THE recent article by Wu and Ma (2005) criticized our article (Luo et al. 2004) on a statistical method for linkage analysis in autotetraploid species by (i) stating that Luo et al.'s linkage analysis is hardly met in practice and (ii) demonstrating the advantages of their method over ours by simulation data. We think it necessary to clarify the misleading criticisms made in their article.
Fisher (1947) and Bailey (1961) considered a two-locus tetrasomic model and exploited the basic features of genetic segregation and linkage of tetrasomic inheritance. They attempted to establish the relationship between the model parameters (the coefficients of double reduction at two linked loci, say α and β, respectively, and recombination frequency between them, r) and frequencies of gamete modes. However, it was clearly pointed out in Bailey (1961, p. 113) that “so far there is no theoretical basis for predicting the frequency of any given mode of gamete formation in terms of, for example, the recombination fraction between the two loci and the two double reduction parameters.” Under exactly the same model, we were able to present the frequency of any given mode of gamete formation in terms of the genetic parameters and to establish an intrinsic relationship between the three parameters in the form given by
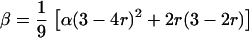 |
(1) |
This relationship is an essential property and natural consequence of the aforementioned model and also the model that Wu and Ma claimed to follow in their research. Thus, it is logically wrong that the model is “strictly based on the assumption that the frequency of double reduction at a marker is determined by the frequency of double reduction at its linked marker and the recombination fraction between these two markers. However, as revealed by cytological and molecular experiments, this assumption that has facilitated Luo et al.'s linkage analysis is hardly met in practice” (Wu and Ma 2005, p. 900). It must be pointed out here that the so-called cytological and molecular experiment evidence that Wu and Ma used to make their criticism is stated in their article as “the values of double reduction are observed to range from 0 to almost 30% (Haynes and Douches 1993) and are likely to be species, chromosome, and position dependent (Butruille and Boiteux 2000)” (Wu and Ma 2005, p. 900). In fact, Equation 1 neither puts any constraint on observed values of double-reduction coefficient nor contradicts the species, chromosome, and position dependency of double reduction. It is clear that these evidences are irrelevant in shaking the basis of Equation 1, and thus their criticism against Equation 1 is baseless and misleading. Given the fact that Wu and Ma's analysis was developed under exactly the same model and they have published actively in this area, we have to suppose that the critical point made in Wu and Ma would be likely due to their failure to understand the model and analysis in our article.
The main difficulties of linkage analysis using genetic marker data (including DNA polymorphic markers) in autotetraploid species lie in two main aspects: systematic segregation distortion due to double reduction and incompleteness of marker data in regard to genotypes at the marker loci. Wu and Ma tried very hard and spent more than half of their page space to defend their previously published article (Wu et al. 2001). However, Wu et al.'s analysis was based entirely on the use of fully informative codominant markers of eight different alleles at each marker between two autotetraploid parents. Under the scenario of using fully informative markers, they avoided all essential theoretical difficulties and their linkage analysis becomes immediately trivial because then both the number of double reduction events and the number of recombinants are directly countable from their offspring genotype data. Built largely on Wu et al. (2001), Wu and Ma presented their analysis of estimating the frequencies of gamete modes, 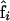 , from gamete data, i.e., the number of different gametes. Estimates of the mode frequencies were then used to calculate α, β, and r through their Equations 6–8, which can be found elsewhere (Bailey 1961, Equation 7.4, p. 113). Having claimed that the gamete-based analysis can be extended to predict the frequencies of gamete mode from marker phenotypic data, the authors surprisingly neglected key steps for justifying how the extension is formulated.
, from gamete data, i.e., the number of different gametes. Estimates of the mode frequencies were then used to calculate α, β, and r through their Equations 6–8, which can be found elsewhere (Bailey 1961, Equation 7.4, p. 113). Having claimed that the gamete-based analysis can be extended to predict the frequencies of gamete mode from marker phenotypic data, the authors surprisingly neglected key steps for justifying how the extension is formulated.
Wu and Ma compared their method to another method, which they claimed to be that developed in our article, by using simulation data. We question here their simulation model and results thus obtained. First, they simulated a case where α = 0.1, β = 0.30, and r = 0.25. Given the claim that Wu and Ma's analysis was under the framework defined by Fisher (1947), the model parameters α, β, and r must follow the relationship given by the above Equation 1 under which β = 0.1833 when α = 0.1 and r = 0.25. Any violation of this relationship contradicts the general model of the autotetrasomic linkage analysis, and simulation thus created is incorrect to make a meaningful comparison. It is clear from Equation 1 that the limit value of β is 0.25 for any given value of α when r takes its upper bound value of 0.75 (please also refer to Sved 1964). Second, Table 2 in their article shows that the two methods were compared using simulation data from the Wu-Ma model and their so-called Luo et al. model. Surprisingly again, they never gave any description about how the Wu-Ma model was simulated and what are the major differences between the two simulation models. The results tabulated in their Table 2 are even more questionable. A comparison of the two analyses under the simulation case of α = 0.1, β = 0.14, and r = 0.10 shows that the two analyses gave similar mean estimates of parameters and their corresponding standard deviations or errors (again not explained) but mean values of the log-likelihood over 200 simulations were −742 under the Wu-Ma analysis and −765 under their so-called Luo et al. model! The difference between the log-likelihood values is 23 (= −765 − (−742)), suggesting that the estimates from the Wu-Ma analysis are ∼1023 times as likely as those from their so-called Luo et al. analysis. This huge difference in the likelihood is absolutely impossible if the comparison was made under a correct setting!
On the basis of the above analyses, we conclude that the criticisms made by Wu and Ma against our article are theoretically wrong and conceptually misleading.
References
- Bailey, N. T. J., 1961. Introduction to Mathematical Theory of Genetic Linkage. Oxford University Press, London.
- Butruille, D. V., and L. S. Boiteux, 2000. Selection-mutation balance in polysomic tetraploids: impact of double reduction and gametophytic selection on the frequency and subchromosomal localization of deleterious mutations. Proc. Natl. Acad. Sci. USA 97: 6608–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1947. The theory of linkage in polysomic inheritance. Philos. Trans. R. Soc. B 233: 55–87. [Google Scholar]
- Haynes, K. G., and D. S. Douches, 1993. Estimation of the coefficient of double reduction in the cultivated tetraploid potato. Theor. Appl. Genet. 85: 857–862. [DOI] [PubMed] [Google Scholar]
- Luo, Z. W., R. M. Zhang and M. J. Kearsey, 2004. Theoretical basis for genetic linkage analysis in autotetraploid species. Proc. Natl. Acad. Sci. USA 101: 1040–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved, J. A., 1964. The relationship between diploid and tetraploid recombination frequencies. Heredity 19: 585–596. [DOI] [PubMed] [Google Scholar]
- Wu, R. L., and C. X. Ma, 2005. A general framework for statistical linkage analysis in multivalent tetraploids. Genetics 170: 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R. L., C. X. Ma, I. Painter and Z-B. Zeng, 2001. A general polyploidy model for analyzing gene segregation in outcrossing autotetraploids. Genetics 159: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]


