Abstract
RNA editing in plant organelles is mediated by site-specific, nuclear-encoded factors. Previous data suggested that the maintenance of these factors depends on the presence of their rapidly evolving cognate sites. The surprising ability of allotetraploid Nicotiana tabacum (tobacco) to edit a foreign site in the chloroplast ndhA messenger RNA was thought to be inherited from its diploid male ancestor, Nicotiana tomentosiformis. Here, we show that the same ndhA editing activity is also present in Nicotiana sylvestris, which is the female diploid progenitor of tobacco and which lacks the ndhA site. Hence, heterologous editing is not simply a result of tobacco's allopolyploid genome organization. Analyses of other editing sites after sexual or somatic transfer between land plants showed that heterologous editing occurs at a surprisingly high frequency. This suggests that the corresponding editing activities are conserved despite the absence of their target sites, potentially because they serve other functions in the plant cell.
Introduction
RNA editing in higher plant plastids and mitochondria changes the genetic information encoded in messenger RNAs by modifying nucleotides at highly specific positions. It is predominantly characterized by C-to-U modifications that restore codons conserved in evolution at a given site (Maier et al, 1992). Unlike the cognate editing sites of animal editing factors APOBEC1 or ADAR1, which are conserved at least in vertebrates (Bass, 2001; Levanon et al, 2005), editing sites in plant organelles are evolutionarily highly dynamic, with even closely related taxa having different sets of sites (Freyer et al, 1995; Schmitz-Linneweber et al, 2002; Sasaki et al, 2003). Indirect evidence suggested that the cognate editing factors are evolving rapidly: a spinach-specific and a maize-specific site introduced artificially into the tobacco plastid chromosome remained unedited (Bock et al, 1994; Reed & Hanson, 1997). In addition, no editing of a tobacco-specific editing site was found in a pea in vitro editing system (Miyamoto et al, 2002). Taken together, these data suggested that an RNA editing site and its cognate site-specific editing factors form an evolutionary unit that is rapidly evolving and is either present or absent in a given species.
The first case reported of a foreign site edited when transferred into plastids of a different species despite the absence of an endogenous homologue was that of the spinach ndhA-189 site by tobacco (Schmitz-Linneweber et al, 2001). Tobacco is an allotetraploid species that originated from a natural interspecific fertilization event that occurred between the progenitors of the modern diploid species Nicotiana sylvestris and Nicotiana tomentosiformis. Site ndhA-189 is present and processed in N. tomentosiformis, but absent from N. sylvestris (Schmitz-Linneweber et al, 2001). On the basis of these results, the authors concluded that the most parsimonious explanation for the ndhA-189 editing activity found in tobacco was that the N. tomentosiformis gene for the factor persisted in the allotetraploid tobacco nuclear genome.
Here we show that the heterologous ndhA editing activity previously identified in tobacco is also present in its female progenitor N. sylvestris. We describe other heterologous editing events in an alloplasmic line and cybrid plants. The unexpectedly high frequency of heterologous editing suggests that there is a subgroup of editing factors that are conserved between plant taxa independently of their target sites. Possibly, such factors are retained because they edit several sites in plastid transcriptomes (Chateigner-Boutin & Hanson, 2002, 2003; Tillich et al, 2005).
Results
N. sylvestris harbours an activity to process ndhA-189
Previously, we used transformation vector pCEE to introduce the spinach ndhA-189 editing site into tobacco chloroplasts (Schmitz-Linneweber et al, 2001). Here, we used the same vector to transform N. sylvestris by particle bombardment (Svab & Maliga, 1993) to test whether this species also possesses an activity to process the foreign editing site ndhA-189, a site widespread in angiosperms (Table 1; Fig 1A). After bombardment of ten leaves, one spectinomycin-resistant line containing the aadA cassette was isolated. From this single transformant, two subclones, designated as SylCEE-2.8 and SylCEE-10.2, were established and maintained by micropropagation in vitro. Correct integration of the transgene was verified by PCR and Southern analysis (Fig 1B,C). Southern analysis further showed that the primary isolates were already homoplastomic for the mutation (Fig 1C).
Table 1.
Editing sites in foreign genetic backgrounds
| The chart summarizes information on editing sites that have been transferred between species. The table is divided by the thick line in editing sites processed in a heterologous nuclear background (above) and those not processed (below). Numbers refer to the codons affected, with the amino acids encoded before (left of the number) and after editing (right of the number) given in one-letter code. Shading: dark grey, sites known to be edited with codon transition and number of codon given (in contrast to previous investigations (Inada et al, 2004), we found ndhD-293 fully edited in pea plants (supplementary Fig 4D online); possibly, differences in growth conditions account for these contradicting observations); white, editing site absent, but capacity to edit the site heterologously was experimentally proven (Schmitz-Linneweber et al, 2005; this study); black, editing site absent, and heterologous editing upon experimental introduction of these sites failed (Bock et al, 1994; Reed & Hanson, 1997; Schmitz-Linneweber et al, 2005); light grey, site absent; hatching, site present but not edited. 1Schmitz-Linneweber et al (2001); 2Fig 1; 3supplementary Fig 4 online; 4Schmitz-Linneweber et al (2002); 5Inada et al (2004); 6Maier et al (1995), Tillich et al (2001); 7Fig 2; 8Hirose et al (1999); 9Sasaki et al (2003); 10Schmitz-Linneweber et al (2005); 11accession no. AJ400848; 12accession no. X86563; 13Bock et al (1994); 14accession no. AB237912; 15accession no. AB240139; 16accession no. AJ316582; 17Reed & Hanson (1997); ND, not determined. |
Figure 1.
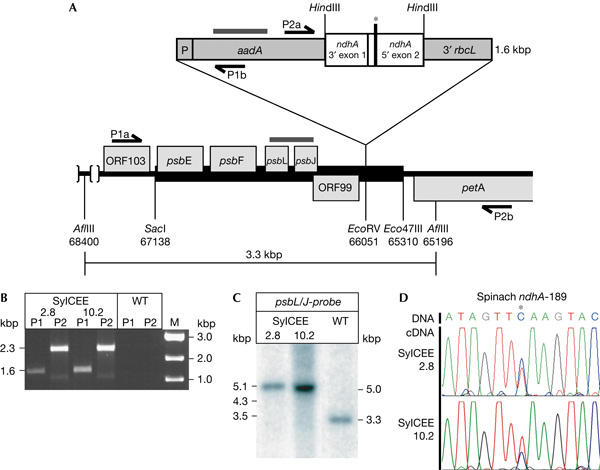
Introduction of a recombinant spinach ndhA construct into the Nicotiana sylvestris plastid chromosome. (A) Schematic representation of parts of the plasmid vector pCEE, as described by Schmitz-Linneweber et al (2001). The thick black bar represents the tobacco plastid DNA fragment used for targeting the recombinant cassette to the petA–psbJ intergenic spacer. The grey bars indicate regions corresponding to probes used in Southern and RNA gel blot analysis. P, 16S ribosomal DNA promoter; 3′rbcL, 3′ stabilizing element. Arrows indicate primers. The asterisk denotes the position of the editing site ndhA-189. Numbers refer to positions in the tobacco plastid chromosome (accession no. Z00044). (B) PCR analysis of correct integration of the transgene into the N. sylvestris plastid chromosome in the two spectinomycine-resistant subclones SylCEE-2.8 and SylCEE-10.2 recovered after transformation. PCR product P1, 1,624 bp; P2, 2,320 bp (see (A) for positions of primers). M, molecular weight marker. (C) Southern analysis of AflIII-digested total genomic DNA (5 μg) of transplastomic lines SylCEE-2.8 and SylCEE-10.2. The probe covers the psbL/J genes (see (A)). Signals obtained correspond to calculated fragment length (indicated on the left; also see (A)). No signal corresponding to wild-type plastid DNA was detected in transplastomic plants, suggesting that they are homoplastomic for the transgene. (D) Complementary DNA sequence analysis of the introduced ndhA editing site in transplastomic lines SylCEE-2.8 and SylCEE-10.2. An excerpt of the corresponding chromatograms is shown with the edited nucleotide in the centre marked by an asterisk. A C-peak corresponding to unedited messages and a T-peak corresponding to edited messages were found in the plant lines analysed (top: corresponding DNA sequence).
Total leaf RNA from SylCEE-2.8 and SylCEE-10.2 was reverse transcribed and transgene-specific complementary DNAs were amplified by PCR and sequenced to assess RNA editing. Surprisingly, the transcripts containing spinach-derived sequences were processed in N. sylvestris plastids (Fig 1D), demonstrating that N. sylvestris harbours an activity to process ndhA-189. RNA editing of this site is only partial, as there is also a signal corresponding to the unedited transcript visible. It is noteworthy that SylCEE-10.2 showed a higher editing efficiency than SylCEE-2.8, which is paralleled by a lower steady-state level of transgenic ndhA transcript in line SylCEE-10.2 (supplementary Fig 1 online), suggesting that the differences in editing of the two transgenic lines may be linked to the differential abundance of target RNAs in these plants, as suggested for similar transformation experiments (Reed & Hanson, 1997; Reed et al, 2001a, 2001b; Chateigner-Boutin & Hanson, 2002).
Why is the ndhA-189 activity maintained in N. sylvestris? Possibly, the underlying editing factors serve a cluster of plastid editing sites. Interestingly, ndhA-189 shares sequence homology with site ndhF-97 (supplementary Fig 2 online), a potential member of a cluster of sites (Chateigner-Boutin & Hanson, 2002, 2003). Processing of this or other sites could be affected in SylCEE plants, because the corresponding factor might be titrated by the competing transgene transcript (Reed et al, 2001a). We therefore tested editing of all described N. sylvestris sites (Sasaki et al, 2003), but did not find any significant reduction in editing anywhere (supplementary Fig 3 online). This is preliminary evidence that the ndhA-189 activity does not serve a further known editing site in the plastid transcriptome.
Heterologous editing in alloplasmic and cybrid plants
To determine whether heterologous editing is a more widespread phenomenon, we analysed plants that harbour foreign plastid genomes. Specifically, we tested the ability of the tobacco nucleus to edit foreign sites introduced into a tobacco nuclear background either by sexual or somatic hybridization.
Nicotiana tabacum Samsoun (NN) is a tobacco variety that carries an introgressed N gene, which confers disease resistance against the tobacco mosaic virus (Holmes, 1938). The N gene was introduced genetically by a backcrossing scheme from the donor species N. glutinosa making N. tabacum Samsoun (NN) effectively an alloplasmic line containing the N. glutinosa plastome (Clausen & Godspeed, 1925; Holmes, 1938; Fig 2A). We confirmed this by sequencing 2,809 bp of the N. glutinosa and N. tabacum Samsoun (NN) plastid chromosomes. They were identical, but had 12 point mutations relative to N. tabacum Petit Havanna. Next, we analysed the N. glutinosa plastid chromosome for the presence of editing sites known to be absent from tobacco but present in other dicots, because such sites could be analysed for heterologous editing in N. tabacum Samsoun (NN). In total, ten editing sites not present in tobacco have been identified in dicots, namely accD-284, clpP-187, matK-219, ndhA-189, ndhB-419, ndhD-293, ndhD-296, petL-2, psbF-26 and rpoB-809 (supplementary Fig 4 online). In addition, we included a maize site, rpoB-206 (codon 208 in tobacco; supplementary Fig 4 online), previously tested for heterologous editing (Reed & Hanson, 1997). The corresponding regions in the N. glutinosa plastid chromosome were amplified and sequenced. This led to the identification of editing site ndhD-293 in N. glutinosa, which was also shown to be present in N. tabacum Samsoun (NN) and represents a widely distributed editing site in angiosperms (Table 1). cDNA analysis showed that this site was processed in N. glutinosa and in N. tabacum Samsoun (NN; Fig 2A). Thus, ndhD-293 is processed after sexual introduction into an N. tabacum nuclear background. Apparently, the tobacco nucleus contains the factor for editing this site despite its absence from the plastid genome, although it cannot be excluded that a gene tightly linked to the N. glutinosa N gene is coding the required activity.
Figure 2.
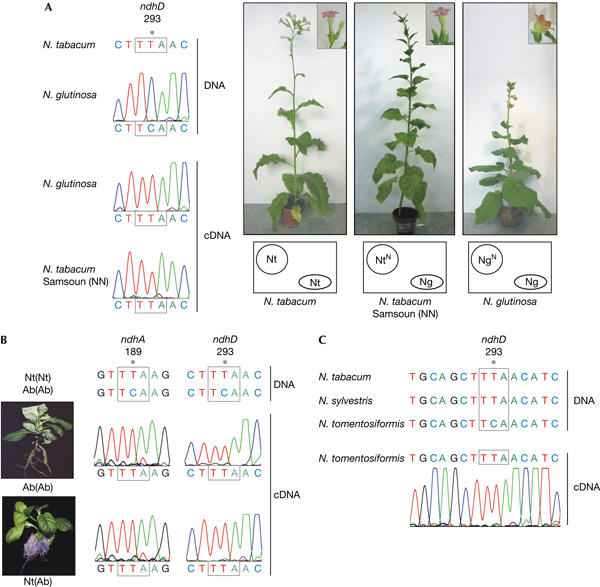
Heterologous processing of editing sites after somatic or sexual transfer into a foreign genetic background. (A) Editing of ndhD-293 in Nicotiana glutinosa, Nicotiana tabacum Petit Havanna and the introgression line N. tabacum Samsoun (NN). Excerpts of the sequencing chromatograms are shown, with the edited position marked by an asterisk and the affected codon boxed. Phenotypes of mature plants are shown, with the inset showing the different flower morphologies. Circles indicate the nuclei and ovals the plastids. Nt, N. tabacum Petit Havanna; Ng, N. glutinosa. Superscript N indicates the presence of the N-gene conferring resistance against tobacco mosaic virus. (B) Complementary DNA sequence analysis of editing sites ndhA-189 and ndhD-293 in nightshade Ab(Ab) and cybrid Nt(Ab) seedlings. Relevant chromatogram excerpts are shown, with the edited position and codons marked as in (A). (C) Nicotiana tomentosiformis possesses and processes editing site ndhD-293, whereas this site is absent in Nicotiana sylvestris and N. tabacum. Partial ndhD DNA sequences for the three Nicotiana species are aligned. Below is an excerpt of a cDNA sequence from N. tomentosiformis together with the corresponding chromatogram. Editing sites and affected codons are marked as in (A).
Apart from alloplasmic lines, a second source for the study of heterologous editing are somatically generated cytoplasmic hybrids (cybrids) that combine plastid editing sites from one species with the nuclear background of another. Nt(Ab) cybrids have a nuclear genome of tobacco, Nt, and a plastid genome of Atropa belladonna (nightshade), (Ab). These cybrids were generated by fusion of somatic cells in vitro, and phenotypically resemble normal wild-type tobacco (Babiychuk et al, 1995). The nightshade plastid genome contains three editing sites not present in tobacco (Schmitz-Linneweber et al, 2002). Two of these heterologous sites, ndhA-189 and ndhD-293, were fully processed in cybrids (Fig 2B), whereas the third site, rpoB-809, remained unedited (Schmitz-Linneweber et al, 2005). This confirms previous findings that site ndhA-189 is processed when brought into a tobacco nuclear background by plastid transformation (Schmitz-Linneweber et al, 2001) and shows that the respective activity present in tobacco chloroplasts is efficiently processing site ndhA-189 when expressed from its natural promoter. This finding further supports that the partial editing observed in transplastomic tobacco (Schmitz-Linneweber et al, 2001) and N. sylvestris lines (Fig 1D) is due to a titration effect of the ndhA-189 activity by overexpression of the target site. The presence of an activity responsible for ndhD-293 editing in the Nt(Ab) cybrid suggests that this activity was not co-introduced with the N gene into N. tabacum Samsoun (NN), but is already encoded in the nuclear genome of tobacco. Interestingly, like site ndhA-189, ndhD-293 is present in tobacco's diploid male progenitor N. tomentosiformis, but absent from its female progenitor and donor of plastids N. sylvestris (Fig 2C; Schmitz-Linneweber et al, 2001). Whether N. sylvestris is able to process this site, similar to the situation for ndhA-189, remains to be determined.
Discussion
This study draws attention to the fact that several plant organellar editing sites are processed after transfer into a foreign nuclear background. Independent transfer events of editing sites between species using three alternative models (transplastomic plants, alloplasmic lines and cybrid lines) prove that editing factors can persist in plant genomes despite the absence of their target site (Figs 1, 2; Schmitz-Linneweber et al, 2001). Four novel heterologous editing events have been found concerning two sites (ndhA-189 in SylCEE and Nt(Ab) and ndhD-293 in Nt SNN and Nt(Ab)). Thus, out of a total of ten different heterologous editing sites analysed here and previously (Bock et al, 1994; Reed & Hanson, 1997; Schmitz-Linneweber et al, 2001, 2005), four are edited. Apparently, the nuclear-encoded activities for these four sites have not been lost after loss of their cognate site, but are maintained. Possibly, such maintenance of editing factors is simply due to chance: the factor is not (yet) lost, because loss of the target site occurred rather recently in evolution. Alternatively, some factors might be maintained by selection, because they are required to fulfil another function.
We assumed earlier that the tobacco activity for ndhA-189 was inherited during allotetraploidization from tobacco's diploid male parent, N. tomentosiformis (Schmitz-Linneweber et al, 2001). Similar arguments have been made for the inheritance of other tobacco editing activities (Sasaki et al, 2003). Here, we show however by a transgenic approach that even though its cognate site is absent, the editing activity for ndhA-189 is present in N. sylvestris. Therefore, the existence of at least this hidden editing activity in tobacco cannot be adequately explained by allotetraploidy alone. Instead, the presence of this activity in both ancestors of tobacco and its survival through the—from a genomic point of view—stressful allotetraploidization event towards tobacco argue for long-term maintenance of the factor and tentatively suggest that some other sort of selective pressure for the stable maintenance of editing activities besides editing the ndhA site guarantees its maintenance. Such a selective pressure for the stable maintenance of editing activities was predicted earlier (Covello & Gray, 1993) and would also help to understand the unexpected high frequency of heterologous editing described here.
What could be the nature of such putative additional selective pressures/functions for editing factors? Most parsimoniously, they could be required for processing a different editing site. This would agree with recent findings that editing sites tend to form evolutionarily and functionally linked clusters, potentially served by group-specific editing factors (Chateigner-Boutin & Hanson, 2003; Tillich et al, 2005) and supports earlier suggestions that editing factors are maintained because they serve not only one target site, but many (Covello & Gray, 1993). Alternatively, the factors could also be involved in a task unrelated to editing. The only specificity factor of plant organelle RNA editing isolated so far belongs to the pentatricopeptide repeat (PPR) protein family (Kotera et al, 2005), members of which have diverse roles in organellar RNA metabolism (Lurin et al, 2004). Further analyses are needed to show whether more editing factors are PPR proteins and whether such factors are exclusively involved in RNA editing or do have several functions, for instance in other aspects of organellar RNA metabolism.
Methods
Preparation of nucleic acids and their analysis. Plants were grown on soil in the green house or on agar-solidified MS medium (Murashige & Skoog, 1962) under sterile conditions in a growth chamber. Total cellular DNA was extracted using a standard cetyltrimethylammonium bromide protocol. Total cellular RNA was isolated using Trizol reagent (Invitrogen, Karlsruhe, Germany). First-strand cDNA synthesis was carried out with Superscript II RNase H− Reverse Transcriptase (Invitrogen). Amplification of DNA fragments by PCR was carried out according to standard protocols (Sambrook et al, 1989). PCR products were sequenced as described previously (Schmitz-Linneweber et al, 2002). All oligonucleotides were bought from MWG-BIOTECH (Ebersberg, Germany) (supplementary Table 1 online). For gel blot analyses, nucleic acids were separated by gel electrophoresis and transferred onto nitrocellulose membranes (Sambrook et al, 1989).
Plastid transformation of Nicotiana sylvestris. Young leaves were harvested from sterile grown N. sylvestris plants and bombarded with plasmid DNA-coated gold particles by using the biolistic PDS-1000/He unit (Bio-Rad, Munich, Germany; Svab & Maliga, 1993). Spectinomycine-resistant shoots were selected on RMOP regeneration medium containing 500 mg/l spectinomycine dihydrochloride (Svab & Maliga, 1993). Plastid transformants resulting from homologous recombination events were identified by PCR, and homoplastomy was verified by Southern blot analysis.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400619-s1.pdf).
Supplementary Material
Supplementary Materials
Acknowledgments
We thank R.M. Maier, who initiated and inspired this work, and Dr U.G. Maier and Dr R.G. Herrmann for support. This work was supported by the Deutsche Forschungsgemeinschaft via SFB-TR1 and a long-term fellowship to C.S.L.
References
- Babiychuk E, Schantz R, Cherep N, Weil JH, Gleba Y, Kushnir S (1995) Alterations in chlorophyll a/b binding proteins in Solanaceae cybrids. Mol Gen Genet 249: 648–654 [DOI] [PubMed] [Google Scholar]
- Bass BL (ed) (2001) RNA Editing. Oxford: Oxford University Press [Google Scholar]
- Bock R, Kossel H, Maliga P (1994) Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J 13: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Hanson MR (2002) Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol Cell Biol 22: 8448–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Hanson MR (2003) Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res 31: 2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen RE, Godspeed TH (1925) Interspecific hybridization in Nicotiana. II. A tetraploid glutinosa–tabacum hybrid, an experimental verification of Winge's hypothesis. Genetics 10: 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello PS, Gray MW (1993) On the evolution of RNA editing. Trends Genet 8: 265–268 [DOI] [PubMed] [Google Scholar]
- Freyer R, Lopez C, Maier RM, Martin M, Sabater B, Kossel H (1995) Editing of the chloroplast ndhB encoded transcript shows divergence between closely related members of the grass family (Poaceae). Plant Mol Biol 29: 679–684 [DOI] [PubMed] [Google Scholar]
- Hirose T, Kusumegi T, Tsudzuki T, Sugiura M (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol Gen Genet 262: 462–467 [DOI] [PubMed] [Google Scholar]
- Holmes FO (1938) Inheritance of resistance to tobacco-mosaic disease in tobacco. Phytopathology 28: 553–561 [Google Scholar]
- Inada M, Sasaki T, Yukawa M, Tsudzuki T, Sugiura M (2004) A systematic search for RNA editing sites in pea chloroplasts: an editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol 45: 1615–1622 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E (2005) Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res 33: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Hoch B, Zeltz P, Kossel H (1992) Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell 4: 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Kossel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251: 614–628 [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Obokata J, Sugiura M (2002) Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol Cell Biol 22: 6726–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Reed ML, Hanson MR (1997) A heterologous maize rpoB editing site is recognized by transgenic tobacco chloroplasts. Mol Cell Biol 17: 6948–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Lyi SM, Hanson MR (2001a) Edited transcripts compete with unedited mRNAs for trans-acting editing factors in higher plant chloroplasts. Gene 272: 165–171 [DOI] [PubMed] [Google Scholar]
- Reed ML, Peeters NM, Hanson MR (2001b) A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res 29: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning—A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sasaki T, Yukawa Y, Miyamoto T, Obokata J, Sugiura M (2003) Identification of RNA editing sites in chloroplast transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol Biol Evol 20: 1028–1035 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Tillich M, Herrmann RG, Maier RM (2001) Heterologous, splicing-dependent RNA editing in chloroplasts: allotetraploidy provides trans-factors. EMBO J 20: 4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM (2002) The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol 19: 1602–1612 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Kushnir S, Babiychuk E, Poltnigg P, Herrmann RG, Maier RM (2005) Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell 17: 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Schmitz-Linneweber C, Herrmann RG, Maier RM (2001) The plastid chromosome of maize (Zea mais): update of the complete sequence and transcript editing sites. Maize Genet Corp News Lett 75: 42–44 [Google Scholar]
- Tillich M, Funk H, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM (2005) Plastid RNA editing in Arabidopsis thaliana ecotypes. Plant J 43: 708–715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials


