Abstract
F1-ATPase is an ATP-driven rotary molecular motor in which the central γ-subunit rotates inside a stator cylinder made of α3β3 subunits. To elucidate the role of rotor-stator interactions in torque generation, we truncated the γ-subunit at its carboxyl terminus, which forms an α helix that penetrates deeply into the stator cylinder. We used an α3β3γ subcomplex of F1-ATPase derived from thermophilic Bacillus PS3 and expressed it in Escherichia coli. We could obtain purified subcomplexes in which 14, 17, or 21 amino-acid residues were deleted. The rotary characteristics of the truncated mutants, monitored by attaching a duplex of 0.49-μm beads to the γ-subunit, did not differ greatly from those of the wild-type over the ATP concentrations of 20 nM–2 mM, the most conspicuous effect being ∼50% reduction in torque and ∼70% reduction in the rate of ATP binding upon deletion of 21 residues. The ATP hydrolysis activity estimated in bulk samples was more seriously affected. The 21-deletion mutant, in particular, was >10-fold less active, but this is likely due to instability of this subcomplex. For torque generation, though not for rapid catalysis, most of the rotor-stator contacts on the deeper half of the penetrating portion of the γ-subunit are dispensable.
INTRODUCTION
F1-ATPase is a water soluble portion of the ATP synthase that catalyzes the synthesis of ATP in mitochondria, chloroplasts, and bacteria in the process of oxidative phosphorylation or photophosphorylation (1–4). When protons flow through the other, membrane-embedded Fo portion of the synthase, ATP is synthesized from ADP and inorganic phosphate in F1. The ATP synthase is a reversible molecular machine, in that ATP hydrolysis in F1 pumps back protons through Fo. The two reactions, the proton flow in Fo and hydrolysis/synthesis in F1, have been proposed to be coupled by mechanical rotation (5,6): both Fo and F1 are rotary motors, Fo being driven by proton flow and F1 by ATP hydrolysis, and the rotary shaft is common to the two motors yet their functional rotary directions are opposite to each other. Thus, when the proton motive force is high and Fo wins, F1 is forcibly rotated in its reverse direction, resulting in ATP synthesis by the reversal of the hydrolysis reaction. When F1 wins, Fo is rotated in reverse and protons flow back. Indeed, isolated F1 has been shown to rotate unidirectionally when it hydrolyzes ATP (7) and reverse rotation by an external force has resulted in ATP synthesis (8,9). Proton-driven rotation of FoF1-ATP synthase has also been demonstrated (10).
The minimal subcomplex of F1 that works as an ATP-driven rotary motor consists of α3β3γ subunits (7), which we refer to here also as F1 unless ambiguity arises. A high-resolution structure of mitochondrial F1 (MF1) has provided a base for mechanistic considerations (11). Three α and three β-subunits are alternately arranged to form a stator cylinder, and an α-helical coiled coil made of the N- and C-termini of the γ-subunit deeply penetrates the cylinder (Fig. 1) to serve as the rotor, as later confirmed (7). Three catalytic sites are located at the interfaces between a β- and an α-subunit, hosted primarily by a β-subunit. In the original structure (11), the three sites contained an ATP analog, ADP, and none, and thus these sites and hosting subunits were designated TP, DP, and E(mpty), respectively (Fig. 1); the central γ-subunit breaks the threefold symmetry seen in the empty α3β3 barrel (12), laying the basis for unidirectional rotation. The remaining three α-β interfaces host, primarily by an α-subunit, noncatalytic nucleotide binding sites which play a role in relieving the enzyme of the so-called MgADP-inhibited state (13–15).
FIGURE 1.
Atomic structure of bovine MF1 ((34); PDB 1E79). α-Subunits are shown in light blue, β-subunits in green, and the γ-subunit in orange. The C-terminus of γ that was truncated in this and previous works is colored according to the scheme in c. Those atoms of α- and β-subunits that are within 0.5 nm from an atom of γ (excluding hydrogens) are colored blue and dark green, respectively. Nucleotides are shown in CPK colors. The black lines in a and the black dot in b represent a putative rotation axis (20). (a) Side views showing the central γ and an opposing α-β pair. The membrane-embedded Fo portion would be above the γ-subunit. (b) Bottom view of the section between the pair of gold lines in a. (c) Amino-acid sequences at the C-terminus of F1 of different origins (CF1, EF1, and MF1 from Nakamoto et al. (39); TF1 from Ohta et al. (40), except that the numbering here starts from Met-1). Colored portions indicate truncations that did not kill the hydrolysis and/or rotation activities: 20 residues in CF1 (23), 12 residues in EF1 (24), and up to 21 residues in TF1 (this work). Note: the amino-acid sequence of the TF1 that we used here is slightly different from the original one described by Ohta et al. (40), and the actual C-terminus is Q285, counting from Met-1 (M. Yoshida, unpublished). Because all differences are outside the C-terminal region shown in c, we adopt the previous, published sequence numbers in this work.
As anticipated from the presence of three catalytic sites, rotation of the γ-subunit occurs in 120° steps, each accompanied by hydrolysis of one ATP molecule (16,17). The asymmetric γ-subunit dictates which of the three basically equivalent sites binds the next ATP molecule (18). ATP binding to that site drives the first 80–90° of a 120° step, and release of a hydrolysis product drives most of the remaining 40–30° (19). The driving force may originate, according to Wang and Oster (20), from a push/pull mechanism: the upper part of a β-subunit bends toward and pushes the curved γ-subunit upon binding of a nucleotide, whereas β retracts and pulls γ upon product release (Fig. 1 a). The bottom of γ (C-terminus shown in yellow to magenta in Fig. 1 a), held in a hydrophobic “sleeve” formed by the α3β3 hexamer (11), would serve as a pivot for the conical rotation. The implication is that steric interactions between β-subunits and the portion of γ that penetrates the α3β3 cylinder would be vital to the torque production. Interactions between the protruding portion of γ and the top surface of β seem less important, because the highly conserved and charged DELSEED sequence that touches γ in the βTP form (the uppermost dark green cluster in Fig. 1 a, left) can all be replaced with alanine without affecting rotational torque (21).
If the penetrating portion of γ is essential for rotation, as the structure suggests (Fig. 1), its truncation should at some point lead to loss of torque. In an early study (22), up to 10 residues at the C-terminus of Escherichia coli F1 (EF1) were truncated, resulting in reduced but significant ATPase activity; deletion of 18 residues gave an inactive enzyme. With chloroplast F1 (CF1), truncation up to 20 C-terminal residues (sea green in Fig. 1 c) did not result in the complete loss of hydrolysis activity (23). Retention of ATPase activity, however, does not necessarily imply active rotation, because rotation and ATP hydrolysis may be decoupled in a defective enzyme. With single-molecule observation, Müller et al. (24) have shown that up to 12 C-terminal residues of EF1 (yellow in Fig. 1, a and c) can be truncated without affecting torque, although hydrolysis activity was gradually diminished at longer deletions. Deletion of 15 or more residues was also attempted, but the enzyme apparently failed to assemble. In this work, we have succeeded in obtaining enzymes with deletion up to 21 residues (magenta in Fig. 1, a and c), owing to higher stability of an F1 derived from thermophilic Bacillus PS3 (TF1). Although most of the contacts at the tip of γ are lost, the mutants produced an appreciable torque.
MATERIALS AND METHODS
ATP, ADP, and phosphoenolpyruvate were from Sigma-Aldrich (St. Louis, MO), and other chemicals were of the highest grade commercially available. In rotation and hydrolysis experiments, Mg2+ was always 2 mM in excess over ATP.
Molecular genetics
In this study, we refer to a mutant (α-C193S, β-His10 at amino terminus, γ-S107C, γ-I210C) α3β3γ subcomplex derived from a thermophilic Bacillus PS3 as wild-type F1 (TF1). The wild-type and deletion mutants were expressed with plasmid pKABG1/HC95 that carries genes for the α- (C193S), β- (His10 at amino terminus), and γ- (S107C, I210C) subunits of TF1 (19,25) in E. coli strain JM103 Δ(uncB-uncD) which has lost the ability to express authentic FoF1-ATPase (26). Plasmids for deletion mutation were prepared in E. coli strain JM109 ((27); obtained from Takara Bio, Otsu, Japan). Culture was in Terrific Broth (28) containing ampicillin (∼50 μg ml−1) or kanamycin (∼50 μg ml−1).
For deletion, BglII-MluI fragment, containing a C-terminal region of the γ-subunit, of plasmid pKABG1/HC95 was transferred to plasmid pET-42b(+) (Novagen, Tokyo, Japan), in which XhoI-EcoRI region had been replaced with a fragment lacking a HindIII site. The resultant plasmid contained a sole HindIII-NheI region, which encompassed the γ C-terminus. Deletions were introduced by polymerase chain reaction, with pKABG1/HC95 as template and with the forward primer 5′-GGAAGCTTCTGCCGCTCACTGAC-3′ that included a HindIII-site. The reverse primer was, for γ-Δ14 with deletion of 14 C-terminal residues, 5′-GGGCTAGCTTATTACGTAATCGCCGCTTGGCGAG-3′ that would eliminate 42 nucleotides (14 amino acid residues) before the γ stop codon and that also contained a NheI-site; for γ-Δ17, 5′-GGGCTAGCTTATTACGCTTGGCGAGCGCGGTTGTAG-3′ that would eliminate 51 nucleotides; for γ-Δ21, 5′-GGGCTAGCTTATTAGCGGTTGTAGGAAAGCGTCAATGTGC-3′ that would eliminate 63 nucleotides; for γ-Δ25, 5′-GGGCTAGCTTATTAAAGCGTCAATGTGCGAATGAGCTCG-3′ that would eliminate 75 nucleotides. The HindIII-NheI portion of the amplified fragment was transferred to the modified pET-42b(+), and its BglII-MluI fragment was introduced into pKABG1/HC95 to produce the expression plasmid. Mutations were confirmed by DNA sequencing (Takara Custom Service, Yokkaichi, Japan).
Doubling time
Growth of E. coli was monitored in 250 ml of Terrific Broth containing ampicillin during 17 h of continuous shaking at 37°C. Turbidity at 600 nm was read every 2 h in a spectrophotometer (UV 3101 PC, Shimadzu, Tokyo, Japan). Doubling time was estimated in the log phase.
Purification of F1-ATPase
Wild-type and mutant F1-ATPases were purified as described (29), with some modifications. Cell lysate was heat treated at 65°C for 8–10 min. After purification with a Ni2+-nitrilotriacetic acid (Ni-NTA) Superflow column (Qiagen, Hilden, Germany), endogenously bound nucleotide was removed on a butyl-Toyopearl column (Tosoh 650M, Tokyo, Japan) by washing with 40 column volumes (400 ml) of buffer C (100 mM potassium phosphate, pH 7.0, and 2 mM EDTA) containing 10% saturated ammonium sulfate. Eluent at 1–0% ammonium sulfate was precipitated in buffer C containing 70% ammonium sulfate and 2 mM dithiothreitol (DTT) and stored at 4°C. Before use, the precipitate was dissolved in buffer C and passed through a size exclusion column (Superdex 200 HR 10/30, Amersham Pharmacia, Piscataway, NJ) preequilibrated with buffer C to remove DTT and possible denatured enzyme. Fractions with the absorbance ratio A280/A260 > 1.95 was selected for the measurement of hydrolysis activity (30). The amount of nucleotide that remained was estimated on a reverse-phase column (TSK-GEL ODS-80Ts, Tosoh) equilibrated with 100 mM sodium phosphate, pH 6.8 (31).
Biotinylation of F1-ATPase
F1 was biotinylated at the two cysteines (γ-107C and γ-210C) by incubation with fourfold molar excess of 6-{N′-[2-(N-maleimide)ethyl]-N-piperazinylamide}hexyl-D-biotinamide (Dojindo, Kumamoto, Japan) for 30 min at room temperature. Unbound biotin was removed on a size exclusion column (Superdex 200 HR 10/30) preequilibrated with buffer C. Biotinylated F1 was frozen with liquid nitrogen and stored at −80°C.
Hydrolysis activity
ATP hydrolysis activity in solution was measured in a spectrophotometer (model U-3100, Hitachi, Tokyo, Japan) thermostated at 23°C. The reaction was started by rapidly adding unbiotinylated F1 in buffer C, at a final concentration of 5–10 nM, into buffer D (10 mM MOPS (3-(N-morpholino)propanesulfonic acid)-KOH, pH 7.0, 50 mM KCl, 2 mM MgCl2) containing a desired amount of MgATP and an ATP regeneration system consisting of 0.2 mM NADH (nicotinamide adenine dinucleotide), 1 mM phosphoenolpyruvate, 250 μg ml−1 pyruvate kinase (rabbit muscle, Roche Diagnostics, Mannheim, Germany), and 50 μg ml−1 lactate dehydrogenase (hog muscle, Roche). Hydrolysis rate was determined from the absorbance decrease of NADH at 340 nm (14).
Ni2+-nitrilotriacetic acid glass preparation
Coverslips (NEO Micro Cover Glass, No. 1, 24 × 32 mm2, Matsunami, Osaka, Japan) to serve as the bottom of an observation chamber were functionalized with Ni-NTA (8). After cleaning in 12 N KOH for 24 h followed by extensive washing in pure water, coverslips held in a ceramic holder were immersed in 0.02% (v/v) acetic acid containing 2% (v/v) 3-mercaptopropyltrimethoxysilane (TSL8380, Toshiba GE silicone, Tokyo, Japan) for 15–20 min with continuous stirring at 60°C. The glass beaker containing the silane solution and coverslips was further incubated for 2 h in an oven at 90°C. After vigorous washing in pure water, 100 mM DTT in 10 mM MOPS-KOH (pH 7.0) was sandwiched between two coverslips and left at room temperature for 20 min to reduce SH groups on the glass surface. After washing with pure water, the paired coverslips were incubated for 30 min at room temperature with 10 mg ml−1 Maleimide-C3-NTA (Dojindo) in 10 mM MOPS-KOH (pH 7.0) in between. The coverslips were again placed on the ceramic holder, washed with pure water, and submerged in 10 mM NiCl2 for 20 min at room temperature. After final washing with pure water, the coverslips were stored in pure water at room temperature; they were used within a few weeks.
Observation of rotation
A flow chamber was constructed of the Ni-NTA coated bottom coverslip above (24 × 32 mm2) and a top uncoated coverslip (18 × 18 mm2) separated by two greased strips of Parafilm cover sheet. Rotation of F1 was observed in buffer D containing an ATP regenerating system consisting of 0.2 mg ml−1 creatine kinase (rabbit muscle, Roche) and 2.5 mM creatine phosphate (Roche). Streptavidin-coated polystyrene beads of diameter 0.49 μm (Bangs Laboratories, Fishers, IN) was centrifuged three times in buffer D containing 5 mg ml−1 bovine serum albumin (BSA) (Fluka, Buchs, Switzerland) to remove preservatives, and the beads were resuspended at 0.1% (w/v) in the same solution. One chamber volume of 500 pM biotinylated F1 was infused into the flow chamber and incubated for 2 min. The chamber was washed three times with 5 mg ml−1 BSA in buffer D and incubated for 5 min. One chamber volume of the washed beads was infused and incubated for 15 min, followed by three washes with buffer D. Finally, five chamber volumes of buffer D containing a desired amount of MgATP was infused, and the chamber was sealed with silicon grease to avoid evaporation.
Rotation was observed under a microscope (IX70, Olympus, Tokyo, Japan) using a Plan Apo 100×/1.4 oil objective. Bead images were captured by a charge-coupled device camera (CCD-72X, Dage-MTI, Michigan, IN) and recorded on a video recorder (EVO-9850, Sony, Tokyo, Japan) at 30 frames s−1. The centroid of bead images was calculated as described (19).
RESULTS
Expression and assembly of the mutant proteins
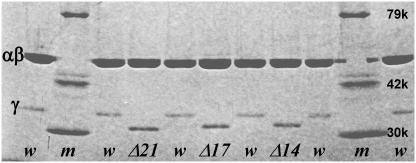
Deletion mutants of TF1, γ-Δ14, γ-Δ17, γ-Δ21, and γ-Δ25 in which 14, 17, 21, or 25 amino acid residues from the C-terminus of the γ-subunit were removed, were expressed in E. coli apparently normally. The doubling time of the E. coli that lacks its authentic F1 genes was almost the same whether a gene encoding wild-type or γ-deleted TF1 was incorporated (Table 1), suggesting that all these mutations were not harmful to the cells. The yield of the assembled subcomplex α3β3γ, the total amount of the subcomplex after final purification versus wet weight of E. coli pellet, did not differ significantly up to γ-Δ21, but we were unable to obtain γ-Δ25, which eluted from the size exclusion column apparently as dissociated subunits (Table 1). As discussed below, γ-Δ21 also appears to be unstable. Deletions were confirmed on the DNA sequence and, except for γ-Δ25, from the sodium dodecyl sulfate (SDS)-gel pattern of purified subcomplexes (Fig. 2). The purified samples contained 0.03–0.07 mol of tightly bound nucleotide per mol of F1.
TABLE 1.
Doubling time, yield, and bound nucleotide for the wild-type and mutants
| Type of F1 | Doubling time* (min) | Yield (weight %) | Bound nucleotide (mol %) |
|---|---|---|---|
| TF1 (wild) | 45 ± 1 | 0.042 | 6.7 |
| TF1 (γ-Δ14) | 54 ± 0 | 0.057 | 5.1 |
| TF1 (γ-Δ17) | 50 ± 2 | 0.039 | 6.5 |
| TF1 (γ-Δ21) | 45 ± 3 | 0.050 | 3.3 |
| TF1 (γ-Δ25) | 55 ± 8 | 0.0 | – |
Mean ± range for two or three samples.
FIGURE 2.
Confirmation of γ-deletions by SDS-polyacrylamide gel electrophoresis (10% gel containing 0.1% SDS; stained with Coomassie brilliant blue G-250). Lanes m, w, Δ14, Δ17, and Δ21, respectively, show molecular weight markers (Wako, Tokyo, Japan), wild-type TF1, the γ-Δ14 mutant, γ-Δ17, and γ-Δ21. α- and β-subunits were unresolved on this gel.
Rotation
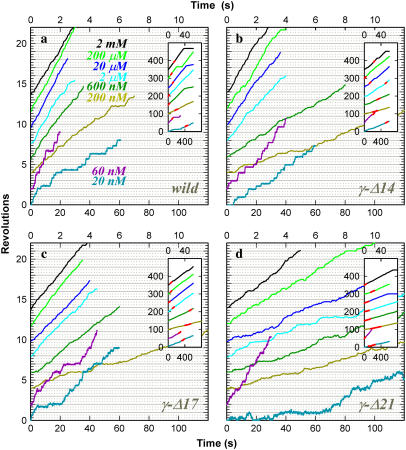
For the analysis of rotation, we chose those F1 molecules that happened to carry a duplex of 0.49-μm beads and that rotated fast without irregular intermissions over at least 25 revolutions. For the estimation of torque, in particular, we tried to choose those duplexes in which the outer bead was slightly above the inner bead, to minimize interaction with the glass surface. Time courses of rotation at various ATP concentrations ([ATP]s) are plotted in Fig. 3, where a fast and smoothly rotating portion over seven revolutions at each [ATP] is selected as shown in the insets. All rotations, whether of wild-type or of truncated mutants, were counterclockwise. At 20 and 60 nM ATP, rotation was clearly stepwise, showing an ATP-waiting dwell every 120° (not very clear in γ-Δ21). In this ATP-limited regime, the mutants showed a somewhat lower stepping rate, consistent with a slower rate of ATP hydrolysis in these mutants (see below). At 2 μM ATP and above, rotation was basically continuous and the speed was independent of [ATP], indicating that the viscous friction on the beads limited the rotary speed. A closer look reveals that, even in this friction-limited regime, the bead duplex tended to fall in occasional short pauses at angles separated by 120°. These pauses likely represent load-dependent stumbling of a chemical reaction at ∼80° ahead of an ATP-waiting angle (31) or short inhibition at a similar angle caused by tight binding of MgADP (15). The rotary speed in the friction-limited regime is determined primarily by the motor torque, and thus the lower speed at longer truncation indicates a lower torque.
FIGURE 3.
Time courses of rotation of a 0.49-μm polystyrene bead duplex attached to the γ-subunit. (a) Wild-type; (b) γ-Δ14; (c) γ-Δ17; and (d) γ-Δ21. Top time axes apply to [ATP] at 200 nM and above, and bottom axes to 20 nM and 60 nM ATP. Records showing the highest average speed over >7 consecutive revolutions are selected and shown. Insets show the whole time courses, where the selected portions are shown in red. Average rotation frequency in Fig. 4 and torque in Fig. 6 were calculated on such selected portions containing >7 consecutive revolutions. Horizontal lines (solid and dotted) are separated by 120°, and each time course has been adjusted such that frequent pauses fall on a horizontal line.
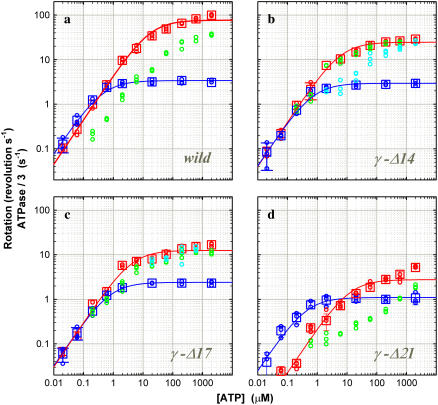
The [ATP] dependence of the time-averaged rotation rate is summarized in Fig. 4, together with the rate of ATP hydrolysis described below. The rotation rate (blue) can be fitted with a Michaelis-Menten kinetics, the saturation being due to the hydrodynamic friction (16). From the fit, the apparent rate of ATP binding at [ATP] below 1 μM is obtained as 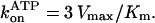 The rate for the wild-type of 2.4 × 107 M−1s−1 agrees with the previous estimates of 2.6 × 107 M−1 s−1 under negligible friction (19) and 1.7 × 107 M−1 s−1 for F1 bearing an actin filament (16). The mutants showed a lower rate: 1.2 × 107 M−1s−1 in γ-Δ14 (52% of the wild-type), 0.99 × 107 M−1s−1 in γ-Δ17 (42%), and 0.77 × 107 M−1s−1 in γ-Δ21 (32%). Because the rate of ATP hydrolysis in solution (red squares in Fig. 4) parallels the rate of rotation at [ATP] below 1 μM, the lower rate of ATP binding in the mutants is genuine, not the result of bead attachment. The extremely low rate of ATP hydrolysis in γ-Δ21 is presumably due to instability of this subcomplex, as discussed below.
The rate for the wild-type of 2.4 × 107 M−1s−1 agrees with the previous estimates of 2.6 × 107 M−1 s−1 under negligible friction (19) and 1.7 × 107 M−1 s−1 for F1 bearing an actin filament (16). The mutants showed a lower rate: 1.2 × 107 M−1s−1 in γ-Δ14 (52% of the wild-type), 0.99 × 107 M−1s−1 in γ-Δ17 (42%), and 0.77 × 107 M−1s−1 in γ-Δ21 (32%). Because the rate of ATP hydrolysis in solution (red squares in Fig. 4) parallels the rate of rotation at [ATP] below 1 μM, the lower rate of ATP binding in the mutants is genuine, not the result of bead attachment. The extremely low rate of ATP hydrolysis in γ-Δ21 is presumably due to instability of this subcomplex, as discussed below.
FIGURE 4.
Comparison of hydrolysis and rotation rates. (a) Wild-type; (b) γ-Δ14; (c) γ-Δ17; and (d) γ-Δ21. For comparison with rotation, hydrolysis rates are divided by three. Circles are from individual time courses, and squares show the average over three points, error bars being a standard deviation larger than the size of the square. [F1] was 5 nM except at 60 and 20 nM ATP, where [F1] was 10 nM. (Cyan) The initial hydrolysis activity estimated at 2–7 s after mixing (2 μM ATP and above), 2–15 s (600 and 200 nM ATP), or from the entire 360-s portion (60 and 20 nM ATP; 200 and 60 nM ATP for γ-Δ21). Values at 60 and 20 nM ATP have been corrected for a small decline in absorbance observed in the absence of F1. (Green) The steady-state hydrolysis activity estimated from the final 200-s portion of a time course; not estimated at 60 and 20 nM ATP (and 200 nM ATP for γ-Δ21) where the activity was low from the beginning. (Red) The maximal hydrolysis activity in a time course: it was equal to the initial activity in the wild-type and γ-Δ21; in γ-Δ14 and γ-Δ17 at intermediate [ATP]s, the maximal activity was observed after an initial lag, and its value was estimated as a slope over a 5-s interval. (Blue) Rotation frequency estimated over >7 consecutive revolutions (see Fig. 3). Red and blue curves show a fit with Michaelis-Menten kinetics: V = Vmax [ATP]/(Km + [ATP]). For hydrolysis (red), Vmax (not divided by three) and Km are 230 s−1 and 17 μM (wild), 75 s−1 and 6.1 μM (γ-Δ14), 38 s−1 and 4.1 μM (γ-Δ17), 8.3 s−1 and 12 μM (γ-Δ21); for rotation (blue), 3.4 s−1 and 0.43 μM (wild), 2.9 s−1 and 0.70 μM (γ-Δ14), and 2.4 s−1 and 0.73 μM (γ−Δ17), 1.1 s−1 and 0.43 μM (γ-Δ21).
Torque
To see whether γ-truncation leads to reduction in the torque, we estimated the torque N from the instantaneous rotary speed ω (in radian s−1) during a 120° step using the following equations (16,31):
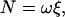 |
(1) |
where ξ is the frictional drag coefficient given, for the case of a duplex of spherical beads, by
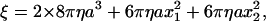 |
(2) |
where a is the bead radius, x1 and x2 are the radii of rotation of the two beads, and η is the viscosity of the medium (∼0.93 × 10−3 N s m−2 at 23°C). The drag in Eq. 2 is likely an underestimate (16,32), but our purpose here is the comparison between the wild-type and mutants.
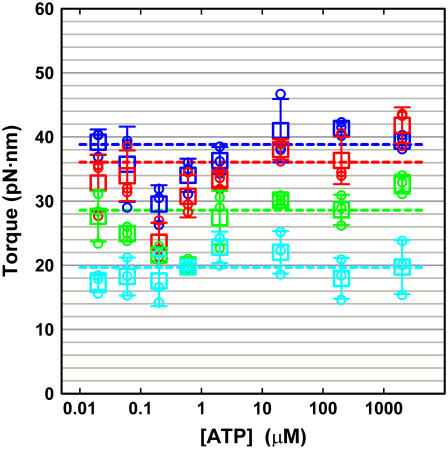
We estimated the angular speed ω by averaging 21 consecutive 120° steps, avoiding the ATP-waiting dwells at low [ATP]s and occasional pauses at high [ATP]s (thick cyan curves in Fig. 5). The slopes, shown in red lines in Fig. 5, indicate that the γ-truncation does not seriously impair torque production in TF1. Torque values estimated as in Fig. 5 are summarized in Fig. 6. Wild-type TF1 gave ∼40 pN·nm of torque over ATP concentrations of 20 nM–2 mM, in agreement with the previous estimate with an actin filament (16,33) and a bead duplex (31). Truncation progressively diminished the torque, but γ-Δ21 still produced ∼20 pN·nm of torque.
FIGURE 5.
Stepping records for torque estimation. (a) Wild-type; (b) γ-Δ14; (c) γ-Δ17; and (d) γ-Δ21. (1) At 2 mM ATP; (2) 200 nM ATP; and (3) 20 nM ATP. Thin lines with dots show 21 consecutive steps selected from a rotation time course, thick cyan line being the average. Individual step records have been shifted vertically by a multiple of 120° to obtain the overlap. Angle zero was chosen such that most of the pauses in the time course fall on a multiple of 120°. Time zero for each step record was assigned by eye to the data point closest to 60°. Straight red lines indicate the slope of the average step record judged by eye.
FIGURE 6.
ATP dependence of the rotary torque. (Blue) Wild-type; (red) γ-Δ14; (green) γ-Δ17; and (cyan) γ-Δ21. Three highest torque values at each [ATP] are plotted in small circles, and the average of the three in a large square, the error bar being ± SD. Dashed lines show the average over all torque values excluding data at 200 nM and 600 nM ATP.
We notice in Fig. 6 that, for both the wild-type and mutants, the torque appears somewhat low at 200 and 600 nM ATP. At this moment we cannot suggest a probable reason, nor are we 100% sure that this trend is statistically meaningful. Rotation at these [ATP]s tends to be irregular (Fig. 5), suggesting susceptibility to surface obstruction.
The small anomaly apart, the major conclusion in this study is that the deletion up to 21 C-terminal residues, or the loss of most of the contacts between the γ-tip and the lower part of the α3β3 cylinder (Fig. 1 a), does not affect the driving torque significantly (at most ∼50%).
Hydrolysis activity
Typical time courses of hydrolysis activity at different [ATP]s are shown in Fig. 7. The slope of the curves is proportional to the rate of hydrolysis. In the wild-type, an initial high activity was gradually replaced with a lower steady-state activity as MgADP inhibition proceeded (13–15). Mutants exhibited peculiar behaviors. In γ-Δ14, a conspicuous initial lag was observed at intermediate [ATP]s, despite the removal of most of the bound nucleotide which, if present in the wild-type, would form MgADP-inhibited enzyme and result in a severe lag. The mutant γ-Δ17 also showed a small lag which is not readily discerned in Fig. 7. The activity of γ-Δ21 was very low and tended to decrease with time as in the wild-type. We did not explore the causes of these different kinetic behaviors. We simply note that, in all the truncation mutants and at all [ATP]s except possibly at 60 nM or below, at least part of the enzyme population was capable of actively hydrolyzing ATP. The rotating F1 molecules that we observed under the microscope must belong to this actively hydrolyzing population.
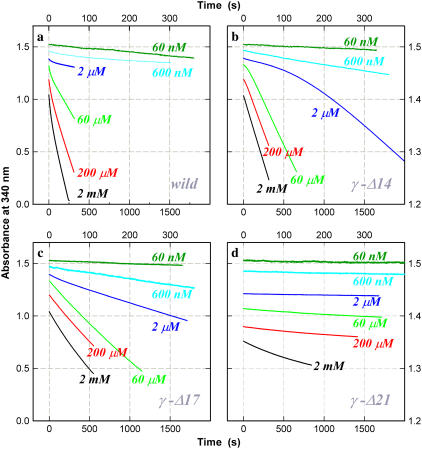
FIGURE 7.
Time courses of ATP hydrolysis at indicated [ATP]. (a) Wild-type; (b) γ-Δ14; (c) γ-Δ17; and (d) γ-Δ21. The decrease in absorbance is proportional to the amount of ATP hydrolyzed. Top and right axes apply to curves for 600 nM and 60 nM ATP. The wild-type showed an initial high activity followed by a steady-state activity (constant slope). At [ATP] ∼10 μM, a low-activity phase appeared before reactivation to the final steady state (see 60 μM). Mutant γ-Δ14 showed an initial lag at and above 2 μM ATP. This was less conspicuous with γ-Δ17, for which a slight initial lag was observed between 600 μM and 20 μM ATP (not clear on the chosen timescale). The activity of γ-Δ21 was very low and was characterized by an initial activity followed by a lower steady-state activity. To avoid overlap, the curves are vertically shifted by an arbitrary amount. [F1] = 5 nM except at 60 nM ATP where [F1] = 10 nM.
In Fig. 4, the maximal hydrolysis activity in a time course, initial in the wild-type and γ-Δ21, near initial in γ-Δ17, and near steady state in γ-Δ14, is plotted in red squares. Except for γ-Δ21, the maximal activity can be approximately fitted with Michaelis-Menten kinetics (red curves), suggesting that these maximal values best represent the activity of an active enzyme. The small deviation seen in the wild-type is due to the carryover of phosphate, which modulates the hydrolysis activity in an [ATP]-dependent manner (R. Shimo-Kon, unpublished): careful removal of phosphate results in simple Michaelis-Menten kinetics (31).
The hydrolysis activity of γ-Δ21 was extremely low, particularly at low [ATP]s where the hydrolysis rate was several times lower than expected from the rotation rate. The implication is that most of the enzyme was inactive from the beginning. With this mutant, the probability of finding a rotating bead duplex was also significantly low at low [ATP]s compared to other mutants. The simplest explanation would be that the γ-Δ21 subcomplex is unstable and tends, for example, to partially dissociate in the absence of nucleotides. In support of this possibility, patterns of native polyacrylamide gel electrophoresis (not shown) indicated a tendency of γ-Δ21 to dissociate into subunits under the electrophoretic conditions, and γ-Δ17 also showed some tendency toward dissociation. A γ-Δ21 subcomplex that happens to retain its integrity would rotate, and addition of ATP might help restore the integrity. We did not pursue further the precise reason the bulk hydrolysis activity of γ-Δ21 was so low. Presumably, the subcomplex of γ-Δ25 was more unstable and could not withstand the purification procedure.
In Fig. 4, except for γ-Δ21, the Michaelis-Menten fit for the maximal hydrolysis activity (red curves) overlaps with that for rotation (blue curves) at low [ATP]s, indicating that three ATP molecules are consumed per turn as expected. In the wild-type, the rotation rate was somewhat higher as in previous studies, suggesting that a portion of the enzyme was already inhibited from the beginning. The F1 in the rotation assay was labeled with streptavidin and beads, but the attachment of beads and/or streptavidin is unlikely to increase the hydrolysis activity. Biotinylation could in principle modify the activity, but biotinylated cysteines are both on the protruding portion of the γ-subunit. At least at 2 mM ATP, biotinylation of either wild-type (R. Shimo-Kon, unpublished) or γ-Δ14 did not change the hydrolysis activity significantly.
As already noted, the apparent rate of ATP binding estimated from either rotation or hydrolysis activity is lower in the mutants than in the wild-type. Larger differences were observed in Vmax, the hydrolysis activity at saturating [ATP] or the rate of catalysis, γ-Δ17 being ∼6-fold less active than the wild-type. These differences in the rates of ATP binding and catalysis are solely related to the structural differences in the γ-subunit: the loss of contacts between γ and α3β3 cylinder somehow affects the hydrolysis kinetics in the catalytic sites remote from the γ-subunit.
DISCUSSION
A crystal structure of the α3β3γ subcomplex of TF1 (Yasuo Shirakihara, Structure Biology Center, National Institute of Genetics, Mishima, Japan, personal communication, 2005) shows close similarity to published structures of MF1 (11,34) and EF1 (35). In particular, positions of the corresponding residues at the C-terminus of the γ-subunit (Fig. 1 c) are essentially the same among these F1 structures although, near the top orifice of the α3β3 cylinder, the γ-subunits of TF1 and EF1 are slightly rotated counterclockwise compared to MF1, making the coiled coil of bacterial γ less twisted than that of MF1.
In Fig. 1, a and b, dark green and blue atoms on the β- and α-subunits, respectively, are those within 5 Å from an atom of γ (hydrogen atoms excluded). The structural similarity above implies that the contacts between the γ-tip and α3β3 cylinder, shown in the dark colors, are essentially the same among MF1, EF1, and TF1. As seen in Fig. 1, a and b, most of the contacts at the γ-tip, below the central cavity in Fig. 1a, should be lost upon deletion of the 21 residues in TF1 (up to and including the residues in magenta). Only a few residues of αE and βE remain within 5 Å from an atom of γ, and Fig. 1 b suggests that the tip of the truncated γ would be allowed a considerable motional freedom in the central cavity. Despite this loose pivot, the γ-Δ21 mutant rotates in the correct direction with few mistakes (Fig. 3 d), and its torque is still 50% of the wild-type (Figs. 5 and 6). It seems that the generation of torque, and of rotary motion, largely depends on mechanical interactions at the top orifice of the α3β3 cylinder.
That the tip of γ is not crucial to mechanical rotation has been suggested by Junge and colleagues: deletion of 12 residues from the C-terminus of the γ-subunit in EF1 (yellow in Fig. 1) did not affect the torque (24), and cross-linking γ-A285, after substitution with cysteine, to a residue on an α-subunit in EF1 did not impair rotation (36). (The cross-linking result, though, might simply imply that the very end of the C-terminal α-helix easily melts into a random coil.) Our γ-Δ21 here is 8 residues shorter than their 12-deletion mutant (Fig. 1 c), confirming and further strengthening the unimportance of the γ-tip.
Previously we have shown that forced, reverse rotation of γ in an α3β3γ subcomplex resulted in ATP synthesis, i.e., the reversal of chemical reactions in the three catalytic sites (8). The simplest interpretation of this reversibility is a γ-dictator mechanism, in which the rotary angle of γ dictates which of the chemical reactions (binding of a substrate, synthesis or hydrolysis of ATP, and release of a product) is to occur in each site or that the equilibrium constants for the chemical reactions are each a function of the γ angle (3). Truncation of the γ-subunit may well impair the communication from γ to catalytic sites, leading to altered kinetics of hydrolysis and rotation, even though the effect on the production of mechanical torque is minimal. Previous reports and this study indicate that the hydrolysis activity diminishes with the extent of truncation, suggesting that contacts at the γ-tip plays some, though not vital, roles in the communication. Thus, for example, membrane-bound EF1 with a γ C-terminal truncation of 4 or 10 residues had 63% and 14% of wild-type activity, respectively (22). Isolated EF1 lacking 3, 6, 9, and 12 C-terminal residues each showed an activity 70%, 50%, 30%, and 24%, respectively, of a control (24). With CF1, Mg-ATPase activity in the presence of an activating anion sulfite was 73%, 45%, 31%, 20%, 13%, and 18% of the wild-type for the deletions of 10, 12, 14, 16, 18, and 20 residues, respectively (23).
Of the contacts at the γ-tip, hydrogen bonds between γ-R254 and βE-D316 (in the MF1 sequence) and between γ-Q255 and βE-T318 (11) have been shown to be important for catalytic activity in a mutation study: when γ-R268 or γ-Q269 of EF1, corresponding to R254 and Q255 in MF1 (Fig. 1 c), was mutated to leucine, the hydrolysis activity dropped to 12% and 1%, respectively, of a control (37). With CF1, though, the mutant with 20-residue deletion including R304 and Q305 was 18% active (23). The tolerance in CF1 is remarkable, in that the N-terminus of γ, the shorter pair of the coiled coil, can also be truncated without complete loss of hydrolysis activity (38). Whether the apparent tolerance in CF1 may result from decoupling, i.e., hydrolysis without rotation, remains to be seen. More important is the question whether γ-truncation in various F1s leads to failure in the γ-dictated synthesis, i.e., clockwise (reverse) rotation failing in ATP synthesis. Further studies are needed to answer these questions.
Acknowledgments
The authors gratefully acknowledge the kind help and cooperation of R. Shimo-Kon, N. Sakaki, Y. Onoue, K. Shiroguchi, and R. Kanda during experiment and preparation of the manuscript. Thanks are also due to M. Fukatsu for encouragement and lab administration. Y. Shirakihara kindly communicated unpublished structural information to us.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Boyer, P. D. 1993. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta. 1140:215–250. [DOI] [PubMed] [Google Scholar]
- 2.Kinosita, K. Jr., R. Yasuda, H. Noji, and K. Adachi. 2000. A rotary molecular motor that can work at near 100% efficiency. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355:473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinosita, K. Jr., K. Adachi, and H. Itoh. 2004. Rotation of F1-ATPase: how an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 33:245–268. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida, M., E. Muneyuki, and T. Hisabori. 2001. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2:669–677. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. D., and W. E. Kohlbrenner. 1981. The present status of the binding-change mechanism and its relation to ATP formation by chloroplasts. In Energy Coupling in Photosynthesis. B. R. Selman and S. Selman-Reimer, editors. Elsevier, Amsterdam. 231–240.
- 6.Oosawa, F., and S. Hayashi. 1986. The loose coupling mechanism in molecular machines of living cells. Adv. Biophys. 22:151–183. [DOI] [PubMed] [Google Scholar]
- 7.Noji, H., R. Yasuda, M. Yoshida, and K. Kinosita Jr. 1997. Direct observation of the rotation of F1-ATPase. Nature. 386:299–302. [DOI] [PubMed] [Google Scholar]
- 8.Itoh, H., A. Takahashi, K. Adachi, H. Noji, R. Yasuda, M. Yoshida, and K. Kinosita Jr. 2004. Mechanically driven ATP synthesis by F1-ATPase. Nature. 427:465–468. [DOI] [PubMed] [Google Scholar]
- 9.Rondelez, Y., G. Tresset, T. Nakashima, Y. Kato-Yamada, H. Fujita, S. Takeuchi, and H. Noji. 2005. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature. 433:773–777. [DOI] [PubMed] [Google Scholar]
- 10.Diez, M., B. Zimmermann, M. Börsch, M. König, E. Schweinberger, S. Steigmiller, R. Reuter, S. Felekyan, V. Kudryavtsev, C. A. M. Seidel, and P. Gräber. 2004. Proton-powered subunit rotation in single membrane-bound FoF1-ATP synthase. Nat. Struct. Mol. Biol. 11:135–141. [DOI] [PubMed] [Google Scholar]
- 11.Abrahams, J. P., A. G. W. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 370:621–628. [DOI] [PubMed] [Google Scholar]
- 12.Shirakihara, Y., A. G. W. Leslie, J. P. Abrahams, J. E. Walker, T. Ueda, Y. Sekimoto, M. Kambara, K. Saika, Y. Kagawa, and M. Yoshida. 1997. The crystal structure of the nucleotide-free α3β3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer. Structure. 5:825–836. [DOI] [PubMed] [Google Scholar]
- 13.Jault, J.-M., C. Dou, N. B. Grodsky, T. Matsui, M. Yoshida, and W. S. Allison. 1996. The α3β3γ subcomplex of the F1-ATPase from the thermophilic bacillus PS3 with the βT165S substitution does not entrap inhibitory MgADP in a catalytic site during turnover. J. Biol. Chem. 271:28818–28824. [DOI] [PubMed] [Google Scholar]
- 14.Matsui, T., E. Muneyuki, M. Honda, W. S. Allison, C. Dou, and M. Yoshida. 1997. Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J. Biol. Chem. 272:8215–8221. [DOI] [PubMed] [Google Scholar]
- 15.Hirono-Hara, Y., H. Noji, M. Nishiura, E. Muneyuki, K. Y. Hara, R. Yasuda, K. Kinosita Jr., and M. Yoshida. 2001. Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. USA. 98:13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda, R., H. Noji, K. Kinosita Jr., and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]
- 17.Adachi, K., R. Yasuda, H. Noji, H. Itoh, Y. Harada, M. Yoshida, and K. Kinosita Jr. 2000. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. USA. 97:7243–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizaka, T., K. Oiwa, H. Noji, S. Kimura, E. Muneyuki, M. Yoshida, and K. Kinosita Jr. 2004. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11:142–148. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda, R., H. Noji, M. Yoshida, K. Kinosita Jr., and H. Itoh. 2001. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 410:898–904. [DOI] [PubMed] [Google Scholar]
- 20.Wang, H., and G. Oster. 1998. Energy transduction in the F1 motor of ATP synthase. Nature. 396:279–282. [DOI] [PubMed] [Google Scholar]
- 21.Hara, K. Y., H. Noji, D. Bald, R. Yasuda, K. Kinosita Jr., and M. Yoshida. 2000. The role of the DELSEED motif of the β subunit in rotation of F1-ATPase. J. Biol. Chem. 275:14260–14263. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto, A., J. Miki, M. Maeda, and M. Futai. 1990. H+-ATPase γ subunit of Escherichia coli: role of the conserved carboxyl-terminal region. J. Biol. Chem. 265:5043–5048. [PubMed] [Google Scholar]
- 23.Sokolov, M., L. Lu, W. Tucker, F. Gao, P. A. Gegenheimer, and M. L. Richter. 1999. The 20 C-terminal amino acid residues of the chloroplast ATP synthase γ subunit are not essential for activity. J. Biol. Chem. 274:13824–13829. [DOI] [PubMed] [Google Scholar]
- 24.Müller, M., O. Pänke, W. Junge, and S. Engelbrecht. 2002. F1-ATPase, the C-terminal end of subunit γ is not required for ATP hydrolysis-driven rotation. J. Biol. Chem. 277:23308–23313. [DOI] [PubMed] [Google Scholar]
- 25.Matsui, T., and M. Yoshida. 1995. Expression of the wild-type and the Cys-/Trp-less α3β3γ complex of thermophilic F1-ATPase in Escherichia coli. Biochim. Biophys. Acta. 1231:139–146. [DOI] [PubMed] [Google Scholar]
- 26.Monticello, R. A., E. Angov, and W. S. A. Brusilow. 1992. Effects of inducing expression of cloned genes for the Fo proton channel of the Escherichia coli F1Fo ATPase. J. Bacteriol. 174:3370–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 33:103–119. [DOI] [PubMed] [Google Scholar]
- 28.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus. 9:12–12. [Google Scholar]
- 29.Adachi, K., H. Noji, and K. Kinosita Jr. 2003. Single molecule imaging of the rotation of F1-ATPase. Methods Enzymol. 361B:211–227. [DOI] [PubMed] [Google Scholar]
- 30.Noji, H., D. Bald, R. Yasuda, H. Itoh, M. Yoshida, and K. Kinosita Jr. 2001. Purine but not pyrimidine nucleotides support rotation of F1-ATPase. J. Biol. Chem. 276:25480–25486. [DOI] [PubMed] [Google Scholar]
- 31.Sakaki, N., R. Shimo-Kon, K. Adachi, H. Itoh, S. Furuike, E. Muneyuki, M. Yoshida, and K. Kinosita Jr. 2005. One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pänke, O., D. A. Cherepanov, K. Gumbiowski, S. Engelbrecht, and W. Junge. 2001. Visco-elastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinosita, K. Jr., R. Yasuda, and H. Noji. 2000. F1-ATPase: a highly efficient rotary ATP machine. Essays Biochem. 35:3–18. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons, C., M. G. Montgomery, A. G. W. Leslie, and J. E. Walker. 2000. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 7:1055–1061. [DOI] [PubMed] [Google Scholar]
- 35.Hausrath, A. C., G. Grüber, B. W. Matthews, and R. A. Capaldi. 1999. Structural features of the γ subunit of the Escherichia coli F1 ATPase revealed by a 4.4-Å resolution map obtained by x-ray crystallography. Proc. Natl. Acad. Sci. USA. 96:13697–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gumbiowski, K., D. Cherepanov, M. Müller, O. Pänke, P. Promto, S. Winkler, W. Junge, and S. Engelbrecht. 2001. F-ATPase: forced full rotation of the rotor despite covalent cross-link with the stator. J. Biol. Chem. 276:42287–42292. [DOI] [PubMed] [Google Scholar]
- 37.Greene, M. D., and W. D. Frasch. 2003. Interactions among γR268, γQ269, and the β subunit catch loop of Escherichia coli F1-ATPase are important for catalytic activity. J. Biol. Chem. 278:51594–51598. [DOI] [PubMed] [Google Scholar]
- 38.Ni, Z.-L., H. Dong, and J.-M. Wei. 2005. N-terminal deletion of the γ subunit affects the stabilization and activity of chloroplast ATP synthase. FEBS J. 272:1379–1385. [DOI] [PubMed] [Google Scholar]
- 39.Nakamoto, R. K., K. Shin, A. Iwamoto, H. Omote, M. Maeda, and M. Futai. 1992. Escherichia coli F0F1-ATPase. Residues involved in catalysis and coupling. Ann. N. Y. Acad. Sci. 671:335–344. [DOI] [PubMed] [Google Scholar]
- 40.Ohta, S., M. Yohda, M. Ishizuka, H. Hirata, T. Hamamoto, Y. Otawara-Hamamoto, K. Matsuda, and Y. Kagawa. 1988. Sequence and over-expression of subunits of adenosine triphosphate synthase in thermophilic bacterium PS3. Biochim. Biophys. Acta. 933:141–155. [DOI] [PubMed] [Google Scholar]