Abstract
The tumor suppressor function of PTEN is strongly linked to its ability to dephosphorylate phosphatidylinositol-3,4,5 trisphosphate and, thereby, control cell growth, survival, and migration. However, the mechanism of action of PTEN in living cells is largely unexplored. Here we use single-molecule TIRF microscopy in living cells to reveal that the enzyme binds to the membrane for a few hundred milliseconds, sufficient to degrade several phosphatidylinositol-3,4,5 trisphosphate molecules. Deletion of an N-terminal lipid-binding motif completely abrogates membrane interaction and in vivo function. Several mechanisms, including C-terminal tail phosphorylations, appear to hold PTEN in a constrained conformation that limits its rate of association with the membrane. The steady-state level of bound PTEN is highest at sites of retracting membrane, including the rear of highly polarized cells. The dynamic membrane association could be modulated temporally or spatially to alter PTEN activity in specific physiological situations and could have important implications for tumor suppressor function.
Keywords: single molecule, membrane binding
PTEN, a lipid phosphatase that dephosphorylates the D3 position of phosphatidylinositol-3,4 bisphosphate and phosphatidylinosiol-3,4,5 trisphosphate (PIP3) (1) antagonizes phosphatidylinositol 3-kinase (PI3K) signaling and is one of the most frequently mutated genes in human cancer (2). Cells that lack PTEN grow faster, are resistant to various apoptotic stimuli, and migrate aberrantly (3–7). The crystal structure of PTEN contains an N-terminal phosphatase domain with the phosphatase signature motif and a C2 domain that binds lipid vesicles (8). Not included in the structure are a C-terminal 50-aa tail with sites for inhibitory phosphorylations and a short N-terminal conserved hydrophobic and polybasic region dubbed “phosphatidylinositol-4,5-bisphosphate-binding domain (PBD)” (9, 10).
PTEN can bind acidic lipid vesicles in vitro and both the C2 domain and the PBD have been shown to contribute to the binding (8, 11); however, in vivo PTEN has been reported to be cytoplasmic and nuclear (12, 13). Only in certain cell lineages and under specific conditions has PTEN been found to translocate to the plasma membrane (14, 15). How then does cytosolic PTEN dephosphorylate its surface membrane substrate? One proposed idea is that PTEN may need to be “activated” to translocate to the membrane and function (reviewed in ref. 16). However, in many cells, loss of PTEN activity results in an increase in basal PIP3, suggesting that it must normally be in a position to keep basal PIP3 levels low.
There are two possible explanations for these disparate findings: Either free cytosolic PTEN can act on the membrane or there is a minor (heretofore undetectable) pool of PTEN that is membrane-associated. The latter scenario raises still more questions: Is this membrane-bound pool stable or dynamic, is it a distinct pool from the cytosolic population, or is it a low-steady-state fraction of the total population? Is little PTEN bound because its affinity for the membrane is low or because the number of binding sites is small? Can the affinity or number of sites be regulated to control PTEN activity temporally or spatially? We developed single molecule imaging of functionally active PTEN-YFP to address these questions. Our results suggest a previously uncharacterized mechanism of action of PTEN in living cells.
Results and Discussion
Single-Molecule Total Internal Reflection Fluorescence Microscopy (TIRFM) Reveals PTEN Membrane Binding.
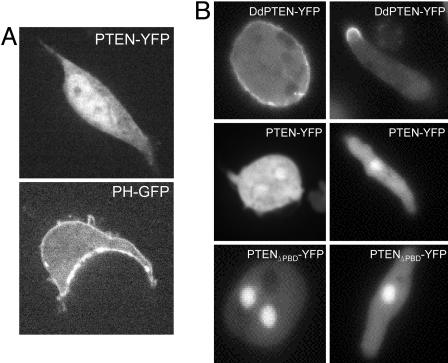
Consistent with previous observations, we found that human PTEN-YFP expressed in human embryonic kidney 293 (HEK293) or HeLa cells and imaged by confocal microscopy does appear cytoplasmic and nuclear (Fig. 1A and data not shown). Although this PTEN-YFP construct is functionally active in maintaining low PIP3 levels in cells (see Fig. 3A and data not shown), there was little evidence for even transient association with the membrane in time-lapse imaging of the fluorescent signal. As a positive control, we show PHAKT-GFP expressed in HEK293, which is membrane and cytoplasmic, as well as the highly homologous Dictyostelium discoideum PTEN-GFP (DdPTEN-GFP), which has been previously reported to be associated with the membrane (17). As shown in Fig. 1B, a portion of DdPTEN is clearly present on the membrane in D. discoideum cells.
Fig. 1.
Confocal microscopy shows cytoplasmic and nuclear localization of human PTEN in HEK293 and D. discoideum cells. (A) HEK293 cells were transiently transfected with the indicated YFP fusion proteins and imaged 48 h later with a confocal laser scanning microscope. (B) D. discoideum cells stably expressing the fusion proteins indicated in the figure were imaged with a confocal microscope. Cells were either under full nutrient conditions (Left) or starved for 5 h to induced them to polarize (Right).
Fig. 3.
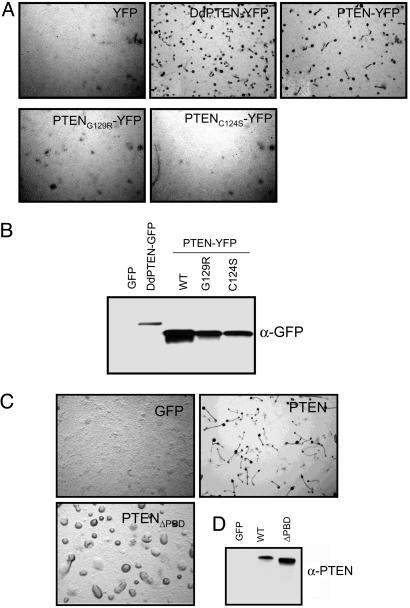
Membrane binding is required for PTEN to complement pten− D. discoideum cells. (A and C) D. discoideum pten− cells stably expressing the indicated constructs were plated in nonnutrient agar to induce development. Images were taken 24 h after starvation. (B and D) Expression levels of the specific proteins were determined by Western blot analyses by using anti-GFP antibody for the YFP-tagged constructs or anti-PTEN antibody for the untagged constructs.
These observations suggest that either PTEN has a low affinity for membranes or mammalian cells have a relatively lower number of membrane-binding sites. To distinguish these possibilities, we expressed PTEN in D. discoideum cells. As shown in Fig. 1B, the differential localization of PTEN and DdPTEN were maintained. In undifferentiated cells, DdPTEN was uniformly associated with the membrane and, as the cells differentiated and adopted a polarized morphology, DdPTEN was concentrated on the membrane at the back of the cells. However, PTEN-YFP was primarily cytoplasmic and nuclear, similar to the localization seen in mammalian cells, whether or not the cells were polarized (Fig. 1B). Despite these apparent differences in the localization, evidence will be presented below that strongly suggests that DdPTEN and PTEN bind to the same sites, indicating that PTEN has a low affinity for the membrane.
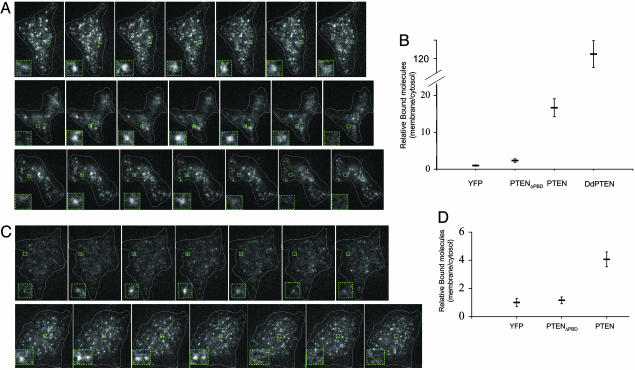
The low level of PTEN on the membrane may be difficult to detect because of epi-fluorescence microscopy (EPI-FM) limitations. We therefore used single-molecule TIRFM to reveal binding of PTEN-YFP to the membrane in living cells. Because with single-molecule TIRFM only one molecule is analyzed at the time, the number of molecules does not influence the sensitivity, and minor events can be visualized (18, 19). Indeed, we detected single molecules of both PTEN and DdPTEN associated with the membrane. We also included in our analysis an allele of PTEN that we previously reported has the C-terminal tail inhibitory phosphorylations mutated to alanine (PTENA4) (20). As will be explained in detail later, this mutant has an enhanced membrane localization. Fig. 2A shows a time series of fluorescence spots representing single molecules of DdPTEN-YFP, PTEN-YFP, and PTENA4-YFP, bound to the plasma membrane in living D. discoideum cells. YFP or PTENΔPBD-YFP, which lacks the N-terminal PBD, displayed very few spots associated with the plasma membrane (see below). To verify that the spots were single molecules, we fixed the cells with formaldehyde and showed that they bleached in a single step (data not shown). To quantify the relative number of molecules, we calculated the steady-state number of single events per μm2 relative to the average fluorescence intensity in the cytosol. As shown in Fig. 2B, the relative number of bound molecules in cells expressing PTEN-YFP was 7- or 16-fold higher than in cells expressing PTENΔPBD-YFP or YFP alone, respectively. However, the relative number was 9-fold higher for DdPTEN-YFP versus PTEN-YFP, consistent with the EPI-FM observations.
Fig. 2.
Single-molecule TIRFM imaging reveals membrane localization of PTEN in Dictyostelium and HEK293 cells. (A) Consecutive frames of cells expressing PTEN-YFP or DdPTEN-YFP observed by TIRFM at 33 ms per frame (see Movies 1–3). Areas outlined within green boxes and magnified insets show appearance and disappearance of individual spots. (B) Quantification of the number of membrane-bound PTEN molecules relative to the average cytosolic fluorescence in D. discoideum cells expressing PTEN, PTENΔPBD-YFP, YFP, or DdPTEN-YFP. The number of spots per μm2 appearing per second detected by TIRFM was divided by the cytosolic fluorescence detected by EPI-FM. Average ± SEM (n ≥ 10). (C) Consecutive frames of cells expressing PTEN or PTENC124S/A4-YFP observed by TIRFM in HEK293 cells (see Movies 4 and 5). Areas outlined within green boxes and magnified insets show appearance and disappearance of individual spots. (D) Quantification of the number of membrane-bound PTEN molecules relative to the average cytosolic fluorescence in HEK293 cells expressing PTEN, PTENΔPBD-YFP, or YFP calculated as in B. Average ± SEM (n ≥ 10).
To corroborate these results, we performed a modified immunofluorescence protocol by using a brief permeabilization of cells. We briefly treated the cells with a detergent that selectively solubilized some of the cytosolic PTEN and enhanced the detection of the membrane-bound PTEN. As seen in Fig. 5, which is published as supporting information on the PNAS web site, a fraction of human PTEN localized to the plasma membrane. Consistent with TIRFM results, no PTENΔPBD was present on the membrane. Thus, immunofluorescense confirms the TIRFM results. Taken together, these results suggest that there is a minor pool of PTEN that associates with the membrane and this binding depends on the PBD.
PTEN Binding to the Membrane Is Dynamic.
To dissect how PTEN interacts with the membrane in living cells, we analyzed the dynamics of the single-bound PTEN molecules. We found that the spots representing DdPTEN, PTEN, and PTENA4 rapidly associated with and dissociated from the membrane (Fig. 2A; see also Movies 1–3, which are published as supporting information on the PNAS web site). Images in Fig. 2A represent consecutive frames at 33-ms intervals. As seen in the insets, the spots representing single molecules appear and disappear in ≈6 frames. Specifically, the lifetimes of individual PTEN-YFP molecules were obtained by counting the time durations between the appearance and the disappearance of the fluorescent spots. When the data were binned in 20-ms intervals, the most frequent event was at 100 ms, suggesting an interesting two-step interaction with the membrane. Thereafter, the probabilities of events of increasing duration dropped off exponentially. The time constant of the exponential decrease in event probability is related to the macroscopic half-time for dissociation. The half-times were ≈310 ms and ≈300 ms for DdPTEN and PTEN, respectively (Table 1). The half-times were independent of expression levels over a 4-fold range (Fig. 6, which is published as supporting information on the PNAS web site).
Table 1.
Kinetics of PTEN membrane binding in living cells
| Cells (temperature, °C) | PTEN allele | Dissociation rate, ms | Diffusion coefficient, μm2/s |
|---|---|---|---|
| D. discoideum(22) | DdPTEN-YFP | 310 (n = 2,040; 6 cells) | 0.15 ± 0.13 (n = 230; 4 cells) |
| PTEN-YFP | 330 (n = 875; 12 cells) | 0.18 ± 0.15 (n = 44; 4 cells) | |
| HEK293 (22) | PTEN-YFP | 180 (n = 828; 12 cells) | 0.18 ± 0.19 (n = 165; 12 cells) |
| PTEN (C124S;A4)-YFP | 290 (n = 657; 4 cells) | 0.34 ± 0.21 (n = 194; 4 cells) | |
| HEK293 (33) | PTEN-YFP | 150 (n = 270; 5 cells) | 0.41 ± 0.60 (n = 138; 5 cells) |
| PTEN (C124S;A4)-YFP | 130 (n = 805, 4 cells) | 0.41 ± 0.39 (n = 107; 4 cells) |
To assess whether dynamic membrane binding is a general property of PTEN, we translated our results to human cells. Stable cell lines expressing PTEN-YPF were derived from HEK293 cells, and single-molecule TIRFM was performed. We analyzed the single molecules of PTEN-YFP and PTENC124s/A4-YFP. The PTENC124s/A4 allele has a point mutation in the catalytic site, cysteine 124 to serine (C124S), as well as the mutations in the phosphorylations sites in the C-terminal tail (A4). This allele has a dramatically increased steady-state number of molecules on the membrane (see below) and was included in the analysis. As shown in Movies 4 and 5, which are published as supporting information on the PNAS web site, and Fig. 2C, PTEN membrane binding in HEK293 cells was also very dynamic with a subsecond exchange rate similar to that observed in D. discoideum cells. The half-times, calculated as described above, were ≈150 ms and ≈130 ms for PTEN-YFP and PTENC124s/A4-YFP, respectively, at 33°C; ≈180 ms and ≈290 ms, respectively, at 22°C (Table 1). YFP or PTENΔPBD-YFP were expressed at equal or higher levels than PTEN-YFP but these molecules displayed very few spots on the membrane. As shown in Fig. 2D, the relative number of bound molecules for PTEN-YFP was 4- and 3.5-fold higher than YFP and PTENΔPBD-YFP, respectively.
These observations suggest that the mechanism of action of PTEN is actually to bind to the membrane, albeit transiently, with a dwell time of <400 ms, to dephosphorylate PIP3. Because the roughly 500–5,000 binding events per sec per cell is lower than the estimated PIP3 turnover rates, it is likely that each binding event comprises multiple catalytic cycles. The membrane and cytosolic pools exchange rapidly, and the bound molecules do not represent a specific pool of PTEN more competent to bind to the membrane.
Dynamic Membrane Binding Is Required for PTEN Activity in Vivo.
Is the low dynamic membrane binding of PTEN, revealed by single-molecule imaging, required for PTEN lipid phosphatase activity in vivo? DdPTEN with intact lipid phosphatase activity is essential for D. discoideum cells to spontaneously polarize and aggregate upon starvation. Human PTEN and PTEN-YFP restored the capacity of D. discoideum pten− cells to migrate efficiently and could complement the pten− phenotype, indicating that it supplied this necessary activity. As expected, phosphatase-dead mutants, PTENC124S-YFP, and PTENG129R-YFP were unable to complement (Fig. 3A). Western blots showed that the human proteins were expressed at comparable levels (Fig. 3B). We next tested whether membrane localization was necessary for functional complementation by PTEN. Deletion of the PBD (PTENΔPBD) completely prevented the human enzyme from rescuing pten− cells even though the levels of expression are even higher than those of PTEN (Fig. 3C). Thus, membrane binding of PTEN is required for its in vivo activity against PIP3. Additionally, these results also suggest that the low levels of PTEN bound to the membrane (Fig. 1B) are sufficient to provide the necessary activity.
PTEN Binding to the Membrane Is Constrained by Several Mechanisms.
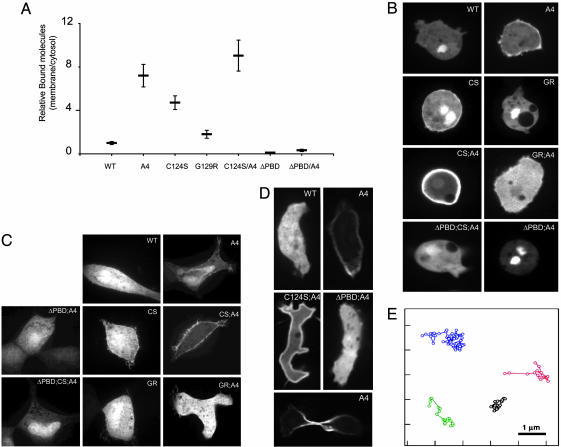
The low affinity of PTEN for the membrane indicates that under normal conditions, the PTEN molecule may be in a constrained conformation. We and others have suggested that the C-terminal tail phosphorylations could inhibit membrane binding of PTEN (21, 22). To test whether phosphorylation interferes with membrane binding, we used the PTENA4 mutant where serine-380, threonine-82, theronine-383, and serine-385 are mutated to alanines. Of note, when human PTEN is expressed in D. discoideum, serine-380 is phosphorylated (data not shown). For each construct, we calculated the number of molecules relative to the average fluorescence intensity in the cytosol as in Fig. 2. Consistently, in D. discoideum cells, we found that PTENA4-YFP has a 7-fold increase in the relative number of molecules bound to the membrane compared to PTEN-YFP (Fig. 4A). This level of membrane binding crossed the threshold apparent by EPI-FM (Fig. 4B). Although we did not quantify the relative number of membrane-bound molecules in HEK293 cells, the EPI-FM was very consistent with the data in D. discoideum cells (Fig. 4C). Thus, phosphorylation of these residues in the C-terminal tail leads to a lower affinity of PTEN for the membrane. It is also possible that the phosphorylation of these sites initiate a chain of phosphorylations or causes a conformational change.
Fig. 4.
Several mutations alter the steady-state number of PTEN membrane-bound molecules. (A) Relative number of membrane-bound PTEN molecules in D. discoideum cells stably expressing PTEN (WT or mutants). The number of spots detected by TIRFM was divided by the relative cytosolic fluorescence detected by EPI-FM calculated as in Fig. 2B. (B) Subcellular localization of PTEN/WT and mutants in undifferentiated D. discoideum cells observed by confocal microscopy. (C) HEK293 cells transiently transfected with the indicated plasmids and imaged 48 h after with a confocal laser scanning microscope. (D) Confocal images of subcellular localization of PTEN in differentiated polarized cells (Top and Middle) and cells undergoing cytokinesis (Bottom). (E) Trajectories of four molecules of PTEN-YFP tracked at 33 frames per s (see also Table 1).
We also noted that a mutation in the catalytic site, C124S, increased the levels of membrane-localized PTEN (Figs. 4 and 5). Because the substrate of PTEN is a membrane lipid, we hypothesized that inactivation of catalytic activity could enhance membrane binding. However, we found that other mutations of the catalytic sites had different consequences. C124S, but not G129R or D92A, enhanced membrane binding 5-fold, to levels that could be detected by EPI-FM (Fig. 4 B and D and data not shown). When we combined the mutations of the tail phosphorylation sites with a C124S mutation, the relative number of spots of PTENC124S/A4-YFP were further increased to 9-fold over PTEN-YFP. However, the G129R/A4 mutation decreased the levels of membrane-bound PTEN and the D92A/A4 mutation did not enhance binding beyond that achieved with PTENA4-YFP (Fig. 4 B and D and data not shown). Because all these mutations (C124S, G129R, and D92A) inactivate the lipid phosphatase activity of PTEN, the lipid phosphatase activity per se does not influence binding. Consistently, neither a decrease in PIP3 levels (generated by treating the cells with the PI3K inhibitor LY294002) nor an increase in PIP3 levels (by deletion of PTEN) influenced the membrane binding of any of the PTEN alleles (Fig. 7, which is published as supporting information on the PNAS web site). Importantly, deletion of the PBD abrogated membrane binding of all alleles, again indicating that all versions of PTEN require this domain for binding. To explain the enhancing effects of the C124S, it was previously suggested that membrane-bound PTEN is subjected to degradation by a mechanism that depends on its phosphatase activity (22). Our data are more consistent with the idea that the phosphatase pocket contributes to membrane binding or that C124S or G129R, respectively, result in an enhanced or decreased exposure of membrane-binding sites. Taken together, the above results suggest that PTEN may be generally in a constrained conformation that can be released by several mechanisms.
The Association of PTEN with the Membrane Is the Critical Regulatory Step.
To dissect the mechanisms of how PTEN affinity for the membrane is modulated, we compared the single-molecule kinetics of the different alleles. The different steady-state levels of membrane-bound PTEN could be the result of altered association or dissociation rates. As noted above, half-times of dissociation for PTEN-YFP and PTENC124S/A4-YFP in HEK293 or between DdPTEN and PTEN in D. discoideum cells were similar (Table 1). The differences in the dissociation rates between the different PTEN alleles are too small to account for the observed relative levels of membrane binding. Furthermore, cells with >5-fold differences in PTEN expression levels displayed nearly identical dissociation rates even though the number of PTEN molecules in the membrane was proportional to the expression levels (Fig. 6). These results indicate that it is the association rate that is altered in each situation and is consistent with a previous report using surface plasmon resonance analysis of PTEN binding to membrane vesicles in vitro (22). These results suggest that the affinity of PTEN for the membrane can be regulated by changes in its association rate and raise the possibility that an increase or decrease in the rate may be a regulatory mechanism in vivo.
Polarized Localization of Membrane-Bound PTEN.
An important question that then remains to be answered is whether the affinity of PTEN for the membrane is regulated in vivo? To begin to explore this possibility, we used D. discoideum cells because the membrane localization of DdPTEN has been shown to be distributed asymmetrically in polarized or dividing D. discoideum cells (23). Interestingly, when the cells were polarized, we found that a fraction PTENA4-YFP was localized at the back of the cells as well as in the furrow of dividing cells (Fig. 4D). These results strongly suggest that PTEN and DdPTEN recognize the same binding sites. In contrast, PTENC124S/A4-YFP was localized uniformly on the membrane, suggesting that a too large increase in the association rate may result in saturation and a failure to detect differences. As shown previously, PTEN-YFP appeared cytosolic, likely because the number of molecules bound in the back was still below detection by EPI-FM (Fig. 4D). These results show that, like DdPTEN, PTEN can be localized asymmetrically in polarized D. discoideum cells and suggest the interesting possibility that polarized membrane distribution of PTEN may be a conserved mechanism.
Changes in the spatial distribution of PTEN could be generated by regulating PTEN diffusion in the membrane or by altering binding and dissociation from different regions. To address this question, we calculated the diffusion coefficients of the membrane bound molecules. The diffusion coefficients were 0.15 ± 0.13 μm2/s and 0.18 ± 0.15 μm2/s for DdPTEN and PTEN, respectively, in D. discoideum cells (at 22°C) and 0.41 ± 0.6 and 0.41 ± 0.39 μm2/s for PTEN-YFP and PTENC124s/A4-YFP, respectively, in HEK293 (at 33°C) (Table 1 and Fig. 4E). These results, taken together with the dissociation times, indicate that PTEN is unlikely to diffuse a significant portion of the cell length during the time is bound (also see Movies 1–4). Thus, the redistributions of bound and, therefore, functional PTEN, are likely to be altered by changing the association rate, or the number of binding sites, in different regions of the membrane.
Conclusions
PTEN maintains basal levels of PIP3 low yet growth factors remain able to induce PIP3 levels via a PI3K-dependent pathway. To explain this observation, it has been proposed that PTEN is either inactivated by growth factors or activated by growth factor depletion (24). Our results suggest an alternative mode of regulation. By maintaining the bulk of PTEN in a conformation with low affinity for the membrane, the bound molecules will be sufficient to keep the basal levels of PIP3 low but not completely oppose increases induced by PI3K stimulation. Because a fraction of PTEN is membrane-associated in the basal state, mutations that inactivate PTEN or interfere with membrane binding will result in the long-term increase in basal PIP3 levels that leads to tumor development.
This strategy allows that in specific biological situations, the constraints are tightened or released to modulate the levels of PTEN in the membrane. We have showed that the membrane binding constraints can be released by blocking phosphorylation of the tail and specific mutations in the catalytic domain. It remains to be determined, however, when in vivo physiological changes in PTEN membrane binding take place. Recently, two reports have shown stimulation of PTEN membrane translocation by ROCK (15) or by shingosine-1-phosphate (25) involving serine/threonine phosphorylation or tyrosine phosphorylation, respectively. How these events alter membrane binding is currently unknown. These results also suggest that increasing membrane binding of PTEN is a potential therapeutic opportunity.
A second important feature of the regulation of PTEN is the asymmetric distribution of the membrane bound molecules in D. discoideum cells. Interestingly, PIP3 and PI3K levels have been shown to be asymmetrically accumulated in a three-dimensional mammary epithelial cells tissue-culture model (26). It would be of interest to investigate the possibility that PTEN localization also may be polarized in epithelial cells and whether the loss of PTEN may contribute, not only to proliferation and survival, but also the loss of polarity seen during tumor progression.
Materials and Methods
Plasmid Construction.
DdPTEN-YFP alleles were constructed by restriction enzyme digestion of inserts from the pJK1 vector and transferring into pJK1 downstream and in frame of YFP (23). PTEN and PTEN mutants were amplified by PCR from the pSG5L-HA-PTEN constructs (3, 20). The A4 mutation contains alanine substitutions of S380, T382, T383, and S385 and has been described in ref. 20. To construct pCDNA3-PTEN-YFP, eYFP and PTEN were amplified with oligonucleotides that have some overlapping sequence. A fusion PCR between the two fragments was generated to obtain PTEN-YFP and subcloned into pCDNA3. For pJK1-PTEN-YFP constructs to generate D. discoideum stable cell lines, PTEN-YFP was amplified from the pCDNA3-PTEN-YFP constructs and cloned into the pJK1 vector. The PBD deletion mutant consisted in the deletion of the first 10 amino acids of PTEN.
D. discoideum Growth and Mammalian Cell Culture.
D. discoideum was described in ref. 17, and mammalian cell culture cells were maintained as described in ref. 20.
Fluorescence Microscopy Analysis on Living Cells.
D. discoideum cells expressing PTEN were plated on eight-well Lab-Tek chambered coverslips (Nunc), allowed to attach for 30 min, and washed before visualization. HEK293 cells were plated and transiently transfected on eight-well Lab-Tek chambered coverslips. Cells were visualized 48 h after transfection in phenol-red-free DMEM. In both types of cells, PTEN cellular localization was visualized with confocal laser scanning microscope (PerkinElmer UltraView) with an inverted microscope (Nikon) with a 63 Å ≈1.4 numerical aperture Apochromat lens.
Immunoblotting.
D. discoideum cells were resuspended at 2 × 107 cells/ml and lysed by the addition of 5× Laemmli buffer (28). Lysates were cleared by centrifugation, and proteins were separated by gel electrophoresis. Immunoblotting was performed as previously described (20).
Single-Molecule Microscopy.
D. discoideum cells were starved for ≈6 h and then washed and suspended. An aliquot was placed on a glass coverslip. After they settled, the cells were overlaid with a sheet of agarose (29). Single molecules of PTEN-YFP were visualized by using an objective-type total internal reflection microscope constructed on an inverted fluorescence microscope (IX70; Olympus) (30). Specimens were illuminated with a 488-nm line of a laser (sapphire 488–20; Coherent, Dieburg, Germany) through an objective lens (PlanApo 60×; numerical aperture 1.45; Olympus). The incident beam from the laser was passed through a beam expander (Sigma Koki, Tokyo), a neutral density filter (Sigma Koki), and quarter-wave plate (WPQ 5900–4M; Sigma Koki), and it was focused on the back focal plane of the objective lens by using focusing lens and a mirror. By adjusting the angle and position of the mirror, the illumination was switched between EPI-FM and TIRFM (31). Fluorescence signals from PTEN-YFP were collected with the objective lens, selected with dichroic mirror (DM2, DM500; Olympus) and emission filter (BA510-550; Olympus), and focused with the optics of IX-70 on a camera. The images were intensified with an image intensifier (GaAsP, C8600-05; Hamamatsu Photonics) and acquired with a charge-coupled device camera (MC681SPD; Texas Instruments).
For observations of HEK293 cells expressing PTEN-YFP or its mutants, the cells were cultured on a coverslip soaked in DMEM containing 10% FBS. After overnight incubation, the coverslip was washed and covered with Hank’s balanced salt solution (HBSS) preheated at 37°C. The single molecules were observed by the same microscope as described above at 33°C.
To prove that a spot-like fluorescence in the obtained image reflects an emission from single YFP molecules, the fluorescent characteristics were examined as described in ref. 30. In brief, the fluorescent intensity of each spot detected in D. discoideum cells expressing PTEN-YFP was ≈1,000 a.u., which is comparable to the intensity of fluorescence emitted from PTEN-YFP molecule adsorbed on a glass surface. The fluorescence of PTEN-YFP molecules on the glass decayed in a single-step fashion, which is a common feature of the bleaching fluorescent molecule.
Data Analysis of Single Molecules.
Single-molecule images were taken at a rate of 33 ms per frame as described above and stored as a stack of frames on a personal computer. Individual fluorescent spots were followed semiautomatically, and the positions (x and y coordinates), the fluorescence intensities, and the frames that appeared were determined. The lifetimes of individual PTEN-YFP molecules were obtained by counting the time durations between the appearance and the disappearance of the fluorescent spots. When the data were binned in 20-ms intervals, the most frequent event was at 100 ms, suggesting an interesting two-step interaction with the membrane. Thereafter the probabilities of events of increasing duration dropped off exponentially. The dissociation rates of PTEN were calculated from the decay curve, exp[−kt], where kis the dissociation rate (30).
Relative Number of Single Molecules.
For the analysis of the relationship between the expression level of PTEN in cytoplasm and the appearance rates of single molecules on membranes, the number of PTEN appearing per second per μm2 on membranes were measured and plotted against the fluorescence intensities of the cytoplasm for each cells. At least 10 cells were measured for each allele of PTEN to obtain the appearance rates.
Diffusion Constants of PTEN.
Diffusion constants of PTEN on membranes were determined as in ref. 32. From the data of x and y coordinates for individual fluorescence spots, the mean square displacements (MSD) were calculated for each time interval (Δt) over a trajectory, and then the diffusion constants were obtained as a slope off the plot of MSD against Δt by least square fitting with an equation, MSD = 4DΔt, where Dis a diffusion constant.
Supplementary Material
Acknowledgments
We thank Mark Landree, Pablo Iglesias, Pere Puigserver, Douglas Robinson, and Jin Zhang for critical reading of the manuscript and the reviewers for thoughtful comments. This work was supported by Department of Defense Grant DAM-17-03-1-0195 (to F.V.), Public Health Service Grants R01-GM28007 and R01-GM34933 (to P.N.D.), and the Ministry of Education, Culture, Sports, Science and Technology’s Leading Project, Bio-Nano-Process (to M.U.).
Abbreviations
- EPI-FM
epi-fluorescence microscopy
- HEK293
human embryonic kidney 293
- PBD
phosphatidylinositol-4,5-bisphosphate binding domain
- PIP3
phosphatidylinosiol-3,4,5 trisphosphate
- PI3K
phosphatidylinositol 3-kinase
- TBS-T
Tris-buffered saline/0.05% Triton X-100
- TIRFM
total internal reflection fluorescence microscopy.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Maehama T., Dixon J. E. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 2.Parsons R. Semin. Cell Dev. Biol. 2004;15:171–176. doi: 10.1016/j.semcdb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R. Proc. Natl. Acad. Sci. USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H., Lesche R., Li D.-M., Liliental J., Zhang H., Gao J., Gavrilova N., Mueller B., Liu X., Wu H. Proc. Natl. Acad. Sci. USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liliental J., Moon S. Y., Lesche R., Mamillapalli R., Li D., Zheng Y., Sun H., Wu H. Curr. Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhu X., Kwon C. H., Schlosshauer P. W., Ellenson L. H., Baker S. J. Cancer Res. 2001;61:4569–4575. [PubMed] [Google Scholar]
- 7.Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 8.Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez F., Sellers W. R. Biochim. Biophys. Acta. 2000;1470:M21–M35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 10.Taylor G. S., Dixon J. E. Methods Enzymol. 2003;366:43–56. doi: 10.1016/s0076-6879(03)66004-0. [DOI] [PubMed] [Google Scholar]
- 11.Walker S. M., Leslie N. R., Perera N. M., Batty I. H., Downes C. P. Biochem. J. 2004;379:301–307. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F., Wagner S., Campbell R. B., Nickerson J. A., Schiffer C. A., Ross A. H. J. Cell Biochem. 2005;96:221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- 13.Lacalle R. A., Gomez-Mouton C., Barber D. F., Jimenez-Baranda S., Mira E., Martinez A. C., Carrera A. C., Manes S. J. Cell Sci. 2004;117:6207–6215. doi: 10.1242/jcs.01545. [DOI] [PubMed] [Google Scholar]
- 14.Sumitomo M., Iwase A., Zheng R., Navarro D., Kaminetzky D., Shen R., Georgescu M. M., Nanus D. M. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Dong X., Wang Z., Liu W., Deng N., Ding Y., Tang L., Hla T., Zeng R., Li L., Wu D. Nat. Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 16.Leslie N. R., Yang X., Downes C. P., Weijer C. J. Biochem. Soc. Trans. 2005;33:1507–1508. doi: 10.1042/BST0331507. [DOI] [PubMed] [Google Scholar]
- 17.Iijima M., Huang Y. E., Luo H. R., Vazquez F., Devreotes P. N. J. Biol. Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 18.Sako Y., Yanagida T. Nat. Rev. Mol. Cell. Biol. 2003;(Suppl.):SS1–SS5. [PubMed] [Google Scholar]
- 19.Douglass A. D., Vale R. D. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez F., Ramaswamy S., Nakamura N., Sellers W. R. Mol. Cell. Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez F., Grossman S. R., Takahashi Y., Rokas M. V., Nakamura N., Sellers W. R. J. Biol. Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 22.Das S., Dixon J. E., Cho W. Proc. Natl. Acad. Sci. USA. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. Dev. Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Cantley L. C., Neel B. G. Proc. Natl. Acad. Sci. USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez T., Thangada S., Wu M.-T., Kontos C. D., Wu D., Wu H., Hla T. Proc. Natl. Acad. Sci. USA. 2005;102:4312–4317. doi: 10.1073/pnas.0409784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Radisky D. C., Wang F., Bissell M. J. J. Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocucci S. M., Sussman M. J. Cell Biol. 1970;45:399–407. doi: 10.1083/jcb.45.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U. K., Molbert E., Showe M., Kellenberger E. J. Mol. Biol. 1970;49:99–113. doi: 10.1016/0022-2836(70)90379-7. [DOI] [PubMed] [Google Scholar]
- 29.Fukui Y., Yumura S., Yumura T. K. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- 30.Ueda M., Sako Y., Tanaka T., Devreotes P., Yanagida T. Science. 2001;294:864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 31.Tokunaga M., Kitamura K., Saito K., Iwane A. H., Yanagida T. Biochem. Biophys. Res. Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 32.Iino R., Koyama I., Kusumi A. Biophys. J. 2001;80:2667–2677. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.