Abstract
Genetic variation in Trypanosoma cruzi is likely a key determinant in transmission and pathogenesis of Chagas disease. We have examined nine loci as markers for the extant T. cruzi strains. Four distinct alleles were found for each locus, corresponding to the sequence classes present in the homozygous discrete typing units (DTUs) I, IIa, IIb, and IIc. The alleles in DTUs IIa and IIc showed a spectrum of polymorphism ranging from DTU I-like to DTU IIb-like, in addition to DTU-specific sequence variation. DTUs IId and IIe were indistinguishable, showing DTU homozygosity at one locus and heterozygosity with DTU IIb and IIc allelic sequences at eight loci. Recombination between the DTU IIb and IIc alleles is evidenced from mosaic polymorphisms. These data imply that two discrete hybridization events resulted in the formation of the current DTUs. We propose a model in which a fusion between ancestral DTU I and IIb strains gave rise to a heterozygous hybrid that homogenized its genome to become the homozygous progenitor of DTUs IIa and IIc. The second hybridization between DTU IIb and IIc strains that generated DTUs IId and IIe resulted in extensive heterozygosity with subsequent recombination of parental genotypes.
TRYPANOSOMA cruzi, the causative agent of Chagas disease, infects 16–18 million people throughout Central and South America. Following acquisition of the parasite, a hemoflagellate protozoan of the order Kinetoplastida, clinical symptoms range from negligible to acute Chagas disease that is characterized by fever, high parasitemia, and ≤5% mortality. The subsequent indeterminate stage may last for decades and is characterized by low parasitemia and the presence of quiescent amastigote forms in muscle tissue. Up to 30% of the individuals in the indeterminate stage develop chronic Chagas disease. Pathologies observed in chronic chagasic patients include digestive tract anomalies (megacolon, megaesophagus) and cardiac enlargement and malfunction (Teixeira 1987), accompanied by integration of parasite minicircle DNA sequences into the host genome (Nitz et al. 2004). Variations in disease have yet to be linked directly to host or parasite genetics; however, both of these factors are likely to play a role (Macedo and Pena 1998; Vago et al. 2000; Macedo et al. 2002, 2004; Campbell et al. 2004).
T. cruzi is classified as a single species although there is substantial genetic and phenotypic diversity among isolates (Dvorak 1984; Tibayrenc and Ayala 1988). Different experimental approaches have yielded varying numbers of genetically and biologically significant subgroups of T. cruzi. Strains can be divided into two main lineages by protein and genetic markers (Miles et al. 1978; Tibayrenc 1995; Souto et al. 1996; Campbell et al. 2004) currently designated T. cruzi I and T. cruzi II (Anonymous 1999) that correspond, respectively, to Miles' Zymodeme I and II and Tibayrenc's discrete typing units (DTUs) I and IIb (Tibayrenc 2003) (see below). Similar approaches provide evidence for a third group, Zymodeme III (Miles et al. 1978), while randomly amplified polymorphic DNA (RAPD) and multilocus isoenzyme electrophoresis (MLEE) analyses support the distinction of six subdivisions referred to as DTUs I, IIa, IIb, IIc, IId, and IIe (Barnabé et al. 2000; Brisse et al. 2000, 2001). The DTU scheme, defined as “sets of stocks that are genetically more related to each other than to any other stock and that are identifiable by common genetic, molecular, or immunological markers called tags” (Tibayrenc 1998; Tibayrenc and Ayala 2002), gives a functional framework that best describes the current understanding of the relationship among the strains.
DTUs IIa and IIc are equivalent to Miles' Zymodeme III and its respective B/A subdivisions (Mendonça et al. 2002; Campbell et al. 2004). Understanding the relationship of DTUs IIa and IIc (Zymodeme III) to other DTUs has been complicated by experimental results yielding contrary associations: MLEE and RAPD assays and HSP60 gene sequences show an overall similarity to DTU IIb (Tibayrenc 1995; Sturm et al. 2003), while individual enzyme profiles and SL RNA and rRNA gene sequences indicate similarity to DTU I (Miles et al. 1978; Coura et al. 2002; Mendonça et al. 2002).
An understanding of the population structure of T. cruzi is critical because of the links to transmission cycles and disease. Broadly, DTU I is associated with the silvatic cycle of transmission and arboreal mammals; DTU II is associated with the domestic cycle and a terrestrial niche (Miles et al. 1978; Fernandes et al. 1998; Zingales et al. 1998; Yeo et al. 2005) and associated triatomine vectors (Gaunt and Miles 2000) in Brazil and other South American countries. This association may vary by region and as the human population encroaches upon the silvatic ecosystem. Genetically distinct organisms can be detected in the heart and esophagus of the same patient (Vago et al. 2000). T. cruzi replication is predominantly clonal (Tibayrenc et al. 1986; Tibayrenc and Ayala 2002; Tibayrenc 2003), although there is evidence for limited genetic exchange among strains (Bogliolo et al. 1996; Carrasco et al. 1996; Souto et al. 1996; Machado and Ayala 2001; Brisse et al. 2003; Sturm et al. 2003). A mechanism for genetic exchange in T. cruzi has been identified experimentally (Gaunt et al. 2003) involving the fusion of parental genotypes, nuclear hybridization, loss of alleles, and homologous recombination. The CL Brener strain, chosen to represent T. cruzi as the first completely sequenced genome (Zingales et al. 1997), is a hybrid strain classified as DTU IIe (Brisse et al. 1998) whose ancestral lineages are represented by DTUs IIb and IIc (Machado and Ayala 2001; Brisse et al. 2003).
On the basis of molecular markers from four strains representing DTUs I, IIb, and IIe we concluded previously that DTUs IIa and IIc and DTUs IId and IIe are two independent hybrid lineages (Sturm et al. 2003). Here we extend our analysis to nine molecular markers, composed of two single-copy coding regions and seven multicopy, noncoding regions, applied to additional strains representing the six T. cruzi DTUs. The current data reveal the genetic makeup of the hybrid strains in greater detail. In this study, four distinct allelic sequences defined five of the six DTUs. Hybrid DTUs IIa and IIc were homozygous at all loci, with mosaic-like sequences sharing similarity to DTU I and/or IIb, their likely ancestral lineages. Hybrid DTUs IId and IIe showed minimal distinction from one another, with allelic heterozygosity at multiple loci and discrete recombination events in specific strains, reflecting a hybridization event between ancestral lineages from DTUs IIb and IIc.
MATERIALS AND METHODS
DNA amplification, cloning, and sequencing:
Most T. cruzi DNA samples (Table 1) were cultured at IRD, Montpellier, France; DNA samples from strains Y from and MT4167 were obtained from Bianca Zingales, USP, Brazil, and Octavio Fernandes, Fiocruz, Brazil, respectively. Primers used to amplify the HSP60, HSP70, 1F8, histones H1, H2A, H2B, and H3 intergenic regions were as described (Sturm et al. 2003). Primers used to amplify the GPI marker were as described (Gaunt et al. 2003). TcMSH2 primers were as described (Augusto-Pinto et al. 2003). Amplification reactions of 50 μl total volume included 1x Red Taq buffer (Sigma), 2 mm MgCl2, 0.4 mm dNTPs, 0.02 μg/μl of each primer, 10 ng total cell DNA, and 1 unit Red Taq (Sigma) polymerase. The reaction was performed with initial denaturation for 2 min at 94°, followed by 25–35 cycles of denaturation for 30 sec at 94°, annealing for 30 sec at 60°, and extension for 1 min at 72°, with a final extension for 10 min at 72°. PCR products were gel purified using the QIAGEN purification kit, cloned using the TOPO-TA cloning kit (Invitrogen), and sequenced commercially by automated fluorescent sequencing (Laragen). We extended our original analysis (Sturm et al. 2003) to include products from at least two strains of each DTU (Table 1). Initially, a single representative cloned PCR product was sequenced on both strands for each examined strain. The number of total strains sequenced for each locus is reflected in the single nucleotide polymorphism (SNP) tables and text; additional clones were sequenced in some cases to characterize precisely heterogeneous loci or in instances of unexpected RFLP results (see below). Nucleotide sequences have been deposited in the GenBank database with the accession nos. AY540638–AY540744 and DQ021887–DQ021899. Some T. cruzi CL Brener strain sequences were retrieved from The Institute for Genomic Research website at http://www.tigr.org.
TABLE 1.
T. cruzi DNA samples used in this study and defined alleles
| Isolatea | DTUb | SL RNAc | D7 rRNAd | Ribocladee | DHFR | TRf | COII-ND1f | PGPg | GPIh | TCPh |
|---|---|---|---|---|---|---|---|---|---|---|
| Cutia | I | I | 2 | A | ||||||
| Silvio X10 cl.1 | I | I | 2 | 2 | A | A | A | A | I | I |
| CANIII | IIa | IIa | 3 | 4ij | D | D | B | IIa | IIa | |
| MT4167 | IIa | IIa | 3 | 4 | ||||||
| CM17 | IIc | “III” | 2 | 3k | B | B | B | |||
| M5631 cl.5 | IIc | IIc | 2 | 3k | ||||||
| M6241 cl.6 | IIc | IIc | 2 | 3k | B | B | B | B | IIc | IIc |
| X9/3 | IIc | 2 | B | |||||||
| CBB cl.3 | IIb | IIb | 1 | C | C | C | ||||
| Esmeraldo cl.3 | IIb | IIb | 1 | C | C | C | C | IIb | IIb | |
| MAS1 cl.1 | IIb | C | IIb | |||||||
| MSC2 | IIb | 1 | C | C | C | |||||
| TU18 cl.2 | IIb | IIb | 1 | C | ||||||
| Y | IIb | IIb | 1 | 1 | ||||||
| GM-0 | IId | |||||||||
| MN cl.2 | IId | IIb | 1/2 | |||||||
| NR | IId | II | 1/2 | 3j | ||||||
| P255 | IId | |||||||||
| SAXP19 | IId | |||||||||
| SC43 cl.1 | IId | IIb | 1 | 3 | IIb, c | I, IIb, c | ||||
| SO3 | IId | II | 1/2 | 3 | B, C | B, C | B | C | IIb | |
| 86/2036 | IIe | B | ||||||||
| CL Brener | IIe | IIb | 1 | 1 | B, C | B, C | B | C | IIb, c | IIb |
| P63 cl.1 | IIe | II | B | |||||||
| Sabcho109a | IIe | 1 | ||||||||
| Tula cl.2 | IIe | II | 1 | B, C | B, C | B | C |
Origin of isolates are described in Brenière et al. (1998); Brisse et al. (2001); Barnabé et al. (2001b).
According to Brisse et al. (2000).
Determined in Souto et al. (1996); Campbell et al. (2004). For consistency with proposed terminology (Anonymous 1999), I and II designations have been reversed from the original publications, e.g., Souto et al. (1996).
1, 125 bp Souto and Zingales (1993); 2, 110 bp Souto and Zingales (1993); 3, 117–120 bp Brisse et al. (2001); Mendonça et al. (2002).
Determined in Kawashita et al. (2001).
Determined in Machado and Ayala (2001).
Determined in Robello et al. (2000)
Determined in Gaunt et al. (2003).
Determined in Brisse et al. (2001).
18S rRNA Kawashita et al. (2001).
Computer analyses:
Sequences were aligned with ClustalX v1.8 (Thompson et al. 1997). Sequence alignments were edited with the BioEdit program (Hall 1999). The assignments of meaningful SNPs were based on their appearance within at least two strains, not necessarily from the same DTU. For phylogenetic analysis, sequence alignments were edited to areas of confident alignment with no gaps or hypervariable regions (alignments available upon request). Phylogenetic trees were determined using MEGA 3.0 software package (Kumar et al. 2004) using the neighbor-joining method. The default analysis settings were used, including the Kimura two-parameter base substitution model. The phylogenetic trees were assessed with 1000 bootstrap replicates. All trees are unrooted, condensed, bootstrap consensus trees displaying topology only, with significant bootstrap values included for the major sequence classes.
RFLP analyses:
Following evidence that the DTU IId and IIe strains were heterozygous hybrids of DTU IIb and IIc (Machado and Ayala 2001; Gaunt et al. 2003), we designed PCR-RFLP analyses (summarized in Table 2) to test the extent of heterozygosity in the DTU IId and IIe strains on the basis of variations between at least two sequenced representatives of the DTU IIb and IIc sequence classes. Restriction digests were performed on PCR products using conditions suggested by the manufacturer (New England Biolabs) from templates representing DTU I (two isolates), DTU IIa (two isolates), DTU IIb (five isolates), DTU IIc (four isolates), DTU IId (seven isolates), and DTU IIe (five isolates). Some products required purification by Qiaquick (QIAGEN) to remove excess salt prior to digestion. Digestion products were run on 1.5 or 3% agarose gels in the presence of ethidium bromide with equal amounts of undigested PCR product run in an adjacent lane. The following enzymes were used for each marker: EcoRV for HSP60; AclI for HSP70; AatII for histone H1; AluI for histone H3; NlaIII for MSH2; HhaI for GPI. When unexpected RFLP patterns were encountered, additional clones were screened and sequenced.
TABLE 2.
RFLP analysis of PCR products
| Marker | PCR (bp) | Enzyme | RFLP SNP(s) | Digestion products (bp) |
|---|---|---|---|---|
| HSP60 | 432–462 | EcoRV | 125 | 314, 148–118 (IIc) |
| HSP70 | 550 | AclI | 362, 363 | 359, 190 (IIb) |
| Histone H1 | 486 | AatII | 116, 118 | 364, 122 (I, IIa, IIc) |
| Histone H3 | 635 | AluI | 216 | 437, 198 (I, IIa, IIb) |
| TcMSH2 | 875 | NlaIII | 2294 | 429, 269, 177 (I, IIc) |
| 429, 177, 168, 101 (IIa, IIb) | ||||
| GPI | 1190 | HhaI | 173 | 817, 447 (I, IIc) |
| 490, 447, 253 (IIa, IIb) |
The RFLP approach serves as a broad survey of particular markers among the T. cruzi strains. Our use of the RFLP technique was primarily as a method to assay for heterozygosity within the loci examined and could be subject to restriction site loss or acquisition in the event of mutation with that restriction site. Specific cases were followed up with sequence analysis. Additional information regarding variability within the strains is likely to be present within the amplified regions (see results).
RESULTS
T. cruzi is considered to be minimally diploid (Gibson and Miles 1986), but aneuploid in hybrid strains (Vargas et al. 2004). In the following sections, we examine the allelic structure of nine loci in the six DTUs of T. cruzi. Examples of variant alleles of the same DTU and the presence of two distinct sequence classes representative of different DTUs were found within particular strains. These two situations are technically heterozygous; however, for the purposes of this study we restricted the use of the term “heterozygous” to represent the latter situation.
Although distinct from one another, both the homozygous DTUs IIa and IIc share common SNPs with DTU I, DTU IIb, or a mixture of these two DTUs (Table 3). In terms of following the genetic flow through the millennia of T. cruzi evolution, the SNP variation between DTUs I and IIb indicated that these two DTUs are most likely representatives of the ancestral T. cruzi I and T. cruzi II sequences from which all other T. cruzi sequences are derived. We present our overall DTU results locus by locus on the basis of the relationship of DTUs IIa/IIc to their progenitor DTUs. Tables of SNPs, phylogenetic analysis, and/or RFLPs are shown for the analyzed loci, two of which are extensions of work previously reported by other groups.
TABLE 3.
Average uncorrected divergence (p-distance) per 100 bp of sequences within and between groups of T. cruzi
| Gene | HSP70 | GPI | H2B | MSH2 | H1 | H3 | HSP60 | 1F8 | H2A | DHFR-TS | TR | NC | CDS | Avg. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Within I | 0.20 | 0.29 | 1.49 | 0.12 | 1.26 | 1.89 | 0.23 | 0.60 | 1.04 | 0.08 | 0.17 | 0.96 | 0.17 | 0.67 |
| Within IIa | 0.98 | 0.67 | 2.48 | 0.12 | 2.32 | 0.63 | 0.94 | 0.60 | 2.60 | 0.20 | na | 1.51 | 0.25 | 1.05 |
| Within IIb | 1.23 | 0.14 | 0.96 | 0.31 | 1.19 | 0.77 | 0.36 | 1.01 | 1.11 | 0.29 | 0.00 | 0.95 | 0.19 | 0.67 |
| Within IIc | 0.59 | 0.37 | 1.39 | 0.18 | 0.74 | 0.71 | 0.59 | 0.37 | 2.11 | 0.12 | 0.12 | 0.93 | 0.20 | 0.66 |
| I vs. IIa | 2.55 | 1.35 | 7.44 | 1.33 | 5.00 | 3.54 | 4.23 | 2.39 | 4.17 | 0.96 | 1.39 | 4.19 | 1.26 | 3.12 |
| I vs. IIb | 4.73 | 2.02 | 7.20 | 1.70 | 3.68 | 4.22 | 3.60 | 3.85 | 5.86 | 1.38 | 1.60 | 4.73 | 1.68 | 3.62 |
| I vs. IIc | 2.16 | 0.93 | 4.22 | 1.50 | 2.79 | 2.95 | 4.73 | 2.57 | 3.68 | 0.73 | 1.03 | 3.30 | 1.05 | 2.48 |
| IIa vs. IIb | 4.80 | 1.95 | 7.75 | 0.89 | 3.96 | 3.54 | 2.85 | 2.69 | 5.30 | 1.04 | 1.63 | 4.41 | 1.38 | 3.31 |
| IIa vs. IIc | 2.75 | 1.20 | 6.10 | 0.91 | 3.76 | 2.79 | 3.72 | 0.78 | 4.80 | 0.64 | 1.44 | 3.53 | 1.05 | 2.63 |
| IIb vs. IIc | 4.98 | 1.82 | 6.66 | 1.37 | 2.86 | 3.28 | 2.85 | 2.87 | 6.01 | 1.04 | 1.72 | 4.22 | 1.49 | 3.22 |
Values underlined are the maximum for each locus, and values in italics are the minimum for each locus. GPI analysis excluded putative recombinant sequences from Hc10, 92.80cl2, Para2, and CL Brener. NC, average of noncoding regions 1F8, Hsp60, Hsp70, H1, H2A, H2B, and H3 values; CDS, average of MSH2, GPI, DHFR-TS, and TR values; Avg, average of all loci.
In individual isolates of DTUs IId and IIe, sequences contained SNPs associated with both parental DTU classes that are referred to as “mosaic” (Gaunt et al. 2003). Mosaics are likely the result of homologous recombination events between alleles of different DTUs present in a single heterozygous individual. For the purposes of our analyses, we chose an arbitrary minimum of three consecutive SNPs from a particular DTU class to constitute a mosaic event when present in the background of a different DTU profile. Using this criterion, mosaic sequences should be distinguished from homoplasy, or the equivalent mutations arising separately in different strains, and will guard our analysis against the intrusion of random PCR infidelity or sequencing errors.
Mosaic sequences were excluded from our phylogenetic analyses and SNP tables for the following reasons: (1) Within a heterozygous background, artifactual mosaic PCR products have occurred (Brakenhoff et al. 1991; Tanabe et al. 2002), and (2) the main point of our sequencing analysis was to determine DTU affiliation of the heterozygous alleles and to assess the loci for useful RFLP sites for extension of the DTU analysis. DTU association is not affected by minor interarray heterogeneity (Thomas et al. 2005). In an attempt to confirm some of the mosaic sequences, the sequencing of additional clones for some loci derived from subsequent PCRs was performed, but the original mosaic sequences were not recovered. Validation of mosaics using PCR could be problematic due to the multicopy nature of most of our loci, with individual reactions selectively amplifying different sequence variants. We used the CL Brener genome database to validate our loci by the presence of mosaics obtained by a PCR-free methodology. In the case of histone H2A, multiple alleles were captured showing affinity with DTUs IIb or IIc or a third class of sequences that were mosaic for SNPs of DTU IIb and IIc (see below). Excluded mosaic sequences were 1F8 and Hsp60 from the NR strain; histones H3, H2A, and H2B from the Tula strain; Hsp60 from SC43; TcMSH2 from the CL Brener strain; histone HI and histone H2A from the M5631 strain; and H2B from the MN strain.
Loci in which DTUs IIa and IIc showed greater affinity to DTU I
HSP70:
The HSP70 gene family is organized in a head-to-tail tandem array of at least seven copies on chromosomes of 1.03 and 1.1 Mbp in CL Brener (Cano et al. 1995; Requena et al. 1988). The HSP70 intergenic region from T. cruzi I, IIb, and IIe strains contains 16 SNPs that correlate with the T. cruzi I/T. cruzi II definition (Sturm et al. 2003).
Alignment of the amplified HSP70 sequences from 13 strains revealed four distinct sequence classes (Table 4). Characteristic SNPs were identified for DTUs I, IIa, IIc, and IIb/IId/IIe. Sequences from 6 of the 7 strains from DTUs IIb, IId, and IIe were identical; SC43 (IId) differed only at nucleotide 271. Phylogenetic analysis of this region revealed robust support for four sequence classes (Figure 1A) with DTU IIb, IId, and IIe sequences in one clade. The restriction enzyme AclI distinguished between DTUs I/IIa/IIc and IIb/IId/IIe and was used for RFLP analysis of the amplified products (Figure 1B). AclI digestion was applied to 15 additional strains previously classified into specific DTUs by RAPD and MLEE analysis (Barnabé et al. 2001a; C. Barnabé and M. Tibayrenc, unpublished results). Strains from DTUs I, IIa, and IIc were not digested by AclI and were characterized by an ∼550-bp product, while strains representing DTUs IIb, IId, and IIe showed complete digestion, yielding ∼360- and ∼190-bp fragments. Data from the CL Brener genome project supported the homozygous nature of the HSP70 locus in this hybrid DTU IIe strain.
TABLE 4.
Positions of SNPs for loci where DTUs IIa/IIc are more similar to DTU I
| Strain | SNP |
|---|---|
| HSP70 | |
| 111111222333333333344a | |
| 688147799379266666779902 | |
| 607435719117101238894662 | |
| Dm28c, Silvio X10 (I) | GGTTTACCCTCATTTGGTTCTTGA |
| CANIII, M4167 (IIa) | GGTTTCCTCATATTTGGATCCCTC |
| CM17, M5631 (IIc) | GGTCTCACCGTATTTGGACCTTGC |
| SC43 (IId) | AACTCCCCTGCGAGGAAATTTTTT |
| Esmeraldo, CBB (IIb), MN (IId), Tula, P63, CL Brener (IIe) | AACTCCCCTGTGAGGAAATTTTTT |
| GPI | |
| 11b | |
| 11111123334445678999900 | |
| 157803337822884496883045913 | |
| 389531483806363976286273262 | |
| X10 (I) | AATTGGCCAGCCTCTCACGTTCCGGCC |
| HC9 (I) | AATTGGCCAGCCTCTCACGGTCCGGCC |
| CanIII (IIa) | ACTTGGCCGACTCCCTATAGTCTGGCC |
| HCL3 (IIa) | ACTTGGCCGACTCCCTACAGTCTGGCT |
| M6241(IIc), RMA34.2 (IIb) | AATTGACCAGCCTCCCATAGTCTGGCT |
| SC43.2 (IId), Chaco2.1 (IId) | AATTGACCAGCCTCCCGTAGGCTGGCT |
| 92.80 cl2.1 (IId) | AATTGACCAGCCCCCCGTAGGCTGGCT |
| Sp14 (IIc) | AATTGGCCAGCCTCCTATAGTCTGGCT |
| Chaco2.2 (IId) | AATTGACCAGCCTCCCATAGGCTAACC |
| CLBrener.2 (IIe) | AATTGGCCAGCGTCCTATAGGTTAATC |
| X9/3, SP13, X109/2.1 (IIc), X109/2.2 (IIc), RMA34.1 (IIb) | AATTGACCAGCCTCCCGTAGTCTGGCT |
| Para2.2 (IId) | TACCAGTTGGAGTCCCGTAGTCTGGCT |
| HC10 (IIb) | TACCAGTTGGAGTTCTACAGTCTGGCC |
| 92.80 cl2.2 (IId) | TACCAGTTGGACCCCTATAGGCTGGCT |
| Esm cl3.2 (IIb), RMA134.1 (IIb) | TACCAGTTGGAGTTCTATATGTTAATC |
| Esm cl3.1 (IIb) | TACCAGTTGGAGTTCTACAGGTTAATC |
| RMA134.2 (IIb), Para2.1 (IId), SC43.1 (IId), CLBrener.1 (IIe) | TACCAGTTGGAGTCCTATAGGTTAATC |
| Histone H2B | |
| 11111112222233334444c | |
| 23666668888999900003343367713490001 | |
| 57013450157257803566763733585643453 | |
| Dm28c, Silvio X10 (I) | TCGCCGGCTTGACCCTCGCTGGCTCAGTTCGACGC |
| M4167 (IIa) | TCAACGATTTTGTTGCTCCTAATCCGGTCCGACGC |
| CANIII (IIa) | TCAACGATTTTGTTGCTCCTAATCCGGTCTGAAGC |
| CM17 (IIc) | TCAACGATTTGACCCTCCTTGGCTTGGTTCGACGC |
| M5631 (IIc) | TCGCCTTTTTGACCCTCCTTAGCTTGGTTCGACGC |
| MN.12 (IId) | TCGCCTTTTTGACCCTCCTTAGCTTGGTTCGACGC |
| TULA.14 (IIe) | TCGCCGATTTGACCCTCCTGAGCTTGGTTCGACGC |
| CL Brener (IIe) | TCGCCTTTTTGACCCTCCTTGGCTTGGTTCGACGC |
| CBB (IIb), Y (IIb) | CTGCAATGACGACCCTCCCAAACTCGTTTTTGAAG |
| Esmeraldo (IIb) | CCGCAATGACGACCCTCCCAAACTCGTTTTTGAAG |
| SC43 (IId) | CTGCAATGACGACCCTCCCAAACTCGTTTTTGAAC |
| MN.11(IId) | CCGCAATGACGACCCTCCCAAACTCGTTTTTGAAC |
| TULA.12, P63.11 (IIe) | CCGCAATGACGACCCTCCCTAACTCGTGTTTGAAC |
| CL Brenerd (IIe) | CTGCAATGACGACCCTCCCAAACTCGTGTTTGAAC |
| MSH2 | |
| 1111111111222222222222e | |
| 6667788999000122223334 | |
| 5575779145567412793680 | |
| 0861212962799034443376 | |
| Silvio, Colombiana, RBI, D7 (I) | CAAACTGGAGTCAGTCAAGGCG |
| 4167, CanIII (IIa) | GAGGCCGGGACTCGCGGAAGCA |
| mt5631 (IIc) | GAAGCCGGAGCCCGCGGAGGCA |
| 115b | GAGGCCGTAGCCCGCGGAGGCA |
| 167a, 226, 231, CL Brener (IIe), Tula, P255(IIe), MN.11 (IId) | GAGGCCGTAGCCCGCGGAAGCA |
| JG, 1005 | GAAGTTAGGGCCCACGACGACA |
| 115a, 167b | GAAGTTGGGGCCCACGACGATA |
| 239a | GGAGTTGGGGCCCACGACGACA |
| 239b | GAAGTTGGGGCCCACGACGACA |
| 577 | GAAGTTGGGGCTCACGACGACA |
| 593 | GGAGTTGGGGCTCACGACGACA |
| CBB (IIb) | GGAGTTGGGGCCCACGACGACA |
| Y (IIb) | GAAGTTAGGGCTCACGACGACA |
| TU18.4 (IIb) | GAAGTTAGGGCCCACGGAGATA |
| TU18 (IIb), CL Brenerf (IIe) | GAAGTTAGGGCTCACGACGATA |
SNP positions relative to CL Brener sequence.
SNP positions relative to amplicons with assignments as reported by Gaunt et al. (2003). For strains with more than one published sequence, different clones were labeled randomly as x.1 and x.2.
SNP positions relative to Dm28c (I) sequence.
The CL Brener IIb-like obtained from TIGR sequence TcTIGR_TCSDP14TR.
SNP positions relative to MSH2 coding region.
CL Brener IIb-like obtained from TIGR sequence TcTIGR-TCHLY26TF.
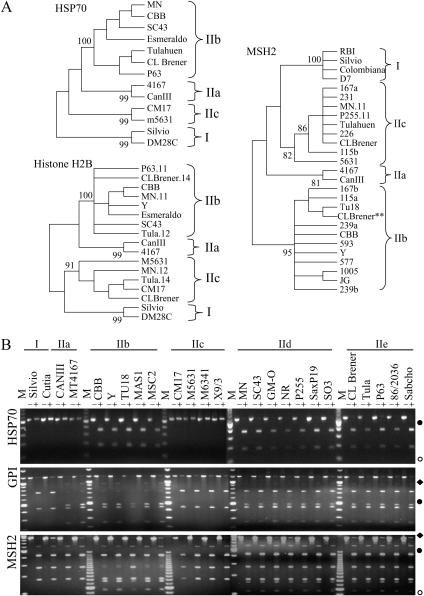
Figure 1.
Loci in T. cruzi where DTU IIa/IIc alleles are more similar to DTU I. (A) Unrooted neighbor-joining trees of the HSP70 and histone H2B intergenic- and MSH2 partial coding-region sequences reveal four DNA sequence classes. **, CL Brener sequences from TIGR. Additional MSH2 sequences were taken from Gaunt et al. (2003). (B) Agarose gel electrophoresis of HSP70, GPI, and MSH2 PCR-RFLP products. The DNA was stained with ethidium bromide. Size markers (M) are either the BRL 1-kb ladder or the 25-bp ladder. (♦) 1-kb bands, (•) 0.5-kb bands, (○) 0.1-kb bands. The SNPs that create the AclI restriction site polymorphism in the HSP70 IIb allele are positions 362 and 363; a SNP at position 173 creates the HhaI polymorphism in the GPI locus; and a SNP at position 2294 creates the NlaIII polymorphism in the MSH2 locus (Table 2).
The presence of four DNA sequence classes, designated I, IIa, IIb, and IIc after their respective DTUs, represents allelic forms of the HSP70 locus. A single sequence class was present in each strain examined, indicating that the locus is homozygous in all tested strains for each DTU. This locus did not allow differentiation of the IIb/IId/IIe DTUs. Selective amplification of sequence classes, loss of the diagnostic restriction site by an isolated SNP, or recombination between alleles of different classes could give a misleading result by PCR-RFLP analysis.
GPI—glucose phosphate isomerase:
The coding region of GPI is heterozygous for CL Brener, with a DTU IIb-like allele and a DTU IIb/IIc mosaic allele (Gaunt et al. 2003). Gaunt et al. (2003) demonstrate the heterozygous nature of DTU IId and IIe strains, with sequences from these strains contained within DTU IIb and IIc clades. We used this knowledge to select an enzyme that would reveal heterozygosity in additional strains originating from a DTU IIb and IIc fusion.
The PCR products of all GPI alleles shared an HhaI restriction site that produced digestion fragments of 817 and 447 bp, as observed in DTUs I and IIc (Figure 1B). Strains from DTUs IIa and IIb have an additional site within the 817-bp fragment, displaying homozygosity with 490-, 447-, and 252-bp products. DTUs IId and IIe showed 817-, 490-, 447-, and 252-bp fragments, indicating that they are all heterozygous.
The GPI locus differentiates among five DTUs of T. cruzi: the four homozygous lines and the heterozygous DTUs IId and IIe that share alleles with DTUs IIb and IIc. The RFLP assay demonstrates the maintenance of heterozygosity in DTUs IId and IIe. However, we can conclude only that these alleles are heterozygous at the site of restriction digestion. This conclusion is also strictly true for all other loci examined by the RFLP assay only.
Histone H2B:
The histone H2B genes are present at ∼18 copies on a single chromosomal band of 3.5 Mbp in CL Brener (García-Salcedo et al. 1994; Henriksson et al. 1995). The intergenic region has four SNP differences between DTU I and DTU IIb/IIe strains (Sturm et al. 2003).
The four classes of sequence variation were resolved by SNP haplotypes (see Table 4). Multiple cloned PCR products from one or two reactions produced the following sequences: for Tula, three DTU IIb and three DTU IIc; for p63, six DTU IIb; for MN, two DTU IIb and three DTU IIc. One representative of each allelic type was included in the phylogenetic analysis, labeled Tula.12, Tula.14, p63.11, MN.11, and MN.12. Mosaic sequences were observed initially from single cloned PCR products from MN, Tula, and P63 strains, but excluded from further analysis. The phylogenetic tree resulting from analysis of the histone H2B intergenic region produced four sequence clades, represented by DTUs I, IIa, IIb, and IIc (Figure 1A). Examination of CL Brener genomic sequence reads from TIGR revealed an ∼2:1 ratio of IIc to IIb allele sequences.
This locus reinforces the DTU clades established by GPI, showing distinction of five of the six DTUs and heterozygosity of DTUs IId and IIe. The heterozygous allelic forms associate with DTUs IIb and IIc.
MSH2—mutS homolog 2:
The single-copy MSH2 DNA mismatch repair gene contains 21 SNPs in a partial open reading frame (Augusto-Pinto et al. 2003) and shows heterozygosity in some strains. We sought to determine the nature of the MSH2 locus in the DTU IId and IIe strains because many of the strains used in the previous study had not been classified by the Tibayrenc rubric and no representatives of DTU IIa were included. To complete the genotypic assessment, we examined the identical partial coding sequence from strains representing the six DTUs.
Neighbor-joining analysis of the MSH2 locus produced four clades represented by DTUs I, IIa, IIb, and IIc (Figure 1A). In RFLP analysis DTUs I/IIa/IIc and IIb can be distinguished by digestion with NlaIII (Figure 1B). DTUs I and IIc were cut at two sites, yielding three fragments, whereas DTU IIb strains had an additional cut site by virtue of which the 269-bp fragment was cleaved to 101 and 168 bp. Strains of DTUs IId and IIe displayed the 269-, 101-, and 168-bp fragments, indicative of allelic heterozygosity. The DTU IIb strain TU18 displayed partial digestion of its 269-bp fragment due to an allelic polymorphism; cloning and sequencing showed that this allele grouped with the DTU IIb clade (Table 4). Originally, a mosaic sequence was observed in a cloned PCR product from MN. Five additional cloned PCR products from a subsequent reaction were all similar to DTU IIc with minor heterogeneity as seen in the SL RNA gene array (Thomas et al. 2005), although the heterozygosity with DTU IIb was evident in RFLP analysis. A single-cloned MN sequence, MN.11, was included in the phylogenetic analysis. Both DTU IIb and IIc allelic forms were found for CL Brener. This coding region follows the same pattern as seen in the previous regions, with four distinct allelic classes and DTU IIb/IIc heterozygosity evident in the DTU IId and IIe strains.
Loci in which DTUs IIa and IIc showed split affinities to DTU I and DTU IIb
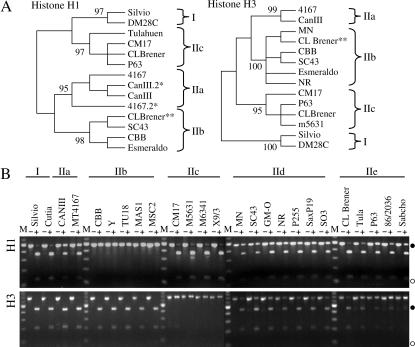
Histone H1:
The 15–20 copies of the histone H1 genes are arrayed tandemly on a chromosomal band of 2.6 Mbp in the CL Brener strain (Åslund et al. 1994; Henriksson et al. 1995). The intergenic region contains five SNPs and a single nucleotide indel among DTUs I and IIb/IIe (Sturm et al. 2003).
Histone H1 sequences from strains representative of the six DTUs displayed four distinct sequence classes (Table 5). Neighbor-joining analysis produced four sequence clades, represented by DTUs I, IIa, IIb, and IIc (Figure 2A). Allelic variants from the CL Brener strain were present in both DTU IIb and DTU IIc clades. Robust bootstrap support was obtained for associating the allelic sequences of DTU IIe strains Tula, CL Brener, and P63 with the DTU IIc clade and the DTU IId strain SC43 and DTU IIe strain P255 with the DTU IIb clade. An AatII restriction site covered SNP positions 116 and 118 and differentiated between DTUs I/IIc and IIb (Figure 2B). After exposure to the restriction enzyme, PCR products from DTU IIb were not digested, while DTUs I and IIc were digested. DTUs IId and IIe displayed a heterozygous phenotype with the presence of the three fragments, as did DTU IIa, which was not anticipated. The misleading result for DTU IIa heterozygosity by RFLP analysis, implying that DTU IIa strains contained DTU IIb-like and IIc-like alleles, was resolved by sequencing both alleles. The DTU IIa strains examined possessed two nearly identical alleles that are polymorphic at the diagnostic RFLP site. Phylogenetic analysis resulted in the formation of a single clade specific to DTU IIa including both alleles. CL Brener and Tula strains showed an unequal intensity of intact bands relative to the digestion fragments, which could be due to unequal PCR amplification, unequal numbers of repeats in homogeneous allelic arrays, or heterogeneous arrays. The CL Brener genome project revealed an ∼3:1 ratio of DTU IIc to DTU IIb histone H1 sequences; therefore, the PCR-RFLP result was consistent with the relative proportion of the two alleles present in the genome of CL Brener. The histone H1 locus again reveals the four allelic types and shows maintenance of heterozygosity for DTUs IId and IIe with alleles similar to DTUs IIb and IIc.
TABLE 5.
Positions of SNPs for loci where DTUs IIa/IIc are split between DTUs I and IIb
| Strain | SNP |
|---|---|
| Histone H1 | |
| 1111111122233344 | |
| 33566890114457948915745 | |
| 78118783685630407000558 | |
| Dm28c (I) | CGGGTCTTCTCCCGTCCCCTTAC |
| Silvio X10 (I) | CGGGTCTTCTCCTGTCCCCTTAC |
| CANIII (IIa),a M4167 (IIa)a | CTGGCTCCTTTTTTGCGTCTCGT |
| CANIII (IIa)b | CTGGTTCCCTTTTTGCGTCTCGT |
| M4167 (IIa)b | CTGGTCTCCTTTTTTCGTCTCGT |
| M5631 (IIc) | CTGGTCTCCTTCTGTCCTCTCAT |
| CM17 (IIc), CL Brener, Tula (IIe) | CTCGTCTCCTTCCGTCCTTTCAT |
| P63 (IIe) | CTCGTCTCCTTCCGTCCTCTCAT |
| SC43 (IId) CL Brenerc (IIe) | TTGATCCCTGTTTGTCCCCCTAT |
| CBB (IIb), Esmeraldo (IIb) | TTGATCCCTGTTTGTTCCCCCAT |
| Histone H3 | |
| 1111111122223333444555555 | |
| 2551445679912570367035001235 | |
| 4054080626767668952556349116 | |
| Dm28c (I), Silvio X10 (I) | ACTTCAGCGCGTTTATCATTGTTTCTTT |
| CANIII, M4167 (IIa) | TCCCCGTTACGTTTACCATCATCCTCTT |
| CM17, M5631 (IIc) | TCTTCGTCGTGAGTGTTATTATTTTTTT |
| CL Brener, P63 (IIe) | TCTTCGTCGTGAGTGTTATTATTTTTTT |
| Esmeraldo (IIb), NR (IIe) | TCTTTGTCCTTTTCATCGCTAGCATTGC |
| CBB (IIb), SC43 (IId) | TATTTGTCCTTTTCATCGCTAGCATTGC |
| MN (IId), CL Brenerd (IIe) | TCTTTGTCCTTTTCATCGCTAGCCTTGC |
SNP positions are relative to CL Brener.
Uncut alleles from DTU IIa strains.
Cut alleles from DTU IIa strains.
CL Brener IIb-like obtained from TIGR sequence TcTSK-TGC107157.
CL Brener IIb-like obtained from TIGR sequence TcTIGR-TCGOI28TF.
Figure 2.
Loci in T. cruzi where the similarity of DTU IIa/IIc alleles are split between DTU I and DTU IIb. (A) Unrooted neighbor-joining trees of histone H1 and histone H3 intergenic sequences reveal four DNA sequence classes. *, the sequence of the uncut allele sequence of IIa strains; **, the CL Brener sequences from TIGR. (B) Agarose gel electrophoresis of PCR-RFLP products from the histone H1 and histone H3 loci of 25 strains. The DNA was stained with ethidium bromide. Size markers (M) are either the BRL 1-kb ladder or the 25-bp ladder. • and ○ represent 0.5- and 0.1-kb bands, respectively. SNPs at positions 116 and 118 create the AatII polymorphism in the H1 locus, and a SNP at position 216 creates the AluI polymorphism in the histone H3 locus (Table 2).
Histone H3:
The histone H3 array contains 10–20 repeats localized to a single chromosomal band of 1.25 Mbp in CL Brener (Bontempi et al. 1994; Henriksson et al. 1995). The intergenic region differs by five SNPs and two single-nucleotide indels between DTUs I and IIb/IIe (Sturm et al. 2003).
The four classes of sequence variation were resolved by sequence alignment (Table 5). The DTU IIb-like sequence from the DTU IId and IIe strains contained sequences similar to the DTU IIb or IIc classes. Phylogenetic analysis provided robust bootstrap support for the four clades (Figure 2A). An AluI site was chosen to examine differential RFLP patterns between DTUs I/IIa/IIb and IIc (Figure 2B). DTU IIc was not cut by AluI, while DTUs I, IIa, and IIb yielded two fragments. All of the representatives of DTUs IId and IIe were heterozygous, displaying the uncut PCR product and the two digestion fragments. Analysis of CL Brener genomic sequence reads from TIGR revealed an ∼1:1 ratio of DTU IIc to IIb allele sequences. The histone H3 region displays maintenance of heterozygosity in DTUs IId and IIe with the same affinities shown in previous heterozygous loci.
Loci in which DTUs IIa and IIc showed greater affinity to DTU IIb
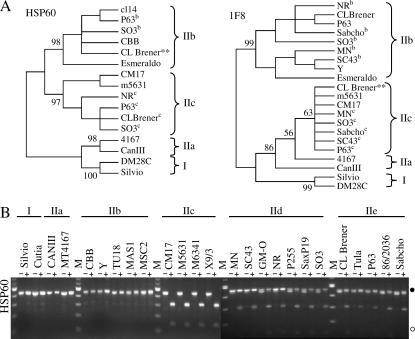
HSP60:
HSP60 is present in an array of ∼10 copies on a single chromosomal band of 3.5 Mbp in CL Brener (Sullivan et al. 1994; Cano et al. 1995). The HSP60 intergenic region possesses 11 SNPs and a 9-bp indel difference between DTUs I and IIb/IIe (Sturm et al. 2003).
Additional sequences obtained by PCR and database mining revealed that this region is heterozygous in some strains, including CL Brener. Four sequence classes were identified, correlating with DTUs I, IIa, IIb, and IIc (Table 6). Strains from DTUs IId and IIe possessed two sequence classes: a DTU IIb-like sequence and a DTU IIc-like sequence. A nonameric microsatellite (ACTATTATT) that corresponded to the originally described 9-bp indel was found in one to five copies in the different strains. Phylogenetic analysis of the HSP60 intergenic region supported the four clades of sequence variation represented by DTUs I, IIa, IIb, and IIc (Figure 3A). The heterozygous strains SO3, P63, and CL Brener were present in both the IIb and IIc DTUs. The length variation due to the microsatellite was evident in the two bands present in the lanes of undigested PCR products from the DTU IId strains (Figure 3B). A SNP at position 125 created an EcoRV restriction site in the DTU IIc-like sequences and allowed facile assessment of genotype by RFLP analysis. PCR products from strains of DTUs I, IIa, and IIb were not cut by EcoRV, whereas DTU IIc products were digested completely. Strains of DTUs IId and IIe showed the heterozygous phenotype, as seen by the combination of 440-, 300- and 140-bp products. In the DTU IId strains with two amplification products, the smaller band corresponding to the DTU IIc-like sequence disappeared upon EcoRV digestion. The ratio of bands in digestions of the DTU IId/IIe strains suggested that the relative amounts of DTU IIb-like and IIc-like products were not equal, possibly reflecting a difference in copy number of the two sequence classes. TIGR CL Brener reads revealed an ∼2:1 ratio of DTU IIc to IIb allele sequences. The HSP60 locus shows heterozygosity among the DTU IId/IIe strains associating with DTUs IIb and IIc.
TABLE 6.
Positions of SNPs for loci where DTUs IIa/IIc are more similar to DTU IIb
| Strain | SNP |
|---|---|
| HSP60 | |
| 1111122222222333333a | |
| 23591234813346799478999 | |
| 38146522463535457932125 | |
| Dm28c, SilvioX10 (I) | GCGTAAATCGCCATAACTAGCGC |
| CANIII (IIa) | GTGTAACCTGTCATAGTGGGAAA |
| M4167 (IIa) | GTGTAACCTGTCGTAGTGGGAAA |
| CM17 (IIc), M5631 (IIc) | ACATGGCTCATCGCGGTGGGCAC |
| NRb (IIe) | ACACGGCTCATCGCGGTGGGAAC |
| CL Brener,b P63b (IIe), SO3b (IId) | ACACGGCTCATCGCGGTGGGCAC |
| CBB, CL14c (IIb), Esmeraldo (IIb), SO3d (IId), P63d (IIe), CL Brenere (IIe) | GCGCAACTCCTTATAGTGGTCAC |
| 1F8 | |
| 1111111222222e | |
| 5890555599223369 | |
| 5735024801374671 | |
| Dm28c, Silvio X10 (I) | CGCGCGGTGGGCTTTT |
| CANIII, M4167 (IIa) | CGCGCGGGGGGCGCGC |
| M5631 (IIc) | CGCGCGGGGTGCGCGC |
| CM17 (IIc), MN,b SC43b (IId) | CGCGCGGGTTGCGCGC |
| SO3b (IId), CL Brener,b P63b, Sabcho109ab (IIe) | CGCGCGGGTTTTGCGC |
| CL Brener,f P63d (IIe) | TATTTTTGGGGCGCGT |
| Y(IIb), NRd (IIe), MNc,b SC43d (IId) | TATGCTTGGGGCGCGT |
| Sabcho109a,d SO3d (IIe) | TATTCTTGGGGCGCGT |
| Esmeraldo (IIb) | TATGCTGCGGGCGCGT |
CL14 is from GenBank X67473.
SNP positions relative to Dm28c (I).
DTU IIb-like allele in hybrid isolates.
CL Brener obtained from TIGR sequence TcTIGR_TCKCA96TR.
DTU IIc-like allele in hybrid isolates.
SNP positions relative to CL Brener (IIe) from GenBank BH843665.
CL Brener IIb-like obtained from TIGR sequence TcTIGR_TCGO128TF.
Figure 3.
Loci in T. cruzi where DTU IIa/IIc alleles are more similar to DTU IIb. (A) Unrooted neighbor-joining trees of HSP60 and 1F8 intergenic sequences reveal four DNA sequence classes. Superscript b or c represent DTU IIb or DTU IIc sequence types, respectively. **, sequence from CL Brener genomic read TCKCA96TR. (B) Agarose gel electrophoresis of PCR-RFLP products from the HSP60 locus of 25 strains. The DNA was stained with ethidium bromide. Size markers (M) are the BRL 1-kb ladders. • and ○ represent 0.5-kb and 0.1-kb bands, respectively. A SNP at position 125 creates the EcoRV polymorphism in the HSP60 locus (Table 2).
1F8—calcium-binding protein:
The 1F8 locus is present in ∼20 tandem copies on chromosome bands of 0.80 and 1.03 Mbp in CL Brener (Gonzalez et al. 1985; Sturm et al. 2003). Eight SNP differences are present between the DTU I and DTU IIb/IIe strains in this 355-bp intergenic region. DTU I and II 1F8 alleles localize to separate chromosome bands in CL Brener (Sturm et al. 2003).
Additional representatives of the 1F8 intergenic region were amplified and sequenced or retrieved from the TIGR database. The SNPs allowed for the discrimination of four sequence classes (Table 6). DTUs IIa and IIc were identical to one another at all but two positions. Both DTUs IId and IIe contained two sequences identified by screening with the DTU I and DTU II discriminating probes (Sturm et al. 2003). Neighbor-joining analysis produced four sequence clades, represented by DTUs I, IIa, IIb, and IIc (Figure 3A). The sequences identified in the DTU IId and IIe strains formed clades with the DTU IIb and IIc sequences. TIGR CL Brener genomic sequence reads revealed an ∼1:1 ratio of DTU IIc to IIb allele sequences, consistent with the relative hybridization intensities (Sturm et al. 2003). The 1F8 locus upholds the developing trend of DTU IId/IIe heterozygosity with DTU IIb and IIc allelic forms.
Mosaic sequences in two DTU IIe strains
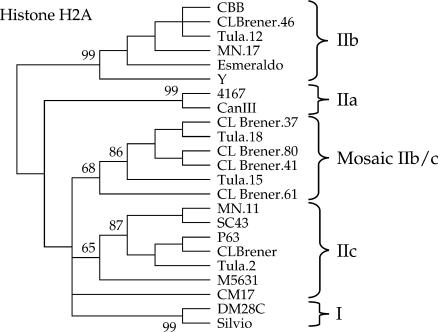
Histone H2A:
The histone H2A genes are present in two arrays in strain Y, containing about six copies of a 1.2-kb repeat and 18–24 copies of a 0.75-kb repeat (Puerta et al. 1994) that localize to two chromosomal bands of 0.7 and 0.9 Mbp in the CL Brener strain (Henriksson et al. 1995). The intergenic region of the 0.75-kb repeat contains 10 SNPs between DTU I and DTUs IIb/IIe (Sturm et al. 2003). In this analysis we included sequences generated by PCR for all DTUs, as well as multiple CL Brener sequences from the genome project. CL Brener sequences 37, 41, 46, 61, and 80 refer to sequences of TIGR-generated genomic reads TCPXO37TF, TCOWB41TR, TCHHN46TR, TCKLU61TF, and TCOUV80TR, respectively.
The histone H2A locus distinguished five sequence classes, including the core DTUs I/IIa/IIb/IIc and a new clade composed of mosaic DTU IIb/IIc variants (Figure 4). Alleles of DTU IId/IIe strains fell within both DTU IIb and IIc sequence clades, demonstrating the heterozygous nature of this locus. Mosaic sequences were observed initially from single cloned PCR products from MN and Tula strains, but excluded from the analysis. Multiple-cloned PCR products yielded the following sequences: from MN, one DTU IIb and five DTU IIc; from Tula, two DTU IIb and two DTU IIc. A single representative from each clade was included in the phylogenetic analysis, labeled MN.11, MN.17, Tula.2, and Tula.12. In addition, two Tula clones from a low-cycle number reaction revealed a new mosaic sequence. The validity of these sequences was strengthened through examination of TIGR CL Brener genomic reads: Four CL Brener histone H2A intergenic region sequences shared elements of a mosaic pattern including DTU IIb/IIc SNPs with the Tula mosaics (Table 7). In CL Brener these sequences were determined to be part of the DTU IIc array on the basis of their clonal mate pairs.
Figure 4.
Mosaicism of DTU IIb and IIc alleles within a multicopy locus for heterozygous DTU IId and IIe strains. Neighbor-joining phylogenetic tree of histone H2A intergenic sequences reveals five sequence classes. Mosaic IIb/IIc sequences from CL Brener and Tula strains form their own class.
TABLE 7.
Positions of histone H2A intergenic region SNPs
| Strain | SNP |
|---|---|
| 1111111111111112222223333333 | |
| 23444561333455566777780014672344445 | |
| 69047049489024847236724833905056788 | |
| Dm28c, Silvio X10 (I) | CAACGCTTGGTTTCTTTCTCGCCACTCCGCGTTTT |
| CANIII (IIa), M4167 (IIa) | CAACACTCGGTTTCTTCCTCGTCACCCCGCACTAC |
| CM17 (IIc), | CAGCGGGTGGTTTCTTCCTTGTCTCCCCGCGTTTT |
| M5631 (IIc) | CAGCGGTTGGTTTCTCCCTTGTCATTCCGCACTAT |
| CL Brener (IIe), P63 (IIe) | CAGCGGTTGGTTTCTTCTCAATTACTTCGCACTAT |
| SC43 (IId) | CAGCGGGTGGTTTCTTCTCAATTACTTCGCGTTTT |
| Tula.2 (IIe) | CAGCGGTTGGTTTCTTCTCAATCATTTCGCGTTTT |
| MN.11 (IId) | CAGCGGTTGGTTTCTTCTCAATCATTTCGCGTTTT |
| Tula.15 (IIe) | TGATACGTGGCTTTCTCCTCGTCTCTCCGCGTTTT |
| Tula.18 (IIe) | TGATACGTGGCTTTCTCCTCGTCTCTCCGCACTAT |
| CL Brener.80 | TAGTTCGTGGCTTTCTCCTCGTCTCTACGCACTAT |
| CL Brener.41, CL Brener.37 | TAGTTCGTGGCTTTCTCCTCGTCTCTCCGCACTAT |
| CL Brener.61 | CGGCCGGTGGCTTTCTCCTCGTCTCTCCGCACTAT |
| MN.17 (IId) | CAACACTCTTTCCTTGCCTCATTGCTCTCTACCAT |
| Y (IIb) | CAACACTCTTTCCTTTCTCCATTGCTCTCTACCAT |
| Esmeraldo (IIb) | CAACACTCTTTCCTTTCCTCATTGCTCTCTACAAT |
| CBB (IIb), Tula.12 (IId), CL Brener.46 (IIe) | CAACACTCTTTCCTTGCCTCATTGCTCTCTACTAT |
SNP position is relative to CL Brener (IIe).
Mosaic sequences are relatively straightforward to identify by SNP haplotypes; however, they can cause ambiguity in standard phylogenetic assignments by lowering bootstrap values or relocating branches and in RFLP analyses by altering restriction sites. The presence of unique SNPs for the CL Brener and Tula mosaic sequences suggests that they have a common origin; their complexity and similarity makes it unlikely that the PCR-generated Tula sequences are due to artifacts of amplification. We have used the example of histone H2A to illustrate a process that is likely to be ongoing and widespread within the heterozygous T. cruzi strains: interallelic recombination as witnessed by mosaic SNPs. This process will generate a new allele.
DTUs I, IIa, IIb, and IIc are homozygous
Our analysis of nine loci above in representatives of all six DTUs revealed a single class of highly conserved sequence at each locus in DTUs I, IIa, IIb, and IIc. Averaged across all loci, DTUs I and IIb were the most divergent from one another (Table 3), indicating that these two DTUs represent the most ancient division within the T. cruzi taxon. Most similar were DTUs I and IIc, followed closely by DTUs IIa and IIc (Table 3), supporting the common ancestry of DTUs IIa and IIc, formerly referred to as Zymodeme 3, and the contribution of DTU I to their hybrid genomes. The recombination of the DTU I and IIb SNPs in the DTU IIa/IIc groups distinguished the latter two DTUs from the first two, and the accumulation of specific mutations within those DTUs distinguished them from one another. The remaining DTUs IId and IIe contained two distinct alleles at eight of the nine loci analyzed, and these DTUs were not substantially different from one another. These data are consistent with previous observations on the heterozygosity of DTUs IId and IIe.
DISCUSSION
A picture of the current population structure of T. cruzi is emerging; however, the comparative genetic structure of all DTUs has been investigated at only a few loci (Machado and Ayala 2001; Gaunt et al. 2003). Here we analyzed an additional nine loci in representatives of the six DTUs. We show that allelic classes present in DTUs I, IIa, IIb, and IIc, as found in previous studies (Machado and Ayala 2001), define DTUs I, IIa, IIb, IIc, and IId/IIe. In all cases, DTUs I, IIa, IIb, and IIc were homozygous at the tested loci. In contrast, DTUs IId and IIe were heterozygous at all but one of the loci tested, possessing sequences similar to IIb and IIc DTUs. Although DTUs IIa and IIc were homozygous at all loci examined, the sequences shared some SNPs in common with DTU I and DTU IIb, indicative of homologous recombination between the parental alleles in a former heterozygous state. Unique DTU-defining SNPs support a substantial evolutionary divergence between DTUs IIa and IIc.
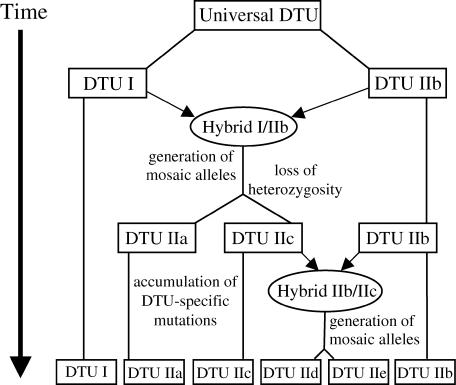
The revelation that T. cruzi can undergo genetic exchange and homologous recombination (Gaunt et al. 2003) provides a mechanism by which to explain the observations of heterozygosity and mosaic SNP haplotypes. The current population structure of T. cruzi reveals the fate of two distinct hybridization events separated by periods of clonal evolution (Figure 5). We hypothesize the initial clonal evolution of two lineages (ancestral DTUs I and IIb) from a common ancestor (universal DTU). The first hybridization event was a fusion between the ancestral DTUs, resulting in a “hybrid progeny” in which mosaic alleles could form (common ancestor of IIa and IIc). The eventual loss of heterozygosity in the progeny was followed by independent clonal evolution to yield DTUs IIa and IIc. It is possible that DTUs IIa and IIc arose independently, but we consider this scenario less likely due to many SNPs shared between the two DTUs. A second hybridization between DTU IIb and DTU IIc generated DTUs IId and IIe. Similar to the proposed evolution of DTUs IIa and IIc, the extensive heterozygosity in DTUs IId and IIe may resolve toward homozygosity. Heterozygosity may be maintained, particularly in genes involved in host-pathogen interactions, if it serves an adaptive advantage such as extending host range or improving cell invasion. DTU fusion provides the opportunity for the formation of an “optimized allele,” for example, in genes that encode housekeeping proteins that are required in general aspects of cell metabolism.
Figure 5.
A schematic of the evolution of T. cruzi groups. Boxes represent extant groups and their ancestral progenitors. The lines connecting boxes represent periods of clonal reproduction. Arrows represent contributions of various DTUs to form new hybrid strains. From a common ancestral universal genotype, two different genotypes arose that are represented today as DTUs I and IIb. Strains from these DTUs subsequently hybridized to produce DTUs IIa and IIc. A second hybridization between strains from DTUs IIb and IIc produced DTUs IId and IIe.
Our hypothesis on the formation of hybrids includes several processes: Two DTUs of T. cruzi undergo a genetic fusion resulting in the creation of a heterozygous DTU with an aneuploid or polyploid genome, such as DTUs IId/IIe; recombination occurs between alleles of the heterozygotes, resulting in mosaic sequences; gene conversion homogenizes loci; chromosome loss restores diploidy resulting in a homozygous diploid organism such as DTUs IIa/IIc. The order of these events is not fixed and could vary for different loci. This model resembles a more complex version of the population structure observed in the haploid protozoan Toxoplasma gondii, where the three predominant extant lineages possess one of two allele classes and appear to be the result of clonal evolution of three siblings from a possible single meiotic event (reviewed in Grigg and Suzuki 2003; Volkman and Hartl 2003).
DTUs IIa and IIc represent the products of a fusion event between a DTU I strain and DTU IIb strain in the distant past. Each of the four reference DTUs are distinct from one another and from the other DTUs at multiple positions (Table 3). The shared SNPs that define DTUs IIa and IIc support their evolutionary association as progeny of the same fusion and resolution events prior to their separation and accumulation of individual defining SNPs. For our evolutionary pedigree, we have adopted the most parsimonious assumption that the hybrids are the result of a single hybridization event based on the presumed rarity of genetic exchange in this species. However, we cannot exclude the possibility of multiple sequential hybridizations and backcrossing with parental strains, a.k.a. reticulate evolution, in the generation of the extant strains.
The identification of the parental lineages of DTUs IIa and IIc resolves the apparently conflicting interpretations of whether these DTUs are closer to DTU I or DTU IIb. In fact, there is no conflict, as both parental DTUs have contributed to DTU IIa and IIc progeny. The recombination of parental genes to generate the current mosaic alleles seen in DTUs IIa and IIc represent a spectrum of allelic intermediates varying from DTU I-like to DTU IIb-like. The existence of heterozygous loci in DTU IIa and IIc strains has not been eliminated.
Heterozygosity and diploidy was indicated by isoenzyme typing and genotyping of various strains of T. cruzi. Heterozygous enzyme activity profiles have been described for phosphoglucomutase, 6-phosphogluconate dehydrogenase, glutamate dehydrogenase NADP+, and malic enzyme (Tibayrenc et al. 1986; Carrasco et al. 1996), while heterozygous phenotypes and genotypes were reported for aldolase, glucose phosphate isomerase and gluteraldehyde 3-phosphatase dehydrogenase (Bogliolo et al. 1996), and DHFR-TS and TR genes (Machado and Ayala 2001). Chromosomal size variation and RFLPs in stocks of hybrid T. cruzi strains have been seen using 18 ESTs (Pedroso et al. 2003), suggesting that many more heterozygous alleles await identification.
DTUs IId and IIe originated from a fusion between a DTU IIc strain and a DTU IIb strain on the basis of this and other studies (Machado and Ayala 2001; Brisse et al. 2003; Gaunt et al. 2003; Elias et al. 2005). The heterozygosity may reflect the relatively recent fusion of the original parentals. Individual strains showed evidence of specific recombination events between the parental alleles, indicating that homologous recombination is an active and ongoing process in the T. cruzi nucleus. We propose that the strains within these heterozygous DTUs are in the process of active reshuffling of their alleles and, following the apparent precedent set by the DTU I/IIb fusion, may subsequently resolve to near or complete homozygosity at all loci. However, the advantages of a heterozygous state are well documented and thus may persist in these strains, presenting an example of hybrid vigor. Indeed, these hybrids are isolated frequently from infected individuals throughout the southern geographical range of the disease (Barnabé et al. 2000, 2001a). Heterozygous hybrids may possess a more successful phenotype that has adapted to, and conquered, a wide range of ecological niches as proposed elsewhere (Widmer et al. 1987).
DTUs IId and IIe were not differentiated in our analysis, indicating that their distinction is subtle, although sufficient to be detected in alternative analyses. The IId and IIe DTUs were distinguished originally on RAPD profiles and MLEE differences for GPI, glucose-6 phosphate dehydrogenase, glutamate dehydrogenase 1 and 2, peptidase 1, and phosphoglucomutase (Tibayrenc and Ayala 1988). DNA sequence variation at these protein-coding loci should allow precise molecular differentiation of the IId and IIe DTUs. Divergence among DTU IId and DTU IIe strains is suggestive of the evolution and loss of heterozygosity of clonal populations from the original hybridization event. In this regard it resembles the beginning of the homogenization process that we have postulated to generate the precursor hybrid of the DTU IIa/IIc branch.
An example of the loss of heterozygosity in the DTU IId/IIe isolates is the rRNA locus. Heterozygous sequences in the D7 region of rRNA were described in DTU IId strain NR, MN, and SO3; however, this locus is homozygous in DTU IId strain SC43 and in DTU IIe strains CL Brener and Tulahuen (Souto et al. 1996). The result is the retention of different alleles in these DTUs, such that CL Brener and Tulahuen are type 1 (DTU IIb) and SC43 is type 2 (DTU IIc) at this locus. The genotype of the D7 rRNA locus in these strains follows the prediction of our hypothesis for the resolution of heterozygosity (summarized in Table 1). The adjacent rRNA promoter region follows a corresponding pattern of DTU affiliation for MN, CL Brener, and SC43 (Brisse et al. 2003).
The T. cruzi genome project reference strain CL Brener is a member of the hybrid DTU IIe. Cumulative analyses indicate the presence of 4 homozygous loci and 13 heterozygous loci in CL Brener (Table 8). However, since the chromosomal location of all the loci are not known, this number cannot be translated into a precise count of homozygous and heterozygous chromosomes. The assignment of alleles with specific chromosome bands (haplotypes) in CL Brener will be problematic given the heterozygous nature of the strain. A reasonable solution to this problem is the sequencing of representatives of the “parental” DTUs, IIb and IIc. Preliminary DNA sequencing (2.5× coverage) of the Esmeraldo strain from DTU IIb has been completed and may provide supporting information to allow further assembly of the CL Brener parental chromosomes. The comparative analysis of these genomes in toto will provide greater details about the resolution process in a much more effective manner than the locus-by-locus strategy that was previously the only avenue; indeed, this study represents but a scratch on the surface of the total genomic information. Additional genome sequencing of representatives from DTUs I, IIa, and IIc will provide the greatest insights into the spectrum of biological diversity encountered in T. cruzi.
TABLE 8.
Summary of genotypes of hybrid groups IId/IIe
| Gene | Chromosomea | Genotype | Parent | Citation |
|---|---|---|---|---|
| SL RNA | XVI, XV | Homozygous | IIb | Souto et al. (1996) |
| LSU D7 rRNA | XIV | Homozygous | IIb | Stolf et al. (2003) |
| Hsp70 | XIII | Homozygous | IIb | This report |
| TSSA | ND | Homozygous | IIb | Campbell et al. (2004); Di Noia et al. (2002) |
| SLA1 snoRNA | XVI, XV | Heterozygous | IIb/IIc | This report |
| Hsp60 | XX | Heterozygous | IIb/IIc | This report |
| Histone H1 | XVII | Heterozygous | IIb/IIc | This report |
| Histone H2A | VII, III | Heterozygous | IIb/IIc | This report |
| Histone H2B | XX | Heterozygous | IIb/IIc | This report |
| Histone H3 | XII, XI | Heterozygous | IIb/IIc | This report |
| 1F8 | XI, V | Heterozygous | IIb/IIc | This report |
| GPI | ND | Heterozygous | IIb/IIc | Gaunt and Miles (2000) |
| TCP | ND | Heterozygous | IIb/IIc | Gaunt and Miles (2000); Robello et al. (2000) |
| TRS | XVIII | Heterozygous | IIb/IIc | Tran et al. (2003) |
| MSH2 | ND | Heterozygous | IIb/IIc | Augusto-Pinto et al. (2003) |
| DHFR-TS | ND | Heterozygous | IIb/IIc | Machado and Ayala (2002) |
| TR | ND | Heterozygous | IIb/IIc | Machado and Ayala (2002) |
ND, not determined.
Chromosome assignments based on CL Brener Cano et al. (1995); Porcile et al. (2003).
Acknowledgments
We thank Michel Tibayrenc (IRD, Montpellier, France), Bianca Zingales and Octavio Fernandes for DNA samples, and Antonio Teixeira (University of Brasilia, Brazil), Robert Hitchcock, Sean Thomas, Jesse Zamudio, Gusti Zeiner, and Carmen Zelaya for helpful discussions and critical reading of the manuscript. S.J.W. is a pre-doctoral trainee of the UCLA Bioinformatics Integrative Graduate Education and Research Traineeship program funded by National Science Foundation (NSF) grant DGE9987641. Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of Trypanosoma cruzi was funded by the National Institute of Allergy and Infectious Disease (NIAID).
References
- Anonymous, 1999. Recommendations from a satellite meeting. Mem. Inst. Oswaldo Cruz 94 (Suppl. 1): 429–432. [DOI] [PubMed] [Google Scholar]
- Åslund, L., L. Carlsson, J. Henriksson, M. Rydåker, G. C. Toro et al., 1994. A gene family encoding heterogenous histone H1 proteins in Trypanosoma cruzi. Mol. Biochem. Parasitol. 65: 317–330. [DOI] [PubMed] [Google Scholar]
- Augusto-Pinto, L., S. M. R. Teixeira, S. D. J. Pena and C. R. Machado, 2003. Single-nucleotide polymorphisms of the Trypanosoma cruzi MSH2 gene support the existence of three phylogenetic lineages presenting differences in mismatch-repair efficiency. Genetics 164: 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabé, C., S. Brisse and M. Tibayrenc, 2000. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology 120: 513–526. [DOI] [PubMed] [Google Scholar]
- Barnabé, C., K. Neibauer, A. Solari and M. Tibayrenc, 2001. a Trypanosoma cruzi: presence of the two major phylogenetic lineages and of several lesser discrete typing units (DTUs) in Chile and Paraguay. Acta Tropica 78: 127–137. [DOI] [PubMed] [Google Scholar]
- Barnabé, C., R. Yaeger, O. Pung and M. Tibayrenc, 2001. b Trypanosoma cruzi: a considerable phylogenetic divergence indicates that the agent of Chagas disease is indigenous to the native fauna of the United States. Exp. Parasitol. 99: 73–79. [DOI] [PubMed] [Google Scholar]
- Bogliolo, A. R., L. Lauria-Pires and W. C. Gibson, 1996. Polymorphisms in Trypanosoma cruzi: evidence of genetic recombination. Acta Tropica 61: 31–40. [DOI] [PubMed] [Google Scholar]
- Bontempi, E. J., B. M. Porcel, J. Henriksson, L. Carlsson, M. Rydåker et al., 1994. Genes for histone H3 in Trypanosoma cruzi. Mol. Biochem. Parasitol. 66: 147–151. [DOI] [PubMed] [Google Scholar]
- Brakenhoff, R. H., J. G. Schoenmakers and N. H. Lubsen, 1991. Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 19: 1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenière, S. F., M.-F. Bosseno, J. Telleria, B. Bastrenta, N. Yacsik et al., 1998. Different behavior of two Trypanosoma cruzi major clones: transmission and circulation in yopung Bolivian patients. Exp. Parasitol. 89: 285–295. [DOI] [PubMed] [Google Scholar]
- Brisse, S., C. Barnabé, A.-L. Bañuls, I. Sidibé, S. Noël et al., 1998. A phylogenetic analysis of the Trypanosoma cruzi genome project CL Brener reference strain by multilocus enzyme electrophoresis and multiprimer random amplified polymorphic DNA fingerprinting. Mol. Biochem. Parasitol. 92: 253–263. [DOI] [PubMed] [Google Scholar]
- Brisse, S., C. Barnabe and M. Tibayrenc, 2000. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int. J. Parasitol. 30: 34–44. [DOI] [PubMed] [Google Scholar]
- Brisse, S., J. Verhoef and M. Tibayrenc, 2001. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int. J. Parasitol. 31: 1218–1226. [DOI] [PubMed] [Google Scholar]
- Brisse, S., J. Henriksson, C. Barnabé, E. J. P. Douzery, D. Berkvens et al., 2003. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect. Genet. Evol. 2: 173–183. [DOI] [PubMed] [Google Scholar]
- Campbell, D. A., S. J. Westenberger and N. R. Sturm, 2004. The determinants of Chagas Disease: Connecting parasite and host genetics. Curr. Mol. Med. 4: 549–562. [DOI] [PubMed] [Google Scholar]
- Cano, M. I., A. Gruber, M. Vazquez, A. Cortés, M. J. Levin et al., 1995. Molecular karyotype of clone CL Brener chosen for the Trypanosoma cruzi genome project. Mol. Biochem. Parasitol. 71: 273–278. [DOI] [PubMed] [Google Scholar]
- Carrasco, H. J., I. A. Frame, S. A. Valente and M. A. Miles, 1996. Genetic exchange as a possible source of genomic diversity in sylvatic populations of Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 54: 418–424. [DOI] [PubMed] [Google Scholar]
- Coura, J. R., A. C. V. Junqueira, O. Fernandes, S. A. S. Valente and M. A. Miles, 2002. Emerging Chagas disease in Amazonian Brazil. Trends Parasitol. 18: 171–176. [DOI] [PubMed] [Google Scholar]
- Di Noia, J. M., C. A. Buscaglia, C. R. De Marchi, I. C. Almeida and A. C. C. Frasch, 2002. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J. Exp. Med. 195: 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J. A., 1984. The natural heterogeneity of Trypanosoma cruzi: Biological and medical implications. J. Cell. Biochem. 24: 357–371. [DOI] [PubMed] [Google Scholar]
- Elias, M. C., N. Vargas, L. Tomazi, A. Pedroso, B. Zingales et al., 2005. Comparative analysis of genomic sequences suggests that Trypanosoma cruzi CL Brener contains two sets of non-intercalated repeats of satellite DNA that correspond to T. cruzi I and T. cruzi II types. Mol. Biochem. Parasitol. 140: 221–227. [DOI] [PubMed] [Google Scholar]
- Fernandes, O., R. P. Souto, J. A. Castro, J. B. Pereira, N. C. Fernandes et al., 1998. Brazilian isolates of Trypanosoma cruzi from humans and triatomines classified into two lineages using mini-exon and ribosomal RNA sequences. Am. J. Trop. Med. Hyg. 58: 807–811. [DOI] [PubMed] [Google Scholar]
- García-Salcedo, J. A., J. L. Oliver, R. P. Stock and A. González, 1994. Molecular characterization and transcription of the histone H2B cluster from the protozoan parasite Trypanosoma cruzi. Mol. Micro. 16: 1033–1043. [DOI] [PubMed] [Google Scholar]
- Gaunt, M., and M. Miles, 2000. The ecotopes and evolution of triatomine bugs (triatominae) and their associated trypanosomes. Mem. Inst. Oswaldo Cruz 95: 557–565. [DOI] [PubMed] [Google Scholar]
- Gaunt, M. W., M. Yeo, I. A. Frame, J. R. Stothard, H. J. Carrasco et al., 2003. Mechanism of genetic exchange in American trypanosomes. Nature 421: 936–939. [DOI] [PubMed] [Google Scholar]
- Gibson, W. C., and M. A. Miles, 1986. The karyotype and ploidy of Trypanosoma cruzi. EMBO J. 5: 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, A., T. J. Lerner, M. Huecas, B. Sosa-Pineda, N. Nogueira et al., 1985. Apparent generation of a segmented mRNA from two separate gene families in Trypanosoma cruzi. Nucleic Acids Res. 13: 5789–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg, M. E., and Y. Suzuki, 2003. Sexual recombination and clonal evolution of virulence in Toxoplasma. Microbes Infect. 5: 685–690. [DOI] [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Henriksson, J., B. Porcel, M. Rydåker, A. Ruiz, V. Sabaj et al., 1995. Chromosome specific markers reveal conserved linkage groups in spite of extensive chromosomal size variation in Trypanosoma cruzi. Mol. Biochem. Parasitol. 73: 63–74. [DOI] [PubMed] [Google Scholar]
- Kawashita, S. Y., G. F. O. Sanson, O. Fernandes, B. Zingales and M. R. S. Briones, 2001. Maximum-likelihood divergence date estimates based on rRNA gene sequences suggest two scenarios of Trypanosoma cruzi intraspecific evolution. Mol. Biol. Evol. 18: 2250–2259. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Macedo, A. M., R. P. Oliveira and S. D. Pena, 2002. Chagas disease: role of parasite genetic variation in pathogenesis. Expert Rev. Mol. Med. 2002: 1–16. [DOI] [PubMed] [Google Scholar]
- Macedo, A. M., C. R. Machado, R. P. Oliveira and S. D. Pena, 2004. Trypanosoma cruzi: genetic structure of populations and relevance of genetic variability to the pathogenesis of chagas disease. Mem. Inst. Oswaldo Cruz 99: 1–12. [DOI] [PubMed] [Google Scholar]
- Macedo, A. M., and S. D. J. Pena, 1998. Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas disease. Parasitol. Today 14: 119–124. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., and F. J. Ayala, 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98: 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. A., and F. J. Ayala, 2002. Sequence variation in the dihydrofolate reductase-thymidylate synthase (DHFR-TS) and trypanothione reductase (TR) genes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 121: 33–47. [DOI] [PubMed] [Google Scholar]
- Mendonça, M. B., N. S. Nehme, S. S. Santos, E. Cupolillo, N. Vargas et al., 2002. Two main clusters within Trypanosoma cruzi zymodeme 3 are defined by distinct regions of the ribosomal RNA cistron. Parasitology 124: 177–184. [DOI] [PubMed] [Google Scholar]
- Miles, M. A., A. Souza, M. Povoa, J. J. Shaw, R. Lainson et al., 1978. Isozymic heterogeneity of Trypanosoma cruzi in the first autochthonous patients with Chagas' disease in Amazonian Brazil. Nature 272: 819–821. [DOI] [PubMed] [Google Scholar]
- Nitz, N., C. Gomes, A. de Cassia Rosa, M. R. D'Souza-Ault, F. Moreno et al., 2004. Heritable integration of kDNA minicircle sequences from Trypanosoma cruzi into the avian genome: insights into human Chagas disease. Cell 118: 175–186. [DOI] [PubMed] [Google Scholar]
- Pedroso, A., E. Cupolillo and B. Zingales, 2003. Evaluation of Trypanosoma cruzi hybrid stocks based on chromosomal size variation. Mol. Biochem. Parasitol. 129: 79–90. [DOI] [PubMed] [Google Scholar]
- Porcile, P. E., M. R. M. Santos, R. T. Souza, N. V. Verbisck, A. Brandão et al., 2003. A refined molecular karyotype for the reference strain of the Trypanosoma cruzi genome project (clone CL Brener) by assignment of chromosome markers. Gene 308: 53–65. [DOI] [PubMed] [Google Scholar]
- Puerta, C., J. Martin, C. Alonso and M. C. López, 1994. Isolation and characterization of the gene encoding histone H2A from Trypanosoma cruzi. Mol. Biochem. Parasitol. 64: 1–10. [DOI] [PubMed] [Google Scholar]
- Requena, J. M., M. C. Lopez, A. Jiminez-Ruiz, J. C. de la Torre and C. Alonso, 1988. A head-to-tail tandem organization of hsp70 genes in Trypanosoma cruzi. Nucleic Acids Res. 16: 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robello, C., F. Gamarro, S. Castanys and F. Alvarez-Valin, 2000. Evolutionary relationships in Trypanosoma cruzi: molecular phylogenetics supports the existence of a new major lineage of strains. Gene 246: 331–338. [DOI] [PubMed] [Google Scholar]
- Souto, R. P., O. Fernandes, A. M. Macedo, D. A. Campbell and B. Zingales, 1996. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 83: 141–152. [DOI] [PubMed] [Google Scholar]
- Souto, R. P., and B. Zingales, 1993. Sensitive detection and strain classification of Trypanosoma cruzi by amplification of a ribosomal RNA sequence. Mol. Biochem. Parasitol. 62: 45–52. [DOI] [PubMed] [Google Scholar]
- Stolf, B. S., R. P. Souto, A. Pedroso and B. Zingales, 2003. Two types of ribosomal RNA genes in hybrid Trypanosoma cruzi strains. Mol. Biochem. Parasitol. 126: 73–80. [DOI] [PubMed] [Google Scholar]
- Sturm, N. R., N. S. Vargas, S. J. Westenberger, B. Zingales and D. A. Campbell, 2003. Evidence for multiple hybrid groups in Trypanosoma cruzi. Int. J. Parasitol. 33: 269–279. [DOI] [PubMed] [Google Scholar]
- Sullivan, M. A., C. L. Olson, A. G. Winquist and D. M. Engman, 1994. Expression and localization of Trypanosoma cruzi hsp60. Mol. Biochem. Parasitol. 68: 197–208. [DOI] [PubMed] [Google Scholar]
- Tanabe, K., N. Sakihama, A. Farnert, I. Rooth, A. Bjorkman et al., 2002. In vitro recombination during PCR of Plasmodium falciparum DNA: a potential pitfall in molecular population genetic analysis. Mol. Biochem. Parasitol. 122: 211–216. [DOI] [PubMed] [Google Scholar]
- Teixeira, A. R. L., 1987. The Stercorarian Trypanosomes, pp. 25–118 in Immune Responses in Parasitic Infections: Immunology, Immunopathology and Immunoprophylaxis, edited by E. J. L. Soulsby. CRC Press, Boca Raton, FL.
- Thomas, S., S. J. Westenberger, D. A. Campbell and N. R. Sturm, 2005. Intragenomic spliced leader RNA array analysis of kinetoplastids reveals unexpected transcribed region diversity in Trypanosoma cruzi. Gene 352: 100–108. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc, M., 1995. Population genetics of parasitic protozoa and other microorganisms. Adv. Parasitol. 36: 47–115. [DOI] [PubMed] [Google Scholar]
- Tibayrenc, M., 1998. Beyond strain typing and molecular epidemiology: Integrated genetic epidemiology of infectious disease. Parasitol. Today 14: 323–329. [DOI] [PubMed] [Google Scholar]
- Tibayrenc, M., 2003. Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evaluation. Kinetoplast. Biol. Dis. 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc, M., and F. J. Ayala, 1988. Isozyme variability in Trypanosoma cruzi, the agent of Chagas' disease: Genetical, taxomonical, and epidemiological significance. Evolution 42: 277–292. [DOI] [PubMed] [Google Scholar]
- Tibayrenc, M., and F. J. Ayala, 2002. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 18: 405–410. [DOI] [PubMed] [Google Scholar]
- Tibayrenc, M., P. Ward, A. Moya and F. J. Ayala, 1986. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc. Natl. Acad. Sci. USA 83: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, A.-N., B. Andersson, U. Pettersson and L. Åslund, 2003. Trypanothione synthetase locus in Trypanosoma cruzi CL Brener strain shows an extensive allelic divergence. Acta Tropica 87: 269–278. [DOI] [PubMed] [Google Scholar]
- Vago, A. R., L. O. Andrade, A. A. Leite, D. d'Avila Reis, A. M. Macedo et al., 2000. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease. Differential distribution of genetic types into diverse organs. Am. J. Pathol. 156: 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, N., A. Pedroso and B. Zingales, 2004. Chromosomal polymorphism, gene synteny and genome size in T. cruzi I and T. cruzi II groups. Mol. Biochem. Parasitol. 138: 131–141. [DOI] [PubMed] [Google Scholar]
- Volkman, S. K., and D. L. Hartl, 2003. Parasitology. A game of cat and mouth. Science 299: 353–354. [DOI] [PubMed] [Google Scholar]
- Widmer, G., J. A. Dvorak and M. A. Miles, 1987. Temperature modulation of growth rates and glucosephosphate isomerase isozyme activity in Trypanosoma cruzi. Mol. Biochem. Parasitol. 23: 55–62. [DOI] [PubMed] [Google Scholar]
- Yeo, M., N. Acosta, M. Llewellyn, H. Sanchez, S. Adamson et al., 2005. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int. J. Parasitol. 35: 225–233. [DOI] [PubMed] [Google Scholar]
- Zingales, B., M. E. S. Pereira, R. P. Oliveira, K. A. Almeida, E. S. Umezawa et al., 1997. Trypanosoma cruzi genome project: biological characteristics and molecular typing of clone CL Brener. Acta Tropica 68: 159–173. [DOI] [PubMed] [Google Scholar]
- Zingales, B., R. P. Souto, R. H. Mangia, C. V. Lisboa, D. A. Campbell et al., 1998. Molecular epidemiology of American trypanosomiasis in Brazil based on dimorphisms of rRNA and mini-exon gene sequences. Int. J. Parasitol. 28: 105–112. [DOI] [PubMed] [Google Scholar]