB087803). These codons encode amino acids between TM1 and TM7 of visual pigments and can modify the absorption spectra of visual pigments (Yokoyama 2000a; Ebrey and Takahashi 2002). Note that among the RH1, SWS1, and M/LWS opsin genes, the first two groups are evolutionarily most closely related (Yokoyama 2000a). Therefore, when we construct the phylogenetic tree of all three groups of opsin genes, each tree topology of orthologous genes is rooted.
Note that fish and mammals diverged ∼400 MY ago (node a, say) and the two mammalian species ∼100 MY ago (node b) (e.g., Kumar and Hedges 1998; Eizirik et al. 2001; Nei et al. 2001; Springer et al. 2003). Using the NG method and these divergence times, we have evaluated ds and dn values and the evolutionary rates of nucleotide substitution. When we consider the 400 MY of zebrafish evolution, the rates of nonsynonymous substitution in the RH1, SWS1, and LWS genes are 0.20 × 10−9, 0.45 × 10−9, and 0.20 × 10−9/site/year, respectively. The respective evolutionary rates of the mammalian ancestor (branch a–b) are significantly lower than those of the orthologous zebrafish genes (Figure 3). Probably more significantly, the SWS1 gene has evolved with the highest rate among the three genes (Figure 3). This result seems to reflect the fact that not only was the amount of λmax-shift that contemporary SWS1 pigments had to achieve the largest among the three pigments but also multiple amino acid changes are needed to achieve the λmax-shift (Shi et al. 2001). Although they are not as strict as in the case of nonsynonymous changes, the evolutionary rate of synonymous substitution tends to be higher in the SWS1 genes as well (Figure 3).
As noted earlier, the proportions of different nucleotides for the RH1, SWS1, and LWS genes between African and Asian elephants are 1/1014, 2/1017, and 1/1059, respectively, which are all synonymous changes. The lack of nonsynonymous nucleotide substitutions shows that the arrhythmic vision of elephants had been established before the separation of the African and Asian elephants. Assuming that the two species diverged 5 MYA (Maglio 1973; see also Eizirik et al. 2001), the evolutionary rates of nucleotide substitution for the three respective opsin genes are (0.10 ± 0.010) × 10−9, (0.20 ± 0.124) × 10−9, and (0.10 ± 0.015) × 10−9. Thus, the evolutionary rates of nucleotide substitution for the three opsin genes have slowed down significantly after the separation of the two elephant species. The cause for these slow evolutionary rates is not immediately clear.
Spectral tuning of mammalian visual pigments:
RH1 pigments:
The λmax-value of the contemporary elephant RH1 pigment is slightly blue shifted by D83N. When a wide range of vertebrates is surveyed, we can also identify several RH1 pigments with blue-shifted λmax-values that are associated with D83N, including marine eel (Anguilla anguilla; λmax = 482 nm; Archer et al. 1996), John Dory (Zeus faber; λmax = 492 nm; Dartnall and Lythgoe 1965), chameleon (Anolis carolinensis; λmax = 491 nm; Kawamura and Yokoyama 1998), bottlenose dolphin (Tursiops truncates; λmax = 488 nm; Fasick and Robinson 1998), and saddleback dolphin (Delphinus delphis; λmax = 489 nm; McFarland 1971; Yokoyama 2000a). Among these, the actual role of D83N in the blue shift in the λmax-value has been experimentally proven only for the bottlenose dolphin (Fasick and Robinson 1998). The biological effects, if any, of the slightly blue-shifted λmax of RH1 pigments on the dim vision in elephants remain to be clarified.
SWS1 pigments:
The λmax-values of UV pigments in a variety of contemporary species have been inherited directly from the vertebrate ancestor (Figure 3). The avian lineage is the exception, where the ancestral pigment acquired violet sensitivity (λmax = 393 nm) by F49V/F86S/L116V/S118A, but some descendants regained UV sensitivity by S90C (Figure 3; Shi and Yokoyama 2003). Curiously, in both elephant and ancestral avian pigments, F86S/L116V is involved in the development of their violet sensitivities. In the ancestral avian pigment, F86S increases the λmax-value by 17 nm and, furthermore, F49V/F86S/S118A and F49V/F86S/L116V/S118A increase the λmax-value by 14 and 33 nm, respectively (Shi and Yokoyama 2003), suggesting that L116V should increase the λmax-value by 19 nm. Our mutagenesis analysis of elephant (P419), however, reveals a very different picture; F86S increases the λmax-value by 51 nm and L116V only by 3 nm (Table 3). Clearly, the effects of these amino acid changes on the λmax-shift are affected strongly by other amino acids. This can be seen by combining the mutagenesis results of the ancestral avian pigment and elephant pigment. That is, F86S/L116V, F49V/S118A, and F49V/F86S/L116V/ S118A increase the λmax-value by 53, −3, and 33 nm, respectively. Consequently, the interaction of the four amino acid changes together decreases the λmax-value by 17 nm.
TABLE 3.
Evolutionary rates of nucleotide substitution (×10−9/site/year)
| Branch | MY | RH1 | Significance | SWS1 | Significance | LWS |
|---|---|---|---|---|---|---|
| a–b | 300 | ds = 0.60 ± 0.077 | ≪ | 1.53 ± 0.256 | ≫ | 0.80 ± 0.103 |
| dn = 0.10 ± 0.013 | ≪ | 0.30 ± 0.026 | ≫ | 0.06 ± 0.010 | ||
| b–human | 100 | ds = 2.00 ± 0.035 | ≫ | 0.50 ± 0.066 | 3.10 ± 1.948 | |
| dn = 0.10 ± 0.013 | ≪ | 0.30 ± 0.026 | ≫ | 0.20 ± 0.020 | ||
| b–elephant | 100 | ds = 2.10 ± 0.531 | 2.20 ± 0.597 | > | 1.00 ± 0.134 | |
| dn = 0.10 ± 0.013 | ≪ | 0.40 ± 0.032 | ≫ | 0.30 ± 0.026 |
Standard errors were computed from [9p(1 − p)/{(3 − 4p)2n}]1/2, where p is the proportion of sites that differ and n is the number of nucleotide sites involved (Kimura and Ohta 1972). The ds and dn values indicate the rates of nucleotide substitution at ∼200 synonymous and ∼616 nonsynonymous sites, respectively. Single and double inequality signs indicate that the differences are significant at 5 and 1% levels, respectively.
Certain amino acid changes at site 86 have played important roles in the evolution of different SWS1 pigments. In addition to the major roles of F86S exhibited in the development of the elephant and avian SWS1 pigments, F86Y in the SWS1 pigment in bovine (Cowing et al. 2002; Fasick et al. 2002) and F86V in the orthologous pigment in guinea pig (Cavia porcellus) (Parry et al. 2004) also increased their λmax-values dramatically. On the other hand, F86L does not shift the λmax-value of the human SWS1 pigment by itself, but it causes the λmax-shift through interactions with other six critical amino acid changes (Shi et al. 2001). In general, therefore, the spectral tuning of SWS1 pigments is based on strong synergistic interactions among ∼10 critical amino acids.
M/LWS pigments:
We have seen that S180A in the elephant LWS pigment has decreased the λmax-value by 6 nm (Table 2). Using mutagenesis and multiple regression analyses, it has been shown that the λmax-value of the M/LWS pigment in the vertebrate ancestor was ∼560 nm and S180A, H197Y, Y277F, T285A, A308S, and S180A/H197Y shift the λmax-values of visual pigments by −7, −28, −8, −15, −27, and +11 nm, respectively (Yokoyama and Radlwimmer 2001; see also Sun et al. 1987). This “five-sites rule” explains the λmax-values of all contemporary and engineered ancestral M/LWS pigments fully (Yokoyama and Radlwimmer 2001). This rule explains the λmax-value of elephant (P552) perfectly.
Parallel evolution:
We have seen that not only has D83N occurred in different RH1 pigments independently but also F86S/L116V occurred in both elephant and avian SWS1 pigments (Figure 3). In fact, the occurrence of such parallel changes is rather common in visual pigments. For example, E122Q has occurred in coelacanth RH1 pigment and decreases the λmax-value by 10 nm (Figure 3). In the coelacanth RH2 pigment, E122Q has also occurred independently and caused a similar λmax-shift (Yokoyama et al. 1999). In addition, identical amino acid changes (A292S) have occurred not only in both coelacanth and dolphin RH1 pigments but also in mouse MWS pigment (A308S) (Figure 3). MWS pigments reveal more extensive parallel evolution; that is, the three identical amino acid replacements (S180A, Y277F, and T285A) have occurred independently in cavefish (Astyanax fasciatus), gecko (Gekko gekko), deer (Odocoileus virginianus), human (Homo sapiens) and macaque (Macacca fascicularis) ancestors, wallaby (Macropus eugenii), and New World monkeys, including squirrel monkey (Saimi boliviensis) (Yokoyama 2000a; Deeb et al. 2003), some of which are shown in Figure 3. Furthermore, in addition to those of elephant and human, S180A caused blue shift in λmax-values of the M/LWS pigments of bovine, goat, deer (O. virginianus), and cat (Yokoyama and Radlwimmer 2001). Molecular analyses of the evolution of the RH1, SWS1, and LWS pigments in elephants provide additional supportive evidence of parallel evolution of visual pigments.
Arrhythmic color vision:
Having the specific RH1, SWS1, and LWS pigments, what do elephants actually see? Vision ultimately depends on many features of the visual nervous system, which are currently unknown for elephants. Furthermore, there is no behavioral measurement on elephant vision. However, by comparing the composition of the visual pigments in elephants to those in other species, we can infer some likely visual capabilities of elephants. We have shown that the RH1, SWS1, and LWS pigments have λmax-values of 496, 419, and 552 nm. Interestingly, these values are virtually identical to those of certain “color-blind” people, known as deuteranopes, who have only RH1, SWS1, and LWS pigments with respective λmax-values of 496 nm (Dartnall et al. 1983), 414 nm (Shi et al. 2001), and 552 nm (Merbs and Nathans 1992). Note that amino acid composition at site 180 of human LWS pigments is highly polymorphic; i.e., S180 and A180 are found in 60 and 40% of a population, respectively (Winderickx et al. 1992).
People with trichromatic color vision see not only four primary colors (blue, green, yellow, and red) but also various intermediate colors between them (Carroll et al. 2001). Instead of seeing four primary colors, however, color-blind people detect only two primary colors (blue and yellow) and do not see intermediate color (Neitz et al. 2001). Thus, when the two primary colors are mixed, the color-blind individuals detect either achromatic, i.e., white or gray, or one of the two basic hues (Jacobs et al. 1993). During the day, therefore, it is highly likely that elephants have the dichromatic color vision of deuteranopes.
What do the elephant and other arrhythmic animals see at night? In a typical human retina, the proportion of a rod-to-cone ratio is 95% with rod-free fovea (Oyster 1999). Many ungulates and carnivores seem to have similar rod/cone ratios of 85–99% (Calderone et al. 2003). Human and arrhythmic mammals, however, have one significant difference; that is, the fovea in the human retina consists of pure cones, but other mammals do not have such a pure cone region in their retina (Oyster 1999). High rod/cone ratios and lack of “rod-free areas” in the retina of many arrhythmic mammals provide an intriguing possibility of an additional dimension in wavelength detection (Jacobs et al. 1994). Note that blue-cone monochromat people are known to distinguish wavelengths in the range of 440–500 nm at twilight by using RH1 rod pigments and SWS1 cone pigments simultaneously (Reitner et al. 1991). The African coelacanths also use rod (RH1) and cone (RH2) pigments to detect a narrow range of wavelengths at ∼480 nm in their habitat (Yokoyama et al. 1999). Therefore, it is highly likely that elephants also use RH1 and SWS1 cone pigments together to discriminate a different range of wavelengths at 420–490 nm at night. Other arrhythmic animals must also have dichromatic color vision during the day and detect wavelengths somewhere between 430 and 500 nm at night, depending on their rhodopsins and specific types of blue pigments.
Acknowledgments
We acknowledge the contributions of those zoos that have submitted elephants to the University of California at Davis Veterinary Medical Teaching Hospital for necropsy. Comments by Ruth Yokoyama are greatly appreciated. This work was supported by National Institutes of Health grant GM-42379 and a start-up fund from Emory University (to S.Y.).
References
- Ali, M. A., and M. A. Klyne, 1985 Vision in Vertebrates. Plenum Press, New York.
- Archer, S. N., A. J. Hope and J. C. Partridge, 1996. The molecular basis for the blue-green sensitivity in the rod visual pigments of the European eel. Proc. R. Soc. Lond. Ser. B 262: 289–295. [DOI] [PubMed] [Google Scholar]
- Asenjo, A. B., J. Rim and D. D. Oprian, 1994. Molecular determinants of human red/green color discrimination. Neuron 12: 1131–1138. [DOI] [PubMed] [Google Scholar]
- Calderone, J. B., B. E. Reese and G. H. Jacobs, 2003. Topography of photoreceptors and retinal ganglion cells in the spotted hyena (Crocuta crocuta). Brain Behav. Evol. 62: 182–192. [DOI] [PubMed] [Google Scholar]
- Carleton, K. L., and T. D. Kocher, 2001. Cone opsin genes of African cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol. Biol. Evol. 18: 1540–1550. [DOI] [PubMed] [Google Scholar]
- Carroll, J., C. J. Murphy, M. Neitz, J. N. Ver Hoeve and J. Neitz, 2001. Photopigment basis for dichromatic color vision in the horse. J. Vision 1: 80–87. [DOI] [PubMed] [Google Scholar]
- Cowing, J. A., S. Poopalasundaram, S. R. Wilkie, P. R. Robinson, J. K. Bowmaker et al., 2002. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem. J. 367: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartnall, H. J. A, and J. N. Lythgoe, 1965. The spectral clustering of visual pigments. Vision Res. 5: 81–100. [DOI] [PubMed] [Google Scholar]
- Dartnall, H. J. A, J. K. Bowmaker and J. D. Mollon, 1983. Human visual pigments: microspectrophotometric results from the eyes of seven persons. Proc. R. Soc. Lond. Ser. B 220: 115–130. [DOI] [PubMed] [Google Scholar]
- Deeb, S. S., M. J. Wakefield, T. Tada, L. Marotte, S. Yokoyama et al., 2003. The cone visual pigments of an Australian marsupial, the Tamar wallaby (Macropus eugenii): sequence, spectral tuning, and evolution. Mol. Biol. Evol. 20: 1642–1649. [DOI] [PubMed] [Google Scholar]
- Ebrey, T., and Y. Koutalos, 2001. Vertebrate photoreceptors. Prog. Retin. Eye Res. 20: 49–94. [DOI] [PubMed] [Google Scholar]
- Ebrey, T., and Y. Takahashi, 2002 Photobiology of retinal proteins, pp. 101–133 in Photobiology for the 21st Century, edited by T. P. Cohil and D. P. Valenzeno. Valdenman, Overland Park, VA.
- Eizirik, E. W., J. Murphy and S. J. O'Brien, 2001. Molecular dating and biogeography of the early placental mammal radiation. J. Hered. 92: 212–219. [DOI] [PubMed] [Google Scholar]
- Fasick, J. I., and P. R. Robinson, 1998. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry 37: 432–438. [DOI] [PubMed] [Google Scholar]
- Fasick, J. I., M. L. Applebury and D. D. Oprian, 2002. Spectral tuning in the mammalian short wavelength sensitive cone pigments. Biochemistry 41: 6860–6865. [DOI] [PubMed] [Google Scholar]
- Gilbert, B., 1990. Once a malcontent, Ruby has taken up brush and palette. Smithsonian 21: 40–46. [Google Scholar]
- Hadjeb, N., and G. A. Berkowitz, 1996. Preparation of T-overhang vectors with high PCR product cloning efficiency. Biotechniques 20: 20–22. [DOI] [PubMed] [Google Scholar]
- Hubbard, R., and A. Kropf, 1958. The action of light on rhodopsin. Proc. Natl. Acad. Sci. USA 44: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, G. H., J. F. Deegan II, M. A. Crognale and J. A. Fenwick, 1993. Photopigments of dogs and foxes and their implications for canid vision. Vis. Neurosci. 10: 173–180. [DOI] [PubMed] [Google Scholar]
- Jacobs, G. H., J. F. Deegan II, J. Neitz, B. P. Murphy, K. V. Miller et al., 1994. Electrophysiological measurements of spectral mechanisms in the retinas of two cervids: white-tailed deer (Odocoileus virginianus) and fallow deer (Dama dama). J. Comp. Physiol. A 174: 551–557. [DOI] [PubMed] [Google Scholar]
- Jacobs, G. H., J. F. Deegan II and J. Neitz, 1998. Photopigment basis for dichromatic color vision in cows, goats, and sheep. Vis. Neurosci. 15: 581–584. [DOI] [PubMed] [Google Scholar]
- Kawamura, S., and S. Yokoyama, 1998. Functional characterization of visual and nonvisual pigments of American chameleon (Anolis carolinensis). Vision Res. 38: 37–44. [DOI] [PubMed] [Google Scholar]
- Khorana, H. G., B. E. Knox, F. Nasi, R. Swanson and D. A. Thompson, 1988. Expression of a bovine rhodopsin gene in Xenopus oocytes: demonstration of light- dependent ionic currents. Proc. Natl. Acad. Sci. USA 85: 7917–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., and T. Ohta, 1972. On the stochastic model for estimation of mutational distance between homologous proteins. J. Mol. Evol. 2: 87–90. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and S. B. Hedges, 1998. A molecular timescale for vertebrate evolution. Nature 392: 917–920. [DOI] [PubMed] [Google Scholar]
- Li, W.-H., 1997 Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Loop, M. S., C. L. Milligan and S. R. Thomas, 1987. Photopic spectral sensitivity of the cat. J. Physiol. 382: 537–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, O., M. Scally, C. J. Douady, D. J. Kao, R. W. Debry et al., 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409: 610–614. [DOI] [PubMed] [Google Scholar]
- Maglio, V. J., 1973. Origin and evolution of the Elephantidae. Trans. Am. Philos. Soc. 63: 1–149. [Google Scholar]
- McFarland, W. N., 1971. Cetacean visual pigments. Vision Res. 11: 1065–1076. [DOI] [PubMed] [Google Scholar]
- Merbs, S. L., and J. Nathans, 1992. Absorption spectrum of human cone pigments. Nature 356: 433–435. [DOI] [PubMed] [Google Scholar]
- Murphy, W. J., E. Eizirik, W. E. Johnson, Y. P. Zhang, O. A. Ryder et al., 2001. Molecular phylogenetics and the origin of placental mammals. Nature 409: 614–618. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nei, M., P. Xu and G. Glazko, 2001. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. USA 98: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitz, J., and G. H. Jacobs, 1989. Spectral sensitivity of cones in an ungulate. Vis. Neurosci. 2: 97–100. [DOI] [PubMed] [Google Scholar]
- Neitz, J., J. Carroll and M. Neitz, 2001. Color vision: almost reason enough for having eyes. Opt. Photon. News 12: 26–33. [Google Scholar]
- Neitz, M., J. Neitz and G. H. Jacobs, 1991. Spectral tuning of pigments underlying red-green color vision. Science 252: 971–974. [DOI] [PubMed] [Google Scholar]
- Nowak, R. M., 1991 Walker's Mammals of the World, Ed. 5. Johns Hopkins University Press, Baltimore.
- Oprian, D. D., R. S. Molday, R. J. Kaufman and H. G. Khorana, 1987. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc. Natl. Acad. Sci. USA 84: 8874–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster, C. W., 1999 The Human Eye: Structure and Function. Sinauer Associates, Sunderland, MA.
- Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima et al., 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289: 739–745. [DOI] [PubMed] [Google Scholar]
- Parry, J. W. L., S. Poopalasundaram, J. K. Bowmaker and D. M. Hunt, 2004. A novel amino acid substitution is responsible for spectral tuning in a rodent violet-sensitive visual pigment. Biochemistry 43: 8014–8020. [DOI] [PubMed] [Google Scholar]
- Radlwimmer, F. B., and S. Yokoyama, 1997. Cloning and expression of the red visual pigment gene of goat (Capra hircus). Gene 198: 211–215. [DOI] [PubMed] [Google Scholar]
- Reitner, A., L. T. Sharp and E. Zrenner, 1991. Is colour vision possible with only rods and blue-sensitive cones? Nature 352: 798–800. [DOI] [PubMed] [Google Scholar]
- Shi, Y., and S. Yokoyama, 2003. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc. Natl. Acad. Sci. USA 100: 8308–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., F. B. Radlwimmer and S. Yokoyama, 2001. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc. Natl. Acad. Sci. USA 98: 11731–11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M. S., W. J. Murphy, E. Eizirik and S. J. O'Brien, 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100: 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., J. P. Macke and J. Nathans, 1997. Mechanisms of spectral tuning in the mouse green cone pigments. Proc. Natl. Acad. Sci. USA 94: 8860–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennesen, M., 1998. Knoxville turns the BIG FIVE-O: a Tennessee zoo marks a milestone. Wildl. Conserv. 101: 66–67. [Google Scholar]
- Walls, G. I., 1942 The Vertebrate Eye and Its Adaptive Radiation. Cranbook Institute of Science, Bloomfield Hills, MI.
- Winderickx, J., D. T. Lindsey, E. Sanocki, D. Y. Teller, A. G. Motulsky et al., 1992. Polymorphism in red photopigment underlies variation in color matching. Nature 356: 431–433. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., and S. Yokoyama, 1990. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc. Natl. Acad. Sci. USA 87: 9315–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., B. E. Knox and S. Yokoyama, 1995. Rhodopsin from the fish, Astyanax: role of tyrosine 261 in the red shift. Invest. Ophthalmol. Vis. Sci. 36: 939–945. [PubMed] [Google Scholar]
- Yokoyama, S., 2000. a Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19: 385–419. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S., 2000. b Phylogenetic analysis and experimental approaches to study color vision in vertebrates. Methods Enzymol. 315: 312–325. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S., and F. B. Radlwimmer, 1999. The molecular genetics of red and green color vision in mammals. Genetics 153: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S., and F. B. Radlwimmer, 2001. The molecular genetics of red and green color vision in vertebrates. Genetics 158: 1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S., and R. Yokoyama, 1996. Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annu. Rev. Ecol. Syst. 27: 543–567. [Google Scholar]
- Yokoyama, S., H. Zhang, F. B. Radlwimmer and N. S. Blow, 1999. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae). Proc. Natl. Acad. Sci. USA 96: 6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]