Abstract
In Arabidopsis thaliana, heterochromatin formation is guided by double-stranded RNA (dsRNA), which triggers methylation of histone H3 at Lys-9 (H3 mK9) and CG plus non-CG methylation on identical DNA sequences. At heterochromatin targets including transposons and centromere repeats, H3 mK9 mediated by the Su(var)3-9 homologue 4 (SUVH4)/KYP histone methyltransferase (MTase) is required for the maintenance of non-CG methylation by the CMT3 DNA MTase. Here, we show that although SUVH4 is the major H3 K9 MTase, the SUVH5 protein also has histone MTase activity in vitro and contributes to the maintenance of H3 mK9 and CMT3-mediated non-CG methylation in vivo. Strikingly, the relative contributions of SUVH4, SUVH5, and a third related histone MTase, SUVH6, to non-CG methylation are locus-specific. For example, SUVH4 and SUVH5 together control transposon sequences with only a minor contribution from SUVH6, whereas SUVH4 and SUVH6 together control a transcribed inverted repeat source of dsRNA with only a minor contribution from SUVH5. This locus-specific variation suggests different mechanisms for recruiting or activating SUVH enzymes at different heterochromatic sequences. The suvh4 suvh5 suvh6 triple mutant loses both monomethyl and dimethyl H3 K9 at target loci. The suvh4 suvh5 suvh6 mutant also displays a loss of non-CG methylation similar to a cmt3 mutant, indicating that SUVH4, SUVH5, and SUVH6 together control CMT3 activity.
INTRODUCTION
Correct maintenance of heterochromatin patterns is essential for gene regulation and genome stability in eukaryotes (reviewed in Lippman and Martienssen, 2004; Matzke and Birchler, 2005). In many eukaryotes, including fission yeast, Drosophila, mammals, and plants, heterochromatin is marked by histone H3 at Lys-9 (H3 mK9). In some species, including mammals and plants, heterochromatin is also marked by cytosine methylation. A major mechanism for guiding H3 K9 and cytosine methylation to appropriate regions of the genome involves double-stranded RNA (dsRNA)–derived species, such as small RNA products of dicer ribonuclease cleavage. For example, in fission yeast, centromere repeat dsRNA is diced into small RNAs, which assemble into the RNA-induced initiation of transcriptional gene silencing effector complex to guide H3 mK9 to centromere repeat regions (Volpe et al., 2003; Verdel et al., 2004). In a potentially related mechanism, small RNAs transfected into human cancer cells can trigger H3 mK9 on identical promoter sequences, resulting in transcriptional silencing of a downstream gene (Ting et al., 2005). Furthermore, in plants, dsRNA-derived species generated from RNA viruses, transcribed inverted repeats, or products of RNA-dependent RNA polymerases can trigger both H3 K9 and cytosine methylation on identical DNA sequences (reviewed in Mathieu and Bender, 2004). However, effector complexes that mediate the connection between RNA and heterochromatin modifications have not yet been characterized in mammalian or plant systems.
Plant RNA-directed DNA methylation occurs in both CG and non-CG contexts. Genetic studies in Arabidopsis thaliana have implicated three structurally distinct cytosine methyltransferases (MTases) in this process: the DRM1/DRM2 cytosine MTases initiate new DNA methylation imprints in response to a dsRNA signal, whereas the MET1 and CMT3 cytosine MTases maintain methylation in CG and non-CG contexts, respectively (reviewed in Mathieu and Bender, 2004). The DRM cytosine MTases also contribute to the maintenance of non-CG methylation at some target regions, including an Arabidopsis SINE transposon, a direct repeat array MEA-ISR, and transgene reporters for RNA-directed DNA methylation (Cao and Jacobsen, 2002; Cao et al., 2003). However, CMT3 is the major MTase that maintains non-CG methylation at centromere repeats and transposons, including DNA transposons and long terminal repeat (LTR) retrotransposons (Bartee et al., 2001; Lindroth et al., 2001; Tompa et al., 2002; Kato et al., 2003; Lippman et al., 2003).
CMT3-mediated non-CG methylation depends on the Su(var)3-9 homologue 4 (SUVH4)/KYP (hereafter referred to as SUVH4) H3 K9 MTase, suggesting that CMT3 is guided to target sequences by the H3 mK9 modification (Jackson et al., 2002; Malagnac et al., 2002). Similarly, H3 mK9 guides cytosine methylation in the fungus Neurospora crassa (Tamaru and Selker, 2001; Tamaru et al., 2003) and in mouse (Lehnertz et al., 2003; Xin et al., 2003). However, at target loci including centromere repeats and transposons, suvh4 mutations confer a weaker loss of non-CG methylation than cmt3 mutations (Jackson et al., 2002; Malagnac et al., 2002; Lippman et al., 2003). The Arabidopsis genome encodes eight other SUVH putative H3 K9 MTases (Baumbusch et al., 2001), suggesting that some of these other SUVHs might contribute to the CMT3 non-CG methylation pathway.
SUVH6 has similar in vitro H3 K9 MTase activity to SUVH4 (Jackson et al., 2004). Therefore, we previously characterized the effects of a suvh6 mutation on H3 K9 and non-CG methylation at CMT3 targets, including transposons, centromere repeats, and the endogenous phosphoribosylanthranilate isomerase (PAI) Trp biosynthetic genes (Ebbs et al., 2005). In the Wassilewskija (Ws) ecotype of Arabidopsis, the PAI genes are arranged as a tail-to-tail inverted repeat of two genes, PAI1-PAI4, and two unlinked singlet genes, PAI2 and PAI3. Transcription through PAI1-PAI4 from a fortuitous unmethylated promoter upstream of PAI1 produces normally polyadenylated PAI1 transcripts and longer species that read into palindromic PAI4 sequences to form dsRNA signals for H3 K9 and cytosine methylation of PAI sequences (Melquist and Bender, 2003; Ebbs et al., 2005). Mutations in CMT3 strongly reduce non-CG methylation at PAI1-PAI4, PAI2, and PAI3 (Bartee et al., 2001). By contrast, mutations in SUVH4 reduce non-CG methylation on PAI2 and PAI3 but have no effect on methylation patterning at PAI1-PAI4 (Malagnac et al., 2002).
We found that double mutation of suvh4 and suvh6 strongly reduced H3 K9 and non-CG methylation on PAI1-PAI4, indicating that SUVH4 and SUVH6 act together to maintain epigenetic modifications at this transcribed inverted repeat locus (Ebbs et al., 2005). However, suvh4 suvh6 retained residual PAI1-PAI4 non-CG methylation relative to cmt3. Furthermore, the suvh6 mutation did not enhance partial demethylation of the Ta3 LTR retrotransposon, the Mu1 DNA transposon, or centromere repeats conferred by the suvh4 mutation relative to cmt3. Here, we show that SUVH5 also has H3 K9 MTase activity in vitro and acts in the CMT3 non-CG methylation pathway in vivo. The suvh4 suvh5 suvh6 triple mutant displays similar strong demethylation of target sequences to a cmt3 mutant at the PAI genes, Ta3 and Mu1 transposons, and centromere repeats, indicating that SUVH4, SUVH5, and SUVH6 together control CMT3-mediated non-CG methylation at these loci.
Importantly, SUVH4, SUVH5, and SUVH6 make different relative contributions to non-CG methylation at different loci. For example, although SUVH4 and SUVH6 control the majority of non-CG methylation at the PAI1-PAI4 transcribed inverted repeat, SUVH4 and SUVH5 control the majority of non-CG methylation at the Ta3 and Mu1 transposons. These results suggest locus-specific variation in the factors that recruit or activate SUVH enzymes at RNA-directed DNA methylation targets. Thus, the Arabidopsis SUVHs can potentially be exploited for genetic and biochemical identification of novel components that connect heterochromatin-modifying enzymes with dsRNA signals.
RESULTS
SUVH5 Has in Vitro H3 K9 MTase Activity
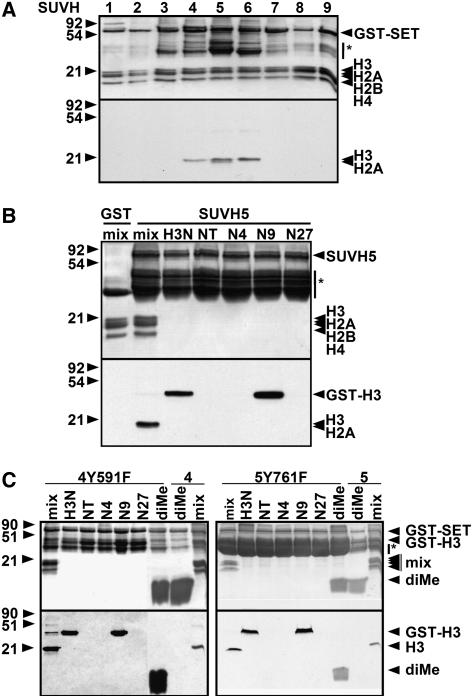
In previous work, we determined that a suvh4 suvh6 double H3 K9 MTase mutant retains residual non-CG methylation on the PAI genes, transposon sequences, and centromere repeats relative to a cmt3 cytosine MTase mutant (Ebbs et al., 2005). To identify H3 K9 MTases that might account for this residual non-CG methylation, we tested the other seven SUVH proteins encoded in Arabidopsis (Baumbusch et al., 2001) for H3 K9 MTase activity in vitro. The SUVH proteins carry a C-terminal catalytic domain (pre-SET, SET, and post-SET motifs), a central conserved YDG domain, and divergent N termini. To assess catalytic activity, we expressed and purified each SUVH protein as a glutathione S-transferase (GST) fusion to the catalytic domain. These recombinant proteins were incubated with a purified bovine histone mix (H1, H2A, H2B, H3, and H4) and S-adenosyl-[14C-methyl]l-Met as the methyl group donor (see Methods). Only SUVH4, SUVH5, and SUVH6 methylated bovine H3 (conserved with Arabidopsis H3; Gendrel et al., 2002) under these conditions (Figure 1A). SUVH5 also methylated bovine H2A (Figure 1A) and the Arabidopsis H2A variants HTA2, HTA7, and HTA13 in vitro (see Supplemental Figure 1 online). Mutational analysis of an HTA13 substrate showed that either of two N-terminal lysines could be modified by SUVH5 (see Supplemental Figure 1 online). SUVH4 did not methylate bovine H2A either in a mix of histones or with H2A as the sole substrate, and it did not methylate Arabidopsis HTA13 in vitro (Figure 1A; data not shown).
Figure 1.
SUVH5 Has in Vitro Histone MTase Activity.
Coomassie blue–stained gels (top panels) and their fluorograms (bottom panels) are shown. GST-SUVH indicates the positions of full-length recombinant SUVH proteins, and asterisks indicate GST-SUVH truncations. Positions of protein molecular mass markers in kilodaltons are shown at left.
(A) In vitro histone MTase assays were performed with a whole histone mix and each of the nine GST-tagged SUVH proteins.
(B) Specificity of SUVH5 for H3 K9. GST-SUVH5 was incubated with a bovine histone mix or a GST-H3 N-terminal peptide as indicated: H3N is the wild type, N4 has K9 and K27 mutated to Arg, N9 has K4 and K27 mutated to Arg, N27 has K4 and K9 mutated to Arg, and NT has K4, K9 and K27 mutated to Arg (Tachibana et al., 2001). GST alone was incubated with the histone mix as a control.
(C) Tyr-to-Phe mutations convert SUVH4 or SUVH5 from an H3 K9 mono-/di-MTase to an H3 K9 tri-MTase. GST-SUVH4Y591F, GST-SUVH4, GST-SUVH5Y761F, and GST-SUVH5 were assayed for activity on bovine histone mix (mix), GST-H3 peptides, or a biotinylated H3 K9 dimethyl peptide (diMe).
To determine the specificity of SUVH5 on H3, we tested for activity against a series of recombinant GST-H3 peptide substrates (residues 1 to 57): wild type (H3N), a mutant with only Lys-4 (N4), a mutant with only Lys-9 (N9), a mutant with only Lys-27 (N27), or a mutant with no Lys residues (NT) (Tachibana et al., 2001). SUVH5 methylated only the H3N and N9 substrates (Figure 1B), indicating specificity for H3 K9.
Histone MTases can add one, two, or three methyl groups to a substrate Lys side chain. Structural analysis of a known histone mono-MTase, the mammalian SET7/9 enzyme, versus a known histone tri-MTase, the Neurospora DIM-5 enzyme, revealed that this difference in activity can be attributed to a conserved active site position that carries a Tyr in the mono-MTase versus a less bulky Phe in the tri-MTase (Zhang et al., 2003; Collins et al., 2005). All of the Arabidopsis SUVH proteins carry a Tyr at this position and thus are predicted to be mono- or di-MTases but not tri-MTases. Consistent with this structural prediction, both SUVH4 and SUVH6 have been shown to add one or two but not three methyl groups to the target Lys on a peptide substrate in vitro (Jackson et al., 2004). Furthermore, immunoblot, immunocytology, and chromatin immunoprecipitation (ChIP) experiments performed with antibodies specific for monomethylated, dimethylated, or trimethylated H3 K9 suggest that Arabidopsis preferentially uses dimethylated H3 K9 as a mark for heterochromatin formation and is deficient in trimethylated H3 K9 (Jackson et al., 2004).
To test the MTase properties of SUVH5 relative to SUVH4, we engineered tri-MTase mutant variants of these enzymes with the conserved Tyr mutated to Phe (SUVH4Y591F and SUVH5Y761F) and assayed their ability to add a third methyl group to an H3 dimethyl K9 peptide substrate. Both mutant enzymes methylated the dimethylated substrate, demonstrating that they have tri-MTase activity, in contrast with the corresponding wild-type enzymes, which did not methylate this substrate (Figure 1C). The tri-MTase mutant enzymes maintained specificity for H3 K9 similar to the wild-type enzymes. These results show that SUVH4 and SUVH5 each can be converted to a tri-MTase by a single mutation, without disruption of overall enzyme function. These results also suggest that, like SUVH4 and SUVH6, SUVH5 is a mono-/di-MTase. Interestingly, besides its primary MTase activity against H3, the SUVH4Y591F mutant displayed low-level secondary MTase activity against one of the other histones in the bovine histone substrate mix (Figure 1C). However, we did not further characterize this secondary activity.
SUVH5 Contributes to PAI Non-CG Methylation and Transcriptional Silencing in the Absence of SUVH4 and SUVH6
To understand the role of SUVH5 in the plant, we analyzed two suvh5 insertion alleles. The suvh5-1 allele is a T-DNA insertion into the catalytic domain–encoding region of the gene isolated in the Ws ecotype, and the suvh5-2 allele is a T-DNA insertion into the catalytic domain–encoding region of the gene isolated in the Columbia (Col) ecotype. Both alleles were crossed into the Ws pai1 reporter background (see below) as single mutations, double mutations with suvh4, or triple mutations with suvh4 and suvh6. Mutant plants were assayed for changes in non-CG methylation and H3 mK9 at representative heterochromatic loci targeted by CMT3, including the PAI Trp biosynthetic genes, the Ta3 LTR retrotransposon, and the Mu1 DNA transposon. PAI and transposon sequences were also tested for transcriptional reactivation.
In wild-type Ws, the PAI1 gene in the PAI1-PAI4 inverted repeat provides the major source of PAI enzyme, as a result of expression from a fortuitous upstream unmethylated promoter. To monitor the transcriptional activity of the functional but silenced PAI2, we isolated a pai1 missense mutation that reduces PAI1 enzyme activity without affecting the dsRNA signal for PAI DNA methylation (Bartee and Bender, 2001). pai1 displays a number of PAI-deficient phenotypes, including blue fluorescence under UV light. These phenotypes can be alleviated by mutations that release the transcriptional silencing of PAI2, for example suvh4 and cmt3 (Bartee et al., 2001; Malagnac et al., 2002). The suvh5-1 and suvh5-2 alleles were crossed into the Ws pai1 background so that we could monitor their effects on PAI2 transcriptional silencing and PAI DNA methylation.
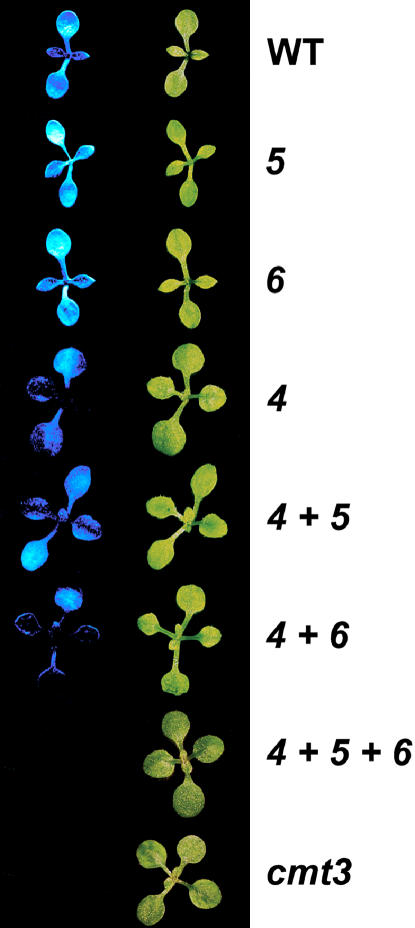
In the blue fluorescence assay for PAI2 silencing, the suvh5 mutation did not affect fluorescence in either the pai1 or pai1 suvh4 background relative to the parental background (Figure 2), as observed previously for the suvh6 mutation (Ebbs et al., 2005). Both pai1 and pai1 suvh5 displayed strong seedling fluorescence, and the pai1 suvh4, pai1 suvh4 suvh5, and pai1 suvh4 suvh6 mutants all displayed reduced but still detectable seedling fluorescence. By contrast, pai1 suvh4 suvh5 suvh6 seedlings were nonfluorescent, similar to pai1 cmt3 seedlings. These fluorescence phenotypes suggest that SUVH5 makes a small contribution to PAI2 silencing in the absence of SUVH4 and SUVH6 and that SUVH6 makes a small contribution to PAI2 silencing in the absence of SUVH4 and SUVH5. The pai1 suvh5, pai1 suvh4 suvh5, and pai1 suvh4 suvh5 suvh6 mutants did not display any obvious morphological defects beyond those conferred by pai1. In addition, the suvh5-1 and suvh5-2 alleles did not confer obvious morphological defects in the wild-type Ws and Col backgrounds, respectively.
Figure 2.
The suvh4 suvh5 suvh6 Mutant Shows Similar PAI2 Transcriptional Reactivation to a cmt3 Mutant in the pai1 Reporter Background.
Representative 2-week-old seedlings are shown photographed under visible light (right row) or short-wave UV light (left row). All mutations assayed were in the Ws pai1 background, with WT indicating wild type, 4 indicating suvh4R302*, 5 indicating suvh5-1, 6 indicating suvh6-1, and cmt3 indicating cmt3illa (Bartee et al., 2001).
In previous work, we determined that non-CG methylation and H3 mK9 of the PAI1-PAI4 inverted repeat are controlled by the combined action of SUVH4 and SUVH6, such that this locus is demethylated in the suvh4 suvh6 double mutant but not in the suvh4 and suvh6 single mutants (Ebbs et al., 2005). However, the suvh4 suvh6 mutant retains residual non-CG methylation on PAI1-PAI4 relative to cmt3.
To determine the effects of the suvh5 mutation on PAI DNA methylation patterning, we performed both DNA gel blot and bisulfite genomic sequencing assays. For DNA gel blot analysis, we used three methylation-sensitive enzymes with cleavage sites within methylated PAI sequences: HpaII (sensitive to methylation of either cytosine in 5′-CCGG-3′), MspI (sensitive to methylation of only the outer CCG cytosine in 5′-CCGG-3′), and HincII (sensitive to methylation of the outermost cytosines in 5′-atGTCAACag-3′, where the enzyme recognition sequence is shown in uppercase). In these assays, DNA samples isolated from the suvh5 single mutant or the suvh4 suvh5 double mutant displayed cleavage patterns similar to those of the parental wild type or suvh4, respectively (Figures 3A and 3B). However, MspI and HincII assays revealed that the residual non-CG methylation present on PAI1-PAI4 in the suvh4 suvh6 mutant relative to cmt3 was lost in suvh4 suvh5 suvh6. Thus, at PAI1-PAI4, SUVH5 makes a small contribution to the maintenance of non-CG methylation in the absence of SUVH4 and SUVH6.
Figure 3.
SUVH4, SUVH5, and SUVH6 All Contribute to PAI1-PAI4 Non-CG Methylation.
(A) and (B) DNA gel blot assays for PAI DNA methylation patterning. Genomic DNA from the indicated mutants was cleaved with HpaII or MspI isoschizomers (A) or HincII (B) and used in DNA gel blot analysis with a PAI1 cDNA probe. P1-P4 indicates PAI1-PAI4, P2 indicates PAI2, and P3 indicates PAI3, with bands diagnostic of methylation on PAI-internal sites denoted with asterisks.
(C) ChIP analysis of H3 2mK4 and H3 2mK9 patterning on the PAI genes. Primer sets specific for PAI1-PAI4 (indicated as PAI1), PAI2, PAI3, or ACTIN were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 3 online), with a representative data set shown. ACTIN is a control unmethylated transcribed gene previously determined to be enriched for H3 2mK4 but not H3 2mK9 in wild-type and DNA methylation–deficient mutant backgrounds (Johnson et al., 2002; Ebbs et al., 2005).
Genotypes are as described for Figure 2.
Bisulfite sequencing analysis of the proximal promoter regions of PAI1 and PAI2 showed that Ws and suvh5 carried similar patterns of CG plus non-CG methylation at PAI1 and PAI2, suvh4 and suvh4 suvh5 carried similar patterns of CG plus non-CG methylation at PAI1 but mainly CG methylation at PAI2, and cmt3, suvh4 suvh6, and suvh4 suvh5 suvh6 carried similar patterns of mainly CG methylation at PAI1 and PAI2 (see Supplemental Figure 2 online). Overall, these patterns are consistent with the PAI DNA gel blot DNA methylation assay results (Figure 3), although the subtle differences in residual non-CG methylation between suvh4 suvh6 and suvh4 suvh5 suvh6 were not within the resolution of the bisulfite sequencing assay.
We used ChIP analysis of the PAI genes to monitor dimethyl H3 K9 (H3 2mK9) and dimethyl H3 K4 (H3 2mK4), a histone modification associated with transcriptional activity (Gendrel et al., 2002; Lippman et al., 2004), in suvh mutant backgrounds. Each PAI locus was monitored with a gene-specific PCR primer pair: the PAI1-PAI4 locus was amplified at the junction between PAI1 and PAI4, PAI2 was amplified across a central intron–exon boundary, and PAI3 was amplified across a central intron–exon boundary (see Supplemental Table 1 online) (Ebbs et al., 2005). The suvh5 single mutant displayed H3 methylation patterns similar to those of wild-type Ws, with all three PAI loci enriched for H3 2mK9 but not H3 2mK4 (Figure 3C; see Supplemental Figure 3 online). The suvh4 suvh5 double mutant displayed H3 methylation patterns similar to those of suvh4: H3 2mK9 was maintained at PAI1-PAI4 but lost from PAI2 and PAI3, and PAI2 and PAI3 gained H3 2mK4. The suvh4 suvh5 suvh6 triple mutant displayed H3 methylation patterns similar to those of suvh4 suvh6: H3 2mK9 was lost from all three PAI loci, and PAI2 and PAI3 gained H3 2mK4. Overall, the PAI H3 2mK9 patterns were consistent with the PAI non-CG methylation patterns (Figure 3). Furthermore, the gain of H3 2mK4 on PAI2 in suvh4 single and multiple mutants (Figure 3C) was consistent with patterns of PAI2 transcriptional activation revealed by fluorescence phenotypes in the pai1 reporter background (Figure 2).
Together, the PAI2 silencing phenotypes and the PAI non-CG methylation patterns (Figures 2 and 3) suggest that SUVH4, SUVH5, and SUVH6 all act at the PAI genes, with the hierarchies SUVH4 > SUVH5 = SUVH6 at the silenced PAI2 and PAI3 target loci and SUVH4 = SUVH6 > SUVH5 at the PAI1-PAI4 transcribed inverted repeat.
We also assayed DNA methylation patterning in a suvh5 suvh6 double mutant at the PAI genes as well as the transposons Ta3 and Mu1, using DNA gel blot assays, and found similar patterns to the wild type at all loci tested (see Supplemental Figure 4 online). These results support the view that SUVH4 is the major H3 K9 MTase involved in the CMT3 DNA methylation pathway.
SUVH5 Contributes to Ta3 and Mu1 Transposon Non-CG Methylation and Transcriptional Silencing in the Absence of SUVH4
The Ta3 and Mu1 transposons are transcriptionally silent elements that carry CG and non-CG methylation and H3 mK9 in wild-type backgrounds (Johnson et al., 2002; Lippman et al., 2003). Ta3 is a single-copy element (Konieczny et al., 1991), whereas Mu1 is one of a group of related elements (Singer et al., 2001) with three sequences detected by DNA gel blot analysis in the Ws ecotype (Ebbs et al., 2005). Both Ta3 and Mu1 display a partial loss of non-CG methylation in suvh4 and a stronger loss of non-CG methylation in cmt3 (Jackson et al., 2002; Lippman et al., 2003).
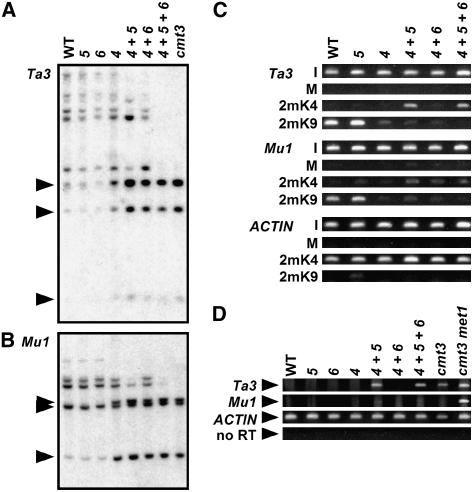
DNA gel blot analysis of Ta3 or Mu1 using an MspI digest that monitors DNA methylation in the non-CG context 5′-CCG-3′ revealed that the suvh5 single mutant showed similar inhibited cleavage patterns to wild-type Ws, diagnostic of dense CCG methylation (Figures 4A and 4B). By contrast, suvh4 suvh5 displayed increased transposon MspI cleavage, diagnostic of reduced CCG methylation relative to suvh4 or suvh4 suvh6. Similar enhanced transposon cleavage patterns were conferred by the suvh5-2 allele in the suvh4 background (see Supplemental Figure 4 online). These results indicate that in the absence of SUVH4, SUVH5 controls residual CCG methylation at the Ta3 element and at Mu1 duplications.
Figure 4.
SUVH5 Contributes to Ta3 and Mu1 Transposon Non-CG Methylation and Ta3 Transcriptional Silencing in the Absence of SUVH4.
(A) and (B) DNA gel blot assays for transposon DNA methylation patterning. DNA from the indicated mutants was cleaved with MspI (A) or HindIII plus MspI (B) and used in DNA gel blot analysis with a Ta3 probe (A) or an Mu1 probe (B). Arrowheads at left indicate the positions of fully cleaved bands.
(C) ChIP analysis of H3 2mK4 and H3 2mK9 patterning on transposons. Primer sets specific for Ta3, Mu1, or ACTIN were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (2mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (2mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 3 online), with a representative data set shown.
(D) Semiquantitative RT-PCR analysis of transposon transcription. Primer sets specific for Ta3, Mu1, or ACTIN were used for RT-PCR analysis of total RNA prepared from 3-week-old plants of the indicated genotypes. The no-RT control is shown for the ACTIN primer set.
Genotypes are as described for Figure 2. The cmt3 met1 mutant (Ebbs et al., 2005) was used as a control for RT-PCR analysis.
The suvh4 suvh5 suvh6 triple mutant displayed a more complete MspI cleavage pattern than suvh4 suvh5 at both Ta3 and Mu1, diagnostic of a loss of residual CCG methylation (Figures 4A and 4B). In both cases, the MspI digestion pattern was similar to that of the cmt3 non-CG MTase. These results suggest that SUVH6 makes a small contribution to the maintenance of non-CG methylation at Ta3 and Mu1 in the absence of SUVH4 and SUVH5.
We used ChIP analysis to monitor H3 2mK9 and H3 2mK4 at Ta3 and Mu1 in suvh mutant backgrounds. The suvh5 single mutant maintained H3 2mK9 similar to wild-type Ws at both transposons (Figure 4C; see Supplemental Figure 3 online), consistent with the maintenance of full DNA methylation at these sequences (Figures 4A and 4B). The suvh4 mutation reduced Ta3 and Mu1 H3 2mK9, such that we could not determine within the sensitivity of the ChIP assay whether this H3 2mK9 depletion was enhanced in the suvh4 suvh5 and the suvh4 suvh5 suvh6 backgrounds (Figure 4C; see Supplemental Figure 3 online). However, at Ta3, the suvh4 suvh5 and suvh4 suvh5 suvh6 mutants displayed a gain of H3 2mK4, diagnostic of transcriptional activation. Furthermore, semiquantitative RT-PCR analysis showed that Ta3 was transcriptionally reactivated in the suvh4 suvh5 and suvh4 suvh5 suvh6 mutants (Figure 4D). Thus, the Ta3 transcriptional activation patterns correlate with non-CG methylation patterns and demonstrate a role for SUVH5 in transposon silencing. Together, these patterns suggest that SUVH4, SUVH5, and SUVH6 all act at Ta3, with the hierarchy SUVH4 > SUVH5 > SUVH6.
Mu1 did not acquire H3 2mK4 or transcriptional activity in any of the suvh mutant backgrounds tested (Figures 4C and 4D), consistent with the previous observation that this element is not transcriptionally activated by defects in the CMT3 non-CG methylation pathway (Lippman et al., 2003). Transcriptional activation of Mu1 was observed only when CG methylation was lost, as in a met1 CG MTase mutant (Figure 4D) (Lippman et al., 2003).
SUVH4, SUVH5, and SUVH6 Together Control the Majority of CMT3-Mediated Repetitive Sequence DNA Methylation
We also examined the effects of suvh mutations on the maintenance of non-CG methylation at the 180-bp centromere (CEN) repeats and at 5S rDNA pericentromeric repeats, using MspI DNA gel blot analysis. At the CEN repeats, the suvh5 and suvh6 single mutants displayed similar cleavage patterns to the wild type (Figure 5A). The ladder of cleaved products was partially shifted downward to a similar extent in the suvh4, suvh4 suvh5, and suvh4 suvh6 mutants, indicative of a partial loss of CCG methylation. The ladder of cleaved products was more strongly shifted downward in the suvh4 suvh5 suvh6 mutant to a similar extent as in cmt3, demonstrating a stronger loss of CCG methylation. Thus, SUVH4, SUVH5, and SUVH6 together control the majority of CMT3-mediated DNA methylation at the CEN repeats, with the hierarchy SUVH4 > SUVH5 = SUVH6.
Figure 5.
SUVH4, SUVH5, and SUVH6 Together Control the Majority of CMT3-Mediated DNA Methylation at Repetitive Sequences.
DNA gel blot assays for repetitive sequences. DNA from the indicated mutants was cleaved with MspI and used in DNA gel blot analysis with a 180-bp centromere repeat probe (A) or a 5S rDNA probe (B). Genotypes are as described for Figure 2.
At the 5S rDNA repeats, the suvh5 and suvh6 mutants had similar MspI cleavage profiles to wild-type Ws, whereas the suvh4, suvh4 suvh5, and suvh4 suvh6 mutants had a similar partial downward shift in the ladder of cleaved products, and the suvh4 suvh5 suvh6 triple mutant had a stronger downward shift (Figure 5B). These results indicate that at the 5S rDNA repeats, as at the CEN repeats, SUVH4, SUVH5, and SUVH6 together control CMT3-mediated DNA methylation, with the hierarchy SUVH4 > SUVH5 = SUVH6. However, in contrast with other loci tested for DNA methylation patterning, the 5S rDNA repeats displayed MspI cleavage patterns diagnostic of residual CCG methylation in suvh4 suvh5 suvh6 relative to cmt3.
SUVH4, SUVH5, and SUVH6 Together Control H3 K9 Monomethylation
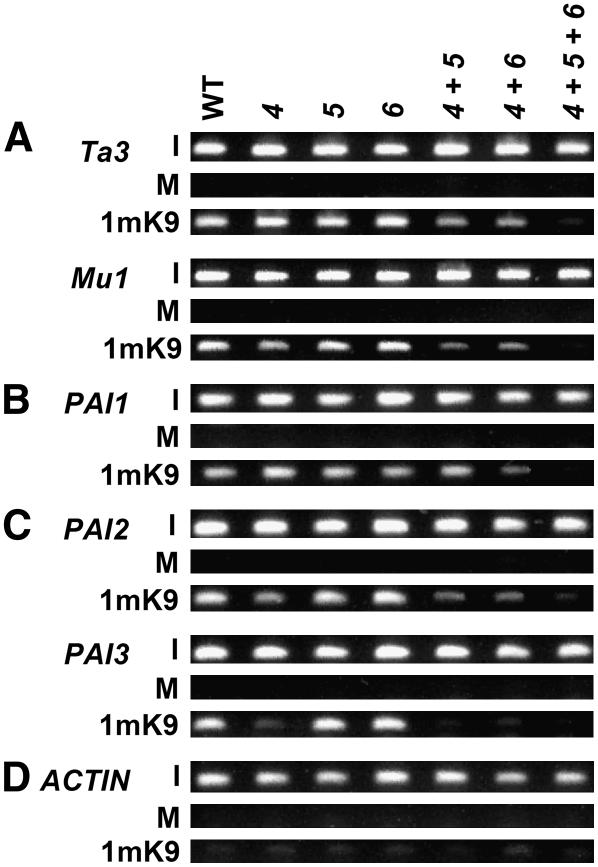
A previous study found that suvh4 maintains similar levels of H3 monomethyl K9 (H3 1mK9) to the wild type at heterochromatin targets, including the Ta3 retrotransposon (Jackson et al., 2004). To determine whether SUVH5 and SUVH6 contribute to H3 1mK9, we performed ChIP analysis for this modification on transposon and PAI gene sequences in suvh mutants (Figure 6; see Supplemental Figure 6 online). We found that SUVH4, SUVH5, and SUVH6 together control H3 1mK9, with different relative contributions at different loci.
Figure 6.
SUVH4, SUVH5, and SUVH6 Together Control H3 1mK9.
ChIP analysis of H3 1mK9 patterning on Ta3 and Mu1 transposons (A), the PAI1-PAI4 transcribed inverted repeat (indicated as PAI1) (B), the silenced PAI2 and PAI3 genes (C), and the unmethylated transcribed control gene ACTIN (D). Primer sets specific for each gene were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), or chromatin immunoprecipitated with H3 anti-monomethyl K9 antibodies (1mK9) from the indicated mutants. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments (see Supplemental Figure 6 online), with a representative data set shown. Genotypes are as described for Figure 2.
At Ta3 and Mu1 transposon sequences, the suvh4, suvh5, and suvh6 single mutants maintained similar levels of H3 1mK9 to the wild type (Figure 6A). However, H3 1mK9 was partially reduced in the suvh4 suvh5 and suvh4 suvh6 double mutants and strongly reduced in the suvh4 suvh5 suvh6 triple mutant, indicating that SUVH4, SUVH5, and SUVH6 together control H3 1mK9 at these loci. At Mu1, the loss of H3 1mK9 in suvh4 suvh5, suvh4 suvh6, and suvh4 suvh5 suvh6 cannot be attributed to transcriptional reactivation induced by the loss of non-CG methylation, because Mu1 is not transcriptionally active in these backgrounds (Figure 4D).
At the PAI1-PAI4 transcribed inverted repeat, the suvh single mutants and the suvh4 suvh5 double mutant maintained similar levels of H3 1mK9 to the wild type (Figure 6B). However, H3 1mK9 was reduced in suvh4 suvh6 and lost in suvh4 suvh5 suvh6. This loss of H3 1mK9 cannot be attributed to transcriptional hyperactivation induced by DNA demethylation, because PAI1-PAI4 maintains a constant level of expression from its upstream unmethylated promoter even when internal non-CG methylation is lost in the cmt3 background (Ebbs et al., 2005). Instead, the loss of H3 1mK9 in suvh4 suvh6 and suvh4 suvh5 suvh6 suggests that SUVH4 and SUVH6 together control the majority of H3 1mK9 at PAI1-PAI4.
At the PAI2 and PAI3 target loci, the suvh4 single mutant displayed reduced H3 1mK9 (Figure 6C), in parallel with the loss of H3 2mK9, non-CG methylation, and transcriptional reactivation (Figures 2 and 3) (Ebbs et al., 2005). At PAI2, residual H3 1mK9 was maintained in suvh4 suvh5 and suvh4 suvh6 but lost in suvh4 suvh5 suvh6, in parallel with patterns of residual transcriptional activation (Figure 2). Therefore, SUVH4 controls the majority of H3 1mK9 at PAI2 and PAI3, but this control could occur indirectly through the maintenance of H3 2mK9, non-CG methylation, and transcriptional silencing. The stronger loss of H3 1mK9 from PAI3 than from PAI2 in suvh4 probably reflects stronger transcriptional reactivation of PAI3 as a result of less extensive promoter PAI sequence identity and DNA methylation (Ebbs et al., 2005).
DISCUSSION
RNA-directed heterochromatin formation is associated with H3 mK9 in fission yeast, Drosophila, mammals, and plants, indicating a fundamental mechanistic connection between RNA signals and H3 K9 MTases (Volpe et al., 2003; Lippman et al., 2004; Pal-Bhadra et al., 2004; Ting et al., 2005). In the plant Arabidopsis, H3 mK9 further guides non-CG methylation mediated by the CMT3 cytosine MTase (Jackson et al., 2002; Malagnac et al., 2002). Here, we show that three Arabidopsis proteins with H3 K9 MTase activity in vitro—SUVH4, SUVH5, and SUVH6—act together to guide H3 K9 monomethylation and dimethylation and CMT3-mediated non-CG methylation. The three SUVH proteins make different relative contributions to the maintenance of H3 K9 and DNA methylation at different loci, suggesting locus-specific mechanisms for their recruitment or activation. The suvh4 suvh5 suvh6 triple mutant displays similar strong non-CG demethylation patterns to a cmt3 mutant at the PAI genes (Figure 3), the Ta3 and Mu1 transposons (Figure 4), and centromere repeats (Figure 5), indicating that the three SUVH proteins control the majority of CMT3-mediated DNA methylation.
At all loci we examined except the PAI1-PAI4 transcribed inverted repeat, the suvh4 single mutation reduced H3 2mK9 and/or non-CG methylation, whereas the suvh5 and suvh6 single mutations had no effect (Figures 3 to 5) (Ebbs et al., 2005). Thus, SUVH4 plays a dominant role relative to SUVH5 and SUVH6 in maintaining heterochromatin-associated modifications at most heterochromatin targets. The dominant role of SUVH4 could be attributable to higher levels of protein expression, intrinsically better MTase activity, or more efficient recruitment to target sequences than SUVH5 or SUVH6.
At PAI1-PAI4, suvh4 suvh6 but not the suvh5 suvh6 or suvh4 suvh5 mutant had reduced H3 mK9 and non-CG methylation (Figures 3 and 6B; see Supplemental Figure 4 online). These results show that either SUVH4 alone or SUVH6 alone can fully maintain heterochromatin-associated modifications at PAI1-PAI4, with only a minor role for SUVH5. By contrast, suvh4 suvh5 displayed a stronger loss of residual non-CG methylation from Ta3 and Mu1 transposons than suvh4 suvh6, and suvh4 suvh5 but not suvh4 suvh6 showed transcriptional reactivation of Ta3 (Figure 4). These data indicate that SUVH5 makes a greater contribution than SUVH6 to heterochromatin modifications at these transposons. The different relative contributions of SUVH5 and SUVH6 at PAI1-PAI4 versus Ta3 and Mu1 transposons suggest that there are locus-specific differences in recruitment or activation of these proteins.
The heterochromatic loci where we observed different relative SUVH activities have different transcriptional activities and derive the dsRNA signal for heterochromatin formation from different sources. The PAI1-PAI4 inverted repeat (SUVH4 = SUVH6 > SUVH5) directly produces dsRNA by constitutive transcription from an upstream promoter, PAI2 and PAI3 (SUVH4 > SUVH5 = SUVH6) are targeted for DNA methylation in trans by PAI1-PAI4 RNA but are not themselves transcribed, and the Ta3 and Mu1 transposons (SUVH4 > SUVH5 > SUVH6) probably generate dsRNA through RNA-dependent RNA polymerase action on transposon-derived transcripts (Melquist and Bender, 2003; Xie et al., 2004). These differences in RNA production and processing could underlie the different SUVH activity patterns. For example, the different sources of dsRNA could feed into different RNA effector complex variants that differentially recruit the SUVH proteins. Alternatively, different RNA signals could promote different patterns of histone modifications on target loci that would lead to differential recruitment of SUVH proteins. It is unlikely that the primary DNA sequence or cytosine methylation pattern on a locus determines the relative activity of the SUVH proteins, because PAI1 and PAI2 are almost identical in sequence and DNA methylation (Luff et al., 1999) and yet have different patterns of dependence on the SUVH proteins for the maintenance of H3 K9 and non-CG methylation.
At the Ta3 and Mu1 transposons, the suvh4 single mutation was sufficient to reduce H3 2mK9 and non-CG methylation, but it did not affect H3 1mK9 levels or confer transcriptional activation (Figures 4 and 6A). These patterns suggest that H3 1mK9 is more easily maintained than H3 2mK9 by the remaining active SUVH enzymes in suvh4. Consistent with this view, a previous in vitro analysis showed that SUVH4 and SUVH6 are better mono-MTases than di-MTases: both enzymes catalyze monomethylation of a peptide substrate within minutes but catalyze dimethylation only after several hours (Jackson et al., 2004). The slow kinetics of SUVH-catalyzed dimethylation in vitro suggests that the relative contribution of a SUVH enzyme to dimethylation of a particular target site in vivo might reflect how stably it is associated with that target. Our finding that SUVH4, SUVH5, and SUVH6 together maintain H3 1mK9 as well as H3 2mK9 and non-CG methylation suggests that in plants the H3 1mK9 modification is part of the CMT3-mediated DNA methylation pathway, rather than serving a novel signaling role. The simplest explanation of our results is that SUVH4, SUVH5, and SUVH6 catalyze both H3 1mK9 and H3 2mK9 in vivo, but we cannot exclude the possibility that these enzymes control H3 mK9 patterns through an indirect mechanism, such as alterations in other histone modification patterns.
The six SUVH proteins that lack in vitro histone MTase activity under our assay conditions (Figure 1A) are all structurally divergent in their catalytic domains from the three active SUVH proteins (Baumbusch et al., 2001). However, in an in vivo setting, with a full complement of interacting factors and substrate histones in a nucleosomal/chromatin context, some of these proteins might also have H3 K9 MTase activity. For example, SUVH2 has H3 K9 MTase activity against nucleosomal substrates in vitro and reduced H3 mK9 in vivo (Naumann et al., 2005). Therefore, the residual DNA methylation detected at the 5S rDNA repeats in the suvh4 suvh5 suvh6 triple mutant relative to cmt3 (Figure 5B) might reflect residual H3 mK9 at these sequences mediated by SUVH2 or other SUVH proteins. It is also possible that some of the other SUVH proteins act as MTases on plant-specific histone variants or nonhistone substrates.
Our finding that SUVH5 can methylate both H3 K9 and Arabidopsis histone H2A variants in vitro (Figure 1; see Supplemental Figure 1 online) suggests that this enzyme might catalyze both H3 K9 and H2A methylation in vivo. However, the lack of DNA methylation phenotypes in the suvh5 single mutant (Figures 3 to 5; see Supplemental Figure 5 online) suggests that SUVH5 H2A MTase activity does not play a role in DNA methylation patterning unless this activity is masked by redundancy with other H2A histone MTases. In addition, a more extensive analysis of the phenotypes conferred by suvh5, especially under nonstandard growth conditions, might reveal a unique pathway controlled by SUVH5 versus SUVH4 or SUVH6. Although the functions of the different Arabidopsis H2A variants remain to be elucidated, H2A variants in fungal and mammalian systems have been shown to be important for specific processes, including telomeric silencing (Wyatt et al., 2003), proper segregation of telomeres during meiotic prophase 1 (Fernandez-Capetillo et al., 2003), protecting euchromatin from heterochromatin spreading (Meneghini et al., 2003), and the nonhomologous end-joining DNA repair pathway (Celeste et al., 2002).
Similar to the cmt3 mutant (Bartee et al., 2001; Lindroth et al., 2001), the suvh4 suvh5 suvh6 triple H3 K9 MTase mutant displays no developmental defects. This observation suggests that the primary function of the three SUVH enzymes is to make histone modifications that guide CMT3-mediated non-CG methylation to transposons and repeated sequences. However, other SUVH proteins might have locus-specific histone methylation functions connected to developmental regulation rather than genome defense. Other plant species, such as maize (Zea mays), also contain multiple SUVH-related genes (Springer et al., 2003), suggesting diversification of SUVH function as a common theme in plant evolution. As we have shown here for SUVH4, SUVH5, and SUVH6, maintaining multiple H3 K9 MTases provides partially redundant modes of protection from different classes of aberrant or invasive DNA sequences.
METHODS
Expression of Recombinant Proteins
The catalytic segment extending from just downstream of the YDG domain through the C terminus of each Arabidopsis thaliana Ws SUVH gene (Baumbusch et al., 2001) was subcloned into either pGEX4T-1 or pGEX4T-2 (Pharmacia) to make an N-terminal GST fusion protein expression construct. Tyr-to-Phe mutations in GST-SUVH4 (residue 591) and GST-SUVH5 (residue 761) were created using oligonucleotide-directed mutagenesis (Kunkel et al., 1987). The GST-H3 peptide plasmids were a gift of Yoichi Shinkai (Tachibana et al., 2001).
Protein expression plasmids were transformed into Escherichia coli strain BL21 Codon Plus RIL (Stratagene). Bacterial cultures for GST-SUVH proteins were grown at 25°C in 1 liter of 2XYT medium supplemented with 100 mg/L ampicillin and 30 mg/L chloramphenicol. Cultures were induced when they reached midlog phase by adding isopropylthio-β-galactoside to a 0.1 mM final concentration and then grown for an additional 4 h. GST-H3 (Figure 1) and GST-HTA (see Supplemental Figure1 online) bacterial expression was performed in a similar manner, except that cultures were grown and induced at 37°C.
Induced cells were lysed in 50 mM Tris, pH 7.5, 0.1 mM EDTA, 0.1% Triton X-100, 1 mg/mL lysozyme (Sigma-Aldrich), 15 units/mL DNase, and a full complement of protease inhibitors (1 mL/4 g wet cell weight; Sigma-Aldrich P-8340) by freeze/thaw. GST fusion proteins were purified from cell lysates using glutathione-coupled Sepharose beads according to the manufacturer's instructions (Amersham Biosciences). Proteins were concentrated using Ultra-free 0.5 concentrators (Millipore), and protein concentration was determined by SDS-PAGE and Coomassie Brilliant Blue R 250 staining.
In Vitro Histone MTase Assays
Calf thymus histone mix (H1, H2A, H2B, H3, and H4) was purchased from Roche Molecular Biochemicals. The biotinylated dimethyl H3 K9 peptide was purchased from Upstate Biotechnology. MTase assays were performed based on a previous protocol (Rea et al., 2000). Briefly, 10 μg of recombinant GST-SUVH protein was incubated with 10 μg of a histone substrate and 300 nCi of S-adenosyl-[methyl-14C]l-Met (100 μM; Amersham) in methylase activity buffer (50 mM Tris, pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, and 250 mM sucrose) at 37°C for 60 min in a total volume of 50 μL. Reactions were separated on an 18% SDS-PAGE gel. Proteins were visualized by Coomassie Brilliant Blue R 250 staining, and the 14C signal was visualized by EN3HANCE (Amersham) fluorography.
In Vitro Analysis of SUVH5 MTase Activity on Arabidopsis H2A Variant Substrates
The Arabidopsis genome encodes 13 histone H2A (HTA) variants, all of which except HTA4 are detectably expressed (Callard and Mazzolini, 1997; Mysore et al., 2000). Although the central sequences of these variants are highly conserved with each other and the mammalian H2A sequences, the N-terminal sequences are divergent (see Supplemental Figure 1A online). To determine whether any of the Arabidopsis HTA proteins could serve as in vitro substrates for methylation by SUVH5, we expressed, purified, and assayed 11 variants as full-length N-terminal GST fusion proteins. Full-length cDNA clones for HTA1 (Nam et al., 1999), HTA2, HTA3, HTA5, HTA7, HTA9 (Callard and Mazzolini, 1997), HTA10, HTA11 (Callard and Mazzolini, 1997), and HTA13 from the Col ecotype were obtained from the ABRC at Ohio State University. HTA6 and HTA8 full-length cDNAs were amplified by RT-PCR from Col RNA. Each of these full-length cDNAs was subcloned into the pGEX* vector (Haldeman et al., 1997) to make an N-terminal GST fusion protein construct.
We found that variants HTA2, HTA7, and HTA13 were used as SUVH5 substrates, with HTA13 being the most robustly methylated (see Supplemental Figure 1B online), whereas HTA1, HTA3, HTA5, HTA6, HTA8, HTA9, HTA10, and HTA11 were not detectably methylated (data not shown). For GST-HTA2, GST-HTA7, and GST-HTA13, both the full-length proteins and truncations of between 31 and 35 kD were observed to be used as SUVH5 substrates (see Supplemental Figure 1B online). Because these truncations are most likely missing C-terminal residues, this finding suggested that SUVH5 recognizes residues in HTA N-terminal tails.
To determine more precisely which HTA residues are methylated, we focused on HTA13 as a model substrate. We made single Lys-to-Arg mutations at residues 13 and 14 of HTA13 (K13R, K14R), counting from the first amino acid after the predicted Met translational start codon as residue 1, and a double mutation of Lys-13 and Lys-14 (KDM). All mutant variants were expressed, purified, and assayed as full-length N-terminal GST fusion proteins. This analysis showed that only the KDM mutation abolished SUVH5-catalyzed methylation, suggesting that SUVH5 can methylate either K13 or K14 in HTA13 (see Supplemental Figure 1C online). We obtained similar results for mutations of the analogous Lys residues in HTA2 (data not shown).
Given the profile of Arabidopsis HTA variants that serve as SUVH5 methylation substrates (see Supplemental Figures 1A and 1B online), the specificity of SUVH5 for Lys-13 and Lys-14 in HTA13 (see Supplemental Figure 1C online), and the specificity of SUVH5 for Lys-9 in histone H3 (Figure 1B), we predicted a recognition sequence context of (V/A)A(R/K*)(R/K*)S for this enzyme, with K* indicating potential substrate Lys residues. To test this prediction, we made a series of context mutations in the HTA13 substrate, including V11A, A12K, K13T, S15A, and S15P. These mutations were chosen based on sequences of HTAs that were not substrates for SUVH5. For example, HTA3, HTA5, and HTA6 all have a T before the K14-analogous residue, whereas HTA9 and HTA11 have a P after the K14-analogous residue (see Supplemental Figure 1A online). Of the context mutations tested, V11A, A12K, K13T, and S15A reduced methylation, whereas only the S15P mutation abolished SUV5-catalyzed methylation (see Supplemental Figure 1C online). Thus, it is likely that particular combinations of amino acid differences in the N termini of nonsubstrate HTAs act together to block SUVH5 activity.
Plant Materials
The Ws suvh5-1 T-DNA insertion mutation was obtained from the FLAGdb/FST mutant collection at the Institute of Agronomic Research in Versailles, France (Samson et al., 2002). The left border insertion junction lies 2071 bp downstream from the ATG translational start codon of SUVH5 in the exonic region that encodes the SET portion of the catalytic sequences. The Col suvh5-2 T-DNA insertion mutation was obtained from the SALK Institute Genomic Analysis Laboratory mutant collection via the ABRC (Alonso et al., 2003). The left border insertion junction lies 2258 bp downstream from the ATG translational start codon of SUVH5 in the exonic region that encodes the SET portion of the catalytic sequences. Each suvh5 allele was crossed with Ws pai1 (Bartee and Bender, 2001), Ws pai1 suvh4R302* (Malagnac et al., 2002), Ws pai1 suvh6-1 (Ebbs et al., 2005), or Ws pai1 suvh4R302* suvh6-1 (Ebbs et al., 2005), and PCR-based genotype markers were used to identify progeny that were homozygous for the three PAI loci from Ws (Luff et al., 1999) and homozygous for each suvh mutation. At least three independent lines were isolated for each suvh mutant, and DNA gel blot assays for PAI and transposon DNA methylation were used to determine that there was no variation in DNA methylation phenotypes among different lines with the same suvh genotype. A representative line of each suvh genotype was used for ChIP analysis (Figures 3C, 4C, and 6) and PAI bisulfite sequencing analysis (see Supplemental Figure 2 online). PCR primers to amplify the suvh5-1 insertion are FSTLB4 (5′-CGTGTGCCAGGTGCCCACGGAATAGT-3′) and SUVH5R2 (5′-GCTTATTCAGACAGAACTGAAC-3′), which yield a 550-bp product. PCR primers to amplify the suvh5-2 insertion are SALKLB (5′-GCGTGGACCGCTTGCTGCAACT-3′) and SUVH5AKFR (5′-ATCACTAAGACAACATCAGTATGATCAAG-3′), which yield a 650-bp product. PCR primers to amplify the intact SUVH5 gene from either suvh5-1 or suvh5-2 are SUVH5R2 and SUVH5GAR2 (5′-GATGGTCTTTGCAATGTTG-3′), which amplify an 844-bp product. PCR-based assays to score the suvh4R302* and suvh6-1 mutations were described previously (Malagnac et al., 2002; Ebbs et al., 2005).
Plant genomic DNA preparations and DNA gel blots were performed as described previously (Melquist et al., 1999).
ChIP Analysis
ChIP assays were performed using a previously described method (Gendrel et al., 2002) starting with 0.7 g of leaf tissue from 3-week-old plants grown in soilless potting mix (Fafard mix 2) under continuous illumination. Chromatin was immunoprecipitated with anti-H3 dimethyl K4 antibodies (Upstate Biotechnology), with anti-H3 dimethyl K9 antibodies (a gift of T. Jenuwein), with anti-H3 monomethyl K9 antibodies (Upstate Biotechnology), or carried through the protocol with no antibody added as a control (mock precipitation). PCR amplification of immunoprecipitated DNA was performed as described previously (Ebbs et al., 2005). PCR-amplified products from ChIP template DNA were visualized on a 2.5% agarose gel stained with GelStar (Cambrex). Each ChIP assay was performed in three independent experiments (see Supplemental Figures 3 and 6 online), with results from representative experiments shown in Figures 3C, 4C, and 6. ChIP primer sequences used in this study are listed in Supplemental Table 1 online.
Semiquantitative RT-PCR
Total RNA was isolated from 4-week-old plants and treated with RNase-free DNase (Promega). RT-PCR was performed as described previously for Mu1 (Singer et al., 2001), Ta3 (Johnson et al., 2002), or ACTIN (Ito et al., 2004). Control reactions without reverse transcriptase were performed to ensure that no background signal was generated from contaminating DNA.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers are as follows: SUVH1 (At5g04940), SUVH2 (At2g33290), SUVH3 (At1g73100), SUVH4 (At5g13960), SUVH5 (At2g35160), SUVH6 (At2g22740), SUVH7 (At1g17770), SUVH8 (At2g24740), SUVH9 (At4g13460), ACTIN (At5g09810), HTA1 (At5g54640), HTA2 (At4g27230), HTA3 (At1g54690), HTA5 (At1g08880), HTA6 (At5g59870), HTA7 (At5g27670), HTA8 (At2g38810), HTA9 (At1g52740), HTA10 (At1g51060), HTA11 (At3g54560), HTA13 (At3g20670), PAI1 (At1g07780), PAI2 (At5g05590), and PAI3 (At1g29410).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SUVH5 Has Histone H2A MTase Activity in Vitro.
Supplemental Figure 2. Bisulfite Genomic Sequencing of DNA Methylation Patterning on the PAI1 and PAI2 Proximal Promoters in suvh Mutants.
Supplemental Figure 3. Replicates of ChIP Analysis of H3 2mK4 and H3 2mK9 Patterning.
Supplemental Figure 4. The suvh5 suvh6 Mutant Does Not Display DNA Methylation Defects.
Supplemental Figure 5. Two suvh5 Alleles Confer Similar Loss of Transposon Non-CG Methylation in the Absence of SUVH4.
Supplemental Figure 6. Replicates of ChIP Analysis of H3 1mK9 Patterning.
Supplemental Table 1. ChIP Primer Sequences.
Supplementary Material
Acknowledgments
We thank Yoichi Shinkai for GST-H3 plasmids; the ABRC for HTA plasmids, the 5S rDNA plasmid pCT4.2, and the Salk collection suvh5-2 T-DNA insertion mutant; the Institute of Agronomic Research for the suvh5-1 T-DNA insertion mutant; and Thomas Jenuwein for H3 anti-dimethyl K9 antibodies. We also thank Cecile Pickart for technical advice and Brandi Rocci for assistance with HTA expression. This work was supported by National Institutes of Health Grant GM-61148 to J.B. and by training grants T32 ES-07141 (National Institute of Environmental Health Sciences) and T32 CA-09110 (National Cancer Institute) to M.L.E.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Judith Bender (jbender@mail.jhmi.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041400.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bartee, L., and Bender, J. (2001). Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 29 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callard, D., and Mazzolini, L. (1997). Identification of proliferation-induced genes in Arabidopsis thaliana. Characterization of a new member of the highly evolutionarily conserved histone H2A.F/Z variant subfamily. Plant Physiol. 115 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Aufsatz, W., Zilberman, D., Mette, M.F., Huang, M.S., Matzke, M., and Jacobsen, S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13 2212–2217. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (suppl. 4), 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste, A., et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R.E., Tachibana, M., Tamaru, H., Smith, K.M., Jia, D., Zhang, X., Selker, E.U., Shinkai, Y., and Cheng, X. (2005). In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 280 5563–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M.L., Bartee, L., and Bender, J. (2005). H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O., Liebe, B., Scherthan, H., and Nussenzweig, A. (2003). H2AX regulates meiotic telomere clustering. J. Cell Biol. 163 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Haldeman, M.T., Xia, G., Kasperek, E.M., and Pickart, C.M. (1997). Structure and function of ubiquitin conjugating enzyme E2-25K: The tail is a core-dependent activity element. Biochemistry 36 10526–10537. [DOI] [PubMed] [Google Scholar]
- Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., Riechmann, J.L., and Meyerowitz, E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 356–360. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Johnson, L., Jasencakova, Z., Zhang, X., PerezBurgos, L., Singh, P.B., Cheng, X., Schubert, I., Jenuwein, T., and Jacobsen, S.E. (2004). Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308–315. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miura, A., Bender, J., Jacobsen, S.E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13 421–426. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., Voytas, D.F., Cummings, M.P., and Ausubel, F.M. (1991). A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. (1987). Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154 367–382. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Ueda, Y., Derijck, A.A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T., Li, E., Jenuwein, T., and Peters, A.H. (2003). Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13 1192–1200. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., and Martienssen, R. (2004). The role of RNA interference in heterochromatic silencing. Nature 431 364–370. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., May, B., Yordan, C., Singer, T., and Martienssen, R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1 E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff, B., Pawlowski, L., and Bender, J. (1999). An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3 505–511. [DOI] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., and Bender, J. (2004). RNA-directed DNA methylation. J. Cell Sci. 117 4881–4888. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Melquist, S., and Bender, J. (2003). Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev. 17 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melquist, S., Luff, B., and Bender, J. (1999). Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M.D., Wu, M., and Madhani, H.D. (2003). Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Nam, J., and Gelvin, S.B. (2000). An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 97 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M.K., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261 429–438. [DOI] [PubMed] [Google Scholar]
- Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., Hause, G., Aurich, A.C., Dorn, R., Jenuwein, T., and Reuter, G. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. (2004). Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669–672. [DOI] [PubMed] [Google Scholar]
- Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., and Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 593–599. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. (2001). Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N.M., Napoli, C.A., Selinger, D.A., Pandey, R., Cone, K.C., Chandler, V.L., Kaeppler, H.F., and Kaeppler, S.M. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132 907–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana, M., Sugimoto, K., Fukushima, T., and Shinkai, Y. (2001). Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276 25309–25317. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., and Selker, E.U. (2001). A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., Zhang, X., McMillen, D., Singh, P.B., Nakayama, J., Grewal, S.I., Allis, C.D., Cheng, X., and Selker, E.U. (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34 75–79. [DOI] [PubMed] [Google Scholar]
- Ting, A.H., Schuebel, K.E., Herman, J.G., and Baylin, S.B. (2005). Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 37 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, R., McCallum, C.M., Delrow, J., Henikoff, J.G., van Steensel, B., and Henikoff, S. (2002). Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12 65–68. [DOI] [PubMed] [Google Scholar]
- Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. (2004). RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. (2003). RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11 137–146. [DOI] [PubMed] [Google Scholar]
- Wyatt, H.R., Liaw, H., Green, G.R., and Lustig, A.J. (2003). Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z., Tachibana, M., Guggiari, M., Heard, E., Shinkai, Y., and Wagstaff, J. (2003). Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 278 14996–15000. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yang, Z., Khan, S.I., Horton, J.R., Tamaru, H., Selker, E.U., and Cheng, X. (2003). Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.