Abstract
Compared to protein-coding sequences, the evolution of noncoding sequences and the selective constraints placed on these sequences is not well characterized. To compare the evolution of coding and noncoding sequences, we have conducted a survey for DNA polymorphism at five randomly chosen loci among a diverse collection of 81 strains of Saccharomyces cerevisiae. Average rates of both polymorphism and divergence are 40% lower at noncoding sites and 90% lower at nonsynonymous sites in comparison to synonymous sites. Although noncoding and coding sequences show substantial variability in ratios of polymorphism to divergence, two of the loci, MLS1 and PDR10, show a higher rate of polymorphism at noncoding compared to synonymous sites. The high rate of polymorphism is not accompanied by a high rate of divergence and is limited to a few small regions. These hypervariable regions include sites with three segregating bases at a single site and adjacent polymorphic sites. We show that this clustering of polymorphic sites is significantly greater than one would expect on the basis of the spacing between polymorphic fourfold degenerate sites. Although hypervariable noncoding sequences could result from selection on regulatory mutations, they could also result from transient mutational hotspots.
PROBABILISTIC models for the molecular evolution of DNA sequences have provided much insight into protein function and evolution (Kimura 1983; Fay and Wu 2003). The power of these models is derived in part from the genetic code, which results in the interspersion of sites with nonsynonymous and synonymous effects on the amino acid sequence of a protein. In contrast to protein-coding sequences, we know relatively little about the function and evolution of cis-regulatory sequences. Although some models have been developed (Moses et al. 2004a,b), a major limitation is the paucity of experimentally identified cis-regulatory sequences.
The examination of polymorphism and divergence in cis-regulatory sequences has shown that while these sequences are constrained, substantial variation exists both within and between species (Jenkins et al. 1995; Ludwig and Kreitman 1995; Ludwig et al. 1998; Tautz and Nigro 1998; Dermitzakis et al. 2003; Moses et al. 2003; Phinchongsakuldit et al. 2004). This variation can be explained under a neutral model since there are degenerate positions within transcription factor binding sites (Moses et al. 2003) and redundant binding sites within an enhancer (Ludwig et al. 1998, 2000). In one study, the DNA sequence variation was found to be inconsistent with a neutral model (Jenkins et al. 1995). However, these studies have been limited to the few regulatory sequences that have been examined in detail, mostly those acting early in Drosophila development.
The genome sequencing of closely related species has provided a wealth of data on the molecular evolution of both coding and noncoding sequences (Cliften et al. 2003; Kellis et al. 2003; Thomas et al. 2003; Richards et al. 2005). One of the main motivations for these projects has been the identification of conserved noncoding sequences, the majority of which likely function in gene regulation. The identification of regulatory sequences by their conservation between species presents a challenge to understanding their evolution since not all regulatory sequences may be tightly conserved. One approach is to study noncoding sequences in their entirety, eliminating any bias in the method used to distinguish functional and nonfunctional sequences.
The examination of polymorphism and divergence in unannotated noncoding sequences has revealed a number of regions showing a higher than expected rate of polymorphism or divergence. The rate of polymorphism but not divergence was found to be greater in the 5′-UTR and intronic sequence of hunchback compared to that in synonymous sites in adjacent hunchback coding sequences (Tautz and Nigro 1998). A small 200-bp region upstream of Attacin C showed a rate of polymorphism 10-fold higher than that found at nearby synonymous sites (Lazzaro and Clark 2001). Divergence and linkage disequilibrium were also much higher in the region. Examination of polymorphism and divergence in 136 5′-UTR sequences in humans revealed a higher ratio of divergence to polymorphism at 5′-UTR compared to that in fourfold degenerate sites found in adjacent coding sequences (Hellmann et al. 2003). The same result was found for noncoding sequences upstream of accessory gland proteins (Kohn et al. 2004). Although there are a number of caveats to comparing variation in noncoding sites to that in synonymous sites, together the data suggest the selective forces acting on coding and noncoding sequences may be quite different.
To compare rates of variation in coding and noncoding sequences, we have surveyed DNA polymorphism at five randomly chosen loci in a diverse collection of 81 strains of Saccharomyces cerevisiae. For each locus we examined 608–845 bp of coding sequence at the 5′ end of the gene and 611–804 bp of noncoding sequence, nearly the entire 5′-intergenic sequence. The five loci include: CCA1, a tRNA nucleotidyltransferase (Aebi et al. 1990); CYT1, which encodes cytochrome c1, a component of the mitrochondrial respiratory chain (Sadler et al. 1984); MLS1, a malate synthase (Hartig et al. 1992); PDR10, an ATP-binding cassette membrane pump involved in pleiotropic drug resistance (Balzi and Goffeau 1995); and ZDS2, known to function in chromatin silencing and cell cycle progression (Bi and Pringle 1996; Roy and Runge 1999). Similar to variation at nonsynonymous sites, all five loci show lower rates of divergence in noncoding compared to synonymous sites. Yet, two genes, MLS1 and PDR10, show higher rates of polymorphism at noncoding compared to synonymous sites.
MATERIALS AND METHODS
Strains:
Strains were obtained from a variety of sources. B1–B6 were obtained from B. Dunn. I14 was collected by J. Fay. CDB and PR were obtained from Red Star Yeast (Oakland, CA). K1–K15 were obtained from N. Goto-Yamamoto and the NODAI culture collection. M1–M34 were provided by R. Mortimer. UC1–UC10 were obtained from the University of California (Davis, CA) Department of Viticulture and Enology culture collection. SB was bought at Whole Foods (Berkeley, CA). Y1–Y12 were provided by C. Kurtzman from the Agriculture Research Service Culture Collection. YJM145–YJM1129 were obtained from J. McCusker. YPS163–YPS1009 were provided by P. Sniegowski.
Polymorphism survey:
Five genes from divergently transcribed intergenic sequences were randomly chosen from the Saccharomyces Genome Database, excluding RNA genes and genes of unknown function. Genes with no clear ortholog in S. paradoxus were not considered. For each gene, the 5′-intergenic sequence and a portion of the coding sequence were amplified by PCR, purified, and both strands were sequenced using BigDye (Perkin Elmer, Boston) termination sequencing. Phred and Phrap were used to call bases and assemble a contiguous sequence for each strain (Ewing and Green 1998). Consed was used to visualize the sequence assemblies and to identify heterozygous sites. Only one of the two haplotypes inferred using PHASE were used in the analyses (Stephens et al. 2001). Sequences were aligned using ClustalW. Population genetic analyses were done using DNASP (Rozas and Rozas 1999). Substitution rates between species were estimated using PAML (Yang 1997).
RESULTS
DNA polymorphism:
DNA polymorphism was surveyed in 81 strains of S. cerevisiae (Table 1), constituting a total of 3561 bp of intergenic sequence and 3671 bp of coding sequence. A total of 191 polymorphic sites were found, constituting 67 unique haplotypes. Four of the polymorphic sites contained 3 segregating bases. Twelve insertions and no deletions were found. The 12 insertions ranged in length from 1 to 3 bp. Of the 12 insertions, 8 were within a string of A or T bases ranging in size from 4 to 11 bp, 1 was within a C4 repeat, 1 consisted of a TA2 repeat, and 1 consisted of a TC5 repeat.
TABLE 1.
Strains studied and their source
| ID | Strain | Location | Source | Date |
|---|---|---|---|---|
| B1 | Lalvin 71B | France | Vineyard (commercial) | NA |
| B2 | Levuline ALS | NA | Vineyard (commercial) | NA |
| B3 | Zymaflore F15 | France | Vineyard (commercial) | NA |
| B4 | Lalvin CY-3079 | NA | Vineyard (commercial) | NA |
| B5 | Lalvin BM45 | NA | Vineyard (commercial) | NA |
| B6 | Zymalfore VL3 | France | Vineyard (commercial) | NA |
| CDB | Côte des Blancs | Germany | Vineyard (commercial) | NA |
| I14 | Italy | Vineyard (soil) | 2002 | |
| K1 | Kyokai no. 1 | Japan | Sake | 1906 |
| K5 | Kyokai no. 5 | Japan | Sake | 1925 |
| K9 | Kyokai no. 9 | Japan | Sake | 1950s |
| K10 | Kyokai no. 10 | Japan | Sake | 1952 |
| K11 | Awamori | Japan | Sake (Shochu) | 1981 |
| K12 | AKU-4011 | Japan | Sake | NA |
| K13 | NRIC 23 | Japan | Sake | NA |
| K14 | NRIC 1413 | Japan | Sake | NA |
| K15 | NRIC 1685 | Japan | Sake | NA |
| M1 | Italy | Vineyard | 1993 | |
| M2 | Italy | Vineyard | 1993 | |
| M3 | Italy | Vineyard | 1993 | |
| M4 | Italy | Vineyard | 1993 | |
| M5 | Italy | Vineyard | 1993 | |
| M6 | Italy | Vineyard | 1993 | |
| M7 | Italy | Vineyard | 1993 | |
| M8 | Italy | Vineyard | 1993 | |
| M9 | Italy | Vineyard | 1993 | |
| M11 | Italy | Vineyard | 1993 | |
| M12 | Italy | Vineyard | 1993 | |
| M13 | Italy | Vineyard | 1993 | |
| M15 | Italy | Vineyard | 1993 | |
| M17 | Italy | Vineyard | NA | |
| M19 | Italy | Vineyard | NA | |
| M20 | Italy | Vineyard | NA | |
| M21 | Italy | Vineyard | NA | |
| M22 | Italy | Vineyard | NA | |
| M24 | Italy | Vineyard | NA | |
| M29 | Italy | Vineyard | 1994 | |
| M30 | Italy | Vineyard | 1994 | |
| M31 | Italy | Vineyard | 1994 | |
| M32 | Italy | Vineyard | NA | |
| M33 | Italy | Vineyard | NA | |
| M34 | Italy | Vineyard | NA | |
| PR | Pasteur red | France | Vineyard (commercial) | NA |
| S288C | California | Nature (fig) | 1937 | |
| SB | S. boulardii | Indonesia | Nature (lychee fruit) | NA |
| UC1 | UCD 51 | France | Vineyard | 1948 |
| UC2 | UCD 175 | Sicily, Italy | Vineyard | 1953 |
| UC4 | UCD 529 | Germany | Vineyard | Pre-1958 |
| UC5 | UCD 612 | Kurashi, Japan | Sake | Pre-1974 |
| UC6 | UCD 765 | Australia | Vineyard | NA |
| UC7 | UCD 781 | Switzerland | Vineyard | NA |
| UC8 | UCD 820 | South Africa | Vineyard | Pre-1988 |
| UC9 | UCD 762 | Italy | Vineyard | Pre-1984 |
| UC10 | UCD 2120 | California | Vineyard | 1998 |
| Y1 | NRRL y390 | Nature (mushroom) | Pre-1940 | |
| Y3 | NRRL y1438 | Africa | Fermentation (palm wine) | Pre-1946 |
| Y4 | NRRL y1532 | Indonesia | Nature (fruit) | Pre-1947 |
| Y5 | NRRL y1546 | West Africa | Fermentation (bili wine) | Pre-1947 |
| Y6 | NRRL yb1952 | French Guiana | NA | Pre-1950 |
| Y8 | NRRL y2411 | Turkey | Vineyard | Pre-1957 |
| Y9 | NRRL y5997 | Indonesia | Fermentation (ragi) | Pre-1962 |
| Y10 | NRRL y7567 | Philippines | Fermentation (coconut) | Pre-1973 |
| Y12 | NRRL y12633 | Ivory Coast | Fermentation (palm wine) | Pre-1981 |
| YJM145 | Segregant YJM128 | Missouri | Clinical | Pre-1989 |
| YJM269 | Fermentation (apple juice) | 1953 | ||
| YJM270 | Europe | Vineyard | Pre-1957 | |
| YJM280 | Segregant YJM273 | United States | Clinical | Pre-1994 |
| YJM308 | United States | Clinical | Pre-1994 | |
| YJM320 | Segregant YJM309 | United States | Clinical | Pre-1994 |
| YJM326 | Segregant YJM310 | United States | Clinical | |
| YJM339 | Segregant YJM311 | United States | Clinical | Pre-1994 |
| YJM421 | Segregant YJM419 | United States | Clinical | Pre-1994 |
| YJM434 | Europe | Clinical | ||
| YJM436 | Europe | Clinical | Pre-1994 | |
| YJM440 | United States | Clinical | Pre-1994 | |
| YJM454 | United States | Clinical | Pre-1994 | |
| YJM627 | Segregant Y55 | France | NA | |
| YJM1129 | NRRL y-567 | Fermentation (distillery) | Pre-1912 | |
| YPS1000 | New Jersey | Nature (oak exudate) | 2000 | |
| YPS1009 | New Jersey | Nature (oak exudate) | 2000 | |
| YPS163 | Pennsylvania | Nature (oak exudate) | 1999 |
ID, identification number; NA, not available.
Heterozygous sites were found in 35 of the 81 strains and at 94 of the 191 polymorphic sites. A chi-square test for Hardy-Weinberg equilibrium identified 45 polymorphic sites with a significant deficit of heterozygous strains (P < 0.001). Because most natural isolates of S. cerevisiae are homothallic diploids (Mortimer et al. 1994), haploid spores are capable of switching mating type and selfing. Thus, loss of heterozygosity is not unexpected. However, of the 35 strains with heterozygous sites, 10 strains were heterozygous at between 11 and 26 sites while the remaining 25 strains were heterozygous at 5 or fewer sites. The strains with high levels of heterozygosity can be explained by a recent mating between two distantly related strains or by loss of their capability to sporulate, a common phenotype found in commercial wine strains (Johnston et al. 2000).
The distinction of haploid and diploid strains is important for allele frequency estimates and other population genetic analyses. Although sporulation is a clear indication of diploidy, the absence of sporulation is uninformative since some diploids sporulate at very low frequencies. To avoid this problem we analyzed only one allele from each strain. For those strains containing heterozygous sites, we inferred haplotypes using the program PHASE (Stephens et al. 2001) and randomly chose one of the two inferred haplotypes. Because 16 of the heterozygous sites are unique variable sites that are present in only a single strain, the random sampling resulted in the loss of 7 polymorphic sites. All subsequent analyses are based on the 184 polymorphic sites that remained (Table 2).
TABLE 2.
Polymorphic sites identified in five genes
| Surveyed sites
|
Polymorphic sites
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Sample size | Noncoding | Coding | Synonymous | NC | N | S | I |
| CCA1 | 73 | 721 | 788 | 179.4 | 20 | 1 | 11 | 2 |
| CYT1 | 67 | 611 | 608 | 149.1 | 15 | 1 | 10 | 2 |
| MLS1 | 77 | 804 | 845 | 186.5 | 41 | 4 | 12 | 0 |
| PDR10 | 75 | 730 | 758 | 177.4 | 26 | 7 | 6 | 8 |
| ZDS2 | 59 | 695 | 672 | 140.7 | 21 | 5 | 8 | 0 |
| Total | 81 | 3561 | 3671 | 833.1 | 123 | 18 | 47 | 12 |
NC, noncoding; N, nonsynonymous; S, synonymous; I, insertion.
Diversity at synonymous sites ranges from 0.33 to 1.32% at the five loci (Table 3), where diversity is measured by the average number of pairwise differences between strains per base pair. The overall average diversity, 0.84%, is higher than that in humans, 0.11–0.15% (Cargill et al. 1999; Halushka et al. 1999), but lower than that in Drosophila melanogaster, 1.41% (Kern and Begun 2005).
TABLE 3.
Population sample statistics from five genes
| π (×100)
|
Kst
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | NC | N | S | Tajima's D | Fu and Li's D | Source | Location |
| CCA1 | 0.39 | 0.03 | 0.76 | −1.08 | −1.54 | 0.28 | 0.28 |
| CYT1 | 0.25 | 0.01 | 1.32 | −1.09 | 0.51 | 0.11 | 0.08 |
| MLS1 | 0.91 | 0.10 | 0.80 | −0.60 | −1.51 | 0.34 | 0.25 |
| PDR10 | 0.55 | 0.15 | 0.33 | −1.05 | −2.46 | 0.46 | 0.37 |
| ZDS2 | 0.51 | 0.16 | 0.99 | −0.69 | 0.37 | 0.23 | 0.20 |
| Average | 0.521 | 0.091 | 0.838 | −0.90 | −0.93 | 0.28 | 0.24 |
π, the average number of pairwise differences between strains per base pair. Kst (Hudson et al. 1992) was measured by source and location as designated in Table 1.
The frequency spectrum is slightly skewed toward rare variants compared to that expected from a randomly mating population of constant size under a Wright-Fisher model. Tajima's D (Tajima 1989) ranges from −0.60 to −1.09 among the five genes, none of which are significant (Table 3). One hundred twenty-eight SNPs have a minor allele frequency of <10% compared to the 99 expected under a Wright-Fisher model (Watterson 1975). Four of the 12 insertions, all found within the promoter of PDR10, have a minor allele frequency of >10%.
Population structure:
We examined population structure stratified by the source from which each strain was obtained and by continent from which each strain was isolated (Table 1). Forty-two strains were from Europe, 14 from Asia, 15 from America, 4 from Africa, and 6 are of unknown origin. Forty-four strains were isolated from grapes, wine fermentations, or commercial wine production. Seven strains were from natural samples, including oak tree exudates, a mushroom, a fig, and various fruits. Eleven strains were obtained from clinical samples of immunocompromised patients. Ten strains were obtained from sake fermentations. Seven strains were obtained from fermentations excluding wine and sake. Two strains were from an unknown source.
Significant population differentiation was found both among sample sources and among sample locations (Table 3, P < 0.001 for all genes). However, the sources and locations from which the strains were isolated are correlated with one another. Most European strains were obtained from vineyards, most North American strains were obtained from clinical samples, and most Asian strains were obtained from fermentation of substrates other than grapes.
Different patterns of variation were found among different groups of strains. A significant reduction in diversity is found within strains from wine and sake compared with diversity within other groups and total diversity (Table 4). In addition to a reduction in diversity, the vineyard strains also show a greater proportion of rare variants, as measured by Tajima's D, compared to that in other groups and to the total (Table 4).
TABLE 4.
Diversity within groups
| Sourcea | Strains | π (×100)b | Rm | TD |
|---|---|---|---|---|
| Sake wine | 9 | 0.10 (0.01) | 3 | 0.839 |
| Grape wine | 23 | 0.14 (0.03) | 5 | −1.57 |
| Clinical | 4 | 0.42 (0.10) | 2 | 0.311 |
| Nature | 5 | 0.50 (0.08) | 3 | −0.31 |
| Fermentation | 4 | 0.55 (0.15) | 0 | 0.563 |
| Total | 45 | 0.42 (0.03) | 31 | −0.57 |
π, the average number of pairwise differences between strains, per base pair; Rm, the minimum number of recombination events; TD, Tajima's D-statistic.
Only strains without missing data are used.
Standard error is shown in parentheses.
Linkage disequilibrium and recombination:
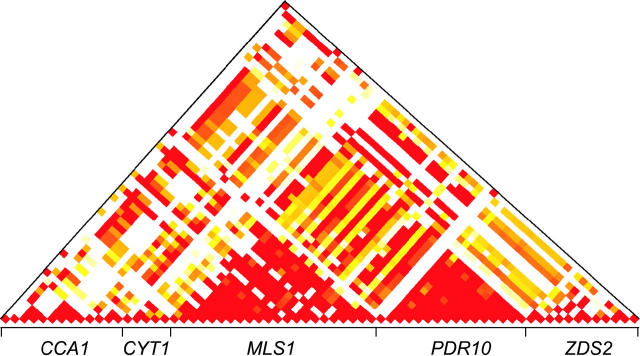
There are significant levels of linkage disequilibrium between unlinked genes (Figure 1). Linkage disequilibrium was measured by the average absolute value of D′ for all polymorphic sites with a minor allele frequency of >10% among the 45 strains for which there was no missing data (Table 5). The average absolute value of D′ from all pairwise comparisons between loci ranges from 0.57 to 0.82. All pairwise comparisons within loci range from 0.71 (ZDS2) to 0.98 (PDR10). The expected absolute value of D′ for unlinked sites is 0.26 and was obtained by resampling of polymorphic sites, keeping the allele frequencies constant. This high rate of linkage disequilibrium can be expected given the population structure found in S. cerevisiae and its ability to reproduce asexually.
Figure 1.—
Linkage disequilibrium measured by |D′|. Only pairs with |D′| > 0.5 and a significant association are shown (Fisher's exact test, P < 0.05). The color indicates |D′| that ranges from 0.5 (yellow) to 1.0 (red).
TABLE 5.
Average pairwise linkage disequilibrium
| Gene | No. sites | CCA1 | CYT1 | MLS1 | PDR10 | ZDS2 |
|---|---|---|---|---|---|---|
| CCA1 | 11 | 0.75 | 0.73 | 0.76 | 0.82 | 0.65 |
| CYT1 | 4 | 0.36 | 0.88 | 0.59 | 0.65 | 0.58 |
| MLS1 | 20 | 0.33 | 0.28 | 0.94 | 0.75 | 0.69 |
| PDR10 | 14 | 0.27 | 0.23 | 0.23 | 0.98 | 0.57 |
| ZDS2 | 10 | 0.34 | 0.33 | 0.30 | 0.26 | 0.71 |
Linkage disequilibrium (LD) within a locus is shown on the diagonal, LD between loci is shown above the diagonal, LD between randomized data is shown below the diagonal. Linkage disequilibrium was measured by the absolute value of D′ for sites with a minor allele frequency of >10%. Only the 45 strains with complete data for all five genes were used.
There is ample evidence of recombination within each of the five loci. Each locus shows evidence of between two and five recombination events by the four-gamete test (Table 4). The absolute value of D′ within a locus is negatively correlated with distance between polymorphic sites (P = 5 × 10−5), but not for the randomized data (Table 6). This could be due to gene conversion or recombination between individuals from the same subpopulation but not from different subpopulations. The recombination mutation ratio, estimated from the ratio of θw from synonymous sites over 4Nc, ranges from 1.4 to 2.7 and the average is 2.1 (Table 7). The mutation rate has been estimated from CAN1 and SUP3 at 2.25 × 10−10 per base pair per generation (Drake 1991). Given that 82% of spontaneous mutations are single-base substitutions (Kang et al. 1992), the point mutation rate is 1.84 × 10−10. The genomic average rate of recombination is 0.34 cM/kbp or 6.8 × 10−6 recombination events per base pair (Cherry et al. 1997). Similar to a previous study (Jensen et al. 2001), the laboratory estimate of the ratio of recombination events to mutation events is four orders of magnitude greater than that inferred from the polymorphism data. This can be explained by higher rates of asexual compared to sexual reproduction as well as by mating-type switching, which enables a cell to mate with its forebear following meiosis.
TABLE 6.
Average absolute value ofD′
| Data | <10 bp | <100 bp | <1000 bp | >1000 bp |
|---|---|---|---|---|
| Observed | 1.00 | 0.96 | 0.87 | 0.87 |
| Resampled | 0.23 | 0.26 | 0.27 | 0.28 |
TABLE 7.
Population sample statistics from 45 strains with data from all five genes
| Gene | θW (×100) | R (×100) | θW/R |
|---|---|---|---|
| CCA1 | 1.28 | 0.48 | 2.66 |
| CYT1 | 1.53 | 1.04 | 1.48 |
| MLS1 | 0.98 | 0.36 | 2.73 |
| PDR10 | 0.65 | 0.27 | 2.40 |
| ZDS2 | 1.30 | 0.94 | 1.38 |
| Average | 1.15 | 0.62 | 2.13 |
θW, a measure of variation based on the number of segregating sites (Watterson 1975); R, a measure of recombination (Hudson 1987).
Selection on synonymous sites:
The detection of selection on nonsynonymous or noncoding sites is greatly facilitated if synonymous sites are effectively neutral. In S. cerevisiae, there is ample evidence that synonymous sites are not neutral (Bennetzen and Hall 1982; Bulmer 1987). However, not all genes and not all synonymous sites may be influenced by selection. For the purposes of detecting selection on nonsynonymous or noncoding sites, synonymous sites may be considered effectively neutral if their substitution rate and pattern of preferred and unpreferred synonymous changes are no different from those in neutral sites.
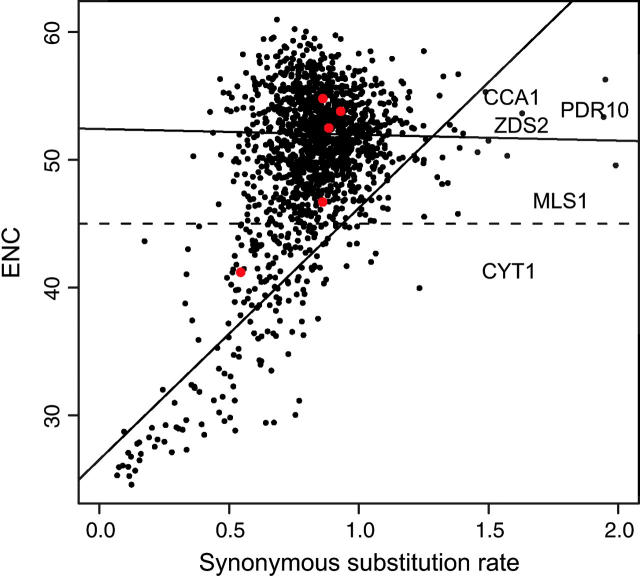
To determine which genes in S. cerevisiae are clearly affected by selection on synonymous sites, we compared the synonymous substitution rate to codon bias (Figure 2). From 1538 genes, there is a clear reduction in the synonymous substitution rate for genes with high codon bias or a small effective number of codons (ENC). However, most genes have a synonymous substitution rate that is not correlated with codon bias. We arbitrarily classified genes as high and low bias, using an ENC cutoff of 45. The 1331 high-bias genes have an average synonymous substitution rate of 0.87 and show no correlation between codon bias and synonymous substitution rate. In contrast, the low-bias genes have an average synonymous substitution rate of 0.60 and show a significant correlation between codon bias and synonymous substitution rate (Pearson's r = 0.74, P < 10−15). With the exception of CYT1, the genes examined in this study have a synonymous substitution rate nearly identical to the average rate and do not have high levels of codon bias.
Figure 2.—
Synonymous substitution rate among S. cerevisiae, S. paradoxus, and S. mikatae in relation to the average codon bias from the three species, as measured by ENC (Wright 1990). The dashed line shows the arbitrary cutoff used to distinguish high- and low-biased genes. The two solid lines are the least-squares fit of a regression of codon bias and synonymous substitution rate for the genes showing high and low codon bias. The five genes examined in this study are shown in red.
The pattern of substitutions at synonymous sites is indicative of whether synonymous sites are at mutation-selection balance. If they are not, the assumption that synonymous sites are effectively neutral is violated. In Drosophila, the relationship between codon bias and synonymous substitution rate is very weak if present (Dunn et al. 2001; Bierne and Eyre-Walker 2003). There is, however, a clear difference in the pattern of preferred and unpreferred synonymous substitutions between D. melanogaster and D. similans (Akashi 1996; Begun 2001). To determine whether patterns of synonymous substitutions in S. cerevisiae show a similar nonequilibrium status, we compared the number of unpreferred and preferred changes along the lineage leading to S. cerevisiae and within strains of S. cerevisiae (Table 8). Both polymorphic and fixed synonymous changes show an equal number of preferred to unpreferred (P → U) and unpreferred to preferred (U → P) changes.
TABLE 8.
Preferred and unpreferred synonymous polymorphism and divergence
| Polymorphic changes
|
Fixed differences
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | P → P | P → U | U → P | U → U | P → P | P → U | U → P | U → U |
| CCA1 | 2 | 3 | 2 | 2 | 3 | 5 | 6 | 5 |
| CYT1 | 2 | 3 | 2 | 1 | 5 | 4 | 2 | 3 |
| MLS1 | 3 | 3 | 4 | 0 | 9 | 8 | 8 | 2 |
| PDR10 | 1 | 0 | 2 | 1 | 3 | 11 | 11 | 5 |
| ZDS2 | 1 | 1 | 2 | 4 | 1 | 5 | 6 | 6 |
| Total | 9 | 10 | 12 | 8 | 21 | 33 | 33 | 21 |
P and U, preferred and unpreferred codons, respectively; e.g., P → P is a synonymous substitution from a preferred to a different preferred codon.
The data show that the synonymous substitution rate and pattern of synonymous substitution in four of the five genes are consistent with those expected for neutral sites. CYT1 has a reduced rate of synonymous substitution, but, interestingly, has the highest rate of synonymous-site diversity (Table 3). The HKA test (Hudson et al. 1987) reveals a lower ratio of synonymous polymorphism to divergence in CYT1 compared to that in PDR10 (P = 0.048), but not in comparison to that in any of the other three genes. After correction for multiple comparisons this difference is not significant.
Selection on nonsynonymous and noncoding sites:
The ratio of nonsynonymous to synonymous substitutions (dN/dS) measures the selective constraint on a protein. In the absence of positive selection or any changes in selective constraint, the dN/dS ratio should be constant across lineages and should not be greater than one (Fay and Wu 2001). None of the five proteins show significant differences in the levels of constraint among the lineages leading to S. cerevisiae, S. paradoxus, and S. mikatae (likelihood-ratio test using PAML, P > 0.05). The combined dN/dS ratios are 0.09, 0.10, and 0.09 for the branch leading to S. cerevisiae, S. paradoxus, and S. mikatae, respectively.
The ratio of noncoding to synonymous substitutions (dNC/dS) measures the selective constraint on noncoding sequences, assuming the mutation rate is the same across the coding and noncoding sequences. All of the promoters show considerable levels of functional constraint. The combined dNC/dS ratios are 0.53, 0.49, and 0.49 for the branch leading to S. cerevisiae, S. paradoxus, and S. mikatae, respectively. This implies that nearly one-half of intergenic sequences are functionally constrained, only slightly >0.52, the median dNC/dS from 2098 genes (Doniger et al. 2005).
Under the same assumptions used to test for branch-specific dN/dS ratios, the ratio of nonsynonymous- to synonymous-site polymorphism (pN/pS) should equal dN/dS. This comparison is the basis for the McDonald-Kreitman (MK) test (McDonald and Kreitman 1991), which compares polymorphic sites and fixed differences rather than estimates of substitution rates. The average pN/pS ratio, 0.11, is nearly identical to that of divergence, 0.09 (Table 9). Similarly, the average pNC/pS ratio of diversity, 0.62, is similar to that of divergence, 0.54 (Table 9).
TABLE 9.
Rates of DNA polymorphism compared to divergence
| Polymorphism (π × 100)
|
Divergence
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | NC | N | S | NC/S | N/S | dNC | dN | dS | dNC/dS | dN/dS |
| CCA1 | 0.39 | 0.03 | 0.76 | 0.52 | 0.04 | 0.13 | 0.01 | 0.21 | 0.59 | 0.02 |
| CYT1 | 0.25 | 0.01 | 1.32 | 0.19 | 0.01 | 0.06 | 0.00 | 0.14 | 0.47 | 0.04 |
| MLS1 | 0.91 | 0.10 | 0.80 | 1.14 | 0.13 | 0.15 | 0.00 | 0.21 | 0.69 | 0.02 |
| PDR10 | 0.55 | 0.15 | 0.33 | 1.65 | 0.46 | 0.13 | 0.05 | 0.28 | 0.45 | 0.18 |
| ZDS2 | 0.51 | 0.16 | 0.99 | 0.52 | 0.16 | 0.13 | 0.04 | 0.26 | 0.49 | 0.15 |
| Average | 0.52 | 0.09 | 0.84 | 0.62 | 0.11 | 0.12 | 0.02 | 0.22 | 0.54 | 0.09 |
Rates of divergence were obtained for the lineage leading to S. cerevisiae using PAML.
The comparison of N/S ratios from polymorphism and divergence can be misleading if positive selection increases the N/S ratio of divergence and negative selection increases the N/S ratio of polymorphism (Fay et al. 2001). The effect of negative selection on the N/S ratio of polymorphism can be examined by comparing low-frequency to common polymorphism. Both Drosophila (Fay et al. 2002) and humans (Fay et al. 2001) show an elevated N/S ratio of rare compared to common polymorphism, indicative of deleterious mutations segregating at low frequency in the population. In S. cerevisiae, the ratio of the rate of nonsynonymous and synonymous polymorphism that is rare, 0.12, is nearly identical to the rate of common polymorphism, 0.11. The ratio of NC/S from rare polymorphism, 0.60, is also very similar to that of common polymorphism, 0.62. This can be explained if most deleterious mutations are recessive and removed from the population following mating-type switching and selfing.
The overall pattern of polymorphism and divergence indicates selective constraint along the lineage leading to S. cerevisiae is similar to that found among extant populations. However, natural selection may influence polymorphism and divergence at individual genes or regions without affecting overall patterns of polymorphism and divergence. Because of the paucity of nonsynonymous polymorphism and divergence, we compared variation only in noncoding to synonymous sites. The NC/S ratio of diversity is larger than that of divergence for MLS1, PDR10, and ZDS2 and lower than that of divergence for CCA1 and CYT1. In addition, the NC/S ratio of polymorphism is greater than unity for both MLS1 and PDR10.
Two tests can be used to assess the significance of the difference between noncoding and synonymous polymorphism and divergence. The MK test can be applied to the number of polymorphic and fixed noncoding and synonymous changes (McDonald and Kreitman 1991). However, the MK test assumes the mutation rate in the two regions is the same, the coalescence time for the two regions is the same, and the number of fixed differences between species can be reliably determined. The first two assumptions are reasonable when the MK test is applied to nonsynonymous and synonymous changes. However, the latter assumption is not justified when divergence is >5–10% because of multiple hits (Templeton 1996). The HKA test can also be applied to noncoding- and synonymous-site polymorphism and divergence (Hudson et al. 1987). The HKA test explicitly accounts for any differences in mutation rates or coalescence times between the two regions, but, like the MK test, does not account for multiple hits. Despite these concerns, we applied an MK and an HKA test to noncoding and synonymous polymorphism and divergence. For the MK test, the number of fixed differences along the lineage leading to S. cerevisiae was estimated by the maximum-likelihood estimate of the synonymous and noncoding substitution rate multiplied by the number of synonymous and noncoding sites. Neither the MK nor the HKA test was significant for any of the genes (P > 0.05).
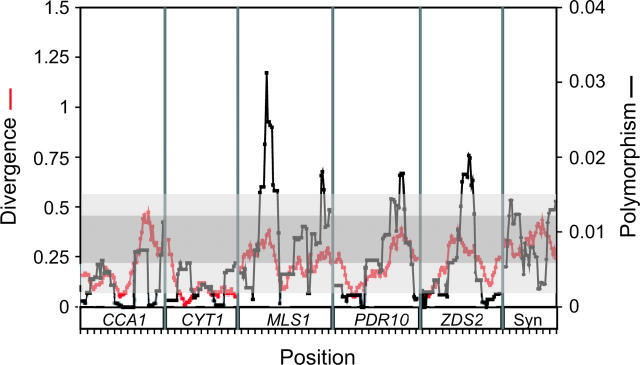
If locus-specific differences in the ratio of noncoding polymorphism to divergence have occurred by chance, the ratio of polymorphism to divergence should be relatively constant across a sliding window of each noncoding sequence. Alternatively, if natural selection has increased or decreased the rate of noncoding polymorphism or divergence, the effect of selection may well be localized to a portion of the noncoding region. To examine diversity in the rate of noncoding polymorphism and divergence, we plotted a sliding window of diversity and substitution rate across the noncoding region from each gene as well as across the concatenated fourfold degenerate synonymous sites from all of the genes (Figure 3). The two y-axes in Figure 3 are scaled such that the average rate of synonymous polymorphism is equal to the average rate of synonymous divergence. Noncoding divergence is more variable and on average lower than synonymous-site divergence, as expected. Noncoding polymorphism is also much more variable than synonymous polymorphism, but in four different regions is greater than the range found at synonymous sites, as shown in Figure 3 (light gray area). Two of the noncoding hypervariable regions lie upstream of MLS1 and the other two are upstream of PDR10 and ZDS2. The two hypervariable regions upstream of MLS1 contain 2 of the 4 sites with 3 segregating bases and 5 additional segregating sites that are within 3 bases of one another (Table 10). The hypervariable region upstream of PDR10 has 10 segregating sites that are within 7 bp of another segregating site (Table 11).
Figure 3.—
Sliding window of polymorphism compared to divergence for noncoding and synonymous sites. Divergence (red) is between S. cerevisiae and S. paradoxus as measured by the Jukes-Cantor model (Jukes and Cantor 1969), and polymorphism (black) is measured by diversity among strains without missing data. The range of synonymous-site divergence (gray) and polymorphism (light gray) is shown as shaded regions. The plot is separated into six regions labeled below the abscissa. The first five regions are from the noncoding sequences 5′ of each gene. The last region (Syn) is the concatenation of fourfold degenerate synonymous sites from the coding sequences of all five genes.
TABLE 10.
Haplotypes from two hypervariable regions upstrem ofMLS1
| Region 1
|
Region 2
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | 201 | 203 | 226 | 233 | 242 | 249 | 253 | 288 | 294 | 295 | 296 | 328 | 637 | 641 | 649 | 661 | 691 | 720 | 721 |
| S. mikatae | C | A | T | T | T | C | — | A | A | G | G | G | N | N | N | N | N | N | N |
| S. paradoxus | C | A | T | A | A | C | A | A | T | A | C | G | T | C | T | A | T | A | A |
| 4 | T | . | . | . | . | . | . | C | . | T | T | . | . | T | . | . | . | C | T |
| K5 | T | . | . | C | . | . | . | C | . | T | T | . | . | T | . | . | . | C | T |
| 7 | T | . | . | . | . | . | . | C | . | . | T | . | . | T | . | . | . | C | T |
| 4 | T | . | . | . | . | . | . | T | . | . | T | . | . | T | . | . | . | C | T |
| 2 | T | T | . | . | C | T | . | . | C | . | . | . | . | T | . | . | . | C | T |
| YJM421 | G | . | . | . | C | T | . | C | C | . | T | . | . | T | . | G | . | C | T |
| YJM440 | T | . | . | . | C | T | . | C | C | . | T | A | . | T | . | . | . | C | C |
| 40 | T | T | . | . | C | T | . | . | C | . | . | . | . | T | C | . | . | T | C |
| Y6 | T | T | . | . | C | T | . | . | C | . | . | . | . | T | . | . | C | T | C |
| Y3 | T | T | . | . | C | T | . | . | C | . | . | . | . | T | . | . | . | T | C |
| 3 | T | G | . | . | C | T | . | T | C | . | T | . | . | T | . | . | . | C | T |
| 2 | . | . | . | . | . | T | . | T | C | . | T | . | C | . | . | . | . | C | C |
| YJM339 | . | . | . | . | . | T | . | T | C | . | T | . | . | . | . | . | . | C | C |
| 3 | . | . | G | . | . | T | G | C | C | . | T | . | . | . | . | . | . | C | C |
| YPS1000 | T | . | . | . | . | T | . | C | C | . | T | . | . | . | . | . | . | C | C |
| YJM454 | T | . | . | . | . | . | . | C | C | . | T | . | . | . | . | . | . | C | C |
| YPS1009 | T | . | . | . | C | T | . | C | C | . | T | . | . | . | . | . | . | C | C |
| 4 | T | T | . | . | C | T | . | C | C | . | T | . | . | . | . | . | . | T | C |
Nucleotide positions are shown in two regions upstream of MLS1. The strains column lists the number of strains with the haplotype shown on the right or the name of the strain if the haplotype is unique.
TABLE 11.
Haplotypes from the hypervariable region upstream ofPDR10
| Nucleotide position
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | 436 | 469 | 472 | 490 | 496 | 533 | 538 | 545 | 577 | 582 | 586 | 605 |
| S. mikatae | — | C | A | — | T | — | — | — | G | T | T | T |
| S. paradoxus | — | G | A | T | T | G | T | G | A | A | T | T |
| 47 | A | . | . | . | . | . | . | A | . | T | G | . |
| Y1 | A | . | . | . | A | . | . | A | . | T | G | . |
| B6 | A | . | . | . | . | C | . | A | . | T | G | . |
| I14 | G | . | . | . | . | . | . | A | . | T | G | . |
| YJM436 | G | . | . | . | . | . | C | . | . | T | G | C |
| YPS1009 | G | . | . | . | . | . | C | . | T | T | G | C |
| Y3 | G | A | C | . | . | . | C | . | . | T | G | . |
| 15 | G | . | C | . | . | . | C | . | . | T | A | C |
| K13 | G | . | C | . | . | . | C | . | . | . | A | C |
| 5 | G | . | C | . | . | . | C | . | . | T | G | C |
| YPS163 | G | . | . | C | . | . | C | . | . | T | G | . |
Nucleotide positions are shown in two regions upstream of PDR10. The strains column lists the number of strains with the haplotype shown on the right or the name of the strain if the haplotype is unique.
To determine whether there are regions with more noncoding polymorphism than can be explained by a neutral model, we examined the distance between segregating sites. The advantage of the distance between segregating sites is that it has well-defined statistical properties compared to a sliding-window analysis, which is dependent on the window length and step size. Assuming a constant rate of polymorphism, p, and given a polymorphic site, the probability of d sites until the next polymorphism is
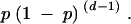 |
Thus, the distance between polymorphic sites is geometrically distributed with parameter p, which can be estimated from the number of polymorphic sites per base pair.
If there is an increase in the rate of polymorphism within a portion of a noncoding region, the distance between segregating sites should be less than that expected under a neutral model. The expected distances were calculated using the geometric distribution with a rate parameter, p, estimated from concatenated fourfold degenerate synonymous sites. To test the goodness-of-fit between the observed and expected distance values we binned the distance between segregating sites in three classes, 0–7 bp, 8–20 bp, and >20 bp, to ensure the expected number of sites in each category is greater than five. Synonymous sites provide a good fit to the geometric distribution using a G-test with Williams' correction (Sokal and Rohlf 1995) (Table 12). Of the five noncoding regions, only MLS1 and PDR10 show a significant deviation from a geometric distribution using the rate parameter from synonymous sites (P = 0.019 and P = 0.049, respectively). Although MLS1 and PDR10 are not individually significant after correction for multiple tests, the combined probability of all five genes is significant (P = 0.007, Fisher's test of combined probabilities) and the sum of the data from all five genes is also significant (P = 0.024, G-test). Furthermore, the G-test is conservative because the overall rate of noncoding polymorphism is less than that of synonymous polymorphism and so the expected distance between segregating noncoding sites should be greater than the distance between concatenated fourfold degenerate sites.
TABLE 12.
Distance between consecutive segregating sites
| Distance
|
||||
|---|---|---|---|---|
| Gene | 0–7 | 8–20 | >20 | P-value |
| CCA1 | ||||
| Obs | 5 | 3 | 11 | 0.103 |
| Exp | 4.9 | 6.0 | 8.0 | |
| CYT1 | ||||
| Obs | 1 | 6 | 7 | 0.079 |
| Exp | 3.6 | 4.4 | 5.9 | |
| MLS1 | ||||
| Obs | 17 | 8 | 15 | 0.019 |
| Exp | 10.4 | 12.7 | 16.9 | |
| PDR10 | ||||
| Obs | 11 | 5 | 9 | 0.049 |
| Exp | 6.5 | 7.9 | 10.6 | |
| ZDS2 | ||||
| Obs | 6 | 6 | 8 | 0.697 |
| Exp | 5.2 | 6.3 | 8.5 | |
| 4d | ||||
| Obs | 4 | 7 | 9 | 0.540 |
| Exp | 5.2 | 6.3 | 8.5 | |
Distance is the number of base pairs to the next polymorphic site. 4d is concatenated fourfold degenerate sites from all five genes. P-value is from a G-test with Williams' correction. Obs, observed; Exp, expected.
The significant clustering of polymorphic sites in noncoding regions suggests that there may be positive or balancing selection on functional noncoding sequences. To determine whether polymorphic sites occur in functional sequences, we compared the number of polymorphic sites in positions conserved among S. cerevisiae, S. paradoxus, and S. mikatae to the number of polymorphic sites in positions that are not conserved. Although a little less than half of the noncoding polymorphic sites are found in conserved positions, the same proportion of fourfold degenerate sites (4d) are found in positions conserved across species (Table 13).
TABLE 13.
Distribution of SNPs in conserved sequences
| Category | Unconserved | Conserved |
|---|---|---|
| Total | 92 | 91 |
| Noncoding | 64 | 54 |
| nd | 3 | 12 |
| 4d | 14 | 8 |
| MLS1 | 12 | 10 |
| PDR10 | 6 | 2 |
Conserved sites have the same base in S. mikatae, S. paradoxus, and S. cerevisiae. MLS1 and PDR10 are the SNPs in hypervariable noncoding sequences. nd, nondegenerate sites; 4d, fourfold degenerate sites.
Previous studies have found reduced levels of variation in experimentally identified functional noncoding sequences (Ludwig and Kreitman 1995; Ludwig et al. 1998; Tautz and Nigro 1998; Dermitzakis et al. 2003; Phinchongsakuldit et al. 2004), but high levels of variation in unannotated noncoding sequences (Tautz and Nigro 1998; Lazzaro and Clark 2001). Of the five intergenic sequences, the promoters of CYT1, MLS1, and COQ5, adjacent to ZDS2, have been identified by deletion constructs. For CYT1, the minimal promoter was delineated to a 209-bp sequence 351 bp upstream of CYT1 (Oechsner et al. 1992). For MLS1, the minimal promoter was delineated to a 190-bp sequence, 474 bp upstream of MLS1 (Caspary et al. 1997). For COQ5, functional promoter elements were found in a 26-bp and a 90-bp sequence starting 400 bp upstream of COQ5 (Hagerman et al. 2002). For CYT1, the rate of polymorphism in the experimentally defined promoter, 6/209, is much less than the total rate found in all synonymous sites, 47/833 (Table 2). In contrast, the experimentally defined promoter of MLS1 has a rate of polymorphism more than twice that of synonymous sites, 14/190, as it encompasses one of the hypervariable regions identified by the sliding-window analyses (Figure 4). While the rate of polymorphism for one of the two COQ5 promoter regions is low, 1/90, the other has a rate of 3/26, four times that of synonymous sites (Figure 4). Thus, while polymorphic sites are not overpresented in conserved positions (Table 13), they tend to be found in experimentally defined promoter sequences (Figure 4).
Figure 4.—
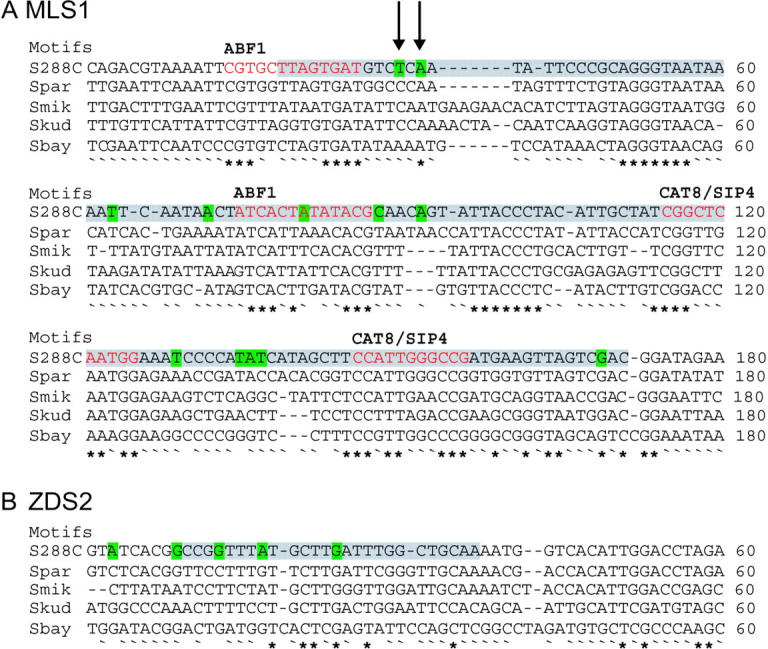
Multiple sequence alignment of experimentally defined promoters (gray) upstream of MLS1 (A) and ZDS2 (B). Polymorphic sites are shown in green and transcription factor-binding sites designated in the original study are shown in red. Arrows show sites with three segregating bases.
DISCUSSION
With short ∼500-bp intergenic sequences, S. cerevisiae provides an excellent opportunity to understand the functional constraints placed on cis-regulatory sequences and their evolution. To address this issue we have compared DNA sequence variation found within and between Saccharomyces species in noncoding and coding sequences. The main result is the observation of noncoding sequences with higher than expected rates of polymorphism. A secondary finding is extensive linkage disequilibrium, even between unlinked loci. Population subdivision, possibly caused by two separate domestication events (Fay and Benavides 2005), is likely a major contributor to linkage disequilibrium.
On average, rates of DNA polymorphism and divergence in noncoding sites are ∼40% lower than those at synonymous sites (Table 9). Yet four small regions within noncoding sequences show rates of polymorphism greater than those at synonymous sites (Figure 3). The clustering of polymorphic sites upstream of MLS1 and PDR10 is significantly greater than that expected on the basis of fourfold degenerate sites.
For a variety of reasons, commonly used statistical tests of neutrality show no significant results for the MLS1 and PDR10 promoter. Neither an MK test nor an HKA test showed any significant differences between rates of polymorphism and divergence at noncoding and synonymous sites. The lack of significance can be explained since both tests measure differences between the rates of polymorphism and divergence between two classes of sites, averaged over the entire region, whereas the hypervariable sequences are limited to small regions within the intergenic sequences. The runs test is designed to detect heterogeneity in the ratio of polymorphism to divergence since polymorphic sites and fixed differences are expected to be evenly interspersed between one another (McDonald 1996). Neither the MLS1 nor the PDR10 noncoding sequences showed any significant departure from neutrality by any of the statistical tests of heterogeneity implemented in DNA Slider (McDonald 1998). It is likely that the power of these tests is limited when rates of divergence are high, since at high divergence the number of runs should become relatively constant for any distribution of polymorphic sites.
There are few explanations for the clustering of polymorphic noncoding sites upstream of MLS1 and PDR10. Our evidence comes from comparing the distance between noncoding polymorphic sites to synonymous polymorphic sites. Thus, a number of factors that affect polymorphism at synonymous sites should be considered. First, a reduced mutation rate at synonymous sites should cause a decrease in diversity and an increase in the distance between polymorphic synonymous sites. However, divergence at synonymous sites is greater than that found at noncoding sites. Second, selection could result in a reduction in diversity at synonymous but not at noncoding sites, thereby increasing the distance between synonymous polymorphic sites. With the exception of PDR10, there is no evidence for a reduction in synonymous-site diversity across the five genes (Table 3). Even if diversity at synonymous sites within PDR10 were reduced, the statistical test for clustering of noncoding sites relies on the combined fourfold degenerate data from all five genes. Finally, some synonymous sites may be functionally constrained, thereby lowering rates of polymorphism and the distance between polymorphic sites. Only CCA1 shows evidence for significant levels of constraint on synonymous sites (Figure 2). Yet instead of showing a reduced rate of polymorphism, CCA1 has the highest level of synonymous-site diversity (Table 9).
Hypervariable noncoding sequences could also be caused by mutation hotspots in noncoding sequences. Yet hotspots should increase both polymorphism and divergence, and only polymorphism appears inflated. Furthermore, there is little evidence for large-scale mutational heterogeneity across the S. cerevisiae genome (Chin et al. 2005). If mutational hotspots are present but not at a fixed location, polymorphic sites should cluster but divergence should average to a uniform distribution over a long enough period of time. Although transient mutational hotspots provide a rather tenuous explanation for the data, two mechanisms are possible. First, a polymorphic site may increase the mutation rate at nearby bases. Second, recombination hotspots have been found to be transient (Ptak et al. 2005; Winckler et al. 2005). If recombination hotspots are transient in yeast and mutagenic, or repair deficient, they may result in transient mutational hotspots. If this is the case, hypervariable noncoding sequences should be observed across the genome. Little or no clustering would be observed in coding sequences since most mutations would be removed by negative selection.
Selection on noncoding sites can both increase and decrease the distance between polymorphic sites. Both changes in selective constraint and the presence of deleterious mutations can influence the ratio of noncoding- to synonymous-site diversity. Yet, the distance between noncoding polymorphic sites should be at the least equal to that found at synonymous sites. Positive selection, diversifying selection, and balancing selection can all increase the rate of polymorphism at noncoding sites above that of synonymous sites. Although positive selection predicts very short sojourn times for variants under selection, population subdivision would inhibit the rapid spread of a selected allele through the entire species (Slatkin and Wiehe 1998). Alternatively, balancing or diversifying selection could account for the excess of noncoding compared to synonymous polymorphism and predicts elevated rates of polymorphism but not of divergence. Although there is no clear way to confidently distinguish these models of selection, one pertinent observation is that the ratio of NC/S is greater than one within some but not all groups of strains categorized by source of isolation.
In conclusion, there are two plausible models that can explain the hypervariable noncoding sequences. First, hypervariable regions could be caused by transient mutational hotspots. Second, hypervariable regions could be caused by some form of natural selection acting on mutations that affect gene expression. With hypervariable regions found in three of the five genes, only two showing significance, it is difficult to determine whether the hypervariable sites are common, and more likely the result of a mutational explanation, or rare, and more likely the result of natural selection.
Acknowledgments
We thank B. Dunn, N. Goto-Yamamoto, R. Mortimer, C. Kurtzman, J. McCusker, and P. Sniegowski for contributing yeast strains; E. Mardis at the Genome and Sequencing Center for use of an ABI 3730xl sequencer; and two anonymous reviewers for useful comments on the interpretation of the data.
References
- Aebi, M., G. Kirchner, J. Y. Chen, U. Vijayraghavan, A. Jacobson et al., 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265: 16216–16220. [PubMed] [Google Scholar]
- Akashi, H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi, E., and A. Goffeau, 1995. Yeast multidrug resistance: the PDR network. J. Bioenerg. Biomembr. 27: 71–76. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., 2001. The frequency distribution of nucleotide variation in Drosophila simulans. Mol. Biol. Evol. 18: 1343–1352. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and B. D. Hall, 1982. Codon selection in yeast. J. Biol. Chem. 257: 3026–3031. [PubMed] [Google Scholar]
- Bi, E., and J. R. Pringle, 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2003. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics 165: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer, M., 1987. Coevolution of codon usage and transfer RNA abundance. Nature 325: 728–730. [DOI] [PubMed] [Google Scholar]
- Cargill, M., D. Altshuler, J. Ireland, P. Sklar, K. Ardlie et al., 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Caspary, F., A. Hartig and H. J. Schuller, 1997. Constitutive and carbon source-responsive promoter elements are involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol. Gen. Genet. 255: 619–627. [DOI] [PubMed] [Google Scholar]
- Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt et al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73. [PMC free article] [PubMed] [Google Scholar]
- Chin, C. S., J. H. Chuang and H. Li, 2005. Genome-wide regulatory complexity in yeast promoters: separation of functionally conserved and neutral sequence. Genome Res. 15: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al., 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E. T., C. M. Bergman and A. G. Clark, 2003. Tracing the evolutionary history of Drosophila regulatory regions with models that identify transcription factor binding sites. Mol. Biol. Evol. 20: 703–714. [DOI] [PubMed] [Google Scholar]
- Doniger, S., J. Huh and J. C. Fay, 2005. Identification of functional transcription factor binding sites using closely related Saccharomyces species. Genome Res. 15: 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88: 7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, K. A., J. P. Bielawski and Z. Yang, 2001. Substitution rates in Drosophila nuclear genes: implications for translational selection. Genetics 157: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Fay, J. C., and J. A. Benavides, 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 2001. The neutral theory in the genomic era. Curr. Opin. Genet. Dev. 11: 642–646. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 2003. Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genomics Hum. Genet. 4: 213–235. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., G. J. Wyckoff and C.-I Wu, 2001. Positive and negative selection on the human genome. Genetics 158: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., G. J. Wyckoff and C.-I Wu, 2002. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415: 1024–1026. [DOI] [PubMed] [Google Scholar]
- Hagerman, R. A., P. J. Trotter and R. A. Willis, 2002. The regulation of COQ5 gene expression by energy source. Free Radic. Res. 36: 485–490. [DOI] [PubMed] [Google Scholar]
- Halushka, M. K., J. B. Fan, K. Bentley, L. Hsie, N. Shen et al., 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239–247. [DOI] [PubMed] [Google Scholar]
- Hartig, A., M. M. Simon, T. Schuster, J. R. Daugherty, H. S. Yoo et al., 1992. Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 20: 5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, I., S. Zollner, W. Enard, I. Ebersberger, B. Nickel et al., 2003. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 13: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50: 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Jenkins, D. L., C. A. Ortori and J. F. Brookfield, 1995. A test for adaptive change in DNA sequences controlling transcription. Proc. R. Soc. Lond. Ser. B Biol. Sci. 261: 203–207. [DOI] [PubMed] [Google Scholar]
- Jensen, M. A., H. L. True, Y. O. Chernoff and S. Lindquist, 2001. Molecular population genetics and evolution of a prion-like protein in Saccharomyces cerevisiae. Genetics 159: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J. R., C. Baccari and R. K. Mortimer, 2000. Genotypic characterization of strains of commercial wine yeasts by tetrad analysis. Res. Microbiol. 151: 583–590. [DOI] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969 Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism III, edited by H. N. Munro. Academic Press, New York.
- Kang, X. L., F. Yadao, R. D. Gietz and B. A. Kunz, 1992. Elimination of the yeast RAD6 ubiquitin conjugase enhances base pair transitions and G.C-T.A transversions as well as transposition of the Ty element: implications for the control of spontaneous mutation. Genetics 130: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren and E. S. Lander, 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kern, A. D., and D. J. Begun, 2005. Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes. Mol. Biol. Evol. 22: 51–62. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1983 The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Kohn, M. H., S. Fang and C. I. Wu, 2004. Inference of positive and negative selection on the 5′ regulatory regions of Drosophila genes. Mol. Biol. Evol. 21: 374–383. [DOI] [PubMed] [Google Scholar]
- Lazzaro, B. P., and A. G. Clark, 2001. Evidence for recurrent paralogous gene conversion and exceptional allelic divergence in the Attacin genes of Drosophila melanogaster. Genetics 159: 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M. Z., and M. Kreitman, 1995. Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 12: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., N. H. Patel and M. Kreitman, 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125: 949–958. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., C. Bergman, N. H. Patel and M. Kreitman, 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., 1996. Detecting non-neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol. Biol. Evol. 13: 253–260. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., 1998. Improved tests for heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol. Biol. Evol. 15: 377–384. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Mortimer, R. K., P. Romano, G. Suzzi and M. Polsinelli, 1994. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10: 1543–1552. [DOI] [PubMed] [Google Scholar]
- Moses, A. M., D. Y. Chiang, M. Kellis, E. S. Lander and M. B. Eisen, 2003. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, A. M., D. Y. Chiang and M. B. Eisen, 2004a Phylogenetic motif detection by expectation-maximization on evolutionary mixtures. Pac. Symp. Biocomput., 324–335. [DOI] [PubMed]
- Moses, A. M., D. Y. Chiang, D. A. Pollard, V. N. Iyer and M. B. Eisen, 2004. b MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 5: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oechsner, U., H. Hermann, A. Zollner, A. Haid and W. Bandlow, 1992. Expression of yeast cytochrome c1 is controlled at the transcriptional level by glucose, oxygen and haem. Mol. Gen. Genet. 232: 447–459. [DOI] [PubMed] [Google Scholar]
- Phinchongsakuldit, J., S. Macarthur and J. F. Brookfield, 2004. Evolution of developmental genes: molecular microevolution of enhancer sequences at the Ubx locus in Drosophila and its impact on developmental phenotypes. Mol. Biol. Evol. 21: 348–363. [DOI] [PubMed] [Google Scholar]
- Ptak, S. E., D. A. Hinds, K. Koehler, B. Nickel, N. Patil et al., 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37: 429–434. [DOI] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, N., and K. W. Runge, 1999. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma 108: 146–161. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sadler, I., K. Suda, G. Schatz, F. Kaudewitz and A. Haid, 1984. Sequencing of the nuclear gene for the yeast cytochrome c1 precursor reveals an unusually complex amino-terminal presequence. EMBO J. 3: 2137–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., and T. Wiehe, 1998. Genetic hitch-hiking in a subdivided population. Genet. Res. 71: 155–160. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995 Biometry: The Principles and Practice of Statistics in Biological Research. W. H. Freeman, New York.
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz, D., and L. Nigro, 1998. Microevolutionary divergence pattern of the segmentation gene hunchback in Drosophila. Mol. Biol. Evol. 15: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R., 1996. Contingency tests of neutrality using intra/interspecific gene trees: the rejection of neutrality for the evolution of the mitochondrial cytochrome oxidase II gene in the hominoid primates. Genetics 144: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. W., J. W. Touchman, R. W. Blakesley, G. G. Bouffard, S. M. Beckstrom-Sternberg et al., 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788–793. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Winckler, W., S. R. Myers, D. J. Richter, R. C. Onofrio, G. J. Mcdonald et al., 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308: 107–111. [DOI] [PubMed] [Google Scholar]
- Wright, F., 1990. The ‘effective number of codons’ used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]