Abstract
The oncoproteins P3k (homolog of the catalytic subunit of class IA phosphoinositide 3-kinase) and Akt (protein kinase B) induce oncogenic transformation of chicken embryo fibroblasts. The transformed cells show constitutive phosphorylation of the positive regulator of translation p70S6 kinase (S6K) and of the eukaryotic initiation factor 4E-BP1 binding protein (4E-BP1), a negative regulator of translation. Phosphorylation activates S6K and inactivates 4E-BP1. A mutant of Akt that retains kinase activity but does not induce phosphorylation of S6K or of 4E-BP1 fails to transform chicken embryo fibroblasts, suggesting a correlation between the oncogenicity of Akt and phosphorylation of S6K and 4E-BP1. The macrolide antibiotic rapamycin effectively blocks oncogenic transformation induced by either P3k or Akt but does not reduce the transforming activity of 11 other oncoproteins. Rapamycin inhibits the kinase mTOR, an important regulator of translation, and this inhibition requires binding of the antibiotic to the immunophilin FKBP12. Displacement of rapamycin from FKBP12 relieves the inhibition of mTOR and also restores P3k-induced transformation. These data are in accord with the hypothesis that transformation by P3k or Akt involves intervention in translational controls.
The two oncoproteins P3k and Akt were originally isolated from tumorigenic retroviruses (1, 2). P3k is the homolog of the catalytic subunit of phosphoinositide (PI) 3-kinase, a lipid kinase that phosphorylates phosphatidylinositol at the D3 position and affects multiple cellular functions, many related to growth and differentiation (3–6). Akt (also called PKB) is a serine–threonine protein kinase; it is a downstream target of PI 3-kinase (7–11). Akt binds to the products of PI 3-kinase, phosphatidylinositol 3,4-biphosphate and phosphatidylinositol 3,4,5-triphosphate, with its pleckstrin homology domain. It then becomes activated by phosphorylation at threonine 308 and serine 473 through the action of the 3-phosphoinositide-dependent kinases PDK1 and PDK2 (12, 13).
Akt affects numerous downstream targets either directly or indirectly (7–11). These can be broadly classified into two groups: (i) survival and death factors and (ii) proteins controlling translation. Among the first group are the pro-apoptotic proteins Bad (14, 15) and caspase 9 (16) and the growth-inhibitory proteins glycogen synthase kinase-3 beta (17) and the forkhead transcription factors FKHR, FKHR-L1, and AFX, all of which are down-regulated by Akt (18–20). Also in this category is the kinase IKK alpha, a positive regulator of NF-κB, which is up-regulated by Akt (21–23). The second category consists of the kinase mTOR (mammalian target of rapamycin, other acronyms: FRAP, RAFT) and its downstream targets p70 S6 kinase (S6K) and the eukaryotic initiation factor 4E binding protein 1 (4E-BP1, also called PHAS-1) (24–27). S6K is activated by mTOR-dependent phosphorylation and controls the translation of 5′TOP mRNAs, so named for the presence of an oligopyrimidine tract at their 5′ termini (28). These messages code for ribosomal proteins and elongation factors; the oligopyrimidine tract mediates coordinate translational regulation in a growth-dependent fashion. 4E-BP is inactivated by mTOR-dependent phosphorylation (29–32). Underphosphorylated 4E-BP binds to the eukaryotic initiation factor 4E (eIF4E, the cap-binding protein) and prevents it from assembling the translation initiation complex at the cap of the mRNA. Phosphorylation abolishes this inhibitory function.
Here, we provide evidence that oncogenic transformation by P3k and Akt is dependent on targets that control translation: transforming activity is correlated with phosphorylation and activation of S6K and is eliminated by the mTOR inhibitor rapamycin. The suggested involvement of translational control in oncogenic transformation is specific for P3k and Akt; transformation induced by numerous other oncoproteins is not inhibited by rapamycin.
Materials and Methods
Cell Culture and Virus Infection.
Primary cultures of chicken embryo fibroblasts (CEF) were prepared from White Leghorn embryos obtained from SPAFAS (Preston, CT). Oncogenicity was assayed by the induction of transformed cell foci according to published techniques (33). The effect of rapamycin (Calbiochem) on focus formation was tested by incorporating the drug at the indicated concentration in the nutrient agar overlay of the infected cells. The cells were fed every other day. Controls received DMSO vehicle instead of rapamycin. The cell cultures were stained with crystal violet on day 20 after infection, and foci of transformed cells were counted. The following previously described viruses and oncogenes were used: PR-A (Prague strain of Rous sarcoma virus) (34), v-src; MH2, v-myc/v-mil; Y73, v-yes (35); PRC (Poultry Research Centre) II, v-fps (36); AEV (Avian erythroblastosis virus, ES4 strain), v-erb A/erb B (37); ASV 1, v-crk (38); S13, v-sea (39); ASV 17, v-jun (40); pRV9-mafQ5H, mutated v-maf (41, 42); NK24, v-fos (43).
The oncogenes v-myc (44), v-mos (45), v-abl, v-ras (46), myr-p3k (13), and myr-akt (12) were expressed with the avian retroviral vector RCAS.
Serum Starvation and Platelet-Derived Growth Factor (PDGF) Stimulation.
For serum starvation, cells were cultured in Ham's F-10 medium with 0.5% FCS and 0.1% chicken serum. After 40 h, the medium was replaced with plain F-10 medium, and the culture was further incubated for 2 h. The cells were then stimulated with 50 ng/ml PDGF (Life Technologies, Grand Island, NY). For rapamycin treatment, rapamycin (10 ng/ml) was added to the culture 2 h before the addition of PDGF.
Western Blots.
Cells were lysed in Nonidet P-40 lysis buffer (20 mM Tris-HCl, pH 7.5/150 mM NaCl/10% glycerol/1% Nonidet P-40/10 mM NaF/1 mM sodium pyrophosphate/1 mM sodium orthovanadate/1 mM microcystin) containing protease inhibitors (Cømplete, Boehringer Mannheim). Lysates containing 60 μg of protein were separated by SDS/PAGE and transferred to Immobilon P membranes (Millipore). The membranes were blocked with 5% nonfat dry milk/Tris-buffered saline/0.05% Tween-20 for 1 h at room temperature. They were then probed with anti-p70s6k (C-19, Santa Cruz Biotechnology) to detect S6K, anti-phospho-p70s6k (Thr-389, Cell Signaling Technology) to detect phosphorylated S6K, anti-4E-BP1 (Santa Cruz Biotechnology) to detect 4E-BP1, anti-phospho-4E-BP1 (Ser-65, Cell Signaling Technology) to detect phosphorylated 4E-BP1, and anti-phospho-Erk (E10, Cell Signaling Technology) to detect phosphorylated extracellular signal-regulated kinase, Erk.
Results
Constitutive Phosphorylation of S6K and 4E-BP1 in CEF Transformed by P3k and Akt.
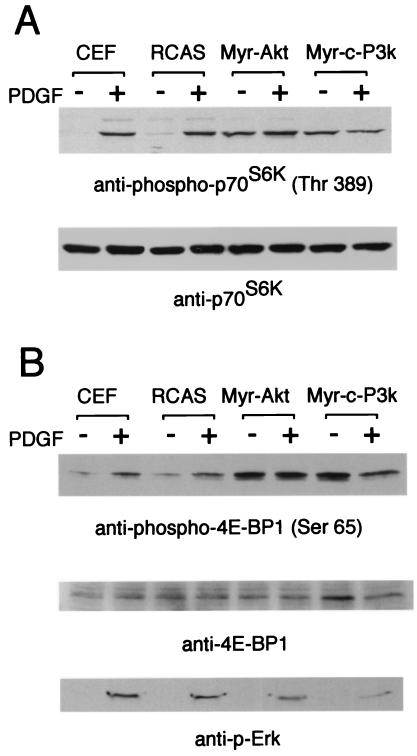
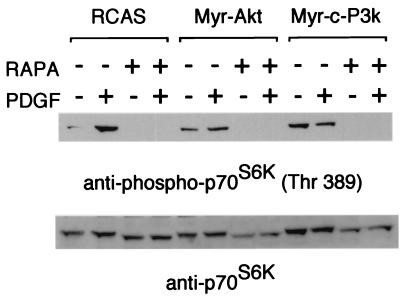
S6K and 4E-BP1 are phosphorylated in a rapamycin-sensitive manner, suggesting a dependence of this process on the mTOR kinase (47–52). Phosphorylation of S6K on the rapamycin-sensitive threonine 389 correlates well with S6K activation in vivo (53). We examined the phosphorylation of S6K in Western blots using a phospho-S6K-specific antibody (Fig. 1A). In serum-starved CEF, uninfected or infected with the RCAS vector alone, phosphorylation of S6K was low, but PDGF induced strong phosphorylation of threonine 389 within 15 min. In contrast, CEF transformed by P3k or Akt showed strong phosphorylation of threonine 389, even under conditions of serum deprivation, suggesting that S6K is constitutively activated in the transformed cells. Control Western blots with phosphorylation-independent antibody against S6K showed that the expression levels of S6K were not altered in P3k or Akt transformation or by stimulation with PDGF. We also investigated the phosphorylation of 4E-BP1 in Western blots using a phospho-specific antibody that recognizes 4E-BP1 phosphorylated on serine 65 (Fig. 1B). This site is phosphorylated in response to growth factors in a rapamycin-sensitive manner (29). Again, in uninfected and RCAS-infected CEF, 4E-BP1 phosphorylation was low under conditions of serum starvation but was strongly induced by PDGF within 15 min. Like S6K, 4E-BP1 was constitutively phosphorylated in P3k- or Akt-transformed CEF, even when the cells were serum deprived. For comparison, phosphorylation of Erk was low in serum-starved P3k- or Akt-transformed cells and was efficiently induced to the level seen in serum-starved CEF stimulated with PDGF.
Figure 1.
Constitutive phosphorylation of S6K and 4E-BP1 in CEF transformed with P3k and Akt. CEF and CEF infected with RCAS, RCAS-Myr-Akt, and RCAS-Myr-c-P3k were serum starved for 40 h and then stimulated with PDGF for 15 min. The cells were lysed, and the lysates were resolved in a 10% (A) or a 15% (B) SDS-polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane. The blot was probed with anti-phospho-S6K (threonine 389), anti-S6K, anti-phospho-4E-BP1 (serine 65), anti-4E-BP1, or anti-phospho-Erk antibody.
The Oncogenic Activity of Akt Is Correlated with the Ability to Induce Phosphorylation of S6K and of 4E-BP1.
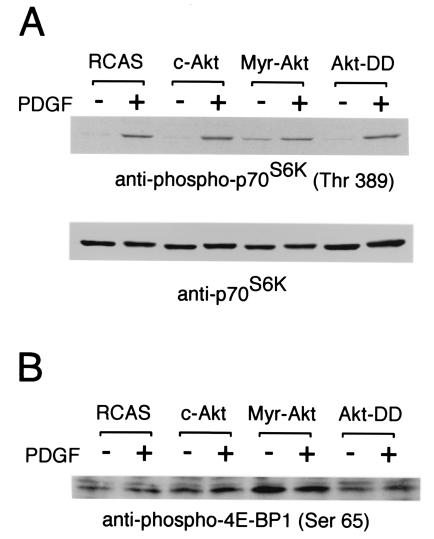
The oncogenicity of Akt requires both membrane localization and specific enzymatic activity (12). Myristylated Akt is highly oncogenic, whereas the construct with the regulatory phosphorylation sites threonine 308 and serine 473 mutated to aspartic acid induces cellular transformation only with a very low efficiency, despite the fact that this mutant retains high kinase activity (12). Western blots of CEF infected with the Akt-DD mutant showed no elevated phosphorylation of S6K, in contrast to Myr-Akt-transformed CEF (Fig. 2A). Similarly, phosphorylation of 4E-BP1 was constitutively increased in Myr–Akt-infected cells but not in Akt–DD-infected cells (Fig. 2B). These results suggest a correlation between oncogenicity and the phosphorylation of S6K and of 4E-BP1. [Thomas and collaborators recently reported that Akt-DD could phosphorylate 4E-BP1 but not S6K (54). The discrepancy with our observations may be due to the mode of Akt-DD expression—transient transfection versus stable transfection—or may reflect the use of different cell types. It is also possible that 4E-BP1 is partially phosphorylated by Akt-DD but not fully inactivated.]
Figure 2.
Correlation between the ability to induce S6k activation and 4E-BP1 inactivation with the ability to induce oncogenic transformation. Cell lysates were prepared as described in the legend to Fig. 1. The lysates were resolved in a 10% (A) or a 15% (B) SDS-polyacrylamide gel. The blot was probed with anti-phospho-S6K (threonine 389), anti-S6K, or anti-phospho-4E-BP1 (serine 65) antibody.
Rapamycin Inhibits Formation of Transformed Cell Foci by P3K in an FKBP12-Dependent Manner.
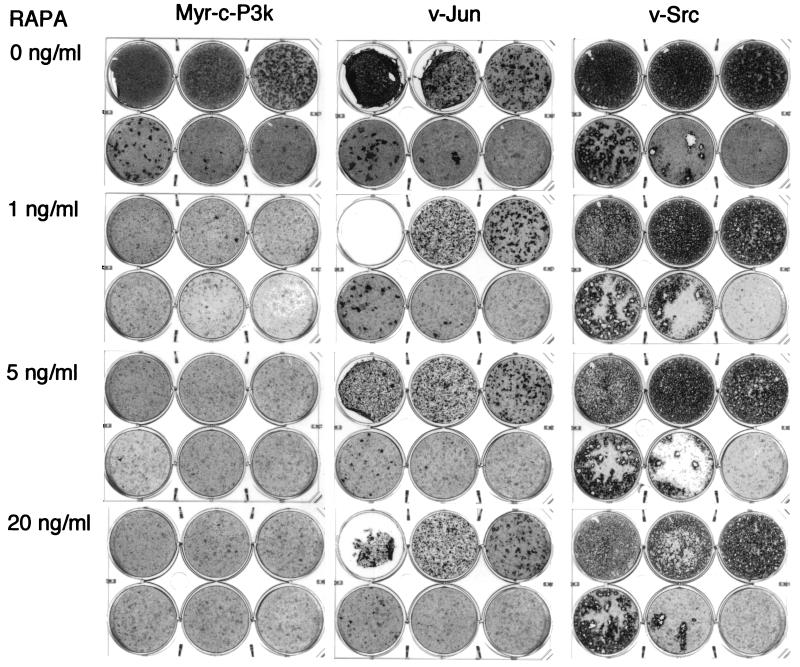
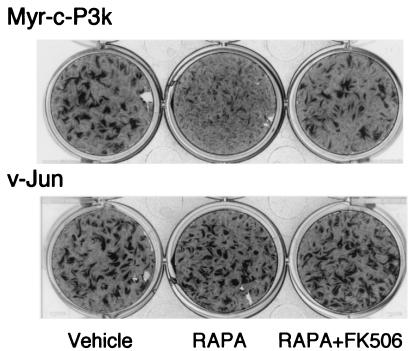
The phosphorylation of S6K is known to be rapamycin-sensitive (55, 56). Since phosphorylation is correlated with oncogenic transformation by P3k and Akt, the target of rapamycin, mTOR, might be involved in cellular transformation induced by P3k or Akt. This possibility was explored by determining the effect of rapamycin on the transforming efficiency of P3k. As shown in Fig. 3, 1 ng/ml rapamycin almost completely eliminated the formation of transformed cell foci by P3k. This inhibition was specific because even 20 ng/ml rapamycin did not significantly influence focus formation by the oncoproteins Src or Jun, although it induced a slight delay in transformation, possibly due to partial growth inhibition caused by the drug in CEF. Rapamycin inhibits mTOR by binding to the immunophilin FKBP12 (54). The interference with P3k transformation probably also involves interaction with FKBP12 because a 100-fold molar excess of FK506, a pharmacological reagent that competes with rapamycin for binding to FKBP12, rescued P3k transformation (Fig. 4).
Figure 3.
Rapamycin inhibits focus formation by P3k but not Jun or Src. CEF were infected with viruses containing the indicated oncoproteins. Each plate was infected with 100 μl of the virus stocks diluted to 10−1 (top left well), 10−2 (top center well), 10−3 (top right well), 10−4 (bottom left well), 10−5 (bottom center well), or with no viruses (bottom right well). The cells were overlaid with nutrient agar containing the indicated concentrations of rapamycin (RAPA) for 17 days and then fixed and stained with crystal violet.
Figure 4.
Rapamycin inhibition of the P3k-induced transformation of CEF is FKBP 12 dependent. CEF were infected with viruses and then overlaid with nutrient agar containing 1 ng/ml rapamycin (Center), 1 ng/ml rapamycin and 100 molar excess of FK506 (Right), or vehicle (DMSO, Left) only.
The Inhibitory Effect of Rapamycin on Oncogenic Transformation Is Specific for P3k and Akt.
The acute sensitivity of P3k-induced oncogenic transformation to inhibition by rapamycin, compared with the refractoriness of Jun and Src, prompted us to test the effect of rapamycin on focus formation caused by other oncoproteins. Akt was the only other oncoprotein that showed sensitivity to rapamycin, reflecting the fact that the Akt protein is part of the PI 3-kinase signaling pathway and suggesting that rapamycin intervenes downstream of Akt. Besides the above-mentioned Src and Jun, the oncoproteins Abl, Crk, ErbB, Fos, Fps, Mos, Qin, Sea, and Yes were not significantly affected in their transforming activity by rapamycin. Surprisingly, the growth-promoting potential of two oncoproteins, Myc and Ras, was strongly enhanced by rapamycin, leading to an increase in the number and size of the transformed cell foci (Table 1).
Table 1.
Effects of rapamycin on transforming activities of various oncogenes in CEF
| Oncoproteins | Rapamycin (ng/ml)
|
||
|---|---|---|---|
| 0 | 1 | 5 | |
| Group 1: inhibited | |||
| Myr-c-P3k | 100 | 0 | 0 |
| Myr-Akt | 100 | 0 | 0 |
| Group 2: unaffected | |||
| v-Src | 100 | 100 | 100 |
| v-Yes | 100 | 75 | 75 |
| v-Sea | 100 | 58 | 83 |
| v-Abl | 100 | 100 | 53 |
| v-Fps | 100 | 102 | 90 |
| v-ErbA/v-ErbB | 100 | 140 | 85 |
| v-Crk | 100 | 78 | 63 |
| v-Mos | 100 | 117 | 133 |
| v-Jun | 100 | 92 | 78 |
| v-Fos | 100 | 117 | 50 |
| Group 3: enhanced | |||
| v-Myc | 100 | 1040 | 480 |
| v-Myc/v-Mil | 100 | 1600 | 1800 |
| v-H-Ras | 100 | 1400 | 800 |
| v-Maf | 100 | 240 | Nd |
Expressed as relative efficiency of focus formation with control cultures set at 100.
Rapamycin Inhibits the Constitutive Phosphorylation of S6K in CEF Transformed by P3k or Akt.
The suggested link between the activating phosphorylation of S6K and oncogenic transformation was strengthened by examining S6K phosphorylation in the presence and absence of rapamycin (Fig. 5). Rapamycin led to an almost complete disappearance of the phosphorylated form of S6K, not only from normal CEF but also from CEF transformed by P3k or Akt. Although transformation by P3k or Akt induces a constitutive phosphorylation of S6K, this activation of S6K is still rapamycin-sensitive as is transformation itself.
Figure 5.
Rapamycin inhibits phosphorylation of S6K. CEF were serum starved for 40 h and treated or not treated with rapamycin (RAPA, 10 ng/ml) for 2 h and then stimulated with PDGF (50 ng/ml) for 15 min. The cell lysates were prepared and resolved in a 10% SDS-polyacrylamide gel. The blot was probed with anti-phospho-S6K or with anti-S6K antibody.
Discussion
The kinase mTOR is at the center of the experiments reported here. mTOR is inhibited by the macrolide antibiotic rapamycin; rapamycin interacts with FKBP12, and this complex binds to mTOR (57–60). FK506 competes with rapamycin in binding to FKBP12 and thus counteracts the inhibition of mTOR (61, 62). Rapamycin is effective at very low concentrations in eliminating mTOR kinase activity (57–60). Since the inhibition of mTOR also abolishes the oncogenic effects of P3k and of Akt, mTOR appears to be an obligatory mediator of the oncogenic signal issued by P3k or Akt. mTOR is phosphorylated, probably directly, by Akt at two carboxyl-terminal sites (63). These two sites are located in a negative regulatory domain of mTOR; phosphorylation may relieve the negative regulation and activate the mTOR kinase (63). Although the complete target spectrum of mTOR remains to be determined, it is clear that mTOR functions as an important regulator of translation. mTOR mediates the phosphorylation of S6K and of 4E-BP1 (47–52, 64). Active S6K is essential for the translation of 5′-TOP mRNAs, which include messages of ribosomal proteins (28). Inactivation of 4E-BP1 facilitates the translation of mRNAs with complex 5′ secondary structures; many of these code for growth-related genes (26, 65).
Our observations suggest that some mTOR-dependent activity, possibly connected to S6K and 4E-BP (and hence linked to eIF4E), is essential for oncogenic transformation induced by P3k or Akt. Transformation by 11 diverse oncoproteins was refractory to inhibition by rapamycin, and transformation by two, Ras and Myc, was enhanced. Some of the rapamycin-refractory oncoproteins, for instance the Src kinase and the adaptor protein Crk, are known to activate PI 3-kinase–Akt signaling (66). Src also induces phosphorylation of S6K (M.A. and P.V., unpublished observations). The resistance to rapamycin indicates that the branch of the PI 3-kinase–Akt signal that is transduced by mTOR is not irreplaceable or essential in the oncogenicity of Src or Crk. In the case of Src, oncogenic signals appear to proceed through two alternative routes, involving mTOR on the one hand and the Ras–mitogen-activated protein kinase pathway on the other. Both must be inhibited to interfere with cellular transformation (67). These comparisons of rapamycin sensitivity and resistance single out P3k and Akt as following a unique mechanism of transformation, not commonly shared by other oncoproteins. A recent publication reported that transformation of an immortalized rat kidney epithelial cell line by the zinc finger transcription factor GLI is also rapamycin sensitive, and this sensitivity is correlated with an inhibition of protein synthesis. Transformation of these same cells by Ras or Myc is not inhibited by the antibiotic (68). The available data support the general statement that translational controls are an essential component of oncogenic transformation by P3k, Akt, or GLI.
Previous studies have suggested an oncogenic potential for several regulators of protein synthesis (for review, see ref. 69). eIF4E was shown to transform NIH 3T3 cells (70, 71), and mutated S6K can affect the morphology of Rat1 cells (72). A cautious interpretation of these data suggests that a gain of function in S6K or of eIF4E may be necessary for certain mechanisms of transformation but is possibly not sufficient. Besides eIF4E, there are two other eukaryotic initiation factors that have shown oncogenic potential, eIF-2A and eIF-4G (73, 74). 4E-BP1 is also hyperphosphorylated and thus inactivated in Src-transformed hamster fibroblasts (75).
There is evidence that PI 3-kinase and Akt act as oncogenic determinants in several human cancers. PIK3CA, the catalytic subunit of class IA PI 3-kinase, is overexpressed in ovarian cancers (76). Loss of function in PTEN, a phosphatase that counteracts PI 3-kinase, is a frequent genetic change in numerous tumors including carcinoma of the prostate and glioblastoma (77–79). mTOR is constitutively phosphorylated in prostate cancer cell lines carrying an inactivating mutation in PTEN or overexpressing Akt3 (63). Akt genes are amplified or overexpressed in several cancers including gastric, ovarian, breast, pancreatic, and prostate cancer (80–85); eIF4E is overexpressed in lymphomas, cancers of the head and neck and in colon carcinomas as well as in cells containing elevated levels of the oncoprotein Myc (86). An importance of S6K in human cancer is suggested by the frequent up-regulation of mRNAs for ribosomal proteins in expression profiles from diverse tumors (87). Several cell lines derived from human tumors, including rhabdomyosarcomas and small cell lung carcinomas, are sensitive to rapamycin (88, 89).
The special mechanisms of transformation suggested here for P3k and Akt may therefore be relevant to human cancer. Akt is an obvious target for the design of novel chemotherapeutic agents, but it might not be desirable to inhibit all of the multiple functions of Akt. mTOR and its subordinate activities may provide opportunities for a chemotherapeutic strategy that is more narrowly aimed and more selective.
Acknowledgments
We thank K. C. Nicolaou, Daniel R Salomon, and Klaus Hahn for valuable comments on the manuscript. Osvaldo Batista and Jeffrey Ludwig provided competent and dedicated technical assistance. This work was supported by National Institutes of Health Research Grants CA42564, CA78230, and CA79616. This is manuscript number 13607 of the Department of Molecular and Experimental Medicine, The Scripps Research Institute.
Abbreviations
- CEF
chicken embryo fibroblasts
- PI 3-kinase
phosphoinositide 3-kinase
- PDGF
platelet-derived growth factor
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011528498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011528498
References
- 1.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 2.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 3.Shepherd P R, Withers D J, Siddle K. Biochem J. 1998;333:471–490. doi: 10.1042/bj3330471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alessi D R, Downes C P. Biochim Biophys Acta. 1998;1436:151–164. doi: 10.1016/s0005-2760(98)00133-7. [DOI] [PubMed] [Google Scholar]
- 5.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 6.Wymann M P, Pirola L. Biochim Biophys Acta. 1998;1436:127–150. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 8.Coffer P J, Jin J, Woodgett J R. Biochem J. 1998;335:1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alessi D R, Cohen P. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 10.Chan T O, Rittenhouse S E, Tsichlis P N. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 11.Datta S R, Brunet A, Greenberg M E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 12.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt P K. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki M, Schetter C, Himly M, Batista O, Chang H W, Vogt P K. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 14.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 15.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 16.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 17.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 18.Biggs W H, III, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Nature (London) 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 20.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 21.Romashkova J A, Makarov S S. Nature (London) 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 22.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. Nature (London) 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 23.Kane L P, Shapiro V S, Stokoe D, Weiss A. Curr Biol. 1999;9:601–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 24.Gray N K, Wickens M. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 25.Dufner A, Thomas G. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- 26.Sonenberg N, Gingras A C. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 27.Raught B, Gingras A C. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- 28.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingras A C, Gygi S P, Raught B, Polakiewicz R D, Abraham R T, Hoekstra M F, Aebersold R, Sonenberg N. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gingras A C, Kennedy S G, O'Leary M A, Sonenberg N, Hay N. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 33.Bos T J, Monteclaro F S, Mitsunobu F, Ball A R, Jr, Chang C H, Nishimura T, Vogt P K. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 34.Duff R G, Vogt P K. Virology. 1969;39:18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
- 35.Kitamura N, Kitamura A, Toyoshima K, Hirayama Y, Yoshida M. Nature (London) 1982;297:205–208. doi: 10.1038/297205a0. [DOI] [PubMed] [Google Scholar]
- 36.Neil J C, Delamarter J F, Vogt P K. Proc Natl Acad Sci USA. 1981;78:1906–1910. doi: 10.1073/pnas.78.3.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai M M, Neil J C, Vogt P K. Virology. 1980;100:475–483. doi: 10.1016/0042-6822(80)90537-1. [DOI] [PubMed] [Google Scholar]
- 38.Tsuchie H, Chang C H, Yoshida M, Vogt P K. Oncogene. 1989;4:1281–1284. [PubMed] [Google Scholar]
- 39.Hayman M J, Kitchener G, Vogt P K, Beug H. Proc Natl Acad Sci USA. 1985;82:8237–8241. doi: 10.1073/pnas.82.23.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maki Y, Bos T J, Davis C, Starbuck M, Vogt P K. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kataoka K, Nishizawa M, Kawai S. J Virol. 1993;67:2133–2141. doi: 10.1128/jvi.67.4.2133-2141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu S, Bottoli I, Goller M, Vogt P K. Proc Natl Acad Sci USA. 1999;96:5716–5721. doi: 10.1073/pnas.96.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishizawa M, Goto N, Kawai S. J Virol. 1987;61:3733–3740. doi: 10.1128/jvi.61.12.3733-3740.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petropoulos C J, Givol I, Hughes S H. Oncogene. 1996;12:2611–2621. [PubMed] [Google Scholar]
- 45.Schmidt M, Oskarsson M K, Dunn J K, Blair D G, Hughes S, Propst F, Vande Woude G F. Mol Cell Biol. 1988;8:923–929. doi: 10.1128/mcb.8.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antczak M, Kung H J. J Virol. 1990;64:1451–1458. doi: 10.1128/jvi.64.4.1451-1458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung J, Kuo C J, Crabtree G R, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 48.Kuo C J, Chung J, Fiorentino D F, Flanagan W M, Blenis J, Crabtree G R. Nature (London) 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 49.Terada N, Lucas J J, Szepesi A, Franklin R A, Takase K, Gelfand E W. Biochem Biophys Res Commun. 1992;186:1315–1321. doi: 10.1016/s0006-291x(05)81549-9. [DOI] [PubMed] [Google Scholar]
- 50.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 51.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 52.Graves L M, Bornfeldt K E, Argast G M, Krebs E G, Kong X, Lin T A, Lawrence J C., Jr Proc Natl Acad Sci USA. 1995;92:7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weng Q P, Kozlowski M, Belham C, Zhang A, Comb M J, Avruch J. J Biol Chem. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]
- 54.Dufner A, Andjelkovic M, Burgering B M T, Hemmings B A, Thomas G. Mol Cell Biol. 1999;19:4525–4534. doi: 10.1128/mcb.19.6.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearson R B, Dennis P B, Han J W, Williamson N A, Kozma S C, Wettenhall R E, Thomas G. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polakiewicz R D, Schieferl S M, Gingras A C, Sonenberg N, Comb M J. J Biol Chem. 1998;273:23534–23541. doi: 10.1074/jbc.273.36.23534. [DOI] [PubMed] [Google Scholar]
- 57.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 58.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature (London) 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 59.Chiu M I, Katz H, Berlin V. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 61.Bierer B E, Mattila P S, Standaert R F, Herzenberg L A, Burakoff S J, Crabtree G, Schreiber S L. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dumont F J, Melino M R, Staruch M J, Koprak S L, Fischer P A, Sigal N H. J Immunol. 1990;144:1418–1424. [PubMed] [Google Scholar]
- 63.Sekulic A, Hudson C C, Homme J L, Yin P, Otterness D M, Karnitz L M, Abraham R T. Cancer Res. 2000;60:3504–3513. [PubMed] [Google Scholar]
- 64.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Nature (London) 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 65.Gingras A C, Raught B, Sonenberg N. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 66.Akagi T, Shishido T, Murata K, Hanafusa H. Proc Natl Acad Sci USA. 2000;97:7290–7295. doi: 10.1073/pnas.140210297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penuel E, Martin G S. Mol Biol Cell. 1999;10:1693–1703. doi: 10.1091/mbc.10.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Louro I D, McKie-Bell P, Gosnell H, Brindley B C, Bucy R P, Ruppert J M. Cell Growth Differ. 1999;10:503–516. [PubMed] [Google Scholar]
- 69.Clemens M J, Bommer U A. Int J Biochem Cell Biol. 1993;31:1–23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 70.Sonenberg N. Curr Opin Cell Biol. 1993;5:955–960. doi: 10.1016/0955-0674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 71.Lazaris-Karatzas A, Montine K S, Sonenberg N. Nature (London) 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 72.Mahalingam M, Templeton D J. Mol Cell Biol. 1996;16:405–413. doi: 10.1128/mcb.16.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donze O, Jagus R, Koromilas A E, Hershey J W, Sonenberg N. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- 75.Tuhackova Z, Sovova V, Sloncova E, Proud C G. Int J Cancer. 1999;81:963–969. doi: 10.1002/(sici)1097-0215(19990611)81:6<963::aid-ijc20>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 76.Shayesteh L, Lu Y, Kuo W L, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills G B, Gray J W. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 77.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ali I U, Schriml L M, Dean M. J Natl Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 79.Maehama T, Dixon J E. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 80.Miwa W, Yasuda J, Murakami Y, Yashima K, Sugano K, Sekine T, Kono A, Egawa S, Yamaguchi K, Hayashizaki Y, Sekiya T. Biochem Biophys Res Commun. 1996;225:968–974. doi: 10.1006/bbrc.1996.1280. [DOI] [PubMed] [Google Scholar]
- 81.Nakatani K, Thompson D A, Barthel A, Sakaue H, Liu W, Weigel R J, Roth R A. J Biol Chem. 1999;274:21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- 82.Ruggeri B A, Huang L, Wood M, Cheng J Q, Testa J R. Mol Carcinog. 1998;21:81–86. [PubMed] [Google Scholar]
- 83.Cheng J Q, Godwin A K, Bellacosa A, Taguchi T, Franke T F, Hamilton T C, Tsichlis P N, Testa J R. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng J Q, Ruggeri B, Klein W M, Sonoda G, Altomare D A, Watson D K, Testa J R. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bellacosa A, de Feo D, Godwin A K, Bell D W, Cheng J Q, Altomare D A, Wan M, Dubeau L, Scambia G, Masciullo V, et al. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 86.De Benedetti A, Harris A L. Int J Biochem Cell Biol. 1999;31:59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 87.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van de Rijn M, Waltham M, et al. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 88.Hosoi H, Dilling M B, Liu L N, Danks M K, Shikata T, Sekulic A, Abraham R T, Lawrence J C, Jr, Houghton P J. Mol Pharmacol. 1998;54:815–824. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- 89.Seufferlein T, Rozengurt E. Cancer Res. 1996;56:3895–3897. [PubMed] [Google Scholar]