Abstract
Evidence in several microorganisms indicates that Amt proteins are gas channels for NH3 and CH3NH2, and this has been confirmed structurally. Chlamydomonas reinhardtii has at least four AMT genes, the most reported for a microorganism. Under nitrogen-limiting conditions all AMT genes are transcribed and Chlamydomonas is sensitive to methylammonium toxicity. All 16 spontaneous methylammonium-resistant mutants that we analyzed had defects in accumulation of [14C]methylammonium. Genetic crosses indicated that 12 had lesions in a single locus, whereas two each had lesions in other loci. Lesions in different loci were correlated with different degrees of defect in [14C]methylammonium uptake. One mutant in the largest class had an insert in the AMT4 gene, and the insert cosegregated with methylammonium resistance in genetic crosses. The other 11 strains in this class also had amt4 lesions, which we characterized at the molecular level. Properties of the amt4 mutants were clearly different from those of rh1 RNAi lines. They indicated that the physiological substrates for Amt and Rh proteins, the only two members of their protein superfamily, are NH3 and CO2, respectively.
BOTH differentiation and metabolism in Chlamydomonas reinhardtii are controlled by the quantity and quality of the nitrogen (N) source. The preferred N source is ammonium. Even in the presence of an alternative N source such as nitrate or arginine, depletion of ammonium leads to substantial changes in transcription of genes whose products are required for acquisition of N (Merchan et al. 2001) and may lead to gametic differentiation (Treier et al. 1989).
Like other microbes, Chlamydomonas has genes coding for ammonium transport proteins (AMT genes). Amt proteins belong to a superfamily that has only one other member, the Rhesus or Rh proteins (Gazzarrini et al. 1999). Chlamydomonas is rare among microbes in having both Amt and Rh proteins (Soupene et al. 2002c) and hence is an organism of choice for discriminating differences in their physiological roles. The best-known Rh proteins compose the Rh blood group substance of humans, a very abundant protein in the red blood cell membrane (Cartron 1999; Avent and Reid 2000).
The substrates for both Amt and Rh proteins have been in dispute. On the basis of evidence in other microbes, we have proposed that Amt proteins are gas channels for NH3 (Soupene et al. 1998, 2001, 2002a,b), whereas others have proposed that they are active transporters for the ion NH+4 (Marini et al. 1997; von Wirén et al. 2000; von Wirén and Merrick 2004). We have provided evidence in Chlamydomonas that Rh proteins are gas channels for CO2 (Soupene et al. 2002c, 2004), whereas others have proposed that they, too, are active transporters for NH+4 (Marini et al. 2000; Westhoff et al. 2002; Hemker et al. 2003; Nakhoul and Hamm 2004). Recently the structure of the AmtB protein from Escherichia coli was determined to an extraordinary 1.35-Å resolution (Khademi et al. 2004). Structures with ligands present confirmed that Amt proteins are gas channels for NH3 or CH3NH2.
To complement our studies of Rh expression and function in Chlamydomonas, we have now used resistance to the toxic ammonium analog methylammonium (we use ammonium to indicate the sum of NH+4 and NH3 and methylammonium to indicate the sum of CH3NH+3 and CH3NH2) to isolate mutant strains with lesions in an AMT gene, AMT4. These strains are particularly useful because it is not yet possible to use homologous recombination to target lesions to particular genes in Chlamydomonas (Lefebvre and Silflow 1999). Properties of the amt4 mutant strains can now be compared to those of RNA interference (RNAi) lines that fail to express RH1 (Soupene et al. 2004; see discussion).
Resistance to methylammonium has been used previously to isolate mutant strains lacking function of Amt proteins (also called Mep or Mea in other microbes) (Arst and Cove 1969; Dubois and Grenson 1979; Marini et al. 1997; Monahan et al. 2002a,b). Franco et al. (1987)(1988) characterized two methylammonium-resistant mutants of C. reinhardtii. They proposed that one strain, called 2170, had a defect in transport of methylammonium and ammonium. Both strains had lesions linked to the NIT1 locus. We show here that AMT4 is not linked to NIT1 and that strain 2170 does not appear to have a lesion in AMT4. Hence the amt4 strains that we describe are different from the methylammonium-resistant strains studied previously. Unexpectedly, we found that a large fraction of them carry transposon-induced lesions and that several of the lesions affected mRNA splicing.
MATERIALS AND METHODS
Strains, culture conditions, and genetic analysis:
Chlamydomonas strains and their sources are listed in Table 1. Strains 4A+ and 4A−, obtained from the laboratories of J. Rochaix and K. K. Niyogi, respectively, are described by Soupene et al. (2004). All strains were maintained on solid Tris-acetate-phosphate (TAP) medium (Harris 1989), supplemented with 5 μg/ml nicotinamide when necessary. The N source was 10 mm NH4Cl, 10 mm KNO3, or 2.5 mm arginine as appropriate. Cells were grown at 25° under continuous light at 25 μE unless otherwise specified. Crosses were performed using standard methods (Levine and Ebersold 1960). Crosses that involved three of the strains were problematic. Strains CR41 and CR45 did not mate readily. At least three matings were required with each strain to obtain sufficient zygotes for analysis. Growth of these strains on plates and in liquid culture was slow and many cells had irregular shapes. Zygotes from CR07 crosses germinated at a low frequency compared with those of other crosses and progeny survival was <50% of survival frequency for other crosses. We failed to obtain complete tetrads only from CR07.
TABLE 1.
Chlamydomonas strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 4A+a | nit1 nit2 mt+ | K. K. Niyogib |
| 4A− | nit1 nit2 mt− | K. K. Niyogib |
| CC-28 | msr-1 ac-17 pyr-1 act-1 can-1 sr-1 nic-13 pf-2 y1 mt+ | CCCc |
| CC-29 | msr-1 ac-17 pyr-1 act-1 can-1 sr-1 nic-13 pf-2 y1 mt− | CCCc |
| CC-124 | nit1 nit2 mt− | CCCc |
| CC-125 | nit1 nit2 mt+ | CCCc |
| CC-1085 | nit1 mt+ | CCCc |
| CC-1086 | nit2 mt+ | CCCc |
| CC-1690 | mt+ | K. K. Niyogib |
| CC-2170 | ma-1 mt+ | E. Fernandezd |
| CC-2290e | mt− | K. K. Niyogib |
4A+ sent to the Chlamydomonas Culture Collection as CC-4051.
Department of Plant and Microbial Biology, University of California, Berkeley, California.
Chlamydomonas Culture Collection, Duke University, Durham, North Carolina.
University of Cordoba, Department of Biochemistry and Molecular Biology, Campus de Rabanales, Edificio Severo Ochoa, Cordoba, Spain.
Also called S1-D2.
Isolation of spontaneous mutants and tests of phenotype:
On TAP/arginine plates strains 4A+ and CC-125 begin to show growth defects at 40 μm methylammonium (but they show no defects up to 1 mm methylammonium when ammonium is the N source). Cells of parental strain 4A+ or CC-125 were grown on TAP/ammonium and then streaked onto solid TAP medium, supplemented with 2.5 mm arginine and various concentrations of methylammonium from 50 μm to 5 mm (see Table 4). After 2–3 weeks incubation in the light, independent resistant mutants were picked from selection plates, purified on TAP/ammonium plates, and maintained on TAP/ammonium. Strains CR05 and CR07, which were among the strains isolated earliest, were subjected to selection twice under the assumption that multiple mutations might be required for methylammonium resistance, as was true in Saccharomyces cerevisiae (Marini et al. 1997). Specifically, two “colonies” picked at 5 mm methylammonium but found to be sensitive were then subjected to selection at 1 mm methylammonium a second time. The second plates were incubated in the light for 10 days and then transferred to the dark for 2 weeks. All 16 mutants that we studied were tested for resistance to methylammonium at a range of concentrations and were also tested for resistance to chloroquine, another weak base. Tests for chloroquine resistance were done at 0.02 mm, 0.1 mm, 0.2 mm, 0.5 mm, and 1.0 mm on solid TAP medium with either arginine or ammonium as N source.
TABLE 4.
Characteristics of parental strains and the three classes of methylammonium-resistant mutants
| Methylammonium (mm)
|
|||
|---|---|---|---|
| Straina | Selected at | Resistant atb | Uptake of [14C]methylammonium (pmol/μg chlorophyll a + b/min)c |
| Wild type | |||
| 4A+ | <0.05 | 80 | |
| CC-125 | <0.05 | 35 | |
| Class 1 | |||
| CR05d | 1 | 1e | <2 |
| CR43 | 0.05 | 1e | <2 |
| Class 2 | |||
| CR02 | 5 | 1 | 3.5 |
| CR03 | 0.1 | 0.1 | 6.0 |
| CR04 | 0.1 | 1 | 2.0 |
| CR07d | 1 | 1 | 3.5 |
| CR39 | 0.05 | 1 | 3.0 |
| CR40 | 0.05 | 1 | 3.5 |
| CR42 | 0.05 | 1 | 4.0 |
| CR46 | 0.05 | 0.1 | 7.0 |
| CR47 | 0.05 | 1 | 3.0 |
| CR48 | 0.05 | 1 | 3.5 |
| CR49 | 0.05 | 1 | 3.5 |
| CR50 | 0.05 | 0.1 | 10 |
| Class 3 | |||
| CR41 | 0.05 | 0.05 | 30 |
| CR45 | 0.05 | 0.05 | 40 |
The parental strain for CR03 and CR04 was CC-125. For all others it was 4A+.
Highest concentration of methylammonium to which the strain was resistant with arginine as N source. The concentrations tested were 0.05, 0.1, and 1 mm.
Rates of uptake were determined from data in Figure 4 and similar experiments. Averages for three experiments are shown except for strains CC-125, CR03, and CR04, which were assayed only once. For parental strains, rates were determined from early time points when <20% of the substrate had been utilized and for mutant strains during the first 15 min.
Subjected to selection twice (see materials and methods).
Grew slowly at 1.0 mm methylammonium.
RNA isolation, Northern hybridization, and RT-PCR:
For Northern hybridizations and RT-PCR, Chlamydomonas cells were grown on an orbital shaker in 20-ml scintillation vials (7 ml medium) or 500-ml flasks (200 ml culture) under continuous light (80 or 17 μE, respectively) to a chlorophyll a + b content of 6–10 μg/ml in TAP/arginine or TAP/ammonium medium. Chlorophyll a + b was determined according to the method of Wintermans and de Mots (1965). RNA was isolated and Northern hybridization was performed according to Gromoff et al. (1989), as described by Soupene et al. (2002c). Poly(A)-tailed RNA was extracted using a GenElute mRNA miniprep kit (Sigma, St. Louis) following instructions of the manufacturer. For Northern hybridization, 3 μg of poly(A)-tailed RNA was separated by electrophoresis and blotted. All probes were generated from genomic DNA and labeled by random-primed labeling with the High-Prime System (Roche) and [α-32P]dCTP following instructions of the manufacturer. The AMT1-specific probe was obtained by PCR amplification of a 600-bp fragment of the last exon (exon 15) using the forward primer AM2 (5′-CGTCCACTGCACCGTTGGTGTG-3′) and the reverse primer AM3 (5′-ACGAATGCAGTTACAATAGGCG-3′). The AMT2-specific probe was obtained by PCR amplification of a 713-bp fragment of the last exon (exon 6) using the forward primer Amt2-122 (5′-TATGCCTATGATCAGTAAGG-3′) and the reverse primer Amt2-123 (5′-ACATTCGGAATATCGTTACAGC-3′). The AMT3-specific probe was obtained by PCR amplification of a 500-bp fragment of the last exon (exon 12) using the forward primer Mep10 (5′-CACGTATGGAAAGCTAAGAGGC-3′) and the reverse primer Mep2a (5′-GGGGCGTACAGTTACAGGATGTCCG-3′). The AMT4-specific probe was obtained by PCR amplification of a 270-bp fragment of exon 5 using the forward primer AMT4-928F and the reverse primer AMT4-1195R (Table 2). The RBCS probe was obtained by PCR amplification of a 664-bp fragment using the forward primer rbc-21 (5′-ATGGCCGCCGTCATTGCCAAG-3′) and the reverse primer rbc-22 (5′-CATCCACCGCCGTTCGTCAGG-3′). Phosphor screens exposed to Northern blots were scanned with a Typhoon 8600 PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
TABLE 2.
PCR and sequencing primers forAMT4
| Primer | Sequence (5′–3′) | Position in DNAa | Position in RNAa |
|---|---|---|---|
| −1577F | ATGCAGCCATGGTGGGAGTGC | −1577–−1557 | |
| ATGF | ATGGCGGACGAGATGGATCC | 1–20 | 1–20 |
| ATGR | TCATGGGATCCATCTCGTCC | 25–6 | 25–6 |
| 500F | TTGCTCAGCTGCAGATCAAACG | 477–498 | |
| 500R | TCGTTTGATCTGCAGCTGAGC | 499–479 | |
| TILL1F | AACAACCCCAACGGCTTTGTG | 700–720 | |
| TILL1R | AAAGCCGTTGGGGTTGTTGC | 717–698 | 360–341 |
| TILL2F | CTCAACTTCAACGCCTACCTC | 1089–1109 | 505–525 |
| TILL2R | TGAGGTAGGCGTTGAAGTTGAGG | 1110–1088 | 526–504 |
| 928F | TGCCTGTCCTGCCTGCTCACC | 1790–1810 | 928–948 |
| 1195R | AGCAGGCCGGCAAAGAACACG | 2058–2038 | 1196–1176 |
| Short1F | ATCGGCATCATCTCCATCTTCG | 2392–2413 | 1303–1324 |
| Short1R | AGATGGAGATGATGCCGATCACC | 2410–2388 | |
| Short2F | ATTGGACTTTCTGCTCATGG | 3484–3503 | 2066–2085 |
| UGA-1R | GTGCTTTCCATCTGAGCACC | 3732–3713 | 2314–2295 |
| EndR | TTTGGACGGCAGACTGTGCG | 4009–3990 | 2591–2572 |
Relative to ATG translational start. Positions of the primers are also shown in Figure 6A.
For RT-PCR, RNA was isolated as described above and treated with DNase I (Pharmacia, Piscataway, NJ) followed by phenol/chloroform extraction. Expression of the four AMT genes was analyzed by RT-PCR using the one step RT-PCR kit (Roche) following instructions of the manufacturer. First-strand DNA was reverse transcribed from 1 μg of total RNA using 0.15 mm of each primer at 60° for 30 min and then cDNA was amplified in the same mixture. For the AMT1, AMT2, and AMT3 genes, the same primer pairs were used as described for Northern analysis. RT-PCR for AMT4 was performed using forward primer AMT4-short2F and reverse primer AMT4-EndR (Table 2).
DNA isolation and PCR:
Cultures were grown in scintillation vials in 7 ml TAP/ammonium medium with constant agitation and continuous light at 80 μE. They were harvested at stationary phase. DNA was isolated using a DNA extraction kit (Stratagene, La Jolla, CA). PCR was performed with the Long Template PCR system (Roche) with 100 ng of genomic DNA in a 100-μl reaction mixture (300 nm of each primer, 500 μm dNTPs, 3 mm MgCl2, and 4 units of Expand Long Template enzyme mix) under the following conditions: 2 min at 95°, 30 cycles of amplification [denaturation (30 sec at 94°)/annealing (1 min at appropriate temperature depending on the primers)/polymerization (appropriate time at 68° depending on the primer sets)], and 10 min at 68°. For large PCR fragments (>4 kb), polymerization time was 7 min and DMSO was added to a final concentration of 5% in the reaction mixture. Primers used to amplify various regions of the AMT4 gene are listed in Table 2.
Methylammonium uptake assays:
Uptake of [14C]methylammonium was determined as described by Soupene et al. (1998). Cells were grown in TAP medium with various N sources to a chlorophyll a + b content of ∼8 μg/ml. They were harvested and subsequently washed and suspended in assay buffer (20 mm HEPES/20 mm acetic acid; pH to 7.2 with KOH). Suspended cells were incubated in a water bath shaker at 25° under lights for 20 min before radiolabeled methylammonium (6 μm; specific activity 6 or 50 Ci/mol) was added. At times between 1 and 60 min, cells were filtered and washed twice with assay buffer, and membranes were counted. Uptake rates were calculated from time points at which ≤20% of the substrate had been utilized.
DNA sequencing of AMT genes:
Sequencing was performed by the University of California, Berkeley Sequencing Facility using the ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Primers for sequencing were the same as those described above for PCR and RT-PCR.
The genome of C. reinhardtii encodes a minimum of four ammonium transport (AMT) genes and may encode as many as eight. Two of the AMT genes were sequenced by Gonzalez-Ballester and Fernandez and deposited in GenBank under accession nos. AF479643, AY058211, and AF530051 for AMT1 cDNA, AMT1 genomic DNA, and AMT2 cDNA, respectively. AMT3 was sequenced in our laboratory and cDNA and genomic DNA sequences were deposited in GenBank as AF509497 and AF509496, respectively. The Joint Genome Institute's (JGI) Chlamydomonas project (Grossman et al. 2003) enabled us to supplement this information with the genomic sequences for AMT2 and AMT4.
Determining the genomic DNA sequence for AMT3 was problematic and we noted that neither the complete sequence for AMT3 nor the adjacent sequence has yet been determined by JGI. Six regions of AMT3 were difficult to sequence using the BigDye Terminator v3.0 kit (Applied Biosystems). These were located in introns 4, 5, 8, 9, 10, and 11, which contain highly repetitive sequences (e.g., [CCA]n, [AGAGGG]n, [AGGGGG]n, and [GT]n). Intron 9, which contains [GT]32, [CA]9, and [CA]5 repeated (microsatellite) sequences, was particularly intractable. Adding DMSO or betaine, or using higher reaction temperatures, did not improve sequencing quality. Genomic DNA sequencing was finally completed by subcloning repetitive motifs and sequencing through them using the dGTP Terminator kit v3.0 (Applied Biosystems). Although this kit permitted sequencing of the repetitive regions, the Standard BigDye kit was required to read into adjacent GC-rich regions due to band compression problems.
Signal peptide predictions:
Peptide cleavage sites and subcellular localization for each of the Amt proteins was determined by the SignalP, ChloroP, TargetP, and PSORT programs (http://us.expasy.org/tools/). Only the Amt2 protein was found to contain a clear signal peptide but the programs did not agree on where it would be targeted.
RESULTS
AMT genes:
The sequences of the four Amt proteins and their closest relative, the Arabidopsis thaliana Amt1;2 protein (Gazzarrini et al. 1999), are compared together in Figure 1A. The regions of highest identity are the middle segments, which, on the basis of predictions for the Arabidopsis Amt proteins and the structure of the E. coli AmtB protein, contain the transmembrane spanning regions (underlined). Polypeptide products of AMT1 and AMT3 are ∼50% identical, as are those of AMT2 and AMT4. When the amino acid sequences of the four proteins are compared, only a 28% identity is found. These similarities fall well short of the degree of identity, 58%, among the five AMT1 family genes of Arabidopsis.
Figure 1.—
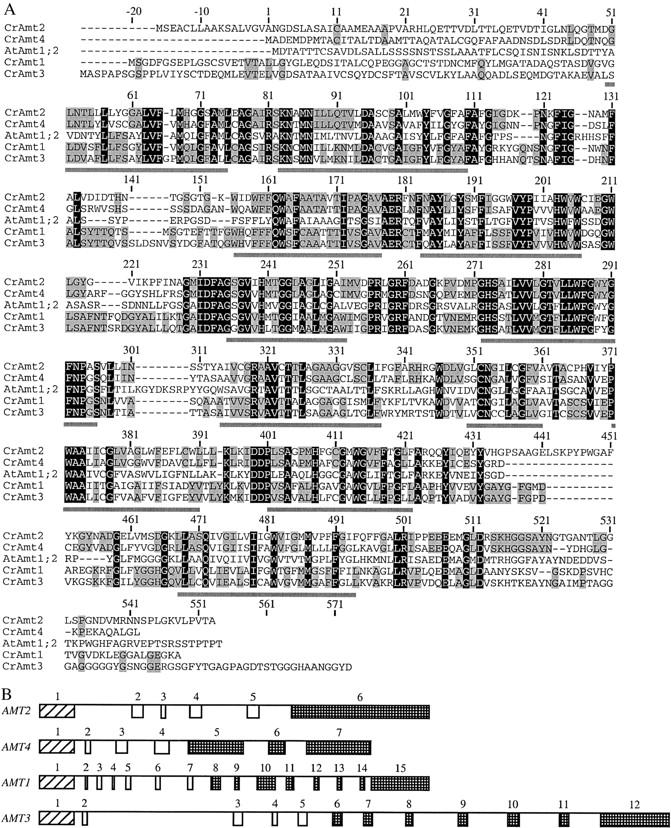
Comparison of Amt proteins and genes. (A) The four Chlamydomonas Amt proteins studied are aligned with the Arabidopsis Amt1;2 protein (Gazzarrini et al. 1999). Numbers are with respect to the putative start site for translation in Chlamydomonas Amt4. Residues identical in the five proteins are on a solid background and residues identical in only Amt2 and Amt4 or Amt1 and Amt3 are shaded. Putative transmembrane segments for the Chlamydomonas proteins, based on comparisons with the E. coli AmtB protein structure, are underlined. (B) Comparison of the structures of the Chlamydomonas AMT1-AMT4 genes. Numbered boxes indicate exons and lines indicate introns. Homologous coding regions of exons are indicated by similar patterns.
Information about the four Chlamydomonas AMT genes (gene size, introns, approximate mRNA length) is summarized in Table 3. There is insufficient information to determine whether any of them might be clustered. Figure 1B summarizes and compares the structures of the four Chlamydomonas AMT genes. The largest intron, intron 2 of AMT3, includes a 370-bp sequence, which is repeated with high similarity 38 times in the Chlamydomonas genome. Fragments of the sequence occur an additional 31 times. In four of the repetitions, the left (268 bp) and right (119 bp) ends of this 370-bp sequence are separated by 3.0–4.0 kb and, in addition, the 370-bp sequence is repeated without interruption at the right end. This arrangement of sequences reflects the organization of the split long terminal repeats (LTRs) of the TOC1 transposon (Day and Rochaix 1991) and suggests that intron 2 of AMT3 at one time may have been a site of insertion for a retrotransposon.
TABLE 3.
C. reinhardtii AMT genes
| Gene | JGI designation (Genome v. 2.0) |
Approximate mRNA lengtha (bases) |
Gene length (bp) |
Intronsb |
|---|---|---|---|---|
| AMT1 | C_110147 | 2.2 × 103 | 4673 | 14 |
| AMT2 | C_456001 | 2.7 × 103 | 4584 | 5 |
| AMT3 | C_2680003 | 2.6 × 103 | 7383 | 11 |
| AMT4 | C_930017 | 2.7 × 103 | 4112 | 6 |
| AMT5 ?? | C_220054 | 10+ | ||
| AMT6 ?? | C_980024 | 6+ | ||
| AMT7 ?? | C_20186 | 9+ | ||
| AMT8 ?? | C_380121 | 6+ |
Based on cDNAs, ESTs, and predicted transcripts from JGI, GenBank, and The Institute for Genomic Research (http://www.tigr.org) databases.
Based on cDNA for AMT1–AMT4 and predictions for the other four putative AMT genes.
The AMT1–AMT4 genes of Chlamydomonas have one splice junction in common: the site before the second transmembrane-spanning segment (W. Inwood, unpublished results). Interestingly, the site of the first splice junction for a variety of RH genes, including Chlamydomonas RH1, Drosophila RH50, and human RhCG and RhAG, is highly conserved and is also located prior to the second predicted transmembrane segment. This conserved intron position could be significant in permitting exchange of leader or N-terminal sequences between Rh or Amt proteins.
The gene prediction program for the JGI C. reinhardtii genome (version 2.0) suggests that there may be an additional four AMT genes in Chlamydomonas (Table 3). Of these, only AMT6 appears to have a strong similarity to the four AMT genes discussed above. Its sequence is only partially complete, so the degree of similarity is not certain. No expressed sequence tag (EST) is associated with any of the four putative AMT genes listed at the JGI database interface. Like the Amt1–Amt4 proteins, the additional hypothetical Amt proteins are most similar to one another and to others in the middle region covering their predicted transmembrane segments. We have not investigated the expression of these other genes and cannot rule out the possibility that they are inactive. They are not considered further.
Expression of AMT1–AMT4:
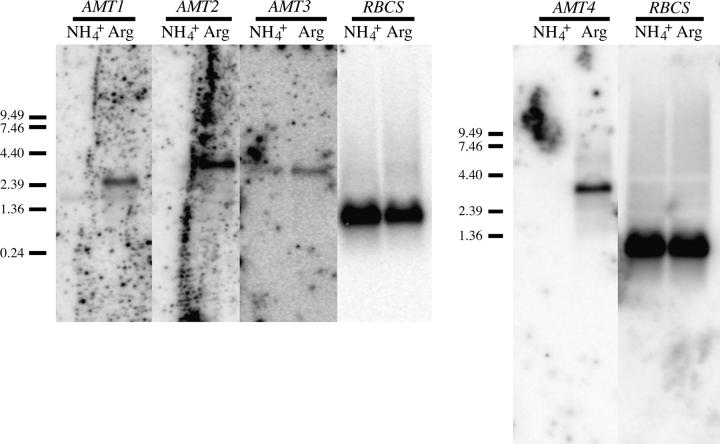
Expression of genes AMT1–AMT4 in strain 4A+ had one of three patterns under the conditions examined. Levels of AMT3 mRNA were relatively low and similar under N-rich and N-limiting conditions (ammonium or arginine as N source, respectively; assessed by Northern hybridization and by RT-PCR; Figures 2 and 3A). Levels of mRNA for AMT1 and AMT2 were undetectable by either assay for cells grown on ammonium, but moderate to high for cells grown on arginine. By RT-PCR, levels of AMT4 mRNA were low but detectable for ammonium-grown cells, but elevated notably for arginine-grown cells. By Northern blot, AMT4 mRNA was detected from arginine- but not ammonium-grown cells.
Figure 2.—
Northern analysis of Amt mRNA levels. Three micrograms of poly(A)-tailed RNA was used for each lane. Cells were grown with ammonium or arginine as N source as indicated. Membranes were hybridized to probes specific to Amt1, Amt2, Amt3, Amt4, and RBCS (see materials and methods).
Figure 3.—
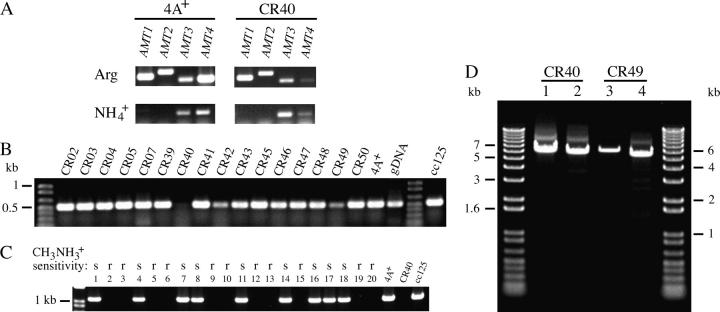
Identification of amt4 mutants by RT-PCR (A and B) and PCR (C and D). (A) Primers specific to the 3′-ends of AMT1-AMT4 (as indicated in materials and methods) were used to amplify mRNA for these genes from strains 4A+ and CR40. RNA was prepared from cells grown on arginine or ammonium as N source and was amplified by RT-PCR. (B) Primers Short2F and EndR (Table 2) were used to amplify the 3′-end of AMT4 mRNA from all 16 methylammonium-resistant strains and parental strains CC-125 and 4A+. RNA was prepared from cells grown on TAP/arginine and was amplified by RT-PCR. The same region was amplified from 4A+ genomic DNA (gDNA). Molecular weight standards are in the unmarked lanes. (C) The ATGF and TILL2R primers (Table 2) were used to amplify the 5′-end of the AMT4 gene by PCR from 20 progeny of a cross between strains CR40 and 4A−. Sensitivity or resistance of these progeny to methylammonium is indicated above the number of the isolate. Strains 4A+, CC-125, and CR40 were used as controls. (D) The AMT4 gene was amplified from DNA (Table 2) isolated from strains CR40 and CR49. Primers and expected fragments from amplification of 4A+ sequences for lanes were: (1) ATGF and 500R, 500 bp; (2) ATGF and 355R, 355 bp; (3) TILL2F and Short1R, 1328 bp; and (4) 928F and Short1R, 627 bp. Long extension times were used to amplify fragments of up to 8 kb (see materials and methods). Molecular weights of standards are indicated next to the unmarked lanes.
Methylammonium-resistant mutants and methylammonium uptake:
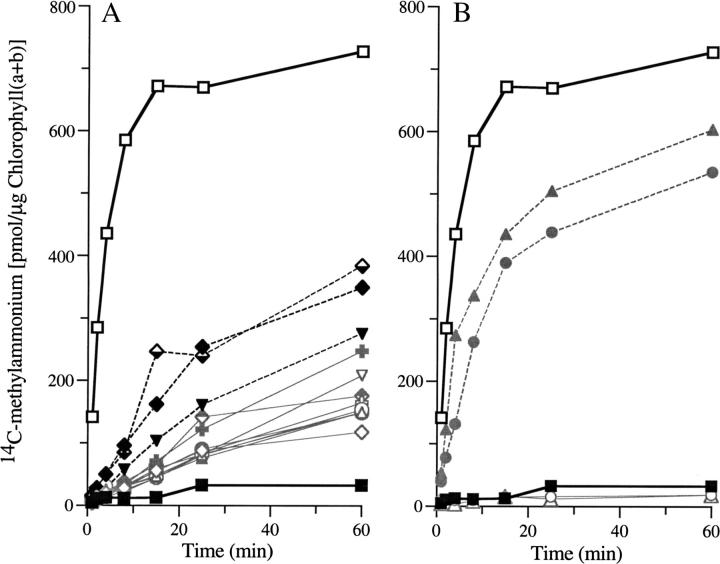
Spontaneous methylammonium-resistant mutants were selected from parental strain 4A+ or CC-125 on TAP/arginine medium supplemented with 0.05, 0.1, 1, or 5 mm methylammonium. Many of the strains were isolated at the lowest concentration of this toxic analog to favor obtaining lesions in single AMT genes of this multigene family and to obtain lesions that might be informative with respect to the mechanism of gas channel function. Sixteen strains were analyzed for their degree of resistance to methylammonium and for their ability to transport it (Table 4 and Figure 4). Figure 4 shows the results for a single experiment and Table 4 presents averages for three experiments, except as noted. For cells grown on TAP/arginine rates of uptake of [14C]methylammonium (6 μm) fell into three distinct classes: class 1, very low (<2% the rate of uptake of strain 4A+; 2 strains); class 2, low (5–10% the rate of 4A+; 12 strains); class 3, intermediate (∼50% the rate of 4A+; 2 strains). Cells grown on ammonium accumulated very little [14C]methylammonium (Figure 4 for strain 4A+ and not shown). When grown on arginine, the class 1 strains showed no greater uptake than did any of the ammonium-grown strains (Figure 4B). The class 1 and 2 strains, those with very low and low uptake, respectively, were able to survive and grow at higher levels of methylammonium than the class 3 strains, which retained intermediate levels of methylammonium uptake (Table 4). Three strains placed in class 2, CR03, CR46, and CR50, were notable for their lower level of resistance to methylammonium compared to the other 9 strains. These 3 strains also had consistently higher methylammonium uptake rates—two to three times the average for the other strains. The relative uptake rates of the other 9 strains in class 2 varied in different experiments.
Figure 4.—
Methylammonium uptake assays. Uptake of [14C]methylammonium (6 μm) by methylammonium-resistant mutants grown on TAP/arginine compared with that of parental strain 4A+ grown on TAP/arginine (open squares) or TAP/ammonium (solid squares). (A) Twelve class 2 mutants (CR02, open diamonds; CR03, solid diamonds, dashed line; CR04, open triangles; CR07, solid triangles; CR39, open circles; CR40, solid circles; CR42, inverted open triangles; CR46, solid inverted triangles, dashed line; CR47, open crosses; CR48, solid crosses; CR49, top-filled diamonds; CR50, bottom-filled diamonds, dashed line). (B) Two class 1 mutants (CR05, open circles; CR43, open triangles) and two class 3 mutants (CR41, solid circles; CR45, solid triangles). Data for strain 4A+ in A and B are the same.
Genetic analysis of methylammonium-resistant mutants:
Our strategy to determine the minimum number of genetic loci represented among the 16 methylammonium-resistant mutant strains involved several steps. First, we crossed each to strain CC-124 or 4A− to show that mutations conferring methylammonium resistance behaved as single lesions in the nuclear genome and segregated 2:2 with respect to mating type. From these crosses mating-type minus methylammonium-resistant strains CR17 and CR181 were obtained from CR02 and CR05, respectively. Each of the remaining 14 mutant strains was then crossed to both of these to determine, by assessing linkage, whether a class might represent a single genetic locus. The results of these crosses are presented in Table 5. The presence of only parental ditype tetrads in crosses between the 2 class 1 strains and between the 12 class 2 strains indicated close linkage or identity of the mutated loci in each class. Finally, CR41 and CR45 were crossed to mating type minus strains obtained from preliminary crosses between them and strain 4A−, and the resulting 22 tetrads were found to be parental ditype (data not shown). Thus the mutations in class 3 strains are closely linked. The classes of mutations defined genetically were the same as those defined by methylammonium uptake assays: CR41 and CR45 (class 3) had intermediate uptake; CR05 and CR43 (class 1) had very low uptake; and the remaining 12 strains (class 2) had low uptake.
TABLE 5.
Summary of genetic crosses
| Strain | CC number | × CR17 tetradsa | × CR181 tetradsa |
|---|---|---|---|
| CR02 | 4052 | Same mutation | ND |
| CR03 | 4036 | 27:0:0 | 7:7:12 |
| CR04 | 4044 | 23:0:0 | 9:9:17 |
| CR05 | 4037 | 3:3:5 | Same mutation |
| CR07 | 4038 | 21:0:0 | 3:4:6 |
| CR39 | 4039 | 27:0:0 | 1:0:2 |
| CR40 | 4040 | 25:0:0 | 2:2:2 |
| CR41 | 4041 | 2:2:2 | 10:7:13 |
| CR42 | 4042 | 15:0:0 | 3:5:8 |
| CR43 | 4043 | 1:3:6 | 15:0:0 |
| CR45 | 4045 | 2:1:3 | 8:4:10 |
| CR46 | 4046 | 14:0:0 | 4:3:6 |
| CR47 | 4047 | 43:0:0 | 5:6:11 |
| CR48 | 4048 | 14:0:0 | ND |
| CR49 | 4049 | 16:0:0 | ND |
| CR50 | 4050 | 12:0:0 | ND |
Parental ditype:nonparental ditype:tetratype (PD:NPD:T). CR17 is a class 2 mutant strain and CR181 is a class 1 mutant strain.
Examination of recombinant progeny from genetic crosses revealed that double-mutant strains, those found in nonparental ditype tetrads, always exhibited the methylammonium resistance level of the more resistant parental strain. In other words, null mutations of class 2 were epistatic to mutations of either of the other classes and mutations of class 1 were epistatic to those of class 3 (see discussion).
Franco et al. (1988) showed that two lesions leading to methylammonium resistance (5 mm) with nitrate as the N source mapped close together and very near the nitrate reductase gene NIT1. Hence, we mapped our three classes of mutations relative to NIT1. A representative of each class was crossed to two strains able to use nitrate as N source, CC-1690 and CC-2290. Methylammonium-resistant strains that were able to grow on nitrate were recovered from these crosses in such proportion to the total progeny as to show that none of our mutated loci was closely linked to either NIT1 or NIT2 (see supplementary Results at http://www.genetics.org/supplemental/). Moreover, we sequenced the AMT4 gene of strain 2170 (see Introduction and Table 1) and found it to be identical to the AMT4 genes of strains CC-1690 and 4A+ (see below). In addition, we showed that transcript levels for all four AMT genes in strain 2170 were the same as those in strains 1690 and 4A+ under N-limiting conditions (arginine or nitrate as N source) and N-excess conditions (RT-PCR; see supplementary Figure 1 at http://www.genetics.org/supplemental/). Thus, expression of the AMT genes in strain 2170 appeared to be normal.
Other phenotypes of methylammonium-resistant mutants:
Class 3-resistant strains proved more sensitive to chloroquine, a weak base, than did other methylammonium-resistant strains. On TAP arginine plates, all strains became somewhat sensitive to chloroquine at 0.5 mm, like parental strains 4A+ and CC-125. On TAP ammonium plates, all strains but two grew well and remained green at all chloroquine concentrations as high as 1 mm. CR41 and CR45 (class 3) were sensitive to chloroquine at 0.5 mm and 1.0 mm and bleached to white after a few days. At 0.2 mm chloroquine, CR45 grew slower than other strains and was lighter green but CR41 was unaffected. In comparison to the other mutant strains and parental strains, CR41 and CR45 grew poorly on hypoxanthine as the N source. Like arginine, hypoxanthine is a limiting N source. These results corroborate findings from genetic crosses (see materials and methods) and methylammonium uptake assays, indicating that CR41 and CR45 are likely to have lesions at the same locus.
Molecular characterization of amt4 mutants—general properties:
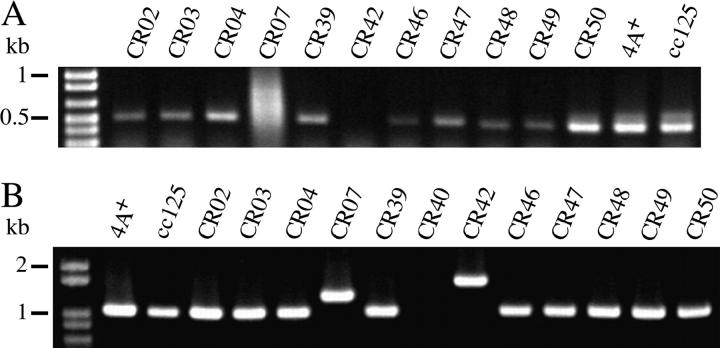
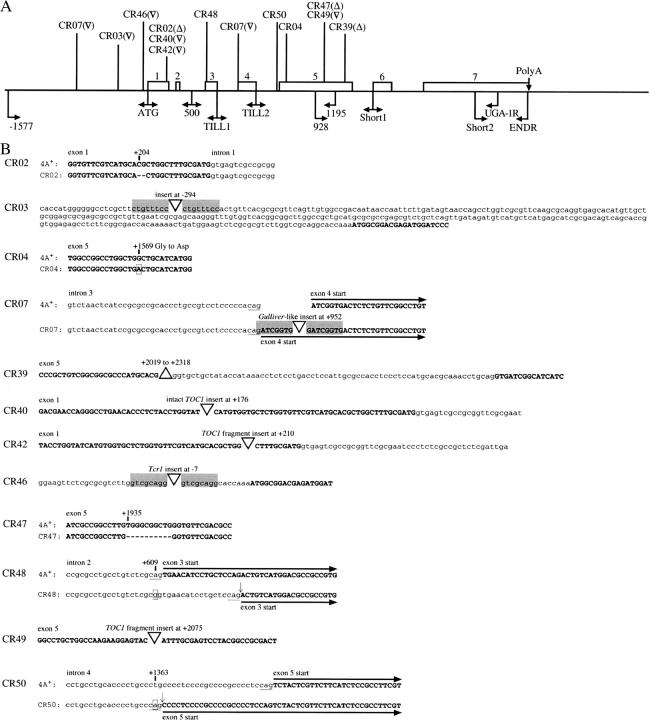
All 16 methylammonium-resistant strains had grossly normal levels of mRNA for AMT1, AMT2, and AMT3 (RT-PCR; see materials and methods) when they were grown on TAP/arginine (data not shown). However, CR40, a class 2 mutant, had little or no transcript for the AMT4 gene on TAP/arginine (3′ probe; Figure 3, A and B). We confirmed with the same RNA preparation from CR40 that levels of transcript for AMT1–AMT3 were normal for both ammonium- and arginine-grown cells and found that mRNA levels for the AMT4 gene were lower than those in 4A+ under both conditions and were not increased on arginine (Figure 3A). We were unable to amplify a 5′ region of AMT4 from CR40 DNA and used this characteristic to determine whether the AMT4 lesion in CR40 was co-inherited with methylammonium resistance in genetic crosses. DNA was isolated from 20 progeny of a cross between CR40 and 4A−, which represented six tetrads, and the 5′ region of AMT4 was amplified. Failure to obtain a PCR product and methylammonium resistance cosegregated in the cross (Figure 3C), indicating that the AMT4 lesion was responsible for methylammonium resistance. The probability that the amt4 lesion was not linked to methylammonium resistance was ∼1/4000. We therefore examined the 5′- and 3′-ends of the AMT4 transcript for each of the other 11 strains in class 2 (RT-PCR; Figures 5A and 3B, respectively). Alterations in some strains reflected gross changes in the DNA at the 5′-end of the gene (Figure 5B). Two changes are most obvious from Figure 5, A and B: First, the 5′-end of the AMT4 transcript from CR07 is not of uniform size and there is a small insert in the DNA. Second, the 5′-end of the AMT4 transcript is absent in CR42 and there is an insert of ∼600 bp in the DNA. These observations led us to the following sequencing strategy to examine other class 2 mutant strains. We divided the AMT4 gene into four parts: upstream and 5′-untranslated region (5′-UTR); 5′ coding region (exons 1–4); middle region (intron 4 and exon 5); 3′ region (intron 5 through exon 7). Because we had observed the inserts shown in Figure 5, we sequenced the 5′ coding region of each mutant first and found changes from the wild-type gene in five strains (CR02, CR07, CR40, CR42, and CR48; Figure 6). We sequenced the middle region next and detected differences in five additional strains (CR04, CR39, CR47, CR49, and CR50). No strain had a change in the 3′ region of the gene, but three strains had changes in the 5′-UTR (CR03, CR46, CR07, second lesion) (Figure 6).
Figure 5.—
Use of RT-PCR and PCR to identify additional macrolesions in and around AMT4. (A) The ATGF and TILL2R primers (Table 2) were used to amplify the 5′-end of AMT4 mRNA from 11 class 2 methylammonium-resistant strains. Cells were grown on arginine and cDNA was amplified from total RNA by RT-PCR. (B) The ATGF and TILL2R primers were used to amplify the 5′-end of the AMT4 gene from 12 class 2 methylammonium-resistant strains. Cells were grown on ammonium and the fragment was amplified from genomic DNA by PCR.
Figure 6.—
amt4 lesions. (A) Locations of amt4 lesions. Mutations in methylammonium-resistant strains are indicated above the line (▵, deletion; ▿, insertion) and primer sites are indicated below. AMT4 exons are indicated as numbered blocks and the map is drawn approximately to scale. (B) Lesions in amt4 mutant strains. Sites of lesions in the DNA are indicated with respect to the translational start as +1. Positions of base-pair changes are ticked and the changes are boxed. Small deletions are indicated by hyphens and large deletions by triangles. Insertions are indicated by inverted triangles and direct repeat sequences are shaded. Exon and intron sequences for strain 4A+ are indicated by boldface uppercase and lowercase letters, respectively. For strains CR07, CR48, and CR50 only, changes in the positions of exons are noted. Conserved splice junction nucleotides are underlined and new splice sites in CR48 and CR50 are indicated by vertical arrows. A minority of splicing events in CR50 occurs at the normal location (see text). Splice sites in CR07 are mentioned in the text and will be discussed in detail elsewhere (K.-S. Kim and W. Inwood, unpublished results). The 4A+ sequence shown with the sequence for CR07 is gapped at the position of the insertion in CR07.
Molecular characterization of amt4 mutants—properties of individual strains:
Spontaneous amt4 lesions fell into three categories (Table 6, Figure 6, and supplementary Results at http://www.genetics.org/supplemental/): single base changes, deletions, and transposon-related events. The single base changes had three consequences: (1) substitution of a charged amino acid, aspartate, for glycine in a putative transmembrane segment, apparently inactivating the Amt4 protein (CR04); (2) destruction of a splice site and consequent deletion of 17 bases of mRNA, leading to a frameshift and protein truncation (CR48); and (3) Creation of an in-frame splice site introducing 24 bases and eight amino acids to increase the length of the protein from 499 to 507 residues (CR50). CR50 retained a moderate level of methylammonium uptake activity (Figure 4 and Table 4) and was less resistant than amt4 null strains to methylammonium. Although the added amino acid residues in the incorrectly spliced transcript, which were inserted in a predicted transmembrane segment, are compatible with a hydrophobic environment, a small amount of normal protein produced from correctly spliced transcript probably accounts for residual uptake activity.
TABLE 6.
Genetic lesions inAMT4 (class 2 mutants)
| Strain | Allele | Type of mutation | Site and effect on protein | Polypeptidea |
|---|---|---|---|---|
| CR02 | amt4-1 | 2-bp deletion | End of exon 1 (truncation) | 121 |
| CR03 | amt4-2 | Insert | 5′-untranslated region | 499 |
| CR04 | amt4-3 | Single base change | Exon 5 (gly to asp) | 499 |
| CR07 | amt4-4 | Gulliver-like insert | Exon 4 (splice change) | 136 |
| CR39 | amt4-5 | 300-bp deletion | Exon 5 and intron 5 (truncation) | 394 |
| CR40 | amt4-6 | TOC1 insert | Exon 1 (truncation) | 80 |
| CR42 | amt4-7 | TOC1 insert | Exon 1 (truncation) | 71 |
| CR46 | amt4-8 | Tcr1 insert | 5′-untranslated region | 499 |
| CR47 | amt4-9 | 10-bp deletion | Exon 5 (truncation) | 367 |
| CR48 | amt4-10 | Single base change | Exon 3 (splice change) | 116 |
| CR49 | amt4-11 | TOC1 insert | Exon 5 (truncation) | 430 |
| CR50 | amt4-12 | Single base change | Intron 4 (splice change) | 507 |
Estimate of polypeptide length assuming normal splicing of AMT4 transcript or, for CR48 and CR50, use of the new splice sites (see text and Figure 6). Amt4 polypeptide in 4A+ and CC-125 is 499 residues.
Deletions of 2, 10, and 300 bp in CR02, CR47, and CR39, respectively, caused frameshifts and translation of truncated proteins of 121, 367, and 394 amino acids, respectively. The three strains were resistant to methylammonium at high concentrations and had similarly low levels of uptake of [14C]methylammonium, leading to the conclusion that all three proteins were inactive. We have not attempted to assess the causes of the three deletions and cannot rule out transposon-related events. The CR02 lesion is a GC deletion between the TOC1 insert sites in strains CR40 and CR42 (see below). The CR39 deletion includes the site of the third TOC1 insertion in CR49.
Transposon-related events probably accounted for the remaining six amt4 mutations. Three were caused by the retrotransposon TOC1 (class I) (Figure 3D), and three were apparently caused by transposons that move by way of DNA intermediates (class II). The TOC1-related lesions (CR40, CR42, and CR49) yielded amt4 null alleles by gross disruption of transcription and transcript processing. A 270-bp remnant of the class II Gulliver-like transposon (Ferris 1989) in CR07 also yielded an Amt4 null phenotype. Like two of the single base-pair changes (see above), the Gulliver-like fragment in CR07, which is located in the splice site between intron 3 and exon 4, disrupts splicing. CR07 also carries an uncharacterized macrolesion upstream of the AMT4 gene that may be a large insertion or a chromosomal rearrangement. The other two lesions apparently caused by class II transposons are located upstream of the translational start for AMT4 (CR03 and CR46). Neither completely disrupts Amt4 function. Details regarding the lesions caused by transposons, which constituted a large fraction of the total spontaneous mutations that we obtained, will be discussed elsewhere (K.-S. Kim and W. Inwood, unpublished results).
DISCUSSION
Studies of methylammonium-resistant mutants of C. reinhardtii allowed us to address the following five issues, discussed below: (1) the molecular biology of amt4 lesions, the largest class; (2) the nature of lesions in the two smaller classes; (3) the relationship of the three classes to methylammonium-resistant mutants of Chlamydomonas studied previously; (4) Chlamydomonas AMT genes and mutations in relation to those in other organisms; and (5) differences in substrate specificity between the Amt4 and Rh1 proteins of Chlamydomonas.
Molecular biology of amt4 lesions:
By selecting for resistance to methylammonium we obtained three classes of mutations in Chlamydomonas that correspond to at least three genetic loci. Mutations in the largest class affect the AMT4 gene, whereas mutations in the other classes do not appear to affect AMT genes or their regulation (see below). Null alleles in AMT4 reduce the uptake of [14C]methylammonium by 90%, and hence Amt4 appears to be a major transporter for methylammonium and, by inference, ammonium. In other organisms, Amt proteins are required for rapid growth at low concentrations of NH3 [e.g. ≤50 nm for enteric bacteria and ≤5 μm for S. cerevisiae (Soupene et al. 1998, 2001)]. At higher concentrations, unmediated diffusion of NH3 is apparently sufficient. Both the AMT4 and AMT2 genes yield prominent transcripts under N-limiting conditions. However, on the basis of computational analysis (see materials and methods), subcellular localization of the two polypeptides appears to be different and hence the Amt2 protein may not be able to compensate for loss of Amt4 in mutants.
With the initial goal of learning more about the details of gas channel function, we characterized all 12 members of the amt4 class molecularly (Figure 6B and supplementary Results at http://www.genetics.org/supplemental/). Figure 6A indicates the locations of lesions in these strains. At least 9 of the lesions are predicted to cause major changes in the protein; 8 of these (in CR02, CR07, CR39, CR40, CR42, CR47, CR48, and CR49) yielded truncated proteins that may also be rapidly degraded and one (CR04) caused a missense mutation likely to result in structural disruption. Two of the remaining lesions (in CR03 and CR46) were upstream of AMT4 and the third (in CR50), which affected splicing, probably allowed synthesis of a small amount of normal protein. Strains carrying all 3 of the latter lesions appeared to retain residual Amt4 function: they had lower resistance to methylammonium and higher residual methylammonium uptake than did the other amt4 strains. Although many of the lesions to methylammonium resistance were selected at the threshold level of sensitivity (50 μm), none of the 12 was revealing about particular amino acid residues in the Amt4 protein required for transport of NH3 gas.
Nature of lesions in the two smaller classes:
Apart from amt4 lesions, we recovered two other classes of mutations that were represented by two strains each. Mutants of class 1 (CR05 and CR43) showed very low residual uptake of [14C]methylammonium (<2%). Nevertheless, lesions in these strains do not appear to be in a gene that regulates AMT4 transcription or transcription of multiple AMT genes because transcript levels for all AMT genes appeared to be grossly normal in these strains. On the basis of our previous studies of [14C]methylammonium transport in other eukaryotic microbes, in which accumulation required energy-dependent acidification of vacuoles (Soupene et al. 2001), we hypothesize that the lesions in class 1 mutants may decrease acidification of vacuoles and/or other acidic compartments. As would be expected if methylammonium is accumulated into acidic compartments in Chlamydomonas, the bulk of the [14C]methylammonium taken up by wild-type (parental) strains CC-125 and 4A+ was not metabolized (E. Feild and W. Inwood, unpublished results). Although others have reported conversion of some [14C]methylammonium to methylglutamine in Chlamydomonas, they employed much higher concentrations than we did (1 mm rather than 6 μm) and used extended incubation times (2 hr rather than 30 min) (Franco et al. 1984). Despite their low uptake of [14C]methylammonium at 6 μm, strains carrying class 1 lesions are not as resistant to 1 mm methylammonium as amt4 null strains. The amt4 null lesions are epistatic to lesions of class 1.
The class 3 mutants, CR41 and CR45, retain intermediate levels of residual [14C]methylammonium uptake, which would make them good candidates for having a lesion in one of the other AMT genes. Their pleiotropic phenotypes mitigate against this: e.g., they grow poorly on hypoxanthine as an N source, are hypersensitive to the weak base chloroquine on ammonium, have an unusual “disorganized” cellular morphology on ammonium, and fail to mate well in genetic crosses. If, however, the function of an Amt protein other than Amt4 is required for gamete formation (see below), the class 3 mutants may have a lesion affecting this protein. Further experiments will be required to determine this and to explain their pleiotropic phenotypes.
Relationship of amt4 mutants to methylammonium-resistant mutants of Chlamydomonas studied previously:
None of our three classes of methylammonium-resistant mutants appears to correspond to the two classes studied previously by Franco et al. (1987)(1988). Unlike the two lesions they studied, which were linked to one another and to NIT1, none of our three classes of mutations was closely linked to NIT1 (see results). The sequence of AMT4 from strain 2170, which is thought to have a defect in uptake of methylammonium and ammonium, was the same as that of AMT4 from its parental strain CC-1690 and from strain 4A+. A 1.5-kb region amplified from upstream of AMT4 also showed no evidence of a large deletion or insertion. Moreover, strain 2170 showed no defect in expression of AMT4 or the other three AMT genes. For a comparison of other properties of strain 2170 to those of our three classes of mutants and to its parental strain CC-1690 (see supplementary Results at http://www.genetics.org/supplemental/).
Chlamydomonas AMT genes and mutations in relation to those in other organisms:
The amt4 mutants of C. reinhardtii can be compared with similar mutants of vascular plants and with mutants of other microorganisms. Just as Amt4 is probably the major transporter of ammonium in Chlamydomonas, a single Amt protein of Arabidopsis appears to be a major transporter in that organism (Kaiser et al. 2002). Loss of function of Amt1;1 resulted in a large defect in uptake of [13N]ammonium upon starvation for N, although Arabidopsis has at least five AMT genes. Loss of function of Amt1;1 also led to increased transcription of other AMT genes.
Like Chlamydomonas, a number of other microbes also have multiple Amt proteins. Those of yeast (called Mep) appear to have different affinities for methylammonium and ammonium (Marini et al. 1997) and one (Mep2) is required for pseudohyphal development of diploids in response to N limitation (Lorenz and Heitman 1998). Likewise, function of one of the three AMT genes of the slime mold Dictyostelium discoideum is required for the culmination of development (Follstaedt et al. 2003). Ammonia gas has long been known to be a major regulator of development in this organism at stages postaggregation (Schindler and Sussman 1977; Bonner 1993). One of the three Amt proteins predicted in the recently completed genome of the marine planctomycete Pirellula (Glöckner et al. 2003) appears to be a hybrid protein of 900 amino acids. The amino terminal half is a typical Amt protein, whereas the carboxy terminal half resembles the well-studied histidine autokinase NtrB. Fusion of Amt to a signal transduction protein suggests that the hybrid protein has a sensory function. It remains to be seen whether any of the Amt proteins of Chlamydomonas will play a role in gamete formation, which occurs in response to N starvation, or in other aspects of sensing and development.
Differences in substrate specificity between the Amt4 and Rh1 proteins:
A principal reason for initiating studies of methylammonium resistance and AMT genes in C. reinhardtii was that this green alga is one of the few microbes to have RH genes in addition to AMT genes. Hence, it is an organism of choice for discriminating between the functions of their protein products. In general, experiments indicating that human Rh proteins transport methylammonium have involved cloning RH genes into microorganisms or cells that do not have them naturally. Our first evidence that the physiological substrates for Amt and Rh proteins differ came from finding that control of expression of the AMT genes of C. reinhardtii differs profoundly from that of its RH1 gene (Soupene et al. 2004). Whereas transcription of three of its four AMT genes is N regulated, transcription of its RH1 gene is highly regulated by availability of CO2. Our second line of evidence for different substrates is that amt4 mutants are resistant to methylammonium and greatly defective in its uptake, whereas RNAi lines lacking expression of RH1 remain sensitive to toxic effects of methylammonium and show no defect in uptake of [14C]methylammonium (Soupene et al. 2004). Rather, they have growth defects specifically at high concentrations of CO2. Results in Chlamydomonas indicate that the substrate for Rh1 is likely to be CO2, whereas that for Amt4 is methylammonium [probably CH3NH2, as we have found in other organisms (Soupene et al. 1998, 2001)] and by inference ammonium (probably NH3).
Recent determination of the X-ray crystal structure of the AmtB protein of E. coli gave a physical face to our physiological characterization of its function and the function of other Amt proteins (Khademi et al. 2004). The extraordinary resolution achieved—1.35 Å—allowed discrimination between charged and uncharged ligands, revealing that it was indeed the gases NH3 and CH3NH2 that were present in the pores or selectivity filters of the channels. (Each monomer of the trimer contains a channel.) The beautiful structures of E. coli AmtB widen opportunities to study the mechanism of protein-mediated gas transport and are the first step in being able to compare directly channels for NH3 and CO2.
Conclusions:
The availability of amt4 mutant lines of C. reinhardtii has allowed discrimination between the substrates for Amt and Rh proteins in one of the few microorganisms to have both naturally. These mutants should facilitate analysis of the role of Amt4 and other Amt proteins in acquisition of ammonium and in sensing and developmental processes controlled by its availability. The amt4 mutants are well behaved in genetic crosses, indicating that Amt4 is not required for gamete formation, and its absence does not appear to have major effects on transcription of other AMT genes. Molecular characterization of the 12 spontaneous amt4 lesions described in this article and a number of others indicated that many were induced by transposition of both class I and class II elements, several of which appear to be novel (K.-S. Kim and W. Inwood, unpublished results).
Acknowledgments
We are grateful to an anonymous reviewer for many helpful suggestions. This work was supported by National Institutes of Health grant GM38361 and a grant from the Torrey Mesa Research Institute, Syngenta Research and Technology, La Jolla, California, to S.K.
References
- Arst, H. N., Jr., and D. J. Cove, 1969. Methylammonium resistance in Aspergillus nidulans. J. Bacteriol. 98: 1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avent, N. D., and M. E. Reid, 2000. The Rh blood group system: a review. Blood 95: 375–387. [PubMed] [Google Scholar]
- Bonner, J. T., 1993. Proteolysis and orientation in Dictyostelium slugs. J. Gen. Microbiol. 139: 2319–2322. [DOI] [PubMed] [Google Scholar]
- Cartron, J. P., 1999. RH blood group system and molecular basis of Rh-deficiency. Baillieres Clin. Haematol. 12: 655–689. [DOI] [PubMed] [Google Scholar]
- Day, A., and J.-D. Rochaix, 1991. A transposon with an unusual LTR arrangement from Chlamydomonas reinhardtii contains an internal tandem array of 76 bp repeats. Nucleic Acids Res. 19: 1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, E., and M. Grenson, 1979. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol. Gen. Genet. 175: 67–76. [DOI] [PubMed] [Google Scholar]
- Ferris, P. J., 1989. Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher plants. Genetics 122: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follstaedt, S. C., J. H. Kirsten and C. K. Singleton, 2003. Temporal and spatial expression of ammonium transporter genes during growth and development of Dictyostelium discoideum. Differentiation 71: 557–566. [DOI] [PubMed] [Google Scholar]
- Franco, A. R., J. Cardenas and E. Fernandez, 1984. Ammonium (methylammonium) is the co-repressor of nitrate reductase in Chlamydomonas reinhardtii. FEBS Lett. 176: 453–456. [Google Scholar]
- Franco, A. R., J. Cardenas and E. Fernandez, 1987. A mutant of Chlamydomonas reinhardtii altered in the transport of ammonium and methylammonium. Mol. Gen. Genet. 206: 414–418. [Google Scholar]
- Franco, A. R., J. Cardenas and E. Fernandez, 1988. Two different carriers transport both ammonium and methylammonium in Chlamydomonas reinhardtii. J. Biol. Chem. 263: 14039–14043. [PubMed] [Google Scholar]
- Gazzarrini, S., L. Lejay, A. Gojon, O. Ninnemann, W. B. Frommer et al., 1999. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot et al., 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100: 8298–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoff, E. D. V., U. Treier and C. F. Beck, 1989. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9: 3911–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez et al., 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E., 1989 The Chlamydomonas Sourcebook. Academic Press, New York.
- Hemker, M. B., G. Cheroutre, R. van Zwieten, P. A. Maaskant-van Wijk, D. Roos et al., 2003. The Rh complex exports ammonium from human red blood cells. Br. J. Haematol. 122: 333–340. [DOI] [PubMed] [Google Scholar]
- Kaiser, B. N., S. R. Rawat, M. Y. Siddiqi, J. Masle and A. D. Glass, 2002. Functional analysis of an Arabidopsis T-DNA “knockout” of the high-affinity NH+4 transporter AtAMT1;1. Plant Physiol. 130: 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi, S., J. O'Connell, III, J. Remis, Y. Robles-Colmenares, L. J. W. Miercke et al., 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305: 1587–1594. [DOI] [PubMed] [Google Scholar]
- Lefebvre, P. A., and C. D. Silflow, 1999. Chlamydomonas: the cell and its genomes. Genetics 151: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, R. P., and W. T. Ebersold, 1960. The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14: 197–216. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., and J. Heitman, 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, A. M., S. Soussi-Boudekou, S. Vissers and B. André, 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 4282–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, A. M., G. Matassi, V. Raynal, B. André, J. P. Cartron et al., 2000. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat. Genet. 26: 341–344. [DOI] [PubMed] [Google Scholar]
- Merchan, F., H. van den Ende, E. Fernandez and C. F. Beck, 2001. Low-expression genes induced by nitrogen starvation and subsequent sexual differentiation in Chlamydomonas reinhardtii, isolated by the differential display technique. Planta 213: 309–317. [DOI] [PubMed] [Google Scholar]
- Monahan, B. J., J. A. Fraser, M. J. Hynes and M. A. Davis, 2002. a Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 1: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan, B. J., S. E. Unkles, I. T. Tsing, J. R. Kinghorn, M. J. Hynes et al., 2002. b Mutation and functional analysis of the Aspergillus nidulans ammonium permease MeaA and evidence for interaction with itself and MepA. Fungal Genet. Biol. 36: 35–46. [DOI] [PubMed] [Google Scholar]
- Nakhoul, N. L., and L. L. Hamm, 2004. Non-erythroid Rh glycoproteins: a putative new family of mammalian ammonium transporters. Pflugers Arch. 447: 807–812. [DOI] [PubMed] [Google Scholar]
- Schindler, J., and M. Sussman, 1977. Ammonia determines the choice of morphogenetic pathways in Dictyostelium discoideum. J. Mol. Biol. 116: 161–169. [DOI] [PubMed] [Google Scholar]
- Soupene, E., L. He, D. Yan and S. Kustu, 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. USA 95: 7030–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., R. M. Ramirez and S. Kustu, 2001. Evidence that fungal MEP proteins mediate diffusion of the uncharged species NH3 across the cytoplasmic membrane. Mol. Cell. Biol. 21: 5733–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., H. Lee and S. Kustu, 2002. a Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc. Natl. Acad. Sci. USA 99: 3926–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., T. Chu, R. W. Corbin, D. F. Hunt and S. Kustu, 2002. b Gas channels for NH3: proteins from hyperthermophiles complement an Escherichia coli mutant. J. Bacteriol. 184: 3396–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., N. King, E. Feild, P. Liu, K. K. Niyogi et al., 2002. c Rhesus expression in a green alga is regulated by CO2. Proc. Natl. Acad. Sci. USA 99: 7769–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., W. Inwood and S. Kustu, 2004. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. USA 101: 7787–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier, U., S. Fuchs, M. Weber, W. W. Wakarchuk and C. F. Beck, 1989. Gametic differentiation in Chlamydomonas reinhardtii: light dependence and gene expression patterns. Arch. Microbiol. 152: 572–577. [Google Scholar]
- von Wirén, N., and M. Merrick, 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Top. Curr. Genet. 9: 95–120. [Google Scholar]
- von Wirén, N., S. Gazzarrini, A. Gojon and W. B. Frommer, 2000. The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 3: 254–261. [PubMed] [Google Scholar]
- Westhoff, C. M., M. Ferreri-Jacobia, D. O. Mak and J. K. Foskett, 2002. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J. Biol. Chem. 277: 12499–12502. [DOI] [PubMed] [Google Scholar]
- Wintermans, J. F., and A. de Mots, 1965. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta 109: 448–453. [DOI] [PubMed] [Google Scholar]